Abstract
Sex pheromone recognition is essential for mating in many insects and plays a major role in maintaining reproductive barriers. A previous study from our lab reported the evolutionary history of the pheromone receptor OR5 in Spodoptera moths. Using heterologous expression in Xenopus oocytes and site-directed mutagenesis, we found that eight amino acid substitutions were sufficient to recapitulate the evolution from an ancestral broadly-tuned to a highly specific receptor. Here, we confirmed this result using expression in Drosophila olfactory neurons. This further confirmed that multiple amino acid changes explain the shift in tuning breadth of Spodoptera OR5 during evolution.
Figure 1. Multiple amino acid changes are responsible for the shift of tuning breadth of a moth ancestral pheromone receptor .
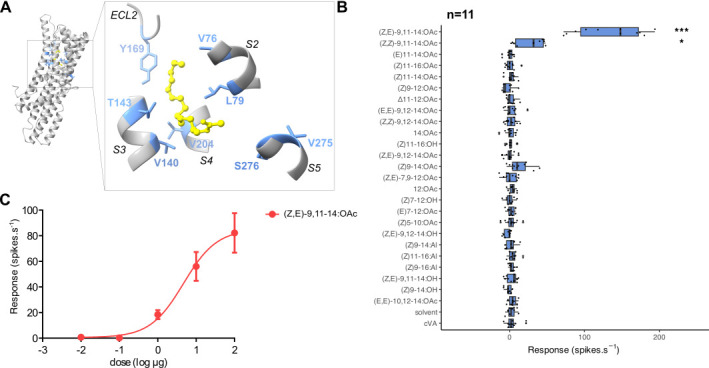
A . 3D model of AncOR5_75_mut8x and its predicted binding pocket. The 8 candidate amino acids are indicated in blue. S2, 3, 4 and 5: transmembrane helices, ECL2: extracellular loop 2. The pheromone compound ( Z,E )-9,11-14:OAc is indicated in yellow. B . Action potential frequency recorded in Drosophila at1 OSNs expressing AncOR5_75_mut8x after stimulation with 25 pheromone compounds (10 µg loaded in the stimulus cartridge). ***: corrected p-value<0.001, *: corrected p-value<0.05, Kruskal-Wallis rank sum test followed by a Wilcoxon pairwise test with a Benjamini-Hochberg correction for multiple testing. C. Dose-response curve of at1 OSNs expressing AncOR5_75_mut8x to different doses of ( Z,E )-9,11-14:OAc.
Description
At the interface between the insect and its environment, olfaction is crucial for reproduction and host selection, and plays an essential role in the adaptation of species to their environment as well as in the speciation process. Insect odorant receptors (ORs) are seven transmembrane domain proteins that detect odorants, with the N-terminus being intracellular and the C-terminus being in the cytosol (Benton et al. 2006) . ORs are expressed at the membrane of the olfactory sensory neurons (OSNs) that are hosted within sensilla of the olfactory organs, mainly the antennae. They follow the classical birth-and-death model of evolution of multigene families (Nei et Rooney 2005) . High rates of tandem duplications are ultimately leading to functional divergence and the emergence of novel response spectra, as well as loss of genes by deletion or pseudogenization events (Sanchez-Gracia et al. 2009). Each species is thus evolving its own OR repertoire. We previously identified a couple of paralogous ORs in the moths Spodoptera littoralis and Spodoptera litura with divergent tuning breadths. In these two sister species, the major component of the sex pheromone is ( Z,E )-9,11-tetradecadienyl acetate (hereafter referred to as ( Z,E )-9,11-14:OAc). While OR5 is able to bind specifically ( Z,E )-9,11-14:OAc in both species, its duplicate OR75 is also a pheromone receptor but exhibits a broader tuning breadth (Li et al. 2023) . Detection of ( Z,E )-9,11-14:OAc is necessary and sufficient to trigger male attraction towards females in S. littoralis and litura . However, this compound is not found in the pheromone blend of other species of the Spodoptera genus, meaning that a major change occurred in the pheromone communication system of a common ancestor of these two species. To identify precisely what changes occurred at the receptor level, we used in this previous work an approach of ancestral gene resurrection and reconstructed the evolutionary trajectory of OR5 and OR75 by studying three different ancestral receptors: AncOR5, AncOR75 (post-duplication) and AncOR5_75 (pre-duplication). These ancestral ORs were resurrected through heterologous expression in Drosophila OSNs and we demonstrated that the specificity likely evolved after duplication only in the OR5 lineage while OR75 remained broadly tuned. More, a combination of 3D modeling of AncOR5_75 and in silico docking allowed to identify 28 amino acid positions potentially interacting with ( Z,E )-9,11-14:OAc, of which eight diverged between AncOR5_75 and AncOR5 (panel A of the figure). To verify whether these eight mutations could have been responsible for the emergence of specificity towards ( Z,E )-9,11-14:OAc, we compared response spectra of AncOR5_75 receptors carrying or not these 8 mutations, using in vitro expression in Xenopus oocytes and stimulation with a panel of six pheromone compounds. By doing so, we could demonstrate that the ancestral receptor carrying the mutations (hereafter referred to as AncOR5_75_mut8x) exhibited the same specificity as AncOR5, suggesting that multiple amino acid changes occurred during evolution to enable the shift of tuning breadth (Li et al. 2023) .
Here, we have endeavored to confirm the shift of specificity of AncOR5_75_mut8x observed in the Xenopus oocyte system by using another expression system and a larger panel of pheromone stimuli. To achieve our goal, we generated a D. melanogaster line expressing AncOR5_75_mut8x in at1 OSNs in place of the endogenous receptor DmelOR67d. These OSNs were then stimulated with air puffs odorized with a large panel of 25 pheromone compounds, and responses were monitored by single-sensillum recordings. ( Z , E )-9,11-14:OAc and its trans isomer ( Z , Z )-9,11-14:OAc significantly activated AncOR5_75_mut8x-expressing OSNs, with a mean response around 130 spikes.s −1 and 28 spikes.s -1 , respectively (panel B of the figure). Such a small response to ( Z , Z )-9,11-14:OAc is not unusual as it was also observed for SlituOR5 in the same heterologous expression system and for AncOR5 as well, although the response was not significant for the latter. Additionally, modest non-significant responses were recorded for ( Z )9-14:OAc. Dose–response analyses revealed that AncOR5_75_mut8x had similar detection thresholds to AncOR5, SlitOR5, and SlituOR5 (Li et al. 2023) . Altogether, these results confirm the specificity of AncOR5_75_mut8x towards ( Z , E )-9,11-14:OAc, which has now been validated in two distinct heterologous expression systems. Compared to the broad tuning breadth that was observed for the non-mutated AncOR5_75 in the same Drosophila OSN setting (Li et al. 2023) , it confirms that the simultaneous change of these 8 amino acid positions lead to a narrowly tuned pheromone receptor and further supports the hypothesis that multiple amino acid changes have been necessary for the alteration of the OR5 tuning breadth as well. This result confirms again the reliability of using Xenopus oocytes and two-electrode voltage clamp as a tool to deorphanize insect odorant receptors (Wang et al. 2016; Hou et al. 2019; Yuvaraj et al. 2021) .
Methods
The AncOR5_75_mut8x mutant full-length open reading frame was optimized for expression in Drosophila , synthetized in vitro and subcloned into the pUAST.attb vector by Synbio Technologies (Monmouth Junction, NJ, USA). The transformant UAS-AncOR5_75_mut8x transformant line was generated by BestGene Inc. (Chino Hills, CA, USA) by injecting the pUAST.attB-AncOR5_75_mut8x plasmid into fly embryos with the genotype y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-51C (Bischof et al. 2007) . The UAS-AncOR5_75_mut8x balanced line was then crossed to the Or67d GAL4 line (Kurtovic, Widmer, et Dickson 2007) to obtain double homozygous flies expressing AncOR5_75_mut8x in at1 OSNs instead of Or67d. Flies were reared on standard cornmeal-yeast-agar medium and kept in a climate- and light-controlled environment (25°C, 12h:12h light:dark cycle).
Twenty-five different pheromone compounds were used to screen the responses of at1 OSNs by using single-sensillum extracellular recordings, as previously described (de Fouchier et al. 2015) . The pheromone compounds were either synthesized in the lab or purchased from Sigma-Aldrich (St Louis, MO, USA) and Pherobank (Wijk bij Duurstede, The Netherlands) and were diluted in hexane (Carlo Erba Reagents, Val de Reuil, France). at1 OSNs were stimulated during 500 ms, using stimulus cartridges made of Pasteur pipettes containing 10 µg of pheromone dropped onto a filter paper. Dose-response analyses were performed using the same methods, with doses ranging from 10 ng to 100 µg of pheromone in the stimulus cartridge. Data were analyzed using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA) and R. Odorants were considered as active if the response they elicited was statistically different from the response elicited by the solvent alone (Kruskal-Wallis followed by a pairwise Wilcoxon test with a Benjamini-Hochberg correction for multiple testing).
Reagents
|
Strain |
Genotype |
Source |
|
UAS-AncOR5_75_mut8x |
yw; UAS-AncOR5_75_mut8x, w + ; + |
BestGene Inc. |
|
Or67d GAL4 |
w + ; +; Or67d GAL4 |
Dickson lab |
|
Plasmid |
Description |
Source |
|
pUAST.attb -AncOR5_75_mut8x |
Plasmid encoding for the ancestral OR carrying eight mutations |
Synbio Technologies |
References
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006 Jan 17;4(2):e20–e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007 Feb 22;104(9):3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fouchier Arthur, Sun Xiao, Monsempes Christelle, Mirabeau Olivier, Jacquin-Joly Emmanuelle, Montagné Nicolas. Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera. Frontiers in Ecology and Evolution. 2015 Aug 12;3 doi: 10.3389/fevo.2015.00095. [DOI] [Google Scholar]
- Hou X, Zhang DD, Yuvaraj JK, Corcoran JA, Andersson MN, Löfstedt C. Functional characterization of odorant receptors from the moth Eriocrania semipurpurella: A comparison of results in the Xenopus oocyte and HEK cell systems. Insect Biochem Mol Biol. 2019 Nov 25;117:103289–103289. doi: 10.1016/j.ibmb.2019.103289. [DOI] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007 Mar 29;446(7135):542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Li Z, Capoduro R, Bastin-Héline L, Zhang S, Sun D, Lucas P, Dabir-Moghaddam D, François MC, Liu Y, Wang G, Jacquin-Joly E, Montagné N, Meslin C. A tale of two copies: Evolutionary trajectories of moth pheromone receptors. Proc Natl Acad Sci U S A. 2023 May 8;120(20):e2221166120–e2221166120. doi: 10.1073/pnas.2221166120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb) 2009 May 13;103(3):208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- Wang B, Liu Y, He K, Wang G. Comparison of research methods for functional characterization of insect olfactory receptors. Sci Rep. 2016 Sep 16;6:32806–32806. doi: 10.1038/srep32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj JK, Roberts RE, Sonntag Y, Hou XQ, Grosse-Wilde E, Machara A, Zhang DD, Hansson BS, Johanson U, Löfstedt C, Andersson MN. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 2021 Jan 26;19(1):16–16. doi: 10.1186/s12915-020-00946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


