Abstract
Traumatic brain injury (TBI) is a leading cause of death and disability in the US. Neural stem/progenitor cells (NSPCs) persist in the adult brain and represent a potential cell source for tissue regeneration and wound healing after injury. The Notch signaling pathway is critical for embryonic development and adult brain injury response. However, the specific role of Notch signaling in the injured brain is not well characterized. Our previous study has established a Notch1CR2-GFP reporter mouse line in which the Notch1CR2 enhancer directs GFP expression in NSPCs and their progeny. In this study, we performed closed head injury (CHI) in the Notch1CR2-GFP mice to study the response of injury-activated NSPCs. We show that CHI induces neuroinflammation, cell death, and the expression of typical TBI markers (e.g., ApoE, Il1b, and Tau), validating the animal model. In addition, CHI induces cell proliferation in GFP+ cells expressing NSPC markers, e.g., Notch1 and Nestin. A significant higher percentage of GFP+ astrocytes and GABAergic neurons was observed in the injured brain, with no significant change in oligodendrocyte lineage between the CHI and sham animal groups. Since injury is known to activate astrogliosis, our results suggest that injury-induced GFP+ NSPCs preferentially differentiate into GABAergic neurons. Our study establishes that Notch1CR2-GFP transgenic mouse is a useful tool for the study of NSPC behavior in vivo after TBI. Unveiling the potential of NSPCs response to TBI (e.g., proliferation and differentiation) will identify new therapeutic strategy for the treatment of brain trauma.
Keywords: Traumatic brain injury, Closed head injury, Neural stem/progenitor cell, Notch1CR2-GFP, Mouse model, GABAergic neuron
1. Introduction
Human traumatic brain injury (TBI) is the leading cause of death and disability in children and young adults in the United States. TBI results in temporary or permanent neurological damage including loss of memory, cognitive function, and motor function. Currently, there is no effective treatment for TBI since little can be done to reverse the tissue damage caused by trauma (Hasan et al., 2017). It is estimated that 5.3 million individuals in the United States are living with disabilities from TBI. TBI is responsible for approximately 282,000 emergency room visits and 56,000 deaths annually (Gardner et al., 2017). TBI is defined as a blow to the head that disrupts brain function and results in temporary or permanent neurological damage including loss of memory, cognitive function, and motor function (Chen et al., 2017). TBI causes cell death from the direct mechanical injury to the head (primary injury) followed by additional cell death from inflammation and swelling (secondary injury). Although the secondary injury is often reduced with anti-inflammatory medicines, cell death from the primary injury is not recoverable.
Several animal models have been established for studying TBI (Ma et al., 2019; Phipps, 2016), including the closed head injury by weight drop (CHI) (Flierl et al., 2009), lateral fluid percussion (LFP) (Van and Lyeth, 2016), a controlled cortical impact (CCI) or impact acceleration models (Campolo et al., 2018). Although no model can fully represent the spectrum of human TBI, the CHI model mimics the majority of clinical cases.
Neural stem/progenitor cells (NSPCs) are multipotent, making them a useful cell source to repair damaged and lost cells after injury (Dixon et al., 2015; Encinas and Fitzsimons, 2017; Ludwig et al., 2018; Weston and Sun, 2018). In the adult brain, endogenous NSPCs are present in the subgranular zone (SGZ) of the hippocampus and the subventricular zone (SVZ) of the lateral ventricles (Encinas and Fitzsimons, 2017; Ming and Song, 2011; Tu et al., 2017). Injury induces neurogenesis in the SVZ of the cerebral cortex (Bohrer and Schachtrup, 2016; Ludwig et al., 2018; Yan et al., 2018) by stimulating NSPC proliferation and differentiation (Wang et al., 2016). NSPCs in the adult SGZ and SVZ have shown to proliferate and differentiate into various mature neural lineages, including astrocytes, oligodendrocytes, and various neuronal cell lineages (Lim and Alvarez-Buylla, 2016). However, the composition of specific cell types derived from injury-induced NSPCs has not been clearly determined (Patel and Sun, 2016; Reis et al., 2017; Zhao et al., 2008).
Several reporter mouse lines (e.g., Nestin-GFP, Pax6-GFP, Sox1-GFP, and Sox2-GFP) have been established to characterize NSPCs in the development of the CNS (Mignone et al. 2004, Barraud et al. 2005, Arnold et al. 2011, Gao et al. 2018). Adding to this pool of NSPC report mouse lines, we have previously established a Notch1CR2-GFP mouse line and characterized GFP+ cells as NSPCs in the developing brain (Tzatzalos et al., 2012) and spinal cord (Li et al., 2016). Notch signaling is highly involved in the development of the central nervous system (CNS) (Louvi and Artavanis-Tsakonas, 2006; Tzatzalos et al., 2012; Yoon and Gaiano, 2005). The evolutionarily conserved Notch signaling pathway is involved in stem cell regulation and differentiation not only during development, but also in the adult brain (Ables et al., 2011; Ables et al., 2010; Woo et al., 2009). Notch1 also regulates adult NSPC quiescence and proliferation, and overexpression of Notch1 has been implicated to promote neural proliferation and self-renewal (Ables et al., 2011; Chapouton et al., 2010; Imayoshi and Kageyama, 2011; Zhou et al., 2010). Notch1 deficiency decreases NSPC number and neurogenesis (Ables et al., 2010). Studies have shown that the astrogliogenic response of the SVZ to injury is accompanied by activation of the Notch signaling pathway (Benner et al., 2013; Carlen et al., 2009; LeComte et al., 2015). Notch signaling is also known to regulate adult endogenous NSPC proliferation and neurogenesis after brain injury (Chojnacki et al., 2003; Puhakka et al., 2017). Notch1 ligands, Jagged1 and Delta1, are co-expressed in the SVZ and SGZ of the injured brain with proliferative NSPCs (Tatsumi et al., 2010; Wang et al., 2009; Wang et al., 2012). In addition, γ-secretase inhibitor of Notch signaling disrupts the maintenance and proliferation of NSCPs (Chojnacki et al., 2003). Different subpopulations of NSPCs exist based on their markers (e.g., Sox2, Nestin, Pax6, and Notch1), neurogenic region, cell state (e.g., quiescent vs. activated), and propensity to differentiate into mature lineages (Artegiani et al., 2017; Dulken et al., 2017). Although NSPCs are heterogeneous, many current studies have either characterized NSPCs as a homogeneous population (e.g., Nestin+). The Notch signaling is a key regulator of NSPCs (Giachino and Taylor, 2014). In our previous established Notch1CR2-GFP reporter line, the Notch1 enhancer CR2 directs GFP expression mainly in interneuron progenitors (Tzatzalos et al., 2012). Thus, this animal model allows the easy tracking of these GFP+ NSPCs in the developing and adult brains.
In this study, we establish a CHI model in Notch1CR2-GFP reporter mouse to examine the behavior of a unique subpopulation of Notch1+ NSPCs in response to TBI. The use of GFP reporter mouse provides the ease of identification and characterization of injury-activated NSPCs during the acute phase of TBI. During the chronic phase of TBI, GFP+ NSPC subpopulation preferentially differentiate into GABAergic interneurons in mice, indicating the preferential lineage potential of Notch1+ NSPCs for GABAergic neuronal differentiation after injury. Thus, our study establishes that Notch1CR2-GFP transgenic mouse is a useful tool for the study of NSPC behavior in vivo after TBI.
2. Materials and methods
All experiments were approved by the Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee at Rutgers University. All animal work was conducted in compliance with the NIH Guide for the Care and Use of Laboratory Animals.
2.1. Transgenic animal
Notch1CR2-GFP (N1CR2) transgenic mouse line was generated and maintained in our lab (Tzatzalos et al., 2012). Animals of 8–12 weeks of age and with equal number of males and females were used for the experiments.
2.2. CHI procedure
Mice 8–12 weeks of age were subjected to CHI using the protocol previously described (Flierl et al., 2009). Briefly, mice were anesthetized with 5% Isoflurane for 3–5 min (min) and kept under 3% Isoflurane for experimental procedure. The parietal bone was exposed by a midline scalp incision after shaving and cleaning the skin with 3 sets of betadine scrub, followed by 70% ethanol. A free-falling rod (327 g weight) with a blunt tip of 3.0 mm diameter dropped onto mouse skull 3.0 mm anterior to the lambda suture and 2.0 mm lateral to the middle line and the falling height was 3.0 cm over the skull to induce injury without cracking the skull. After surgery, mice were allowed to recover on a heating pad until fully awake.
2.3. Behavior
Mice were evaluated beginning at 1 h (hr) post-CHI using mNSS behavior tests by two blinded, trained observers (Flierl et al., 2009).
2.4. Tissue preparation
At designated time points after injury, animals were euthanized, and brains were removed for tissue processing. Brains were washed in 1× PBS, and fixed with 4% (w/v) PFA for 1 day. Fixed brains were then washed four times with 1× PBS before being added to 30% (w/v) sucrose for 1–2 days. Next, the brain tissue was embedded in cryopreservation solution (Tissue Tek OCT compound) and frozen at −80 °C until cryosectioning.
2.5. Immunohistochemistry (IHC)
Frozen brain tissue was cryosectioned at 12 μm using a cryostat (Thermo Shandon Cryostat Cryotome) and air dried. Tissues sections were frozen at −80 °C until staining. Before staining, tissue sections were removed and stored at ambient room temperature for 30 min until beginning procedure. Sections were antigen retrieved with methanol for 10 min at room temperature, blocked and permeabilized for 1 h at room temperature in blocking buffer (10% donkey serum, 0.1% Triton-100, 0.1% Tween 20), then incubated with primary antibody overnight at 4 °C. The next day, samples were washed with 1× PBS, then incubated with corresponding fluorophore-conjugated secondary antibodies for 1 h at room temperature. Slides were washed with PBS, DAPI was added, then slides were dried before adding Cytoseal20 mounting media prior to fluorescent imaging. The primary antibodies used include: GFP (Abcam ab5450), Ki67 (Abcam ab15580), Nestin (Abcam ab6142), Sox2 (Millipore MAB4343), Caspase3 (Cell Signaling 661S), CD68 (Millipore mab1435), Notch1 (Abcam ab8925), DCX (Cell Signaling 4604S), NG2 (Millipore MAB5384), S100b (Abcam ab52642), Olig2 (Millipore AB9610), NeuN (Millipore MAB377), vGlut2 (Abcam ab79157), and GABA (Abcam ab8891).
2.6. Imaging and cell counting
Horizontal brain sections were imaged with a Zeiss LSM800 microscope for confocal imaging or a Leica DMi8 motorized fluorescence microscope with LED light source for tiled imaging to show a whole or large brain area. Cell counting was performed manually to quantify and compare fluorescence intensity of GFP and cell markers in CHI and sham brain sections. For each marker, 3–5 sections from at least 3 animals of each gender were counted at each time point. Since some markers were nuclear and others cytoplasmic, DAPI nuclei staining was necessary to confirm co-expression of markers. All images, under multiple conditions, were taken using the same optical parameters to avoid saturation. The images were acquired and analyzed blindly by a separate analyzer. Data was presented as dot plots, where each dot represents an animal with the average result of ≥3 quantified cryosections from that animal. Statistics was performed on each group and error bars are represented as Mean ± SEM. For statistical significance, students t-test was performed with * = P < .05 significance, ** = P < .01 significance, *** = P < .0005 significance, and **** = P < .0001.
2.7. Quantitative real-time PCR (qPCR)
For qPCR, total RNA was extracted from mouse SVZ, hippocampus, and cerebral cortex at specified time points with Tri Reagent. RNA was isolated, and cDNA was synthesized using qSCRIPT cDNA SuperMix. qPCR was performed using the Roche 480 LightCycler platform with SYGR Green FastMix and primers for genes of interest and GAPDH as a reference gene. Analysis was performed using the Livak method (Livak and Schmittgen, 2001).
3. Results
3.1. CHI induces substantial inflammation and cell death in the cerebral cortex and hippocampus
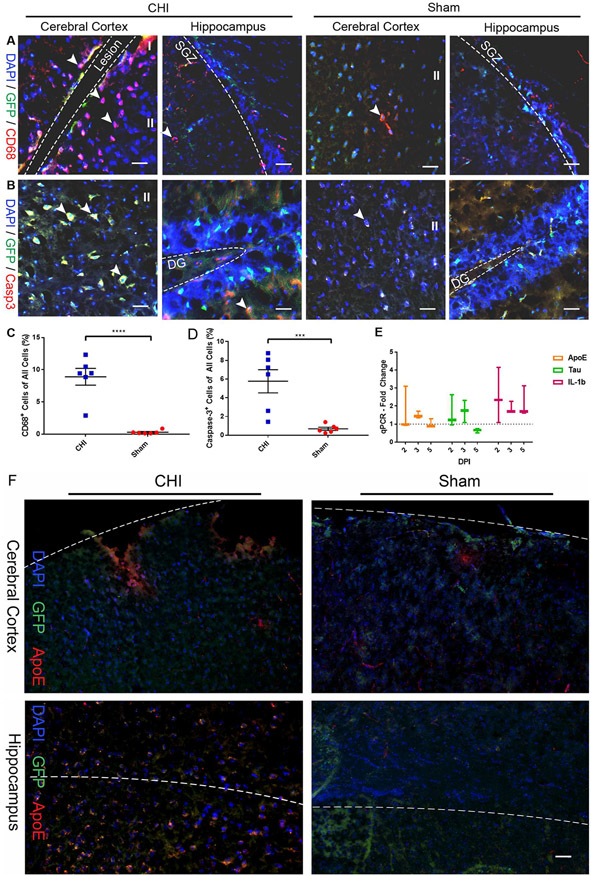
We performed a closed head injury (CHI) on Notch1CR2-GFP transgenic mice using a weight-drop device (Chen et al., 1996; Flierl et al., 2009) with a drop weight of 327 g at a height of 3.0 cm (Chen et al., 1996; Evanson et al., 2018; Khalin et al., 2016; Leinhase et al., 2006; Sun et al., 2018) to determine the effects of TBI on inflammation, cell death, and NSPC activation and differentiation after injury (Fig. 1). It has been established that TBI induces substantial neuroinflammation and cell death (Hsieh et al., 2013; Schwab et al., 2002; Stoica and Faden, 2010; Turtzo et al., 2014). Neuroinflammation involves the activation of glia (e.g., microglia and astrocytes) to release inflammatory mediators within the brain, and the subsequent recruitment of peripheral immune cells (Chiu et al., 2016; Schwab et al., 2002; Stoica and Faden, 2010). An increase in the number of macrophages and cell death has been observed in the cerebral cortex near the injury site after TBI (Ray et al., 2002). We thus examined neuroinflammation and cell death to confirm the extent of injury and validate the cellular response in the cerebral cortex and hippocampus by immunohistochemistry (IHC) analysis. Marker gene expression for neuroinflammation (CD68, Fig. 2A) and cell death (Caspase-3 or Casp3, Fig. 2B) was observed in injured and sham brain tissues at 2 days post injury (DPI). The increased number of CD68+ cells were found mostly in the ipsilateral hemisphere of the injured brain compared to the contralateral hemisphere of the brain. In the injured brain tissues, a majority of CD68+ cells were found in the ipsilateral cerebral cortex of the CHI brain at or near the site of impact compared to the sham brain (Fig. 2A). A majority of Casp3+ cell death was found in the ipsilateral hemisphere of the cerebral cortex and hippocampus of the CHI brain compared to the sham brain (Fig. 2B). Both GFP+ cells and non-GFP+ cells express cell death marker Casp3 near the injury site in both the cerebral cortex and hippocampus (Fig. 2A) indicating that the injury is substantial. The increase in the number of cells with inflammation marker CD68 (8.91 ± 1.30% in CHI (n = 6) vs. 0.30 ± 0.12% in sham (n = 6); p < .0001; Fig. 2C) and cell death marker Casp3 (5.77 ± 1.24% in CHI (n = 6) vs. 0.68 ± 0.17% in sham (n = 6); p < .0005; Fig. 2D) was significant in injury brain tissues at 2 DPI. Antibody staining confirms that there was an increase of markers for cell death and inflammation after CHI compared to the sham animals. The minimal expression of these markers in the sham animals confirms that the surgical procedure did not induce injury in the brain. To further confirm the CHI model, we performed qPCR analysis on TBI and inflammation markers, e.g., ApoE, Tau, and IL-1b, in the injured brain. When compared to sham animals, we found that there was an increase for TBI markers ApoE (1.67 ± 0.71 fold increase at 2 DPI) and Tau (1.72 ± 0.36 fold increase at 3 DPI) after injury (Fig. 2E). The expression level of Tau was similar to a previous study in brain tissue (Pluta et al., 2018). Additionally, we also observed an increase of inflammation marker IL-1b (2.52 ± 0.89 fold increase at 2 DPI) after injury (Fig. 2E). The expression level of these inflammation markers are consistent to the previous published data (Newell et al., 2018); (Kumar et al., 2016; Ma et al., 2017). In addition to Casp3, we verified the expression of TBI marker ApoE using immunohistochemistry analysis on 14 DPI samples. A higher level of ApoE signal was observed in the CHI brain compared to the sham brain (Fig. 2F). Thus, our results confirm that the CHI model induces substantial cell death and neuroinflammation.
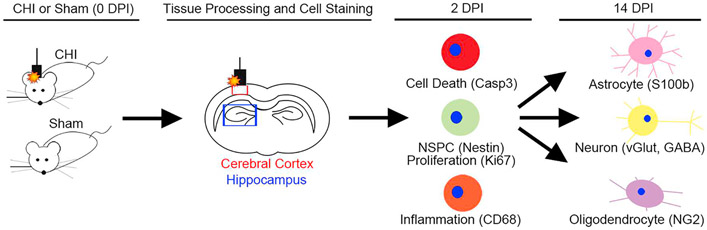
Fig. 1. Experimental Design.
Closed head injury (CHI) was induced by weight-drop impact in Notch1CR2-GFP mice after scalp skin opening. Sham mice did not receive weigh-drop impact injury but received an incision on the scalp to expose the skull. At 2 days post injury (DPI) and 14 DPI, brain tissue was harvested and cryosectioned in coronal orientation. At 2 dpi, markers for cell death, inflammation, cell proliferation, and TBI were examined by IHC and qPCR analysis. At 14 dpi, neural lineage markers were analyzed to determine the cell fate for injury-induced GFP+ cells. Imaging analysis was performed in both the cerebral cortex (red box) at the injury site and hippocampus (blue box) beneath the injured cerebral cortex.
Fig. 2. CHI induces substantial inflammation and cell death in the cerebral cortex and hippocampus.
Injury-induced inflammation and cell death in the cerebral cortex and hippocampus were determined by immunostaining and qPCR. Cells expressing macrophage marker CD68 (A) and cell death marker Caspase-3 (Casp3) (B) were detected in coronal sections at 2 days post injury (DPI). Quantification shows a significant increase in the number of CD68+ macrophages (p < .0001) (C) and Caspase-3+ cells (p < .0005) (D) in the cerebral cortex of injured animals as compared to the sham animals. Mean ± SEM, n = 6. (E) qPCR analysis shows fold-change increase in markers of TBI (ApoE, Tau) and inflammation (IL-1b) at 2, 3, and 5 DPI compared to sham animals. (F) Representative photomicrographs of TBI marker ApoE staining. Scale bars = 30 μm.
3.2. CHI increases the number of GFP+ cells in the cerebral cortex and hippocampus
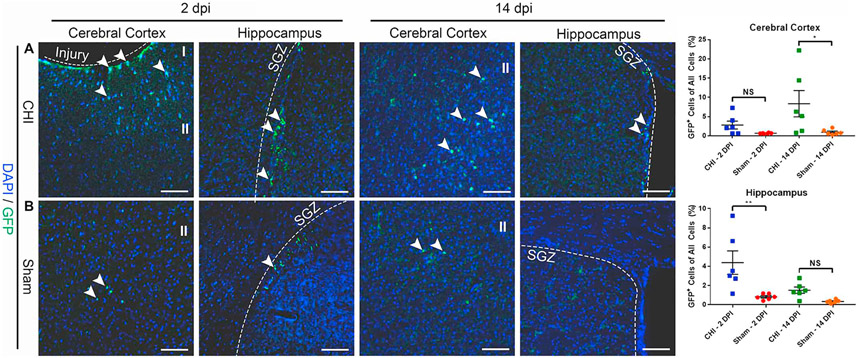
With confirmation of the CHI model by markers of cell death, inflammation, and TBI, we next performed cellular and molecular characterization of GFP+ cell behavior after injury. Our previous study has shown that GFP+ cells label interneuron progenitor cells in Notch1CR2-GFP animals (Tzatzalos et al., 2012). It has been also shown that injury induces NSPCs in the SGZ of the hippocampus and the SVZ in the cerebral cortex (Bohrer and Schachtrup, 2016; Ludwig et al., 2018; Yan et al., 2018). To identify brain regions with GFP+ cells in the Notch1CR2-GFP transgenic mice, we performed IHC in the ipsilateral cerebral cortex and hippocampus. We compared the CHI (n = 6, Fig. 3A) and sham (n = 6, Fig. 3B) mice after CHI. A majority of GFP+ cells were located in the ipsilateral cerebral cortex and hippocampus, with an increased GFP+ cell expression after injury. At 2 DPI, there was an increased number of GFP+ cells at cerebral cortex (injury site) and the SGZ, with a significant increase of GFP+ cells located in the SGZ (p < .01), a known region of NSPCs in the adult brain. By 14 DPI, there was still increased GFP+ cells, but with a significant increase of GFP+ cells located in the ipsilateral cerebral cortex (p < .05), indicating injury-activated NSPCs either at the site of injury or migrating to the site of injury are increased after CHI. IHC confirms that injury consistently induces GFP+ cell activation in the ipsilateral cerebral cortex and the hippocampus of the CHI animal compared to the sham animal.
Fig. 3. CHI increases the number of Notch1CR2-GFP+ cells in the cerebral cortex and hippocampus.
Injury-induced GFP+ cells in the cerebral cortex and hippocampus were determined by immunostaining in a mouse model of closed head injury (CHI). GFP+ cells in the injured cerebral cortex and hippocampus (SGZ) were detected in coronal sections of the brain at 2 DPI and 14 DPI. An increased number of GFP+ cells was observed in the injured hippocampus at 2 DPI (p < .01) and in injured cerebral cortex at 14 DPI (p < .05) (A) as compared to sham mice (B). Scale bar = 50 μm, Mean ± SEM, n = 6.
3.3. GFP+ cells express Notch1 in the injured brain
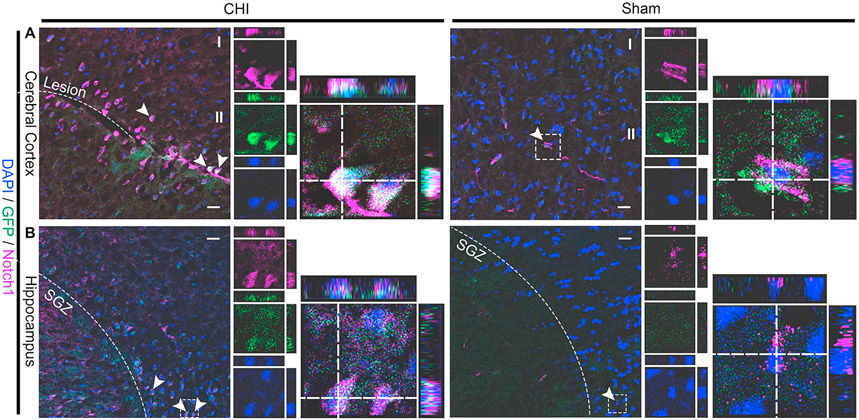
To determine whether CHI upregulates Notch1 signaling in injured brain tissue, we performed IHC on tissue sections from the cerebral cortex (Fig. 4A) and hippocampus (Fig. 4A) using antibody against Notch1 at 2 DPI. We found there was in increased number of Notch1+ cells after CHI in injured mice (n = 6) compared to sham mice (n = 6). A majority of GFP+ cells co-label with Notch1, confirming the increase of Notch1 signaling in GFP+ cells after CHI. To determine the identity of CHI-induced GFP+ cells, we next analyzed the early response to injury (i.e., proliferation and cell death) and the lineage development (i.e., astrocytes, neurons, oligodendrocytes).
Fig. 4. CHI increases the number of Notch1+ cells in the cerebral cortex and hippocampus.
Injury-induced Notch1+ cells in the cerebral cortex (A) and hippocampus (B) were determined by immunostaining in a mouse model of closed head injury (CHI). Notch1+ cells in the injured cerebral cortex and hippocampus (SGZ) were detected in coronal sections of the brain at 2 DPI. An increased number of Notch1+ cells was observed in the injured hippocampus and cerebral cortex at 2 DPI as compared to sham mice. Scale bar = 50 μm.
3.4. CHI induces cell proliferation in the hippocampus
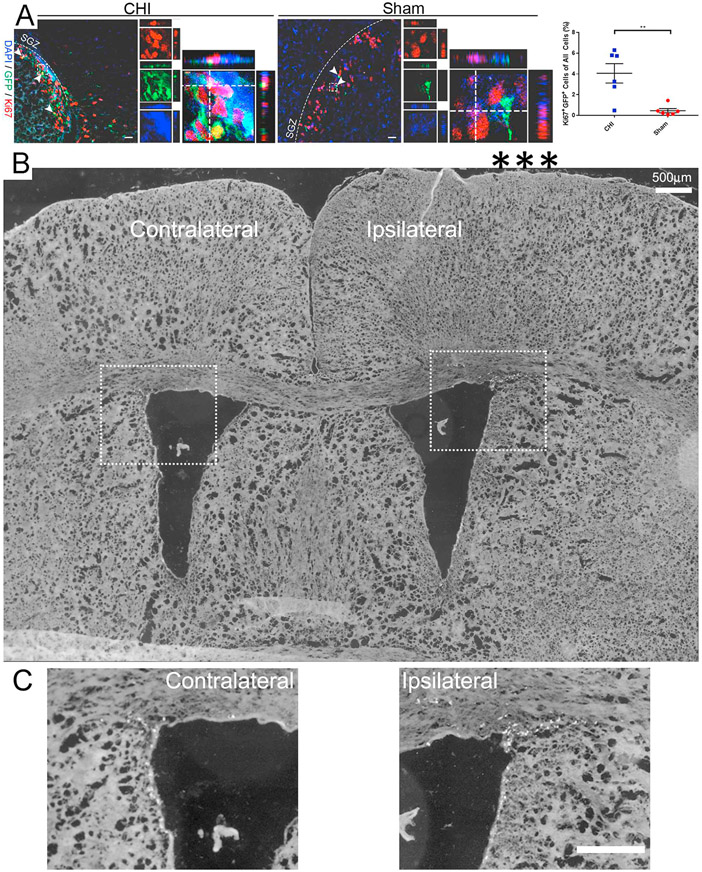
To analyze the cellular response and GFP+ cell identity after injury, we performed IHC using cell proliferation marker Ki67 at 2 DPI in the SGZ (Fig. 5A). The expression of Ki67 in the SGZ of the injured mouse brain was detected, a known region of NSPC proliferation (Lim and Alvarez-Buylla, 2016; Sibbe et al., 2012). Increased number of Ki67+ cells was observed in the ipsilateral hippocampus of injured mice compared to the contralateral hippocampus of injured mice and the hippocampus of sham mice. Ki67+/GFP+ co-expression was detected in all tissue sections, with a majority of the signal observed in the SGZ. Ki67+/GFP+ co-labeled cells are significantly increased in the ipsilateral hippocampus at 2 DPI (4.06 ± 0.93% in CHI (n = 6) vs. 0.45 ± 0.21% in sham (n = 6); p < .01; Fig. 5A) based on quantification of co-labeling merged fluorescence images. The most profound increase in Ki67 expression is in the SGZ, a known region where active NSPCs reside. Ki67 staining in GFP+ cells indicate injury induces cell proliferation.
Fig. 5. CHI increases cell proliferation in the hippocampus.
(A) Injury-induced GFP+ cells in the hippocampus were determined by immunostaining in a mouse model of closed head injury (CHI). Confocal images show GFP+ cells expressing cell proliferation marker Ki67 (red) in coronal sections of the hippocampus at 2 DPI. Arrowheads indicate co-labeled cells. Dash line boxed region shows an example of Ki67+/GFP+ co-labeled cells in separate image channels and in a higher magnification with orthogonal views of Z-stack. Quantification shows a significant increase in the number of Ki67+/GFP+ cells (4.06 ± 0.93% in CHI vs. 0.45 ± 0.21% in sham; p < .01) out of all the cells in the hippocampus of injured mice as compared to sham mice. Scale bar = 20 μm, Mean ± SEM, n = 6. (B) tiled imaging shows both contralateral and ipsilateral brain region. A higher magnification of the wall in the lateral ventricles within the dotted boxes is shown in (C). Scale bars in (B–C) = 500 μm.
We also performed a low power tiled imaging analysis of brain regions containing both the contralateral and ipsilateral sites with each of the zoomed in panels to show the location of impact site of CHI (indicated by the three asterisks) and Ki67 labeled cells (Fig. 5B-C). In the closed head injury (CHI) model, there was no obvious tissue damage (Fig. 5B). However, CHI significantly increased the percentage of Ki67+ cells along the wall of the lateral ventricle (LV) in the ipsilateral site of the brains compared to that of the contralateral site (white dots are Ki67 labeled cells in Fig. 5C).
3.5. CHI increases the number of GFP+ NSPCs in the hippocampus
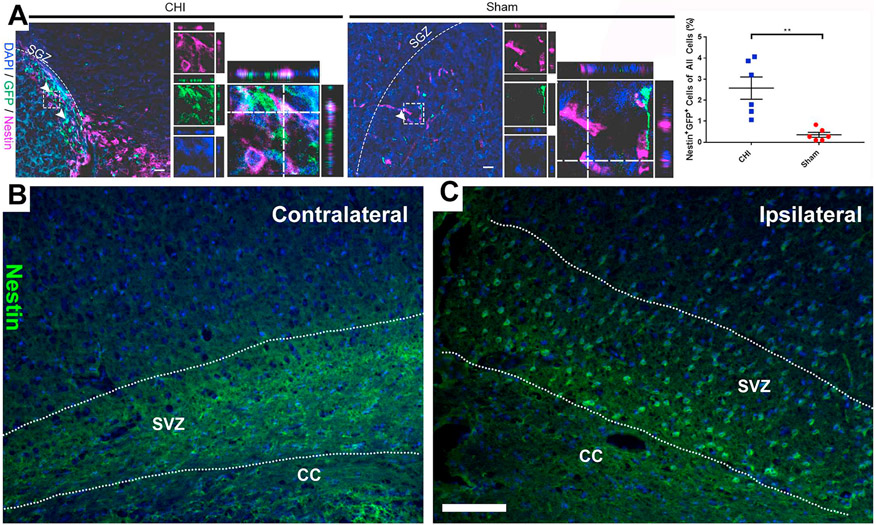
To determine whether the injury-induced GFP+ cells are NSPCs, we examined the co-expression of GFP with a NSPC marker Nestin by IHC at 2 DPI in the SGZ of the hippocampus. Quantification showed that CHI significantly increased the percentage of Nestin+/GFP+ co-labeled cells in the ipsilateral hippocampus at 2 DPI (2.58 ± 0.53% after CHI (n = 6) vs. 0.36 ± 0.12% in sham (n = 6); p < .01; Fig. 6A). This result indicates that injury-induced GFP+ cells were NSPCs. It is known that TBI induces the activation of NSPCs in the SVZ of the cerebral cortex (Chang et al., 2016). Thus, we also examined the expression of NSPC marker Nestin in the SVZ. We found that CHI dramatically increased the Nestin+ cells in the SVZ of the ipsilateral cerebral cortex compared to that of the contralateral cerebral cortex (Fig. 6B-C). The increased number of Nestin+/GFP+ cells indicates that CHI activates NSPCs.
Fig. 6. CHI induces Nestin+ cells in the hippocampus.
(A) Injury-induced GFP+ cells in the hippocampus were determined by immunostaining in a mouse model of closed head injury (CHI). Confocal images show GFP+ cells co-labeled with NSPC marker Nestin (purple) in the hippocampus in coronal sections at 2 DPI. Arrowheads indicate co-labeled cells. Dash line boxed region shows an example of Nestin+/GFP+ co-labeled cells in separate image channels and in a higher magnification with orthogonal views of Z-stack. Quantification shows an increase in the number of Nestin+/GFP+ cells (2.58 ± 0.53% in CHI vs. 0.36 ± 0.12% in sham; p < .01) out of all the cells in the ipsilateral cerebral cortex of injured mice compared to sham mice. Mean ± SEM, n = 6. Examples of photomicrographs of anti-Nestin antibody staining in the SVZ of the contralateral (B) and ipsilateral (C) brain regions of the CHI animal. Scale bars = 20 μm.
3.6. Injury-induced GFP+ NSPCs differentiate into astrocytes in the cerebral cortex
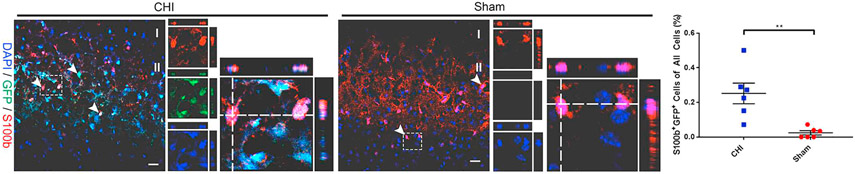
Studies have shown that TBI−/stroke-activated NSPCs mainly give rise to astrocytes (Carlen et al., 2009; Givogri et al., 2006; LeComte et al., 2015; Makara et al., 2003; Shimada et al., 2011). It is also found that Notch signaling plays a role in astrocyte proliferation after stroke/ischemic injury (LeComte and Spees, 2016; Zhang et al., 2015). However, the extent of astrogliogenesis in injured brain is not well examined. We next determined the percentage of astrocytes express GFP by IHC at 14 DPI using astrocyte marker S100b. S100b is an astrocyte-specific CNS protein upregulated after TBI (Kim et al., 2018). IHC analysis shows that an increased number of GFP+ cells co-labeled with astrocyte marker S100b was observed in the injury site of the cerebral cortex compared to sham mice (0.25 ± 0.06% in CHI (n = 6) vs. 0.02 ± 0.01% in sham (n = 6); p < .01) at 14 DPI (Fig. 7). The most profound increase in S100b expression was in the cerebral cortex surrounding the injury site. The increased number of S100b+/GFP+ co-labeled astrocytes indicate injury induced enhancer Notch1CR2 activation in NSPCs, and Notch1CR2 plays an important role astrogliosis in injured brain tissue.
Fig. 7. CHI increases GFP+ astrocytes in the cerebral cortex.
The lineage adoption of injury-induced Notch1CR2-GFP+ cells in the cerebral cortex was determined by immunostaining in a mouse model of closed head injury (CHI). Confocal images show GFP+ cells expressing astrocyte lineage marker S100b in coronal sections of the cerebral cortex at 14 DPI. Arrowheads indicate co-labeled cells. Dash line boxed region shows an example of S100b + GFP+ cells in separate image channels and in a higher magnification with orthogonal views of Z-stack Quantification shows a significant increase in the number of S100b+/GFP+ (co-labeled) astrocytes (0.25 ± 0.06% in CHI vs. 0.02 ± 0.01% in sham; p < .01) out of all the cells in the cerebral cortex of injured mice compared to sham mice. Scale bar = 20 μm, Mean ± SEM, n = 6.
3.7. Injured-induced GFP+ NSPCs preferentially differentiate into GABAergic neurons in the cerebral cortex
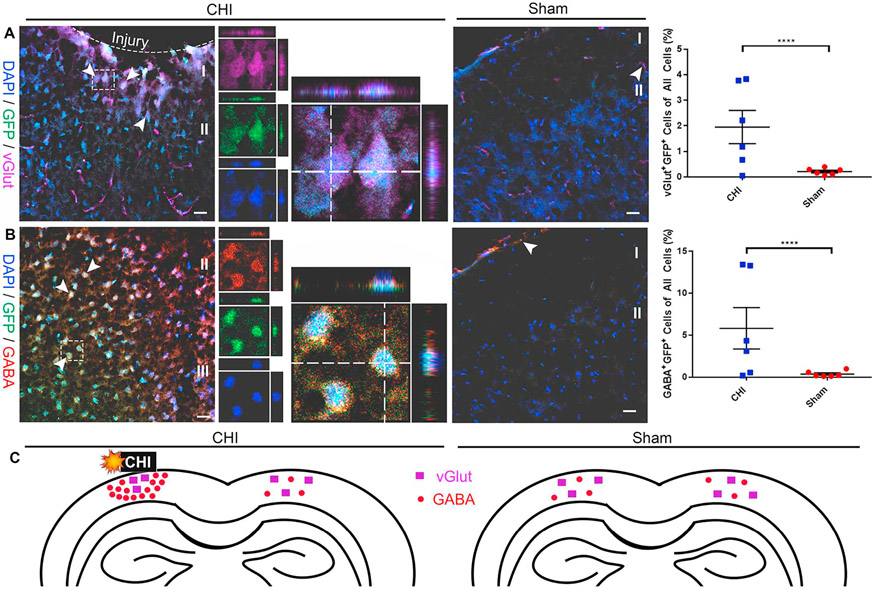
The extent of injury-induced neuronogenesis has not been well characterized. Thus, the potential of injury-activated GFP+ NSPCs to differentiate into neuronal lineages was examined by IHC using various markers for neurons (GABA and vGlut2). GABA+/GFP+ and vGlut2+/GFP+ cells at 14 DPI were analyzed by confocal imaging and quantification. We found that co-labeled cells were mainly localized near the injury site in the cerebral cortex (Fig. 8). The percentage of vGlut2+/GFP+ neurons (1.95 ± 0.65% in CHI (n = 6) vs. 0.21 ± 0.05% in sham (n = 6); p < .0001; Fig. 8A) and GABA+/GFP+ neurons (5.82 ± 2.47% in CHI (n = 6) vs. 0.37 ± 0.14% in sham (n = 6); p < .0001; Fig. 8B) was significantly higher in CHI animals as compared to sham group (Fig. 8A).
Fig. 8. CHI-induced GFP+ cells contain increased number of glutamatergic and GABAergic neurons in the cerebral cortex.
Lineage adoption of injury-induced Notch1CR2-GFP+ cells in the cerebral cortex was determined by immunostaining in a mouse model of closed head injury (CHI). Confocal images show GFP+ cells expressing excitatory neuronal marker vGlut2 (A) and inhibitory neuronal marker GABA (B) in coronal sections of the cerebral cortex at 14 DPI. Arrowheads indicate co-labeled cells. Dash line boxed region shows an example of vGlut2+/GFP+ (A) and GABA+/GFP+ cells (B) in separate image channels and in a higher magnification with orthogonal views of Z-stack. Quantification shows a significant increase in the number of vGlut2+/GFP+ neurons (1.95 ± 0.65% in CHI vs. 0.21 ± 0.05% in sham; p < .0001) and GABA+/GFP+ neurons (5.82 ± 2.47% in CHI vs. 0.37 ± 0.14% in sham; p < .0001) out of all the cells in the ipsilateral cerebral cortex of injured brains compared sham brains. The approximate distribution of vGlut2+ excitatory neurons and GABA+ inhibitory neurons (C) in the injured and sham brain is shown. Scale bar = 20 μm, Mean ± SEM, n = 6.
3.8. CHI does not affect GFP+ oligodendrocyte differentiation
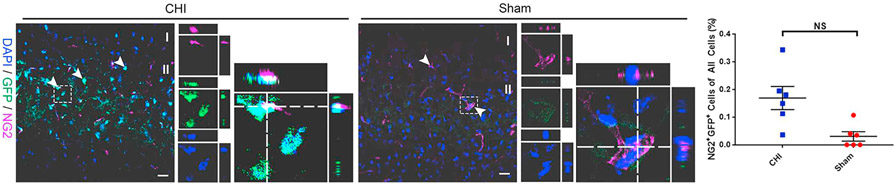
To evaluate the potential for oligodendrocyte lineage development of injury-induced GFP+ cells, we determined the fate of GFP+ cells in the ipsilateral cerebral cortex. Co-expression of NG2+/GFP+ cells was determined by IHC at 14 DPI using an oligodendrocyte progenitor cell marker NG2. GFP+ cells were found to be co-labeled with NG2 in the ipsilateral cerebral cortex after CHI (Fig. 9). Although co-labeling of GFP+ oligodendrocyte progenitors was observed after injury, the number of NG2+/GFP+ co-labeled cells (0.17 ± 0.04% in CHI (n = 6) vs. 0.03 ± 0.02% in sham (n = 6); Fig. 9) was not statistically significant compared to the sham at 14 DPI. These results indicate that Notch1-activated NSPCs (GFP+) have the potential to differentiate into oligodendrocytes in the injured and uninjured cerebral cortex, but injury-response does not alter oligodendrocyte lineage adoption.
Fig. 9. CHI does not alter the number of GFP+ oligodendrocyte progenitors in the cerebral cortex.
The lineage adoption of injury-induced Notch1CR2-GFP+ cells in the cerebral cortex was determined by immunostaining in a mouse model of closed head injury (CHI). Confocal images show GFP+ cells expressing oligodendrocyte progenitor marker NG2 in coronal sections of the cerebral cortex at 14 DPI. Arrowheads indicate co-labeled cells. Dash line boxed region shows an example of NG2+/GFP+ cells in separate image channels and in a higher magnification with orthogonal views of Z-stack. Quantification shows no significant (NS) increase in the number of NG2+/GFP+ (0.17 ± 0.04% in CHI vs. 0.03 ± 0.02% in sham; NS) oligodendrocyte progenitors out of all the cells in the cerebral cortex of injured animals compared to sham animals. Scale bar = 20 μm, Mean ± SEM, n = 6.
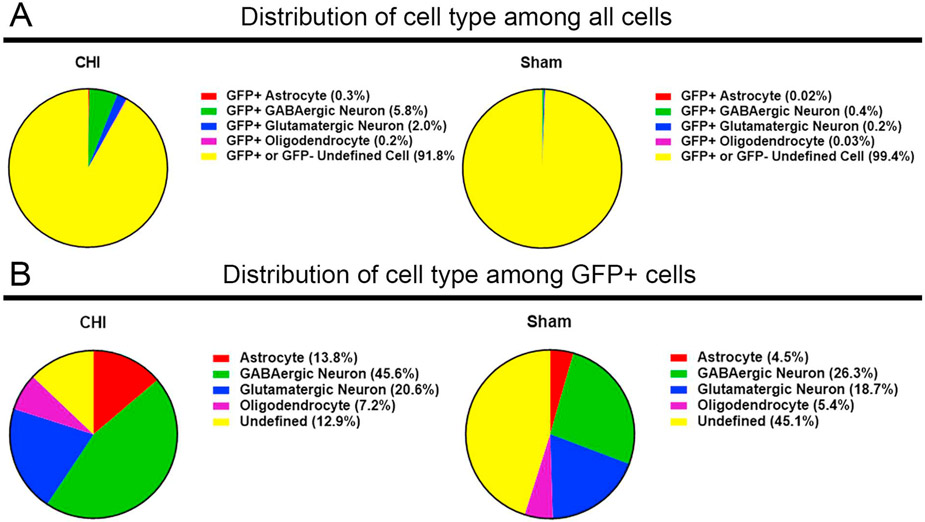
3.9. Differences in cell type distribution of the GFP+ cells
Although the number of GFP+ cells is increased after CHI, the GFP+ cells represent a small cell population compared to the total cells in the brain (Fig. 10A). Focusing only on the GFP+ cell population, we wonder whether the cellular distribution of GFP+ cells is different between the CHI and sham animal groups. Since the cellular identities (e.g., astrocytes, GABAergic neurons, glutamatergic neurons, and oligodendrocytes) of all GFP+ cells were determined by IHC, we then calculated the percentage of GFP+ cells in each different cell type (Fig. 10B). Compared with the sham group, there was a significant difference in the cellular distribution of the GFP+ cells in CHI group. Notablly, there was a significantly higher percentage of GFP+ astrocytes (13.8% in CHI vs. 4.5% in sham) and GFP+ GABAergic neurons (45.6% in CHI vs. 26.3% in sham) at the injury site in mice with CHI.
Fig. 10. Distribution of the GFP+ cell population in CHI and sham brains.
Pie charts (A) show distribution of various sham and injury-induced GFP+ neural cell populations out of all cells in CHI and sham transgenic mouse brains at 14 DPI. There is an increase of GFP+ cell types after CHI compared to sham animals. Pie charts (B) show distribution of various sham and injury-induced GFP+ neural cell populations out of all GFP+ cells in CHI and sham transgenic mouse brains at 14 DPI. In injured brain regions, there was an increased proportion of GFP+ astrocytes, GABAergic neurons, and glutamatergic neurons after CHI compared to sham animals.
Although there was no major increase in the total number of GFP+ glutamatergic neurons between CHI (20.6%) and sham (18.7%), the location of these cells was different with a majority of the glutamatergic neurons being centralized at the injury site after injury (Fig. 8C). The difference in the distribution of GFP+ glutamatergic neurons suggests a possibility that the microenvironment at the injury site may favor the glutamatergic neurons vs. GABAergic neurons.
There was no significant changes in the number of GFP+ oligodendrocytes between the CHI brain (7.16%) and the sham (5.40%) brain. The number of NG2+/GFP+ co-labeled oligodendrocyte progenitors were 7.16% in the brain with CHI compared to 5.40% in sham animals, indicating that Notch1CR2 activity may not have an effect on NSPC differentiation to the oligodendrocyte lineage. The undefined GFP+ cells may be representative of quiescent populations of NSPCs or other specific neural lineages not evaluated in this study.
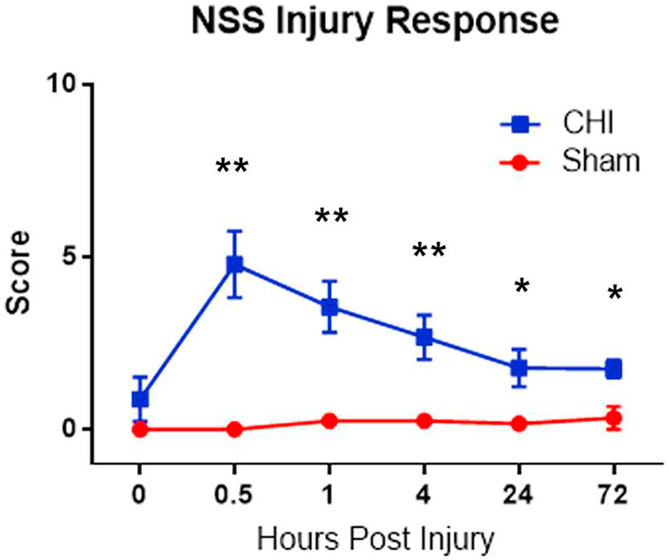
3.10. CHI causes neurological impairments
To assess the neurological impairment after CHI, we obtained neurological severity scores (NSS) (Chen et al., 1996; Flierl et al., 2009) The score consists of 10 individual clinical parameters, including tasks on motor function, alertness and physiological behavior. Compared with the sham, CHI increased the NSS scores at various stages post-injury (Fig. 11).
Fig. 11. Overall scores of injury response using NSS tests.
The NSS score consists of 10 individual clinical parameters, including tasks on motor function, alertness and physiological behavior. Values are expressed as means ± SD. Statistical analysis was carried out using Student-t-test for each timepoint (n = 6). Asterisk (*) indicates statistically significant differences (** P < .01 and * P < .05) in neurological deficit caused by CHI.
4. Discussion
In the Notch1CR2-GFP mouse line, the reporter GFP marks NSPCs for interneurons (Li et al., 2016; Tzatzalos et al., 2012). Using this reporter line, we characterize GFP+ cells in the young adult mouse after TBI. Injury induces NSPC proliferation in response with an expansion of Notch1CR2-GFP+ NSPCs in the acute phase of injury at 2 DPI. During a subchronic phase of injury at 14 DPI, GFP+ cells constitute a higher percentage of GABAergic neurons and astrocytes around the impact site (mainly in the SVZ and SGZ). Since injury is known to induce astrocytes, it is thus suggested that GFP+ NSPCs preferentially differentiate into GABAeregic neurons.
The increased percentage of GFP+ GABAergic neurons in the injured brain correlates well with the increased Notch signaling. This supports our previous finding that Notch signaling is preferentially active in GABAergic progenitors (Li et al., 2016; Tzatzalos et al., 2012). Thus, our study establishes that Notch1CR2-GFP transgenic animal is a valuable model for the study of NSPC behavior after brain injury. Since our published data show that manipulating transcription factors (e.g., Nkx6.1) regulates Notch1 signaling (Li et al., 2016), our mouse line would make it simple to monitor this important signaling pathway by tracking the GFP activity during normal and pathological conditions.
The Notch signaling pathway is essential for continuous production of NSPCs/neuroblasts and neural differentiation during development (Ables et al., 2011; Louvi and Artavanis-Tsakonas, 2006; Yoon and Gaiano, 2005). It has been shown that increased Notch1 signaling activity increases NSPC proliferation, whereas inhibiting Notch1 signaling results in a reduction of proliferating cells in the SVZ (Benner et al., 2013; Oya et al., 2009; Tatsumi et al., 2010; Wang et al., 2009). Transient activity of Notch signaling regulates proliferation and differentiation of NSPCs was observed in the injured brain (Tatsumi et al., 2010). However, its specific role in neuroregeneration after injury still needs to be investigated.
Injury-induced Notch signaling is known to contribute mainly to astrogliosis in mammals (Benner et al., 2013; Chojnacki et al., 2003; Givogri et al., 2006; LeComte et al., 2015; Tanigaki et al., 2001; Zhang et al., 2015). Consistent with the role of Notch signaling in astrogliosis, we found that CHI increases S100b+/GFP+ astrocytes. The increased number of astrocytes could be due to the following two processes: 1) injury-induced reactive astrogliosis; and/or 2) NSPC differentiation after injury.
In adult brain, NSPCs in the SVZ and SGZ generate neuroblasts that migrate and integrate into olfactory bulb circuitry or dentate gyrus (Braun and Jessberger, 2014; Ming and Song, 2011). One study in zebrafish shows that the injury-induced NSPCs differentiated into Tbr1+ neurons in the regions surrounding the injury site (Kishimoto et al., 2012). However, the injury-induced neurons in mammalian brain have not been well characterized. In this study, CHI increases the number of GABAergic neurons at the injury site in Notch1CR2-GFP animals. This finding suggests that Notch signaling promotes the generation of neurons in the injured brain and it provides a potential target for the development of TBI therapeutics/regenerative medicine.
The Notch1CR2-GFP mouse model is complementary to other animal models (e.g., Nestin-GFP) which identify the subset of Nestin+ NSPCs (Gao et al., 2009; Wang et al., 2016). Our previous studies established that GFP marks NSPCs for interneurons in the developing mouse brain (Tzatzalos et al., 2012) and spinal cord (Li et al., 2016). The higher percentage of GFP+ GABAergic neurons in the injured animals has not been reported in Nestin-GFP animals. It is not clear to what extent the cellular composition of the Notch1CR2-GFP+ cells are different from that of the Nestin-GFP+ cells. Thus, Notch1CR2-GFP mouse line provides a useful model for the study of interneurons and other neural types (e.g., glutamatergic neurons and astrocytes) in the adult brain in normal and pathological conditions.
In summary, our cellular and molecular analysis of the injured Notch1CR2-GFP animals identify a subpopulation of NSPCs that respond to injury and the extent of the Notch1-activated cellular response (e.g., activation, lineage adoption). Future studies should further investigate this response by examining the molecular mechanisms driving the injury response in specific subtypes of NSPCs that benefit functional and behavioral recovery after TBI.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Cai lab for helpful discussion and the Maribel Vasquez lab for imaging analysis. This work was supported by the grants from the New Jersey Commission on Spinal Cord Research [08–3074-SCR-E0; 10–3091-SCR-E-0; 15IRG006] and Busch Biomedical Award [659218]. J.A. and M.N.P. are fellows of the NIH Biotechnology Training Program [NIH T32 GM00839]. M.P. is also a recipient of the U.S. Department of Education GAANN Precision and Personalized Medicine Pre-Doctoral Training Fellowship (P200A150131).
Abbreviations:
- ApoE
Apolipoprotein E
- Casp3
Caspase-3
- CHI
Closed head injury
- CNS
Central nervous system
- DPI
days post injury
- GFP
green fluorescent protein
- Il1b
Interleukin 1 beta
- NSPC
neural stem/progenitor cell
- SGZ
subgranular zone
- SVZ
subventricular zone
- TBI
traumatic brain injury
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.expneurol.2019.113119.
References
- Ables JL, Decarolis NA, Johnson MA, Rivera PD, Gao Z, Cooper DC, Radtke F, Hsieh J, Eisch AJ, 2010. Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J. Neurosci 30, 10484–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ables JL, Breunig JJ, Eisch AJ, Rakic P, 2011. Not(ch) just development: notch signalling in the adult brain. Nat. Rev. Neurosci 12, 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K, 2011. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9 (4), 317–329. 10.1016/j.stem.2011.09.001. 21982232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegiani B, Lyubimova A, Muraro M, van Es JH, van Oudenaarden A, Clevers H, 2017. A single-cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep. 21, 3271–3284. [DOI] [PubMed] [Google Scholar]
- Barraud P, Thompson L, Kirik D, Bjorklund A, Parmar M, 2005. Isolation and characterization of neural precursor cells from the Sox1-GFP reporter mouse. Eur J Neurosci 22 (7), 1555–1569. 10.1111/j.1460-9568.2005.04352.x.16197496. [DOI] [PubMed] [Google Scholar]
- Benner EJ, Luciano D, Jo R, Abdi K, Paez-Gonzalez P, Sheng H, Warner DS, Liu C, Eroglu C, Kuo CT, 2013. Protective astrogenesis from the SVZ niche after injury is controlled by notch modulator Thbs4. Nature 497, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrer C, Schachtrup C, 2016. ID(ealizing) control of adult subventricular zone neural stem/precursor cell differentiation for CNS regeneration. Neurogenesis (Austin) 3, e1223532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun SM, Jessberger S, 2014. Adult neurogenesis: mechanisms and functional significance. Development 141, 1983–1986. [DOI] [PubMed] [Google Scholar]
- Campolo M, Esposito E, Cuzzocrea S, 2018. A controlled cortical impact preclinical model of traumatic brain injury. Methods Mol. Biol 1727, 385–391. [DOI] [PubMed] [Google Scholar]
- Carlen M, Meletis K, Goritz C, Darsalia V, Evergren E, Tanigaki K, Amendola M, Barnabe-Heider F, Yeung MS, Naldini L, Honjo T, Kokaia Z, Shupliakov O, Cassidy RM, Lindvall O, Frisen J, 2009. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat. Neurosci 12, 259–267. [DOI] [PubMed] [Google Scholar]
- Chang EH, Adorjan I, Mundim MV, Sun B, Dizon ML, Szele FG, 2016. Traumatic brain injury activation of the adult subventricular zone neurogenic niche. Front. Neurosci 10, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P, Skupien P, Hesl B, Coolen M, Moore JC, Madelaine R, Kremmer E, Faus-Kessler T, Blader P, Lawson ND, Bally-Cuif L, 2010. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci 30, 7961–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Constantini S, Trembovler V, Weinstock M, Shohami E, 1996. An experimental model of closed head injury in mice: pathophysiology, histopathology, and cognitive deficits. J. Neurotrauma 13, 557–568. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhong X, Smith DK, Tai W, Yang J, Zou Y, Wang LL, Sun J, Qin S, Zhang CL, 2017. Astrocyte-specific deletion of Sox2 promotes functional recovery after traumatic brain injury. Cereb. Cortex 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CC, Liao YE, Yang LY, Wang JY, Tweedie D, Karnati HK, Greig NH, Wang JY, 2016. Neuroinflammation in animal models of traumatic brain injury. J. Neurosci. Methods 272, 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacki A, Shimazaki T, Gregg C, Weinmaster G, Weiss S, 2003. Glycoprotein 130 signaling regulates Notch1 expression and activation in the self-renewal of mammalian forebrain neural stem cells. J. Neurosci 23, 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon KJ, Theus MH, Nelersa CM, Mier J, Travieso LG, Yu TS, Kernie SG, Liebl DJ, 2015. Endogenous neural stem/progenitor cells stabilize the cortical microenvironment after traumatic brain injury. J. Neurotrauma 32, 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A, 2017. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 18, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas JM, Fitzsimons CP, 2017. Gene regulation in adult neural stem cells. Current challenges and possible applications. Adv. Drug Deliv. Rev 120, 118–132. [DOI] [PubMed] [Google Scholar]
- Evanson NK, Guilhaume-Correa F, Herman JP, Goodman MD, 2018. Optic tract injury after closed head traumatic brain injury in mice: a model of indirect traumatic optic neuropathy. PLoS One 13, e0197346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl MA, Stahel PF, Beauchamp KM, Morgan SJ, Smith WR, Shohami E, 2009. Mouse closed head injury model induced by a weight-drop device. Nat. Protoc 4, 1328–1337. [DOI] [PubMed] [Google Scholar]
- Gao X, Enikolopov G, Chen J, 2009. Moderate traumatic brain injury promotes proliferation of quiescent neural progenitors in the adult hippocampus. Exp. Neurol 219, 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhang W, Ma L, Li X, Wang H, Li Y, Freimann R, Yu Y, Shuai L, Wutz A, 2018. Derivation of haploid neural stem cell lines by selection for a Pax6-GFP reporter. Stem Cells Dev 27 (7), 479–487. 10.1089/scd.2017.0193. 29471728. [DOI] [PubMed] [Google Scholar]
- Gardner AJ, Shih SL, Adamov EV, Zafonte RD, 2017. Research Frontiers in traumatic brain injury: Defining the Injury. Phys. Med. Rehabil. Clin. N. Am 28, 413–431. [DOI] [PubMed] [Google Scholar]
- Giachino C, Taylor V, 2014. Notching up neural stem cell homogeneity in homeostasis and disease. Front. Neurosci 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givogri MI, de Planell M, Galbiati F, Superchi D, Gritti A, Vescovi A, de Vellis J, Bongarzone ER, 2006. Notch signaling in astrocytes and neuroblasts of the adult subventricular zone in health and after cortical injury. Dev. Neurosci 28, 81–91. [DOI] [PubMed] [Google Scholar]
- Hasan A, Deeb G, Rahal R, Atwi K, Mondello S, Marei HE, Gali A, Sleiman E, 2017. Mesenchymal stem cells in the treatment of traumatic brain injury. Front. Neurol 8, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL, Kim CC, Ryba BE, Niemi EC, Bando JK, Locksley RM, Liu J, Nakamura MC, Seaman WE, 2013. Traumatic brain injury induces macrophage subsets in the brain. Eur. J. Immunol 43, 2010–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R, 2011. The role of notch signaling in adult neurogenesis. Mol. Neurobiol 44, 7–12. [DOI] [PubMed] [Google Scholar]
- Khalin I, Jamari NL, Razak NB, Hasain ZB, Nor MA, Zainudin MH, Omar AB, Alyautdin R, 2016. A mouse model of weight-drop closed head injury: emphasis on cognitive and neurological deficiency. Neural Regen. Res 11, 630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Tsao JW, Stanfill AG, 2018. The current state of biomarkers of mild traumatic brain injury. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto N, Shimizu K, Sawamoto K, 2012. Neuronal regeneration in a zebrafish model of adult brain injury. Dis. Model. Mech 5, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Alvarez-Croda DM, Stoica BA, Faden AI, Loane DJ, 2016. Microglial/macrophage polarization dynamics following traumatic brain injury. J. Neurotrauma 33, 1732–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeComte MD, Spees JL, 2016. Notch1-STAT3-ETBR signaling in brain injury and cancer. Cytokine 80, 64–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeComte MD, Shimada IS, Sherwin C, Spees JL, 2015. Notch1-STAT3-ETBR signaling axis controls reactive astrocyte proliferation after brain injury. Proc. Natl. Acad. Sci. U. S. A 112, 8726–8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinhase I, Holers VM, Thurman JM, Harhausen D, Schmidt OI, Pietzcker M, Taha ME, Rittirsch D, Huber-Lang M, Smith WR, Ward PA, Stahel PF, 2006. Reduced neuronal cell death after experimental brain injury in mice lacking a functional alternative pathway of complement activation. BMC Neurosci. 7, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tzatzalos E, Kwan KY, Grumet M, Cai L, 2016. Transcriptional regulation of Notch1 expression by Nkx6.1 in neural stem/progenitor cells during ventral spinal cord development. Sci. Rep 6, 38665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Alvarez-Buylla A, 2016. The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harb. Perspect. Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S, 2006. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci 7, 93–102. [DOI] [PubMed] [Google Scholar]
- Ludwig PE, Thankam FG, Patil AA, Chamczuk AJ, Agrawal DK, 2018. Brain injury and neural stem cells. Neural Regen. Res 13, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma MW, Wang J, Dhandapani KM, Brann DW, 2017. NADPH oxidase 2 regulates NLRP3 Inflammasome activation in the brain after traumatic brain injury. Oxidative Med. Cell. Longev 2017, 6057609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Aravind A, Pfister BJ, Chandra N, Haorah J, 2019. Animal models of traumatic brain injury and assessment of injury severity. Mol. Neurobiol 5332–5345. 10.1007/s12035-018-1454-5. 30603958. [DOI] [PubMed] [Google Scholar]
- Makara JK, Rappert A, Matthias K, Steinhauser C, Spat A, Kettenmann H, 2003. Astrocytes from mouse brain slices express ClC-2-mediated cl- currents regulated during development and after injury. Mol. Cell. Neurosci 23, 521–530. [DOI] [PubMed] [Google Scholar]
- Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G, 2004. Neural stem and progenitor cells in nestin-GFP transgenic mice. J Comp Neurol 469 (3), 311–324. 10.1002/cne.10964. 14730584. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H, 2011. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EA, Todd BP, Mahoney J, Pieper AA, Ferguson PJ, Bassuk AG, 2018. Combined blockade of interleukin-1alpha and -1beta signaling protects mice from cognitive dysfunction after traumatic brain injury. eNeuro 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya S, Yoshikawa G, Takai K, Tanaka JI, Higashiyama S, Saito N, Kirino T, Kawahara N, 2009. Attenuation of notch signaling promotes the differentiation of neural progenitors into neurons in the hippocampal CA1 region after ischemic injury. Neuroscience 158, 683–692. [DOI] [PubMed] [Google Scholar]
- Patel K, Sun D, 2016. Strategies targeting endogenous neurogenic cell response to improve recovery following traumatic brain injury. Brain Res. 1640, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps HW, 2016. Systematic review of traumatic brain injury animal models. Methods Mol. Biol 1462, 61–88. [DOI] [PubMed] [Google Scholar]
- Pluta R, Bogucka-Kocka A, Ulamek-Koziol M, Bogucki J, Januszewski S, Kocki J, Czuczwar SJ, 2018. Ischemic tau protein gene induction as an additional key factor driving development of Alzheimer’s phenotype changes in CA1 area of hippocampus in an ischemic model of Alzheimer’s disease. Pharmacol. Rep 70, 881–884. [DOI] [PubMed] [Google Scholar]
- Puhakka N, Bot AM, Vuokila N, Debski KJ, Lukasiuk K, Pitkanen A, 2017. Chronically dysregulated NOTCH1 interactome in the dentate gyrus after traumatic brain injury. PLoS One 12, e0172521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SK, Dixon CE, Banik NL, 2002. Molecular mechanisms in the pathogenesis of traumatic brain injury. Histol. Histopathol 17, 1137–1152. [DOI] [PubMed] [Google Scholar]
- Reis C, Gospodarev V, Reis H, Wilkinson M, Gaio J, Araujo C, Chen S, Zhang JH, 2017. Traumatic brain injury and stem cell: pathophysiology and update on recent treatment modalities. Stem Cells Int. 2017, 6392592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab JM, Beschorner R, Meyermann R, Gozalan F, Schluesener HJ, 2002. Persistent accumulation of cyclooxygenase-1-expressing microglial cells and macrophages and transient upregulation by endothelium in human brain injury. J. Neurosurg 96, 892–899. [DOI] [PubMed] [Google Scholar]
- Shimada IS, Borders A, Aronshtam A, Spees JL, 2011. Proliferating reactive astrocytes are regulated by Notch-1 in the peri-infarct area after stroke. Stroke 42, 3231–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbe M, Haussler U, Dieni S, Althof D, Haas CA, Frotscher M, 2012. Experimental epilepsy affects Notch1 signalling and the stem cell pool in the dentate gyrus. Eur. J. Neurosci 36, 3643–3652. [DOI] [PubMed] [Google Scholar]
- Stoica BA, Faden AI, 2010. Cell death mechanisms and modulation in traumatic brain injury. Neurotherapeutics 7, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Brady RD, van der Poel C, Apted D, Semple BD, Church JE, O’Brien TJ, McDonald SJ, Shultz SR, 2018. A concomitant muscle injury does not worsen traumatic brain injury outcomes in mice. Front. Neurol 9, 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T, 2001. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron 29, 45–55. [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Okuda H, Makinodan M, Yamauchi T, Makinodan E, Matsuyoshi H, Manabe T, Wanaka A, 2010. Transient activation of notch signaling in the injured adult brain. J. Chem. Neuroanat 39, 15–19. [DOI] [PubMed] [Google Scholar]
- Tu M, Zhu P, Hu S, Wang W, Su Z, Guan J, Sun C, Zheng W, 2017. Notch1 signaling activation contributes to adult hippocampal neurogenesis following traumatic brain injury. Med. Sci. Monit 23, 5480–5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtzo LC, Lescher J, Janes L, Dean DD, Budde MD, Frank JA, 2014. Macrophagic and microglial responses after focal traumatic brain injury in the female rat. J. Neuroinflammation 11, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatzalos E, Smith SM, Doh ST, Hao H, Li Y, Wu A, Grumet M, Cai L, 2012. A cis-element in the Notch1 locus is involved in the regulation of gene expression in interneuron progenitors. Dev. Biol 372, 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van KC, Lyeth BG, 2016. Lateral (parasagittal) fluid percussion model of traumatic brain injury. Methods Mol. Biol 1462, 231–251. [DOI] [PubMed] [Google Scholar]
- Wang X, Mao X, Xie L, Greenberg DA, Jin K, 2009. Involvement of Notch1 signaling in neurogenesis in the subventricular zone of normal and ischemic rat brain in vivo. J. Cereb. Blood Flow Metab 29, 1644–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo F, Pan C, Lou Y, Zhang P, Guo S, Yin J, Deng Z, 2012. Effects of low temperatures on proliferation-related signaling pathways in the hippocampus after traumatic brain injury. Exp. Biol. Med. (Maywood) 237, 1424–1432. [DOI] [PubMed] [Google Scholar]
- Wang X, Seekaew P, Gao X, Chen J, 2016. Traumatic brain injury stimulates neural stem cell proliferation via mammalian target of rapamycin signaling pathway activation. eNeuro 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston NM, Sun D, 2018. The potential of stem cells in treatment of traumatic brain injury. Curr. Neurol. Neurosci. Rep 18, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SM, Kim J, Han HW, Chae JI, Son MY, Cho S, Chung HM, Han YM, Kang YK, 2009. Notch signaling is required for maintaining stem-cell features of neuroprogenitor cells derived from human embryonic stem cells. BMC Neurosci. 10, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Kong L, Xia Y, Liang W, Wang L, Song J, Yao Y, Lin Y, Yang J, 2018. Osthole promotes endogenous neural stem cell proliferation and improved neurological function through notch signaling pathway in mice acute mechanical brain injury. Brain Behav. Immun 67, 118–129. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N, 2005. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci 8, 709–715. [DOI] [PubMed] [Google Scholar]
- Zhang Y, He K, Wang F, Li X, Liu D, 2015. Notch-1 signaling regulates astrocytic proliferation and activation after hypoxia exposure. Neurosci. Lett 603, 12–18. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH, 2008. Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660. [DOI] [PubMed] [Google Scholar]
- Zhou ZD, Kumari U, Xiao ZC, Tan EK, 2010. Notch as a molecular switch in neural stem cells. IUBMB Life 62, 618–623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.