Abstract
GCN5, a putative transcriptional adapter in humans and yeast, possesses histone acetyltransferase (HAT) activity which has been linked to GCN5’s role in transcriptional activation in yeast. In this report, we demonstrate a functional interaction between human GCN5 (hGCN5) and the DNA-dependent protein kinase (DNA-PK) holoenzyme. Yeast two-hybrid screening detected an interaction between the bromodomain of hGCN5 and the p70 subunit of the human Ku heterodimer (p70-p80), which is the DNA-binding component of DNA-PK. Interaction between intact hGCN5 and Ku70 was shown biochemically using recombinant proteins and by coimmunoprecipitation of endogenous proteins following chromatography of HeLa nuclear extracts. We demonstrate that the catalytic subunit of DNA-PK phosphorylates hGCN5 both in vivo and in vitro and, moreover, that the phosphorylation inhibits the HAT activity of hGCN5. These findings suggest a possible regulatory mechanism of HAT activity.
Gene activity is normally repressed due to the organization of DNA in chromatin. Prior to transcription, the repressive chromatin structure is remodeled to allow access to DNA binding sites (reviewed in references 33, 34, 59, and 72). Following processes that alter DNA structure, such as replication (60) and DNA repair (27), the chromatin structure is rebuilt. A number of proteins that remodel chromatin have been identified (for reviews, see references 63 and 82), and they exhibit evolutionary conservation, suggesting a fundamental role in eukaryotes.
Several factors shown by biochemical means to alter chromatin structure were initially identified as transcriptional regulatory factors in the yeast Saccharomyces cerevisiae, suggesting that modification of chromatin contributes to gene regulation. For example, components of the SWI/SNF complex both alter chromatin structure and are involved in transcriptional regulation (17, 63). The SWI/SNF complex (61) disrupts nucleosome-DNA interactions in vitro, promoting the binding of transcription factors to chromatin (22, 43). SWI2 contains ATPase activity that is required for its role in both transcription and nucleosome disruption (43, 44). Components of SWI/SNF were isolated in genetic screens in yeast (56, 71), and mutations in them down-regulated transcription (46, 62). The yeast complex is functionally related to multiple similar complexes in both yeast (13) and higher eukaryotes (40, 81).
A second link between factors required in transcription and chromatin alteration has emerged recently. Components of the ADA complex were identified by genetic selections in yeast for reduced function of the herpes simplex virus activator, VP16 (7). Several genes were identified (52, 53, 64, 66) and their products were shown to mutually interact in vitro and in vivo (14, 37). Two of them, ADA2 and GCN5, are evolutionarily conserved (15). ADA factors interact with specific activators (6, 20, 70) and with the general factor TBP (TATA-binding protein) (6), suggesting that an ADA complex potentiates transcription in vivo through these associations. Recently, GCN5 was shown to possess histone acetyltransferase (HAT) activity (12). Since hyperacetylation of amino-terminal tails of core histones correlates with activity of certain genes (11, 23, 50, 83), the HAT activity of GCN5 suggests a link between nucleosome acetylation and transcriptional activation. Moreover, since ADA components physically associate with transcription factors, targeted histone acetylation may have a direct role in transcriptional activation. Supporting this notion, deletion (16) and substitution (80a) mutations in GCN5 have defined a HAT domain in vitro whose integrity of function corresponds closely to the ability of GCN5 to potentiate activated transcription in vivo.
Many factors that regulate chromatin structure have a conserved domain of unknown function, called a bromodomain (BrD) (36). The BrD motif is present in nearly all HAT-associated transcriptional cofactors, including yeast and human GCN5 (12, 15, 29, 80), p300/CBP-associated factor (85), CBP/p300 (3, 5, 21, 58), and human (hTAFII250) and Drosophila TAFII230 (55, 69). The BrD also is present in the SWI2/SNF2 protein of the SWI/SNF chromatin remodeling complex and in other members of the SWI2/SNF2 family (36, 74). Secondary structure prediction suggests that the BrD may form a surface for protein-protein contacts (36).
Phosphorylation is a common mechanism for regulation of various cellular processes, including DNA-related activities such as transcription, replication, and DNA repair. A well-characterized kinase that specifically requires association with DNA for its activity (18, 48) is DNA-dependent protein kinase (DNA-PK) (19). The DNA-PK holoenzyme consists of a 450-kDa catalytic subunit (DNA-PKcs) (35), which phosphorylates serine/threonine, and a DNA-binding component known as Ku autoantigen (31). Ku is a heterodimer comprised of 70-kDa (65) and 80-kDa (84) subunits, and the 70-kDa subunit possesses DNA helicase activity (76). Ku and DNA-PK have been implicated in transcriptional repression (30, 41), DNA repair (49, 73), and immunoglobulin gene rearrangements [V(D)J recombination (8, 26)]. These last two processes are mechanistically linked via the DNA-PK holoenzyme, since mutations in DNA-PKcs cause the mouse SCID phenotype (8) and mutations in Ku cause sensitivity to ionizing radiation brought about by defects in DNA repair (73). Moreover, the phenotype of a mouse bearing a Ku80 disruption includes both radiation sensitivity and V(D)J recombination defects (57). Importantly, although several substrates of DNA-PKcs have been identified in vitro (reviewed in references 2 and 19), physiological targets are still obscure.
In this report, we describe a functional interaction between the BrD of the histone acetyltransferase hGCN5 and the DNA-PK holoenzyme. The apparent result of this interaction, both in vitro and in vivo, is phosphorylation of hGCN5 and inhibition of its HAT activity. These data are the first report of regulation of HAT activity within this new class of chromatin modifying agents.
MATERIALS AND METHODS
Two-hybrid analysis.
The LexA fusions of hBrD.Pro (amino acids [aa] 339 to 363) and hBrD.HT (aa 364 to 421) (see Table 1, footnote a, for definitions) were generated as PCR products, bearing BamHI and BglII restriction sites, cloned in BamHI site of BTM116 (10). The carboxyl-terminal fragment of Ku70 fused to the GAL4 activation domain (GAL4AD) was recovered from a HeLa cDNA library (MatchMaker; Stratagene) in the two-hybrid screen (25) using yeast strain L-40 (78) with LexA-BrD of hGCN5. β-Galactosidase assays (68) were carried out in yeast strain PSY316 (ade2-101 Δhis3-200 leu2-3,112 lys2 Δtrp1::hisG ura3-52) transformed with a reporter bearing eight LexA binding sites upstream of the lacZ gene (15).
TABLE 1.
Interaction between LexA DNA-binding domain fusions and the carboxyl terminus of Ku70 in the yeast two-hybrid assay
| LexADBD fusiona | β-Galactosidase activity (U/mg of protein)
|
Fold inductionb | |
|---|---|---|---|
| GAL4AD | KuC-GAL4AD | ||
| rho | 10 ± 6 | 17 ± 1 | ∼2 |
| hGCN5.BrD | 9 ± 1 | 489 ± 58 | 54 |
| hGCN5.BrD.HT | 7 ± 4 | 91 ± 21 | 13 |
| hGCN5.BrD.Pro | 6 ± 2 | 19 ± 10 | ∼3 |
| CBP.BrD | 7 ± 1 | 120 ± 21 | 17 |
| TAF250.BrD | 8 ± 4 | 147 ± 38 | 18 |
Shown are LexA DNA-binding domain (LexADBD) fusion proteins derived from rho, the hGCN5 BrD, the helix-turn-helix motif from the hBrD (BrD.HT; aa 364 to 421), the proline-rich region from the hBrD (BrD.Pro; aa 339 to 363), the CBP BrD, and the TAF250 BrDs. The LexA fusions were cotransformed into yeast with either the GAL4AD or the carboxyl terminus of Ku70 (aa 279 to 610) fused to the GAL4AD.
Ratio of each LexA fusion interaction with GAL4AD-KuC to interaction with the GAL4AD alone.
GST interactions.
Human Ku70 and hGCN5 were cloned as PCR products bearing BamHI sites into expression vector pGEX3, opened with BamHI. The full-length hADA2 was cloned as a PCR product, bearing BamHI and EcoRI restriction sites in the same sites of expression vector pGEX5. The glutathione S-transferase (GST) deletion mutants of the BrD of hGCN5, as well as the BrD of CBP, were cloned as PCR products with BamHI and BglII restriction sites into the BamHI site of pGEX3. Expression and purification of GST fusion proteins were performed as described previously (6). The same PCR products of Ku70 and hGCN5 genes were cloned into pSP64 vector for translation in vitro. hGCN5ΔBrD was cloned in pSP64 cleaved with BamHI as a PCR fragment bearing BamHI restriction sites. In vitro translation of proteins in rabbit reticulocytes was performed as instructed by the manufacturer (TNT kit; Promega). Aliquots (25 μl) of TNT extracts were incubated for 1.5 h at 4°C with 25 μl of beads carrying 5 to 7 μg of fusion protein in binding buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 10 mM MgCl, 12% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF]). After extensive washes with binding buffer, the beads were washed twice with binding buffer containing 0.25 M NaCl and 0.1% Nonidet P-40 (NP-40). Material remaining on beads was eluted with elution buffer (20 mM reduced glutathione, 100 mM Tris-Cl [pH 8.0], 0.1% NP-40) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was fixed, dried, and exposed to X-ray film.
Fractionation and coimmunoprecipitation.
Nuclear extract was prepared as described previously (1). Nuclear extract (0.5 g [100 ml]) was loaded onto a 100-ml phosphocellulose (P11) column (Whatman) in 0.1 M KCl DB (20 mM HEPES [pH 7.9], 10% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM PMSF, leupeptin [1 μg/ml], pepstatin A [1 μg/ml], apoprotinin [5 μg/ml]). The flowthrough (FT; 100 ml) was loaded onto a 25-ml DE-52 column (Whatman) in 0.1 M KCl DB. Proteins were eluted in a 0.1 M to 0.5 M KCl DB gradient over 100 ml and collected in 5-ml fractions. Fractions containing GCN5 and Ku proteins, as judged by Western blotting, were pooled (0.3 M KCl), and 0.5 ml of this pool was loaded onto a 20-ml Superose 6 gel filtration column (Pharmacia).
In the immunoprecipitation experiments, 25 μl of fraction was used in each reaction. Proteins were incubated with either 2 μl of antihemagglutinin (α-HA) epitope or 2 μl of α-Ku80 monoclonal serum for 2 h on ice in binding buffer (10 mM HEPES [pH 7.6], 300 mM potassium acetate, 0.1% NP-40, 1 mM PMSF), following by addition of 40 μl of protein G slurry. Then 0.5 μg of sonicated salmon sperm DNA was added to the immunoprecipitation reaction using α-Ku80 monoclonal antibody, to test the influence of DNA. Antibodies were allowed to bind to protein G beads for 30 min and then were washed extensively with binding buffer. Proteins were eluted with 1 M NaCl–0.2% SDS and immunoblotted with anti-hGCN5 (α-hGCN5) serum.
In vitro kinase assay.
DNA-PKcs and Ku proteins were purified to near homogeneity from HeLa cells, as described previously (18), with the following modifications. Enzyme from the phosphocellulose column was separated from Ku by fast protein liquid chromatography (FPLC) on a Superose 12 gel filtration column at 0.4 M KCl. Fractions containing DNA-PKcs which were devoid of Ku were pooled and further chromatographed by FPLC on MonoQ. The Ku-containing fractions, devoid of DNA-PKcs as judged by silver staining and Western analysis, were pooled and dialyzed against B/0.1 (18). Purified DNA-PKcs (1.5 μl [100 μg/ml]) and an equal amount of Ku heterodimer (when used) were incubated with 100 ng of purified recombinant hGCN5 at 30°C for 30 min in reaction buffer (HEPES [pH 7.6], 50 mM NaCl, 10 mM magnesium chloride, 8% glycerol, 1 mM dithiothreitol, 12.5 μM [γ-32P]ATP) in the presence of 100 ng of sonicated salmon sperm DNA (where indicated). When hGCN5 was phosphorylated for subsequent testing in the HAT assay, 0.5 mM cold ATP was used instead of radioactive [γ-32P]ATP. Reactions were stopped by addition of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholate); hGCN5 was immunoprecipitated with hGCN5 antiserum or preimmune serum and subjected to SDS-PAGE. Phosphorylated proteins were visualized by autoradiography of the dried gel.
HAT assay.
The HAT assay was performed as described elsewhere (12). Purified recombinant hGCN5, or immunoprecipitated hGCN5 on protein A-agarose beads, was incubated with DNA-PKcs, ATP, Ku, and DNA for phosphorylation (as described above) and then incubated for 30 min at 30°C with free calf thymus type IIA histones (Sigma) in the presence of 0.1 mCi of 3H-labeled acetyl coenzyme A (Sigma). Acetylated histones were subjected to SDS-PAGE (15% gel). The gel was fixed, soaked in Intensify liquid scintillant (Du Pont), dried, and exposed to X-ray film. In parallel, 10-ml aliquots of reaction mixtures were spotted on P-81 Whatman filters, washed four times in 50 mM sodium bicarbonate buffer (pH 9.0), and measured in a liquid scintillation counter. To measure HAT activity of GCN5 immunoprecipitated from MO59K or MO59J cells, loaded beads were incubated for 30 min at 30°C with rotation.
Immunodepletion.
Fifty-microliter portions of the FT after P11 column chromatography were incubated with α-DNA-PKcs or α-HA monoclonal antibody overnight on ice. Immunodepletion of hGCN5 was done with antigen-purified hGCN5 polyclonal antiserum. To each immunodepletion reaction, 40 μl of protein G beads preblocked in 5% bovine serum albumin (BSA) was added, and the mixtures were incubated for an additional hour. Beads were removed by centrifugation, and supernatants were subsequently used in the kinase assay. In the add-back experiment, 100 ng of semipurified DNA-PKcs (18) was added to the DNA-PKcs-depleted sample.
Affinity depletion and competition.
Equalized amounts of glutathione-Sepharose beads bearing either GST or GST-BrD were incubated with 100-μl portions of P11 FT overnight with rotation. In competition experiments, equalized amounts of eluted GST or GST-BrD were used. Beads were removed by centrifugation. Proteins bound to beads were subjected to SDS-PAGE and Western blot analysis with α-Ku70 monoclonal antibody. Affinity-depleted supernatants were used in kinase assays, followed by immunoprecipitation using preimmune or α-hGCN5 serum. Immunoprecipitates were analyzed for phosphorylation of hGCN5 by SDS-PAGE and autoradiography. As a control for equal amounts of hGCN5, 20-μl aliquots of affinity-depleted supernatants were analyzed by immunoblotting using α-hGCN5 polyclonal serum.
Phosphatase treatment.
Phosphorylated hGCN5 was immunoprecipitated from P11 FT fractions either immunodepleted for DNA-PK or mock treated with α-HA monoclonal antibody. Following washes, equal portions of protein G beads bearing hGCN5 were either treated with 50 U of calf intestinal phosphatase at 37°C for 30 min or mock treated. Immunoprecipitates were then either tested for HAT activity or subjected to SDS-PAGE and exposed to X-ray film.
In vivo labeling.
Human MO59K or MO59J glioblastoma cells that had reached 70 to 80% confluence were transfected with GCN5 expression vector by using Lipofectin (Boehringer). Prior to labeling, transfected cells were washed with either Met/Cys-free or phosphate-free medium. For labeling with [35S]Met, cells were incubated for 2 h in Met/Cys-free Dulbecco modified Eagle’s medium (DMEM) supplemented with 2% dialyzed fetal bovine serum prior to addition of 0.5 mCi of [35S]Met-Cys (ICN). Cells were labeled for 1 h, washed with cold phosphate-buffered saline and lysed with RIPA buffer supplemented with protease inhibitors. For labeling with 32P, cells were incubated for 3 h in phosphate-free DMEM supplemented with 2% dialyzed fetal bovine serum to exhaust the pool of endogenous phosphates. Cells were labeled in 4 ml of medium with 1 mCi of 32Pi (DuPont) overnight and lysed with RIPA buffer supplemented with 50 mM NaF and protease inhibitors. Extracts were centrifuged at 40,000 rpm, and supernatants were incubated with either preimmune or GCN5 antiserum for 3 h on ice following incubation with protein G beads for 1 h. Beads were washed 8 to 10 times with cold RIPA buffer and subjected to SDS-PAGE.
RESULTS
In vivo and in vitro interactions between the BrD of hGCN5 and Ku70.
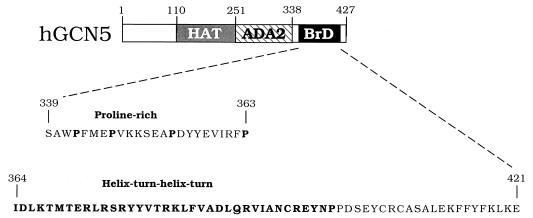
The BrD is a conserved protein motif of unknown function present in proteins that functionally interact with chromatin. The BrD derived from hGCN5 (Fig. 1) was used in a yeast two-hybrid screen (25) with a HeLa cDNA library fused to the yeast GAL4AD. We isolated six clones out of 106 primary transformants that interacted with LexA-hGCN5.BrD but not with other unrelated fusion proteins such as LexA-protein kinase C, LexA-lamin, and LexA-rho, using a LacZ filter assay (data not shown). All six clones contained overlapping cDNA fragments coding for the carboxyl terminus of the 70-kDa subunit of human Ku autoantigen (Ku70C-GAL4AD). Two independent clones were obtained (aa 279 to 499 and 349 to 602), and therefore the likely region of interaction encompasses aa 349 to 499. The largest clone (aa 349 to 602) was used in subsequent experiments, because it interacted more strongly with LexA-hGCN5.BrD. Recently, Ku70 also has been identified in a two-hybrid screen as a protein interacting with the proto-oncogene p95vav (67), and an overlapping region of Ku70 was found to interact with both p95vav and hGCN5.
FIG. 1.
hGCN5 domain structure. The domains of GCN5 are aa 1 to 110 (nonessential function) (14), aa 110 to 251 (the HAT domain) (16), aa 251 to 338 (the ADA2 interaction domain) (14), and aa 338 to 427 (the BrD motif) (53). Sequences within the BrD are aa 339 to 363 (the proline-rich sequence [hBrD.Pro]) and aa 364 to 421 (the helix-turn-helix-turn motif [hBrD.HT]). The prolines and helix-turn-helix-turn region are shown in boldface.
To quantitate the strength and specificity of interaction, β-galactosidase activities of various LexA fusions and Ku70C-GAL4AD were determined (Table 1). The interaction between LexA-hGCN5.BrD and Ku70C-GAL4AD was strong, showing 54-fold induction over interaction with the GAL4AD alone. In contrast, an unrelated LexA fusion (LexA-rho) showed less than twofold induction. BrDs derived from human TAFII250 (aa 1400 to 1608) (69) and CBP (aa 1107 to 1247) (21) were also able to interact with Ku70C-GAL4AD (Table 1), albeit somewhat more weakly than did LexA-hGCN5.BrD. Interaction between the BrDs derived from either hGCN5 or CBP was tested in vitro. Full-length in vitro-translated [35S]Ku70 bound to both GST-hGCN5.BrD and GST-CBP.BrD (Fig. 2B). These interactions indicate a general affinity between Ku70 and BrDs, although the significance of the CBP and TAFII250 interactions with Ku70 has not been further tested.
FIG. 2.
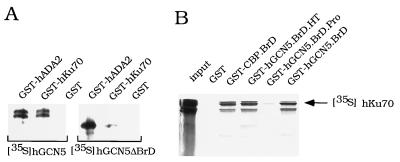
In vitro interactions between hGCN5 and Ku70. (A) hGCN5 binding to GST-hKu70. Equal amounts of GST-hADA2, GST-hKu70, or GST beads were incubated with either 35S-labeled hGCN5 or hGCN5ΔBrD translated in vitro. After washing, the material remaining on beads was eluted with 20 mM reduced glutathione and subjected to SDS-PAGE and autoradiography. (B) hKu70 binding to GST-BrDs. Equal amounts of GST, GST-CBP.BrD, GST-hGCN5.BrD.HT, GST-hGCN5.BrD.Pro, GST-hGCN5.BrD, and GST beads were incubated with in vitro-translated 35S-labeled hKu70. The material bound to beads was treated the same as for panel A. hKu70 input represented 50% of the amount used in each reaction. See Table I, footnote a, for explanation of GST fusion protein abbreviations.
Biochemical interaction between hGCN5 and Ku70 was further tested using GST fused to full-length Ku70 and [35S]hGCN5 translated in vitro. hGCN5 bound to GST-Ku70 beads but did not bind to control GST beads (Fig. 2A, left). The BrD of hGCN5 was required for interaction with Ku70, since hGCN5 lacking the BrD (hGCN5ΔBrD) bound poorly to GST-Ku70 (Fig. 2A, right). In a control experiment, both hGCN5 and hGCN5ΔBrD interacted with GST-hADA2 (Fig. 2A). The GCN5-ADA2 interaction previously was shown to be independent of the BrD (14, 53).
To further analyze the interaction between Ku70 and the BrD of hGCN5, we identified the region within the BrD of interaction with Ku70. Secondary structure prediction reveals in all BrDs the presence of two amphipathic α-helices followed by reverse turns (BrD.HT) (36) (Fig. 1). Also, the adjacent region of the BrD contains several highly conserved prolines (BrD.Pro) (Fig. 1). To address the question of whether the BrD.HT or the BrD.Pro is required for interaction with Ku70, two BrD deletion derivatives were fused to either LexA or GST to test for interaction in vivo and in vitro. In the two-hybrid assay, LexA-hGCN5.BrD.HT interacted with Ku70C-GAL4AD, but LexA-hGCN5.BrD.Pro interacted poorly (Table 1). Similarly, in the GST pull-down assay, GST-hGCN5.BrD.HT interacted with hKu70, but GST-hBrD.Pro bound poorly to hKu70 (Fig. 2B). These data indicate that interaction between Ku70 and hGCN5 requires the helix-turn-helix-turn region within the BrD of hGCN5.
Cofractionation of hGCN5 and Ku70/80.
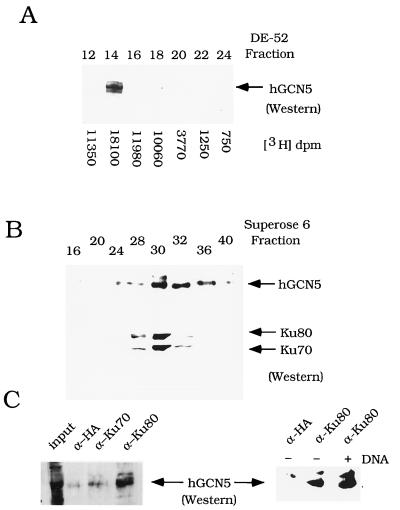
To determine whether hGCN5 and Ku70/80 interacted in vivo, HeLa nuclear extract was fractionated over three chromatographic columns and the elution pattern of the proteins was monitored. Using Western blot analysis, hGCN5 was found largely in the P11 column FT (data not shown). Following separation of the P11 FT over DE-52 ion-exchange resin, hGCN5 eluted in one sharp peak, which coincided with the peak of histone acetylation activity (Fig. 3A). The peak of Ku70/80 elution coincided with the peak of hGCN5 elution (fraction 14) on the DE-52 column (data not shown). The peak fractions of hGCN5-Ku70/80 immunological staining in the DE-52 fractions were pooled and size fractionated over Superose 6 resin. hGCN5 and the Ku heterodimer coeluted from the sizing column with an apparent molecular mass of approximately 350 kDa (Fig. 3B).
FIG. 3.
Association of hGCN5 and Ku70/80 in HeLa nuclear extract. (A) Cofractionation of hGCN5 and HAT activity over DE-52 ion-exchange chromatography. Western blot analysis was performed on DE-52 column fractions, using α-hGCN5 polyclonal serum. The arrow designates the position of hGCN5 protein. HAT activity was quantitated by liquid scintillation counting, and the values are shown for each fraction. (B) Cofractionation of Ku70/80 and hGCN5 over Superose 6 sizing chromotography. Western blot analysis was performed on Superose 6 column fractions. Upper panel, hGCN5 immunoblot; lower panel, Ku70/80 immunoblot. The column was calibrated with size marker proteins: dextran blue (2,000 kDa) fraction 15; thyroglobulin (669 kDa) fraction 24; ferritin (440 kDa) fraction 28; aldolase (158 kDa) fraction 32. (C) Coimmunoprecipitation analysis of Ku and hGCN5. Western blot analysis of hGCN5 was performed on immunoprecipitates from fraction 14 after DE-52 column chromatography (A). The monoclonal antibodies used in immunoprecipitation were α-HA epitope, α-Ku70, and α-Ku80. The input lane (left panel) represents 15% of the material used in each reaction. The right panel shows the effect of adding sonicated salmon sperm DNA. Silver staining of immunoprecipitates with anti-Ku80 or anti-HA serum indicated no differences in the overall pattern of proteins bound by each antiserum (data not shown).
To determine whether hGCN5 and Ku were physically associated, coimmunoprecipitation of the proteins was tested. Ku70 and Ku80 monoclonal antibodies were added to fraction 14 after the DE-52 column chromatography, which contained hGCN5 (Fig. 3A). hGCN5 was detected in the α-Ku80 precipitate, at approximately 15% of input (Fig. 3C). hGCN5 was not detected in the α-Ku70 precipitate (Fig. 3C, left). Since the Ku70 monoclonal antibody is directed against its carboxyl terminus (79), and since the carboxyl terminus of Ku70 is likely to be the region of interaction with hGCN5, the monoclonal antibody is likely to disrupt the Ku70-hGCN5 interaction. A control monoclonal antibody (α-HA epitope) also failed to precipitate hGCN5 (Fig. 3C). Ethidium bromide had no effect on coimmunoprecipitation of the proteins (data not shown), suggesting that contact between hGCN5 and Ku is mediated by mutual interaction rather than by coassembly on DNA. The influence of DNA on stability of the Ku70/80-hGCN5 interaction was tested, since Ku binds to the ends of double-stranded DNA (54). The addition of DNA did not affect the coimmunoprecipitation of Ku and hGCN5 (Fig. 3C, right). Thus, hGCN5 cofractionates and coimmunoprecipitates with Ku from HeLa nuclear extract, indicating an association between the proteins.
In vitro phosphorylation by DNA-PKcs lowers HAT activity of recombinant hGCN5.
These data show interaction between, and cofractionation of, hGCN5 and Ku70/80. Since human Ku autoantigen is the DNA-binding component of the DNA-PK holoenzyme (31), this raised the possibility that DNA-PKcs was also physically associated with hGCN5 and Ku. We examined the column fractions described above by immunostaining for DNA-PKcs and Ku70/80 proteins. Although cofractionating with Ku70/80 proteins over P11 and DE-52 columns (Fig. 4A), DNA-PKcs eluted from the Superose 6 column in fractions 24 to 26 (data not shown), distinct from the peak fraction of hGCN5 and Ku70/80 (Fig. 3B and 4A). These results are consistent with observations of others (24, 31) that DNA-PKcs and the Ku heterodimer can be separated from each other by gel filtration chromatography and, apparently, assemble into a complex only in the presence of DNA.
FIG. 4.
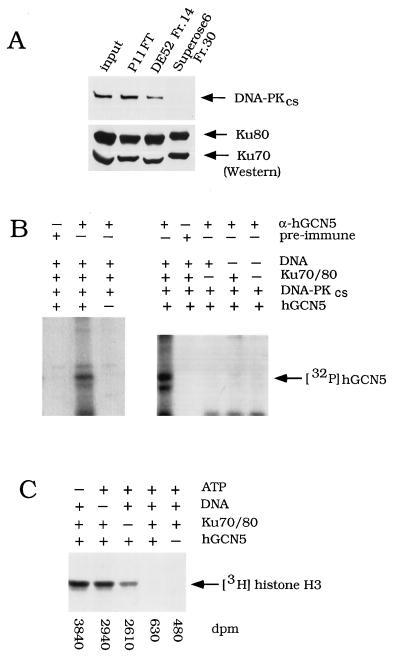
Effect of Ku/DNA-PKcs on recombinant hGCN5. (A) Western blot analysis of DNA-PKcs in sequential column fractions. Input, HeLa nuclear extract; P11 FT, FT from the P11 column; DE-52 Fr.14, fraction 14 (0.3 M KCl elution) (Fig. 3A); Superose 6 Fr.30, fraction 30 (Fig. 3B). Equal volumes of fractions described above were subjected to SDS-PAGE and transferred onto nitrocellulose. Blots were incubated with either DNA-PKcs monoclonal antibody (upper panel) or a mixture of Ku70 and p80 monoclonal antibodies (lower panel). (B) Phosphorylation of hGCN5 by DNA-PKcs in vitro. Recombinant hGCN5 was incubated with sonicated salmon sperm DNA, purified Ku70/80, and purified DNA-PKcs, as indicated. In the control experiment in the left panel, recombinant hGCN5 was not added to the lane marked −. [γ-32P]ATP was added and, following the kinase reaction, samples were immunoprecipitated with α-hGCN5 or preimmune serum and subjected to SDS-PAGE and autoradiography. (C) HAT activity assay of phosphorylated hGCN5. Recombinant hGCN5 and purified DNA-PKcs were incubated with ATP, sonicated salmon sperm DNA, and Ku70/80, as indicated. Following the HAT assay, to visualize 3H-histones, samples were subjected to SDS-PAGE and fluorography. One-fourth of the sample volume was quantitated by liquid scintillation counting, and these values are shown below the gel.
As a next step, we tested whether purified recombinant hGCN5 can be phosphorylated by DNA-PKcs purified from HeLa extract (18). DNA-PKcs phosphorylated hGCN5, as judged from control reactions using preimmune serum or lacking recombinant hGCN5 (Fig. 4B, left). The absence of Ku70/80 and/or DNA in the reaction abolished phosphorylation (Fig. 4B, right), consistent with previous observations that maximal activity of DNA-PKcs requires Ku and free DNA ends (31, 47). Thus, the phosphorylation of hGCN5 was likely to be mediated by DNA-PKcs and not by a nonspecific kinase activity present in the enzyme preparation.
The phosphorylation of hGCN5 by DNA-PKcs shown above prompted us to test whether phosphorylation affected hGCN5’s HAT activity. Recombinant hGCN5 acetylated free histone H3 (Fig. 4C) (55, 80), and, strikingly, phosphorylation by DNA-PKcs decreased HAT activity six- to eightfold (Fig. 4C). In control reactions, where hGCN5 was poorly phosphorylated because either ATP, Ku70/80, or DNA was omitted (Fig. 4A), hGCN5’s HAT activity was affected only slightly (Fig. 4C). This indicated that the inhibition of HAT activity was caused by the kinase activity of the DNA-PK holoenzyme and not by nonspecific effects of the reagents. The data above indicate that recombinant hGCN5 interacted with the Ku heterodimer and was a target of phosphorylation by DNA-PKcs, resulting in decreased HAT activity of hGCN5.
Phosphorylation by DNA-PKcs affects the HAT activity of endogenous hGCN5.
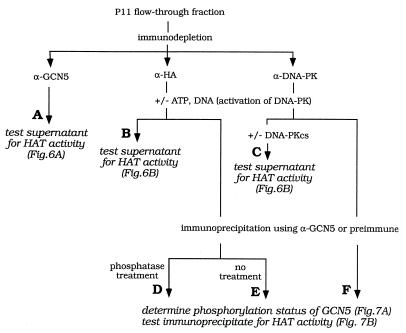
Next we examined the physiological relevance of these observations. We performed a series of experiments using the P11 column FT after HeLa extract chromatography. The P11 FT is a crude fraction and contains hGCN5 (see Fig. 6A, lower panel), Ku70/80 (Fig. 4A), and DNA-PKcs (Fig. 4A; see also Fig. 6B, lower panel). These experiments, outlined in flow diagrams (Fig. 5 and 8A), were designed to test whether endogenous hGCN5’s HAT activity is affected by DNA-PKcs in these native conditions (Fig. 6) and, if so, whether hGCN5 is a phosphorylation substrate for DNA-PKcs (Fig. 7) and, finally, if Ku70/80 interaction with the BrD of hGCN5 is required for this phosphorylation (Fig. 8).
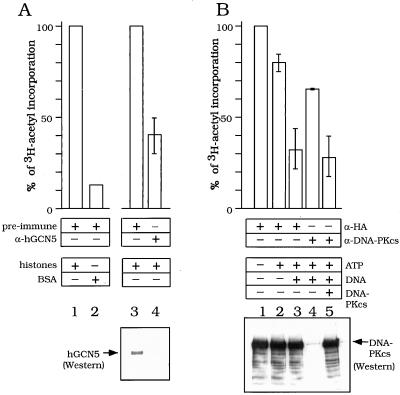
FIG. 6.
Effect of immunodepletion of endogenous hGCN5 or DNA-PKcs on HAT activity in HeLa cell extracts. (A) HAT activity of endogenous hGCN5. P11 FT fraction was preincubated with preimmune (lanes 1 to 3) or hGCN5 (lane 4) antiserum. Proteins bound to antibodies were depleted by incubation with protein G beads followed by centrifugation. Supernatants were incubated with either BSA (lane 2) or free histones (lanes 1, 3, and 4) and subjected to HAT assay; 100% activity represented the incorporation of the 3H-acetyl moiety (dpm) mediated by the P11 FT fraction after mock immunodepletion with preimmune antiserum (lanes 1 and 3). Activities of other samples were calculated as a percentage of the mock-treated values. The standard deviations are based on results of two independent experiments. (B) Effect of DNA-PKcs on HAT activity in P11 FT. P11 FT was preincubated with HA-epitope monoclonal antibody (lanes 1 to 3) or DNA-PKcs monoclonal antibody (lanes 4 and 5) and immunodepleted as for panel A. Prior to the HAT assay, immunodepleted samples were incubated with ATP (lanes 2 to 5) and nonspecific DNA (lanes 3 to 5). Purified DNA-PKcs was added to the DNA-PKcs-immunodepleted sample (lane 5). Following the kinase reaction, samples were subjected to HAT assay; 100% activity represented the incorporation of 3H-acetyl moiety (dpm) mediated by the P11 FT fraction after mock immunodepletion with HA epitope monoclonal antibodies (lane 1). Activities of other samples were calculated as a percentage of the mock-treated values. The standard deviations are based on results of three independent experiments.
FIG. 5.
Outline of experiments testing the effect of DNA-PK on endogenous hGCN5. HeLa nuclear extract was fractionated on a P11 column, and the FT was tested for hGCN5-dependent HAT activity (column A) as well as DNA-PKcs-dependent effects on hGCN5’s HAT activity (columns B and C) and DNA-PKcs-dependent phosphorylation of hGCN5 (columns D to F). The experimental results are shown in Fig. 6 and 7.
FIG. 8.
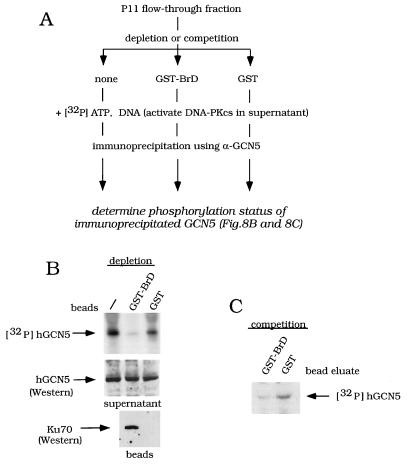
Effect of BrD affinity depletion or affinity competition on hGCN5 phosphorylation in the P11 FT fraction. (A) Outline of experiments testing effect of GST-BrD on hGCN5 phosphorylation. P11 column FT was tested for Ku70-dependent effects on hGCN5 phosphorylation, following either affinity depletion of Ku70 or affinity competition for Ku70, using GST-BrD. The experimental results are shown in panels B (depletion) and C (competition). (B) Effect of affinity depletion of Ku70 on phosphorylation of hGCN5. The P11 FT was either not treated (−) or incubated with GST-BrD beads or GST beads. Proteins bound to the beads were depleted by centrifugation. Upper panel, [γ-32P]ATP and sonicated salmon sperm DNA were added to the supernatants to activate DNA-PKcs. Following the kinase reaction, samples were immunoprecipitated using α-hGCN5 and subjected to SDS-PAGE and autoradiography. Middle and lower panels, following bead depletion, either the supernatants were assayed for hGCN5 (middle) or the beads were assayed for Ku70 (lower), using Western analysis. (C) Effect of affinity competition for Ku70 on phosphorylation of hGCN5. GST-BrD and GST were eluted from the glutathione-Sepharose beads, using reduced glutathione. The P11 FT was incubated with the bead-eluted GST-BrD or GST, and a kinase assay was performed as for panel B.
FIG. 7.
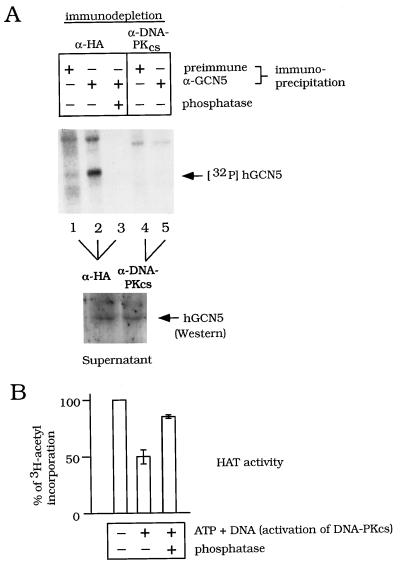
Effect of immunodepletion of DNA-PKcs and phosphatase treatment on phosphorylation and HAT activity of endogenous hGCN5. (A) Effect on phosphorylation of hGCN5. The P11 FT was preincubated with HA epitope monoclonal antibody (lanes 1 to 3) or DNA-PKcs monoclonal antibody (lanes 4 and 5) and immunodepleted as for Fig. 6A. The kinase reaction was done in the immunodepleted samples in the presence of [γ-32P]ATP and sonicated salmon sperm DNA. Following the kinase reaction, hGCN5 was immunoprecipitated and phosphatase was added to the reaction in lane 3, as indicated. The phosphorylation status of hGCN5 was determined by SDS-PAGE and autoradiography (upper panel). To show that the amount of hGCN5 input in any of the lanes in the upper panel was the same following immunodepletion by α-HA (lanes 1 to 3) or α-DNA-PKcs (lanes 4 and 5), hGCN5 input was analyzed by Western blotting (lower panel). (B) Effect of phosphatase treatment on HAT activity of hGCN5. The P11 FT was preincubated with HA epitope monoclonal antibody as for panel A. The kinase reaction was performed with nonradiolabeled ATP and sonicated salmon sperm DNA where indicated. Following the kinase reaction, samples were immunoprecipitated with α-hGCN5, and HAT activity was determined; 100% activity represented the incorporation of 3H-acetyl moiety (dpm) mediated by the P11 FT fraction without activation of DNA-PKcs (lane 1). Activities of other samples were calculated as a percentage of the mock-treated values. Standard deviations are based on results of three independent experiments.
The FT was tested for GCN5-dependent HAT activity (Fig. 6). Immunodepletion of hGCN5 (Fig. 5, column A) resulted in a 50% decrease in HAT activity (Fig. 6A, lanes 3 and 4). The remaining HAT activity is likely to be due to other, unknown HATs in the FT, since hGCN5 was not detected by Western analysis following the immunodepletion (Fig. 6A, lower panel, lanes 3 and 4). In a control experiment, the acetyltransferase activity of the FT was largely specific for exogenous histones, since the activity was 10-fold higher for free histones than for BSA (Fig. 6A, lanes 1 and 2).
Next, a possible role for DNA-PKcs in modulation of the HAT activity in the FT was investigated (Fig. 5, columns B and C). The HAT activity was lowered 25% by adding ATP (Fig. 6B, lane 2) and was reduced an additional 50% by adding both ATP and DNA (Fig. 6B, lane 3), which are effectors of DNA-PKcs kinase activity. To determine whether this latter reduction of HAT activity was due to repression by DNA-PKcs, DNA-PKcs was immunodepleted prior to the HAT assay, as confirmed by Western analysis (Fig. 6B, lower panel; compare lanes 3 and 4). DNA-PKcs immunodepletion significantly alleviated the HAT repression (Fig. 6B; compare lanes 2, 3, and 4). Furthermore, add-back of purified DNA-PKcs once again repressed HAT activity (Fig. 6B, lane 5). Taken together, the data in Fig. 6 suggest that DNA-PKcs, in the presence of its effectors, repressed the HAT activity of endogenous hGCN5.
We then determined whether hGCN5 was indeed a phosphorylation substrate of DNA-PKcs in the FT and, if so, attempted to establish a correlation between GCN5’s phosphorylation state and the level of its HAT activity (Fig. 5, columns D to F; Fig. 7). The FT was immunodepleted by using α-DNA-PKcs or was mock treated with α-HA, and the supernatant was used in a kinase reaction. The phosphorylation state of endogenous hGCN5 was determined after immunoprecipitation using α-GCN5 or preimmune serum as a control. As shown in Fig. 7A (upper panel), hGCN5 was phosphorylated in the supernatant following mock immunodepletion (lane 2) but not following immunodepletion of DNA-PKcs (lane 5). As a control, the amount of hGCN5 was comparable after immunodepletion by the two antibodies, as shown by Western analysis (Fig. 7A, lower panel). Significantly, phosphatase treatment of the mock-immunodepleted sample both reversed the DNA-PKcs-dependent phosphorylation of hGCN5 (Fig. 7A; compare lanes 2 and 3) and, importantly, largely alleviated the DNA-PKcs-dependent repression of HAT activity of immunoprecipitated hGCN5 (Fig. 7B). In the experiments shown in Fig. 7, histones were added subsequent to the kinase reaction, indicating that phosphorylation of histones themselves is unlikely to cause changes in HAT activity. In summary, endogenous hGCN5 is apparently a phosphorylation substrate for DNA-PKcs, and this phosphorylation reversibly affects hGCN5’s HAT activity.
The BrD is required for phosphorylation of hGCN5 by the DNA-PK holoenzyme.
We then determined whether hGCN5-Ku70 interaction, and specifically the BrD of hGCN5, is required for DNA-PKcs-mediated phosphorylation in the P11 FT fraction. The experimental strategy, outlined in Fig. 8A, is based on the previous observation that GST-BrD interacts with Ku70 in vitro (Fig. 2).
To test the importance of Ku70 for phosphorylation of endogenous hGCN5, GST-BrD beads were used to affinity deplete the FT fraction prior to the kinase reaction. Affinity depletion of the FT with GST-BrD beads reduced phosphorylation of hGCN5, while a mock depletion with GST alone had little effect (Fig. 8B, upper panel). The amounts of hGCN5 in the supernatants after GST-BrD or GST affinity depletion were equivalent (Fig. 8B, middle panel). GST-BrD precipitated Ku70, while GST alone interacted poorly with Ku70 (Fig. 8B, lower panel), showing that GST-BrD interaction with Ku70 competed with the normal association between hGCN5 and Ku70. These data illustrate that phosphorylation of endogenous hGCN5 requires the native Ku70-containing complex and thus extend our previous observation of the role of Ku in vitro (Fig. 4).
We then addressed the specific role of the BrD in the phosphorylation of hGCN5. In contrast to the experiment using GST-BrD beads (shown in Fig. 8B), GST-BrD was eluted from the beads and incubated with the FT fraction (Fig. 8C). Thus, in this experiment Ku70 was not depleted prior to the kinase reaction. However, phosphorylation of hGCN5 was still reduced in the presence of excess eluted GST-BrD but not GST (Fig. 8C). Thus Ku70 was still present during the kinase reaction in Fig. 8C but was titrated out by GST-BrD, thus being unable to participate in the phosphorylation of full-length GCN5. These data indicate that physical interaction between the BrD of GCN5 and Ku70 is required for phosphorylation of hGCN5 by DNA-PKcs.
Activity of hGCN5 is inhibited by DNA-PKcs in vivo.
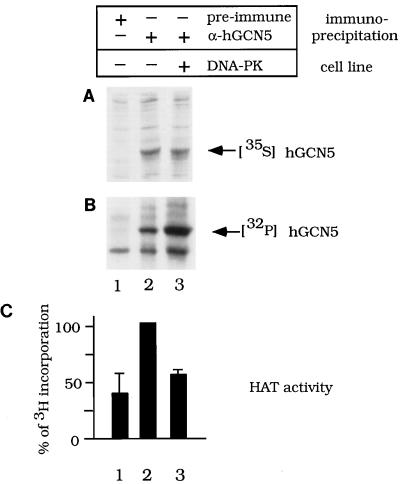
To further address the question of whether DNA-PKcs inhibits activity of GCN5 in vivo, we examined the state of phosphorylation and levels of HAT activity of hGCN5 in cells expressing DNA-PKcs or lacking DNA-PKcs. hGCN5 was transfected into the human glioblastoma cell line MO59K (DNA-PKcs+) or MO59J (DNA-PKcs−) (49). hGCN5 was expressed equally in both cell lines, as judged by [35S]Met labeling (Fig. 9A). Phosphorylation of immunoprecipitated hGCN5 was determined following 32Pi labeling of cells and was approximately two times higher in DNA-PKcs+ cells than in DNA-PKcs− cells (Fig. 9B). Strikingly, hyperphosphorylated hGCN5 from DNA-PKcs+ cells possessed lower HAT activity compared to hGCN5 from DNA-PKcs− cells (Fig. 9C). Thus, observations in both cell lines (Fig. 9) and cell fractions (Fig. 6 to 8) strongly support in vitro evidence (Fig. 4) that GCN5 is phosphorylated by DNA-PKcs, causing inhibition of GCN5’s HAT activity.
FIG. 9.
Phosphorylation levels and HAT activity of hGCN5 in DNA-PKcs+ and DNA-PKcs− glioblastoma cells. MO59K cells (DNA-PK+) (lane 3) or MO59J cells (DNA-PK−) (lanes 1 and 2) were transfected with hGCN5 expressed from the cytomegalovirus promoter. The cells were lysed, and samples were immunoprecipitated with hGCN5 antiserum (lanes 2 and 3) or preimmune serum as a control (lane 1). Transfected cells were labeled with either [35S]Met (A) to determine hGCN5 expression levels in the two cell lines or 32Pi (B) to detect the levels of hGCN5 phosphorylation. (C) Histogram of the HAT activity detected in immunoprecipitates from unlabeled cells that were similarly transfected with hGCN5. Standard deviations are based on results of two independent experiments. The high background of HAT activity in lane 1 derived from a combination of nonspecific precipitation of HAT activity by the protein G-Sepharose beads, as well as by the preimmune serum (data not shown). Total levels of incorporation of 32Pi into proteins from the two cell lines were comparable, as judged by SDS-PAGE analysis (data not shown).
DISCUSSION
The BrD is present in proteins that modify or remodel chromatin, and the motif is conserved in primary structure through evolution. Therefore, it is striking that no function has yet been attributed to the BrD. Because of the presence of the BrD in proteins of similar overall function, i.e., chromatin alteration, we initiated a two-hybrid genetic screen, using the BrD derived from human GCN5. We identified a physical interaction between hGCN5 and the Ku70/80 heterodimer, both in vitro and in vivo, which resulted in phosphorylation of hGCN5 by DNA-PKcs. The consequence of the phosphorylation was repression of hGCN5’s HAT activity, which is the first report of modulation of HAT activity. Hence, this is a potential regulatory mechanism of HAT activity.
Role of the BrD in HATs.
The BrD is found commonly in transcription factors having a role in histone acetylation, including GCN5 (12), hTAFII250 and Drosophila TAFII230 (55), and CBP (5, 58). We and others previously have shown that the BrD is dispensable for catalytic HAT activity (16, 55, 58), which suggested that it may have an ancillary or regulatory function related to HAT activity. Our current results suggest that the BrD may play a role in negative modulation of HAT activity.
Generally, deletion of the BrD causes either no effect or a modest effect on overall function. No phenotypic effect was observed by deletion of the BrD in SWI2/SNF2 (45) or in the related Drosophila melanogaster protein, brahma (74). Similarly, deletion of the BrD in the yeast transcription factor SPT7, belonging to the TBP-related group of SPT proteins, caused no detectable loss of function (28). For yeast GCN5, deletion of the BrD caused a slight reduction of the ability of the protein to complement growth or transcription (16, 53), although the stability of the truncated protein was reduced (16). Our observation that the BrD confers a negative effect on hGCN5 activity may explain the absence of a clear function for the BrD in previous studies. Since only potential positive effects of the BrD were tested previously, it may be necessary to examine the role of the BrD in negative regulation of protein activity.
We have shown that interaction with Ku70 requires the helix-turn-helix motif in the BrD of hGCN5 but not the proline-rich region located upstream (Fig. 1). Since both motifs are evolutionarily conserved in BrDs (36), it remains to be determined whether the proline-rich region is required for other, yet unknown functions of the BrD. For example, proline-rich regions are present in transcriptional activation domains (75) and thus are likely to be regions of protein-protein interaction.
BrDs derived from the HAT enzymes TAFII250 and CBP also interacted with Ku70 in the current study. TAFII250 is associated with TBP and constitutes part of the TFIID complex that is required for activated transcription (reviewed in reference 77). CBP interacts with and potentiates transcription of several transcriptional activators, including CREB and steroid receptors (reviewed in reference 39). In addition, DNA-PKcs and Ku70/80 were shown to be components of an RNA polymerase II holoenzyme purified from HeLa cell nuclear extracts (51). Thus, the interaction that we have observed between these other BrDs and Ku70 raises the intriguing possibility that the DNA-PK holoenzyme may inhibit the activity of these other transcriptionally relevant HATs, which is currently under investigation.
hGCN5 may be a physiological substrate of DNA-PK.
After obtaining Ku70 as a yeast two-hybrid partner of the BrD of hGCN5, we tested the role of this interaction in three contexts: in vitro using recombinant proteins, in HeLa cell nuclear extracts using endogenous proteins, and in vivo using DNA-PK+/− cell lines. In all three contexts hGCN5 was seen to be phosphorylated by DNA-PK, in a Ku70- and BrD-dependent manner, resulting in inhibition of HAT activity. Taken together, these data constitute strong evidence supporting a physiologically significant role for the hGCN5-Ku interaction and phosphorylation by DNA-PKcs.
Various proteins previously were shown to be phosphorylated by DNA-PKcs in vitro; however, few of these have been shown to be physiological targets. For example, many DNA-binding transcriptional activators, such as Oct1, SP1, p53, and glucocorticoid receptor (GR), are phosphorylated by DNA-PK (reviewed in reference 19), but none have been reported to be modified in vivo. Other in vitro targets include the carboxyl-terminal tail of the largest subunit of RNA polymerase II (24) and the 34-kDa subunit of the replication factor RPA (9). As we have shown for hGCN5, RPA is phosphorylated in vivo (9). Importantly, the DNA-PKcs-dependent phosphorylation of hGCN5 has a functional consequence, i.e., down-regulation of HAT activity both in vitro and in vivo.
The consensus motifs for DNA-PKcs kinase activity in vitro have been determined. DNA-PKcs is a Ser/Thr kinase and recognizes -SQ-, -TQ-, -PS-, or -PT- sites (reviewed in references 2 and 19) surrounded by acidic residues (4). Since only RPA was phosphorylated in vivo (9), and those sites have not been characterized, it remains to be determined what sites are used in vivo. Similarly, it will be important to identify sites in hGCN5 utilized by DNA-PKcs in vitro and to determine whether the same sites are phosphorylated in vivo in a DNA-PK-dependent manner. Our preliminary data suggest that the phosphorylation of recombinant hGCN5 occurs within the amino terminus (5a), and within this region reside two potential phosphorylation sites. Currently, we are testing the role of these sites in phosphorylation of hGCN5 and function in vitro and in vivo.
Given the wide range of phosphorylation substrates, it is possible that DNA-PK holoenzyme targets a large number and variety of transcription factors, including DNA binding activators as well as coactivators or HATs, to widely repress transcription at sites of repair or recombination (see below for further discussion).
Potential role of inhibition of GCN5’s HAT activity by DNA-PKcs.
What might be the functional consequence of phosphorylation of hGCN5 and inhibition of HAT activity? The DNA-PK holoenzyme is known to play a role in DNA repair and V(D)J recombination (38). One possibility is that during processes that alter the DNA template, transcription of nearby genes must be repressed. A second possibility is that following transcriptional activation associated with increased acetylation of histones, DNA-PK plays a role in inactivating HATs. As discussed above, the presence of DNA-PK in the human RNA polymerase II holoenzyme (51) lends support to the idea that modulation of HAT activity by the DNA-PK holoenzyme may be involved in transcriptional regulation.
There are several examples of Ku70/80 and/or DNA-PKcs-dependent repression of transcription. For example, Ku mediates repression of GR-dependent transcription of the mouse mammary tumor virus long terminal repeat in vivo (30). DNA-PK does indeed phosphorylate GR in vitro (30); however, it is not yet known whether phosphorylation occurs in vivo. Thus, the connection between DNA-PK-mediated phosphorylation and transcriptional repression remains to be established.
RNA polymerase I transcription is repressed in vitro in two ways by DNA-PK holoenzyme. First, Ku autoantigen was seen to compete with the positive regulator of RNA polymerase I transcription, UBF (upstream binding factor) (42), for binding to DNA. Moreover, phosphorylation of the RNA polymerase I cofactor, SL1, by DNA-PKcs prevented formation of the preinitiation complex in vitro and lowered the overall level of transcription (41). The relevance of this mechanism in vivo has not been reported.
Repression of hGCN5 activity via the DNA-PK holoenzyme might operate at several levels. First, Ku70/80 may bind to hGCN5 via the bromodomain interaction and may sequester hGCN5 in nonfunctional complexes. Interestingly, hGCN5 was found in a relatively small complex (320 kDa) from HeLa cells, compared to yeast GCN5, which is present in larger ADA protein-associated complexes (800 kDa and 1.8 MDa) (32). A second level of repression may be phosphorylation of hGCN5 by DNA-PKcs, through DNA-PKcs interaction with Ku, resulting in inhibition of HAT activity of hGCN5. Future experiments will elucidate the role of DNA-PK-mediated modulation of hGCN5 activity.
ACKNOWLEDGMENTS
We thank T. Carter for DNA-PKcs monoclonal antibodies and for help in purification of Ku70/80 DNA-PKcs; W. H. Reeves for α-Ku monoclonal antibodies; D. Jensen for help in the two-hybrid screen; N. Malik for help in purification of Ku and DNA-PKcs; and T. Carter, P. Lieberman, G. Moore, X. Nui, J. Ozer, F. J. Rauscher III, D. Reinberg, and R. Sheikhatter for valuable discussions and critical reading of the manuscript.
The work was supported by grants from the NSF and The Council for Tobacco Research to S.L.B.; grants from the NIGMS and the NSF to J.L.W.; and a Cancer Core grant from the NIH and a grant from the Pew Charitable Trust to The Wistar Institute. T.O.-H. was the recipient of an EMBO long-term fellowship; S.L.B. is the recipient of an ACS Junior Faculty Research Award, and J.L.W. is a Leukemia Society Scholar.
REFERENCES
- 1.Abmayr S M, Workman J L. Preparation of nuclear and cytoplasmic extracts from mammalian cells, units 12.1.1–12.1.9. In: Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. [DOI] [PubMed] [Google Scholar]
- 2.Anderson C W. DNA damage and the DNA-activated protein kinase. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- 3.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 4.Bannister A, Gottlieb T, Kouzarides T, Jackson S. c-Jun is phosphorylated by the DNA-dependent protein kinase in vitro; definition of the minimal kinase recognition motif. Nucleic Acids Res. 1993;21:1289–1295. doi: 10.1093/nar/21.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannister A, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5a.Barlev, N., and S. L. Berger. Unpublished observations.
- 6.Barlev N, Candau R, Wang L, Darpino P, Silverman N, Berger S. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 7.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 8.Blunt T, Finnie N, Taccioli G, Smith G, Demengeot J, Gottlieb T, Mizuta R, Varghese A, Alt F, Jeggo P, Jackson S. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 9.Boubnov N V, Weaver D T. scid cells are deficient in Ku and replication protein A phosphorylation by the DNA-dependent protein kinase. Mol Cell Biol. 1995;15:5700–5706. doi: 10.1128/mcb.15.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J, Allis C. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 12.Brownell J, Zhou J, Ranalli T, Kobayashi R, Edmondson D, Roth S, Allis C D. Tetrahymena histone acetyltransferase A: a transcriptional co-activator linking gene expression to histone acetylation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 13.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 14.Candau R, Berger S L. Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 15.Candau R, Moore P, Wang L, Barlev N, Ying C, Rosen C, Berger S. Identification of functionally conserved human homologs of the yeast adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candau R, Zhou J, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson M, Laurent B C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 18.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter T H, Anderson C W. The DNA-dependent protein kinase, DNA-PK. Prog Mol Subcell Biol. 1996;12:37–58. [Google Scholar]
- 20.Chiang Y, Komarnitsky P, Chase D, Denis C. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 21.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 22.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 23.Csordas A. On the biological role of histone acetylation. Biochem J. 1990;265:23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvir A, Stein L Y, Calore B L, Dynan W S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA PolII. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 25.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 26.Finnie N J, Gottlieb T M, Blunt T, Jeggo P, Jackson S P. DNA-PK activity is absent in xrc-6 cells; implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaillard P-H L, Martini E M-D, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 28.Gansheroff L, Dollard C, Tan P, Winston F. The S. cerevisiase SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giffin W, Tarrance H, Rodde D J, Pope L, Hache R J G. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 31.Gottlieb T, Jackson S. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 32.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast GCN5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an ADA complex and the SAGA (SPT/ADA) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 33.Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990;6:395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- 34.Hager G, Smith C, Svaren J, Horz W. Initiation of expression: remodelling genes. In: Elgin S C R, editor. Chromatin structure and gene expression. Vol. 9. Oxford, England: IRL Press; 1995. pp. 89–103. [Google Scholar]
- 35.Hartley K, Gell D, Smith G, Zhang H, Divecha N, Connelly M, Admon A, Lees-Miller S, Anderson C, Jackson S. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 36.Haynes S, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 39.Janknecht R, Hunter T. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 40.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn A, Gottlieb T, Jackson S, Grummt I. DNA-dependent protein kinase: a potent inhibitor of transcription by RNA pol I. Genes Dev. 1995;9:193–203. doi: 10.1101/gad.9.2.193. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn A, Stefanovsky V, Grummt I. The nucleolar transcription activator UBF relieves Ku antigen-mediated repression of mouse ribosomal gene transcription. Nucleic Acids Res. 1993;21:2057–2063. doi: 10.1093/nar/21.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 44.Laurent B C, Carlson M. Yeast SNF2/SWI2, SNF5 and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 45.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 46.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lees-Miller S, Anderson C W. DNA-activated protein kinase, DNA-PK: a potential coordinator of nuclear events. Cancer Cells. 1991;3:341–345. [PubMed] [Google Scholar]
- 48.Lees-Miller S P, Chen Y-R, Anderson C W. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day III R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1186. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 50.Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- 51.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 52.Marcus G, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcus G, Silverman N, Berger S, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2—putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mimori T, Hardin J. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 55.Mizzen C, Yang X-J, Kokubo T, Brownell J, Bannister A, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 56.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in S. cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nussenzweig A, Chen C, Soares V C, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 58.Ogryzko V, Schlitz R, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 59.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryotic Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 60.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 61.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 63.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. . (Review.) [DOI] [PubMed] [Google Scholar]
- 64.Piña B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reeves W H, Sthoeger Z M. Molecular cloning of cDNA encoding the p70 (Ku) lupus autoantigen. J Biol Chem. 1989;264:5047–5052. [PubMed] [Google Scholar]
- 66.Roberts S, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romera F, Dargemont C, Poso F, Reeves W, Camonis J, Gesselbrecht S, Fischer S. p95vav associates with the nuclear protein Ku70. Mol Cell Biol. 1996;16:37–44. doi: 10.1128/mcb.16.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose M, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 69.Ruppert S, Wang E H, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 70.Silverman N, Agapite J, Guarente L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 72.Svaren J, Horz W. Regulation of gene expression by nucleosomes. Curr Opin Genet Devel. 1996;6:164–170. doi: 10.1016/s0959-437x(96)80046-3. [DOI] [PubMed] [Google Scholar]
- 73.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 74.Tamkun J, Deuring R, Scott M, Kissinger M, Pattatucci A, Kaufman T, Kennison J. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 75.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 76.Tuteja N, Tuteja R, Ochem A, Taneja P, Huang N, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J, Zhang J, Wang S, Pongor S, Falaschi A. Human DNA helicase II: a novel DNA unwinding enzyme identified as the Ku autoantigen. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verrijzer C, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 78.Vojtek A, Hollenberg S, Cooper J. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 79.Wang J, Satoh M, Pierani A, Schmitt J, Chou C-H, Stunnenberg H G, Roeder R, Reeves W H. Assembly and DNA binding of recombinant Ku (p70/80) autoantigen defined by a novel monoclonal antibody specific for p70/80 heterodimers. J Cell Sci. 1994;107:3223–3233. doi: 10.1242/jcs.107.11.3223. [DOI] [PubMed] [Google Scholar]
- 80.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S. Histone acetyltransferase activity is conserved between yeast and human GCN5 and required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80a.Wang, L., L. Liu, and S. L. Berger. Unpublished observations.
- 81.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kaplana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 82.Wolffe A, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 83.Wolffe A P. Transcription: in tune with the histones. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 84.Yaneva M, Wen J, Ayala A, Cook R. cDNA-derived amino acid sequence of the 86-kDa subunit of the Ku antigen. J Biol Chem. 1989;264:13407–13411. [PubMed] [Google Scholar]
- 85.Yang X, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral E1A oncoprotein. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]