Abstract
We report the characterization of clone 1.9.2, a gene expressed in mineralizing osteoblasts. Remarkably, clone 1.9.2 is the murine homolog of the alpha chain of the nascent polypeptide-associated complex (α-NAC). Based on sequence similarities between α-NAC/1.9.2 and transcriptional regulatory proteins and the fact that the heterodimerization partner of α-NAC was identified as the transcription factor BTF3b (B. Wiedmann, H. Sakai, T. A. Davis, and M. Wiedmann, Nature 370:434–440, 1994), we investigated a putative role for α-NAC/1.9.2 in transcriptional control. The α-NAC/1.9.2 protein potentiated by 10-fold the activity of the chimeric activator GAL4/VP-16 in vivo. The potentiation was shown to be mediated at the level of gene transcription, because α-NAC/1.9.2 increased GAL4/VP-16-mediated mRNA synthesis without affecting the half-life of the GAL4/VP-16 fusion protein. Moreover, the interaction of α-NAC/1.9.2 with a transcriptionally defective mutant of GAL4/VP-16 was severely compromised. Specific protein-protein interactions between α-NAC/1.9.2 and GAL4/VP-16 were demonstrated by gel retardation, affinity chromatography, and protein blotting assays, while interactions with TATA box-binding protein (TBP) were detected by immunoprecipitation, affinity chromatography, and protein blotting assays. Based on these interactions that define the coactivator class of proteins, we conclude that the α-NAC/1.9.2 gene product functions as a transcriptional coactivator.
In addition to the sequence-specific DNA-binding transcription factors and the components of the basal transcriptional machinery, the control of gene expression requires another class of proteins that are involved in activated transcription. These molecules have been ascribed different names, such as mediators, adaptors, and coactivators (13). While further studies may reveal that these semantic distinctions reflect true mechanistic differences, the various names have been used indiscriminately in various contexts. The term “coactivator” was first ascribed to the TAFs (TATA-binding protein [TBP]-associated factors), a class of proteins that contact TBP and interact with one or several sequence-specific transcription factors to potentiate activator-dependent transcription (9). We have utilized these characteristics to define the function of coactivators herein.
These proteins provide interfaces to link the sequence- specific factors to the basal transcriptional machinery (13, 33). Recent studies have identified specific enzymatic functions for some coactivators, indicating that activated transcription does not merely result from protein-protein contacts but also requires covalent modification of the factors involved (2, 8, 12).
The TAFs represent a group of proteins tightly associated with TBP so that they copurify with it in the chromatographic fraction TFIID (9). Several human and Drosophila TAFs have now been cloned (reference 4 and references therein). Recent genetic evidence in yeast may question whether or not TAFs are universally required for transcription (25, 35), but the yeast results can perhaps be explained by the relative simplicity of the yeast genome (33). Indeed, recent evidence suggests that specific TAFs are implicated in the control of developmentally restricted gene expression and tissue-specific transcriptional activation in multicellular organisms (29). Coactivators distinct from the TFIID fraction have also been identified and cloned (reviewed in reference 13).
Several of the coactivators characterized to date share functional properties. However, some of them also exhibit very unique functions. For example, the Drosophila TAF with a molecular weight of 150,000 (dTAFII150) has been shown to possess specific DNA binding affinity (34), while hTAFII250 was shown to act as a protein serine kinase (8). The CBP and p300 coactivators possess histone acetyltransferase activity (2). Thus a diverse range of structural and functional domains in coactivator proteins have been characterized.
We have used the technique of differential display of mRNA amplified by PCR (differential display PCR) (19) to compare genes expressed in mineralizing osteoblasts with those expressed in dedifferentiated, nonmineralizing cells of the same lineage. We report the characterization of clone 1.9.2, a 794-bp cDNA corresponding to a transcript expressed in differentiated osteoblasts. Remarkably, clone 1.9.2 is the murine homolog of the alpha chain of the nascent polypeptide-associated complex (α-NAC) (GenBank accession no. X80909 [unpublished data]) and shall hereafter be referred to as α-NAC/1.9.2. NAC has been previously purified as a heterodimeric complex binding the newly synthesized polypeptide chains as they emerge from the ribosome (37). The β-NAC subunit has been identified as BTF3b (39), a protein involved in regulating transcription in yeast (15) and in higher eukaryotes (39). Based on sequence similarities between α-NAC/1.9.2 and transcriptional regulatory proteins and the identification of the heterodimerization partner of α-NAC as the transcription factor BTF3b (37), we investigated a putative role for α-NAC/1.9.2 in transcriptional control. Additional work from our laboratory characterizing the function of the muscle-specific isoform of α-NAC/1.9.2, skNAC, as a sequence-specific DNA-binding transcription factor and demonstrating that α-NAC/1.9.2 has inherent DNA binding activity in vitro (38) further supports a role for α-NAC/1.9.2 in the regulation of gene transcription.
We present evidence that the α-NAC/1.9.2 protein can potentiate the activity of the chimeric activator GAL4/VP-16 in vivo. Specific protein-protein interactions between α-NAC/1.9.2 and the activator were demonstrated. The α-NAC/1.9.2 protein was also shown to interact with TBP. Based on these interactions that define the coactivator class of proteins, we conclude that the α-NAC/1.9.2 gene product functions as a transcriptional coactivator.
MATERIALS AND METHODS
Cloning of 1.9.2 cDNA.
Differential display PCR of mRNA isolated from terminally differentiated osteoblasts (10) and passaged, dedifferentiated bone cells was performed as described previously (19). Clone 1.9.2 was identified as a band specific for differentiated osteoblasts, excised, reamplified, and used to clone the full-length 1.9.2 cDNA from a primary osteoblast library prepared in lambda-ZAPII (Stratagene, La Jolla, Calif.).
Immunolocalization of α-NAC/1.9.2.
Exponentially growing or serum-starved MC3T3-E1 osteoblastic cells (32) were fixed in 4% paraformaldehyde. Immunochemistry was performed according to standard protocols (36) with the anti-αNAC antibody (38) and a secondary goat anti-rabbit antibody conjugated to fluorescein isothiocyanate (TAGO Immunologicals, Burlingame, Calif.). Antibody dilutions were 1:1,000 and 1:50 for the primary and secondary antibodies, respectively.
Transient transfection assays.
All vectors were constructed by standard molecular biology procedures, and full details and sequences are available on request. For assays using GAL4/VP-16, P19 EC cells (24) were transfected with 0.8 μg of the reporter plasmid 5Gal4-E1b-CAT (20), 0.1 μg of an expression plasmid for GAL4/VP-16 (pSGVP [26]), and 2 μg of either the BTF3b vector (39) or the cytomegalovirus (CMV)-1.9.2 expression vector. Fifty nanograms of the luciferase expression plasmid pGL2-control (Promega Corp., Madison, Wis.) was also included as a reference to monitor for variations in transformation efficiency. All transfections were performed with 5 μl of the Lipofectamine reagent (Gibco BRL, Gaithersburg, Md.) according to the instructions of the manufacturer, and cells were harvested 24 h posttransfection. Chloramphenicol acetyltransferase (CAT) and luciferase activities were assayed as previously described (31).
Northern blot assay.
Total RNA from transiently transfected cells was probed with a PCR-labeled fragment from the reporter CAT gene by conventional protocols. The membrane was subsequently stripped and rehybridized with a probe directed against the α-tubulin transcript to monitor for variations in the quality and quantity of the loaded mRNA.
Analysis of GAL4/VP-16 half-life.
We used the cycloheximide-Western protocol of Maheswaran et al. (22) to estimate the half-life of the GAL4/VP-16 activator. P19 EC cells (24) were transfected as described above with 0.5 μg of pSGVP (26) together with 0.5 μg of either an inert plasmid (pBKCMV; Stratagene Corp.) or the CMV-1.9.2 expression vector. Transfection efficiency was monitored with 0.1 μg of the luciferase expression plasmid pGL2-control. Transfected cells were pooled 24 h posttransfection and split in the appropriate number of dishes. Cycloheximide (30 μg/ml) was added to each dish 36 h posttransfection, and cells were harvested at intervals. Protein lysates were prepared, and 700 μg of total protein per sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with an anti-GAL4/VP-16 antibody (Upstate Biotechnology, Inc., Lake Placid, N.Y.).
Gel mobility shift assay.
The 17-bp GAL4 binding site oligonucleotide (20) was labeled by Klenow filling in and incubated with the purified recombinant proteins in binding buffer (12 mM HEPES-NaOH [pH 7.9], 4 mM Tris-Cl [pH 7.9], 60 mM KCl, 1 mM CaCl2, 1 mM dithiothreitol, 12% glycerol) for 30 min at room temperature. The final reaction volume was 20 μl. The amounts of protein used were 50 ng for GAL4/VP-16, 100 ng for GAL4(DBD-1–147 [DNA binding domain residues 1 to 147]), and 500 ng for α-NAC/1.9.2. The preimmune sera and anti-αNAC antibody (38) were used at a final dilution of 1:400; binding reaction mixtures with antibodies were preincubated on ice for 30 min prior to addition of the probe. Bound probe was separated from free oligonucleotide on 6% gels in binding buffer supplemented with 1 mM EDTA. Samples were migrated at 150 V for 3 h with buffer recirculation. The gels were then dried and autoradiographed.
Affinity chromatography.
Crude nuclear extracts from serum-starved P19 EC cells (24) were prepared as described by Dignam et al. (7). Crude lysates from bacteria expressing GAL4/VP-16 or GAL4/VP16ΔFP442 (1) were obtained by sonication. The nuclear extracts (2 mg of total protein) or the bacterial lysates (15 mg of total protein) were precleared on glutathione-Sepharose 4B beads (Pharmacia Canada, Baie d’Urfé, Quebec, Canada) and then passed on a glutathione-Sepharose 4B column previously loaded with glutathione S-transferase (GST)–1.9.2 (300-μl final bed volume). Bound proteins were eluted with a step gradient of NaCl, precipitated with trichloroacetic acid, and analyzed by Western blot assay (11). The antibodies directed against GAL4/VP-16 and TBP were obtained from Upstate Biotechnology, Inc. Recombinant human TBP was obtained from Upstate Biotechnology, Inc.
Far-Western protein blot assay.
The α-NAC/1.9.2 cDNA was subcloned into the pGEX-4T-3 vector (Pharmacia Canada), and the GST-1.9.2 fusion protein was purified according to the manufacturer’s instructions and then was biotinylated with the protein biotinylation system (Canadian Life Technologies, Burlington, Ontario, Canada) based on the protocol supplied. The far-Western blot assay was performed as described by Inostroza et al. (17), except that the blocked membrane was exposed to the biotinylated protein for only 1 h. GST-TBP was purchased from Santa Cruz BioTechnology (Santa Cruz, Calif.), while purified GAL4/VP-16 was a generous gift of James T. Kadonaga (University of California—San Diego, La Jolla, Calif.). The amount of purified protein loaded in each lane was 0.5 μg.
Immunoprecipitation.
Immunoprecipitation reactions were performed according to standard protocols (28). Four hundred microliters of crude nuclear extract from serum-starved P19 embryonal carcinoma cells (4 μg/μl) was incubated with 4 μl of antibodies. The subsequent immunoblotting step was performed by standard protocols (11). The anti-RNA polymerase II antibody was from Santa Cruz BioTechnology (Santa Cruz, Calif.). The antibody dilutions used were 1:1,000 for the anti-αNAC antibody; 1:250 for the anti-TBP, anti-RNA polymerase II, and preimmune sera; and 1:7,500 for the alkaline phosphatase-conjugated second antibody.
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this paper is U22151.
RESULTS
α-NAC/1.9.2 localizes to the nucleus in G0-phase cells.
The α-NAC/1.9.2 protein sequence features a putative nuclear targeting sequence (RSEKKARK) located at residues 71 to 78 (not shown). This sequence motif, combined with the observation that the dimerization partner of α-NAC has been identified as the transcriptional regulatory protein BTF3b (37), prompted us to investigate the putative nuclear localization of α-NAC/1.9.2 as well as its possible involvement in the regulation of gene transcription.
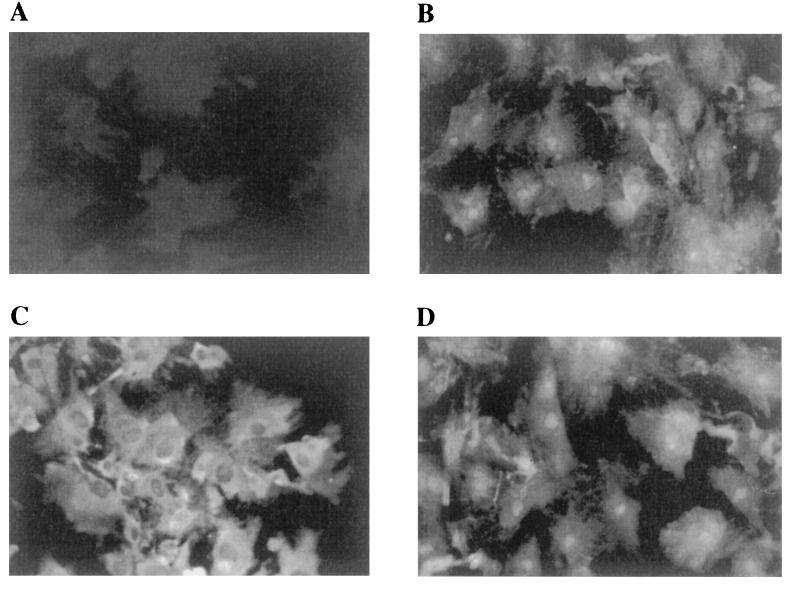
Immunocytochemical analysis with anti-αNAC antibodies (38) revealed that the nuclear localization of the protein was cell cycle dependent: in cells arrested at the G0/G1 border by serum deprivation, the α-NAC/1.9.2 protein was detected in the nucleus of all cells, as well as in the cytoplasm (Fig. 1B). When the cells were challenged with serum for 4 h, the protein mostly localized to the cytoplasm, although some nuclear staining remained evident (Fig. 1C). When serum-stimulated cells were subsequently serum deprived, the α-NAC/1.9.2 protein relocalized to the nucleus (Fig. 1D). Control staining with preimmune sera showed a complete absence of signal (Fig. 1A).
FIG. 1.
α-NAC/1.9.2 localizes to the cytoplasm and nucleus in serum-deprived osteoblasts. MC3T3-E1 osteoblastic cells were stained with polyclonal antibodies raised against the fusion GST-1.9.2 protein. (A) Control staining with preimmune sera. (B to D) Staining for α-NAC/1.9.2. A strong signal was observed in the cytoplasm and the nucleus of serum-starved cells (B). Following stimulation with fetal calf serum for 4 h, α-NAC/1.9.2 localized mainly to the cytoplasm (C). Removal of serum after the 4-h stimulation provoked reentry of α-NAC/1.9.2 into the nucleus (D).
α-NAC/1.9.2 acts as a transcriptional coactivator with GAL4/VP-16.
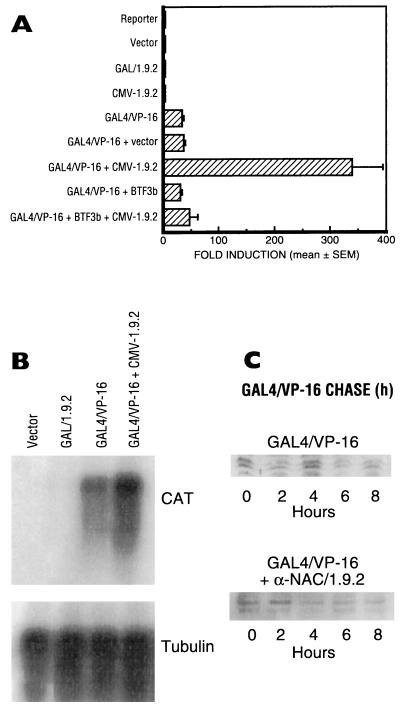
In order to assess the putative transcriptional activation function of α-NAC/1.9.2, the full-length α-NAC/1.9.2 protein was linked in frame to the DBD of the yeast GAL4 transcription factor (amino acids 1 to 147) (27) and assayed for its capacity to activate the transcription of the 5Gal4-E1b-CAT reporter plasmid (20). Under conditions in which the control chimeric activator GAL4/VP-16 (26) strongly stimulated the transcription of the reporter gene (Fig. 2A and B), we found no evidence of increased transcription mediated through the GAL4/α-NAC/1.9.2 fusion protein (Fig. 2A and B), suggesting that α-NAC/1.9.2 could not transactivate. It is interesting that a C-terminal fragment of α-NAC/1.9.2 fused to GAL4 [construct GAL4-1.9.2(111–215)] moderately increased transcription of the 5Gal4-E1b-CAT reporter plasmid (about threefold) in ROS 17/2.8 osteosarcoma cells, but not in P19 embryonal carcinoma cells or C2C12 myoblasts (data not shown). We surmise that this may be due to the interaction of α-NAC/1.9.2 with TBP (see below).
FIG. 2.
α-NAC/1.9.2 acts as a transcriptional coactivator. Transient transfection assays of the reporter plasmid 5Gal4-E1b-CAT (20) with expression vectors for GAL4/VP-16, GAL4/1.9.2, α-NAC/1.9.2, and BTF3b alone or in combination. (A) Relative fold induction levels of the CAT reporter gene in transfected cells. Vector represents the parental pBK-CMV vector in which the α-NAC/1.9.2 cDNA was subcloned to generate CMV-1.9.2. The expression level detected in cells transfected with the reporter alone was arbitrarily ascribed a value of 1. Results are expressed as mean fold induction ± standard error of four independent transfections. (B) Northern blot assays of total RNA from the transfected cells. The blot was first hybridized to a CAT probe (upper panel) and then was stripped and reprobed with an α-tubulin probe (lower panel). (C) Half-life of the GAL4/VP-16 protein in the presence or absence of α-NAC/1.9.2. P19 EC cells (24) were transfected with the GAL4/VP-16 expression vector alone or in combination with the α-NAC/1.9.2 expression vector. Cycloheximide was added to each dish, and cells were harvested at the indicated intervals. Samples were analyzed by Western blotting with an anti-GAL4/VP-16 antibody.
Expression of the α-NAC/1.9.2 cDNA under the control of the CMV promoter (construct CMV-1.9.2) also had no effect on the expression of the 5Gal4-E1b-CAT reporter gene (Fig. 2A). These results are in agreement with our previous findings showing that despite its capacity to bind the same DNA sequence as the muscle-specific skNAC isoform, α-NAC/1.9.2 could not stimulate transcription from promoters containing the skNAC response element (38).
However, when GAL4/VP-16 was cotransfected with construct CMV-1.9.2, a 10-fold enhancement of the response mediated by GAL4/VP-16 alone was observed (Fig. 2A; (compare the 33.5 ± 3.5-fold activation [mean ± standard error] observed with GAL4/VP-16 to the 339 ± 55-fold activation measured with GAL4/VP-16 together with CMV-1.9.2). The enhanced expression generated by the coexpression of GAL4/VP-16 and α-NAC/1.9.2 was also observed at the level of the mRNA of the reporter gene (Fig. 2B), demonstrating that the potentiation occurred at the transcriptional level and not through an increase in the translation, stability, or folding of the nascent CAT protein. The observed effect was specific to the α-NAC/1.9.2 protein, because the empty vector had no influence on the activity of the GAL4/VP-16 activator (Fig. 2A). Moreover, the coactivation function of α-NAC/1.9.2 was not observed when reporter templates devoid of GAL4 binding sites were used or when α-NAC/1.9.2 was cotransfected with vectors expressing only the DBD of GAL4 (data not shown).
β-NAC (37), identified as BTF3b (37, 39), was also tested in this system. While it had no effect on the transcriptional activation function of GAL4/VP-16, BTF3b inhibited the stimulation of transcription mediated by α-NAC/1.9.2 when coexpressed with α-NAC/1.9.2 and GAL4/VP-16 (Fig. 2A).
Similar levels of expression were achieved for each recombinant protein (data not shown). Moreover, we used cycloheximide treatment to estimate the half-life of the GAL4/VP-16 activator in the presence or absence of α-NAC/1.9.2. Figure 2C shows that the coexpression of α-NAC/1.9.2 did not affect the level of expression of the GAL4/VP-16 protein (leftmost lane, zero hour time point), nor did it influence the stability of the chimeric activator.
α-NAC/1.9.2 interacts with GAL4/VP-16.
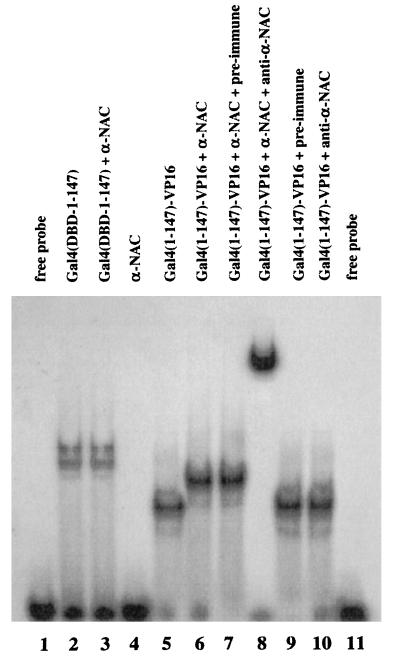
Considering the results presented above, we analyzed whether α-NAC/1.9.2 could directly interact with GAL4/VP-16 as well as components of the basal transcriptional machinery. We first used gel mobility shift assays with purified recombinant proteins to assess direct contacts between α-NAC/1.9.2 and GAL4/VP-16. As shown in Fig. 3, both the GAL4 DBD [GAL4(DBD-1–147); lane 2] and GAL4/VP-16 (lane 5) bound the labeled GAL4 probe. GAL4(DBD-1–147) is a fusion with GST, thus explaining the reduced mobility of the bound complex compared to that of GAL4/VP-16. When α-NAC/1.9.2 was added to the binding reaction mixture, the migration of the bound GAL4/VP-16 complex was further retarded (lane 6), thus demonstrating an interaction of α-NAC/1.9.2 with the GAL4/VP-16 protein bound to the GAL4 probe. Control reaction mixtures established that α-NAC/1.9.2 interacted with the VP-16 moiety of the GAL4/VP-16 fusion protein: α-NAC/1.9.2 did not bind the labeled probe by itself (lane 4) and did not interact with GAL4(DBD-1–147) (lane 3). The migration of the bound complex in reaction mixtures containing GAL4/VP-16 together with α-NAC/1.9.2 was further altered by antibodies directed against α-NAC/1.9.2 (supershifting; see lane 8), further confirming the presence of α-NAC/1.9.2 in the bound complex. The anti-αNAC antibodies had no effect on the binding of GAL4/VP-16 alone (lane 10), and preimmune sera did not influence the migration of any of the bound complexes (lanes 7 and 9). These data demonstrate direct protein-protein interactions between α-NAC/1.9.2 and the acidic activator VP-16.
FIG. 3.
α-NAC/1.9.2 interacts with GAL4/VP-16. Results from gel mobility shift assays with purified recombinant proteins with a labeled 17-mer GAL4 probe are shown. GAL4(DBD-1–147) is a fusion protein between GST and the DBD (residues 1 to 147) of GAL4. GAL4(1–147)-VP-16 is the same as GAL4/VP-16. α-NAC denotes the purified full-length α-NAC/1.9.2 protein. Preimmune sera (lanes 7 and 8) and antibodies raised against α-NAC/1.9.2 (anti-α-NAC [lanes 8 and 10]) were added to some binding reaction mixtures.
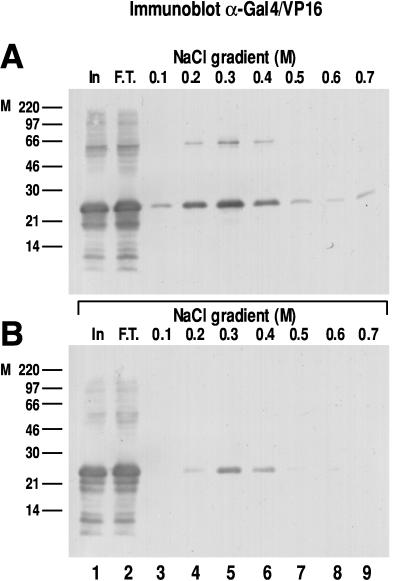
We estimated the strength of the interaction between α-NAC/1.9.2 and GAL4/VP-16 by using affinity chromatography with step gradient elution. As depicted in Fig. 4A, bacterially expressed GAL4/VP-16 was retained on a GST-α-NAC/1.9.2-loaded glutathione-Sepharose column. The recombinant GAL4/VP-16 did not interact with a control GST resin (not shown). The interaction of the bound GAL4/VP-16 protein with α-NAC/1.9.2 was disrupted with increasing concentrations of salt, and the bulk of the bound protein eluted in the 0.3 and 0.4 M fractions (Fig. 4A, lanes 5 and 6). These results demonstrate that the binding between α-NAC/1.9.2 and the chimeric activator is a strong interaction. A significant proportion of the input protein was recovered in the flowthrough fraction (compare lanes 1 and 2), because these experiments were intentionally planned with an excess of GAL4/VP-16 to ascertain that the affinity column was completely saturated.
FIG. 4.
Interactions between α-NAC/1.9.2 and wild-type and mutant GAL4/VP-16. (A) Affinity chromatography of crude lysates from bacteria transformed with an expression vector for wild-type GAL4/VP-16. Bound proteins were eluted with a step gradient of NaCl, and the immunoblot (lanes 1 to 9) was probed with an anti-GAL4/VP-16 antibody. (B) Affinity chromatography of crude lysates from bacteria expressing GAL4/VP16ΔFP442 eluted with a salt step gradient. In, input (1/100 of total material loaded on column); F.T., flowthrough. Molecular mass (kilodaltons) markers (M) are indicated to the left of each panel.
We also tested the interaction of α-NAC/1.9.2 with a transcriptionally compromised mutant of GAL4/VP-16, GAL4/VP16ΔFP442 (1, 5, 21). While the mutant recombinant protein still bound to the α-NAC/1.9.2 affinity resin (Fig. 4B), significantly less protein was retained on the column, and most of it was eluted with 0.3 M salt (lane 5). It is particularly striking that no GAL4/VP16ΔFP442 protein could be detected in the 0.5 to 0.7 M fractions, whereas some of the wild-type GAL4/VP-16 fusion protein was recovered in those fractions (compare lanes 7 to 9, Fig. 4A and B). Equal amounts of recombinant proteins were loaded on each column (lane 1). Similar results were obtained in three separate experiments and with bacterial vectors that expressed only the VP-16 or VP16ΔFP442 moiety (not shown). Thus α-NAC/1.9.2 interacts more strongly with transcriptionally competent VP16 than with transcriptionally compromised mutants, supporting our interpretation that the potentiating effect of α-NAC/1.9.2 on GAL4/VP-16-dependent gene expression is mediated at the level of gene transcription.
α-NAC/1.9.2 interacts with TBP.
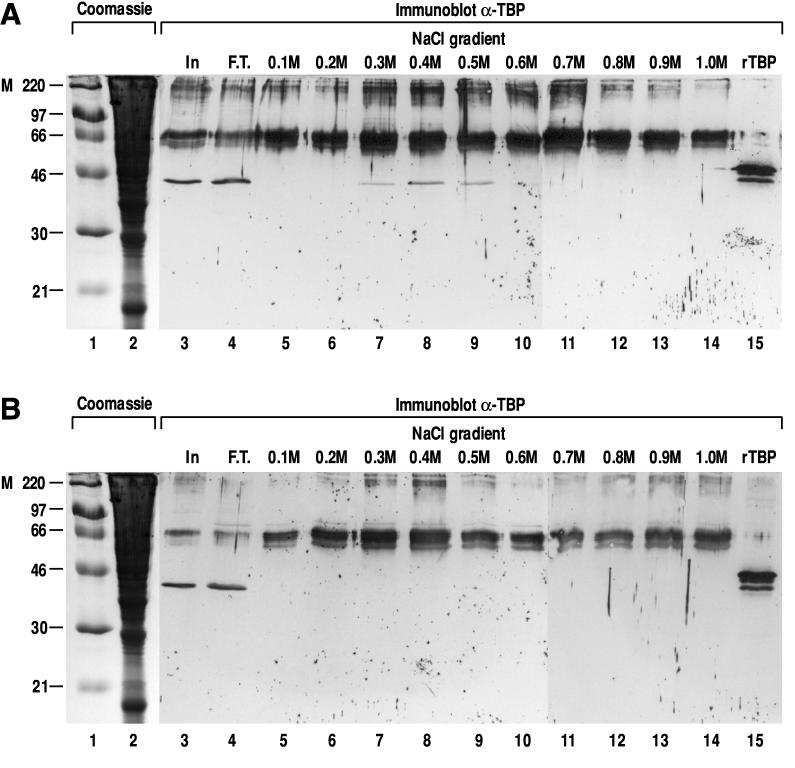
We also examined whether α-NAC/1.9.2 could interact with components of the general transcriptional machinery by performing affinity chromatography of crude nuclear extracts on a glutathione-Sepharose column loaded with recombinant GST-α-NAC/1.9.2 fusion protein, followed by immunoblotting of the step gradient eluate. The data presented in Fig. 5A show that TBP bound to the column. Testing for interactions between α-NAC/1.9.2 and other components of the basal transcriptional apparatus revealed that α-NAC/1.9.2 did not interact with RNA polymerase II, TFIIB, TFIIA, RAP30, RAP74, TAFII55, and TAFII70 (data not shown). As illustrated in Fig. 5A, native TBP was retained on the GST-α-NAC/1.9.2 affinity column and eluted with a peak in the 0.4 M salt fraction (lane 8), confirming the strength of the interaction between α-NAC/1.9.2 and TBP. The identity of the protein identified in the fractions from the column was ascertained with two different anti-TBP antibodies (not shown) as well as by running a control sample of purified recombinant human TBP (lane 15 [the murine protein appears to migrate slightly faster than the recombinant human homolog]). TBP did not bind to a control column loaded with the GST moiety alone (Fig. 5B), confirming the specificity of the observed interaction. Polypeptides in the range of 66 and 220 kDa were detected in every fraction eluted from the GST-α-NAC/1.9.2 affinity columns (Fig. 5A). These protein bands were also revealed by immunoblotting of the fractions eluted from the control GST column (Fig. 5B), demonstrating that they correspond to nonspecific interactions of abundant proteins with the GST moiety of the fusion protein, most likely detected as background by the alkaline phosphatase-conjugated secondary antibody.
FIG. 5.
Interactions between α-NAC/1.9.2 and TBP. (A) Affinity chromatography of crude nuclear extracts eluted with a salt step gradient. The immunoblot (lanes 3 to 15) was probed with the anti-TBP antibody. Recombinant human TBP (rTBP) was migrated in lane 15. (B) Control affinity chromatography column loaded with the GST moiety alone. The same crude nuclear extract used in panel A was passed through the column. Immunoblotting was with the anti-TBP antibody. In, input (1/100 of total material loaded on column); F.T., flowthrough. Lanes 1 and 2 contained Coomassie blue-stained molecular mass (kilodaltons) markers (M) and input material, respectively.
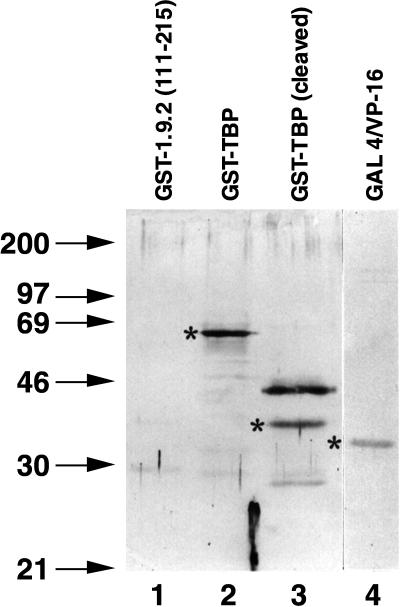
The results shown in Fig. 4 and 5 demonstrate strong protein-protein interactions between α-NAC/1.9.2 and the activator GAL4/VP-16, as well as between α-NAC/1.9.2 and TBP. Direct protein contacts were also detected with far-Western protein blotting assays with purified recombinant proteins. Figure 6, lane 4, shows that α-NAC/1.9.2 could interact with GAL4/VP-16 in the absence of DNA. The interaction was specific and was not mediated through the GST portion of the fusion protein, since GST-α-NAC/1.9.2 did not interact with GST-1.9.2(111–215), a fusion protein with an Mr of 39,400, which comprises the C-terminal half of α-NAC/1.9.2 fused to the DBD of GAL4 (Fig. 6, lane 1). GST-α-NAC/1.9.2 also failed to bind the DBD of GAL4 in far-Western assays (not shown). A direct interaction between GST-α-NAC/1.9.2 and GST-TBP or its cleaved TBP moiety (Fig. 6, lanes 2 and 3) was also detected by the far-Western assay.
FIG. 6.
Protein-protein interactions between α-NAC/1.9.2 and GAL4/VP-16 and between α-NAC/1.9.2 and TBP. Results of a far-Western protein blot assay show the interaction of biotinylated GST-1.9.2 with TBP (lanes 2 and 3) and GAL4/VP-16 (lane 4). The fusion protein GST-1.9.2(111–215) (Mr, 39,400) served as a negative control in this assay (lane 1). Asterisks point to the proper molecular size of GST-TBP (lane 2), cleaved TBP (lane 3), and GAL4/VP-16 (lane 4). Molecular mass (kilodaltons) markers are indicated in the left-hand lane.
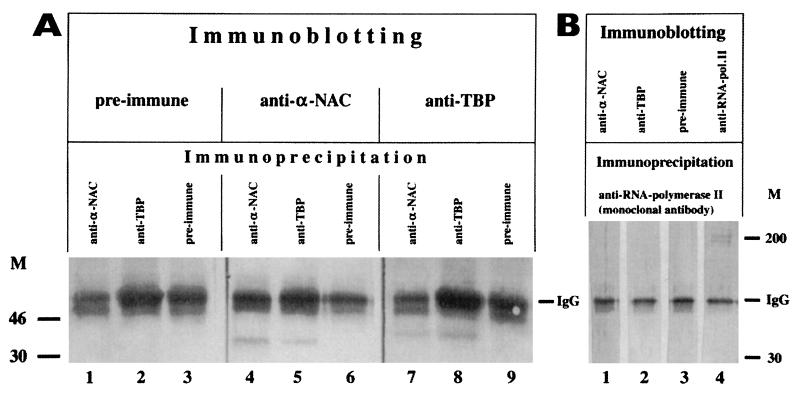
Immunoprecipitation experiments with crude nuclear extracts from serum-starved cells further established that α-NAC/1.9.2 is associated with TBP in vivo. Immunoprecipitation reactions were performed with anti-αNAC or anti-TBP antibodies, and the immunocomplexes were then analyzed by immunoblotting with the same antibodies. Preimmune sera and antibodies directed against the large subunit of RNA polymerase II served as controls in these experiments. Figure 7A shows that complexes immunoprecipitated with the anti-α-NAC/1.9.2 antibody contained both α-NAC/1.9.2 (lane 4) and TBP (lane 7). Conversely, immunocomplexes obtained with the anti-TBP antibody contained TBP (lane 8), TAFII55 and TAFII70 (not shown), as well as α-NAC/1.9.2 (lane 5). The specificity of these protein-protein interactions was demonstrated with antibodies directed against a different component of the basal transcriptional machinery, namely RNA polymerase II. The anti-RNA polymerase II antibody failed to coimmunoprecipitate α-NAC/1.9.2 (Fig. 7B, lane 1) or TBP (lane 2). Preimmune sera had no effect in these assays (Fig. 7A, lanes 1 to 3, 6, and 9; and B, lane 3).
FIG. 7.
α-NAC/1.9.2 is associated with TBP in vivo. Immunoprecipitation and immunoblotting assays demonstrating the specific interaction of α-NAC/1.9.2 with TBP in crude nuclear extracts. Immunoprecipitation reactions were performed with preimmune sera [lanes 3, 6, and 9]), anti-TBP antibodies (A [lanes 2, 5, and 8]), anti-αNAC antibodies (A [lanes 1, 4, and 7]), or anti-RNA polymerase II (pol. II) antibodies (B [lanes 1 to 4]). The immunocomplexes were then analyzed by immunoblotting with the antibodies indicated in the upper portion of each panel. TBP was specifically associated with the anti-αNAC immunocomplexes (A, lane 7). Similarly, α-NAC/1.9.2 was specifically associated with the anti-TBP immunocomplexes (A [lane 5]). M, molecular mass (kilodaltons) markers (M). IgG, immunoglobulin G.
These data show that α-NAC/1.9.2 specifically interacted with GAL4/VP-16 and TBP, resulting in enhanced transcriptional activation. We interpret these results to mean that α-NAC/1.9.2 acted as a transcriptional coactivator.
DISCUSSION
We have shown that clone 1.9.2 corresponds to α-NAC, a heterodimeric complex previously implicated in binding nascent polypeptide chains as they emerge from the ribosome (37). Based on sequence similarity with DNA binding proteins and the identification of the β-NAC dimerization partner as the transcription factor BTF3b, we have examined the possibility that α-NAC/1.9.2 might participate in transcriptional control. These studies were also prompted by our previous observations that the muscle-specific isoform of α-NAC/1.9.2, skNAC, acts as a DNA-binding transcription factor (38). We have shown that α-NAC/1.9.2 performs protein-protein interactions with TBP and the chimeric transcriptional activator GAL4/VP-16. Transient transfection assays have revealed that α-NAC/1.9.2 acts as a coactivator to enhance activated transcription. Considering the apparent dual function of the protein, we propose a different meaning for the α-NAC acronym: nascent-polypeptide-associated complex and coactivator alpha.
Our data support the role of α-NAC/1.9.2 as a transcriptional coactivator. Previous results from our laboratory have shown that recombinant α-NAC/1.9.2 can bind DNA with sequence specificity in vitro (38). However, the protein does not appear to contain an intrinsic activation domain. This fact is supported by the following observations. (i) We could not demonstrate transcriptional activation by α-NAC/1.9.2 through GAL4 binding sites when full-length α-NAC/1.9.2 was expressed as a fusion protein with the DBD of GAL4 (Fig. 2). (ii) α-NAC/1.9.2 also failed to activate transcription from promoter templates containing its cognate response element (38). Thus, the transcriptional role of α-NAC/1.9.2 is more consistent with the function of a coactivator than a site-specific transcription factor. We have observed, however, that the C-terminal domain of α-NAC/1.9.2 (residues 111 to 215), when fused to GAL4, could moderately increase transcription from a GAL4-dependent reporter template (about threefold) in a subset of bone cell lines (not shown). A possible explanation for this result would be that the C-terminal domain of α-NAC/1.9.2 is the portion of the molecule that contacts TBP. It has been shown that protein-protein interactions with TBP are sufficient to activate transcription (18). The apparent specificity of this response might be due to the relative amount of TBP available to interact with α-NAC/1.9.2 in particular cell types.
It remains to be determined if the DNA binding function of α-NAC/1.9.2 is expressed or if it is masked in vivo. Interestingly, the simian virus 40 large T antigen, a known DNA binding protein, has recently been shown to function as a TAF and to interact with TBP (6). A possible role for a specific DNA binding function within a coactivator molecule would be to target it to particular promoters (12, 34), thus generating an increased degree of specificity to the transcriptional response to α-NAC/1.9.2 during development.
The interaction between VP16 and the transcriptional machinery is highly specific: a phenylalanine-to-proline substitution at position 442 of VP16 (VP16ΔFP442) abolishes transcriptional activity in vivo and correspondingly decreases binding of VP16 to TBP and TFIIB in vitro (5, 16, 21). Similarly, the GAL4/VP16ΔFP442 fusion protein is transcriptionally compromised regarding GAL4 binding site-dependent transcriptional activation of reporter genes both in vivo and in vitro (1). The interaction between GAL4/VP16ΔFP442 and the ADA5 transcriptional coactivator is accordingly weakened (23). The weaker interaction observed between GAL4/VP16ΔFP442 and α-NAC/1.9.2 (Fig. 4), combined with the stimulation of reporter gene mRNA synthesis and the lack of effect on the half-life of the activator (Fig. 2B and C, respectively), demonstrates that α-NAC/1.9.2 potentiates GAL4/VP-16-dependent activation at the transcriptional level and confirms our interpretation that α-NAC/1.9.2 functions as a transcriptional coactivator.
The α-NAC/1.9.2 protein shares several functional features with the previously characterized TAF class of transcriptional coactivators. The coimmunoprecipitation data presented in Fig. 7 raise the question as to why α-NAC/1.9.2 has not been previously identified as a component of the multisubunit TFIID complex. The most likely possibility is related to the fact that we observed nuclear localization of α-NAC/1.9.2 only when cells were starved and arrested in their cell cycle progression (Fig. 1). Nuclear extracts are normally prepared from cycling, serum-fed cells, and the amounts of nuclear α-NAC/1.9.2 complexed to TBP are probably undetectable under those conditions.
The α-NAC/1.9.2 protein is most abundant in the nucleus of cells arrested at the G0/G1 border of the cell cycle. Terminally differentiated cells are postmitotic; however, this does not mean that they are transcriptionally inactive. Postmitotic neurons, for example, can respond to a variety of signals by the induction of new programs of gene expression (30). Thus, the observation that α-NAC/1.9.2 localizes to the nucleus mostly in noncycling cells is not contradictory to a role in transcriptional regulation. We have initiated experiments aimed at defining the signalling events involved in the subcellular distribution of α-NAC/1.9.2.
It is difficult to reconcile the previously proposed role for α-NAC/1.9.2 in the binding of nascent chains as they emerge from the ribosome (37) with the data presented herein. A similar conflict has recently been described for the yeast TAFII30 gene, Tfg3, which has been shown to be a component of the SWI/SNF complex as well as the TFIIF and TFIID transcription complexes (3), but was initially identified as ANC1, a gene implicated in cytoskeletal function (14).
NAC has been purified as a heterodimeric protein (37). It is conceivable, although unlikely, that the heterodimeric form of NAC is implicated in protein translation while the α-NAC/1.9.2 monomer is implicated in transcriptional control. Indeed, cotransfection of the CMV-1.9.2 vector with a BTF3b expression vector (kindly supplied by J.-M. Egly) inhibited the coactivation function of α-NAC/1.9.2 (Fig. 2). This observation is in accord with a recently reported inhibitory role of yeast BTF3b in the transcription of various yeast genes (15). Several parameters could modulate the association of α-NAC/1.9.2 with BTF3b. First, we have found evidence of posttranslational modification of the protein in the form of variable phosphorylation (not shown). Second, the subcellular localization of α-NAC/1.9.2 is modulated during the cell cycle (Fig. 1), suggesting that α-NAC/1.9.2 may only be available to interact with BTF3b during certain phases of the cycle. Finally, we have observed that the levels of α-NAC/1.9.2 mRNA are far more abundant than the levels of BTF3b transcripts in bone cells (not shown). Thus, the possible stoichiometric excess of α-NAC/1.9.2 over BTF3b may leave a proportion of α-NAC/1.9.2 molecules unassociated with BTF3b and able to exert its coactivation function. Additional work will be required to reconcile the transcriptional and putative translational regulatory activities of α-NAC/1.9.2.
ACKNOWLEDGMENTS
W.V.Y. and A.M. participated equally in this study and should both be considered first authors.
We express our gratitude to the following investigators for their generous gift of plasmids or reagents: Michael Green (5Gal4-E1b-CAT), Mark Ptashne (pSGVP), James T. Kadonaga (purified GAL4/VP-16 fusion protein), Robert Tjian (pSV6tkCAT), Jean-Marc Egly (BTF3b), and Steven J. Triezenberg (GAL4/VP16ΔFP442). We thank Edwin Wan for oligonucleotide synthesis and Jane Wishart and Mark Lepik for preparing the figures.
This work was supported by a grant from the Shriners of North America. W.V.Y. was a Shriners’ Fellow, and R.S.-A. is a Chercheur-Boursier from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 2.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 3.Cairns B R, Henry N L, Kornberg R D. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J-L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 5.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 6.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 7.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 9.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 10.Ecarot-Charrier B, Glorieux F H, van der Rest M, Pereira G. Osteoblasts isolated from mouse calvaria initiate matrix mineralization in culture. J Cell Biol. 1983;96:639–643. doi: 10.1083/jcb.96.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher S, Winston S E, Fuller S A, Hurrell J G R. Immunoblotting and immunodetection. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1993. pp. 10.8.1–10.8.17. [Google Scholar]
- 12.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 13.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 14.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 15.Hu G-Z, Ronne H. Yeast BTF3 protein is encoded by duplicated genes and inhibits the expression of some genes in vivo. Nucleic Acids Res. 1994;22:2740–2743. doi: 10.1093/nar/22.14.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 17.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 18.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 19.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 20.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y-S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 22.Maheswaran S, Inglert C, Bennett P, Heinrich G, Haber D A. The WT1 gene product stabilizes p53 and inhibits p53-mediated apoptosis. Genes Dev. 1995;9:2143–2156. doi: 10.1101/gad.9.17.2143. [DOI] [PubMed] [Google Scholar]
- 23.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBurney M W. P19 embryonal carcinoma cells. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- 25.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 26.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sauer F, Wassarman D A, Rubin G M, Tjian R. TAFIIs mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 30.Sheng M, Greenberg M E. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 31.St-Arnaud R, Moir J M. Wnt-1-inducing factor-1: a novel G/C box-binding transcription factor regulating the expression of Wnt-1 during neuroectodermal differentiation. Mol Cell Biol. 1993;13:1590–1598. doi: 10.1128/mcb.13.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudo H, Kodama H-A, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tansey W P, Herr W. TAFs: guilt by association? Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 34.Verrijzer C P, Yokomori K, Chen J-L, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 35.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 36.Watkins S. Immunohistochemistry. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. pp. 14.6.1–14.6.13. [Google Scholar]
- 37.Wiedmann B, Sakai H, Davis T A, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 38.Yotov W V, St-Arnaud R. Differential splicing-in of a proline-rich exon converts αNAC into a muscle-specific transcription factor. Genes Dev. 1996;10:1763–1772. doi: 10.1101/gad.10.14.1763. [DOI] [PubMed] [Google Scholar]
- 39.Zheng X M, Black D, Chambon P, Egly J M. Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature. 1990;344:556–559. doi: 10.1038/344556a0. [DOI] [PubMed] [Google Scholar]