Abstract
The relationship between gastrointestinal tract infection, the host immune response, and the clinical outcome of disease is not well understood in COVID-19. We sought to understand the effect of intestinal immune responses to SARS-CoV-2 on patient outcomes including the magnitude of systemic antibody induction. Combining two prospective cohort studies, International Severe Acute Respiratory and emerging Infections Consortium Comprehensive Clinical Characterisations Collaboration (ISARIC4C) and Integrated Network for Surveillance, Trials and Investigations into COVID-19 Transmission (INSTINCT), we acquired samples from 88 COVID-19 cases representing the full spectrum of disease severity and analysed viral RNA and host gut cytokine responses in the context of clinical and virological outcome measures. There was no correlation between the upper respiratory tract and faecal viral loads. Using hierarchical clustering, we identified a group of fecal cytokines including Interleukin-17A, Granulocyte macrophage colony-stimulating factor, Tumor necrosis factorα, Interleukin-23, and S100A8, that were transiently elevated in mild cases and also correlated with the magnitude of systemic anti-Spike-receptor-binding domain antibody induction. Receiver operating characteristic curve analysis showed that expression of these gut cytokines at study enrolment in hospitalised COVID-19 cases was associated negatively with overall clinical severity implicating a protective role in COVID-19. This suggests that a productive intestinal immune response may be beneficial in the response to a respiratory pathogen and a biomarker of a successful barrier response.
INTRODUCTION
Whilst SARS-CoV-2 is primarily considered a respiratory pathogen, a growing body of evidence shows that the virus is also capable of directly infecting gut mucosal tissue. The susceptibility of gut mucosal tissues to SARS-CoV-2 infection was hypothesized based on observations of high levels of expression of SARS-CoV-2 entry receptors angiotensin-converting enzyme 2 (ACE2) and tpye II transmembrane serine protease (TMPRSS) on duodenal and ileal enterocytes1, 2. Indeed, histological evidence confirmed SARS-CoV-2 entry into ACE-2-expressing small intestine enterocytes from hospitalized patients with COVID-19 early in the pandemic3. Since then, small intestine organoid models and cultured gut explants have shown the high level of sensitivity of human gut tissue to SARS-CoV-2 infection4, 5, 6. Furthermore, the presence of replication-competent virus in human fecal samples provides evidence of the potential for active viral replication in the gastro intestinal (GI) tract in vivo7.
Although it is now well-established that GI infection with SARS-CoV-2 is a feature of COVID-19, the clinical implications of gut infection remain less well-defined. Although GI symptoms including diarrhoea, nausea, and vomiting are common in COVID-198, 9, 10, 11 it remains unclear whether these are directly associated with gut infection or are the result of other phenomena such as effects of infection-induced systemic cytokines. Existing literature on the subject is somewhat paradoxical, with studies showing a lack of correlation between detection of viral RNA in fecal samples and both the presence of GI symptoms and overall clinical outcome10, 12. Furthermore, there are conflicting reports on the relationship between presence of GI symptoms and overall clinical outcome of infection13, 14. Prolonged viral shedding in fecal samples has been observed after resolution of symptoms and clearance of virus from the upper respiratory tract (URT), suggesting a complex, non-linear association between presence of virus in the GI tract and symptomatology15, 16, 17.
The systemic immune response has been shown to be a key determinant in the clinical outcome of SARS-CoV-2 infection, with dysregulated serum cytokines responses associating with the most severe outcomes18, 19, 20. However, the relationship between the gut immune response, severity of GI symptoms and overall clinical, and virological outcomes of infection remain to be determined. Here we leverage two prospective human cohorts to study a group of 88 individuals representing the full spectrum of COVID-19 outcomes to determine whether gut mucosal immune responses are associated with favorable clinical outcomes and optimal induction of systemic antibody responses.
RESULTS
Cohort and clinical data
We obtained paired clinical data and faecal samples from 88 COVID-19 cases with varying degrees of severity, ranging from asymptomatic to fatal infection. Of these, 45 were community cases recruited via the Integrated Network for Surveillance, Trials and Investigations into COVID-19 Transmission (INSTINCT) study, two of which were admitted to hospital during the study. In total, 43 were hospitalised patients recruited via the International Severe Acute Respiratory Infection Comprehensive Clinical Characterisation Collaboration (ISARIC4C) study. In INSTINCT, the National Health Service Test and Trace contact tracing system and UK Health Security Agency infrastructure were used to recruit recently symptomatic COVID-19 cases and their household contacts. INSTINCT samples were collected from May 2020–March 2021 when the pre-alpha and alpha (B.1.1.7) strains were predominant in the UK. Longitudinal URT, blood, and faecal samples were collected over a period of 28 days from the day of enrolment. Faecal samples were collected on day 7 and day 28 post-enrolment (Fig. 1A). Samples from asymptomatic, persistently polymerase chain reaction (PCR)-negative, and seronegative cohabitants of COVID-19 cases were recruited in INSTINCT and used as uninfected controls. Faecal samples from the 43 hospitalised patients in the ISARIC4C study were collected on the day of hospitalisation. ISARIC4C samples were collected from March 2020–August 2020, in this time the pre-alpha strain was the predominant circulating variant in the UK. Time between symptom onset and day of collection of first faecal sample (INSTINCT D7, ISARIC4C D0) was not significantly different between cohorts allowing us to compare these samples across cohorts. INSTINCT mean duration from symptom onset to sample collection = 8.8 days (IQR 2.0). ISARIC4C = 6.9 days, (IQR 5.0).
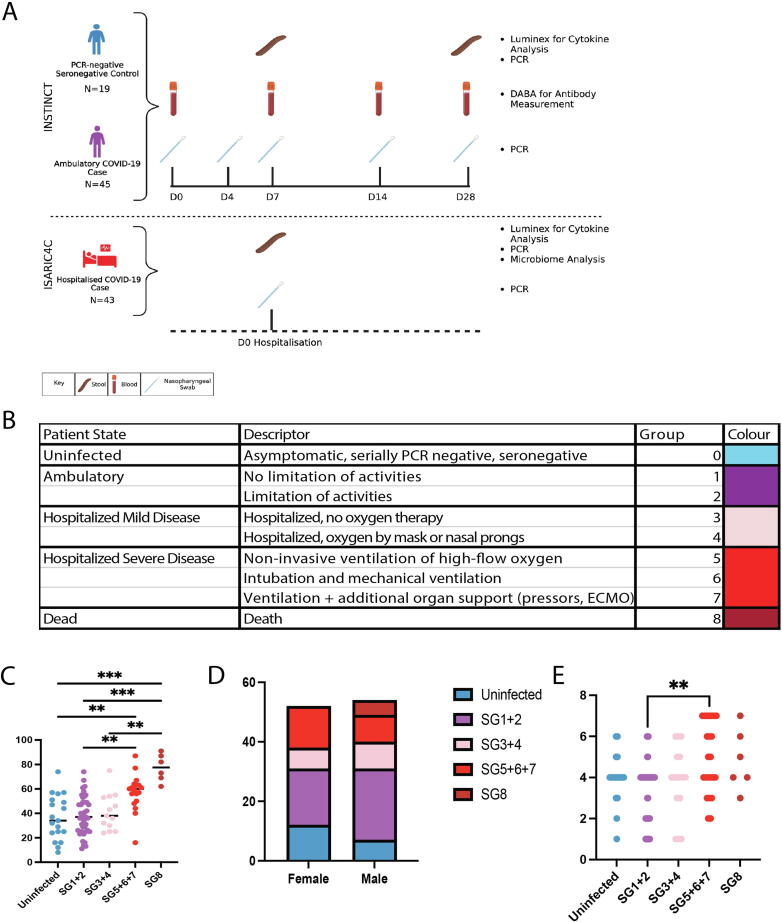
Fig. 1.
INSTINCT (n = 45) and ISARIC4C (n = 43) study sample collection schedules and cohort demographics. (A) PCR-negative controls and ambulatory community COVID-19 cases were collected via INSTINCT. Serial faecal, blood and nasopharyngeal swabs were collected over 28 days. Moderate and severely ill COVID-19 cases were recruited via ISARIC4C. Faecal samples and nasopharyngeal swabs were collected at enrolment which occurred immediately following hospitalisation. ISARIC4C and INSTINCT sample collection schedules are aligned to show that comparisons are made between faecal samples collected at D0 and D7 respectively. (B) Table showing patient state and descriptor for each severity group. (C) Participant age and (D) Biological sex and (E) Bristol stool score stratified by symptom severity group. Colours represent Severity Groups. Results of Kruskal Wallis tests with Dunn’s multiple comparison tests are displayed *p < 0.05, **p < 0.01, ***p < 0.001. ECMO = extracorporeal membrane oxygenation.
Faecal samples from both cohorts were processed in the same manner for downstream cytokine profiling and viral E gene PCR. Severity groups (SG) were assigned based on the WHO ordinal scale for clinical improvement as described by Thwaites et al. (Fig. 1B)19. Serially PCR negative, seronegative control participants were assigned to severity group 0 (SG0), ambulatory community cases were assigned SG1 and 2 and the hospitalised cases span SG3-8 wherein SG3 equates to minimal treatment and SG8 equates to death. The INSTINCT cohort comprises SG0,1+2 with two participants in SG3. ISARIC4C comprises the remainder of SG3-8. Unsurprisingly, the highest SG contained significantly older patients (Fig. 1C). However, severity stratification by biological sex showed a similar number of participants in each severity group, suggesting that age but not sex is an important factor in the severity grouping in these cohorts (Fig. 1D). Bristol stool scores for fecal consistency varied widely across the cohorts; higher scores tended to be observed in the highest SG (Fig. 1E).
Mild COVID-19 cases have a pronounced pro-inflammatory gut mucosal cytokine response
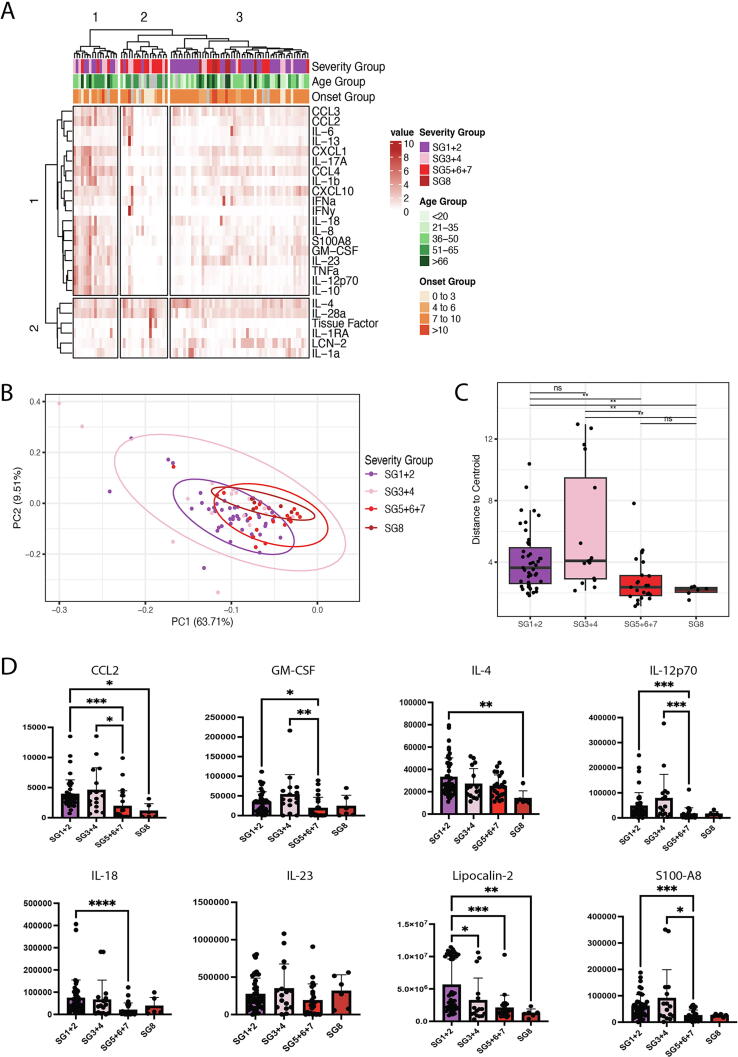
Luminex-based cytokine profiling was used to investigate the relationship between gut cytokine production and clinical outcomes of SARS-CoV-2 infection. Hierarchical clustering based on concentrations of 25 cytokines measured in faecal samples yielded three defined patient clusters (Fig. 2A). Cluster 1 consists of participants with mild COVID-19 in the lower SG that have a pronounced faecal cytokine response. Cluster 2 contains primarily participants who are hospitalised and have severe disease. Cluster 3 consists primarily of individuals with asymptomatic or mild COVID-19. Contingency testing showed that SG were not independent of cluster assignment (χ2, p < 0.05) showing that the distribution of SG was not equal in each of the three clusters. On the other hand, onset groups were independent of cluster assignment (χ2, p > 0.05), indicating that cluster assignment is not driven by number of days from symptom onset to sample collection (data not shown). Non-clustered normalised cytokine expression in gut samples acquired from INSTINCT and ISARIC stratified by individual symptom group supports the conclusion that gut cytokine expression level varies by COVID-19 disease severity (Supplementary Fig.1).
Fig. 2.
Mild COVID-19 cases have a pronounced pro-inflammatory gut mucosal cytokine response. (A) Hierarchical clustered heatmap of cytokines from faecal samples in both INSTINCT (n = 45) and ISARIC4C (n = 43) cohorts. Further annotated with severity group, age group and time from symptom onset to sample acquisition group. (B) Principal component analysis showing clustering of cytokines according to severity group (n = 88). Cytokine loadings associated with PCA shown in Supplementary Fig. 6 (C) Distance of cytokines from the clustered severity group centroid (n = 88). (D) Concentrations of 8 cytokines in faecal samples stratified by severity groups. Results of Kruskal Wallis tests with Dunn’s multiple comparison tests are displayed *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Principal component analysis of cytokine data from early-timepoint ISARIC4C and INSTINCT shows tight clustering of SG5-8 cases and sparse distribution of SG1-4 cases, indicating a heterogenous gut cytokine response in mild COVID-19 disease (Fig. 2B). This is supported empirically by Permutational multivariate analysis of variance (PERMANOVA) of cytokine data against SG, which showed that the variance is not uniform between SG (f-statistic = 3.051, r2 = 0.097, SSQ = 210.81, df = 3, p < 0.05). Analysis of dispersion of severity groups from the group centroid (Fig. 2C) showed that both SG1+2 and SG3-4 had significantly higher distance to centroid values compared to SG5, 6+7. SG3+4 also had a significantly higher distance to centroid than SG8. Investigation of individual cytokines confirmed that pro-inflammatory cytokines were evident at higher concentrations in the gut of mildly ill cases compared to severely ill cases (Fig. 2D and Supplementary Fig. 2). Cytokines present in mildly ill cases include chemokine (C-C) motif ligand 2 (CCL2), Granulocyte macrophage colony-stimulating factor (GM-CSF), which are associated with myeloid recruitment and differentiation, as well as lipocalin and S100A8, which are generally used as measures of intestinal inflammation clinically.
To understand whether the lack of cytokines present in severe cases may be associated with a particular microbiome signature, we analysed the microbial composition of fecal samples from the ISARIC4C cohort. SG 5–8 had significantly lower alpha diversity than the milder cases (Supplementary Figs. 3 and 4A) though interpretation of this observation is confounded by the potential effect of antibiotic use. Indeed, a contingency test of inpatient antibiotic use and SG 5–8 showed that the two variables were not independent of one another (Fisher’s Exact Test, p < 0.05) (Supplementary Fig. 4B). Longitudinal data detailing antibiotic use prior to admission was limited and hence we were unable to conclusively demonstrate a causative role of antibiotic use on the differences in microbiome composition between groups. In order to study whether the intestinal cytokine response observed across both ISARIC4C and INSTINCT cohorts without potential complications from antibiotic use in the most severe patients, downstream analysis focused on the INSTINCT cohort.
Viral burden in the URT and gut are non-correlated
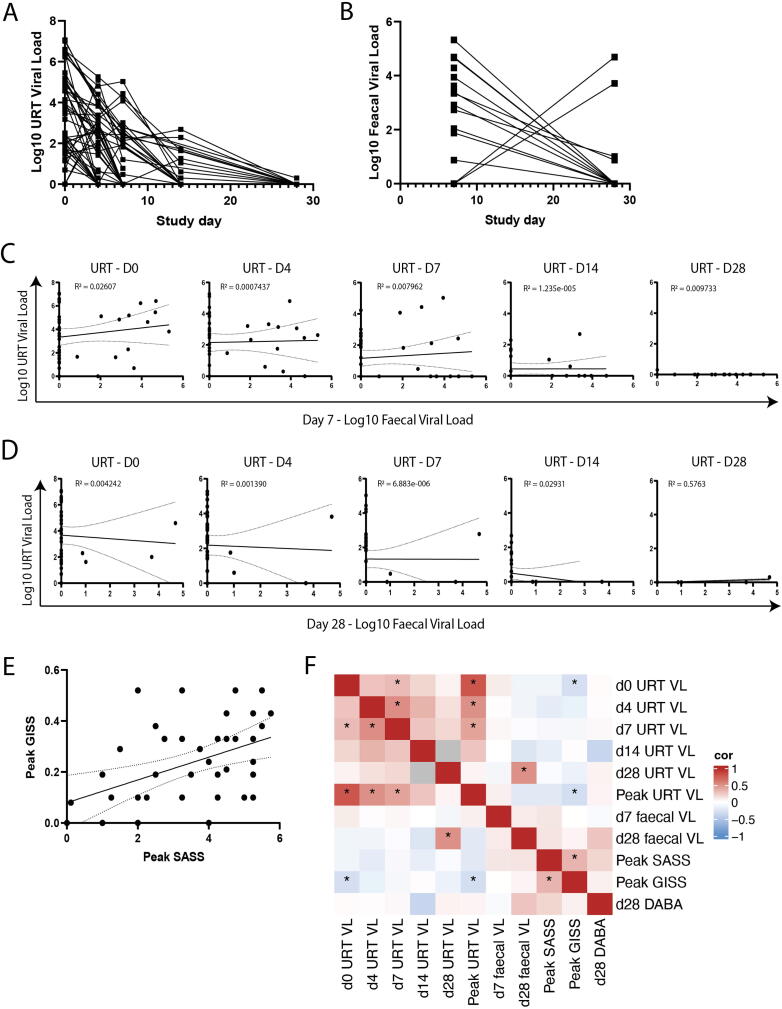
Having observed a pro-inflammatory gut cytokine response in individuals with mild COVID-19 disease, we sought to understand how SARS-CoV-2 infection in the intestine related to upper respiratory tract (URT) infection and clinical outcomes in these individuals. Viral load (VL) trajectories in mild disease were measured from the upper respiratory tract and faecal samples with peak URT VL generally occurring at day 0 and abating to mostly undetectable levels by day 28, suggestive of induction of an appropriate immune response to effectively control the virus (Fig. 3A). Similar trajectories were observed in the gut VL for the majority of samples with a greater VL measured at day 7 compared to mostly undetectable levels at day 28. However, some samples had a diminished but persistent VL at day 28 and a number of samples displayed undetectable virus in the gut at day 7 with elevated VLs at day 28 perhaps indicating some temporal variation in the viral immune response in the gut (Fig. 3B). VL in the URT and gut were compared at each of their respective sample timepoints which evidenced no correlation between VLs at later time points (day 14–28), and weak correlation (r < 0.3) at early timepoints, suggesting discordance between the URT and the gut, particularly at early stages of symptom onset (Figs. 3C and D).
Fig. 3.
Viral burden in the upper respiratory tract and gut are non-correlated (n = 45). (A) URT and (B) Faecal viral load were quantified by PCR targeting SARS-CoV-2 E gene. Viral load trajectories are plotted over 28 days. (C) Viral load between the URT sample timepoints versus faecal viral load at day 7 and (D) Day 28. (E) Peak symptom associated severity score (SASS) versus peak GI symptom score (GISS). (F) Correlation heatmap between URT viral loads, faecal viral loads, peak SASS, peak GISS and D28 serum DABA. Correlation measured by Kendall rank correlation coefficient, tau. Correlations that are statistically different than zero (p < 0.05) are marked with an asterisk (*).
To stratify and quantitate the clinical outcomes of infection within the mild end of the spectrum of COVID-19 disease, a COVID-19 severity associated symptom score (SASS) was assigned based on symptoms recorded in self-collected participant symptom diaries as previously described21, 22. Peak SASS correlated with peak GI symptom score (GISS) (Fig. 3E). Kendall correlation matrix analysis was performed to investigate associations between these clinical outcome measures, longitudinal URT and fecal VLs, and systemic antibody induction (Fig. 3F). Unsurprisingly, URT VL measures at D0 and D4 correlated with one another as well as with peak URT VL. In accordance with the data displayed in Figs. 3C and D, URT VL did not correlate with faecal VL at early timepoints. A positive association between D28 faecal VL and D28 URT VL was observed though it was driven by few data points. No significant positive associations were observed between any VL measure and SASS, GISS or antibody induction. To the contrary, a significant negative association between peak URT VL and peak GISS was observed. Overall, these data are consistent with neither URT VL nor faecal VL strongly driving systemic or GI-specific symptoms.
Longitudinal cytokine analysis shows elevated early gut cytokine production in mild COVID-19 disease
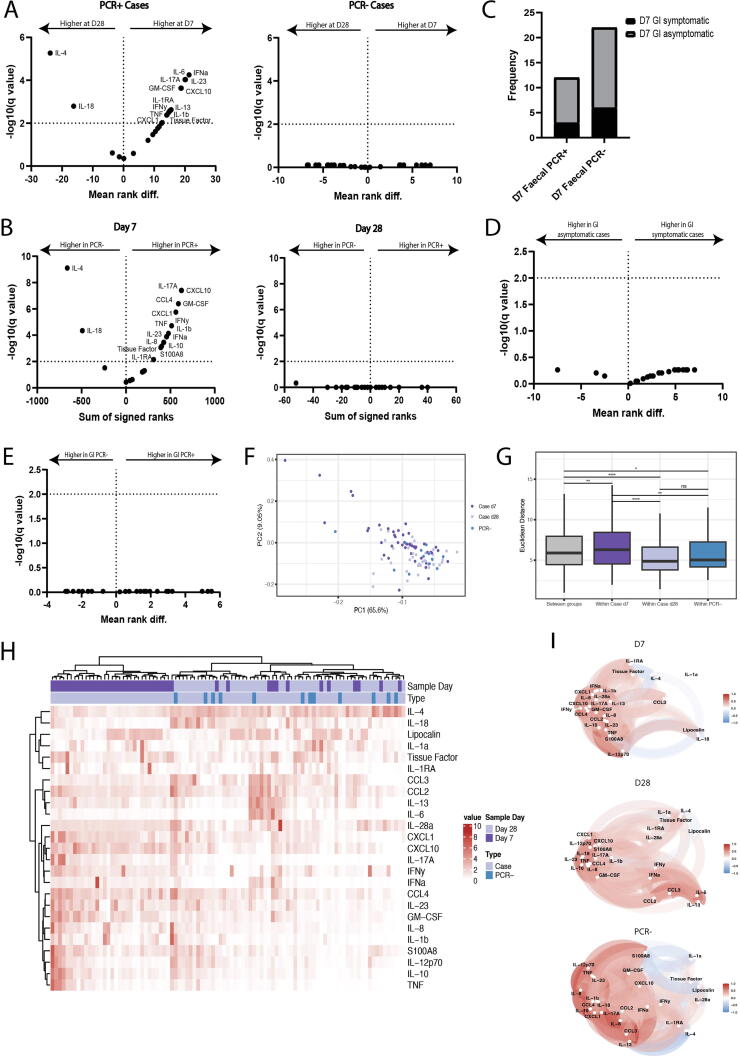
Having established that symptom score, day 28 antibody levels and VL from these longitudinal faecal samples were not correlated, we investigated longitudinal cytokine data to determine whether specific gut immune responses correlated with these outcome measures. A majority of cytokines measured were elevated in mild cases at day 7. Conversely, interleukin (IL)-4, IL-18 and lipocalin-2 (LCN-2) were observed at relatively higher concentrations at d28 (Fig. 4A). The cytokine concentrations in d7 samples from uninfected and infected cases were compared, revealing elevated levels of IL-4 and IL-18 in uninfected contacts compared to infected cases. These findings suggest that the expression of these cytokines is suppressed during early infection (Fig. 4B).
Fig. 4.
Longitudinal cytokine analysis shows elevated early gut cytokine production in mild COVID-19 disease (PCR- n = 12, PCR+ Day 7 n = 45, PCR+ Day 28 n = 38). (A) Mean rank difference of faecal cytokines measured at day 7 and day 28 in PCR+ and PCR- cases. (B) Sum of signed ranks of faecal cytokines measured in PCR+ and PCR- cases at day 7 and day 28. (C) Frequency of participants experiencing GI symptoms who have a measurable viral load (PCR+) or not (PCR-) at day 7 (p > 0.999). (D) Mean rank difference of faecal cytokines in participants with or without GI symptoms at day 7 and (E) in faecal PCR+ and PCR- participants. (F) PCA of faecal cytokines measured in samples collected from PCR+ cases at D7 or D28 or from PCR- cases. Cytokine loadings associated with PCA are shown in Supplementary Fig. 6. (G) Euclidian distance of pairwise samples between and within PCR+ cases at D7, PCR+ cases at day D28 and PCR- cases. (H) Heatmap of cytokines measured in faecal samples from PCR- control individuals and PCR+ cases at day 7 and day 28. (I) Correlation networks of cytokines from faecal cytokines measured in PCR+ cases at day 7 samples, PCR+ cases at day 28 samples and in PCR- individuals where the correlation is shown by proximity of points and direction and strength of the correlation between cytokines is indicated by color (red = positive correlation, blue = negative correlation). Correlation measured by Kendall rank correlation coefficient, tau. Only correlations ≥0.3 are shown.
Previous studies have revealed incongruous results when looking at GI symptoms and clinical outcome12, 13, 14. Stratifying samples from participants with mild disease based on the VL detected in faecal samples and presence of GI symptoms shows that presence of virus is not related to GI symptoms experienced (p > 0.999) (Fig. 4C). Furthermore, neither GI symptoms nor VL in faecal samples accounts for the gut cytokine signature observed in these participants (Figs. 4D–E).
Analysis of cytokines detected in longitudinal faecal samples from participants with mild COVID-19 disease and PCR-negative controls show that cytokine measurements from faecal samples at day 28 cluster closer to samples from PCR-negative controls (Fig. 4F). Cytokine responses between PCR-negative controls, mild cases at day 7, and mild cases at day 28 are statistically distinct as measured by PERMANOVA (f-statistic = 4.38, r2 = 0.087, SSG = 204.374, p < 0.05). The PCR-negative controls and case groups have distinguishable cytokine profiles as measured by their within-group variance relative to the between group variances. Increased cytokine expression is seen in day 7 samples with a more heterogeneous response in samples from this timepoint (Fig. 4H), as demonstrated by a greater within-group similarity (based on pairwise sample distances) for mild cases at day 7 (p < 0.05) as compared mild cases at day 28 and PCR-negative individuals (Fig. 4G). Correlation network analysis shows a cluster of cytokines closely correlated at day 7 of infection, which returns to a correlation pattern similar to uninfected controls at day 28 (Fig. 4I).
Gut cytokine profile following recent SARS-CoV-2 infection associates with systemic anti-SARS-CoV-2 antibody induction
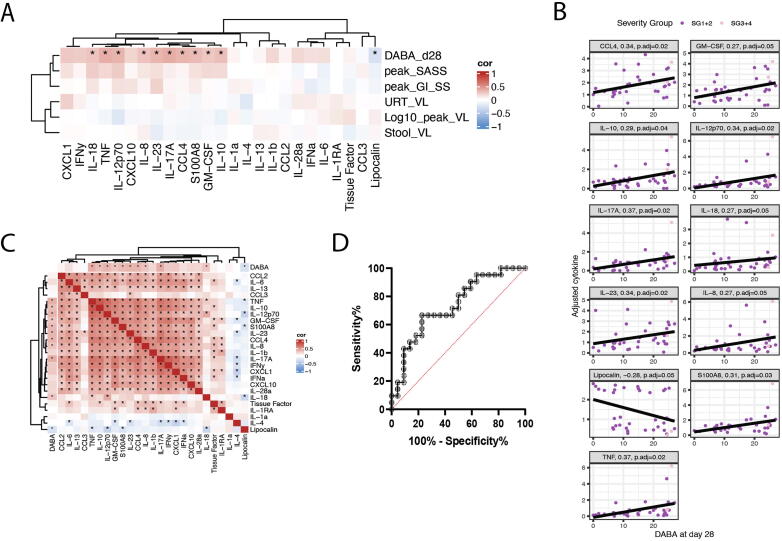
To understand whether these striking differences in faecal cytokine levels over time were associated with outcomes, we investigated associations between faecal cytokines and VL, symptom scores, and serum antibody levels at day 28. Gut cytokine levels at day 7 in mild cases positively correlated with day 28 serum Double antigen binding assay (DABA) levels (p < 0.05), but not with symptom scores (peak SASS, peak GISS), log10 peak URT VL, nor faecal and URT VLs at day 7 (Table 1, Fig. 5A). Cytokine concentrations at day 28 were also non-correlated with peak symptom scores, log10 peak URT VLs and faecal and URT VLs at day 28 and did not correlate with d28 serum DABA (Supplementary Table 1). Serum cytokine levels do not show a significant correlation with day 28 serum DABA levels (Supplementary Fig. 5A). Critically, serum and faecal cytokines were not significantly correlated (Supplementary Fig. 5B), suggesting the faecal cytokine measures represent the mucosal immune response and not systemic cytokine levels.
Table 1.
PERMANOVA of faecal cytokines from PCR+ cases at day 7 (n = 45) against day 28 DABA levels, peak SASS, peak GISS, log10 of peak URT viral load (VL) in URT, URT viral load at day 7, fecal viral load at day 7.
| Df | Sum of Sqs | R2 | F | P value | P value (adjusted) | |
|---|---|---|---|---|---|---|
| D28 DABA | 1 | 82.53 | 0.084 | 3.198 | 0.006 | 0.036 |
| Peak SASS | 1 | 37.76 | 0.029 | 1.275 | 0.212 | 0.636 |
| Log10 (Peak URT VL) | 1 | 13.71 | 0.010 | 0.454 | 0.897 | 0.897 |
| Peak GISS | 1 | 13.84 | 0.011 | 0.459 | 0.868 | 0.897 |
| D7 URT VL | 1 | 23.26 | 0.018 | 0.776 | 0.547 | 0.897 |
| D7 Faecal VL | 1 | 16.56 | 0.013 | 0.550 | 0.787 | 0.897 |
DABA = Double antigen binding assay; GISS = GI symptom score; PCR = polymerase chain reaction; PERMANOVA = permutational multivariate analysis of variance; SASS = severity associated symptom score; Sqs = XXX; URT = upper respiratory tract.
Fig. 5.
Gut cytokine profile following recent SARS-CoV-2 infection associates with systemic anti-SARS-CoV-2 antibody induction. (A) Correlation between faecal cytokines measured from day 7 samples (n = 45) and day 28 serum DABA levels, peak SASS, peak GISS, URT viral load at day 7, faecal viral load at day 7, and log 10 of peak viral load in the URT. Correlation measured by Kendall rank correlation coefficient, tau. Correlations that are statistically different than zero (p < 0.05) are marked with an asterisk (*). (B) Individual correlation graphs between cytokines that significantly correlate with D28 DABA measurements. (C) Correlation matrix of faecal cytokines measured from PCR+ day 7 samples (n = 45). Correlation of cytokines against D28 serum DABA measurements are shown along the top and left axes. Correlation measured by Kendall rank correlation coefficient, tau. Correlations that are statistically different than zero (p < 0.05) are marked with an asterisk (*). (D) A receiver operating characteristic (ROC) graph showing the sensitivity and specificity with which the antibody-associated cytokine profile score predicted the need for invasive ventilation in the ISARIC4C cohort (n = 43).
Concentrations of 10 of 24 measured cytokines (CCL4, GM-CSF, IL-17A, IL-18, tumor necrosis factor (TNF)α, IL-23, IL-8, IL-10, IL-12p70, S100A8) in faecal samples correlated positively with serum anti-Spike RBD antibody as measured by DABA (Fig. 5B–C). This suggests an association between gut mucosal cytokine response and antibody induction. Having previously observed a trend towards higher levels of inflammatory gut cytokines occurring in milder COVID-19 cases across the INSTINCT and ISARIC4C cohorts, we aimed to determine whether the serum antibody-associated gut cytokine profile identified in the INSTINCT cohort predicted clinical outcome of COVID-19 in the ISARIC4C cohort. An “antibody-associated cytokine profile” score was calculated for each individual recruited in the ISARIC4C study i.e., SG3-8 by Z-scoring concentrations of each of these 10 cytokines and calculating the mean of these concentrations. The sensitivity and specificity with which this “antibody-associated cytokine profile” score predicted the need for invasive ventilation was assessed using Receiver operating characteristic analysis. 21 of 43 hospitalised COVID-19 cases required invasive ventilation and the area under the receiver operating characteristic curve was 0.734 (95% CI 0.584-0.884. p = 0.0087) (Fig. 5D). Together these findings suggest that SARS-CoV-2 infection induces production of pro-inflammatory cytokines in the gut which associate with both systemic antibody induction and favourable clinical outcomes.
DISCUSSION
A considerable body of evidence shows that GI symptoms are a common feature of COVID-19 disease, and that SARS-CoV-2 can directly infect gut tissue4, 7, 9, 10; however, the importance of the intestinal immune response during SARS-CoV-2 infection has not been fully explored. Here we leveraged two patient cohorts that span the severity spectrum of COVID-19 in order to understand the relationship between SARS-CoV-2 infection of the GI tract, the gut cytokine response, and clinical and serological outcomes.
We found discordance between URT and gut VLs in ambulatory COVID-19 cases. Where URT and faecal samples were collected contemporaneously SARS-CoV-2 VLs did not correlate across the two sites. Cases in which URT SARS-CoV-2 positivity was observed in the absence of detectable gut infection as well as the inverse were observed, suggesting compartmentalisation of infection in these two anatomical sites, consistent with observations of different rates of resolution of infection in the two compartments16. Interestingly, presence of detectable virus in the gut did not predict presence of GI symptoms, nor did it associate strongly with a particular gut cytokine profile. It is possible that direct infection of myeloid cells within the intestine may result in a local viral reservoir and response that could not be detected by fecal qPCR23, 24.
We identified a dynamic gut cytokine response associated with favourable clinical outcomes in mild ambulatory COVID-19 patients who, by definition, mounted immune responses that appropriately contained infection and disease. Pro-inflammatory cytokines including GM-CSF and type I, II and III Interferons as well as chemokines including chemokine (C-X-C) motif ligand 10 (CXCL10) and CCL4 were transiently elevated in mild cases whilst resolution-associated cytokine IL-4 and the alarmin IL-18 were downregulated. The downregulation of IL-18, in contrast to other inflammatory cytokines, may point to the unique role of this cytokine in intestinal homeostasis25. The differences between IL-18 and IL-1β, both inflammasome activated cytokines, may also point to infection and response by different immune cell populations within the intestine. The observed cytokine signature is consistent with the rapidly resolving inflammatory response seen systemically in mild COVID-19 cases18, 20; however, we did not observe a correlation between intestinal and systemic cytokine levels. This dichotomy between serum and gut is consistent with compartmentalisation of both infection and immune response to COVID-19 infection. In contrast to observations made in serum samples across the COVID-19 disease spectrum, level of expression of these acute infection-associated pro-inflammatory gut cytokines did not increase with disease severity level19. To the contrary, we observed generally lower levels of these cytokines in the gut of more severely ill patients suggesting that a pro-inflammatory gut response may be a component of an effective immune response to curtail SARS-CoV-2 infection.
The composition of the gut microbiome likely has an important role to play in COVID-19-induced gut immune responses and clinical outcomes of infection. Changes in the gut microbiota could contribute to intestinal permeability, inflammation and could correlate to GI symptoms. Recent data shows that the composition of the microbiome of COVID-19 cases changes throughout the course of disease and is associated with overall disease severity26. However, analysis of the microbial composition in the severe, hospitalised ISARIC4C cohort was prevented due to the confounding effect of antibiotic use demonstrated by contingency table analyses showing non-independence of COVID-19 severity and antibiotic use.
In contrast to serum cytokine levels, we observed a positive correlation between concentration of a subset of inflammatory cytokines in the gut early after infection and serum concentration of anti-Spike RBD antibodies at day 28 in ambulatory cases. Elevated expression of the same cytokines in the hospitalised ISARIC4C cohort predicted better clinical outcome, suggesting that production of these antibody-associated cytokines in the gut is a protective phenomenon. Because the intestine harbours a large population of immune cells, it has a great potential to impact systemic responses and responses at other mucosal sites. Because of limited access to clinical data from hospitalized patients, we were unable to look for other possible confounders, including underlying diseases.
Based on data suggesting intestinal infection can persist beyond respiratory infection and that the humoral immune response to SARS-CoV-2 continues to evolve after the infection is resolved27, 28, it is tempting to propose the intestine as a key site of antibody maturation and production. Alternatively, pro-inflammatory cytokine production at the gut mucosa could affect barrier function of the tissue and increase antigen load systemically, impacting antibody production29. Levels of GM-CSF and CCL2 in the intestine can affect monocyte differentiation and recruitment30. Recruitment of inflammatory cells to the intestine may act as a shunt away from the lungs or may be a biomarker of a productive respiratory immune response.
A thorough understanding of the complex interplay between presence of virus, the immune response, and symptomology will require further work, likely involving animal models. This work was limited by the availability of tissue samples due to health and safety concerns during the early stages of the pandemic. In addition, we were unable to link other mucosal immune data from these cohorts due to limited donor overlap. Elucidating the cellular sources of the cytokines measured in this study and how these compare to their counterparts in the lung will be a key factor in understanding how the gut immune response is associated with outcomes. By understanding the local signals that lead to optimal systemic antibody levels, so far the best correlate of protection, it might be possible to tune the immune response in severe patients toward a more protective response and mimic these signals when designing the next generation of vaccines.
METHODS
Study design and participants
The INSTINCT study is a prospective longitudinal community cohort study. Samples and participant data were obtained between May 2020 and November 2021 under IRAS: 282820 (REC ref. 20/NW/0231). Community COVID-19 cases were identified through Public Health England (PHE) and the National Test and Trace programme in the United Kingdom. Research nurses obtained informed consent for home visits and sampling, having obtained consent to contact potential participants about the study via PHE. Participants received no compensation for participation. A total of 45 PCR-confirmed COVID-19 cases and 19 uninfected household contacts (defined by serial negative PCR and serological tests) were included in this study based on availability of longitudinal URT PCR data, clinical data and availability of at least one stool sample. 7 PCR-positive cases and two uninfected controls received a single dose of a COVID-19 vaccination (produced by either Pfizer or Oxford/Astra Zeneca). Multiple Mann-Whitney U tests showed no significant differences in concentrations of any measured gut cytokine nor D28 DABA value between vaccinated and unvaccinated INSTINCT participants (data not shown).
The ISARIC4C study is a prospective hospitalised cohort study. Samples and participant data were obtained between March 2020 and August 2020. Identified patients hospitalised during the SARS-COV-2 pandemic were recruited into the International Severe Acute Respiratory and Emerging Infection Consortium World Health Organization Clinical Characterisation Protocol UK (IRAS260007 and IRAS126600) (REC ref. 13/SC/0149). Informed consent was obtained from all participants. Faecal samples were collected from a total of 46 COVID-19 cases at time of admission. Additional longitudinal data was collected as part of the ISARIC4C study with clinical outcomes used in this study. Clinical data could not be linked to samples from three participants; hence data from 43 individuals was used in downstream analyses.
Potential sources of bias were minimized at the level of cohort selection, data collection and data analysis. Participant inclusion was dependent on availability of all data types used in downstream analysis including demographic, clinical and immunological data. Standardization of methods within cohorts and between cohorts where feasible, including methods for faecal cytokine quantification. Regular quality control checks at the level of data analysis were performed to ensure accurate reporting of data. Analysis was limited to participants with clinical endpoint data available hence bias introduced by loss to follow up was minimised.
Sample processing
Samples were collected on days indicated and placed in –80 storage. Samples were thawed at room temperature for downstream aliquoting. Using a sterile disposable spatula (VWR International Ltd, Leicestershire, UK), 100 mg ± 10 mg of faecal sample was placed into a ZR BashingBead (TM) lysis tube (0.1 and 0.5 mm beads) (Cambridge Bioscience Ltd, Cambridge, UK) containing 700 µl of qiagen buffer (10% VXL in AVL) (QIAGEN Ltd, Manchester, UK) for further viral RNA isolation. 200 mg ± 20 mg of faecal sample was placed into a ZR BashingBead (TM) lysis tube (0.1 and 0.5 mm beads) containing 750 µl of DNA/RNA shield (Cambridge Bioscience Ltd, Cambridge, UK) for further DNA isolation. 200 mg ± 20 mg of faecal sample was placed into a ZR BashingBead (TM) lysis tube (2 mm beads) containing 1mL of antibody isolation buffer for further analysis. 200 mg ± 20 mg of faecal sample was placed into a ZR BashingBead (TM) lysis tube (2 mm beads) containing 1 mL of protease inhibitor buffer for further analysis.
All tubes containing faecal samples in buffer were shaken for 10 minutes to homogenize the faecal samples. The tubes containing the faecal samples in qiagen buffer and DNA/RNA shield were shaken for another 10 minutes and then stored at −80. The tubes containing the faecal samples in antibody isolation buffer or protease inhibitor buffer (Merck Life Science UK Limited, Darmstadt, Germany) were spun down for 10 minutes at 10XG and then stored at –80°C.
Luminex
Luminex (Bio-Techne, Minneapolis, USA) was carried out using 50 µl of faecal sample homogenized in antibody isolation buffer or 50 µl of standard, reconstituted according to the manufacturers protocol, was added to their respective wells, with each sample and standard being done in duplicate. 50 µl of microparticle cocktail resuspended as per the instructions were added to each well. The plate was then covered with a foil seal and incubated for 2 hours at room temperature on a horizontal orbital microplate shaker set at 800 rpm. Using a microplate magnetic device to align the microparticle beads, the plate was washed by removing the liquid from the bottom of the well and adding 100 µl of PBS-tween 20, leaving for 1 minute before removing the liquid. This was repeated three times.
The remaining steps were performed according to the manufacturers protocol and the microparticle beads were resuspended in 50 µl of PBS-tween 20 and incubated for 2 minutes on the microplate shaker at room temperature before reading on the BioRad Bio-Plex 200 systems (Bio-Rad Laboratories Ltd, Watford, UK).
VL quantification
Nose and throat samples were collected using flocked swabs placed in COPAN Universal Transport Medium (Copan Diagnostics, Murrieta, CA, USA) in participant’s homes and stored at 2–8°C for up to four days. Viral RNA was extracted using the innuPREP Virus TS RNA 2.0 Kit on a CyBio Felix (Analytik Jena, Jena, Germany), following the manufacturer’s instructions. Faecal samples were collected and processed as previously described. Faecal samples were aliquoted into homogenisation tubes (Zymo, Irvine, CA, USA) containing VXL buffer and homogenised with a vortex genie with vortex adaptor (QIAGEN Ltd, Manchester, UK). Viral RNA was extracted using the QIAmp viral RNA mini kit (QIAGEN Ltd, Manchester, UK) according to manufacturer’s instructions. TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) was used in a triplex PCR targeting viral E and N genes and host RNAse P. Samples with adequate RNAse P RNA and an E gene Ct <36.5 (which equates to 5 RNA copies per PCR reaction) were considered PCR-positive.
DABA
10mL of serum was collected, centrifuged at 2000g for 10 minutes and aliquoted into 500 μl aliquots, which were frozen at –20°C. The serological samples were processed at the Molecular Diagnostic Unit (MDU), Imperial College London. Antibody (Immunoglobulin M and Immunoglobulin G) to SARS-CoV-2 receptor binding domain (anti-RBD) was measured using a two-step double antigen binding assay (DABA) (Imperial College London, London, UK) with recombinant S1 antigen on the solid-phase and labeled recombinant RBD as detector in the fluid-phase as previously described [31]. UK Patent Application No. 2011047.4 for “SARS-CoV-2 antibody detection assay” has been filed.
Faecal DNA extractions
Faecal samples were extracted using the ZymoBIOMICS DNA/RNA miniprep kit (Zymo, Irvine, CA, USA) according to the manufacturer’s instructions. Sample homogenization was performed using a Qiagen Vortex adapter coupled to a vortex genie 2 for 20 minutes at maximum speed. Initial DNA quantification was performed on a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA).
16S ribosomal (r)RNA gene sequencing
The Ion Torrent 16S Metagenomics Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to amplify 16S rRNA genes of all the samples. 2uL of each patient DNA sample was used for amplification of the 16S rRNA gene. For amplification of the 16S hypervariable regions, PCR was performed in two pools, each containing a different primer set (Pool 1 contained primers targeting the V2-4-8 regions, pool 2 contained primers targeting the V3-6, 7–9 regions). Following PCR amplification, equal volumes of PCR products from pools 1 and 2 of each sample were combined. In total, 30µl of each combined PCR product was purified using the Agencourt AmpureXP kit according to the manufacturer’s instructions (Beckman Coulter, Brea, CA, USA).
DNA was quantified using the Promega quantifluor one dsDNA Quantification kit (Promega, Madison, WI, USA) and DNA library was prepared using Ion Plus Fragment Library Kit (Thermo Fisher Scientific, Waltham, MA, USA) and Ion Xpress Barcodes Adapters 1–25 Kit according to the manufacturer’s protocol. The DNA library was then purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). Library concentrations were determined using the Ion Universal Library Quantification Kit (Thermo Fisher Scientific, Waltham, MA, USA). The libraries were then serially diluted to a specified concentration and equal volumes of each library were pooled.
The sequencing was performed on the Ion Chef instrument (Thermo Fisher Scientific, Waltham, MA, USA), using 530 sequencing chips. Analysis of results was performed using the Ion Reporter Software (Thermo Fisher Scientific, Waltham, MA, USA) on the Ion 16S Metagenomics Kit analysis module. Reads were mapped to three reference 16S rRNA databases; Greengenes, MicroSEQ ID and SILVA database. The default parameters for processing raw sequence data by Ion Reporter Software were applied. Operational taxonomic unit analysis was performed using Ion Reporter Software, which runs Quantitative Insights into Microbial Ecology. Alpha indices txt tables were then extracted and read into R for further analysis. Data was rarefied and average read depth was 21078 per sample and samples with >7150 sequences per sample were retained for analysis.
Sequences that did not have at least 50 reads were removed for analyses. Sequence read counts were analysed at the genus level. Read counts were normalised using centred-log ratio transformation using the R package “ALDEx2”, which imputed values using Monte-Carlo instances from a Dirichlet distribution.
Statistical analyses
Luminex cytokine data points above or below the limit of detection of the assay were assigned the highest or lowest values on standard curves for each for the 25 measured analytes respectively. Values were adjusted based on the per gram mass of faecal samples and converted to pg/ml units. These cytokine concentrations were subsequently normalised by standard deviation. Adjusted normalised cytokines were used in all statistical and multivariate analyses. Principal Component Analysis (PCA) was performed using R function “prcomp”. Heatmaps dendrograms were made with hierarchical clustering using distances based on Kendall correlation and clustered with Ward D2 linkage. PERMANOVA was implemented using the R function “adonis2” from the R package “vegan.” PERMANOVA of adjusted cytokines against PCR-, case day 7, and case cay 28 sample groups was applied with restricted permutation to account for repeated measures in Case individuals. Cytokines of Case individuals were analysed cross-sectionally, at day 7 and at day 28. Cross-sectional analysis included correlation and PERMANOVA of cytokines against outcome measures DABA (at day 28), peak SASS, log10 of peak URT VL, peak GISS, faecal VL, and URT VL. Correlations were calculated using the R function “cor.test”, performing non-parametric Kendall correlation. Correlation networks of cytokines within PCR-, PCR+ case day 7 and PCR+ case day 28 sample groups were made using R package “corrr” where proximity of network nodes was determined by multidimensional scaling of the correlation matrix. Only correlations ≥0.3 are shown in the correlation networks. Pairwise sample similarity was measured using Euclidean distance. Sample similarity comparisons were organised into between group comparisons or within group comparisons. For case-control analysis sample groups were PCR-, PCR+ case day 7, and PCR+ case day 28. For severity analysis, sample groups were dictated by severity group. Dispersion of sample groups (e.g. SG or case-control groups) were calculated using R function “betadisper” from R package “vegan,” which measured the distance to the centroid for each group. Pairwise statistical testing was performed using Wilcoxon Rank Sum test. Correction for multiple testing was performed using the Benjimani-Hochberg method. Contingency test of SG across heatmap clusters were performed using χ2 test with simulated p-values. Contingency test of SG and antibiotic use was performed by Fisher’s Exact Test.
AUTHOR CONTRIBUTIONS
D.C. designed and performed experiments, analysed data, and wrote the manuscript. J.F. designed study and experiments, analysed data, and wrote the manuscript. SY advised on biostatistics, analysed data, and wrote the manuscript. N.I. and K.M. advised on biostatistics and analysed data. S.B., J.H., W.G., E.C., A.K., I.J., M.H., A.B., E.P., C.R., performed and facilitated experiments. K.J. designed study and experiments. M.M., R.T., G.T., Designed and led projects related to virological and serological assays. K.B., M.S., P.O. Designed and led the ISARIC4C study and consortium and reviewed the manuscript. C.P. designed and performed experiments and assisted in obtaining funding. J.J. designed and facilitated experiments, assisted in obtaining funding, and reviewed the manuscript. A.L. designed study and designed and facilitated experiments, obtained funding, and reviewed the manuscript. E.T. designed study, designed, performed, and facilitated experiments, obtained funding, and wrote the manuscript.
DECLARATION OF COMPETING INTEREST
JF, DC, ET have no competing interests to declare. PO has been a member of scientific advisory boards for Janssen, Cephid, Seqirus, Pfizer, GSK, Moderna and Sanofi outside the scope of this work.
FUNDING
This work is supported by the NIHR Health Protection Research Unit (HPRU) in Respiratory Infections (NIHR200927), in partnership with the UK Health Security Agency; University of Oxford COVID-19 Research Response Fund; MRC HIU core grant MC_UU_00008, Nuffield Department of Medicine. The development of the hybrid DABA assay used for quantification of SARS-CoV-2 anti-Spike RBD antibodies was supported by MRC grant: MC_PC_19078: nCoV: Serological detection of past SARS-CoV-2 infection by non-invasive sampling for field epidemiology and quantitative antibody detection.
This work uses Data/Material provided by patients and collected by the NHS as part of their care and support #DataSavesLives. The Data/Material used for this research were obtained from ISARIC4C. The COVID-19 Clinical Information Network (CO-CIN) data was collated by ISARIC4C investigators. Data and Material provision was supported by grants from: the National Institute for Health Research (NIHR; award CO-CIN-01), the Medical Research Council (MRC; grant MC_PC_19059), and by the NIHR Health Protection Research Unit (HPRU) in Emerging and Zoonotic Infections at University of Liverpool in partnership with Public Health England (PHE), (award 200907), NIHR HPRU in Respiratory Infections at Imperial College London with PHE (award 200927), Liverpool Experimental Cancer Medicine Centre (grant C18616/A25153), NIHR Biomedical Research Centre at Imperial College London (award IS-BRC-1215-20013), and NIHR Clinical Research Network providing infrastructure support.
ACKNOWLEDGMENTS
This work is supported by the NIHR Health Protection Research Units (HPRU) in Respiratory Infections (NIHR200927), in partnership with the UK Health Security Agency. The Data/Material used for this research were obtained from ISARIC4C and INSTINCT. Data/Material collected as part of ISARIC4C was provided by patients and collected by the NHS as part of their care and support #DataSavesLives. The COVID-19 Clinical Information Network (CO-CIN) data was collated by ISARIC4C investigators. Data/ Material collected as part of INSTINCT was generously provided by members of households in London and collected by INSTNCT study members. We would like to thank Dr Zoe Christodoulou, the WIMM Biological Safety Officer, for facilitating work in the MRC Weather Institute of Molecular Medicine CL3 facility. We would also like to thank Wilna Oosthuyzen and Hayley Hardwick for facilitating work with the ISARIC4C cohort.
ETHICS STATEMENT
INSTINCT Study: COVID-19 cases and household contacts were recruited into the INSTINCT study protocol (IRAS282820). Written informed consent was obtained from all participants. Ethical approval was given by the North West Greater Manchester East Research Ethics Committee in England (REC reference 20/NW/0231).
ISARIC4C study: Identified patients hospitalised during the SARS-COV-2 pandemic were recruited into the International Severe Acute Respiratory and Emerging Infection Consortium World Health Organization Clinical Characterisation Protocol UK (IRAS260007 and IRAS126600). Written informed consent was obtained from all patients. Ethical approval was given by the South Central–Oxford C Research Ethics Committee in England (reference: 13/SC/0149), Scotland A Research Ethics Committee (reference: 20/SS/0028) and World Health Organization Ethics Review Committee (RPC571 and RPC572l; 25 April 2013).
COLLABORATORS
Prof Maria Zambon, Prof. Jake Dunning and their teams at UKHSA collaborated with NHS Test and Trace to identify and report COVID-19 cases to the INSTINCT study team
PATIENT AND PUBLIC INVOLVEMENT
Central research questions posed in the INSTINCT and ISARIC studies were designed to answer pressing public health priorities during the early stages of the COVID-19 pandemic. In INSTINCT, clinical research nurses interacted with members of the public in their homes, directly providing information pertinent to the project to participants. Reciprocally, feedback from participants informed amendment of INSTINCT study documents including Participant Information Sheets and Case Record Forms. The hospitalised portion of the study was designed and performed early in the COVID19 pandemic, and therefore did not include public involvement. Patient involvement was needed for primary data collection but was not involved in design, conduct, reporting, or dissemination of this work.
Footnotes
Supplementary material to this article can be found online at https://doi.org/10.1016/j.mucimm.2023.11.005.
Contributor Information
Joe Fenn, Email: jfenn@ic.ac.uk.
Emily E. Thornton, Email: emily.thornton@imm.ox.ac.uk.
Appendix A. Supplementary material
The following are the Supplementary material to this article:
DATA AVAILABILITY STATEMENT
Anonymised data is available upon reasonable request. Data generated by the ISARIC4C consortium is available for collaborative analysis projects through an independent data and materials access committee at isaric4c.net/sample access.
REFERENCES
- 1.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Pover. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao F., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamers M.M., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zang R., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu H., et al. SARS-CoV-2 induces a more robust innate immune response and replicates less efficiently than SARS-CoV in the human intestines: an ex vivo study with implications on pathogenesis of COVID-19. Cell. Mol. Gastroenterol. Hepatol. 2021;11:771–781. doi: 10.1016/j.jcmgh.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo T., et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276–284. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 9.Mao R., et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redd W.D., et al. Prevalence and characteristics of gastrointestinal symptoms in patients with severe acute respiratory syndrome coronavirus 2 infection in the United States: a multicenter cohort study. Gastroenterology. 2020;159:765–767.e2. doi: 10.1053/j.gastro.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marasco G., et al. Prevalence of gastrointestinal symptoms in severe acute respiratory syndrome coronavirus 2 infection: results of the prospective controlled multinational GI-COVID-19 study. Am. J. Gastroenterol. 2022;117:147–157. doi: 10.14309/ajg.0000000000001541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britton G.J., et al. Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS-CoV-2-specific IgA in patients with acute COVID-19. Sci. Rep. 2021;11:13308. doi: 10.1038/s41598-021-92740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livanos A.E., et al. Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 2021;160:2435–2450.e34. doi: 10.1053/j.gastro.2021.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shehab M., Alrashed F., Shuaibi S., Alajmi D., Barkun A. Gastroenterological and hepatic manifestations of patients with COVID-19, prevalence, mortality by country, and intensive care admission rate: systematic review and meta-analysis. BMJ Open Gastroenterol. 2021;8:e000571. doi: 10.1136/bmjgast-2020-000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wölfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morone G., et al. Incidence and persistence of viral shedding in COVID-19 post-acute patients with negativized pharyngeal swab: a systematic review. Front. Med. (Lausanne) 2020;7:562. doi: 10.3389/fmed.2020.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergamaschi L., et al. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity. 2021;54:1257–1275.e8. doi: 10.1016/j.immuni.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thwaites R.S., et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Valle D.M., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakki S., et al. Onset and window of SARS-CoV-2 infectiousness and temporal correlation with symptom onset: a prospective, longitudinal, community cohort study. Lancet Respir. Med. 2022;10:1061–1073. doi: 10.1016/S2213-2600(22)00226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houston H., et al. Broadening symptom criteria improves early case identification in SARS-CoV-2 contacts. Eur. Respir. J. 2022;60:2102308. doi: 10.1183/13993003.02308-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sefik E., et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 2022;606:585–593. doi: 10.1038/s41586-022-04802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junqueira C., et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;606:576–584. doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison O.J., et al. Epithelial-derived IL-18 regulates Th17 cell differentiation and Foxp3+ Treg cell function in the intestine. Mucosal Immunol. 2015;8:1226–1236. doi: 10.1038/mi.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., et al. Robust cross-cohort gut microbiome associations with COVID-19 severity. Gut Microbes. 2023;15:2242615. doi: 10.1080/19490976.2023.2242615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natarajan A., et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med. 2022;3:371–387.e9. doi: 10.1016/j.medj.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teyssou E., et al. Long-term evolution of humoral immune response after SARS-CoV-2 infection. Clin. Microbiol. Infect. 2022;28:1027.e1–1027.e4. doi: 10.1016/j.cmi.2022.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews C., McLean M.H., Durum S.K. Cytokine tuning of intestinal epithelial function. Front. Immunol. 2018;9:1270. doi: 10.3389/fimmu.2018.01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Däbritz J., et al. Reprogramming of monocytes by GM-CSF contributes to regulatory immune functions during intestinal inflammation. J. Immunol. 2015;194:2424–2438. doi: 10.4049/jimmunol.1401482. [DOI] [PubMed] [Google Scholar]
- 31.Rosadas C., et al. Detection and quantification of antibody to SARS CoV 2 receptor binding domain provides enhanced sensitivity, specificity and utility. J Virol Methods. 2022;302:114475. doi: 10.1016/j.jviromet.2022.114475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymised data is available upon reasonable request. Data generated by the ISARIC4C consortium is available for collaborative analysis projects through an independent data and materials access committee at isaric4c.net/sample access.