Abstract
A tripartite domain of the murine immunoglobulin μ heavy-chain enhancer contains the μA and μB elements that bind ETS proteins and the μE3 element that binds leucine zipper-containing basic helix-loop-helix (bHLH-zip) factors. Analysis of the corresponding region of the human μ enhancer revealed high conservation of the μA and μB motifs but a striking absence of the μE3 element. Instead of bHLH-zip proteins, we found that the human enhancer bound core binding factor (CBF) between the μA and μB elements; CBF binding was shown to be a common feature of both murine and human enhancers. Furthermore, mutant enhancers that bound prototypic bHLH-zip proteins but not CBF did not activate transcription in B cells, and conversely, CBF transactivated the murine enhancer in nonlymphoid cells. Taking these data together with the earlier analysis of T-cell-specific enhancers, we propose that ETS-CBF is a common composite element in the regulation of antigen receptor genes. In addition, these studies identify the first B-cell target of CBF, a protein that has been implicated in the development of childhood pre-B-cell leukemias.
The immunoglobulin μ heavy-chain (IgH) gene enhancer (μ enhancer), located in the JH-Cμ intron, is necessary for IgH gene expression in B lymphocytes (17, 23). The μ enhancer has also been shown to play a key role in the initiation of IgH gene rearrangements in the most immature B-cell precursors (2, 30, 32, 42). These observations indicate that detailed analysis of the μ enhancer will provide insights into the general problem of enhancer function as well as early regulatory events in B lymphopoiesis.
Studies using the murine μ enhancer have shown that the enhancer contains binding sites for several nuclear factors that mediate its transcription-activating function (6). μ enhancer binding proteins can be broadly classified into two groups: those whose expression is tissue restricted such as the μA, μB, and octamer motif binding proteins; and those whose expression is more ubiquitous, such as the basic helix-loop-helix (bHLH) family of transcription factors that bind the μE1 to μE5 motifs. How these two kinds of protein factors collaborate to produce a functional, cell-specific enhancer is unknown. Furthermore, mutation of individual motifs within the enhancer does not significantly affect enhancer activity, indicating a degree of functional redundancy among the various motifs that have been identified (16).
To simplify the analysis of this enhancer, we have previously described a minimal domain of the murine μ enhancer containing the μA, μB, and μE3 motifs that is active in B cells (24). Based on the observation that minimal enhancer activity depends on all three motifs, we proposed that this domain contains no redundant elements. The μA and μB elements bind the ETS domain proteins Ets-1 and PU.1, respectively, whereas the μE3 element binds several members of the bHLH-zip (leucine zipper-containing bHLH) family, such as TFE3 and USF. Thus, like the full μ enhancer, the minimal enhancer is composed of binding sites for tissue-restricted (PU.1) and ubiquitously expressed (TFE3 and USF) factors, suggesting that it is a good model in which to examine the mechanism of enhancer function. To strengthen the proposed importance of the minimal enhancer, in this study we examined the corresponding region of the intronic μ enhancer from the human IgH locus (11).
We found that the sequences of the μA and μB sites, as well as the spacing between them, were highly conserved between the two enhancers. Consistent with this observation, Ets-1 and PU.1 proteins bound to these sites. However, the intervening μE3 element was less well conserved between the two enhancers, and we detected no binding of either of two prototypic bHLH-zip proteins, TFE3 and USF, to the human enhancer. Because transcriptional activity of the minimal murine enhancer requires an intact μE3 site, we predicted that the lack of a μE3-like element in the human enhancer would render a corresponding minimal human enhancer fragment inactive in transfection assays. This was not the case. A μA/μB-containing region of the human enhancer was as active as the minimal murine enhancer in S194 plasma cells. Mutagenic analysis further showed that sequences between the μA and μB elements were necessary for enhancer activity, suggesting that the minimal human μ enhancer also required an element in addition to the ETS protein binding sites. We found that the intervening element bound the transcription factor CBF (core binding factor; also known as PEBP2 or AML1 [15, 39]), and binding was disrupted in all mutants that were inactive in transfection assays. These observations identify the first B-cell-specific target of CBF, a factor that has previously been implicated in the activation of several T and myeloid cell-specific promoters and enhancers (7, 12, 27, 35, 41, 44), and demonstrate that ETS-CBF is a common composite element in antigen receptor gene enhancers.
MATERIALS AND METHODS
Mammalian and bacterial expression plasmids.
The PU.1 (pEVRF-PU.1), Ets-1 (pEVRF-Ets-1), and CBFα2451 [pcDNA/CBFα2(451)] expression vectors have been previously described (5, 43). The bacterial expression plasmids His-PU.1 and His-ETS(Ets-1) are described in reference 25.
The bacterial expression plasmids GST (glutathione S-transferase)-TFE3 (an NcoI-EcoRI fragment of the TFE3 cDNA filled in with Klenow enzyme, ligated into pGEX2T [Pharmacia Biotech, Inc.] cut with SmaI) and GST-USF were a gift of K. Calame, Columbia University, New York, N.Y. All expression plasmids were sequenced to ensure that the appropriate reading frame was maintained.
Construction of reporter plasmids.
The μ70 dimer reporter was described previously (24). The murine and human μ51 wild-type (mWT and hWT) dimer reporters were constructed by ligating two tandem repeats of the annealed oligonucleotides 5′ TCG ACC TGG CAG GAA GCA GGT CAT GTG GCA AGG CTA TTT GGG GAA GGG AAC 3′ and 5′ TCG AGT TCC CTT CCC CAA ATA GCC TTG CCA CAT GAC CTG CTT CCT GCC AGG 3′ (mWT) and 5′ TCG ACC TGG CAG GAA GCA GGT CAC CGC GAG AGT CTA TTT TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT AAA ATA GAC TCT CGC GGT GAC CTG CTT CCT GCC AGG 3′ (hWT) into the Δ56CAT enhancerless reporter plasmid digested with SalI. The human μ51 mutant (hM1 to hM6) and murine μC− (mμC−) reporters were constructed similarly with the following annealed oligonucleotides: hM1, 5′ TCG ACC TGG CAG GAA GCA ttg CAC CGC GAG AGT CTA TTT TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT AAA ATA GAC TCT CGC GGT Gca aTG CTT CCT GCC AGG 3′; hM2, 5′ TCG ACC TGG CAG GAA GCA GGT ata CGC GAG AGT CTA TTT TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT AAA ATA GAC TCT CGC Gta tAC CTG CTT CCT GCC AGG 3′; hM3, 5′ TCG ACC TGG CAG GAA GCA GGT CAt atg GAG AGT CTA TTT TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT AAA ATA GAC TCT Cca taT GAC CTG CTT CCT GCC AGG 3′; hM4, 5′ TCG ACC TGG CAG GAA GCA GGT CAC Ctc tAG AGT CTA TTT TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT AAA ATA GAC TCT aga GGT GAC CTG CTT CCT GCC AGG 3′; hM5, 5′ TCG AGT TGG CAG GAA GCA GGT CAC CGC GAG gta CTA TTT TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT AAA ATA Gta cCT CGC GGT GAC CTG CTT CCT GCC AGG 3′; hM6, 5′ TCG ACC TGG CAG GAA GCA GGT CAC CGC GAG AGT CTA ccc TAG GAA GCA AAC 3′ and 5′ TCG AGT TTG CTT CCT Agg gTA GAC TCT CGC GGT GAC CTG CTT CCT GCC AGG 3′; and mμC−, 5′ TCG ACC TGG CAG GAA GCA GGT CAT GTG GaA AGG CTA TTT GGG GAA GGG AAC 3′ and 5′ TCG AGT TCC CTT CCC CAA ATA GCC TTt CCA CAT GAC CTG CTT CCT GCC AGG 3′. The mutated nucleotides are in lowercase and underlined. All reporter plasmids were sequenced to ensure that the appropriate mutations were introduced.
Cell culture, transfections, and CAT assays.
S194 cells were grown in RPMI medium supplemented with 5% newborn serum, 5% inactivated fetal calf serum, and 50 μg each of penicillin and streptomycin per ml. M12 cells were grown in RPMI medium supplemented with 10% inactivated fetal calf serum and 50 μg each of penicillin and streptomycin per ml. Murine and human dimeric enhancer-containing chloramphenicol acetyltransferase (CAT) reporter plasmids (5 μg) were transfected into S194 and M12 cells by the DEAE-dextran method as previously described (24); 40 to 48 h after transfection, whole-cell extracts were prepared by three rounds of freeze-thawing, and the levels of CAT protein in the extracts were determined by CAT enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim Corp.) according to the manufacturer’s instructions.
HeLa cells were grown in Dulbecco modified Eagle medium supplemented with 10% newborn serum and 50 μg each of penicillin and streptomycin per ml. μ70 wild-type, μA−, μE3−, and μB− dimeric enhancer-containing CAT reporter plasmids (2 μg) were cotransfected with PU.1 (1 or 2 μg), Ets-1 (1 or 2 μg), or CBFα2 (2 μg) expression vectors into HeLa cells by the calcium phosphate method. Plasmid pEVRF2 (18) was included as a carrier to maintain a total of 6 μg of DNA per transfection. Briefly, 6 × 105 cells were split into individual plates 2 to 4 h before transfection, and the DNA-containing calcium phosphate precipitate was gently dropped on the medium. The cells were washed with fresh medium at 16 h; after harvesting at 40 to 48 h, whole-cell extracts were prepared by three rounds of freeze-thawing, and the level of CAT protein in 100 μg of extract was determined by CAT ELISA (Boehringer Mannheim) according to the manufacturer’s instructions.
In vitro protein expression, EMSAs, and supershifts.
Full-length PU.1 and the ETS domain of Ets-1 [Ets-1(ETS)] proteins were expressed as hexahistidine-tagged proteins (25). Full-length TFE3 and USF proteins were expressed as GST fusion proteins and were purified as described by the manufacturer (Pharmacia Biotech, Inc.) The CBFα241-190 and CBFα241-214 proteins containing the DNA binding Runt domain of CBFα2 were prepared as previously described (3). For electrophoretic mobility shift assays (EMSAs), either bacterially expressed and purified proteins or 4, 8, and 16 μg of S194 nuclear extracts were incubated with 32P-labeled oligonucleotide DNA probes (20,000 cpm) in the presence of 25 ng of poly(dI-dC) · (dI-dC) (1 μg for extracts), 70 mM NaCl, and 10% glycerol for 10 min at room temperature, and reactions were resolved on a 4% polyacrylamide gel. Wild-type and mutant probes in each experiment were of comparable specific activity. EMSAs in Fig. 5, 6, 8, and 9 were performed with annealed double-stranded oligonucleotides, whereas those in Fig. 7 were performed with PstI-BamHI fragments from the murine enhancer (bp 380 to 433) as described elsewhere (24). The CBF high-affinity consensus probe used for Fig. 8 was obtained by annealing two oligonucleotides, 5′-AAT TCG AGT ATT GTG GTT AAT ACG-3′ and 5′-AAT TCG TAT TAA CCA CAA TAC TGG-3′. Supershifts were performed by incubating 20 μg of S194 or 27 μg of 70Z extracts with 32P-labeled oligonucleotide DNA probes (50,000 cpm) for 20 min at room temperature, followed by incubation with the specified antibodies for an additional 30 min on ice (21, 22).
FIG. 5.
DNA binding of PU.1 and Ets-1 to the human enhancer mutants. EMSAs were carried out with bacterially expressed and purified His-Ets-1(ETS) (lanes 1 to 6) and His-PU.1 (lanes 7 to 12) proteins and hWT and mutant enhancer probes as indicated above the lanes. The mutant probes are numbered as in Fig. 4B. Specific nucleoprotein complexes are indicated by arrows 1 [His-Ets-1(ETS)–DNA] and 2 (His-PU.1–DNA). A lower-mobility complex in the lanes with the PU.1 protein is due to the double occupancy of the μB and μA sites.
FIG. 6.
CBFα2 binds the human IgH μ enhancer in vitro. (A) EMSAs were carried out with bacterially expressed and purified CBFα241-190, which contains the DNA binding Runt domain of CBFα2, and hWT and mutant enhancer probes as indicated. Specific nucleoprotein complexes in lanes 1, 2, 5, and 6 are indicated by an arrow. (B) In vitro competition assays with CBFα241-190 bound to the human WT probe. EMSAs were carried out as described in the text, with 25-, 125-, and 250-fold molar excesses of competitor DNA fragments indicated by triangles. Competitor DNA was excised as dimeric fragments from reporter plasmids used for transfections in Fig. 4C and contain wild-type or mutated human enhancer sequences as indicated.
FIG. 8.
The murine and human μ enhancers bind CBF in B-cell nuclear extracts. (A) In vitro competition assays with CBF bound to a high-affinity consensus binding probe. EMSAs were carried out with 20 μg of S194 nuclear extracts with 32P-labeled CBF oligonucleotide DNA probes (50,000 cpm), in the presence of no competitor DNA (lane 1), 5-, 25-, and 133-fold molar excesses of murine μ enhancer DNA (lanes 2 to 4) and human μ enhancer DNA (lanes 5 to 7), and 8- and 33-fold molar excesses of self (lanes 8 and 9) and 16-, 32-, and 66-fold molar excesses of nonspecific (lanes 10 to 12) competitor oligonucleotides, indicated by triangles. Specific nucleoprotein complexes are indicated by an arrow. The nonspecific competitor is a high-affinity binding site for Ets-1 (28). (B) Supershift EMSAs were carried out with 20 μg of S194 plasma cell and 27 μg of 70Z pre-B-cell extracts and a CBF high-affinity consensus binding site probe. Lanes 1 to 5 show complexes formed by the incubation of S194 extracts with CBF probes followed by no antiserum (lane 1), antiserum specific for AML-1, -2, and -3 (lanes 2, 3, and 4, respectively), and normal rabbit serum (NRS) (lane 5). Lanes 6 to 10 show complexes formed by the incubation of 70Z extracts with CBF probes followed by no antiserum (lane 6), antiserum specific for AML-1, -2, and -3 (lanes 7, 8, and 9, respectively), and normal rabbit serum (lane 10). Specific complexes are indicated by an arrow on the left.
FIG. 9.
CBFα2 binding correlates with minimal μ enhancer activity in B-cell lines. (A) Sequences of murine and human wild-type enhancers compared to sequences of two mutants in the murine (mμC−) and human (hM3) enhancers. The overlapping μE3 (bHLH-zip protein binding) and μC (CBF binding) sites in the murine enhancer are overlined and underlined, respectively. The murine μC− mutation alters the single nonoverlapping base between the two motifs. This position in the CBFα2 consensus binding site has been shown to be important for binding (19). The μC motif in the human motif is also underlined and is shown to be mutated in the hM3 mutation, which introduces an E-box motif into the human enhancer sequence that is absent in the wild-type sequence. (B) EMSAs were carried out with bacterially expressed and purified GST-TFE3, CBFα241-214, and S194 plasma cell extracts and the indicated murine and human probes. Lanes 1 to 4 show complexes formed by the incubation of approximately 50 ng of GST-TFE3 with the indicated probes. Specific complexes in lanes 1, 2, and 4 are indicated by the top arrow on the left, and a nonspecific complex is indicated by an asterisk. Lanes 5 to 8 show complexes formed by the incubation of 100 ng of CBFα241-214 and the indicated probes. Specific nucleoprotein complexes in lanes 5 and 7 are indicated by the bottom arrow on the left. Lanes 9 to 11 and 12 to 14 show complexes formed by the incubation of 4, 8, and 16 μg of S194 extracts, respectively, with the mWT and mμC− probes. The μE3 binding complex is indicated by the arrow on the right. (C) Transcriptional activity of the murine enhancer. Reporter plasmids (5 μg) containing dimeric murine wild-type [WT or (μ70)2] and mutant (μC−) enhancers or no enhancer (Δ56) were transfected into the M12 B-cell line, and CAT assays were performed by ELISA as described in the Materials and Methods. CAT enzyme activity is shown on the y axis as the percentage of the activity of the reporter plasmid containing the wild-type murine (μ70)2 enhancer. Results shown are the averages of at least two transfections carried out in duplicate. Error bars indicate the average deviations of the data.
FIG. 7.
CBFα2 binds the murine IgH μ enhancer in vitro. EMSAs were carried out with bacterially expressed and purified CBFα241-190, which contains the DNA binding Runt domain of CBFα2, and the mWT μ enhancer and μA, μE3, and μB mutant enhancer probes as indicated.
RESULTS
The human μ enhancer does not contain a μE3 element.
To extend our ongoing characterization of the murine IgH μ enhancer, we examined the organization of the human μ enhancer. Of the several bHLH protein binding sites known in the murine enhancer, μE1, μE2, and μE4 were easily recognizable in the human enhancer (Fig. 1). Although the μE5 and μE3 sites showed regions of similarity, they were significantly less conserved (Fig. 1). Specifically, the regions corresponding to both the μE3 and μE5 sites lacked one half of the minidyad that is characteristic of the μE elements. In contrast, the lymphoid cell-restricted elements μA, μB, and octamer were highly conserved between the two enhancers, as was the spacing between the μA and μB elements (Fig. 1). Thus, two of the three elements present in the minimal murine enhancer (consisting of μA, μB, and μE3 elements) were conserved in the human sequence.
FIG. 1.
Alignment of IgH μ enhancer sequences. Sequences of IgH μ enhancer from four different species (GenBank accession no. V01523 for mouse, M13799 for rat, K01901 for human, and X13700 for rabbit) were aligned by using the Pileup program in the Wisconsin package version 8.1 (Genetics Computer Group, Madison, Wis.). Elements containing previously identified recognition motifs are overlined, and positions where the nucleotides from all species are identical are indicated by asterisks.
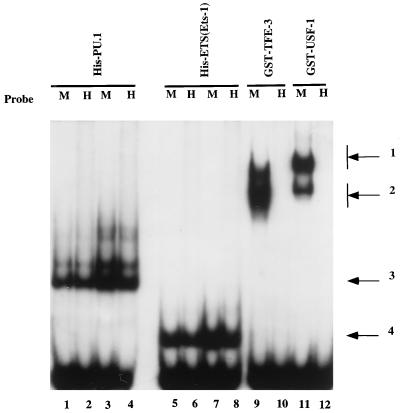
We examined the binding of μA and μB binding proteins to the human enhancer to directly establish the validity of the sequence comparisons. For these experiments, full-length PU.1 (μB binding protein) and Ets-1(ETS) (μA binding protein) were expressed as hexahistidine-tagged proteins in bacteria. The proteins were purified by adsorption to nickel chelate resins and used in EMSAs. Prototypic μE3 binding proteins, TFE3 and USF, were expressed as GST fusion proteins. Both murine and human enhancer sequences bound His-tagged PU.1 and Ets-1(ETS) comparably (Fig. 2, lanes 1 to 8). TFE3 and USF bound well to the murine probe but not at detectable levels to the human probe (Fig. 2, lanes 9 to 12). We conclude that the five nucleotides of the putative human μE3 element that are identical to the murine sequence at the 5′ end of the site (Fig. 1) do not allow efficient binding of either of these bHLH-zip proteins to the human enhancer.
FIG. 2.
DNA binding analysis of PU.1, Ets-1, TFE3, and USF1 to the murine and human enhancers. The human (H) and murine (M) μ enhancer probes were used in binding assays with the following: lanes 1 and 2, His-PU.1 (80 ng); lanes 3 and 4, His-PU.1 (160 ng); lanes 5 and 6, Ets-1(ETS) (100 ng); lanes 7 and 8, Ets-1(ETS) (200 ng); lanes 9 and 10, GST-TFE3 (50 ng); and lanes 11 and 12, GST-USF1 (40 ng). Arrows: 1 and 2, USF and TFE3 binding to the murine probe only; 3, PU.1-DNA complex; 4, Ets-1(ETS)–DNA complex. EMSAs were performed as described previously (5).
Functional analysis of the minimal human μ enhancer.
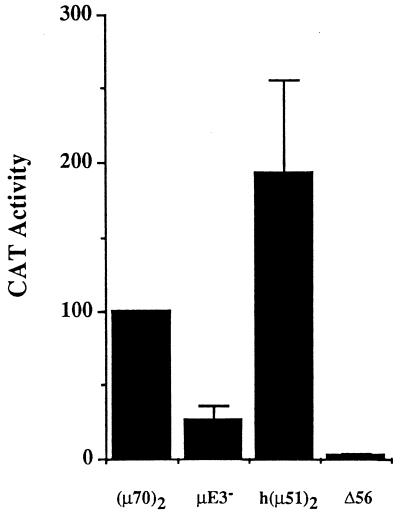
Our earlier analysis of the minimal murine μ enhancer showed that mutation of the μE3 site significantly decreased enhancer activity in B cells. Absence of a μE3-like element in the human μ enhancer suggested that a corresponding fragment of this enhancer would have very low enhancer activity, similar to that of the μE3 mutated murine enhancer. To check if this was so, we tested the transcription activation properties of a μA/μB-containing fragment of the human enhancer. Synthetic oligonucleotides encompassing the μA/μB elements from the human enhancer were cloned as dimers 5′ of a CAT reporter gene transcribed from a c-fos gene promoter and assayed by transient transfection in S194 plasma cells. Surprisingly, this human μ enhancer fragment was as active as the minimal murine μ70 enhancer in S194 plasma cells (Fig. 3) but not in nonlymphoid cells (data not shown). As expected, high-level activity of the murine enhancer was dependent on the μE3 site because a μE3 mutation significantly decreased activity. We conclude that despite the absence of a recognizable μE3-like element that can bind bHLH-zip proteins such as TFE3 and USF, a fragment of the human enhancer spanning the μA and μB sites is a functional B-cell-specific enhancer, which we shall refer to as the minimal human enhancer.
FIG. 3.
The minimal murine and human IgH μ enhancers activate transcription comparably. Reporter plasmids (5 μg) containing dimeric murine μ70 [(μ70)2], murine μE3 mutant (μE3−), and human [h(μ51)2] enhancers were transfected into S194 plasma cell lines, and CAT assays were performed by ELISA as described in Materials and Methods. CAT enzyme activity is shown on the y axis as the percentage of the amount of CAT enzyme obtained with the dimeric murine μ70 reporter. Results show the averages of at least two transfections carried out in duplicate. Δ56 refers to an enhancerless reporter. Error bars indicate the average deviations of the data.
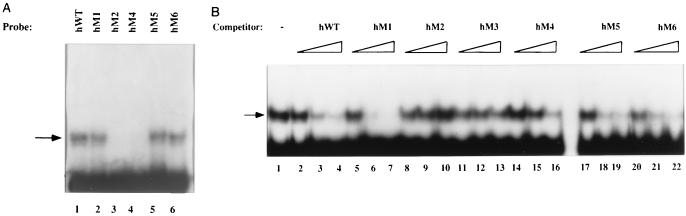
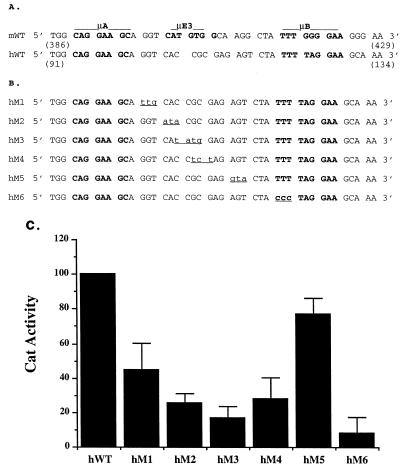
Activity of the minimal human enhancer may be due only to the μA and μB motifs and their respective binding proteins, or it may require additional factors. To distinguish between these possibilities, we analyzed a panel of mutations that introduced changes in the sequence between the μA and μB sites (Fig. 4A and B). All mutant fragments were obtained as synthetic oligonucleotides, cloned as dimers in the fos-CAT vector, and assayed by transient transfection into S194 cells (Fig. 4C). Mutation of the conserved nucleotides just 3′ of the μA site (hM1) reduced but did not eliminate enhancer activity. In vitro binding assays suggested that reduced binding of Ets-1 to the μA element may be partly responsible for the observed decrease. However, mutations in the nonconserved region further 3′ resulted in diminished enhancer activity similar to the μE3− mutation in the murine enhancer (Fig. 4C). These observations are consistent with the requirement for an additional factor, other than ETS proteins at μA and μB sites, for activity of the minimal human enhancer in B cells. As observed previously with the murine enhancer, mutation of the μB element in the human enhancer (hM6) abolished enhancer activity. We propose that the minimal human enhancer is also activated by three closely positioned factors.
FIG. 4.
Identification of an element in the human IgH enhancer in the region corresponding to the murine μE3 motif. (A) Comparison of the murine and human enhancers indicating the absence of a μE3 motif in the human enhancer. The μA, μE3, and μB motifs are indicated in boldface, and the nucleotide numbers of the murine and human enhancers are from references 8 and 11, respectively. (B) Sequences of a panel of mutants in the human enhancer (hM1 to hM6) corresponding to the region spanning the murine μE3 motif. The altered sequence in each mutant is indicated in lowercase and underlined. (C) Transcriptional activities of the minimal human mutant enhancers. Reporter plasmids (5 μg) containing dimeric wild-type hWT and mutant (hM1 to hM6) enhancers were transfected into S194 plasma cell lines, and CAT assays were performed by ELISA as described in Materials and Methods. CAT enzyme activity is shown on the y axis as the percentage of the activity of the reporter plasmid containing the hWT enhancer. Results shown are the averages of at least two transfections carried out in duplicate. Error bars indicate the average deviations of the data.
To ensure that mutations hM1 to M6 did not disrupt DNA-protein interaction at the μA or μB site, we used EMSA to study the binding of PU.1 and Ets-1 to the mutant enhancers. All of the human enhancer derivatives bound Ets-1(ETS) (Fig. 5, lanes 1 to 6), and all except hM6 bound PU.1 (Fig. 5, lanes 7 to 12), indicating that the functional effects described above were not due to mutations in the μA or μB site. These results strengthen the conclusion that a third, unidentified factor is required for activity of the minimal human enhancer.
Analysis of proteins binding to the human μ enhancer.
Close examination of the human sequence revealed a similarity to the recognition site of CBF, the consensus binding site for which is PyGPyGGT (15, 19, 37). On the noncoding strand of the human enhancer, the nucleotides corresponding to the murine μE3 motif are 5′-CGCGGT-3′ (Fig. 1). We therefore tested whether CBFα2 (the DNA binding subunit of CBF) bound to the human enhancer fragment.
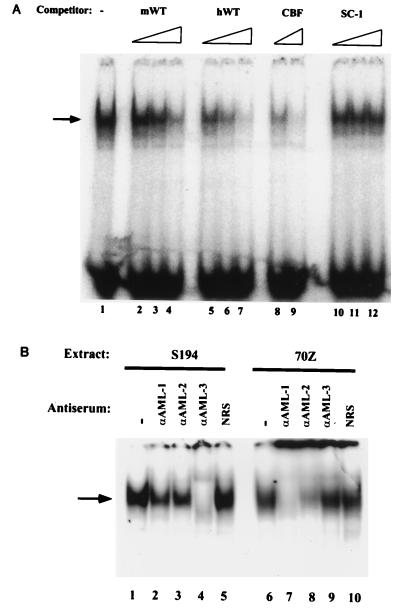
Bacterially expressed DNA binding (Runt) domain from CBFα2 (AML1) formed a discrete nucleoprotein complex with the human enhancer probe (Fig. 6A, lane 1). To strengthen the conjecture that the activity of the human enhancer may be mediated by CBF, we also analyzed the panel of intervening site mutants that were tested by transient transfection. The inactive mutants, hM2 and hM4, did not bind CBFα2 efficiently (Fig. 6A, lanes 3 and 4), whereas the transcriptionally active mutants hM1 and hM5 bound CBFα2 in vitro (Fig. 6A, lanes 2 and 5). The transcriptionally inactive μB mutant, hM6, also retained CBFα2 binding (Fig. 6A, lane 6), whereas the inactive mutant hM3 did not bind CBFα2 (see Fig. 9). The close correspondence between CBFα2 binding and functionally active intervening site mutants suggested that the human enhancer is activated by a combination of ETS proteins and CBF.
The wild-type and mutated human enhancer sequences were further characterized by in vitro competition assays (Fig. 6B). The CBFα2 Runt domain and a wild-type human enhancer probe were incubated in the presence of increasing amounts of competitor oligonucleotides. The hWT sequence as well as hM1, hM5, and hM6 competed efficiently for CBFα2 binding, whereas hM2 to hM4 competed inefficiently even at the highest concentrations tested. Subtle variations were seen between different competitors; for example, in several experiments hM1 competed more efficiently than hWT, and hM4 retained some protein binding as shown by detectable competition at its highest concentration. The increased affinity of hM1 may reflect a dependence on flanking sequences beyond the core recognition site for CBFα2 binding, and the weakness of hM4 may be because all the critical guanosine residues are left unaltered in this mutation (Fig. 4B). We note that the transcriptional activities of mutations hM2 to hM4 in B cells partially recapitulate the relative affinities of these sequences for CBFα2 in vitro. For example, in hM3, three of the four guanosines in the core CBFα2 recognition site have been altered, and this sequence has the least transcriptional activity. In contrast, hM4, which is the most transcriptionally active of the three mutations, also retains more CBFα2 binding ability.
The murine enhancer also binds CBF.
The results presented above were consistent with the idea that the minimal domain of the human enhancer examined here is activated by ETS domain proteins binding to μA and μB sites and a CBF family member binding to the intervening sequence. The human enhancer has two features that are similar to the murine enhancer: it requires μA and μB binding proteins, and three protein binding sites are required for transcriptional activity. The major difference between the two is that the murine enhancer is believed to be activated by bHLH-zip proteins such as TFE3, whereas the human enhancer does not bind TFE3 and may be activated by CBF. Although it was possible that ETS domain proteins combined with different factors to activate the two enhancers, we tested whether CBF binding was a common feature of both enhancers. Fragments of the wild-type murine enhancer and mutations thereof were assayed for CBF binding by EMSA. The wild-type sequence as well as mutants μA and μB bound CBFα2 in vitro (Fig. 7, lanes 1, 2, and 4); however, mutant μE3 did not (Fig. 7, lane 3). We conclude that the murine enhancer contains a CBF binding site between the μA and μB elements, which is lost in mutant μE3−. Thus, CBF binding is a common feature of both enhancers.
The μ enhancers bind CBF present in B-cell nuclear extracts.
To detect CBF DNA binding activity in B-cell extracts, we used a high-affinity CBF binding site in EMSA. In S194 plasma cell extracts, a discrete nucleoprotein complex was detected with this probe (Fig. 8A, lane 1). This complex was specific, because it could be competed with the self oligonucleotide (Fig. 8A, lanes 8 and 9) but not with a high-affinity Ets-1 binding site (Fig. 8A, lanes 10 to 12). The murine and human enhancer sequences also competed this complex (Fig. 8A, lanes 2 to 4 and 5 to 7, respectively), although approximately fourfold-higher levels were required compared to the self competitor. Furthermore, a murine enhancer fragment containing a mutated μE3 element did not compete for CBF binding in S194 extracts (data not shown). We conclude that the murine and human Ig enhancer sequences bind endogenous B cell CBF with comparable affinities.
To further characterize the CBF proteins present in B-cell lines, we performed antibody supershift experiments using S194 and 70Z pre-B-cell nuclear extracts. The levels of CBF binding were comparable in the two extracts (Fig. 8B, lanes 1 and 6), indicating that CBF proteins are expressed at early stages of B-cell differentiation. EMSAs were carried out in the presence of antibodies specific for the three AML proteins, AML-1 (CBFα2), AML-2 (CBFα3), and AML-3 (CBFα1) (21, 22). The CBF complex in S194 cells was reduced significantly only with an anti-AML-3 antibody (Fig. 8B, lane 4), while the complex in 70Z cells was most sensitive to the anti-AML-1 antibody (Fig. 8B, lane 7). Consistent with earlier observations of Meyers et al. (21, 22), we observed a similarly migrating complex in Jurkat T-cell nuclear extracts, which was affected by the anti-AML-1 antibody (data not shown). These results demonstrate that different CBF proteins are expressed during B-cell development; however, the functional significance of this difference is unclear at present. AML-1 mRNA was also detected in two pre-B-cell lines, a B-cell line and several T-cell lines (data not shown).
CBF binding correlates with both murine and human enhancer activities.
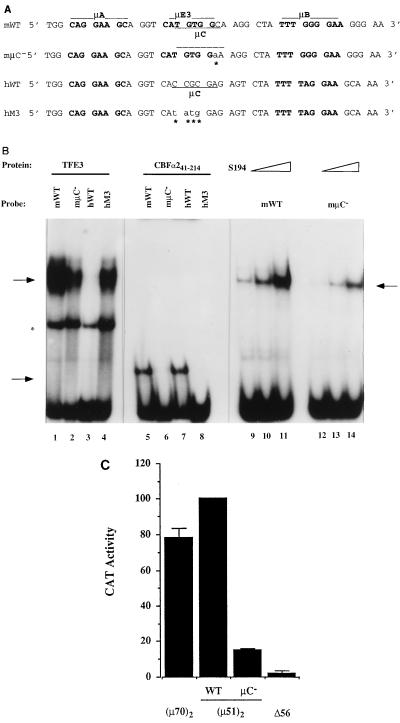
Analysis of the human enhancer suggested that ETS domain proteins plus CBF are sufficient to generate transcriptional activity in B cells. Furthermore, the murine enhancer was found to contain a CBF binding site that overlapped the μE3 site. It is likely that CBF binds to the TGTGG motif of the murine enhancer, which is also a part of the CATGTGG recognition site of bHLH-zip proteins. Because the μE3 mutation also eliminated CBF binding, we could not ascertain whether CBF and bHLH-zip proteins (such as TFE3) could both provide the third essential component to the tripartite enhancer. Ideally we wanted to analyze an enhancer that bound TFE3, but not CBF, to determine whether bHLH-zip proteins could confer the observed properties of this enhancer. In an attempt to distinguish between CBF and TFE3 binding to the murine enhancer, we mutated the C residue 3′ of the CBF core recognition site to an A (Fig. 9A), because previous studies showed that TGTGGA was recognized poorly by CBFα2 (37). This mutation, we anticipated, would substantially reduce affinity for CBF binding while retaining TFE3 binding. Fortuitously, an analogous situation was created in the hM3 human enhancer mutation, where the sequence CATATGG is similar to the murine μE3 site and may therefore bind TFE3 (Fig. 9A).
Protein binding to the mutated enhancers was assayed by EMSA. Recombinant TFE3 bound strongly to the murine enhancer probe (Fig. 9B, lane 1) but not to the wild-type human enhancer sequence (Fig. 9B, lane 3). The single-base-mutated murine sequence μC− retained TFE3 binding, though binding affinity was reduced approximately twofold (Fig. 9B, lane 2); however, CBFα2 binding was undetectable (Fig. 9B, lanes 5 and 6). The hM3 human sequence, in sharp contrast to its wild-type counterpart, gained significant TFE3 binding (Fig. 9B, lane 4) while losing the ability to bind CBFα2 (Fig. 9B, lanes 7 and 8). Effect of the murine μC− mutation was also assayed in S194 extracts. As seen above with recombinant TFE3, the μC− probe bound a factor in S194 extracts with approximately twofold-reduced affinity (Fig. 9B, lanes 9 to 13). We conclude that μC− and hM3 sequences do not bind CBFα2 in vitro but bind bHLH-zip proteins, albeit with reduced affinity compared to that of the wild-type murine μE3 sequence.
Inactivity of the hM3 mutant enhancer in S194 cells suggested that the residual TFE3 binding was insufficient to confer transcriptional activity. We further tested the murine μC− mutation by transient transfection. Compared to the wild-type murine enhancer, the μC− mutant was a much poorer enhancer in M12 cells (Fig. 9C). Indeed, the reduced activity was reminiscent of the μE3− mutation that abolished both CBFα2 and TFE3 binding (24). These results indicate that two enhancer derivatives that bind TFE3, but not CBFα2, are not efficient transcriptional enhancers. Though we cannot rule out the possibility that reduced TFE3 binding is partly responsible for the lack of activity of the μC− enhancer, we favor the interpretation that TFE3 or TFE3-like bHLH-zip proteins do not activate this domain of the enhancer in B cells. Therefore, the requirement for the sequences between μA and μB for enhancer function likely represents a role for CBF in the activation of both the murine and human enhancers.
CBFα2 plus Ets-1 activate the μ enhancer in nonlymphoid cells.
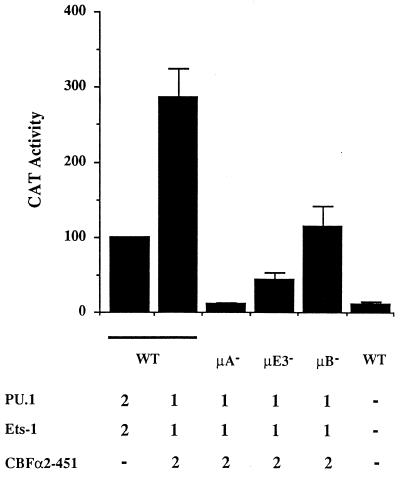
The preceding analysis suggested that CBF may work together with ETS proteins to activate the minimal μ enhancer. To obtain further supporting evidence, we assayed the ability of CBFα2 to transactivate the μ enhancer fragment in nonlymphoid cells. In HeLa cells, the μ70 enhancer was activated about 10-fold by coexpression of PU.1 and Ets-1 (Fig. 10, compare first and last bars). In the presence of PU.1, Ets-1, and CBFα2, significantly higher transcriptional activity was observed (Fig. 10, bar 2) which required all three elements in the enhancer to be intact (Fig. 10, bars 3 to 5). Of the three mutations, the μB mutation had the weakest effect, presumably because exogenously expressed Ets-1 and CBFα2 (μA and μE3 binding proteins, respectively) partially reduced the requirement for the μB site in the cotransfection assay. Similar effects were observed when only Ets-1 and TFE3 were coexpressed, resulting in μB-independent activation of μ70 (unpublished observations). Consistent with the mutational analysis, Ets-1 plus CBFα2 transactivated the μ70 enhancer, whereas PU.1 and CBFα2 did not (data not shown). These results indicate that CBFα2 may be a functional μ enhancer binding protein.
FIG. 10.
CBFα2 activates the IgH μ enhancer in nonlymphoid cells in cooperation with Ets-1. HeLa cells were transfected with reporter plasmids containing the (μ70)2 enhancer along with expression plasmids for PU.1 (2 μg) and Ets-1 (2 μg) (bar 1) and for PU.1 (1 μg), Ets-1 (1 μg), and CBFα2 (2 μg) (bar 2) or with an empty pEVRF2 expression plasmid as carrier DNA (bar 6). The transcription activation abilities of PU.1 (1 μg), Ets-1 (1 μg), and CBFα2 (2 μg) were also tested by cotransfecting these reporter plasmids with binding site mutant versions of the (μ70)2 enhancer, μA− (bar 3), μE3− (bar 4), and μB− (bar 5). Cells were harvested 2 days after being transfected, and CAT analysis was performed by ELISA. Results shown are the averages of at least two transfections carried out in duplicate. Error bars indicate the average deviations of the data.
DISCUSSION
We have previously defined a functional domain of the murine IgH gene enhancer that contains the μA and μB motifs that bind ETS domain proteins and the μE3 element that binds several ubiquitously expressed bHLH-zip proteins. All three elements are required for activity of this domain in transient transfection assays. Here we describe the analysis of the corresponding region of the human IgH enhancer. Both μA and μB sites were highly conserved between the two enhancers; however, the intervening μE3 element was not. Lack of sequence similarly in this region was reflected in the inability of prototypic bHLH-zip proteins TFE3 and USF to bind to the human enhancer. However, in striking contrast to the μE3 mutated murine enhancer that is inactive in transfection assays, the human enhancer domain was an active transcriptional enhancer in B cells. Nucleotides between the μA and μB motifs were necessary for enhancer activity and were found to bind the transcription factor CBF. Furthermore, we also found that the μE3 element of the murine enhancer was a CBF binding site. These observations highlight the similarity in organization of the murine and human enhancers, particularly with respect to the core enhancer domain. The CBF binding site in the Ig μ enhancers will be referred to as the μC element.
Although it has been assumed that the murine μE3 sequence works by binding bHLH-zip proteins, our observation that this element is also a CBF binding site raised the question of whether TFE3-like proteins or CBF family members were the functional μE3 binding proteins. Based on the analysis of the murine and human enhancers and several mutants thereof and transactivation studies, we propose that CBFα2 is likely to be the common, functional protein required to generate transcriptional activity in B cells. Is TFE3 binding to the murine sequence, then, a complete coincidence, or do these proteins also activate the enhancer under some circumstances? Reexamination of the early in vivo footprint experiments of Ephrussi et al. (4) provides some interesting ideas. They observed protections over the bHLH protein binding sites of the murine μ enhancer, including the μE3 motif, in plasmacytoma cells, which represent the terminal stage of B-cell differentiation. Indeed, the residues that scored in the assay are reminiscent of TFE3-μE3 interactions and unlike the expected pattern for CBFα2. Specifically, only two of three guanosines on the coding strand were protected by dimethyl sulfate in vivo and by TFE3 proteins in vitro, whereas methylation interference assays with CBFα2 would be expected to identify all three guanosines. Furthermore, no protections were seen over the μA and μB elements in the in vivo footprinting studies, even though these sites are crucial for enhancer activity in transfection experiments. The recent analysis of TFE3-deficient mice in which serum Ig levels are reduced indicates a defect in terminal B-cell differentiation (20). One possibility is that activation of the enhancer requires ETS and CBF proteins at earlier stages of B-cell differentiation, and in later stages of differentiation such as plasma cells, enhancer activity is maintained by bHLH proteins such as TFE3. Our observation that TFE3 and Ets-1 can interact directly as well as transactivate the μ enhancer is consistent with the possibility that bHLH-zip proteins also play a role in μ enhancer regulation (unpublished data).
CBF has been previously implicated in the regulation of T and myeloid cell-specific genes (1, 27, 36, 44). Since its identification as the protein that confers T-cell tropism to transformation by Moloney murine leukemia virus (34), CBF binding sites have been found in the enhancers of all the T-cell receptor (TCR) genes (7, 10, 12, 14, 41). Interestingly, CBF binding sites in both TCRα and -β enhancers are close to sites that bind ETS proteins. T-cell-specific activity of the TCRα enhancer depends on an ATF/CREB site, a LEF binding site, and a composite ETS-CBF element (7). It is likely that T-cell specificity is largely determined by the LEF site which also binds TCF-1, a T-cell-restricted factor (38, 40). In the TCRβ enhancer, two ETS-CBF elements have been identified, and our recent experiments suggest that both elements plus an additional element between them are necessary for enhancer activity in T-cell lines (1a). The elements that confer T-cell specificity to the TCRβ enhancer are not known. Our identification of an ETS-CBF composite motif in the Ig μ enhancer suggests that this is a common element that regulates both B- and T-cell antigen receptor genes. An interesting possibility is that the ETS-CBF motif is a hematopoietic cell-specific element whose activity is further modulated in a lineage (or developmental stage)-specific manner by other factors.
Two differences may be noted in the organization of ETS-CBF motifs in the TCR enhancers compared to the IgH μ enhancer. First, the ETS and CBF binding sites in both TCRα and -β enhancers are close together, whereas the μA and μC motifs of the μ enhancer are well separated. For example, the ETS-CBF motif in the βE4 element of the TCRβ enhancer has the sequence GGATGTGG, and the μA/μC sequence is shown in Fig. 1. Second, both TCR enhancers contain a second CBF site very close to the ETS-CBF element (this results in an ETS/CBF/CBF element in the TCR enhancers), whereas the μ enhancer contains a second ETS site (μB), making it an ETS-CBF/ETS-dependent regulatory sequence. It is likely that the second ETS site in the μ enhancer confers cell specificity by binding the B-cell- and macrophage-specific transcription factor PU.1. We have recently shown that the μA/μE3/μB enhancer activates transcription in B cells as well as macrophage cell lines (26) but not in T cells (unpublished observations), strengthening the idea that the μB site may specify transcriptional activation within hematopoietic cell types.
One of the genes encoding DNA binding α subunits of CBF (also known as PEBPA2B or AML1) is targeted in the most prevalent form of chromosomal translocation, t(12;21), identified in childhood acute lymphoblastic leukemias (9, 31, 33). The resulting TEL-AML1 fusion protein contains an N-terminal region derived from the ETS domain gene, TEL, which is fused to AML1 coding sequences that include the DNA binding Runt homology domain. Thus, the oncoprotein retains the ability to bind to CBF binding sites, and it is hypothesized that dysregulation of CBF-dependent gene regulation is a major factor in the development of disease (13). Because the leukemia induced by the t(12;21) translocation is one of immature B cells (9, 31, 33), CBF is likely to be important in early B-cell gene expression. Furthermore, in mice carrying a targeted disruption of the Cbfa2 (murine AML1) gene, both B- and T-cell development is blocked, reemphasizing the importance of this factor in lymphopoiesis (29) (unpublished observations). However, no CBF-regulated B-cell genes had been previously identified. Our studies are the first to identify a B-cell-specific enhancer that may be a target of CBF and highlight the combinatorial mechanisms by which ETS-CBF composite elements may be used to regulate B- and T-cell-specific gene expression.
ACKNOWLEDGMENTS
Anti-AML antisera were generously provided by S. Hiebert (Vanderbilt University Cancer Center, Nashville, Tenn.).
We thank W. Dang and G. Tian for discussions, assistance with plasmid constructions, and gifts of purified proteins.
This work was supported by NIH grants GM38925 to R.S. and CA58343 to N.A.S.
REFERENCES
- 1.Cameron S, Taylor D S, TePas E C, Speck N A, Mathet-Prevot B. Identification of a critical regulatory site in the human interleukin-3 promoter by in vivo footprinting. Blood. 1994;83:2851–2859. [PubMed] [Google Scholar]
- 1a.Carvajal, I., and R. Sen. Unpublished observations.
- 2.Chen J, Young F, Bottaro A, Stewart V, Smith R K, Alt F. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crute B, Lewis A, Wu Z, Bushweller J, Speck N. Biochemical and biophysical properties of the core-binding factor alpha2 (AML1) DNA-binding domain. J Biol Chem. 1996;271:26251–26260. doi: 10.1074/jbc.271.42.26251. [DOI] [PubMed] [Google Scholar]
- 4.Ephrussi A, Church G M, Tonegawa S, Gilbert W. B lineage specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 5.Erman B, Sen R. Context dependent transactivation domains activate the immunoglobulin mu heavy chain gene enhancer. EMBO J. 1996;15:4565–4575. [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst P, Smale S T. Combinatorial regulation of transcription II: the immunoglobulin heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 7.Giese K, Kingsley C, Kirshner J, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 8.Gillies S D, Morrison S L, Oi V T, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 9.Golub T R, Barker G F, Bohlander S K, Hiebert S, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallberg B, Thornell A, Holm M, Grundstrom T. SEF1 binding is important for T cell specific enhancers of genes for T cell receptor-CD3 subunits. Nucleic Acids Res. 1992;20:6495–6499. doi: 10.1093/nar/20.24.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayday A C, Gillies S D, Saito H, Wood C, Wiman C, Hayward W S, Tonegawa S. Activation of a translocated human c-myc gene by an enhancer in the immunoglobulin heavy-chain locus. Nature. 1984;307:334–340. doi: 10.1038/307334a0. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Munain C, Krangel M. c-Myb and core-binding factor/PEBP2 display functional synergy but bind independently to adjacent sites in the T-cell receptor delta enhancer. Mol Cell Biol. 1995;15:3090–3099. doi: 10.1128/mcb.15.6.3090. . (Erratum, 15:4659.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiebert S W, Sun W, Davis N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussel M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiang Y H, Spencer D, Wang S, Speck N A, Raulet D H. The role of viral “core” motif-related sequences in regulating T cell receptor γ and δ gene expression. J Immunol. 1993;150:3905–3916. [PubMed] [Google Scholar]
- 15.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenardo M, Pierce J W, Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987;236:1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- 17.Libermann T A, Baltimore D. Transcriptional regulation of immunoglobulin gene expression. Mol Aspects Cell Regul. 1990;6:399–421. [Google Scholar]
- 18.Matthias P, Müller M M, Schreiber E, Rusconi S, Shaffner W. Eukaryotic expression vectors for the analysis of mutant proteins. Nucleic Acids Res. 1989;17:6418. doi: 10.1093/nar/17.15.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melnikova I, Crute B, Wang S, Speck N. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrell K, Wells S, Henderson A, Gorman J, Alt F, Stall A, Calame K. The absence of the transcription activator TFE3 impairs activation of B cells in vivo. Mol Cell Biol. 1997;17:3335–3344. doi: 10.1128/mcb.17.6.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers S, Downing J, Hiebert S. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyers S, Lenny N, Sun W, Hiebert S. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 23.Nelsen B, Sen R. Regulation of immunoglobulin gene transcription. Int Rev Cytol. 1992;133:121–149. doi: 10.1016/s0074-7696(08)61859-8. [DOI] [PubMed] [Google Scholar]
- 24.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 25.Nikolajczyk B, Nelsen B, Sen R. Precise alignment of sites required for mu enhancer activation in B cells. Mol Cell Biol. 1996;16:4544–4554. doi: 10.1128/mcb.16.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikolajczyk B S, Cortes M, Feinman R, Sen R. Combinatorial determinants of tissue-specific transcription in B cells and macrophages. Mol Cell Biol. 1997;17:3527–3535. doi: 10.1128/mcb.17.7.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuchprayoon I, Meyers S, Scott L, Suzow J, Hiebert S, Friedman A. PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2β/CBFβ proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol Cell Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nye J A, Tersen J, Gunther C V, Jonsen M D, Graves B J. Interaction of murine Ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 29.Okuda T, van Deursen J, Hiebert S, Grosveld G, Downing J. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 30.Oltz E M, Alt F W, Lin W, Chen J, Taccioli G, Desiderio S, Rathbun G. A V(D)J recombinase-inducible B-cell line: role of transcriptional enhancer elements in directing V(D)J recombination. Mol Cell Biol. 1993;13:6223–6230. doi: 10.1128/mcb.13.10.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romana S P, Poirel H, Leconiat M, Flexor M-A, Mauchauffe M, Jonveaux P, Macintyre E A, Berger R, Bernard O A. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 32.Serwe M, Sablitzky F. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 1993;12:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shurtleff S A, Buijs A, Behm F G, Rubnitz J E, Raimondi S C, Hancock M L, Chan G C-F, Pui C-H, Grosveld G, Downing J R. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 34.Speck N A, Renjifo B, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 35.Sun W, Graves B, Speck N. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by CBF and ETS requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae S-C, Lu J, Maruyama M, Zhang Y W, Oka H, Arai N, Arai K, Ito Y. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor (GM-CSF) promoter activity by AML1-related transcription factor, PEBP2. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 37.Thornell A, Hallberg B, Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988;8:1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Wang Q, Crute B, Melnikova I, Keller S, Speck N. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterman M, Fischer W, Jones K. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 41.Wotton D, Ghysdael J, Wang S, Speck N, Owen M. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yancopoulos G D, Alt F W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 43.Zaiman A, Lewis A, Crute B, Speck N, Lenz J. Transcriptional activity of core binding factor alpha (AML1) and beta subunits on murine leukemia virus enhancer cores. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D, Hetherington C, Meyers S, Rhoades K, Larson C, Chen H, Hiebert S, Tenen D. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]