Abstract
Gastric cancer (GC) is a prominent contributor to global cancer-related mortalities, and a deeper understanding of its molecular characteristics and tumor heterogeneity is required. Single-cell omics and spatial transcriptomics (ST) technologies have revolutionized cancer research by enabling the exploration of cellular heterogeneity and molecular landscapes at the single-cell level. In the present review, an overview of the advancements in single-cell omics and ST technologies and their applications in GC research is provided. Firstly, multiple single-cell omics and ST methods are discussed, highlighting their ability to offer unique insights into gene expression, genetic alterations, epigenomic modifications, protein expression patterns and cellular location in tissues. Furthermore, a summary is provided of key findings from previous research on single-cell omics and ST methods used in GC, which have provided valuable insights into genetic alterations, tumor diagnosis and prognosis, tumor microenvironment analysis, and treatment response. In summary, the application of single-cell omics and ST technologies has revealed the levels of cellular heterogeneity and the molecular characteristics of GC, and holds promise for improving diagnostics, personalized treatments and patient outcomes in GC.
Keywords: gastric cancer, single-cell omics, spatial transcriptomics, cellular heterogeneity, tumor microenvironment
1. Introduction
Gastric cancer (GC) is the fourth leading cause of cancer-related mortality (1). According to the World Health Organization and the International Agency for Research on Cancer, it is the fifth most frequently diagnosed cancer worldwide, with >1 million new cases reported each year (2). The incidence rates of gastric cancer show regional variations. In East Asia, particularly in countries such as Japan, China and South Korea, there is a notably higher prevalence of this disease compared with in other regions (3,4). GC is also among the cancer types with the highest mortality, with the fourth highest cancer mortality rate globally (1). Its late diagnosis, often due to initially subtle symptoms, results in poor treatment outcomes for numerous patients (5–8). Known risk factors for GC include infection with Helicobacter pylori, alcohol consumption, smoking, age, high salt intake, and diets low in fruits and vegetables (9,10). H. pylori infection, in particular, can progress from gastritis to gastroduodenal ulcers, gastric carcinoma and lymphoma (11).
The diagnosis of GC involves histological examination through endoscopic biopsy and staging with CT, endoscopic ultrasound, positron emission tomography and laparoscopy (10). GC is anatomically classified as true gastric adenocarcinoma (gADC; refers specifically to cancer originating in the stomach itself) or gastroesophageal-junction adenocarcinoma (refers to cancer that occurs at the junction where the esophagus meets the stomach), with histological categorization into diffuse and intestinal types (12). Intestinal-type GC, mainly caused by H. pylori infection, is characterized by tubular or glandular structures, and is associated with intestinal metaplasia (13). Diffuse-type GC is characterized by poorly differentiated tumor cells with decreased adhesion, leading to infiltration of the stroma as small subgroups or cellular forms (13). Adenocarcinoma accounts for the majority of GC cases (90–95%), followed by less frequent types such as lymphoma (4%), gastrointestinal stromal tumors (<1%), carcinoid tumors (3%) and hereditary diffuse GC (1–3%) according to the American Cancer Society (9).
GC treatment necessitates a multidisciplinary approach. For early-stage cases with low lymph node metastasis (LNmet) risk, endoscopic therapy or surgery alone can be curative (14). Innovations such as sentinel lymph node biopsy can improve the quality of life without compromising oncologic outcomes, yet their use outside East Asia is limited and long-term studies are ongoing (14). Later-stage localized GC requires extensive lymphadenectomy and multimodality therapy to prevent the occurrence of nodal and distant metastases (14). Targeted therapies have also been implemented for treatment, including the use of trastuzumab, an anti-HER2 antibody, and ramucirumab, a VEGFR-2 antibody (12). Despite these advancements, the prognosis and recurrence rates of GC remain discouraging due to its complex heterogeneity and the specific tumor microenvironment (TME) that promotes tumor progression and metastasis (15).
In 2009, Tang et al (16) first introduced single-cell RNA sequencing (scRNA-seq) technology to address the issue of cellular heterogeneity present in bulk RNA sequencing. Since then, single-cell sequencing (SCS) technologies, in conjunction with other bulk and/or single-cell omics technologies, have revolutionized the exploration of tumor heterogeneity at the single-cell level, providing insights into transcriptional, genomic, proteomic, epigenomic, metabolic and multi-omics characteristics of individual cells (17–25). Notably, single-cell omics biotechnology, such as scRNA-seq, enables molecular expression profiling of individual cells, allowing the mechanisms underlying tumor development to be assessed, and revealing the molecular characteristics of the TME (26–33). The present review aims to provide an overview of the most notable single-cell omics technologies, platforms and their applications in GC studies. Additionally, spatial transcriptomics (ST) technologies have emerged to construct spatial tissue atlases and characterize the spatiotemporal heterogeneity of cancers, offering the potential to profile spatial heterogeneity in tumors (34–39). Therefore, ST technologies and their applications in GC studies are also discussed. As no current ST method offers as comprehensive a coverage of the transcriptome as scRNA-seq, there is a growing demand to integrate single-cell and spatial data (40–45). Finally, studies and applications that involve integrating scRNA-seq with ST in GC research were explored.
2. Single-cell omics technologies
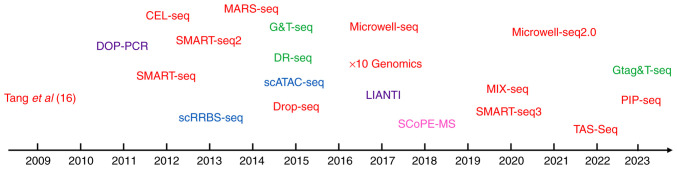
The field of single-cell omics has experienced remarkable advancements since its inception. Technologies of milestone significance in the field of single-cell sequencing are discussed subsequently. In 2009, the first single-cell mRNA sequencing method was proposed (16), followed by its application to human cancer cells in 2011 (46). Subsequently, in 2012, the first single-cell exon was sequenced (47). Building upon these developments, Picelli et al (48) introduced Smart-seq2 in 2013, a method that improved coverage and sensitivity compared with previous techniques. In 2017, Zheng et al (49) introduced a novel scRNA-seq method referred to as 10× Genomics, which revolutionized the study of cellular communication, the TME and tumor heterogeneity (Fig. 1). These advancements in single-cell omics technologies have paved the way for investigating genomic, transcriptomic and epigenomic heterogeneity at the single-cell level. Table I presents an overview of single-cell technologies and their respective characteristics.
Figure 1.
Timeline of the development of multiple single-cell omics methods. Red, scRNA sequencing; purple, scGenome sequencing; blue, scEpigenome sequencing; pink, scProteomics; green, scMulti-Omics. CEL-seq, cell expression by linear amplification and sequencing; DOP-PCR, degenerate oligonucleotide-primed PCR; DR-seq, DNA and RNA sequencing; Drop-seq, droplet-sequencing; G&T-seq, single cell genome and transcriptome sequencing; Gtag&T-seq, genome-tag and transcriptome sequencing; LIANTI, linear amplification via transposon insertion; MARS-seq, massively parallel RNA single cell sequencing; Microwell-seq, Microwell-sequencing; MIX-seq, multiplexed interrogation of gene expression through single-cell RNA sequencing; PIP-seq, protein interaction profile sequencing; sc, single-cell; scATAC-seq, single-cell assay for transposase-accessible chromatin with high-throughput sequencing; SCoPE-MS, single-cell proteomics by mass spectrometry; scRRBS-seq, single-cell reduced representation bisulfite sequencing; SMART-seq, switching mechanism at 5′end of RNA template sequencing; TAS-Seq, terminator-assisted solid-phase cDNA amplification and sequencing.
Table I.
Summary of single-cell technologies and their respective characteristics.
| Omics | Characteristics | Methods |
|---|---|---|
| Single-cell genome | Detecting single-cell SNV, CNV and other genomic sequence or structure variations | DOP-PCR, 2011 (46), MDA, 2019 (104), MALBAC, 2012 (66), LIANTI, 2017 (67), META-CS, 2021 (68) |
| Single-cell transcriptome | Detecting mRNA expression in single cells and identifying cell clusters | STRT-seq, 2011 (170), Smart-seq, 2012 (171), CEL-seq, 2012 (52), InDrop, 2015 (172), Drop-seq66, 10× Chromium Genomics, 2017 (49), MARS-seq, 2019 (62), Seq-Well, 2017 (60), Microwell-seq, 2018 (173), SPLit-seq, 2018 (61), Quartz-seq, 2013 (174), C1-CAGE, 2019 (63), RamDa-seq, 2018 (175) |
| Single-cell epigenome | Detecting the epigenomic status of cells, such as DNA methylation, histone modification and chromatin states | scRRBS, 2016 (70), WGBS, 2015 (176), CGI-seq, 2017 (177), ATAC-seq, 2015, 2017 (57,178), DNase-seq, 2013 (179), ChIP-seq, 2009 (73),Drop-ChIP, 2015 (180), scBS-seq, 2020 (23)scAba-seq, 2016 (181), CUT&Tag, 2019 (182), Single-cell Hi-C, 2013 (183) |
| Single-cell protein mass spectrometry | Use of mass spectrometry or flow cytometry instead of sequencing to measure the protein expression patterns | SCoPE-MS, 2018 (76), SCoPE2, 2021 (77), sc-CyTOF, 2022 (75) |
| Single-cell CRISPR sequencing | Combination of single-cell toolkits and CRISPR screening | Perturb-seq, 2016 (79), CRISP-seq, 2016 (80), CROP-seq, 2017 (81), Mosaic-seq,2017 (82), Direct-capture Perturb-seq, 2020 (83) |
| Single-cell multiomics | Combining analyses of genome, epigenome and transcriptome | Trio-seq, 2016 (86) |
| Combining surface proteins with transcriptome | CITE-seq, 2017 (87), REAP-seq, 2017 (184) | |
| Combining genome with transcriptome | G&T-seq, 2015 (185), DR-seq, 2015 (186) | |
| Combining DNA methylation with transcriptome | scM&T-seq, 2016 (88) | |
| Combining protein-DNA contacts with transcriptome | scDam&T-seq, 2019 (187) | |
| Combining open chromatin with TCR | T-ATAC-seq, 2018 (188) | |
| Combining open chromatin with transcriptome | SNARE-seq, 2019 (189), scCAT-seq, 2019 (190) |
CNV, copy number variation; SNV, single nucleotide variant; TCR, T-cell receptor.
scRNA-seq
scRNA-seq offers a deeper understanding of cellular heterogeneity compared with traditional bulk cell analysis (50). It provides insights into gene expression at the single-cell level. The process involves isolating and lysing individual cells, followed by reverse transcribing mRNA and then amplifying it (50). However, previous single-cell isolation methods, such as manual picking (31,51–53), FACS-sorting (54–56) and integrated microfluidic circuits (57–59), have limitations in scalability due to cost, time and labor constraints. To address these limitations, advancements have been made to enhance efficiency and reduce costs. For example, Seq-Well (60), developed in 2017, is a portable and straightforward platform for massively-parallel scRNA-seq. Its working principle involves confining mRNA capture beads with unique barcodes and cells within small pores (pinholes), followed by sealing with a semipermeable membrane. This setup is conducive to efficient cell lysis and mRNA capture. After lysis, the beads are removed for parallel sequencing. The unique barcode on each bead allows for the identification of the originating cell of each transcript. Due to its simplicity and portability, Seq-Well can be implemented in a variety of settings, making it a versatile tool for single-cell genomics and transcriptomics research. It uses selective chemical functionalization (mRNA capture beads) to enable rapid cell lysis and efficient transcript capture while minimizing cross-contamination. Another scalable method, split-pool ligation-based transcriptome sequencing (SPLiT-seq), was proposed in 2018. SPLiT-seq enables efficient sample multiplexing without the need for specialized equipment, and is compatible with fixed cells or nuclei (61). In 2019, massively parallel RNA single cell sequencing (version 2.0) was introduced, combining sub-microliter reaction volumes, optimized enzymatic mixtures and an enhanced analytical pipeline (62). These methods substantially reduced costs, improved reproducibility and decreased well-to-well contamination (62). Unlike most single-cell transcriptomic profiling methods that focus on the 3′-end of polyadenylated transcripts, C1 Cap Analysis of Gene Expression, developed in 2019, detects transcript 5′-ends using an original sample multiplexing strategy in the C1TM microfluidic system. Analyzing transcript 5′-ends enhances the understanding of gene expression by allowing precise mapping of transcription start sites, which sheds light on the complexity of promoter usage and regulatory mechanisms. This approach not only reveals the diversity of transcript isoforms, contributing to protein variability, but also improves the accuracy of gene expression profiling across different conditions and cell types. Consequently, focusing on 5′-ends is crucial for unraveling the intricacies of gene regulation and the functional diversity within cellular processes (63).
Single-cell genome sequencing
Single-cell genome sequencing has allowed greater examination of genetic diversity, making it easier to analyze both de novo germline and somatic mutations in both normal and cancerous cells (23,64). In 2001, a simple method using rolling circle amplification was introduced for the amplification of vector DNA from single colonies or plaques, removing the requirement for lengthy growth periods and conventional DNA isolation techniques (65). In 2011, Navin et al (46) used flow-sorted nuclei, whole genome amplification and next-generation sequencing to precisely measure genomic copy numbers within individual nuclei. This method was used to explore the population structure and evolutionary dynamics of tumors in human breast cancer cases. Another method, multiple annealing and looping based amplification cycles, introduced quasi-linear preamplification in 2012 (66), reducing biases associated with nonlinear amplification (whole genome amplification). Furthermore, linear amplification via transposon insertion, proposed in 2017 (67), overcame limitations of current whole genome amplification methods, including amplification bias, amplification errors and limited resolution for detecting variations, enabling micro-copy number variation detection with kilobase resolution, while minimizing amplification biases and errors. Multiplexed end-tagging amplification of complementary strands (METACs) (68), developed in 2021, improved single-cell whole-genome amplification by leveraging the complementary strands of double-stranded DNA to filter out false positives and reduce sequencing costs, achieving high accuracy in detecting single-nucleotide variations and other genomic variants, which improves single-cell whole-genome amplification by leveraging both strands of the DNA. Unique tags are added to DNA ends before amplification, allowing for the pairing of complementary strands during sequencing. This method helps filter out false positives and reduces sequencing costs by requiring less sequencing depth for high accuracy. METACs is particularly effective in detecting single-nucleotide variations and other genomic variants, differing from traditional methods that often amplify only one DNA strand and may have higher error rates. It is applicable in fields such as single-cell genomics, clinical diagnostics and population genetics (68).
Single-cell epigenome sequencing
Single-cell epigenome analysis provides valuable insights into DNA methylation, histone modification and chromatin states, which influence cellular activity (69). In 2013, the first single-cell method for methylome analysis, single-cell reduced representation bisulfite sequencing, was introduced (70,71). This technique enables the measurement of the methylation state in ~10% of CpG sites through enrichment of CpG dense regions (70,71). These sites predominantly cover most promoters, yet a limitation is their relatively poor coverage of a number of crucial regulatory regions, such as enhancers (70). The post-bisulfite adapter tagging method involves bisulfite conversion prior to library preparation, ensuring that DNA degradation does not compromise the adapter-tagged fragments. This allows for the measurement of methylation at up to 50% of CpG sites in individual cells (53). Chromatin immunoprecipitation (ChIP) followed by sequencing demonstrates improved data compared with chromatin immunoprecipitation combined with DNA microarray (ChIP combined with DNA microarray technology is a method used to identify DNA regions that interact with specific proteins, by precipitating protein-DNA complexes and hybridizing the extracted DNA onto microarrays) by providing higher resolution, greater genomic coverage, increased sensitivity and cost-effectiveness, leading to more precise and comprehensive analysis of DNA-protein interactions, allowing genome-wide profiling of DNA-binding proteins, histone modifications and nucleosomes (72,73). Another method, single cell assay for transposase-accessible chromatin with sequencing, utilizes a transposase enzyme to insert sequencing adapters into open chromatin regions, revealing which parts of the genome are active or accessible in each cell. This technology is widely applied in epigenetics to understand cell-to-cell variability, identify regulatory elements such as enhancers and promoters, and explore the mechanisms of gene regulation in diverse cell types (57).
Single-cell proteomics (SCoPE)
Single-cell protein mass spectrometry allows for comprehensive measurement of protein expression patterns in a cell (74). Cytometry by time of flight (75), a mass cytometry-based method, has been used to analyze surface and intracellular proteins using metal-labeled antibodies labeled with heavy metal isotopes, allowing simultaneous detection of multiple proteins in cells with minimal overlap and higher precision compared with fluorescent labels used in traditional flow cytometry. SCoPE by mass spectrometry (76), a high-throughput method, was developed based on liquid chromatography-tandem mass spectrometry techniques, and is a high-throughput method for single-cell proteomic analysis that allows for the isolation, enzymatic digestion, and subsequent identification and quantification of proteins from individual cells. This technique provides detailed insights into the protein composition of single cells, revealing cellular functions and heterogeneity at the proteomic level. Subsequent improvements led to the development of SCoPE2 (77), which offers enhanced quantitative accuracy, proteome coverage, sample preparation ease and cost-effectiveness.
Single-cell CRISPR sequencing
Single-cell CRISPR sequencing is a cutting-edge technique that integrates CRISPR-Cas9, a powerful gene-editing tool, with SCS methods. This innovative approach primarily focuses on executing targeted gene edits at the single-cell level and then analyzing the consequent changes in the cell transcriptome using SCS, thereby allowing researchers to directly observe the effects of specific genetic alterations on gene expression in individual cells (78). Through this approach, a deeper understanding of gene functions, cellular networks and disease mechanisms is attainable. These techniques allow the use of compiled CRISPR libraries for collective cellular interventions, followed by high-throughput phenotypic analysis by using collective cellular interventions via CRISPR libraries to simultaneously edit multiple genes, followed by high-throughput phenotypic analysis, allowing for a comprehensive study of the resulting changes in cellular behavior and characteristics, revealing complex gene functions and interactions within cellular networks (78). To date, ≥30 different single-cell CRISPR techniques have been developed; the present review focuses on introducing a few representative technologies. Perturb-seq (79) and CRISP-seq (80) were among the first single-cell CRISPR techniques to be developed. Perturb-seq combines CRISPR-mediated gene perturbation with scRNA-seq for large-scale gene function screening and studying the impact of gene expression changes on cellular states (79). CRISP-seq, similar to Perturb-seq, focuses on studying individual genes or a small numbers of genes, assessing how specific gene-editing events affect cell function (80). By introducing specific gene alterations via CRISPR, CRISPR droplet sequencing (81), followed by scRNA-seq evaluates the influence of gene perturbations on cellular states and behaviors, which combines CRISPR technology with droplet-based single-cell sequencing, allowing simultaneous editing and gene expression profiling in individual cells. Unlike traditional CRISPR techniques that focus primarily on gene editing, CRISPR droplet sequencing integrates gene editing with detailed, single-cell level transcriptomic analysis, revealing how edits affect cellular functions (78). Mosaic-seq (82) generates cellular mosaics (a collection of cells in which each cell has a distinct genetic alteration) with various genetic perturbations, and analyzes the combined effects of these disruptions using scRNA-seq. This method is used to study the interactions between different genes and their impact on cellular functions. Direct-capture Perturb-seq (83), a variant of Perturb-seq, improves data quality and analysis efficiency by directly capturing and sequencing CRISPR-guided RNA, facilitating a more precise association between gene-editing events (deliberate alterations made to the genome using CRISPR-Cas9 technology, such as knocking out, knocking in or modifying specific gene sequences) and transcriptomic changes.
Single-cell CRISPR sequencing and its derivative technologies hold potential in cancer research. These techniques assess the complex molecular networks within tumor cells and aid the understanding of the TME, drug responses and mechanisms of treatment resistance (79). For instance, Jun et al (84), using in vitro experiments, explored all cytosine-to-thymine mutations in the exon regions of three genes (MAP2K1, KRAS and NRAS), revealing the insertions and deletions and transcriptomic markers contributing to melanoma drug resistance. Roth et al (85) developed pooled knockin sequencing (PoKI-seq), a technology that measures cell abundance and state both ex vivo and in vivo. This method facilitates the barcoding and tracking of targeted integrations of large non-viral DNA templates in primary human T cells. The technology notably identified a novel TGF-β R2-41BB chimeric receptor, enhancing the clearance of solid tumors. PoKI-seq enables the parallelized rewriting of endogenous genetic sequences, accelerating the identification of effective knockin programs for cell therapies. However, specific applications of single-cell CRISPR technology in GC research have yet to be discovered.
Integration of multi-omics
Notable advancements have been made in integrating single cell multiple-omics analyses. For instance, Trio-seq allows for the simultaneous analysis of the genome sequence, epigenome and transcriptome in a single cell (86). Another technique, cellular indexing of transcriptomes and epitopes by sequencing, combines surface protein analysis with transcriptome sequencing (87), while Single Cell Methylome and Transcriptome Sequencing enables the simultaneous analysis of both the epigenome (specifically DNA methylation patterns) and transcriptome (gene expression profiles) at the single-cell level, and combines DNA methylation analysis with transcriptome sequencing within a single cell (88).
In conclusion, single-cell omics technologies, encompassing SCS, single-cell proteomics and multi-omics have undergone advancements, enabling researchers to explore the intricate details of cellular heterogeneity across various omics layers. These technologies offer insights into gene expression, genetic variations, epigenomic modifications and protein expression patterns at the single-cell level, enhancing the understanding of cellular dynamics and disease mechanisms.
3. Application of single-cell omics technologies in GC
Single-cell omics technologies have transformed the comprehension of GC by revealing cellular heterogeneity and molecular landscapes. Numerous studies have used SCS methods to investigate GC, providing insights into tumor heterogeneity, metastasis, genetic alterations, diagnosis, treatment response and the TME (47,89–92) (Fig. 2). The present review summarizes key findings of these studies, highlighting the contributions of different SCS technologies in advancing the knowledge of GC. Additionally, a summary of SCS technology applications in GC is presented in Table II.
Figure 2.
Application of single-cell sequencing technologies in research of GC, including five aspects: Tumor heterogeneity, genetic alterations, tumor diagnosis, tumor metastasis and treatment response. GC, gastric cancer; NK, natural killer.
Table II.
Summary of the applications of single-cell sequencing technologies in GC.
| First author/s, year | Sample | Method | Findings | (Refs.) |
|---|---|---|---|---|
| Andor et al, 2020 | GC cell lines. | Droplet-based reagent delivery system | Integrating single-cell genomes and transcriptomes in GC cell lines revealed substantial genetic and transcriptional diversity. | (93) |
| Peng et al, 2019 | 34 cancerous cells and 9 healthy cells from a patient with GC. | Agilent SureSelect Platform | 24 significant mutated genes were identified. | (94) |
| Wang et al, 2021 | 3 patients with GC with primary cancer tissues and matched metastatic lymph node cancer samples. | SMART-seq2 | Marker genes (ERBB2, CLDN11 and CDK12) for lymph node metastasis and potential evolution-driving genes (FOS and JUN) were identified. | (98) |
| Sathe et al, 2020 | 7 patients diagnosed with GC and 1 patient with intestinal metaplasia. | 10× Genomics | TME variations, including increased stromal cell and Treg numbers, unique transcriptional cell states in DCs and exhausted | (105) |
| CTL subclasses, and specific extracellular matrix composition in TME stromal cells. | ||||
| Zhang et al, 2021 | 9 samples of tumor tissues and 3 samples of non-tumor tissues. | 10× Genomics | Identified a set of biomarkers capable of distinguishing between benign and malignant epithelium. | (99) |
| Wang et al, 2021 | 15 patients with gastric adenocarcinoma. | 10× Genomics | Peritoneal carcinomatosis specimens were categorized into two distinct subtypes based on their prognostic implications, accompanied by the identification of a 12-gene prognostic signature. | (100) |
| Eum et al, 2020 | 5 specimens from 4 individuals with GC, consisting of 4 peritoneal ascites samples and 1 cerebrospinal fluid sample. A total of 3 samples were procured from donors without cancer, peritonitis, bacterial infection or hepatitis B/C virus. | SMART-seq2 | Tumor-associated macrophages in the malignant ascites of patients with GC exhibited pronounced M2-like characteristics. Furthermore, the presence of this M2-like phenotype in TAMs was associated with a poor prognosis in patients with GC. | (101) |
| Meyer et al, 2020 | Carcinogen-induced mouse model. | SMART-seq2 | Increased group 2 innate lymphoid cell levels in stomach tissues of patients with spasmolytic polypeptide-expressing metaplasia suggested their involvement in coordinating the metaplastic response to severe gastric injury. | (191) |
| Fu et al, 2020 | Cancer tissue samples from 2 patients with GC. Peripheral blood samples were collected from 3 patients with GC before surgery. A total of 2 normal samples of blood were obtained. | 10× Genomics | The results revealed down-regulation of the IRF8 transcription factor in CD8+ tumor-infiltrating lymphocytes from GC tissues. Furthermore, decreased IRF8 levels in blood CD8+ T cells suggested an advanced disease stage. | (106) |
| Kwon et al, 2021 | 19 patients with metastatic GC. | 10× Genomics | A diverse TCR repertoire was associated with prolonged progression-free survival in patients treated with pembrolizumab. Additionally, increased PD-1 and CD8+ T cell levels were associated with durable clinical benefits. | (192) |
| Li et al, 2022 | 9 untreated patients with non-metastatic GC. | 10× Genomics | The GC tissues exhibited enrichment of immune suppression-related gene expression in Treg cells. No distinct exhausted CD8+ T cell cluster was identified, and low expression levels of exhaustion markers PDCD1, CTLA4, HAVCR2, LAG-3 and TIGIT were observed. Furthermore, specific ACKR1 expression in tumor endothelial cells was associated with poor prognosis. | (102) |
| Huang et al, 2023 | 35 patients across four medical centers (with/without GC peritoneal metastasis). | 10× Genomics | Elevated monocyte-like DCs during GC progression were associated with poor prognosis. Additionally, high-plasticity GC, characterized by autophagy-related genes MARCKS and TXNIP, was associated with a worse prognosis. | (107) |
| Kang et al, 2022 | Tumors and matched normal tissue of 24 treatment-naïve patients with GC. | 10× Genomics | Tumors exhibited prominent overrepresentation of activated fibroblasts and endothelial cells. Additionally, immuno-suppressive myeloid cell subgroups and Tregs were pivotal in creating an immuno-suppressive microenvironment, which was associated with an unfavorable prognosis and resistance to anti-programmed cell death 1 treatment in patients. | (91) |
| Kim et al, 2022 | A total of 12 chemotherapy-naïve Korean patients with metastatic advanced GC | 10× Genomics | Identified the defining characteristics linked to a positive response to platinum-based chemotherapy. | (111) |
| Huang et al, 2022 | 9 patients were chosen, each providing both primary tumor and normal tissue samples for the study. | 10× Genomics | A comprehensive transcriptomic landscape of malignant epithelial cells in GC was constructed. Prognostic signatures for predicting overall survival in patients with GC were developed by combining the scRNA-seq data with bulk RNA sequencing datasets. | (103) |
| Sundar et al, 2022 | A total of 53 tumor samples, including 13 APB-high, 27 APB-int and 13 APB-low samples. | Illumina Hiseq sequencer | scRNA-seq analysis validated the presence of distinct immunological populations and revealed decreased proportions of T cells in tumors characterized by high APB levels. | (108) |
| Zhou et al, 2023 | A total of 14 tissue samples were collected, including 1 normal mucosa tissue, 6 DGC tissues, 6 PDGC tissues and 1 NEC tissue. | 10× Genomics | In both malignant cells and the immune microenvironment, there were distinctive molecular characteristics observed in DGC, PDGC and NEC. The analysis indicated a progressive reduction in interferon pathway responses as cells transitioned from DGC to NEC, leading to an increased capacity for immune evasion. | (193) |
| Yang et al, 2022 | 3 patients with synchronous gastric and colorectal cancer. | BD® Single-Cell Multiplexing Kit | The differential prognoses and drug responses observed in GC and colorectal cancer could be primarily attributed to the variations in the TME, which were influenced by the mutational landscape and microbiome components. | (109) |
| Li et al, 2022 | 10 GC specimens collected before and after neoadjuvant treatment with camrelizumab in combination with mFOLFOX6. | 10× Genomics | Conducted single-cell RNA sequencing on 10 GC samples both before and after neoadjuvant treatment. The study highlighted that high expression of interferon-γ in CD8+ T cells was associated with enhanced responses to this combination therapy, indicating an immunological impact on the tumor environment. | (112) |
| Zhang et al, 2019 | Tumor tissue of 13 patients. | 10× Chromium platform | Identification of OR51E1 as a marker for distinctive endocrine cells in early-malignant lesions, and the potential utility of HES6 in identifying metaplasia at an early stage was revealed. | (104) |
| Jiang et al, 2022 | 10 human tissue samples from 6 patients. | 10× Genomics | Using scRNA-seq, the study assessed primary tumors and different metastases (liver, peritoneum, ovary and lymph node) in GC, and aimed to analyze intra- and intertumoral heterogeneity of carcinoma cells and the TME and understood organ-specific metastatic patterns, with particular emphasis on lymphatic metastases. | (194) |
| Chen et al, 2021 | 111 patients with advanced GC. | Illumina HiSeq X Ten system | Patients exhibited a high prevalence of mutations in the KRAS and Rap1 signaling pathway, particularly in 3 baseline small CTCs displaying trisomy 8. Additionally, distinctive mutations in the MET/PI3K/AKT pathway and the SMARCB1 gene were detected in the patients, specifically in >6 large multiploid CTCs. | (95) |
| Nagaoka et al, 2020 | C57BL/6 mice inoculated with two GC cell lines (YTN16 and YTN2). | 10× Genomics | In the murine GC model, the combination of anti-IL-17 and anti-PD-1 monoclonal antibodies was associated with robust tumor regression. | (113) |
| Zhang et al, 2020 | 13 mucosa biopsies from 9 patients. These patients included individuals with wild superficial gastritis (non-atrophic gastritis, with three biopsies), which served as the normal control group, chronic atrophic gastritis (with three biopsies), intestinal metaplasia (with six biopsies) and EGC (with one biopsy). | 10× Genomics | A panel of specific signatures was identified for EGC, which hold clinical significance for accurate diagnosis of EGC | (104) |
| Li et al, 2022 | Eight pairs of GC and adjacent mucosal samples. | 10× Genomics | The analysis focused on examining the characteristics of different cancer-associated fibroblast subsets and their role in regulating the dynamic communication between cancer-associated fibroblasts (CAFs) within the TME and other cells. | (110) |
APB, alternate promoter burden; CTCs, circulating tumor cells; CTL, cytotoxic T cell; DCs, dendritic cells; DGC, differentiated gastric cancer; EGC, early gastric cancer; GC, gastric cancer; IRF8, interferon regulatory factor 8; mFOLFOX6, 5-fluorouracil, leucovorin and oxaliplatin; NEC, neuroendocrine carcinoma; PDGC, poorly differentiated gastric cancer; scRNA-seq, single-cell RNA sequencing; SMART-seq, single molecule amplification and re-sequencing technology for sequencing; TCR, T-cell receptor; TME, tumor microenvironment; Treg, regulatory T cell.
Genetic alterations of single cells
Early applications of SCS in GC focused on transcriptome and single-cell genome analysis of GC cell lines and reported marked genetic and transcriptional diversity (93). In one study, the identification of 24 notable mutated genes among tumor cells demonstrated the genetic alterations underlying GC and potential therapeutic targets (94). Analysis of circulating tumor cells (CTCs) from patients with advanced GC demonstrated numerous mutations in the genes associated with the KRAS and Rap1 pathways, as well as mutations in the genes associated with the MET/PI3K/AKT pathway and the SMARCB1 gene in patients with large multiploid CTCs (95), leading to the development of resistance to either chemotherapy alone (96) or chemotherapy combined with targeted therapy (97) in patients with GC.
Tumor diagnosis and prognosis
In a study on GC lymph node metastasis, scRNA-seq was performed on primary and metastatic tissues from 3 patients, revealing intratumoural heterogeneity and distinct carcinoma profiles. The results identified a subgroup of cells indicating a transitional state in the metastasis process, and also revealed potential marker genes (ERBB2, CLDN11 and CDK12) and genes driving gastric cancer evolution (FOS and JUN), offering insights for GC treatment (98). A panel of biomarkers was identified for discriminating between benign and malignant epithelial tissues, potentially aiding in early detection and diagnosis of GC (99). Using scRNA-seq analysis, subtypes of peritoneal carcinomatosis samples from patients with GC were classified, and a 12-gene prognostic signature was identified (100). Investigation of metastatic GC using peritoneal ascite samples and cerebrospinal fluid revealed that poor prognosis was associated with M2-like characteristics in tumor-associated macrophages (101). In a study of patients with non-metastatic GC, immunosuppressive gene expression patterns were enriched in regulatory T cells (Tregs) within gastric tumor tissues, indicating an immunosuppressive TME. The absence of a separate exhausted CD8+ T cell cluster and low expression levels of exhaustion markers were also observed, and ACKR1 was identified as a potential marker associated with poor prognosis (102). Using scRNA-seq, a broad spectrum of GC subtypes was assessed to create a transcriptomic map of biomarkers from malignant epithelial cells for the prediction of overall survival in patients with GC (103). OR51E1 has been identified as a key marker gene for unique endocrine cells in early-malignant lesions of gastric cancer, offering a potential avenue for early detection of malignancy (103). Simultaneously, HES6 has been recognized for its potential utility in identifying metaplasia at an early stage, demonstrating its importance in the early diagnosis and intervention of precancerous gastric conditions (104). Furthermore, a panel of early GC-specific signatures was identified using mucosa biopsies, which may be used in clinical applications for early diagnosis (104).
TME analysis
Analysis of the TME in patients with GC revealed increased stromal cells and Tregs, unique transcriptional cell states in dendritic cells (DCs), exhausted cytotoxic T lymphocytes and a specific extracellular matrix composition not found in normal tissue (105). Furthermore, in more advanced disease stages, a downregulation of interferon regulatory factor 8 in CD8+ tumor-infiltrating lymphocytes has been reported, revealing changes in the immunological landscape in GC and its potential implications for disease progression (106).
Another study demonstrated that monocyte-like DCs and autophagy-related genes marking high-plasticity (ability of certain cancer cells to adapt and change in response to different environments or therapeutic pressures) GC were associated with poor prognosis during GC peritoneal metastasis progression (107). In tumors with a high alternate promoter burden (APB-high), characterized by increased use of alternative gene promoters, distinct immunological populations were observed along with a reduced proportion of T cells. These findings shed light on the immunological aspects of GC and how the APB-high status influences tumor progression and the immune response (108).
Comparisons between GC and colorectal cancer revealed distinct mutational landscapes and microbiomes, contributing to differences in the TME, and thus, disease prognosis (109). Furthermore, the communication between cancer-associated fibroblasts (CAFs) within the TME and other cells provides insights into their regulatory functions (110).
Treatment response
Activated fibroblasts, endothelial cells, immunosuppressive myeloid cell subsets and Tregs present in the TME were associated with an unfavorable prognosis and resistance to anti-programmed cell death 1 therapy in patients GC (91). Characteristics linked to a positive response to platinum-based chemotherapy were defined, aiding personalized treatment decisions. For example, response was associated with on-treatment TME remodeling, including natural killer cell recruitment, decreased tumor-associated macrophages, M1-macrophage repolarization and increased effector T-cell infiltration (91). In non-responders to chemotherapy, Kim et al (111) observed low or no programmed death-ligand 1 expression, an increase in Wnt signaling and B-cell infiltration, a higher presence of lymphocyte activating 3-expressing T cells, and a reduction in dendritic cells. This suggests a distinct pattern of immune changes associated with chemotherapy resistance.
To assess the effects of combination therapy with camrelizumab and 5-fluorouracil, leucovorin and oxaliplatin on GC and its impact on the TME, Li et al (112) conducted single-cell RNA sequencing on 10 GC samples both before and after neoadjuvant treatment. This study highlighted that high expression of interferon-γ in CD8+ T cells was associated with enhanced responses to this combination therapy, indicating an immunological impact on the tumor environment (112). Additionally, a murine model suggested the potential therapeutic approach of combining anti-IL-17 and anti-programmed death-1 monoclonal antibodies for GC tumor regression (113).
In conclusion, these studies (112,113) demonstrated the wide-ranging application and use of single-cell omics technologies in GC research. By analyzing single cells, these studies have provided insights into intratumoral heterogeneity, the TME, immune and treatment responses, prognostic markers, and potential therapeutic targets. In addition, these techniques contribute to the understanding of GC biology and hold promise for improved diagnostics and personalized treatments.
4. ST technologies
ST techniques have notably improved the understanding of cellular function within multicellular organisms by revealing the precise location of cells in tissue sections. These techniques can be broadly categorized into two main types: Imaging-based methods and sequencing-based methods (114). Imaging-based methods include in situ hybridization (ISH) and in situ sequencing (ISS), while sequencing-based methods include laser capture microdissection (LCM) and in situ barcoding (ISB) (115). Table III provides an overview of the key characteristics of ST technologies.
Table III.
Summary of ST technologies.
| Category | ST methods | Samples | Characteristic |
|---|---|---|---|
| LCM | LCM-seq, 2016 (131), tomo-seq, 2014 (133) Geo-seq, 2017 (134), PIC, 2021 (195), NICHE-seq (132), | Formalin fixed paraffin-embedded/fresh frozen | Uses a laser to precisely dissect and capture targeted cells or areas, enabling downstream RNA analysis to obtain transcriptomic information specific to the isolated cells or regions. |
| ISB | NanoString Technologies, Inc. DSP, 2020 (137), HDST, 2019 (138), Visium, 2016 (139), Slide-seq, 2019 (141), Stereo-seq (140) | Formalin fixed paraffin-embedded/fresh frozen | Involves barcoding individual cells within intact tissue sections using DNA-barcoded antibodies or oligonucleotide-conjugated antibodies, facilitating subsequent identification and spatial localization during RNA sequencing analysis. |
| ISH | Sm FISH, 1998 (116), Seq FISH+, 2019 (119), MERFISH, 2015 (196), SABER, 2019 (197), seqFISH (117,118), multiplexed error-robust FISH (120,121) and RollFISH (122) | Formalin fixed paraffin-embedded/fresh frozen | Uses complementary DNA or RNA probes labeled with fluorescent or chromogenic markers to detect and visualize target RNA molecules in their original spatial context. |
| ISS | ExSeq, 2021 (126), FISSEQ, 2014 (127), ISS, 2013 (198), STAR map, 2018 (128), BaristaSeq 2018 (127) | Formalin fixed paraffin-embedded/fresh frozen/cell cultures | Involves iterative cycles of sequential hybridization of spatially barcoded oligonucleotides, followed by imaging and nucleotide incorporation, to determine the RNA sequence and its spatial location. |
ISB, in situ barcoding; ISH, in situ hybridization; ISS, in situ sequencing; LCM, laser capture microdissection; ST, spatial transcriptomics.
ISH techniques, such as single-molecule RNA fluorescence ISH (FISH) (116), seqFISH (117,118), seqFISH+ (119), multiplexed error-robust FISH (120,121) and RollFISH (122), use labeled probes to detect and visualize specific RNA molecules within tissue sections. These techniques involve hybridization of labeled probes to complementary target RNA sequences, followed by signal detection and localization. However, ISH is limited by the need for prior probe design knowledge and its inability to provide transcriptome-wide coverage, thereby constraining its applications (117,123,124).
ISS-based ST techniques, including fluorescent ISS (125), expansion sequencing (126), BaristaSeq (127) and spatially-resolved transcript amplicon readout mapping (128), enable direct sequencing of RNA molecules in their tissue context, thereby offering spatially resolved transcriptomic information. While ISS-based techniques offer subcellular resolution, their use is impacted by limitations, including a restricted number of targeted genes or low detection efficiency (125–130).
LCM-based ST techniques, such as laser capture microdissection sequencing (131), NICHE-seq (132), tomo-seq (133) and Geo-seq (134) facilitate precise assessment of specific cell populations or defined spatial regions of interest within tissues. These techniques allow for the examination of gene expression patterns within complex tissue architecture. However, LCM-based techniques are labor-intensive and low-throughput, rendering them impractical for processing samples in large batches (135,136).
ISB-based ST techniques provide transcriptome-wide resolution at the cellular and subcellular levels, enabling investigations into gene expression patterns within the tissue context. Notable examples of ISB-based techniques include NanoString Technologies, Inc. digital spatial profiling (DSP) (137), High-Definition ST (138), Visium (139), Stereo-seq (140) and Slide-seq (141). These techniques enable simultaneous detection of multiple genes and offer valuable insights into cellular spatial organization and tissue heterogeneity (138,140,142,143).
By using ST techniques, researchers can gain a comprehensive and detailed understanding of the spatial distribution of gene expression within tissues. These advancements have notable implications for various fields, including developmental biology, disease research and regenerative medicine.
5. Application of ST technologies in GC
Previous research has demonstrated the diverse applications of ST technologies. These applications encompass in situ cell typing (144,145), spatial gene expression pattern acquisition (139), tumor trajectory mapping (146), exploration of tumor pathogenesis (147–150), investigation of the TME (33,151–155) and prediction of disease prognosis (156,157). In Table IV, a summary of ST technology applications in GC is presented. For instance, Kumar et al (90) pinpointed specific B-cell sublineages exhibiting increased proportions in diffuse-type gastric cancer and highlighted KLF12 expression in epithelial cells as a potential driver of plasma cell recruitment. Furthermore, a stepwise accumulation of CAF subpopulations characterized by high co-expression of INHBA and FAP was identified.
Table IV.
Summary of spatial transcriptomics technology applications in GC.
| First author/s, year | Sample | Method | Findings | (Refs.) |
|---|---|---|---|---|
| Kumar et al, 2022 | 10 tumor (patients diagnosedwith gastric adenocarcinoma) and 3 normal samples. | NanoString Technologies, Inc. GeoMx | Increased plasma cells and stage-dependent accumulation of cancer-associated fibroblasts in diffuse-type gastric tumors, supported by spatial transcriptomics. | (90) |
| Jeong et al, 2021 | 5 patients with diffuse-type GC. | IHC and ISH | IHC and duplex ISH unveiled the spatial distribution of different cell types, along with the presence of CCL2-expressing endothelial cells and fibroblasts, signifying tumor invasion. | (158) |
| Sundar et al, 2021 | Each assay encompassed a total of 64 primary GC samples and their corresponding regional lymph node metastases, all originating from patients with locally advanced, resectable GC. | NanoString Technologies, Inc. ‘PanCancer Progression Panel’ | In GC, metastases to regional lymph nodes are typically rooted in the deeper subregions of the primary tumor. Upcoming trials for novel targeted treatments should focus on evaluating these deep tumor areas, as key genes relevant to therapy may show unique changes there. | (159) |
| Yamasaki et al, 2022 | Three gastric neoplasia tumor organoids from gastric neoplasia mice (GAN-WT, GAN-p53 and GAN-KP). | Visium, 10× Genomics | Potential involvement of hypoxia and MAPK signaling in the progression of KRAS-mutated GC, independent of Wnt signaling. Potential use of trametinib as a therapeutic candidate for suppressing hypoxia-induced tumor-stroma interactions and inhibiting metastatic progression. | (160) |
| Grosser et al, 2022 | 6 SARIFA-positive and 6 SARIFA-negative cases. | NanoString Technologies, Inc. GeoMx DSP | DSP demonstrated that SARIFA-positive cases exhibited upregulation of genes associated with triglyceride catabolism and endogenous sterols. Specifically, differential expression of COL15A1, FABP2 and FABP4 was observed in the positive cases. | (161) |
DSP, digital spatial profiling; GAN, gastric neoplasia; GAN-WT, gastric neoplasia; GAN-p53, gastric neoplasia with p53 knockout; GAN-KP, KRASG12V-expressing gastric neoplasia-p53KO; GC, gastric cancer; IHC, immunohistochemistry; ISH, in situ hybridization; LNmet, lymph node metastasis; PTdeep, primary tumor deep; SARIFA, stroma areactive invasion front areas.
Furthermore, immunohistochemistry (IHC) and duplex ISH techniques were used to evaluate the distribution of major cell types, and identified CCL2-expressing endothelial cells and fibroblasts, thereby providing evidence of tumor invasion (158). Utilizing the NanoString Technologies, Inc. ‘PanCancer Progression Panel’, Sundar et al (159) conducted a differential gene expression analysis and revealed that only 16% of genes exhibited significant differences between primary tumor deep (PTdeep) areas and corresponding LNmet samples. Notably, both LNmet and PTdeep samples exhibited increased expression of several genes with potential therapeutic significance, such as IGF1, PIK3CD and TGFB1, compared with superficial primary tumors (159). In a separate study using 10× Genomics Visium, Yamasaki et al (160) demonstrated the role of hypoxia signaling in the metastatic progression of KRASG12V-expressing gastric neoplasia-p53KO tumors, highlighting trametinib as a promising therapeutic approach to curb metastasis in KRAS-mutated GC. Furthermore, the application of DSP revealed upregulation of genes related to triglyceride catabolism and endogenous sterols, such as COL15A1, FABP2 and FABP4, particularly in cases positive for stroma-reactive invasion front areas (161).
To summarize, the applications of ST technologies in GC have yielded valuable insights into cell types, spatial gene expression patterns, tumor invasion, metastatic progression and potential therapeutic targets. These findings contribute to the understanding of GC pathogenesis and open avenues for improved treatment strategies.
6. Integration of scRNA-seq and ST
scRNA-seq is a tool for identifying cell subpopulations within tissues. However, it is unable to capture the spatial arrangement of cells and the immediate networks of intercellular communication in their native locations (41). ST technologies have not yet achieved the same level of resolution as scRNA-seq in transcriptomic maps of tissues (117). Therefore, integrating both single-cell and ST data can provide a comprehensive understanding of cell-type distribution and the potential mechanisms of intercellular communication underlying tissue architecture (41).
Li et al (110) conducted scRNA-seq combined with multi-staining registration of 16 samples from cancer and adjacent mucosa tissues (8 patients). This study reported four subsets of CAFs with distinct properties in GC.
Kumar et al (90) compared single-cell profiles between patient-derived organoids and primary tumors, highlighting similarities and differences within and between lineages. These findings were compared with ST using in vitro and in vivo models, providing a high-resolution molecular resource for intra- and interpatient lineage states across distinct GC subtypes.
Jeong et al (158) performed scRNA-seq on tissue samples from different layers of diffuse-type GC and validated the results using IHC and ISH on formalin-fixed paraffin-embedded tissues. This study revealed spatial reprogramming of the TME that may contribute to the invasive tumor potential in diffuse-type GC.
Sundar et al (159) investigated spatial intratumoral heterogeneity in primary GC and matched these to LNmet using transcriptomic profiles, DNA copy number profiles and histomorphological phenotypes. These findings suggested that regional lymph node metastases likely originate from deeper subregions of the primary tumor.
Jia et al (162) performed scRNA-seq and IHC on samples from patients with gADC to characterize the immune cell population in the TME. The results provided novel insights into the immune and tumor cell signatures in the TME of gADC with tertiary lymphoid structures and highlighted the potential role of IgA-mediated humoral immunity in these patients.
Using simultaneous single-cell and spatial analysis, Xie et al (163) demonstrated that secreted phosphoprotein 1 was expressed at high levels in GC and was associated with macrophage infiltration, advanced tumor stage and increased mortality in patients with advanced GC. Enrichment of tumor-specific macrophages in the deep layer of GC tissue was identified, emphasizing their role in the disease.
In summary, integrating single-cell and ST data holds great potential for unraveling the complex architecture of tissues and understanding intercellular communication networks. The aforementioned studies (90,110,158,159) provide valuable insights into the cellular heterogeneity, immune response and spatial organization of GC, which contributes to the knowledge of GC and demonstrates future research and therapeutic strategies.
7. Conclusion and future direction
GC is an invasive disease associated with high morbidity and characterized by notable heterogeneity. Single-cell omics technologies and analytical tools have been identified as resources for elucidating the complexity of the TME, and intra- and intertumoral heterogeneity. In the present review, an overview of current single-cell omics technologies and their applications in GC research was provided. Discussing the rapidly advancing field of ST and understanding the spatial organization of tumors is crucial for evaluating tumorigenesis and disease progression, and how it can be used with single-cell omics to gain deeper insights into the characteristics of GC. This combined approach provides a method to construct spatial histology information and assess the spatial structure of tumors. By integrating these complementary approaches, previously unknown mechanisms of tumor heterogeneity can be assessed. This integrative effort holds great promise for defining disease subtypes, predicting prognosis and enabling targeted therapies to be delivered, based on the spatial distribution of specific cell subtypes. It also allows the identification of ligands and receptors involved in their mechanism of action. However, SCS and ST research on GC has not yet been applied to the clinical practice of treating GC. Ultimately, this comprehensive approach will further the understanding of tumorigenesis and allow the development of novel techniques for precision therapy in the future.
8. Prospects for integrating single-cell omics and ST with artificial intelligence
In cancer research, the combination of high-resolution data from single-cell omics and ST with the analytical capabilities of artificial intelligence (AI) offers insights into cellular heterogeneity and intercellular interactions (25,164,165). AI, especially deep learning, efficiently processes vast and complex data, automatically identifying cell states and subgroups (166). This integration not only allows the combination of results from a number of studies and data classification, but also reveals intratumoral cell communication and interactions, aiding in predicting tumor progression pathways, identifying novel drug targets and providing decision support for precision therapy and personalized strategies (167). This interdisciplinary fusion markedly advances the exploration of tumor complexity, accelerates research progress and promotes therapeutic innovation, marking a notable advancement in the field of cancer medicine (168,169).
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Natural Science Foundation of Sichuan Province (grant nos. 2022NSFSC1610, 2023NSFSC0678 and 2023NSFSC0569).
Availability of data and materials
Not applicable.
Authors' contributions
Visualization of the data was performed by LR, LN, PC and YiZ. Writing of the original draft of the manuscript was performed by LR and DH. Reviewing and editing of the manuscript was performed by LR, DH, HoL, LN, PC, XY, YaZ, HaL, JS, NL and YiZ, and validation of the manuscript and supervision was provided by HaL, NL and YiZ. Data authentication is not applicable. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21:4012. doi: 10.3390/ijms21114012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong MCS, Huang J, Chan PSF, Choi P, Lao XQ, Chan SM, Teoh A, Liang P. Global incidence and mortality of gastric cancer, 1980–2018. JAMA Netw Open. 2021;4:e2118457. doi: 10.1001/jamanetworkopen.2021.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.GBD 2017 Stomach Cancer Collaborators, corp-author. The global, regional, and national burden of stomach cancer in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease study 2017. Lancet Gastroenterol Hepatol. 2020;5:42–54. doi: 10.1016/S2468-1253(19)30328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham D, Starling N, Rao S, Iveson T, Nicolson M, Coxon F, Middleton G, Daniel F, Oates J, Norman AR, Upper Gastrointestinal Clinical Studies Group of the National Cancer Research Institute of the United Kingdom Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med. 2008;358:36–46. doi: 10.1056/NEJMoa073149. [DOI] [PubMed] [Google Scholar]
- 7.Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M, Miyashita K, Nishizaki T, Kobayashi O, Takiyama W, et al. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): A phase III trial. Lancet. Oncol. 2008;9:215–221. doi: 10.1016/S1470-2045(08)70035-4. [DOI] [PubMed] [Google Scholar]
- 8.Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, Hironaka S, Sugimoto N, Lipatov O, Kim TY, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): A double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–1235. doi: 10.1016/S1470-2045(14)70420-6. [DOI] [PubMed] [Google Scholar]
- 9.Salvatori S, Marafini I, Laudisi F, Monteleone G, Stolfi C. Helicobacter pylori and Gastric cancer: Pathogenetic mechanisms. Int J Mol Sci. 2023;24:2895. doi: 10.3390/ijms24032895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 11.Polk DB, Peek RM., Jr Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 13.Onoyama T, Ishikawa S, Isomoto H. Gastric cancer and genomics: Review of literature. J Gastroenterol. 2022;57:505–516. doi: 10.1007/s00535-022-01879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li GZ, Doherty GM, Wang J. Surgical management of gastric cancer: A review. JAMA Surg. 2022;157:446–454. doi: 10.1001/jamasurg.2022.0182. [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z, Shi L, Dong Y, Zhang Y, Yang F, Wei J, Huo M, Li P, Liu X. Effect of crosstalk among conspirators in tumor microenvironment on niche metastasis of gastric cancer. Am J Cancer Res. 2022;12:5375–5402. [PMC free article] [PubMed] [Google Scholar]
- 16.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 17.Ou Z, Lin S, Qiu J, Ding W, Ren P, Chen D, Wang J, Tong Y, Wu D, Chen A, et al. Single-nucleus RNA sequencing and spatial transcriptomics reveal the immunological microenvironment of cervical squamous cell carcinoma. Adv Sci (Weinh) 2022;9:e2203040. doi: 10.1002/advs.202203040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun D, Guan X, Moran AE, Wu LY, Qian DZ, Schedin P, Dai MS, Danilov AV, Alumkal JJ, Adey AC, et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat Biotechnol. 2022;40:527–538. doi: 10.1038/s41587-021-01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casado-Pelaez M, Bueno-Costa A, Esteller M. Single cell cancer epigenetics. Trends Cancer. 2022;8:820–838. doi: 10.1016/j.trecan.2022.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Hu W, Zeng H, Shi Y, Zhou C, Huang J, Jia L, Xu S, Feng X, Zeng Y, Xiong T, et al. Single-cell transcriptome and translatome dual-omics reveals potential mechanisms of human oocyte maturation. Nat Commun. 2022;13:5114. doi: 10.1038/s41467-022-32791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye J, Yang C, Xia L, Zhu Y, Liu L, Cao H, Tao Y. Protoplast preparation for algal single-cell omics sequencing. Microorganisms. 2023;11:538. doi: 10.3390/microorganisms11020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu T, Hu X, Wang M, Wang J, Zou B, Tan P, Cui T, Dou Y, Ning L, et al. CellCall: Integrating paired ligand-receptor and transcription factor activities for cell-cell communication. Nucleic Acids Res. 2021;49:8520–8534. doi: 10.1093/nar/gkab638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashima Y, Sakamoto Y, Kaneko K, Seki M, Suzuki Y, Suzuki A. Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med. 2020;52:1419–1427. doi: 10.1038/s12276-020-00499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Wang J, Zhao Y, Wang H, Liu T, Li Y, Cui T, Li W, Feng Y, Luo J, et al. cncRNAdb: A manually curated resource of experimentally supported RNAs with both protein-coding and noncoding function. Nucleic Acids Res. 2021;49:D65–D70. doi: 10.1093/nar/gkaa791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YF, Wang YH, Gu ZF, Pan XR, Li J, Ding H, Zhang Y, Deng KJ. Bitter-RF: A random forest machine model for recognizing bitter peptides. Front Med (Lausanne) 2023;10:1052923. doi: 10.3389/fmed.2023.1052923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Z, Kan C, Sun M, Yang F, Wong M, Wang S, Zheng H. Mapping breast cancer microenvironment through single-cell omics. Front Immunol. 2022;13:868813. doi: 10.3389/fimmu.2022.868813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao B, Jiang B, Xing W, Xie Z, Luo Z, Zou W. Discovery and application of postnatal nucleus pulposus progenitors essential for intervertebral disc homeostasis and degeneration. Adv Sci (Weinh) 2022;9:e2104888. doi: 10.1002/advs.202104888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moline DC, Zenner ML, Burr A, Vellky JE, Nonn L, Vander Griend DJ. Single-cell RNA-Seq identifies factors necessary for the regenerative phenotype of prostate luminal epithelial progenitors. Am J Clin Exp Urol. 2022;10:425–439. [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, An G, Wang H, Wu X, Ping P, Hu L, Chen Y, Fan J, Cheng CY, Sun F. Human obstructive (postvasectomy) and nonobstructive azoospermia-Insights from scRNA-Seq and transcriptome analysis. Genes Dis. 2022;9:766–776. doi: 10.1016/j.gendis.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanemoto S, Sujino T, Miyamoto K, Moody J, Yoshimatsu Y, Ando Y, Koya I, Harada Y, Tojo AO, Ono K, et al. Single-cell transcriptomics of human gut T cells identifies cytotoxic CD4+CD8A+ T cells related to mouse CD4 cytotoxic T cells. Front Immunol. 2022;13:977117. doi: 10.3389/fimmu.2022.977117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning L, Abagna HB, Jiang Q, Liu S, Huang J. Development and application of therapeutic antibodies against COVID-19. Int J Biol Sci. 2021;17:1486–1496. doi: 10.7150/ijbs.59149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ning L, Liu M, Gou Y, Yang Y, He B, Huang J. Development and application of ribonucleic acid therapy strategies against COVID-19. Int J Biol Sci. 2022;18:5070–5085. doi: 10.7150/ijbs.72706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Pan X, Shi T, Gu Z, Yang Z, Liu M, Xu Y, Yang Y, Ren L, Song X, et al. P450Rdb: A manually curated database of reactions catalyzed by cytochrome P450 enzymes. J Adv Res. 2023 Oct 21; doi: 10.1016/j.jare.2023.10.012. doi: 10.1016/j.jare.2023.10.012 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 34.Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14:68. doi: 10.1186/s13073-022-01075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson AC, Yanai I, Yates LR, Wang L, Swarbrick A, Sorger P, Santagata S, Fridman WH, Gao Q, Jerby L, et al. Spatial transcriptomics. Cancer Cell. 2022;40:895–900. doi: 10.1016/j.ccell.2022.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Chen D, Song D, Liu X, Zhang Y, Xu X, Wang X. Clinical and translational values of spatial transcriptomics. Signal Transduct Target Ther. 2022;7:111. doi: 10.1038/s41392-022-00960-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsson L, Bergenstråhle L, He M, Andrusivova Z, Lundeberg J. SnapShot: Spatial transcriptomics. Cell. 2022;185:2840–2840.e1. doi: 10.1016/j.cell.2022.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu T, Wang J, Zou B, Li L, Yao L, Chen K, Ning L, Wu B, Zhao X, Wang D. Cellinker: A platform of ligand-receptor interactions for intercellular communication analysis. Bioinformatics: btab036. 2021 doi: 10.1093/bioinformatics/btab036. doi: 10.1093/bioinformatics/btab036 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren L, Ning L, Yang Y, Yang T, Li X, Tan S, Ge P, Li S, Luo N, Tao P, Zhang Y. MetaboliteCOVID: A manually curated database of metabolite markers for COVID-19. Comput Biol Med. 2023;167:107661. doi: 10.1016/j.compbiomed.2023.107661. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed R, Zaman T, Chowdhury F, Mraiche F, Tariq M, Ahmad IS, Hasan A. Single-Cell RNA sequencing with spatial transcriptomics of cancer tissues. Int J Mol Sci. 2022;23:3042. doi: 10.3390/ijms23063042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longo SK, Guo MG, Ji AL, Khavari PA. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 2021;22:627–644. doi: 10.1038/s41576-021-00370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kijima Y, Evans-Yamamoto D, Toyoshima H, Yachie N. A universal sequencing read interpreter. Sci Adv. 2023;9:eadd2793. doi: 10.1126/sciadv.add2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren L, Xu Y, Ning L, Pan X, Li Y, Zhao Q, Pang B, Huang J, Deng K, Zhang Y. TCM2COVID: A resource of anti-COVID-19 traditional Chinese medicine with effects and mechanisms. Imeta. 2022:e42. doi: 10.1002/imt2.42. doi: 10.1002/imt2.42 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Liu C, Liu M, Liu T, Lin H, Huang CB, Ning L. Attention is all you need: Utilizing attention in AI-enabled drug discovery. Brief Bioinform. 2023;25:bbad467. doi: 10.1093/bib/bbad467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren L, Pan X, Ning L, Gong D, Huang J, Deng K, Xie L, Zhang Y. Construction of a combined hypoxia-related genes model for hepatocellular carcinoma prognosis. Curr Comput Aided Drug Des. 2023;19:150–161. doi: 10.2174/1573409919666221223123610. [DOI] [PubMed] [Google Scholar]
- 46.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Z, Li J, Huang P, Zhang Y, Yang J, Liu K, Jiang Y. Applications and achievements of single-cell sequencing in gastrointestinal cancer. Front Oncol. 2022;12:905571. doi: 10.3389/fonc.2022.905571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picelli S, Björklund Å K, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 49.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang L, Yu J, Li J, Li N, Liu J, Xiu L, Zeng J, Wang T, Wu L. Integration of scRNA-Seq and bulk RNA-Seq to analyse the heterogeneity of ovarian cancer immune cells and establish a molecular risk model. Front Oncol. 2021;11:711020. doi: 10.3389/fonc.2021.711020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, II, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 59.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gierahn TM, Wadsworth MH, II, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. Seq-Well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keren-Shaul H, Kenigsberg E, Jaitin DA, David E, Paul F, Tanay A, Amit I. MARS-seq2.0: An experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat Protoc. 2019;14:1841–1862. doi: 10.1038/s41596-019-0164-4. [DOI] [PubMed] [Google Scholar]
- 63.Kouno T, Moody J, Kwon AT, Shibayama Y, Kato S, Huang Y, Böttcher M, Motakis E, Mendez M, Severin J, et al. C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution. Nat Commun. 2019;10:360. doi: 10.1038/s41467-018-08126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyu T, Lin Y, Wu K, Cao Z, Zhang Q, Zheng J. Single-cell sequencing technologies in bladder cancer research: Applications and challenges. Front Genet. 2022;13:1027909. doi: 10.3389/fgene.2022.1027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, Xie XS. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI) Science. 2017;356:189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xing D, Tan L, Chang CH, Li H, Xie XS. Accurate SNV detection in single cells by transposon-based whole-genome amplification of complementary strands. Proc Natl Acad Sci USA. 2021;118:e2013106118. doi: 10.1073/pnas.2013106118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weichenhan D, Lipka DB, Lutsik P, Goyal A, Plass C. Epigenomic technologies for precision oncology. Semin Cancer Biol. 2022;84:60–68. doi: 10.1016/j.semcancer.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. Single-cell epigenomics: Powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016;17:72. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo H, Zhu P, Wu X, Li X, Wen L, Tang F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 73.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tajik M, Baharfar M, Donald WA. Single-cell mass spectrometry. Trends Biotechnol. 2022;40:1374–1392. doi: 10.1016/j.tibtech.2022.04.004. [DOI] [PubMed] [Google Scholar]
- 75.Iyer A, Hamers AAJ, Pillai AB. CyTOF(®) for the Masses. Front Immunol. 2022;13:815828. doi: 10.3389/fimmu.2022.815828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Budnik B, Levy E, Harmange G, Slavov N. SCoPE-MS: Mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:161. doi: 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Specht H, Emmott E, Petelski AA, Huffman RG, Perlman DH, Serra M, Kharchenko P, Koller A, Slavov N. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 2021;22:50. doi: 10.1186/s13059-021-02267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bock C, Datlinger P, Chardon F, Coelho MA, Dong MB, Lawson KA, Lu T, Maroc L, Norman TM, Song B, et al. High-content CRISPR screening. Nat Rev Methods Primers. 2022;2:9. doi: 10.1038/s43586-021-00093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. A multiplexed Single-Cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. Dissecting immune circuits by linking CRISPR-Pooled screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 81.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol Cell. 2017;66:285–299.e5. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 83.Replogle JM, Norman TM, Xu A, Hussmann JA, Chen J, Cogan JZ, Meer EJ, Terry JM, Riordan DP, Srinivas N, et al. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat Biotechnol. 2020;38:954–961. doi: 10.1038/s41587-020-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jun S, Lim H, Chun H, Lee JH, Bang D. Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Commu Biol. 2020;3:154. doi: 10.1038/s42003-020-0888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roth TL, Li PJ, Blaeschke F, Nies JF, Apathy R, Mowery C, Yu R, Nguyen MLT, Lee Y, Truong A, et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell. 2020;181:728–744.e21. doi: 10.1016/j.cell.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu X, Wen L, Tang F, Huang Y, Peng J. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood S, Ponting CP, Voet T, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deng G, Zhang X, Chen Y, Liang S, Liu S, Yu Z, Lü M. Single-cell transcriptome sequencing reveals heterogeneity of gastric cancer: Progress and prospects. Front Oncol. 2023;13:1074268. doi: 10.3389/fonc.2023.1074268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M, Yasuda T, Koh V, Huang KK, Tay ST, et al. Single-Cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12:670–691. doi: 10.1158/2159-8290.CD-21-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kang B, Camps J, Fan B, Jiang H, Ibrahim MM, Hu X, Qin S, Kirchhoff D, Chiang DY, Wang S, et al. Parallel single-cell and bulk transcriptome analyses reveal key features of the gastric tumor microenvironment. Genome Biol. 2022;23:265. doi: 10.1186/s13059-022-02828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zulfiqar H, Guo Z, Ahmad RM, Ahmed Z, Cai P, Chen X, Zhang Y, Lin H, Shi Z. Deep-STP: A deep learning-based approach to predict snake toxin proteins by using word embeddings. Front Med (Lausanne) 2024;10:1291352. doi: 10.3389/fmed.2023.1291352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andor N, Lau BT, Catalanotti C, Sathe A, Kubit M, Chen J, Blaj C, Cherry A, Bangs CD, Grimes SM, et al. Joint single cell DNA-seq and RNA-seq of gastric cancer cell lines reveals rules of in vitro evolution. NAR Genom Bioinform. 2020;2:lqaa016. doi: 10.1093/nargab/lqaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng L, Xing R, Liu D, Bao L, Cheng W, Wang H, Yu Y, Liu X, Jiang L, Wu Y, et al. Characterization and validation of somatic mutation spectrum to reveal heterogeneity in gastric cancer by single cell sequencing. Sci Bull (Beijing) 2019;64:236–244. doi: 10.1016/j.scib.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 95.Chen Y, Li Y, Qi C, Zhang C, Liu D, Deng Y, Fu Y, Khadka VS, Wang DD, Tan S, et al. Dysregulated KRAS gene-signaling axis and abnormal chromatin remodeling drive therapeutic resistance in heterogeneous-sized circulating tumor cells in gastric cancer patients. Cancer Lett. 2021;517:78–87. doi: 10.1016/j.canlet.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 96.Li Y, Zhang X, Ge S, Gong J, Lu M, Zhang Q, Cao Y, Wang DD, Lin PP, Shen L. Clinical significance of phenotyping and karyotyping of circulating tumor cells in patients with advanced gastric cancer. Oncotarget. 2014;5:6594–6602. doi: 10.18632/oncotarget.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y, Zhang X, Liu D, Gong J, Wang DD, Li S, Peng Z, Li Y, Wang X, Lin PP, et al. Evolutionary expression of HER2 conferred by chromosome aneuploidy on circulating gastric cancer cells contributes to developing targeted and chemotherapeutic resistance. Clin Cancer Res. 2018;24:5261–5271. doi: 10.1158/1078-0432.CCR-18-1205. [DOI] [PubMed] [Google Scholar]
- 98.Wang B, Zhang Y, Qing T, Xing K, Li J, Zhen T, Zhu S, Zhan X. Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq. Sci Rep. 2021;11:1141. doi: 10.1038/s41598-020-80881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang M, Hu S, Min M, Ni Y, Lu Z, Sun X, Wu J, Liu B, Ying X, Liu Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut. 2021;70:464–475. doi: 10.1136/gutjnl-2019-320368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang R, Dang M, Harada K, Han G, Wang F, Pool Pizzi M, Zhao M, Tatlonghari G, Zhang S, Hao D, et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat Med. 2021;27:141–151. doi: 10.1038/s41591-020-1125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eum HH, Kwon M, Ryu D, Jo A, Chung W, Kim N, Hong Y, Son DS, Kim ST, Lee J, et al. Tumor-promoting macrophages prevail in malignant ascites of advanced gastric cancer. Exp Mol Med. 2020;52:1976–1988. doi: 10.1038/s12276-020-00538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Y, Hu X, Lin R, Zhou G, Zhao L, Zhao D, Zhang Y, Li W, Zhang Y, Ma P, et al. Single-cell landscape reveals active cell subtypes and their interaction in the tumor microenvironment of gastric cancer. Theranostics. 2022;12:3818–3833. doi: 10.7150/thno.71833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang Z, Wu C, Liu X, Lu S, You L, Guo F, Stalin A, Zhang J, Zhang F, Wu Z, et al. Single-Cell and bulk RNA sequencing reveal malignant epithelial cell heterogeneity and prognosis signatures in gastric carcinoma. Cells. 2022;11:2550. doi: 10.3390/cells11162550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang P, Yang M, Zhang Y, Xiao S, Lai X, Tan A, Du S, Li S. Dissecting the Single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 2019;27:1934–1947.e5. doi: 10.1016/j.celrep.2019.04.052. [DOI] [PubMed] [Google Scholar]
- 105.Sathe A, Grimes SM, Lau BT, Chen J, Suarez C, Huang RJ, Poultsides G, Ji HP. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin Cancer Res. 2020;26:2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fu K, Hui B, Wang Q, Lu C, Shi W, Zhang Z, Rong D, Zhang B, Tian Z, Tang W, et al. Single-cell RNA sequencing of immune cells in gastric cancer patients. Aging (Albany NY) 2020;12:2747–2763. doi: 10.18632/aging.102774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang XZ, Pang MJ, Li JY, Chen HY, Sun JX, Song YX, Ni HJ, Ye SY, Bai S, Li TH, et al. Single-cell sequencing of ascites fluid illustrates heterogeneity and therapy-induced evolution during gastric cancer peritoneal metastasis. Nat Commun. 2023;14:822. doi: 10.1038/s41467-023-36310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sundar R, Huang KK, Kumar V, Ramnarayanan K, Demircioglu D, Her Z, Ong X, Bin Adam Isa ZF, Xing M, Tan AL, et al. Epigenetic promoter alterations in GI tumour immune-editing and resistance to immune checkpoint inhibition. Gut. 2022;71:1277–1288. doi: 10.1136/gutjnl-2021-324420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang W, Zhao Y, Ge Q, Wang X, Jing Y, Zhao J, Liu G, Huang H, Cheng F, Wang X, et al. Genetic mutation and tumor microbiota determine heterogenicity of tumor immune signature: Evidence from gastric and colorectal synchronous cancers. Front Immunol. 2022;13:947080. doi: 10.3389/fimmu.2022.947080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li X, Sun Z, Peng G, Xiao Y, Guo J, Wu B, Li X, Zhou W, Li J, Li Z, et al. Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics. 2022;12:620–638. doi: 10.7150/thno.60540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim R, An M, Lee H, Mehta A, Heo YJ, Kim KM, Lee SY, Moon J, Kim ST, Min BH, et al. Early tumor-immune microenvironmental remodeling and response to first-line fluoropyrimidine and platinum chemotherapy in advanced gastric cancer. Cancer Discov. 2022;12:984–1001. doi: 10.1158/2159-8290.CD-21-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li S, Li K, Tian F, Li H, Xia Q, Li T, Dong B, Li D, Yu J, Zhang J, et al. A high interferon gamma signature of CD8+ T cells predicts response to neoadjuvant immunotherapy plus chemotherapy in gastric cancer. Front Immunol. 2022;13:1056144. doi: 10.3389/fimmu.2022.1056144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nagaoka K, Shirai M, Taniguchi K, Hosoi A, Sun C, Kobayashi Y, Maejima K, Fujita M, Nakagawa H, Nomura S, Kakimi K. Deep immunophenotyping at the single-cell level identifies a combination of anti-IL-17 and checkpoint blockade as an effective treatment in a preclinical model of data-guided personalized immunotherapy. J Immunother Cancer. 2020;8:e001358. doi: 10.1136/jitc-2020-001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu Q, Jiang M, Wu L. Spatial transcriptomics technology in cancer research. Front Oncol. 2022;12:1019111. doi: 10.3389/fonc.2022.1019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 117.Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays. 2020;42:e1900221. doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 118.Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, Cai L. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xia C, Fan J, Emanuel G, Hao J, Zhuang X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci USA. 2019;116:19490–19499. doi: 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang G, Moffitt JR, Zhuang X. Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Sci Rep. 2018;8:4847. doi: 10.1038/s41598-018-22297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu C, Simonetti M, Rossell C, Mignardi M, Mirzazadeh R, Annaratone L, Marchiò C, Sapino A, Bienko M, Crosetto N, Nilsson M. RollFISH achieves robust quantification of single-molecule RNA biomarkers in paraffin-embedded tumor tissue samples. Commun Biol. 2018;1:209. doi: 10.1038/s42003-018-0218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dar D, Dar N, Cai L, Newman DK. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science. 2021;373:eabi4882. doi: 10.1126/science.abi4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goh JJL, Chou N, Seow WY, Ha N, Cheng CPP, Chang YC, Zhao ZW, Chen KH. Highly specific multiplexed RNA imaging in tissues with split-FISH. Nat Methods. 2020;17:689–693. doi: 10.1038/s41592-020-0931-8. [DOI] [PubMed] [Google Scholar]
- 125.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alon S, Goodwin DR, Sinha A, Wassie AT, Chen F, Daugharthy ER, Bando Y, Kajita A, Xue AG, Marrett K, et al. Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science. 2021;371:eaax2656. doi: 10.1126/science.aax2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen X, Sun YC, Church GM, Lee JH, Zador AM. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 2018;46:e22. doi: 10.1093/nar/gkx1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361:eaat5691. doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y, Liu B, Zhao G, Lee Y, Buzdin A, Mu X, Zhao J, Chen H, Li X. Spatial transcriptomics: Technologies, applications and experimental considerations. Genomics. 2023;115:110671. doi: 10.1016/j.ygeno.2023.110671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Elhanani O, Ben-Uri R, Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell. 2023;41:404–420. doi: 10.1016/j.ccell.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 131.Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q, Hedlund E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun. 2016;7:12139. doi: 10.1038/ncomms12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li H, Iannacone M, Shulman Z, Amit I. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science. 2017;358:1622–1626. doi: 10.1126/science.aao4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159:662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 134.Chen J, Suo S, Tam PP, Han JJ, Peng G, Jing N. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 135.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 136.Nichterwitz S, Benitez JA, Hoogstraaten R, Deng Q, Hedlund E. LCM-Seq: A method for spatial transcriptomic profiling using laser capture microdissection coupled with PolyA-Based RNA sequencing. Methods Mol Biol. 2018;1649:95–110. doi: 10.1007/978-1-4939-7213-5_6. [DOI] [PubMed] [Google Scholar]
- 137.Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, Hoang M, Jung J, Liang Y, McKay-Fleisch J, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38:586–599. doi: 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- 138.Vickovic S, Eraslan G, Salmén F, Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 140.Chen A, Liao S, Cheng M, Ma K, Wu L, Lai Y, Qiu X, Yang J, Xu J, Hao S, et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 2022;185:1777–1792.e1721. doi: 10.1016/j.cell.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 141.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cho CS, Xi J, Si Y, Park SR, Hsu JE, Kim M, Jun G, Kang HM, Lee JH. Microscopic examination of spatial transcriptome using Seq-scope. Cell. 2021;184:3559–3572.e22. doi: 10.1016/j.cell.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell. 2019;178:473–490.e26. doi: 10.1016/j.cell.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, Thennavan A, Wang C, Torpy JR, Bartonicek N, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, Guo MG, George BM, Mollbrink A, Bergenstråhle J, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:497–514.e22. doi: 10.1016/j.cell.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 147.Saviano A, Henderson NC, Baumert TF. Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J Hepatol. 2020;73:1219–1230. doi: 10.1016/j.jhep.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sharma A, Seow JJW, Dutertre CA, Pai R, Blériot C, Mishra A, Wong RMM, Singh GSN, Sudhagar S, Khalilnezhad S, et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–394.e21. doi: 10.1016/j.cell.2020.08.040. [DOI] [PubMed] [Google Scholar]
- 149.Massalha H, Bahar Halpern K, Abu-Gazala S, Jana T, Massasa EE, Moor AE, Buchauer L, Rozenberg M, Pikarsky E, Amit I, et al. A single cell atlas of the human liver tumor microenvironment. Mol Syst Biol. 2020;16:e9682. doi: 10.15252/msb.20209682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ben-Moshe S, Shapira Y, Moor AE, Manco R, Veg T, Bahar Halpern K, Itzkovitz S. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab. 2019;1:899–911. doi: 10.1038/s42255-019-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, Li J, Li F, Tan HB. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 152.Andersson A, Larsson L, Stenbeck L, Salmén F, Ehinger A, Wu SZ, Al-Eryani G, Roden D, Swarbrick A, Borg Å, et al. Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat Commun. 2021;12:6012. doi: 10.1038/s41467-021-26271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nerurkar SN, Goh D, Cheung CCL, Nga PQY, Lim JCT, Yeong JPS. Transcriptional Spatial Profiling of Cancer Tissues in the Era of Immunotherapy: The potential and promise. Cancers (Basel) 2020;12:2572. doi: 10.3390/cancers12092572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grauel AL, Nguyen B, Ruddy D, Laszewski T, Schwartz S, Chang J, Chen J, Piquet M, Pelletier M, Yan Z, et al. TGFβ-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nat Commun. 2020;11:6315. doi: 10.1038/s41467-020-19920-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, Hajdu CH, Simeone DM, Yanai I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 156.Svedlund J, Strell C, Qian X, Zilkens KJC, Tobin NP, Bergh J, Sieuwerts AM, Nilsson M. Generation of in situ sequencing based OncoMaps to spatially resolve gene expression profiles of diagnostic and prognostic markers in breast cancer. EBioMedicine. 2019;48:212–223. doi: 10.1016/j.ebiom.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, Cheng Y, Huang S, Liu Y, Jiang S, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at Single-cell level. Cancer Discov. 2022;12:134–153. doi: 10.1158/2159-8290.CD-21-0316. [DOI] [PubMed] [Google Scholar]
- 158.Jeong HY, Ham IH, Lee SH, Ryu D, Son SY, Han SU, Kim TM, Hur H. Spatially distinct reprogramming of the tumor microenvironment based on tumor invasion in Diffuse-type gastric cancers. Clin Cancer Res. 2021;27:6529–6542. doi: 10.1158/1078-0432.CCR-21-0792. [DOI] [PubMed] [Google Scholar]
- 159.Sundar R, Liu DH, Hutchins GG, Slaney HL, Silva AN, Oosting J, Hayden JD, Hewitt LC, Ng CC, Mangalvedhekar A, et al. Spatial profiling of gastric cancer patient-matched primary and locoregional metastases reveals principles of tumour dissemination. Gut. 2021;70:1823–1832. doi: 10.1136/gutjnl-2020-320805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yamasaki J, Hirata Y, Otsuki Y, Suina K, Saito Y, Masuda K, Okazaki S, Ishimoto T, Saya H, Nagano O. MEK inhibition suppresses metastatic progression of KRAS-mutated gastric cancer. Cancer Sci. 2022;113:916–925. doi: 10.1111/cas.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Grosser B, Glückstein MI, Dhillon C, Schiele S, Dintner S, VanSchoiack A, Kroeppler D, Martin B, Probst A, Vlasenko D, et al. Stroma AReactive invasion front areas (SARIFA)-a new prognostic biomarker in gastric cancer related to tumor-promoting adipocytes. J Pathol. 2022;256:71–82. doi: 10.1002/path.5810. [DOI] [PubMed] [Google Scholar]
- 162.Jia L, Wang T, Zhao Y, Zhang S, Ba T, Kuai X, Wang B, Zhang N, Zhao W, Yang Z, Qiao H. Single-cell profiling of infiltrating B cells and tertiary lymphoid structures in the TME of gastric adenocarcinomas. Oncoimmunology. 2021;10:1969767. doi: 10.1080/2162402X.2021.1969767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie W, Cheng J, Hong Z, Cai W, Zhuo H, Hou J, Lin L, Wei X, Wang K, Chen X, et al. Multi-transcriptomic analysis reveals the heterogeneity and tumor-promoting role of SPP1/CD44-mediated intratumoral crosstalk in gastric cancer. Cancers (Basel) 2022;15:164. doi: 10.3390/cancers15010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yang Y, Gao D, Xie X, Qin J, Li J, Lin H, Yan D, Deng K. DeepIDC: A prediction framework of injectable drug combination based on heterogeneous information and deep learning. Clin Pharmacokinet. 2022;61:1749–1759. doi: 10.1007/s40262-022-01180-9. [DOI] [PubMed] [Google Scholar]
- 165.Han YM, Yang H, Huang QL, Sun ZJ, Li ML, Zhang JB, Deng KJ, Chen S, Lin H. Risk prediction of diabetes and pre-diabetes based on physical examination data. Math Biosci Eng. 2022;19:3597–3608. doi: 10.3934/mbe.2022166. [DOI] [PubMed] [Google Scholar]
- 166.Chen J, Xu H, Tao W, Chen Z, Zhao Y, Han JDJ. Transformer for one stop interpretable cell type annotation. Nat Commun. 2023;14:223. doi: 10.1038/s41467-023-35923-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ma Q, Xu D. Deep learning shapes single-cell data analysis. Nat Rev Mol Cell Biol. 2022;23:303–304. doi: 10.1038/s41580-022-00466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yuan Z, Yao J. Harnessing computational spatial omics to explore the spatial biology intricacies. Semin Cancer Biol. 2023;95:25–41. doi: 10.1016/j.semcancer.2023.06.006. [DOI] [PubMed] [Google Scholar]
- 169.Liu J, Fan Z, Zhao W, Zhou X. Machine intelligence in Single-cell data analysis: Advances and new Challenges. Front Genet. 2021;12:655536. doi: 10.3389/fgene.2021.655536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Islam S, Kjällquist U, Moliner A, Zajac P, Fan JB, Lönnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 174.Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR. Quartz-Seq: A highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, Nikaido I. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun. 2018;9:619. doi: 10.1038/s41467-018-02866-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schönegger A, Klughammer J, Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Han L, Wu HJ, Zhu H, Kim KY, Marjani SL, Riester M, Euskirchen G, Zi X, Yang J, Han J, et al. Bisulfite-independent analysis of CpG island methylation enables genome-scale stratification of single cells. Nucleic Acids Res. 2017;45:e77. doi: 10.1093/nar/gkx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Litzenburger UM, Buenrostro JD, Wu B, Shen Y, Sheffield NC, Kathiria A, Greenleaf WJ, Chang HY. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol. 2017;18:15. doi: 10.1186/s13059-016-1133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mooijman D, Dey SS, Boisset JC, Crosetto N, van Oudenaarden A. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol. 2016;34:852–856. doi: 10.1038/nbt.3598. [DOI] [PubMed] [Google Scholar]
- 182.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 185.Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 186.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rooijers K, Markodimitraki CM, Rang FJ, de Vries SS, Chialastri A, de Luca KL, Mooijman D, Dey SS, Kind J. Simultaneous quantification of protein-DNA contacts and transcriptomes in single cells. Nat Biotechnol. 2019;37:766–772. doi: 10.1038/s41587-019-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Satpathy AT, Saligrama N, Buenrostro JD, Wei Y, Wu B, Rubin AJ, Granja JM, Lareau CA, Li R, Qi Y, et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat Med. 2018;24:580–590. doi: 10.1038/s41591-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen S, Lake BB, Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol. 2019;37:1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu L, Liu C, Quintero A, Wu L, Yuan Y, Wang M, Cheng M, Leng L, Xu L, Dong G, et al. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat Commun. 2019;10:470. doi: 10.1038/s41467-018-08205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Meyer AR, Engevik AC, Madorsky T, Belmont E, Stier MT, Norlander AE, Pilkinton MA, McDonnell WJ, Weis JA, Jang B, et al. Group 2 innate lymphoid cells coordinate damage response in the stomach. Gastroenterology. 2020;159:2077–2091.e8. doi: 10.1053/j.gastro.2020.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kwon M, An M, Klempner SJ, Lee H, Kim KM, Sa JK, Cho HJ, Hong JY, Lee T, Min YW, et al. Determinants of response and intrinsic resistance to PD-1 blockade in microsatellite instability-high gastric cancer. Cancer Discov. 2021;11:2168–2185. doi: 10.1158/2159-8290.CD-21-0219. [DOI] [PubMed] [Google Scholar]
- 193.Zhou X, Yang J, Lu Y, Ma Y, Meng Y, Li Q, Gao J, Jiang Z, Guo L, Wang W, et al. Relationships of tumor differentiation and immune infiltration in gastric cancers revealed by single-cell RNA-seq analyses. Cell Mol Life Sci. 2023;80:57. doi: 10.1007/s00018-023-04702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jiang H, Yu D, Yang P, Guo R, Kong M, Gao Y, Yu X, Lu X, Fan X. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell RNA Sequencing. Clin Transl Med. 2022;12:e730. doi: 10.1002/ctm2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Honda M, Oki S, Kimura R, Harada A, Maehara K, Tanaka K, Meno C, Ohkawa Y. High-depth spatial transcriptome analysis by photo-isolation chemistry. Nat Commun. 2021;12:4416. doi: 10.1038/s41467-021-24691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kishi JY, Lapan SW, Beliveau BJ, West ER, Zhu A, Sasaki HM, Saka SK, Wang Y, Cepko CL, Yin P. SABER amplifies FISH: Enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods. 2019;16:533–544. doi: 10.1038/s41592-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.