Abstract
The alpha chain of the nascent polypeptide-associated complex (α-NAC) coactivator was shown to potentiate the activity of the homodimeric c-Jun activator, while transcription mediated by the c-Fos/c-Jun heterodimer was unaffected. The use of deletion mutants in pull-down assays revealed that α-NAC interacted with amino acids 1 to 89 of the c-Jun protein and that the coactivator could interact with both the unphosphorylated and the serine 73-phosphorylated form of c-Jun. N-terminal-deleted c-Jun protein failed to interact with α-NAC in mammalian two-hybrid assays, while mutant c-Jun proteins lacking the leucine zipper or the basic domain retained interaction with α-NAC in vivo. Kinetics studies with purified c-Jun homodimer and recombinant α-NAC proteins allowed determination of the mechanism of coactivation by α-NAC: the coactivator stabilized the AP-1 complex formed by the c-Jun homodimer on its DNA recognition sequence through an eightfold reduction in the dissociation constant (kd) of the complex. This effect of α-NAC was specific, because α-NAC could not stabilize the interactions of JunB or Sp1 with their cognate binding sites. Interestingly, the expression of α-NAC was first detected at 14.5 to 15 days postconception, concomitantly with the onset of ossification during embryogenesis. The α-NAC protein was specifically expressed in differentiated osteoblasts at the centers of ossification. Thus, the α-NAC gene product exhibits the properties of a developmentally regulated, bone-specific transcriptional coactivator.
Tissue-specific gene expression is regulated by a combination of sequence-specific DNA-binding transcriptional activators, general or basal transcription initiation factors, and associated cofactors. The sequence-specific transcription factors can be divided into several classes or families based on either the activation domain they possess or the protein motif they use for DNA binding. In addition to these site-specific proteins, accurate transcription initiation by RNA polymerase II requires at least six general transcription initiation factors: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (reviewed in references 16 and 29).
While the general initiation factors are sufficient for basal-level transcription, enhancement of transcription by transcriptional activator proteins bound to enhancer elements requires the presence of additional mediator proteins, known as transcriptional coactivators (12, 16, 19, 38). Coactivators are defined functionally by their ability to selectively potentiate the stimulatory activity of specific subsets of enhancer binding transcriptional activators. Among the best-characterized coactivators are the TAFs (TATA box-binding protein [TBP]-associated factors) (12), which are subunits of TFIID. Purification and molecular cloning of several TAFs have confirmed that they provide protein interfaces to link the sequence-specific factors to the basal transcriptional machinery, and some exhibit specific enzymatic functions essential for activated gene transcription (11, 27, 28). Moreover, coactivators distinct from those associated with TBP have also been identified and cloned (reviewed in reference 19).
We have recently demonstrated that the alpha chain of the nascent polypeptide-associated complex (α-NAC) protein, previously thought to be involved in some aspects of translational control (41), could translocate to the nucleus, where it was shown to function as a transcriptional coactivator in conjunction with the chimeric activator GAL4/VP-16 in vivo (43a). Specific interactions between α-NAC and TBP were detected. Examination of the expression pattern of α-NAC in adult tissues allowed us to identify and characterize a muscle-specific isoform of α-NAC, skNAC, also involved in the regulation of gene transcription (43). In this report, we have examined the expression pattern of α-NAC during embryogenesis and observed that it was selectively expressed in developing bone.
In an attempt to identify endogenous transcriptional activators that could interact with α-NAC to regulate gene transcription in differentiating bone cells, we examined the possibility that AP-1 proteins, known modulators of bone development in vivo (17), might represent natural targets for α-NAC. The AP-1 proteins are formed through the heterodimerization of Fos family members and Jun family members through a structural motif called the leucine zipper (30). The heterodimer can then bind DNA at a consensus site termed the AP-1 site and act as a transcription factor to modulate the expression of AP-1-responsive genes (30). All of the Jun family members can also homodimerize to exert the same function (30).
Interestingly, α-NAC was shown to potentiate the activity of the homodimeric c-Jun activator, while transcription mediated by the c-Fos/c-Jun heterodimer was unaffected. We have delineated the domain of the c-Jun protein that interacts with α-NAC and measured the influence of α-NAC on kinetic parameters of c-Jun binding to AP-1 sites. These observations have allowed us to propose a model for the α-NAC-mediated coactivation of c-Jun-dependent transcription. The observed restricted expression pattern of α-NAC during embryogenesis and the identification of its interaction with c-Jun suggest new perspectives in the study of the regulation of gene transcription during osteoblastic differentiation.
MATERIALS AND METHODS
Northern blot assay.
The commercially available mouse embryo Northern blot (Clontech Laboratories, Palo Alto, Calif.) was probed with the full-length, PCR-labeled α-NAC cDNA. The membrane was subsequently stripped and rehybridized with a probe directed against the S28 ribosomal protein mRNA to monitor for variations in the quality and quantity of the loaded mRNA.
Immunolocalization of α-NAC.
Embryos were dissected, embedded in Tissue-Tek O.C.T. compound (Miles, Inc., Elkhart, Ind.), frozen, and sectioned at 6 μm. Immunohistochemistry was performed according to standard protocols (40) with the anti-αNAC antibody (43) and a secondary goat anti-rabbit antibody conjugated to fluorescein isothiocyanate (TAGO Immunologicals, Burlingame, Calif.). The antibody dilutions were 1:1,000 and 1:50 for the primary and secondary antibodies, respectively. Counterstaining of the embryos was with toluidine blue.
Transient transfection assays.
All vectors were constructed by standard molecular biology procedures, and full details and sequences are available on request. C3H10T1/2 embryonic fibroblasts (39) were cotransfected with 0.1 μg of a reporter vector (AP-1-tk-luc) in which a canonical AP-1 binding site (5′-TGACTCA-3′) was subcloned upstream of the minimal herpes simplex virus thymidine kinase promoter driving the luciferase reporter gene together with 0.7 μg of cytomegalovirus (CMV)-driven expression vectors for c-fos (8), c-jun, or α-NAC, alone or in combination. Variations in transfection efficiency were monitored with 0.1 μg of the expression plasmid pSV6tkCAT (7).
For mammalian two-hybrid assays, ROS 17/2.8 osteoblastic cells (24) were cotransfected with combinations of simian virus 40-driven expression vectors for the GAL4 DNA binding domain (GAL4-DBD) (0.1 μg) (32) or the GAL4-α-NAC fusion protein (0.1 μg) and the jun and fos expression vectors (in increasing amounts from 0.1 to 0.4 μg). The reporter gene used was 5Gal4-E1b-CAT (0.1 μg) (22), and transfection efficiency was monitored with the luciferase vector pGL2control (Promega Corp., Madison, Wis.). Similar levels of expression were achieved for each recombinant protein (data not shown).
All transfections were performed with 5 μl of the Lipofectamine reagent (Gibco BRL, Gaithersburg, Md.) according to the instructions of the manufacturer, and cells were harvested 24 h posttransfection. Chloramphenicol acetyltransferase (CAT) and luciferase activities were assayed as previously described (34).
GST pull-down assays.
Pull-down assays were performed essentially as described by Folkers and van der Saag (13) with Promega’s TNT in vitro transcription and translation kit in the presence of [35S]methionine. In vitro-transcribed and -translated plasmids included pSP65-c-fos (8), Jac7 (c-jun cDNA cloned into pGEM-7 [31]), pT7-GAL4/VP-16 (in which the GAL4/VP-16 fusion cDNA was subcloned into the GPP-73 vector [37]), and the control plasmid T7-luciferase supplied with the TNT kit (Promega). For pull-down assays with deleted c-Jun fusion protein, glutathione-Sepharose beads loaded with glutathione S-transferase (GST)–jun(1–89) were purchased from New England Biolabs (Mississauga, Ontario, Canada) and phosphorylated with serum-induced whole-cell extracts from P19 embryonal carcinoma cells (25) according to the protocol supplied by the manufacturer. Binding and washes were performed as recommended with the buffers supplied. The anti-GST antibody was from Pharmacia (Baie d’Urfé, Quebec, Canada).
Affinity chromatography.
Crude nuclear extracts from serum-stimulated MC3T3-E1 cells (36) were prepared as described by Dignam et al. (10). The nuclear extracts (2 mg of total protein) were precleared on glutathione-Sepharose 4B beads (Pharmacia) and then passed on a glutathione-Sepharose 4B column previously loaded with GST-α-NAC (300-μl final bed volume). Bound proteins were eluted with a step gradient of NaCl, precipitated with trichloroacetic acid, and analyzed by Western blot assay (15). The antibodies directed against the serine 73-phosphorylated form of c-Jun were obtained from New England Biolabs.
On and off rate measurements.
Electrophoretic mobility shift assays were performed as described previously (34), with the exception that binding reactions were carried out at 4°C in a total volume of 180 μl. Commercially available purified c-Jun and Sp1 protein (estimated at 7 × 10−8 M, final concentration) (Promega Corporation), in vitro-translated JunB protein, and purified recombinant GAL4/VP-16 and α-NAC protein (3 × 10−7 M, final concentration) were used in the binding assays. Twenty-microliter aliquots were removed at intervals and loaded on the gels. Binding reactions and calculations were as described by Chodosh et al. (5). The probes used were end-labeled oligonucleotides corresponding to the AP-1 site from the hMT-IIA promoter (21), the canonical GAL4 binding site (22), or the consensus Sp1 DNA recognition sequence (3a) (each estimated at around 2 × 10−7 M, final concentration).
RESULTS
α-NAC is specifically expressed in bone during development.
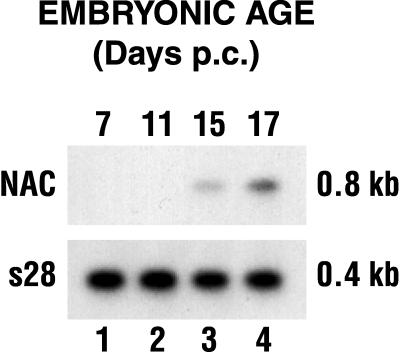
We used both Northern blot assays and immunohistochemistry to address the tissue specificity and developmental regulation of the expression of α-NAC. Figure 1 shows that the α-NAC mRNA could not be detected in poly(A)+ RNA isolated from 7- and 11-day-postconception (p.c.) whole mouse embryos. However, the α-NAC transcript was readily detected in mouse embryos at 15 and 17 days p.c. (Fig. 1). Expression of the α-NAC mRNA was widespread in the adult mouse (43).
FIG. 1.
Expression pattern of α-NAC mRNA during mouse embryogenesis. Whole mouse embryo mRNA was probed with the full-length α-NAC cDNA in a Northern blot assay. The membrane was subsequently stripped and rehybridized with a probe directed against the S28 ribosomal protein.
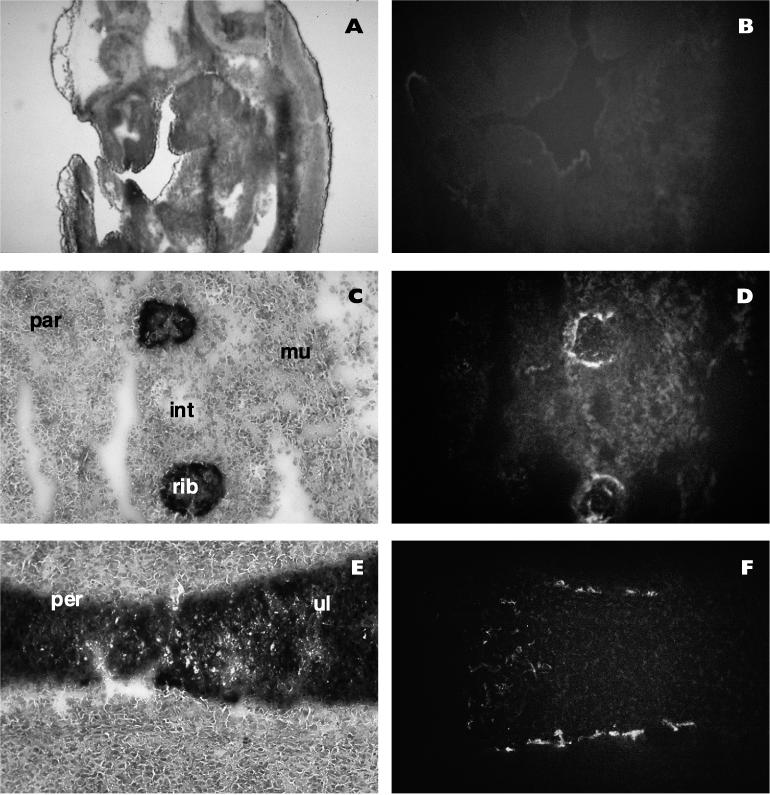
We next studied the pattern of expression of the α-NAC protein during mouse development. Confirming the results obtained with Northern blot assays (Fig. 1), we could not detect the α-NAC protein prior to 14 days p.c. (Fig. 2B and data not shown). The onset of mineralization is first detected at 14 to 14.5 days p.c. (20); the ribs, for example, show evidence of endochondral ossification (Fig. 2C). Ossification of the limbs also begins at the same time, and the primary ossification centers of long bones can be observed along the periosteum in the midshaft region (Fig. 2E) (20). The α-NAC protein was specifically detected in the nucleus of differentiated osteoblasts in the primary ossification centers of ribs and long bones at 14.5 days p.c. (Fig. 2D and F). No signal was detected in other tissues, as evidenced by the absence of staining in the tissue flanking the ribs in Fig. 2C and D, and control staining with preimmune sera gave no signal (not shown). This specific pattern of expression was maintained until birth; postnatally, expression of the α-NAC protein was detectable in other tissues (43).
FIG. 2.
The α-NAC protein is specifically expressed in mineralizing bone during development. Cryosections of embryos at various developmental ages were probed with the anti-α-NAC antibody and fluorescein isothiocyanate-conjugated secondary antibody. (A, C, and E) Phase-contrast photomicrographs of the toluidine blue-stained sections. (B, D, and F) Indirect fluorescence photomicrographs. (A and B) Sagittal section from a 12-day-p.c. embryo. No detectable signal could be observed in any field examined. (C and D) Parasagittal section from a 14.5-day-p.c. embryo showing ossification of the ribs and expression of α-NAC in differentiated osteoblasts. (E and F) Longitudinal section of the ulna from a 14.5-day-p.c. embryo. The ossification center is in the midshaft region and corresponds to the area of expression of α-NAC. Par, panniculus carnosus (cutaneous muscle of the trunk); int, intercostal muscles; mu, muscle; per, periosteum; ul, ulna.
The AP-1 c-Jun homodimer may be a physiological target for α-NAC.
The tissue-restricted pattern of expression observed for α-NAC during development prompted us to search for transcriptional activators implicated in the control of gene expression during osteoblastic differentiation that could represent natural targets for the α-NAC transcriptional coactivation function. Considering the large body of experimental evidence demonstrating that the AP-1 transcription factor plays a key role in the regulation of bone tissue metabolism (17), we tested for a possible interaction between AP-1 family members and α-NAC. The coactivation function of α-NAC was tested with both the c-Fos/c-Jun heterodimer and the c-Jun homodimer.
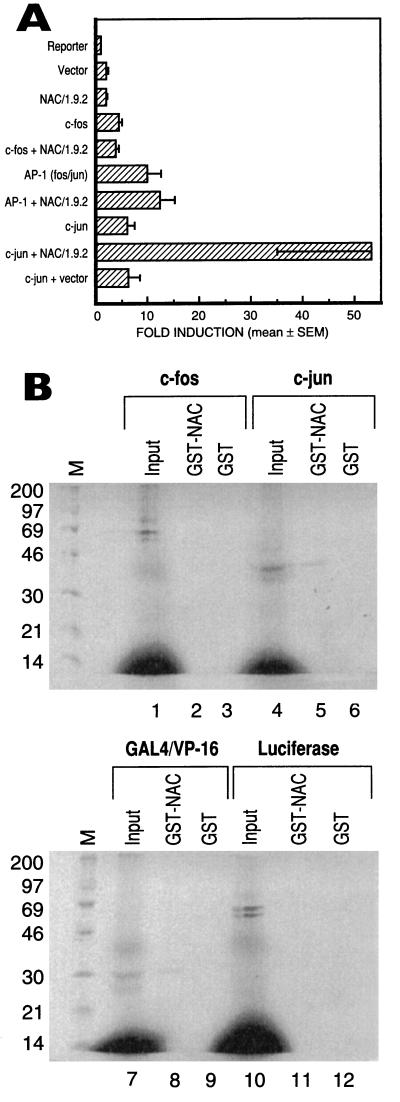
As shown in Fig. 3A, α-NAC and c-Fos, alone or in combination, had no effect on the expression of a luciferase reporter gene under the control of a single AP-1 binding site. The AP-1 Fos/Jun heterodimer stimulated the expression of the reporter gene by about 10-fold; however, α-NAC had no effect on the expression levels driven by the heterodimer. The c-Jun homodimer also stimulated the transcription of the luciferase reporter; mean fold induction levels were measured as 6.2 ± 1.4 (mean ± standard error). Interestingly, α-NAC potentiated the c-Jun-mediated transcriptional induction by a further ninefold, to 53.1 ± 18.0. Similar levels of expression were achieved for each recombinant protein (data not shown).
FIG. 3.
α-NAC interacts with c-Jun to potentiate c-Jun-mediated transcription. (A) Transient transfection assays with expression vectors for c-fos, c-jun, and α-NAC, alone or in combination. The expression level detected in cells transfected with the reporter alone (AP-1-tk-luc) was arbitrarily ascribed a value of 1. Results are expressed as mean fold induction ± standard error of four independent transfections. (B) Pull-down protein-protein interaction assays. [35S]methionine-labeled in vitro-translated proteins, identified above each panel, were incubated with the fusion GST-α-NAC protein or the free GST moiety. Input represents 1/10 of the total input of in vitro-translated protein. M, molecular mass (kilodaltons) markers.
We next tested for specific protein-protein interactions between α-NAC and AP-1 family members by using the protein pull-down assay. As demonstrated in Fig. 3B, lane 5, α-NAC interacted with the c-Jun protein. The interaction was specific, because the GST moiety of the fusion GST-NAC protein did not bind c-Jun (lane 6). No interactions were observed between α-NAC and the monomeric c-Fos protein (lanes 1 to 3). The luciferase and GAL4/VP-16 proteins served as negative and positive controls for the pull-down assay, respectively (lanes 7 to 12).
α-NAC interacts with the N terminus of c-Jun.
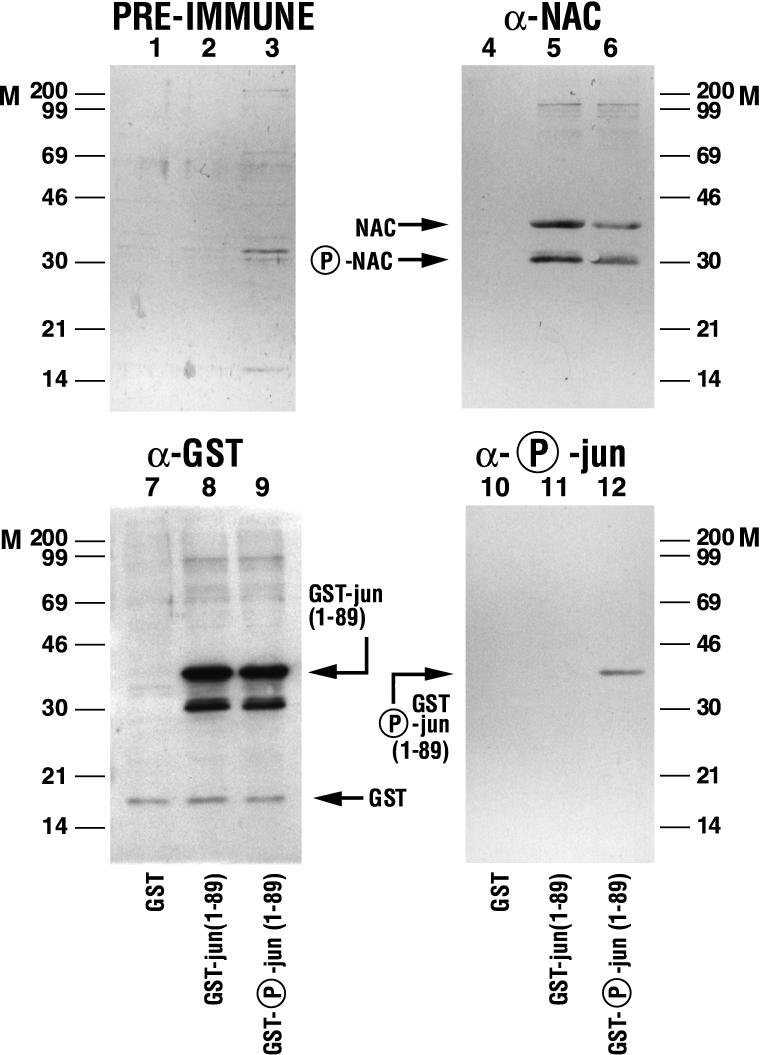
A deletion mutant of c-Jun fused to GST [GST-jun(1–89)] was utilized in pull-down protein interaction assays to map the domain of c-Jun interacting with α-NAC. The GST-jun(1–89) fusion protein phosphorylated on the serine residue at position 73 by the specific action of the JNK kinase (9) was also tested in these assays. As shown in Fig. 4, lane 5, α-NAC interacted with GST-jun(1–89), demonstrating that the N terminus of the c-Jun protein is involved in the binding of c-Jun to α-NAC. Two forms of the α-NAC protein were detected by the anti-α-NAC antibody in these interaction assays (lanes 5 and 6); treatment of nuclear extracts from bone cells with protein phosphatases prior to immunoblotting with the anti-αNAC antibody eliminated the protein form showing higher electrophoretic mobility (data not shown), suggesting that it represents a phosphorylated form of α-NAC.
FIG. 4.
α-NAC interacts with the N terminus of c-Jun. Pull-down protein interaction assays were performed with the phosphorylated or unphosphorylated GST-jun(1–89) fusion protein. Crude cell extracts from P19 embryonal carcinoma cells were incubated with glutathione-Sepharose beads loaded with the GST fusion proteins or the negative control GST moiety. Following binding, centrifugation, and washes, the bound proteins were analyzed by immunoblotting with anti-α-NAC (lanes 4 to 6), anti-GST (lanes 7 to 9), or anti-phospho-Ser 73 (lanes 10 to 12) antibodies. Background staining was assessed with preimmune sera (lanes 1 to 3). M, molecular mass (kilodaltons) markers.
The c-Jun protein must be phosphorylated at positions 63 and 73 to become transcriptionally active (9). Interestingly, α-NAC was shown to interact with both the phosphorylated and nonphosphorylated forms of c-Jun (Fig. 4, lanes 5 and 6). The observed interactions were specific because the cellular α-NAC proteins were not pulled down by the GST moiety of the GST-jun(1–89) fusion protein (lane 4). The amounts of fusion protein loaded in the pull-down assays were monitored with an anti-GST antibody (lanes 7 to 9), while the specificity of the phosphorylation of GST-jun(1–89) by the JNK kinase (9) was assessed with an antiphosphoserine 73 antibody (lanes 10 to 12). Preimmune sera did not detect any significant cross-reactivity of the secondary antibodies used in the immunodetection (lanes 1 to 3).
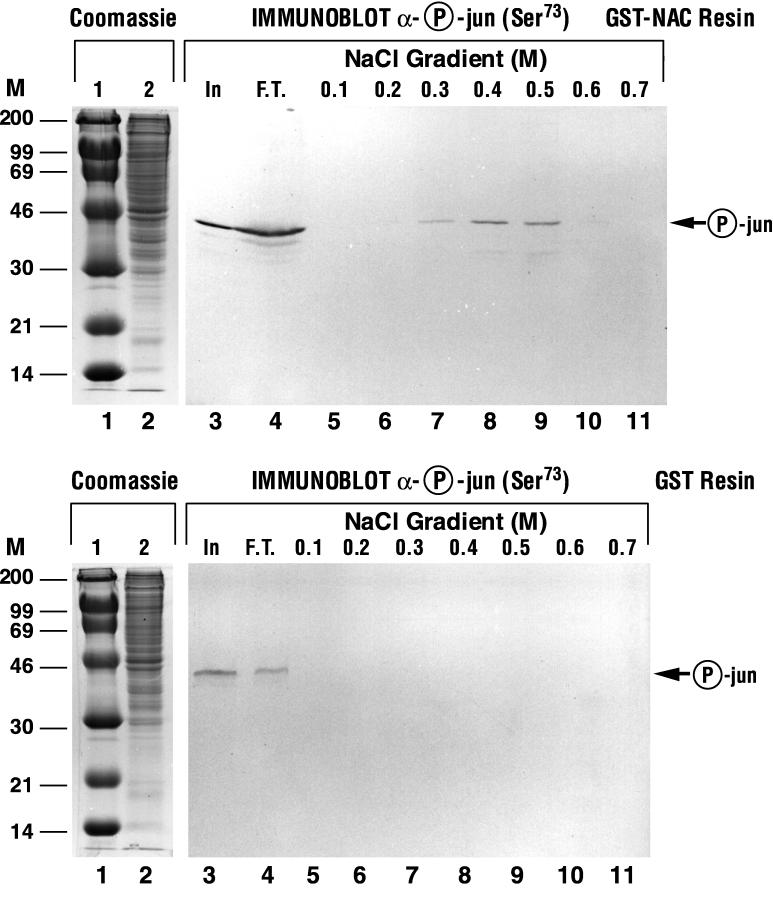
The strength of the interaction between α-NAC and phospho-c-Jun was next assessed by performing affinity chromatography of crude nuclear extracts from serum-stimulated osteoblasts on a glutathione-Sepharose column loaded with recombinant GST-α-NAC fusion protein, followed by immunoblotting of the step gradient eluate with the antiphosphoserine 73 antibody. As illustrated in Fig. 5, upper panel, the native phosphorylated c-Jun protein was retained on the GST-α-NAC affinity column, and eluted with a peak in the 0.5 M salt fraction (lane 9), confirming the strength of the interaction between α-NAC and phospho-c-Jun. The phosphorylated c-Jun did not bind to a control column loaded with the GST moiety alone (Fig. 5, lower panel).
FIG. 5.
Binding between α-NAC and the phosphorylated form of c-Jun is a strong interaction. (A) Affinity chromatography on a GST-α-NAC column. The recombinant GST-α-NAC protein was immobilized on a glutathione-Sepharose column. A nuclear extract from serum-stimulated MC3T3-E1 osteoblastic cells was passed through the column, allowing specific interactions between α-NAC and nuclear proteins to occur. Bound proteins were eluted with a step gradient of salt and detected by Western blotting with a monoclonal antibody directed against the phosphorylated form of c-Jun. Note that bound phospho-c-Jun elutes at 0.4 to 0.5 M salt. (B) The same nuclear extract was passed through a control GST column. Molecular mass (kilodaltons) markers (M) are indicated to the left. In, input protein; F.T., flowthrough; lanes 1 and 2, Coomassie blue-stained molecular size markers and input material, respectively.
α-NAC and c-Jun interact in vivo.
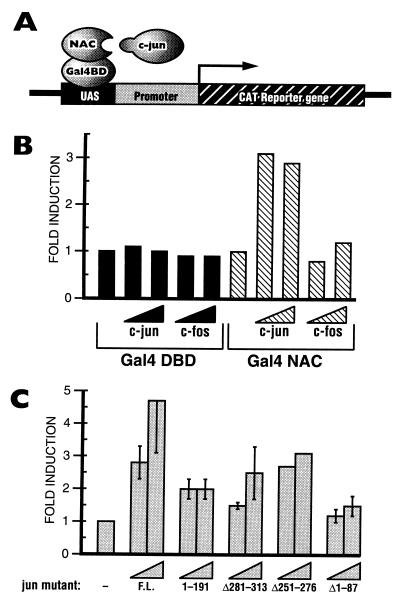
We performed mammalian two-hybrid assays (Fig. 6A) in order to confirm that c-Jun interacts with α-NAC in vivo and to test the specificity of this interaction. The full-length α-NAC cDNA was cloned in frame with the yeast GAL4-DBD and transfected in ROS 17/2.8 osteoblastic cells (24), together with a GAL4-dependent reporter construct and expression vectors for AP-1 family members. A GAL4-DBD expression vector was used as a control in these experiments. As illustrated in Fig. 6B, c-Jun and c-Fos did not interact with the GAL4-DBD. Moreover, the GAL4-α-NAC fusion protein did not significantly influence the expression of the reporter gene, confirming our previous observations that α-NAC does not possess an intrinsic activation domain (43, 43a).
FIG. 6.
α-NAC binds the c-Jun N terminus in vivo. (A) Schematic representation of the mammalian two-hybrid assay. (B) ROS 17/2.8 osteoblastic cells were transfected with the expression vectors for the various proteins mentioned. The expression level detected in cells transfected with the reporter and the GAL4 NAC vector was arbitrarily ascribed a value of 1. The c-Jun and c-Fos proteins did not interact with the GAL4-DBD. The fusion protein GAL4 NAC, which is transcriptionally inert, did not bind c-Fos; however, the interaction of c-Jun with α-NAC tethers the c-Jun activation domain to the GAL4-dependent promoter and results in the expression of the reporter gene. (C) The mammalian two-hybrid assay was performed with c-Jun deletion mutants. The expression level detected in cells transfected with the reporter and the GAL4 NAC vector was arbitrarily ascribed a value of 1. F.L., full-length c-Jun protein; 1–191, a truncated form of c-Jun in which the C-terminal domain was deleted; Δ281–313, leucine zipper deletion mutant; Δ251–276, basic domain deletion mutant; Δ1–87, N-terminal deletion mutant.
The expression of the reporter gene was stimulated when an expression vector for c-Jun was coexpressed with the GAL4-α-NAC fusion protein (Fig. 6B). This result confirms the interaction of the c-Jun protein with α-NAC in vivo, which tethers the c-Jun activation domain (1) to the GAL4-dependent promoter and allows transcription of the reporter gene. As previously observed in vitro (Fig. 3B), the c-Fos protein did not bind the GAL4-α-NAC fusion and did not activate the expression of the reporter construct (Fig. 6B).
We also tested c-Jun deletion mutants in the mammalian two-hybrid assay (Fig. 6C). As observed previously, the full-length c-Jun protein interacted with the GAL4-α-NAC fusion (Fig. 6C, F.L.). Several c-Jun mutant proteins also bound GAL4-α-NAC in this assay, leading to the activation of the reporter gene in the range of two- to threefold (Fig. 6C): a truncated c-Jun comprising only the N-terminal region (amino acids 1 to 191 [1–191]), a leucine zipper deletion mutant (Δ281–313), and a basic domain deletion mutant (Δ251–276). A mutation deleting the first 87 amino acids of the protein prevented the interaction of c-Jun with GAL4-α-NAC, resulting in expression levels of the reporter gene that did not exceed 1.5-fold over those of the control (Fig. 6C, Δ1–87). These in vivo results corroborate the data obtained with the in vitro pull-down protein interaction assays where the domain of the c-Jun protein interacting with the α-NAC coactivator was identified within the N-terminal amino acids region of residues 1 to 89 (Fig. 4).
α-NAC stabilizes the binding of the c-Jun homodimer to the AP-1 site.
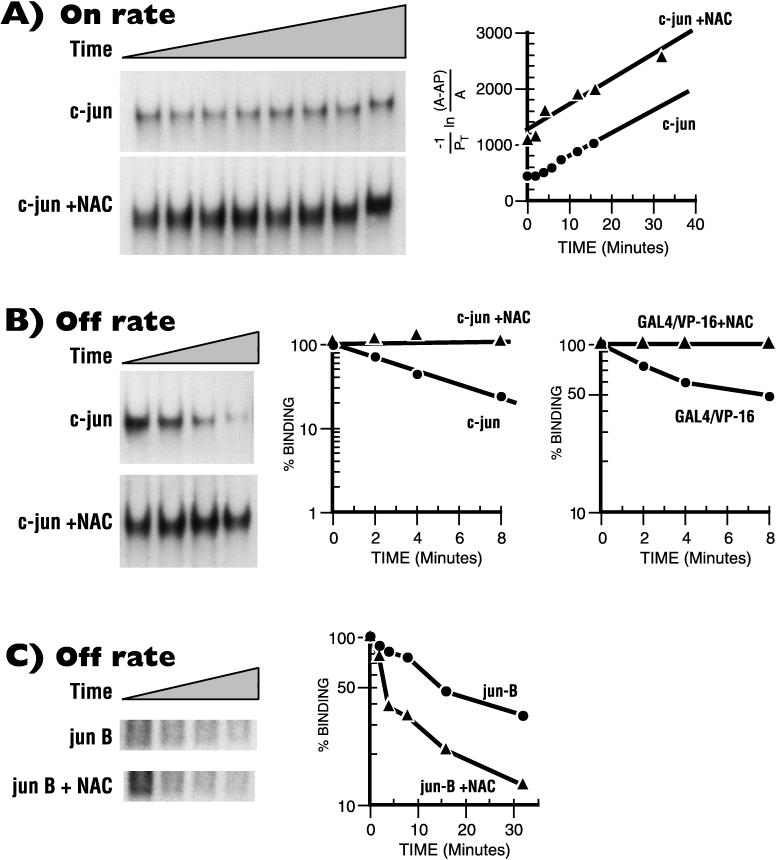
In order to gain further understanding of the mechanism of α-NAC-mediated potentiation of c-Jun-activated transcription, we measured kinetic parameters of the c-Jun interaction with its cognate AP-1 binding site in the presence or absence of α-NAC by using the electrophoretic gel mobility shift assay. α-NAC did not influence the on rate of association of the c-Jun homodimer with the AP-1 recognition sequence (Fig. 7A). However, α-NAC significantly affected the off rate of c-Jun from its DNA binding site (Fig. 7B), thus dramatically stabilizing the AP-1 complex formed by the c-Jun homodimer. In three independent experiments, α-NAC caused, on average, an eightfold reduction in the dissociation constant (kd) of the complex (Table 1).
FIG. 7.
α-NAC stabilizes the binding of c-Jun to the AP-1 site. Binding reactions and electrophoretic mobility shift assays were performed with purified c-Jun and α-NAC proteins. (A) On rate measurements. Samples were removed at intervals and analyzed for DNA binding. Bound probe was measured by cutting out the bands on the gel and counting in a gamma counter; calculations were done as described by Chodosh et al. (5). Note that the slope of the binding curve, which represents the on rate, is not affected by α-NAC. A, free c-Jun; AP, c-Jun–DNA complex; PT, total DNA. (B) Off rate measurements. The bound complexes were formed as described, and a 50-fold excess of unlabeled AP-1 or GAL4 oligonucleotides was subsequently added to the binding reaction mixture. Samples were removed at intervals and processed as described above. The slope allows calculation of kd, which is reduced in the presence of α-NAC, thus stabilizing the c-Jun AP-1 complex and the GAL4/VP-16 complex. (C) Off rate measurement of the JunB AP-1 complex: in vitro-translated JunB protein was used as described for c-Jun in panel B. Note that α-NAC does not stabilize the JunB AP-1 complex.
TABLE 1.
Effect of α-NAC on the kd of c-Jun binding to an AP-1 site
| Expt |
kd (s−1)
|
Fold decrease in kd | |
|---|---|---|---|
| c-Jun | c-Jun + α-NAC | ||
| 1 | 1.70 × 10−4 | 2.93 × 10−5 | 5.8 |
| 2 | 7.30 × 10−4 | 6.67 × 10−5 | 10.9 |
| 3 | 2.28 × 10−4 | 5.15 × 10−5 | 4.4 |
| Mean (± SE) | 3.76 × 10−4 | 4.92 × 10−5 | 7.6 ± 2.7 |
We tested the specificity of the stabilizing effect of α-NAC on DNA binding. While α-NAC stabilized the binding of GAL4/VP-16 to its cognate site (Fig. 7B, right panel), it had no effect on the stability of binding of JunB (Fig. 7C) and Sp1 (data not shown) to their respective DNA recognition sequences. These data suggest that α-NAC only stabilizes the binding of factors with which it interacts to potentiate the transcriptional activation function.
These experiments demonstrate a direct interaction between α-NAC and the c-Jun homodimer, resulting in increased transcriptional activation from an AP-1-responsive promoter. Combined with the restricted expression pattern observed for α-NAC during development, these data suggest that α-NAC may be implicated in the regulation of gene transcription during osteoblastic differentiation.
DISCUSSION
We have shown that the c-Jun AP-1 homodimer is a natural target for the coactivation function of α-NAC. Moreover, our data revealed that α-NAC represents one of the rare examples of a tissue-specific, developmentally regulated transcriptional coactivator. These results provide novel perspectives for our understanding of the specificity of AP-1-dependent gene transcription and the regulation of osteoblast-specific gene expression.
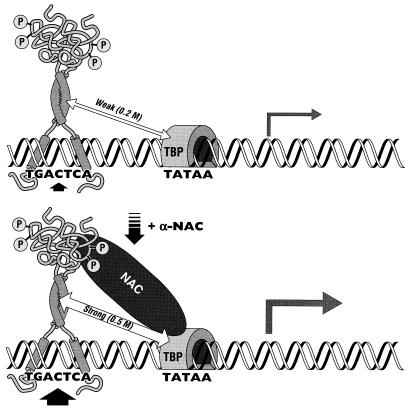
In a search for proteins interacting with c-Jun and implicated in c-Jun-mediated transcriptional activation, Franklin et al. (14) have shown that the C terminus of the c-Jun protein associates directly with TBP and TFIIB in vitro. While these interactions may contribute to the molecular mechanisms regulating c-Jun-driven transcription, they were measured as relatively weak interactions (c-Jun dissociated from TBP in 0.2 M salt) (14). The high affinity of α-NAC for c-Jun (Fig. 5) and TBP (43a) is thought to stabilize the interaction of the c-Jun homodimer with the basal transcriptional machinery. Moreover, α-NAC stabilized the AP-1 complex formed by the c-Jun homodimer through an eightfold reduction in the kd of the complex (Table 1). We thus propose the following model for the potentiation of c-Jun-mediated transcription by α-NAC: the rate of transcription from a c-Jun-dependent promoter is increased in the presence of the α-NAC coactivator through the stabilization of the c-Jun dimer on its binding site and through the recruitment of the basal transcriptional machinery by α-NAC, which stabilizes the interaction between c-Jun and TBP (Fig. 8).
FIG. 8.
Model of the α-NAC potentiation of c-Jun-activated transcription. (Upper panel) The c-Jun homodimer binds the AP-1 site and interacts weakly with TBP. (Lower panel) The α-NAC coactivator interacts with the N-terminal domain of c-Jun. This interaction leads to a stabilization of the interaction of the c-Jun homodimer with the AP-1 site through a reduction in the dissociation rate (kd). Moreover, since α-NAC interacts strongly with both c-Jun and TBP, it strengthens the contacts between c-Jun and the basal transcriptional machinery, resulting in enhanced transcription from the c-Jun dimer (represented in the form of the larger arrow than that in the upper panel). For simplicity, only the phosphorylated form of c-Jun is depicted in the model; however, in vitro protein interaction assays demonstrated that α-NAC can also interact with the unphosphorylated form of c-Jun.
Coactivator proteins that potentiate the transcriptional response to the c-Jun homodimer have been reported previously. The interaction of c-Jun with CBP (CREB binding protein) has been described previously (2, 3). More recently, the JAB-1 coactivator which selectively potentiates the activity of c-Jun and JunD, but not JunB or v-Jun, was identified with the yeast two-hybrid protein interaction assay (6). It is interesting to note the similarities between the mechanisms of action of JAB-1 and α-NAC, despite the complete absence of sequence homology between them. Both proteins interact with the N-terminal portion of c-Jun and can bind the phosphorylated as well as the unphosphorylated forms of the transcription factor (6). Moreover, the two coactivators appear to act by stabilizing the interaction of the c-Jun homodimer to its cognate DNA binding site (6). However, the target contacts of JAB-1 with the basal transcriptional machinery have not been identified, and its tissue distribution has not been described. It also remains unclear whether JAB-1 can interact with Fos family members or affect the transcriptional response to the Fos/Jun AP-1 heterodimer. Therefore, several distinct coactivators may contribute to the tissue specificity of target gene activation by AP-1 proteins. Additional specificity may also be provided by posttranslational modifications; CBP, for example, interacts only with the phosphorylated form of c-Jun (2).
There is a growing body of experimental evidence showing that AP-1 is essential for proper bone development (reviewed in reference 17). While most of these studies have focused on the c-fos component of the AP-1 family, it is well known that the Fos protein needs a dimerizing partner in order to bind DNA and activate transcription (1, 30). We have previously shown that c-jun is expressed in osteoblasts (4); indeed, expression of c-jun in bone cells has been reported both during early embryogenesis (42) and at all stages of osteoblastic differentiation (26). Thus, it is probable that the c-Jun activator plays a physiological role in the control of gene expression in bone cells.
Genetic and biochemical evidence has recently demonstrated that transcriptional coactivators such as the TAFIIs mediate transcriptional activation during development (33). Moreover, coactivators have been shown to be involved in tissue-specific gene expression. For example, the coactivation function of the small subunit of TFIIA (TFIIA-S) has been shown to be required for the Ras-mediated determination of photoreceptor cells during Drosophila development (44). Several groups have also identified a B-cell-specific coactivator (18, 23, 35). These studies have led to the cloning and characterization of OBF-1/Bob1, a coactivator interacting with some members of the Oct family of octamer motif-binding proteins (18, 35). This coactivator appears responsible for restricting the activity of the immunoglobulin promoter to B cells (18, 35), thus converting ubiquitously expressed transcription factors to cell-type-specific activators. Since coactivators interact with one or several distinct transcriptional activators, it is plausible that the developmental regulation of the expression of a particular coactivator could lead to the coordinate induction of a number of genes in a given tissue. We propose that the restricted expression of α-NAC to bone cells during development serves such a function. The identification of additional endogenous transcription factors interacting with α-NAC, together with the identification of α-NAC-responsive genes in bone, should further our understanding of the molecular mechanisms regulating osteoblastic differentiation and function.
ACKNOWLEDGMENTS
A.M. and W.V.Y. participated equally in this study and should both be considered first authors.
We thank the following investigators for their generous gift of plasmids or reagents: Michael Green (5Gal4-E1b-CAT), Mark Ptashne (pSGVP), Robert Tjian (pSV6tkCAT), Daniel Nathans (Jac7), Frank J. Rauscher III (CMV-c-fos and pSP65-c-fos), and Michael Karin and Trang Hoang (c-Jun deletion mutants). We thank Edwin Wan for oligonucleotide synthesis and Jane Wishart and Mark Lepik for preparing the figures.
This work was supported by a grant from the Shriners of North America. W.V.Y. was a Shriners’ Fellow, and R.S.-A. is a Chercheur-Boursier from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 2.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 4.Candeliere G A, Prud’homme J, St-Arnaud R. Differential stimulation of Fos and Jun family members by calcitriol in osteoblastic cells. Mol Endocrinol. 1991;5:1780–1788. doi: 10.1210/mend-5-12-1780. [DOI] [PubMed] [Google Scholar]
- 5.Chodosh L A, Carthew R W, Sharp P A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986;6:4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claret F-X, Hibi M, Dhutt S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- 7.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 8.Curran T, Gordon M B, Rubine K L, Sambucetti L C. Isolation and characterization of the c-fos (rat) cDNA and analysis of post-translational modification in vitro. Oncogene. 1987;2:79–84. [PubMed] [Google Scholar]
- 9.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 12.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 13.Folkers G E, van der Saag P T. Adenovirus E1A functions as a cofactor for retinoic acid receptor β (RARβ) through direct interaction with RARβ. Mol Cell Biol. 1995;15:5868–5878. doi: 10.1128/mcb.15.11.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin C C, McCulloch A V, Kraft A S. In vitro association between the Jun protein family and the general transcription factors, TBP and TFIIB. Biochem J. 1995;305:967–974. doi: 10.1042/bj3050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher S, Winston S E, Fuller S A, Hurrell J G R. Immunoblotting and immunodetection. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1993. pp. 10.8.1–10.8.17. [Google Scholar]
- 16.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 17.Grigoriadis A E, Wang Z-Q, Wagner E F. Fos and bone cell development: lessons from a nuclear oncogene. Trends Genet. 1995;11:436–441. doi: 10.1016/s0168-9525(00)89142-8. [DOI] [PubMed] [Google Scholar]
- 18.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 19.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman M H. The atlas of mouse development. London, United Kingdom: Academic Press; 1992. [Google Scholar]
- 21.Lee W, Haslinger A, Karin M, Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- 22.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, Fujii H, Gerster T, Roeder R G. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- 24.Majeska R J, Rodan G A. Parathyroid hormone-responsive clonal cell lines from rat osteosarcoma. Endocrinology. 1980;107:1494–1503. doi: 10.1210/endo-107-5-1494. [DOI] [PubMed] [Google Scholar]
- 25.McBurney M W. P19 embryonal carcinoma cells. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- 26.McCabe L R, Kockx M, Lian J, Stein J, Stein G. Selective expression of fos- and jun-related genes during osteoblast proliferation and differentiation. Exp Cell Res. 1995;218:255–262. doi: 10.1006/excr.1995.1154. [DOI] [PubMed] [Google Scholar]
- 27.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 30.Ransone L J, Verma I M. Nuclear proto-oncogenes fos and jun. Annu Rev Cell Biol. 1990;6:539–557. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 31.Ryder K, Nathans D. Induction of protooncogene c-jun by serum growth factors. Proc Natl Acad Sci USA. 1988;85:8464–8467. doi: 10.1073/pnas.85.22.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauer F, Wassarman D A, Rubin G M, Tjian R. TAFIIs mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 34.St-Arnaud R, Moir J M. Wnt-1-inducing factor-1: a novel G/C box-binding transcription factor regulating the expression of Wnt-1 during neuroectodermal differentiation. Mol Cell Biol. 1993;13:1590–1598. doi: 10.1128/mcb.13.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 36.Sudo H, Kodama H-A, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanese N, Pugh B F, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 38.Tansey W P, Herr W. TAFs: guilt by association? Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 39.Taylor S M, Jones P A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 40.Watkins S. Immunohistochemistry. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. pp. 14.6.1–14.6.13. [Google Scholar]
- 41.Wiedmann B, Sakai H, Davis T A, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson D G, Bhatt S, Ryseck R-P, Bravo R. Tissue-specific expression of c-jun and junB during organogenesis in the mouse. Development. 1989;106:465–471. doi: 10.1242/dev.106.3.465. [DOI] [PubMed] [Google Scholar]
- 43.Yotov W V, St-Arnaud R. Differential splicing-in of a proline-rich exon converts αNAC into a muscle-specific transcription factor. Genes Dev. 1996;10:1763–1772. doi: 10.1101/gad.10.14.1763. [DOI] [PubMed] [Google Scholar]
- 43a.Yotov W V, Moreau A, St-Arnaud R. The alpha chain of the nascent polypeptide-associated complex functions as a transcriptional coactivator. Mol Cell Biol. 1998;18:1303–1311. doi: 10.1128/mcb.18.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeidler M P, Yokomori K, Tjian R, Mlodzik M. Drosophila TFIIA-S is up-regulated and required during Ras-mediated photoreceptor determination. Genes Dev. 1996;10:50–59. doi: 10.1101/gad.10.1.50. [DOI] [PubMed] [Google Scholar]