Abstract
The TATA-binding protein (TBP) is common to the basal transcription factors of all three RNA polymerases, being associated with polymerase-specific TBP-associated factors (TAFs). Simian virus 40 large T antigen has previously been shown to interact with the TBP-TAFII complexes, TFIID (B. Damania and J. C. Alwine, Genes Dev. 10:1369–1381, 1996), and the TBP-TAFI complex, SL1 (W. Zhai, J. Tuan, and L. Comai, Genes Dev. 11:1605–1617, 1997), and in both cases these interactions are critical for transcriptional activation. We show a similar mechanism for activation of the class 3 polymerase III (pol III) promoter for the U6 RNA gene. Large T antigen can activate this promoter, which contains a TATA box and an upstream proximal sequence element but cannot activate the TATA-less, intragenic VAI promoter (a class 2, pol III promoter). Mutants of large T antigen that cannot activate pol II promoters also fail to activate the U6 promoter. We provide evidence that large T antigen can interact with the TBP-containing pol III transcription factor human TFIIB-related factor (hBRF), as well as with at least two of the three TAFs in the pol III-specific small nuclear RNA-activating protein complex (SNAPc). In addition, we demonstrate that large T antigen can cofractionate and coimmunoprecipitate with the hBRF-containing complex TFIIIB derived from HeLa cells infected with a recombinant adenovirus which expresses large T antigen. Hence, similar to its function with pol I and pol II promoters, large T antigen interacts with TBP-containing, basal pol III transcription factors and appears to perform a TAF-like function.
The three eukaryotic DNA-dependent RNA polymerases each associate with a unique set of transcription factors that direct basal transcription from their respective promoters. However, one factor, the TATA-binding protein (TBP), is common to the basal transcription factors of all three polymerases (10, 52). In each case, TBP is found associated with a polymerase-specific set of TBP-associated factors (TAFs). Specifically, polymerase I (pol I) transcription requires TBP to be associated with three pol I TAFs which together form the selectivity factor 1 (SL1) complex (9). Polymerase II (pol II) transcription utilizes a complex of proteins called TFIID which consists of TBP associated with at least eight different pol II TAFs (14, 54). In the case of polymerase III (pol III) transcription, TBP is associated with two pol III transcription complexes, TFIIIB and small nuclear RNA (snRNA)-activating protein complex (SNAPc) (6, 36, 42). TFIIIB plays an essential role in pol III transcription and is thought to contact the polymerase directly (30). Yeast TFIIIB is a complex of three proteins: TBP, TFIIB-related factor (BRF), and TFC5 (6, 17, 29). Mammalian TFIIIB is not clearly defined but can be separated by ion-exchange chromatography into two fractions, designated 0.38M-TFIIIB and 0.48M-TFIIIB (36). Mital et al. (38) have isolated a TBP-associated protein from the 0.38M-TFIIIB fraction which is homologous to yeast BRF and designated human BRF (hBRF). In addition, Wang and Roeder (49) have isolated a TBP-associated protein that they call TFIIIB90, which is similar to hBRF. A second TBP-containing pol III complex, SNAPc, also contains three TAFs: SNAP 43, SNAP 45, and SNAP 50 (25). Unlike the other TBP-TAF complexes, the SNAPc complex has been implicated in both pol II and pol III transcription of the promoters of snRNA genes (42).
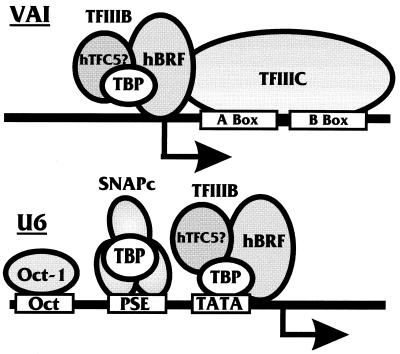
RNA pol III directs RNA synthesis from a diverse array of genes including the tRNA gene family, the 5S, 7SK, and 7SL ribosomal genes, the U6 snRNA gene, and the adenovirus type 2 (Ad2) VAI gene. Promoters of these pol III genes fall into three classes based on structure and the factors that they bind (Fig. 1). Class 1 and 2 promoters, e.g., 5S RNA and tRNA promoters, respectively, are intragenic; i.e., their promoter elements are located downstream of the initiation site, in contrast to class 3 promoters, which are extragenic.
FIG. 1.
Pol III transcription initiation complexes formed on the Ad2 VAI and U6 promoters. Transcription factor TFIIIC binds the A and B boxes on the intragenic promoter of the adenovirus VAI gene and recruits transcription factor TFIIIB, which is comprised of TBP and hBRF and, by analogy to the yeast system, may also recruit the human TFC5 factor. The human U6 snRNA promoter is similar to pol II promoters lying upstream of the coding sequence and containing a TATA box located approximately 25 nucleotides upstream of the initiation site. A PSE and an octamer binding site are located upstream of the TATA element. The transcription factor TFIIIB utilizes TBP to interact with the TATA box. A second TBP-containing transcription complex, SNAPc, binds to the PSE. In addition, the Oct-1 transcription factor is capable of binding the upstream octamer motif.
The mammalian U6 snRNA promoter is a class 3 promoter (Fig. 1). Similar to a pol II promoter, it has a TATA box located 25 nucleotides upstream of the +1 transcription start site that serves as a binding site for TBP (34, 35). Upstream of the TATA box lies an octamer motif and a proximal sequence element (PSE) (Fig. 1). The octamer motif binds the Oct-1 transcription factor, while the PSE binds the SNAPc complex. Although TBP is a component of the SNAPc complex, it does not seem to directly bind the DNA at the PSE; binding here is probably mediated by one or more of the TAFs present in the SNAPc complex (42).
Transcription from the U6 promoter has been shown to be independent of TFIIIA and TFIIIC (39, 48). These components of the pol III machinery are primarily used by the intragenic class 1 and 2 promoters. An example of such a promoter is the adenovirus VAI promoter (a class 2, pol III promoter), which has no TATA box but has two sequence elements termed A and B boxes that lie downstream of the initiation site (Fig. 1). These elements bind two distinct polypeptides that constitute the TFIIIC transcription factor (44). The TBP-containing TFIIIB complex is recruited to the VAI promoter through interactions with the TFIIIC complex (5, 28, 30). For all classes of pol III promoters, it is the TFIIIB complex that directly contacts the pol III enzyme and recruits it to the promoter (30).
Cells transformed with simian virus 40 (SV40) have been found to have increased levels of pol III transcripts (1, 41, 43, 51). In addition, SV40-transformed cells show an increase in the levels of TFIIIC and changes in its phosphorylation state (51). The mechanism by which SV40 enhances pol III transcription has not been elucidated; however, given the known functions of the SV40 early protein, large T antigen, it would be reasonable to suspect that it is involved.
Large T antigen (hereafter T antigen) is a very promiscuous transcriptional activator capable of activating pol II promoters of both viral and cellular genes (23, 31, 40). Previous work from this laboratory has shown that T antigen requires only a simple pol II promoter structure for activation, i.e., a TATA element and one upstream transcription factor (e.g., SP1 or Tef-1) binding site. In order for activation to occur, T antigen must interact with both the basal transcription complex (TFIID) and the upstream-bound transcription factor (22, 24). These data and more recent findings from our laboratory (12) have shown that T antigen stably associates with the TFIID complex and functions in a TAF-like manner. Coimmunoprecipitation studies suggest that the association of T antigen with TFIID is mediated by interactions with TBP and TAFs (12, 24). The fact that TBP is a transcription factor utilized by all three DNA-dependent RNA polymerases suggested to us that T antigen may influence transcription mediated by other polymerases through its interaction with TBP.
In agreement with this idea, recent studies of pol I transcriptional activation have shown that T antigen interacts with the TBP-TAFI complex, SL1 (53), and that this interaction is critical for transcriptional activation. In this work, we show that T antigen can also affect pol III transcription through interactions with the pol III basal factors containing TBP. We find that T antigen can transcriptionally activate the U6 pol III promoter which contains a TATA box and an upstream PSE but cannot activate the TATA-less intragenic VAI promoter. Mutants of large T antigen previously shown to be unable to activate pol II promoters also fail to activate the U6 promoter. We provide in vitro evidence that T antigen can interact with the TBP-containing pol III transcription factor hBRF as well as with at least two of the three TAFs in the SNAPc complex. To confirm the relevance of these in vitro interactions, we demonstrate that T antigen can cofractionate and coimmunoprecipitate with the hBRF-containing complex TFIIIB derived from cells infected with a recombinant adenovirus expressing SV40 T antigen. Hence, similar to the pol I and II situations, T antigen appears to interact with basal pol III transcription factors and appears to perform a TAF-like function.
MATERIALS AND METHODS
Plasmids and viruses.
Plasmid pRSV-Tex contains the SV40 large T antigen cDNA under the control of the Rous sarcoma virus long terminal repeat (37). The matching control plasmid, pRSV3BgIII, was generated by cleaving pRSV-Tex with BglII and religation of the vector, which effectively removes the cDNA. Plasmid p6-1dl contains the SV40 genomic fragment encoding both large T and small t antigens under the control of the SV40 early promoter (31). The control plasmid for p6-1dl is pL16HX, which contains only the SV40 early promoter. Plasmids T2811 and pT(Rb-) (pSG5-K1) encode for T antigens that, respectively, cannot bind p53 and Rb, under the control of the SV40 early promoter (3, 27). Mutants inA2803, inA2807, inA2815, inA2817, inA2831, inA2835, and inA2420 have been previously described (55).
Plasmid pSP72U6-sense was used to study activation of the U6 promoter. It was constructed by cleaving pU6/Hae/RA.2 (35) with SacI and BamHI to release the U6 promoter fragment attached to a β-globin coding element. The fragment was then inserted into a pSP72 (Promega) vector cleaved with SacI and BamHI. Plasmid pSP72-VAI was used for analysis of the transcriptional activation of the VAI gene; it contains the Ad2 VAI gene promoter and gene sequence. To make this construct, pBSM13+VAI (35) was cleaved with SacI and HindIII and ligated to a pSP72 vector fragment cleaved with SacI and HindIII. Plasmid pα4X(A+C) (46) contains an α-globin mRNA expressed from an α-globin promoter and was used as an internal control for transfection experiments.
The glutathione S-transferase (GST)-hBRF-expressing plasmid contains a cDNA copy of the hBRF gene fused to sequences encoding the glutathione binding moiety of GST and has been described previously (38). The plasmid expressing GST fused to full-length T antigen (GST-T) contains a cDNA copy of the T antigen coding region fused to the GST moiety (12). Plasmids for expression of the in vitro transcription and translation of the SNAP proteins, pCITE SNAPc43, pCITE SNAPc 45, pCITE SNAPc 50, have been described previously (25).
A recombinant adenovirus vector expressing SV40 T antigen, kindly provided by Alan Wildeman (11), was used to infect HeLa cells in suspension for the purification of hBRF-containing complexes from T-antigen-producing cells.
Transfections and infections.
CV-1 cells (3 × 106) were plated in 100-mm-diameter tissue culture dishes and grown overnight. Monolayers at approximately 80% confluency were transfected with the appropriate plasmids by the calcium phosphate procedure as previously described for similar experiments (23).
HeLa cells were propagated and maintained in suspension in Iscove’s medium supplemented with 5% fetal calf serum. For infection, cells were grown in spinner flasks and infected with the adenovirus vector at a multiplicity of infection of 10.
Protein purifications.
HeLa cells infected as described above were harvested 18 h postinfection, and nuclear extracts were prepared according by the method of Dignam et al. (13). To purify hBRF from the infected cells, the nuclear extract was chromatographed first on a phosphocellulose column and then on a HiLoad Sepharose-Q column as previously described (38). Fractions collected from the Sepharose column were analyzed for the presence of hBRF and TBP by in vitro activity assays and Western analysis. The fractions were tested for the presence of T antigen by Western analysis.
RNA preparation and analysis.
Total cellular RNA was prepared and RNase protection assays were performed as described previously (23). For the antisense U6 probe, the U6-sense plasmid was linearized with KpnI and antisense RNA was transcribed by using SP6 polymerase. For the VAI antisense probe, plasmid pSP72-VAI was linearized with SacI and antisense RNA was transcribed with SP6 polymerase. Results were quantitated with a PhosphorImager.
Coimmunoprecipitation and Western analysis.
Monoclonal antibodies used for our experiments included Pab419, an anti-T-antigen monoclonal antibody, and CSH407, an anti-hBRF antibody. Immunoprecipitations were performed as described by Damania and Alwine (12). Fractions from the HiLoad Sepharose-Q column were allowed to incubate with either Pab419 or CSH407 and protein-A agarose beads overnight, and the beads were subsequently washed four times with radioimmunoprecipitation buffer. The eluted proteins were separated by polyacrylamide gel electrophoresis (PAGE) on a sodium dodecyl sulfate–12% polyacrylamide gel and transferred to nitrocellulose. Specific proteins were detected by incubating the blot with the appropriate antibody followed by visualization using an enhanced chemiluminescence kit (Amersham) or by 125I-labeled secondary antibody.
Protein binding assay.
Expression and purification of GST-T and GST-hBRF were done as previously described (24). The amounts of GST proteins used in the binding reactions were normalized by semiquantitative comparison of protein bands on a silver-stained gel.
GST protein binding assays were performed at 4°C on a nutator. Test proteins were prepared by in vitro transcription and translation (Promega) and labeled with [35S]methionine. The radiolabeled proteins were incubated for 1 h with the GST fusion proteins attached to glutathione beads in NETN buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK]) plus 3% bovine serum albumin, which served as a blocking agent. The beads were then washed five times with NETN buffer, and the bound proteins were eluted from the beads by using Laemmli buffer. The eluted proteins were separated by PAGE on an SDS–10% polyacrylamide gel and visualized by autoradiography.
RESULTS
T antigen can activate the human U6 promoter but not the Ad2 VAI promoter.
Our previous findings have suggested that T antigen is capable of activating simple pol II promoters consisting of a TATA box and one upstream transcription factor binding site (22). As shown in Fig. 1 and discussed above, the pol III promoter of the U6 gene is structurally analogous to many pol II promoters. In contrast, the Ad2 VAI promoter is structurally distinct (Fig. 1). To determine whether T antigen could transcriptionally activate pol III promoters, we transfected CV-1 cells with reporter plasmids containing either the U6 promoter attached to a β-globin gene reporter or the VAI promoter as it exists within the VAI gene (Fig. 1). Activation was tested by cotransfection with either the T-antigen-expressing plasmid pRSV-Tex or a control plasmid, pRSV3BglII.
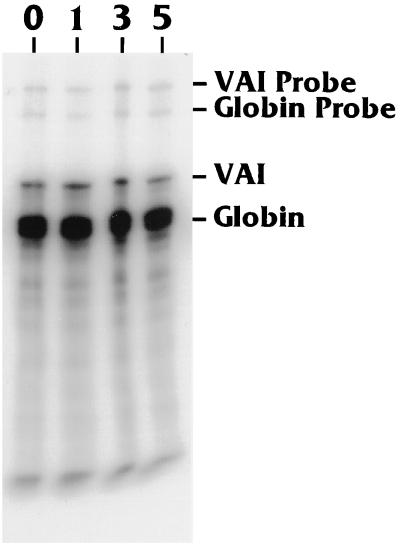
Figure 2 shows a nuclease protection analysis of VAI RNA produced in CV-1 cells transfected with the Ad2 VAI gene plus increasing amounts (0 to 5 μg) of pRSV-Tex. Each transfection also contained an α-globin mRNA-expressing plasmid [pα4X(A+C) (46); α-globin RNA production was used as a transfection efficiency control. Each transfection was performed in duplicate, and the experiment was repeated four times. The data show that the levels of VAI RNA produced in the absence and in the presence of T antigen are equivalent (this was verified by PhosphorImager analysis [not shown]), suggesting that T antigen has neither a positive nor a negative effect on VAI promoter activity.
FIG. 2.
T antigen does not activate transcription of the VAI gene. CV-1 cells were transfected with a VAI RNA-expressing plasmid, pSP72-VAI, together with 0, 1, 3, or 5 μg of a plasmid expressing SV40 T antigen (pRSV-Tex). Input amounts of DNA were maintained by using a filler plasmid, pRSV3BglII. Two micrograms of pα4X(A+C), which produces α-globin mRNA, was also cotransfected as an internal transfection efficiency control. At 42 h after transfection, the total RNA was harvested, and VAI RNA and α-globin RNA production was analyzed by RNase protection as described in Materials and Methods. The protected VAI and α-globin RNAs were 161 and 132 nucleotides, respectively. The undigested VAI (316 nucleotides) and α-globin (220 nucleotides) antisense probes are also shown.
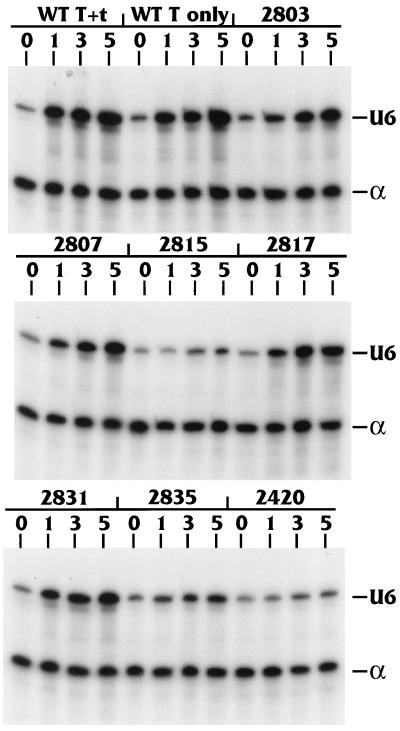
Figure 3 shows similar analysis of transfections using the reporter plasmid containing the U6 promoter (pU6-sense), plus the α-globin control plasmid (pα4xA+C), cotranfected with increasing amounts (0 to 5 μg) of various plasmids which produce T antigen or specific mutants of T antigen. In the top panel, the lanes labeled “WT T+t” indicate transfections using p6-1dl, a plasmid which encodes the entire early region of SV40 and produces both large T and small t antigens. The lanes labeled “WT T only” indicate transfections using pRSV-Tex, a plasmid containing a cDNA encoding wild-type (WT) T antigen that produces only large T antigen. In contrast to the VAI data, both T-antigen-expressing plasmids clearly activated U6 transcription. Quantitation of the data showed that T antigen alone increased RNA production from the U6 promoter at least fivefold, whereas the presence of both large T and small t antigens increased RNA production approximately sevenfold. These data indicate that T antigen alone is responsible for the U6 promoter activation and that the presence of small t antigen had little, if any, additional effect. In agreement with this conclusion, the expression of small t antigen alone did not activate the U6 promoter (data not shown).
FIG. 3.
T antigen activates transcription of the U6 RNA gene. CV-1 cells were transfected with the U6 RNA producing plasmid pSP72-U6 sense, along with 0, 1, 3, or 5 μg of plasmids expressing full-length large T antigen and small t antigen (WT T+t), full-length T antigen alone (WT T only), or various mutants of T antigen (Table 1). The α-globin RNA producing plasmid pα4X(A+C) was used as the internal control. At 42 h posttransfection, the total RNA was harvested and U6 RNA and α-globin RNA production was analyzed by RNase protection as described in Materials and Methods. The protected U6 (184 nucleotides) and α-globin (132 nucleotides) bands are indicated.
Effects of transcriptional activation mutants of T antigen on activation of the U6 promoter.
We next tested the activation of the U6 promoter by mutants of T antigen that have been previously characterized for transactivation of pol II promoters (12, 55). The mutants used were all small in-frame deletions and insertions in large T antigen (Table 1). All of the mutants are in the context of the entire early region; therefore, both large T and small t antigens are produced. The expression of these mutant T antigens has previously been shown to be equivalent to WT expression (12). Each transfection was performed in duplicate and the experiment was repeated four times. Mutants inA2803, inA2807, inA2817, and inA2831 were capable of activating the U6 promoter to levels similar to that of WT T antigen (Table 2). However, mutants inA2815, inA2835, and inA2420 were defective for the transcriptional activation of the U6 promoter (Table 2). The effects of each mutant on the U6 pol III promoter were analogous to their previously measured effects on pol II promoters (12), suggesting that T antigen uses a common mechanism to activate transcription in both cases (see Discussion).
TABLE 1.
Large-T-antigen mutants tested
| Mutant | Type of mutation | Region or domain affected |
|---|---|---|
| inA2803 | Insertion at aa 34 | N terminus |
| inA2807 | Insertion at aa 302 | Zinc finger |
| inA2815 | Insertion at aa 168 | DNA binding domain |
| inA2817 | Insertion at aa 219 | DNA binding domain |
| inA2831 | Deletion of aas 4 to 34 | N terminus |
| inA2835 | Insertion at aa 85 and 86 | N terminus |
| inA2420 | Truncation after aa 138 | Missing C-terminal 569 aa |
TABLE 2.
Activation of U6 promoter by large-T-antigen mutantsa
| WT or mutant | % of WT level | SD |
|---|---|---|
| WT | 100 | 3.0 |
| inA2803 | 67 | 4.5 |
| inA2807 | 68 | 1.5 |
| inA2815 | 20 | 2.5 |
| inA2817 | 75 | 2.0 |
| inA2831 | 79 | 2.0 |
| inA2835 | 40 | 6.5 |
| inA2420 | 26 | 3.0 |
Quantitation was taken from the points for 5 μg of plasmid input in Fig. 3.
Chesnokov et al. (8), White et al. (50), and Larminie et al. (33) reported that p53 and Rb both inhibit pol III-directed transcription from the U6 promoter. Since p53 and Rb have been shown to have antagonistic effects with respect to T antigen on many pol II promoters, we examined the effect of T-antigen mutants which were defective for p53 and Rb binding. The results (not shown) suggested that the ability or inability to interact with p53 and Rb had no effect on T antigen’s ability to activate the U6 promoter.
Interactions of SNAPc components and hBRF with T antigen.
The data presented above show that T antigen can activate the U6 promoter but not the VAI promoter in CV-1 cells. The U6 promoter contains binding sites for the Oct-1 transcription factor, the TBP-containing SNAPc complex, as well as the TBP-hBRF (TFIIIB) complex. Our lab has previously shown that T antigen is not capable of interacting with the Oct-1 protein in a protein-protein binding assay and cannot activate simple pol II promoters that contain an Oct-1 binding site (24). However, we have demonstrated T antigen’s ability to bind TBP and pol II TAFs and be a component of TFIID (12, 24). Therefore, we tested whether T antigen could interact with any of the remaining transcription factors that associate with the U6 promoter: hBRF (TFIIIB), SNAP 43, SNAP 45, and SNAP 50.
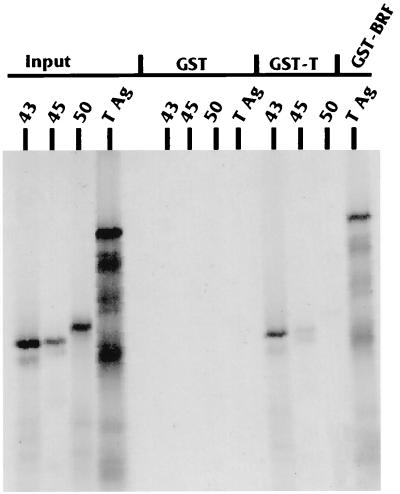
To determine whether T antigen can interact with components of the SNAPc complex, we used the GST-T fusion protein to assay binding to [35S]Met-labeled SNAP 43, SNAP 45, and SNAP 50 produced by in vitro transcription and translation. Figure 4 shows the results of the binding analysis. T antigen interacted strongly with SNAP 43, moderately with SNAP 45, and very little with SNAP 50. The SNAP proteins did not bind the control GST moiety. Figure 4 also shows the result of a binding experiment done with a GST fusion of full-length hBRF and [35S]Met-labeled T antigen produced by in vitro transcription and translation. T antigen interacted very well with hBRF but not with the control GST moiety. GST binding was not altered in the presence of 200 mg of ethidium bromide per ml, showing that the binding was attributable to protein-protein interactions rather than tethering due to binding a common piece of DNA (reference 32 and data not shown).
FIG. 4.
T antigen interacts in vitro with components of SNAPc and hBRF. The three SNAPc components SNAP 43, 45, and 50 were produced by in vitro transcription and translation; the input lanes indicate 20% of the total amount of in vitro-transcribed and -translated 35S-labeled proteins used in each binding reaction. Each protein was tested for in vitro binding with the GST moiety alone or with a GST fusion with full-length T antigen as described in Materials and Methods. In addition, in vitro-transcribed and -translated T antigen was tested for binding to the GST moiety alone and to a GST fusion with full-length hBRF.
Cofractionation of T antigen with a pol III transcription factor, hBRF, from infected HeLa cells.
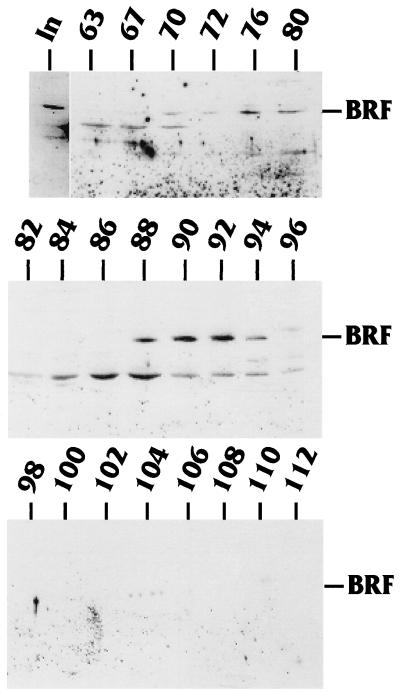
The GST binding assay described above showed that T antigen can interact very well in vitro with hBRF as well as SNAP 43 and to a lesser extent with SNAP 45. The fact that T antigen could interact with components of the SNAPc complex is interesting because the SNAPc complex is necessary for the activation of both pol II snRNA genes like U1 and U2 and the pol III snRNA U6 gene (42). However, what was most interesting was that T antigen could interact with hBRF, a factor which, unlike the SNAPc complex, is uniquely involved in pol III transcription. To test whether T antigen’s ability to bind hBRF in vitro held true in vivo, HeLa spinner cells were infected with a recombinant adenovirus which produced WT T antigen. The infected cells were harvested 18 h postinfection, and nuclear extracts were prepared by the method described by Dignam et al. (13). The extracts were fractionated on a phosphocellulose column followed by a HiLoad Sepharose-Q column (see Materials and Methods). Fractions from the Sepharose-Q column were fractionated by SDS-PAGE, and the gel was transferred to nitrocellulose. Western analysis was performed with an anti-hBRF antibody to detect the presence of hBRF; bands were visualized with an enhanced chemiluminescence kit. Figure 5 shows the fractions that were tested. hBRF eluted from the Sepharose-Q column in two peaks: fractions 70 through 80 and fractions 88 through 96. In additional analyses (not shown), the fractions were tested for the presence of TBP. TBP was found to cofractionate with both peaks of hBRF. These findings are consistent with previous reports (30, 36, 45).
FIG. 5.
Purification of hBRF. Nuclear extract from HeLa cells infected with a recombinant adenovirus vector expressing T antigen was fractionated on a phosphocellulose and Sepharose-Q column (see Materials and Methods). Fractions from a Sepharose-Q column, previously shown to contain TFIIIB activity, were analyzed for hBRF by Western analysis using an anti-hBRF antibody. The hBRF elutes in two peaks (fractions 70 to 80 and fractions 88 to 96). In, input.
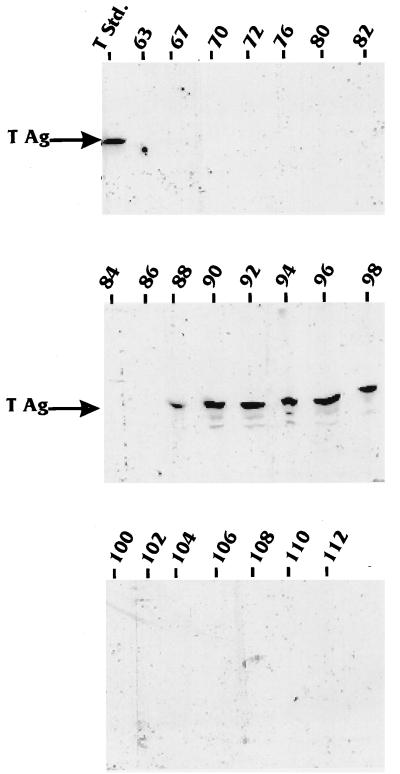
We next tested the same fractions to determine whether T antigen cofractionated with hBRF from the infected HeLa cell nuclear extracts. The Western blot was probed with a monoclonal anti-T-antigen antibody, and the bands were visualized by using an 125I-labeled secondary antibody. Figure 6 shows that T antigen eluted from the Sepharose-Q column in one peak extending from fractions 88 through 98. These results suggest that T antigen cofractionates with one of the two hBRF peaks.
FIG. 6.
T antigen cofractionates with hBRF. The hBRF-containing fractions examined in Fig. 5 were examined for the presence of T antigen (T Ag) by Western blotting using anti-T antigen antibody Pab419. Large T antigen eluted in one peak from the Sepharose-Q column (fractions 88 to 98). Std., standard.
Large T antigen and hBRF coimmunoprecipitate.
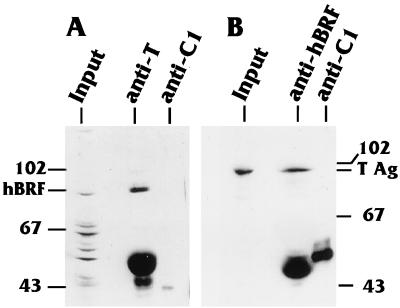
The data presented above strongly suggest that T antigen cofractionates with TFIIIB which contains both TBP and hBRF. To confirm that a true interaction was occurring between T antigen and hBRF, coimmunoprecipitations were performed with Sepharose-Q fractions 88 to 96, in which both proteins cofractionated. The pooled fractions were dialyzed into buffer D (see Materials and Methods) and coimmunoprecipitated with an anti-hBRF or anti-T antigen antibody, and the precipitates were analyzed by Western blotting. Figure 7A shows that hBRF coimmunoprecipitated with T antigen using an anti-T antibody; conversely, Fig. 7B shows that T antigen coimmunoprecipitated with hBRF using anti-hBRF (Fig. 7B). The nonspecific control antibody against the hnRNPC1 protein immunoprecipitated neither protein. Previous experiments had shown that the anti-T antibody did not cross-react with hBRF and that the anti-hBRF antibody did not cross-react with T antigen. The overall data indicate that large T antigen cofractionates and coimmunoprecipitates with hBRF, suggesting that T antigen exists in a stable complex with TFIIIB in HeLa cells infected with a recombinant adenovirus expressing SV40 T antigen.
FIG. 7.
T antigen and hBRF coimmunoprecipitate. Fractions 88 to 96 from the Sepharose-Q column, containing both hBRF and T antigen, were pooled and dialyzed into 0.1 M KCl buffer D. The samples were then subjected to immunoprecipitation reactions. (A) Immunoprecipitation reaction using an anti-T antigen antibody or an anti-hnRNPC1 (anti-C1) control antibody. The immunoprecipitates were resolved on a gel and subjected to Western analysis using an anti-hBRF antibody. (B) Immunoprecipitation reaction using an anti-hBRF antibody or an anti-hnRNPC1 control antibody. The immunoprecipitates were resolved on a gel and subjected to Western analysis using an anti-T antigen (T Ag) antibody.
DISCUSSION
Eukaryotic RNA pol III is responsible for the transcription of small nuclear and cytoplasmic RNAs of both cellular and viral origin (21). Transcription of these genes is enhanced during viral infection by the expression of viral early proteins like the pseudorabies virus immediate-early protein and the adenovirus E1A and/or E1B proteins (4, 19, 26) and is suppressed by poliovirus infection (16). In addition, cells that have been transformed with SV40 show an increase in pol III RNA transcripts (41, 51) and, like cells that have been transformed with adenovirus, show an increase in the level of the pol III transcription factor TFIIIC (44, 51). Although the effects of SV40 infection and transformation on pol III transcription have been well studied, the mechanism underlying this process remained undefined. However, given the known transcriptional activation functions of large T antigen, it seemed likely that it may be directly involved in the activation of pol III promoters. Indeed, the identification and characterization of the key factors involved in pol III transcription has now made it possible to investigate the mechanism by which SV40 large T antigen activates some pol III promoters.
Previous studies in our laboratory and others have suggested that T antigen can activate cellular transcription through protein-protein interactions (22, 24). Recently we have shown that T-antigen-mediated activation of pol II promoters involves its ability to form a stable complex with TFIID (12), where it performs a TAF-like function interacting with upstream-bound transcription factors. The interaction with TFIID is mediated at least in part by a direct interaction with TBP. Since TBP is a common factor in the basal apparatuses of all three mammalian RNA polymerases, it seemed reasonable to expect that T antigen may also exist in the TBP-containing transcription complexes used by pol I and III. In accordance with this suggestion, Zhai et al. (53) have recently reported that SV40 T antigen binds to the TBP-TAFI complex, SL1, and coactivates rRNA transcription. Zhai et al. (53) specifically show that the activation of a rRNA promoter by T antigen is dependent on its ability to interact with TBP as well as the two pol I TAFs, TAFI110 and TAFI48. Similarly, the data presented herein, examining pol III promoter activation, establish a similar mechanism. As in the case of pol I and II, T antigen appears to mediate pol III transcriptional activation through its ability to interact with TBP-containing pol III transcription factors.
Our data demonstrate that large T antigen can interact with multiple components of the pol III transcription family. Using a protein-protein interaction assay, with a biochemical fractionation scheme and coimmunoprecipitation analysis, we have established that T antigen is able to bind to three pol III transcription factors in addition to TBP: SNAP 43, SNAP 45, and hBRF. Despite these interactions, we found that T antigen does not directly activate all classes of pol III promoters. While it failed to activate the TATA-less intragenic adenovirus VA1 promoter (class 2), it readily activated the class 3 U6 promoter, which, like pol II promoters, contains a TATA box and upstream transcription factor binding sites. Thus with pol I, II, and III promoters, the interaction involving T antigen and TBP, or a TBP-containing complex, is very significant. Further, a TAF-like function for T antigen in affecting activated transcription is indicated in each case. Our data also suggest that activation of the U6 promoter is not a function of T antigen’s ability to interact with tumor suppressors like p53 and Rb. T-antigen mutants defective in p53 and Rb binding maintain their abilities to activate the U6 promoter. This agrees with previous data which suggested that T-antigen-mediated activation of pol II promoters is independent of p53 and RB (47, 55).
Three mutants of T antigen, inA2815, inA2835, and inA2420, were found to be defective for transactivation of the U6 promoter. Mutant inA2835 has an insertion at amino acid (aa) 85, and mutant inA2420 is a frameshift mutation that produces a truncated 138-aa product. Both mutations map to the amino terminus of T antigen. Previous data from our laboratory have indicated that the N-terminal 172 aa of T antigen interact with TBP (24). This could explain the inability of both of these mutants to activate the U6 promoter. Mutant inA2815 has an insertion at aa 168 which corresponds to the DNA binding domain. However, it is unlikely that the loss of ability to bind DNA is the reason for the defect in transactivation since the ability of T antigen to bind DNA has never been correlated with transcriptional activation (2, 18, 31). Instead it is likely that this mutation prevents the protein from folding correctly and activating transcription (7). Mutants inA2815 and inA2835 have previously been shown to be defective in transcriptional activation of pol II promoters. In the case of mutant inA2815, this has been shown to be due to its inability to bind the TBP-containing TFIID complex (12). Hence, the inability of this mutant to transactivate the U6 promoter may analogously be linked to an inability to interact with the TBP-containing complexes TFIIIB and/or SNAPc.
We found that T antigen was not able to activate the Ad2 VAI promoter. This finding supports previous data suggesting that the ability of T antigen to activate a gene is, in part, dependent on promoter structure (22, 23). The VAI promoter is intragenic, with the A and B boxes binding the transcription factor TFIIIC, which then recruits TFIIIB to the promoter. Although T antigen is capable of interacting with components of TFIIIB, it appears that this interaction is not sufficient to drive transcription from the VAI promoter. This may be related to the direction and placement of the basal complex on the internal promoter, causing it to hinder T antigen’s ability to access or interact with specific components of the transcription machinery. An intriguing finding has been that SV40-transformed cells show an upregulation in the abundance of the TFIIIC transcription factor (51). TFIIIC plays a key role in the activation of class 1 and 2 pol III promoters but is dispensable for class 3 promoters (20). Thus, it appears that SV40 has found different ways to activate pol III transcription in the cell. One mechanism is direct and requires T antigen to interact with TFIIIB and/or SNAPc to activate some pol III genes. The other mechanism appears to be indirect, most likely through activation of pol II transcription causing an increase in the expression of the pol III transcription factor TFIIIC.
SV40 T antigen has long been established as a very promiscuous transcriptional activator capable of activating many viral and cellular promoters. This function of T antigen has been attributed to the fact that in order to replicate its genome, the virus must push the infected cell into the cell cycle. This may be accomplished, in part, by T antigen’s ability to activate many cellular genes whose products may be needed for cellular growth and proliferation. The TBP-TAF complexes in the cell appear to be functional targets for T antigen. Through association with either the pol I-associated factor SL1 (55), the pol II-associated factor TFIID (12), or the pol III-associated factors TFIIIB and SNAPc, T antigen may be able to activate a diverse array of genes transcribed by any of the three mammalian RNA polymerases.
ACKNOWLEDGMENTS
We thank Nouria Hernandez for pol III promoters as well as SNAPc and hBRF expression plasmids and technical advise; we thank Charles Cole for mutant T-antigen-expressing plasmids. In addition, we thank Noam Harel for critical comments on the manuscript and other members of the Alwine laboratory for their help and support.
This work was supported by Public Health Service grant CA28379 awarded to J.C.A. by the National Cancer Institute.
REFERENCES
- 1.Aufiero B, Schneider R J. The hepatitis B virus X-gene product transactivation both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard P, Bruggmann H. Control of transcription in vitro from simian virus 40 promoters by proteins from viral minichromosomes. Curr Top Microbiol Immunol. 1989;144:47–54. doi: 10.1007/978-3-642-74578-2_6. [DOI] [PubMed] [Google Scholar]
- 3.Bentivoglio C, Zhu J, Cole C N. Mechanisms of interference with simian virus 40 (SV40) DNA replication by trans-dominant mutants of SV40 large T antigen. J Virol. 1992;66:2551–2555. doi: 10.1128/jvi.66.7.4209-4219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger S L, Folk W R. Differential activation of RNA polymerase III-transcribed genes by the polyoma virus enhancer and adenovirus E1A gene products. Nucleic Acids Res. 1985;13:1413–1428. doi: 10.1093/nar/13.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun B R, Riggs D L, Kassavetis G A, Geiduschek E P. Multiple states of DNA-protein interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc Natl Acad Sci USA. 1989;86:2530–2534. doi: 10.1073/pnas.86.8.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology with TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 7.Casaz P, Rice P W, Cole C N, Hansen U. A TEF-1-independent mechanism for activation of the simian virus 40 (SV40) late promoter by mutant SV40 large T antigen. J Virol. 1995;69:3501–3509. doi: 10.1128/jvi.69.6.3501-3509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokov I, Chu W, Botchan M, Schmid C. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol Cell Biol. 1996;16:7084–7088. doi: 10.1128/mcb.16.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 10.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 11.Coulombe J, Berger L, Smith D B, Hehl R K, Wildeman A G. Activation of simian virus 40 transcription in vitro by T antigen. J Virol. 1992;66:4591–4596. doi: 10.1128/jvi.66.7.4591-4596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 13.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associates with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 15.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:15–16. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 16.Fradkin L G, Yoshinaga S K, Berk A J, Dasgupta A. Inhibition of host cell RNA polymerase III-mediated transcription by poliovirus: inactivation of specific transcription factors. Mol Cell Biol. 1987;7:3880–3887. doi: 10.1128/mcb.7.11.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrielsen O S, Marzouki N, Ruet A, Sentenac A, Fromageot P. Two polypeptide chains in yeast transcription factor to interact with DNA. J Biol Chem. 1989;264:7505–7511. [PubMed] [Google Scholar]
- 18.Gallo G J, Gruda M C, Manuppello J R, Alwine J C. Activity of simian DNA-binding factors is altered in the presence of simian virus 40 (SV40) early proteins: characterization of factors binding to elements involved in the activation of the SV40 late promoter. J Virol. 1990;64:173–184. doi: 10.1128/jvi.64.1.173-184.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaynor R B, Feldman L T, Berk A J. Transcription of class III genes activated by viral immediate early proteins. Science. 1985;230:447–450. doi: 10.1126/science.2996135. [DOI] [PubMed] [Google Scholar]
- 20.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 21.Geiduschek E P, Tocchini-Valentini G P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 22.Gilinger G, Alwine J C. Transcriptional activation by simian virus 40 large T antigen: requirement for simple promoter structures containing either TATA or initiator elements with variable upstream factor binding sites. J Virol. 1993;67:6682–6688. doi: 10.1128/jvi.67.11.6682-6688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruda M, Alwine J C. Simian virus 40 T-antigen transcriptional activation mediate through the Oct/SPH region of the SV40 late promoter. J Virol. 1991;65:3553–3558. doi: 10.1128/jvi.65.7.3553-3558.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruda M, Zabolotny J, Xiao J, Davidson I, Alwine J. Transcriptional activation by SV40 large T antigen: interaction with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry R, Sadowski C, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 26.Hoeffler W K, Roeder R G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988;53:907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 27.Kaelin W G, Jr, Ewen M, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interaction in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2556. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression and function of TFC5, the gene encoding the B” component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassavetis G A, Braun B R, Nguyen L H, Geidoschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 31.Keller J M, Alwine J C. Analysis of an activatable promoter. Sequences in the SV40 late promoter required for T-antigen-mediated trans activation. Mol Cell Biol. 1985;5:1859–1869. doi: 10.1128/mcb.5.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai J, Herr W. Ethidium bromide provides a simple tool for establishing DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larminie C, Cairns C, Mital R, Martin K, Kouzarides T, Jackson S, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobo S, Lister J, Sullivan M, Hernandez N. The cloned RNA polymerase II transcription factor IID selects RNA polymerase III to transcribe the human U6 gene in vitro. Genes Dev. 1991;5:1477–1489. doi: 10.1101/gad.5.8.1477. [DOI] [PubMed] [Google Scholar]
- 35.Lobo S, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 36.Lobo S M, Tanaka M, Sullivan M L, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 37.Loeken M, Khoury G, Brady J. Stimulation of the adenovirus E2 promoter by simian virus 40 T antigen and E1a occurs by different mechanisms. Mol Cell Biol. 1986;6:2020–2026. doi: 10.1128/mcb.6.6.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy R. Transcription of a mouse U6 small nuclear RNA gene in vitro by RNA polymerase III is dependent on transcription factors different from transcription factors TFIIIA, TFIIIB, and TFIIIC. J Biol Chem. 1988;263:15980–15984. [PubMed] [Google Scholar]
- 40.Rice P W, Cole C N. Efficient transcriptional activation of many simple modular promoters by simian virus 40 large T antigen. J Virol. 1993;67:6689–6697. doi: 10.1128/jvi.67.11.6689-6697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rigby P J, La Thangue N B, Murphy D, Skene B. The regulation of cellular transcription by simian virus 40 large T antigen. Proc R Soc Lond Ser B. 1985;226:15–23. doi: 10.1098/rspb.1985.0076. [DOI] [PubMed] [Google Scholar]
- 42.Sadowski C L, Henry W, Lobo S, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 43.Singh K, Carey M, Saragosti S, Botchan M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985;314:553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- 44.Sinn E, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of a TFIIIC2 subunit (TFIIICB) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- 45.Taggart A K P, Fisher T S, Pugh B F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 47.Trifilis P J, Picardi J, Alwine J C. SV40 T antigen can transcriptionally activate and mediate DNA replication in cells which lack retinoblastoma susceptibility gene product. J Virol. 1990;64:1345–1347. doi: 10.1128/jvi.64.3.1345-1347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldschmidt R, Wanandi I, Seifart K H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;10:2595–2603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionary conserved and contains both TFIIB and high mobility group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White R, Trouche D, Martin K, Jackson S, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 51.White R, Stott D, Rigby P J. Regulation of RNA polymerase III transcription in response to simian virus 40 transformation. EMBO J. 1990;9:3713–3721. doi: 10.1002/j.1460-2075.1990.tb07584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White R J, Jackson S P, Rigby P W J. A role for the TATA-box-binding protein component of transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci USA. 1992;89:1949–1953. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhai W, Tuan J, Comai L. SV40 large T antigen binds to the TBP-TAF complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Q, Lieberman P, Boyer T, Berk A. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J, Rice P W, Chamberlain M, Cole C N. Mapping the transcriptional transactivation function of simian virus 40 large T antigen. J Virol. 1991;65:2778–2790. doi: 10.1128/jvi.65.6.2778-2790.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]