Abstract
We propose a novel model for the regulation of the p85/p110α phosphatidylinositol 3′-kinase. In insect cells, the p110α catalytic subunit is active as a monomer but its activity is decreased by coexpression with the p85 regulatory subunit. Similarly, the lipid kinase activity of recombinant glutathione S-transferase (GST)-p110α is reduced by 65 to 85% upon in vitro reconstitution with p85. Incubation of p110α/p85 dimers with phosphotyrosyl peptides restored activity, but only to the level of monomeric p110α. These data show that the binding of phosphoproteins to the SH2 domains of p85 activates the p85/p110α dimers by inducing a transition from an inhibited to a disinhibited state. In contrast, monomeric p110 had little activity in HEK 293T cells, and its activity was increased 15- to 20-fold by coexpression with p85. However, this apparent requirement for p85 was eliminated by the addition of a bulky tag to the N terminus of p110α or by the growth of the HEK 293T cells at 30°C. These nonspecific interventions mimicked the effects of p85 on p110α, suggesting that the regulatory subunit acts by stabilizing the overall conformation of the catalytic subunit rather than by inducing a specific activated conformation. This stabilization was directly demonstrated in metabolically labeled HEK 293T cells, in which p85 increased the half-life of p110. Furthermore, p85 protected p110 from thermal inactivation in vitro. Importantly, when we examined the effect of p85 on GST-p110α in mammalian cells at 30°C, culture conditions that stabilize the catalytic subunit and that are similar to the conditions used for insect cells, we found that p85 inhibited p110α. Thus, we have experimentally distinguished two effects of p85 on p110α: conformational stabilization of the catalytic subunit and inhibition of its lipid kinase activity. Our data reconcile the apparent conflict between previous studies of insect versus mammalian cells and show that p110α is both stabilized and inhibited by dimerization with p85.
Phosphatidylinositol (PI) 3′-kinases constitute a family of enzymes that mediate intracellular signaling initiated by receptor tyrosine kinases and heterotrimeric G-protein-coupled receptors. Activation of PI 3′-kinase leads to increases in the intracellular levels of PI[3,4]P2 and PI[3,4,5]P3, which are presumed second messengers (4). PI 3′-kinases have been implicated in the control of proliferation, cytoskeletal organization, apoptosis, and vesicular trafficking (6, 16, 20, 31, 46).
A classification of PI 3′-kinases has been described by Zvelebil and coworkers (48). The class I enzymes are heterodimeric proteins that are composed of separate regulatory and catalytic subunits and that utilize PI, PI[4]P, and PI[4,5]P2 as substrates. Class I enzymes include the p85/p110 PI 3′-kinase, which is activated by binding to phosphotyrosyl proteins, and the p101/p120 PI 3-kinase-γ isoform, which is activated by βγ subunits from trimeric G proteins (11, 13, 38, 39). Class II PI 3′-kinases contain C-terminal C2 domains and preferentially utilize PI and PI[4]P as substrates (24, 26, 43). Class III enzymes include the yeast VPS34 and its mammalian homolog (35, 44) and recognize PI but not higher-order phosphoinositides as substrates.
Regulation of the p85/p110 PI 3′-kinase is complex. Two homologous p85 regulatory subunits have been identified (9, 27, 36). Each contains an N-terminal SH3 domain followed by a proline rich domain, a breakpoint cluster region (BCR) homology domain, a second proline-rich domain, and two SH2 domains. Shorter forms (p55) lacking the SH3 and BCR homology domains have also been cloned (1, 29). All the p85/p55 proteins contain putative coiled-coil domains that mediate stable dimerization with the p110 catalytic subunit (7). The three known isoforms of p110 catalytic subunits (p110α, p110β, and p110δ) contain closely spaced N-terminal binding sites for p85 and p21ras-GTP, as well as C-terminal kinase domains (12, 14, 18, 32, 42). In vitro studies have shown that the p85 SH3 domain binds to proline-rich peptides, the proline-rich domains bind to SH3 domains from Fyn and Lyn, the BCR homology domain binds to GTP-loaded CDC42, and the SH2 domains bind to tyrosyl phosphopeptides (3, 17, 37, 41, 47). Each of these binding events activates p85/p110α PI 3′-kinase in vitro, although it is not clear yet whether these distinct activating interactions are redundant or additive (2, 10, 28, 47). p110α binding to p21ras-GTP was also found to increase PI 3′-kinase activity in an in vitro assay using lipid vesicles as the substrates (32). However, this increase in activity may reflect the targeting of p110α to the lipid vesicles, since nonisoprenylated GTP-ras binds to p110 but does not activate it in vitro.
The p85 regulatory subunit of PI 3′-kinase has been generally viewed as an activator of p110α. Several groups have reported that p110α is catalytically inactive as a monomer in Cos cells and requires coexpression with p85 for activity (11, 18). In fact, the p110α binding domain from p85 (the inter-SH2 [iSH2] domain) has been tethered to the N terminus of p110α to produce a constitutively active PI 3′-kinase (15). However, p110α is active when expressed in baculovirus-infected Sf-9 cells (11). In addition, p110β is active as a monomer in HEK 293T cells (13). Interestingly, the studies reporting that p110α is inactive as a monomer in mammalian cells used C-terminal tags or C-terminally directed antibodies, whereas the study reporting active monomeric p110β used an N-terminal tag inserted between residues 30 and 31 (11, 13, 15, 18).
In this study, we have examined the mechanism by which p85 regulates p110α activity. In insect cells, p110α is an active monomer that is reversibly inhibited by stable binding to p85. Activation of recombinant p85/p110α by phosphotyrosyl proteins reflects a disinhibition of the p85/p110α dimer. In contrast, monomeric p110 had little activity in HEK 293T cells, and its activity was increased 15- to 20-fold by coexpression with p85. However, this requirement can be completely supplanted by either the addition of an N-terminal glutathione S-transferase (GST) tag or culture of the mammalian cells at 30°C. Under conditions that stabilize monomeric p110α in mammalian cells (GST-p110α in cells grown at 30°C), coexpression of GST-p110α with p85 causes an inhibition of activity similar to that seen in insect cells. Our data show that p85 acts by both stabilizing and inhibiting the p110α catalytic subunit.
MATERIALS AND METHODS
Antibodies and Western blotting.
Affinity-purified rabbit antibodies against residues 324 to 721 of human p85 have been previously described (2). Antibodies against p110α (AE-40) were produced by immunizing rabbits with a GST fusion protein containing residues 1 to 140 of bovine p110α. The antibodies were affinity purified with a column made by coupling the same GST fusion protein to Affigel (Bio-Rad). All Western blots were visualized with primary and secondary antibodies (where required) and 125I-protein A (New England Nuclear) and quantitated with a Molecular Dynamics PhosphorImager.
Mutagenesis of p85 and p110.
Construction of N-terminally hemagglutinin (HA)-tagged p85 has been previously described (33). Mutation of Ser-608 to alanine was performed by the method of Kunkel et al. (23). Bovine p110α (provided by M. Waterfield, Ludwig Institute for Cancer Research) was mutated with the pALTER system (Promega). A new XhoI site was inserted between the first and second codons of the p110α cDNA, which allowed the insertion of double-stranded cassettes between an upstream BamHI site and the XhoI site. The cassettes contained an idealized Kozak sequence (21, 22) followed by the initial ATG and the various epitope tags. The myc tag was inserted with anticomplementary synthetic oligonucleotides. The Tris-HA tag was subcloned from a vector provided by E. Skolnik, New York University, by PCR. The GST tag was subcloned from the pGEX-2T vector (Pharmacia) by using PCR. The resulting sequences, inserted immediately N-terminal to Pro-2 of p110α, were as follows: p110α/N-myc-p110: MAEEQKLISEEDLRRG; 3HA-p110α: MPRGGGRIFYPYDVPDTAGYPYDVPDYAGSTPYDVPDYAAQCGRARG; GST-p110α: M-(GST)-LVPRGSRG. To produce C-terminally myc-tagged p110α, a new BglII site was inserted immediately 5′ to the stop codon of the p110α cDNA, and anticomplementary synthetic nucleotides were used to insert the sequence RSDLGEQKLISEEDLG-STOP. All constructions were confirmed by sequencing. The p110α cDNA constructs were subcloned into the pSG5 expression vector (Stratagene) for expression in mammalian cells; the HA-p85 cDNA construct was subcloned into the expression vector pCMVhis (40). All plasmids were purified by equilibrium centrifugation in CsCl.
Preparation of recombinant PI 3′-kinase in Sf-9 cells.
The p85 and p110α gene constructs were subcloned into pBluebacIII (Invitrogen) and cotransfected into Sf-9 cells with Baculogold (Pharmingen) linearized baculovirus DNA, which contains a lethal mutation rescued by recombination with the baculovirus transfer vector. Recombinant virus was amplified 2 or 3 times to produce high-titer stocks. To produce the recombinant proteins, Sf-9 cells were grown in six-well dishes and infected with recombinant baculovirus. After 3 days in culture, the cells were washed in ice-cold phosphate-buffered saline and lysed by cycles of freezing and thawing in 10 mM Tris (pH 7.4)–150 mM NaCl–1 mM EDTA–100 μg of aprotinin per ml–1 μg of leupeptin per ml–350 μg of phenylmethylsulfonyl fluoride per ml (baculolysis buffer). After removal of particulate material by centrifugation at 12,000 × g, the lysates were assayed directly for PI 3′-kinase activity with sonicated bovine liver PI (200 μg/ml) and ATP (45 μM) as described by Ruderman et al. (34).
Purification of GST-p110.
Sf-9 cells, grown in two 15-cm-diameter dishes, were infected with GST-p110α baculovirus. After 48 h, the cells were lysed in 20 mM Tris (pH 7.5)–137 mM NaCl–1 mM CaCl2–1 mM MgCl2–1% Nonidet P-40 (NP-40)–10% glycerol (NP-40 lysis buffer). After centrifugation at 12,000 × g to remove particulate material, the lysate was passed twice over glutathione-Sepharose, washed with 20 column volumes of 10 mM Tris (pH 7.4)–150 mM NaCl, and eluted with 50 mM Tris (pH 7.4) containing 10 mM glutathione. The eluate was assayed directly for lipid kinase activity; control experiments showed that this buffer did not affect the activity of recombinant p110α. Peak fractions were assayed in the absence or presence of p85 as described above.
p85 and p110α mixing and coexpression experiments.
Lysates from Sf-9 cells expressing wild-type p85 or mutant p85 or lysates from uninfected Sf-9 cells (approximately 20 μg of protein) were mixed with lysates from Sf-9 cells expressing wild-type or tagged p110α (approximately 60 μg of total protein). After 30 min on ice, the samples were assayed for PI 3′-kinase activity as described below. Where indicated, the p85 was first immunopurified by absorption onto protein A-Sepharose with an antibody that recognizes the C-terminal SH2 domain of p85 antibody; we have previously shown that this antibody does not affect p85/p110 activity (25). p110 activity was measured in the presence of the washed p85/protein A beads. Alternatively, Sf-9 cells were coinfected with baculovirus coding for p110α and p85. Duplicate samples were assayed for PI 3′-kinase activity as previously described (34); assays used mixtures including 10 mM MgCL2, 40 μM ATP containing 20 μCi of [32P]ATP/assay, and 200 μM PI. Background lipid kinase activity present in lysates from uninfected Sf-9 cells was subtracted. Parallel samples were assayed for p110α expression by immunoblotting with anti-p110α antibodies. For analysis of PI[4]P and PI[4,5]P2 phosphorylation, assays contained sonicated mixtures of 10 μg of phosphatidylserine, 5 μg of PI, and 10 μg of either PI[4]P or PI[4,5]P2 and were conducted with 500 μM ATP. After extraction in acidic chloroform-methanol (1:1), the organic phase was washed with 1 volume of synthetic upper phase and analyzed by thin-layer chromatography in 1-propanol–2 M acetic acid (65:35).
PI 3′-kinase kinetic analysis.
Recombinant p110α monomers were incubated in the absence or presence of wild-type or mutant p85 as described above and then incubated for an additional 60 min in the absence or presence of a 1 μM concentration of a bisphosphopeptide derived from p85 binding sequences in IRS-1 [DD(P)YMPMSPGAGAGAGAGAGNGD(P)YMPMSPKS] (33). For the kinetic analysis with variable lipid concentrations (Fig. 2 and Table 1), the mixtures were assayed in a solution containing 10 mM MgCl2, 1 mM ATP, 20 μCi of [32P]ATP per assay, and 0 to 1,000 μM PI. For the kinetic analysis with variable ATP, the mixtures were assayed in a solution containing 10 mM MgCl2, 20 μCi of [32P]ATP per assay, 400 μM phospholipid, and 0 to 600 μM ATP. After 10 min at 22°C, the lipids were extracted and lipid kinase activity was measured (34). Incorporation of [32P]ATP into the phospholipid was quantitated with a Molecular Dynamics PhosphorImager, and the result was converted to counts per minute by quantifying serial dilutions of [32P]ATP with the PhosphorImager. All determinations were made in duplicate. Kinetic analysis was performed with Kaleidograph software.
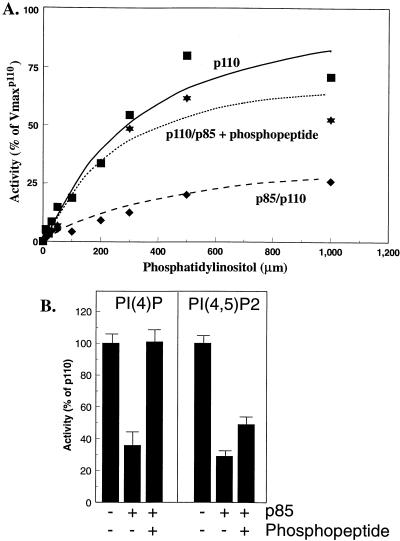
FIG. 2.
Lipid kinase activities of p110α monomers and p85/p110α dimers. (A) N-myc p110α monomers were incubated for 30 min at 4°C in the absence or presence of p85 and then incubated for an additional 60 min at 4°C in the absence or presence of 1 μM bisphosphopeptide. The mixtures were assayed in the presence of 0 to 1,000 μM PI and 1 mM ATP. After 10 min at 22°C, the lipids were extracted and analyzed as described in Materials and Methods. All determinations were performed in duplicate, and the data are the means from three separate experiments. Curves represent the best Michaelis-Menten fit and were generated with Kaleidograph software. (B) N-myc-p110 monomers or p85/N-myc-p110 dimers were incubated in the absence or presence of phosphopeptide as described above. The samples were assayed with sonicated mixtures of PI,PS and either PI[4]P or PI[4,5]P2 as described in Materials and Methods. The data are the means ± standard errors of the means for two (PIP) or three (PIP2) experiments.
TABLE 1.
Kinetic analysis of p110 activitya
| Protein | Kinetic data for assay with variable amounts of:
|
|||||
|---|---|---|---|---|---|---|
| PI
|
ATP
|
|||||
| Km (μM) | Vmax (relative) | Vmax/Km (relative) | Km (μM) | Vmax (relative) | Vmax/Km (relative) | |
| p110 | 415 ± 161 | 100 | 0.24 | 99 ± 23 | 100 | 1.01 |
| p110/p85 | 512 ± 151 | 20.5 ± 8.3 | 0.04 | 130 ± 19 | 48.5 ± 2.0 | 0.37 |
| p110/p85 + phosphopeptide | 492 ± 117 | 89.9 ± 18 | 0.18 | 58 ± 12 | 54.5 ± 11 | 0.95 |
p110 monomers were incubated in the absence or presence of p85 and then incubated for an additional 60 min in the absence or presence of 1 μM bisphosphopeptide. The mixtures were assayed in the presence of 0 to 1,000 μM PI (in the presence of 1 mM ATP) or 0 to 600 μM ATP (in the presence of 400 μM phospholipid). After 10 min at 22°C, the lipids were extracted and analyzed as described in Materials and Methods. The relative Vmax values in each experiment were determined by defining the Vmax of p110 as 100%. All determinations were made in duplicate, and the data are the means ± standard errors of the means of the Km and relative Vmax values from each of 3 or 4 experiments with each substrate. Incorporation of [32P]ATP into the phospholipid was quantitated with a Molecular Dynamics PhosphorImager and normalized to serial dilutions of [32P]ATP. The actual Vmax of p110 was 0.43 ± 0.07 nmol/min/mg, based on an estimation of p110 from silver staining. The kinetic analysis was performed with Kaleidograph software.
Analysis of p85 serine phosphorylation.
Sf-9 cells were infected with p85 baculovirus. After 48 h, the cells were washed into phosphate-free Graces medium and incubated with [32P]orthophosphate (0.5 mCi/ml) for 4 h. The cells were lysed as described above in baculolysis buffer containing 100 mM NaF, 10 mM sodium pyrophosphate, and 100 μM orthovanadate. Alternatively, the cells were lysed in buffer without phosphatase inhibitors and then incubated for 1 h at 30°C in 2 mM dithiothreitol–2 mM MnCl2–0.5 μg of recombinant protein phosphatase 1 (provided by Z.-Y. Zhang, Albert Einstein College of Medicine). The proteins were diluted to 0.5 ml, immunoprecipitated with anti-p85 antibody and protein A-Sepharose, eluted in sample buffer, and separated by reducing sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis (SDS–7.5% PAGE). Proteins were visualized by autoradiography, and 32P incorporation was quantitated with a Molecular Dynamics PhosphorImager.
Production of recombinant PI 3′-kinase in HEK 293T cells.
Confluent HEK 293T cells (provided by J. Krolewski, Columbia University) were split 1:10 24 h prior to transfection, preincubated in 25 μM chloroquine, and transfected with 30 to 45 μg of expression vectors for N-myc-p110α, C-myc-p110α, 3HA-p110α, GST-p110α, and p85 or with empty vector by a calcium-phosphate precipitation method. After 48 h the cells were solubilized, and PI 3′-kinase was immunoprecipitated with monoclonal anti-myc antibody (9E10; Oncogene Science), monoclonal anti-HA antibody (12CA5), or polyclonal anti-GST antibody (Pharmacia). After absorption of the lysate to protein G-Sepharose beads (Pharmacia), the beads were washed and lipid kinase activity was determined as described by Ruderman et al. (34), with quantitation with a Molecular Dynamics PhosphorImager. All determinations were made in triplicate. Immunoprecipitates from parallel samples were blotted with anti-p110α or anti-p85 antibodies.
Metabolic labeling experiments.
HEK 293T cells were transfected with expression vectors for C-myc-p110 in the absence or presence of p85 as described above. Thirty-six hours after transfection, the cells were incubated in methionine-free medium containing 10% fetal bovine serum and 0.1 mCi of [35S]methionine for 16 h. The cells were then washed with phosphate-buffered saline and lysed or incubated for various times in complete medium supplemented with 10× methionine. The cells were lysed, and proteins were immunoprecipitated with anti-myc antibodies and protein G-Sepharose beads eluted, separated by SDS-PAGE, and visualized by autoradiography.
Heat inactivation.
Recombinant wild-type p110 was produced in Sf-9 cells and incubated in buffer without or with recombinant p85 for 1 h at 4°C. The samples were then shifted to the indicated temperatures for 30 min, chilled on ice, and assayed for lipid kinase activity as described above.
RESULTS
p110α is active as a monomer in insect cells and is inhibited by coexpression or reconstitution with p85.
We examined the effects of p85 on p110α activity by using recombinant proteins expressed in baculovirus-infected Sf-9 cells. p110α showed significant activity in the absence of p85, consistent with previous reports (11) (Fig. 1A). Coexpression with p85 increased the amount of p110α protein in Sf-9 lysates (Fig. 1B) but decreased its activity dramatically (Fig. 1A), suggesting that p110α was inhibited by p85.
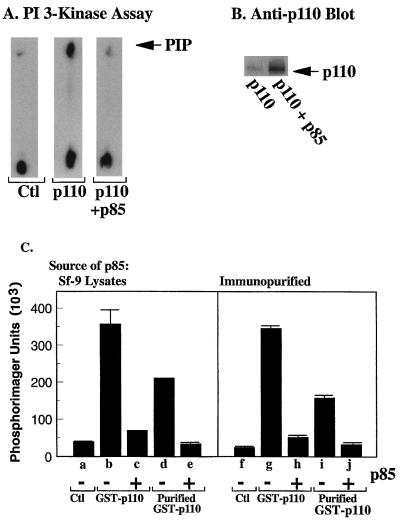
FIG. 1.
Expression of p110α in Sf-9 cells. (A) Control Sf-9 cells or p110α baculovirus-infected Sf-9 cells were additionally infected without the p85 baculovirus or not infected. The cells were lysed, and PI 3′-kinase activities in the lysate were measured. (B) Western blot of parallel samples with anti-p110α antibody. (C) Left panel: control Sf-9 lysates, lysates from cells expressing GST-p110α, or glutathione-Sepharose-purified GST-p110α was mixed with lysates from cells expressing p85 as indicated. Lipid kinase activity was then measured. Right panel: control Sf-9 lysates, lysates from cells expressing GST-p110α, or glutathione-Sepharose-purified GST-p110α was mixed with immunopurified p85 as indicated, and PI 3′-kinase activities were measured. All determinations were made in triplicate, and the data are the means ± standard errors of the means from three experiments.
We reconstituted the p85-induced inhibition of p110α by producing the proteins separately in Sf-9 cells and then mixing them in vitro; p110α could be immunoprecipitated with anti-p85 antibodies under these conditions, demonstrating that the two formed a complex (data not shown). Incubation of GST-p110α with HA-tagged p85 caused an 80% decrease in lipid kinase activity (Fig. 1C, lane c). To show that the inhibition was not due to contaminating factors in the Sf-9 cytosol, we purified an identical amount of GST-p110α by absorption on glutathione-Sepharose beads. Total p110α activity was reduced slightly, reflecting the loss of some GST-p110α from the beads during the washes (lane d). However, the purified GST-p110α was still inhibited by incubation with p85 (lanes e and j). Similarly, we purified p85 by absorption with a highly specific anti-p85 antibody and showed that the immunopurified p85 inhibited GST-p110α activity in Sf-9 lysates (lane h) and in glutathione-Sepharose-purified GST-p110α (lane j). These data show that the inhibition of p110α by p85 does not require coexpression, as has been previously suggested (45). Moreover, purification of p110α and p85 had no effect on the inhibition of p110α by p85. Subsequent experiments were performed with p110α and p85 in Sf-9 lysates, as purified GST-p110α was unstable and lost activity rapidly with storage (data not shown).
We measured the activity of p110α and p85/p110α dimers in the absence or presence of a bisphosphopeptide that activates heterodimeric PI 3′-kinase in vitro (33). Substrate velocity curves, measured in the presence of 1 mM ATP or 400 μM sonicated PI, were obtained for PI and ATP, respectively. A representative curve for PI is shown in Fig. 2, and the calculated kinetic data are shown in Table 1. The addition of p85 to p110α significantly decreased the utilization of both PI and ATP as substrates. Relative Vmax/Km values for PI and ATP decreased by 85 and 60%, respectively, compared to those seen with p110α monomers. The addition of bisphosphopeptide (Fig. 2A; Table 1) restored the relative Vmax/Km values for lipid and ATP to 75 and 95%, respectively, of those seen with p110α monomers. However, the activity of p85/p110α dimers in the presence of activating phosphopeptide was never greater than the activity of an equivalent amount of monomeric p110α. The activity of monomeric p110α was not affected by phosphopeptide (data not shown). The inhibition of p110α by p85 was confirmed by using PI[4]P and PI[4,5]P2 as the substrates (Fig. 2B). The addition of p85 inhibited p110α activity by 50 and 70% with PI[4]P and PI[4,5]P2, respectively. The addition of bisphosphopeptide increased the activity of p85/p110α dimers to 108 and 50%, respectively, of that seen with p110α monomers.
These data suggest that p85 inhibits the activity of p110α. Subsequent activation of p85/p110α dimers by IRS-1 or phosphopeptides reflects a disinhibition of p110α rather than a true increase in activity relative to p110α monomers.
Inhibition of p110α by p85 is independent of serine or threonine phosphorylation.
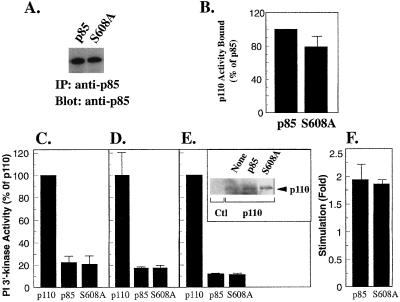
Dhand et al. identified Ser-608 in the iSH2 domain of p85 as an inhibitory regulatory site (8). To determine whether the observed inhibition of p110α could be due to phosphorylation of Ser-608, we mutated the residue to alanine. The p85-S608A mutant was expressed in Sf-9 cells as an 85-kDa polypeptide (Fig. 3A) and showed no differences in binding to p110α when compared to wild-type p85 (Fig. 3B). p85-S608A was indistinguishable from wild-type p85 with regard to inhibition of p110α; this was observed when monomeric p110α was mixed in vitro with either p85 from Sf-9 lysates (Fig. 3C) or immunopurified p85 (Fig. 3D). Similarly, coinfection of Sf-9 cells with either wild-type p85 or p85-S608A increased p110α expression (Fig. 3E, inset) but decreased p110α activity (Fig. 3E). p85-S608A/p110α dimers could be activated by phosphopeptides to the same extent as wild-type p85/p110α dimers (Fig. 3F). Although it is possible that phosphorylation of Ser-608 has an additional inhibitory function in the regulation of p110α, these data show that the inhibition observed here does not depend on the phosphorylation of this residue.
FIG. 3.
Inhibition of p110α by p85-S608A. (A) Expression of wild-type p85 and p85-S608A in Sf-9 lysates was measured by blotting with anti-p85 antibody. (B) Wild-type p85 and p85-S608A were immobilized on anti-p85–protein A-Sepharose beads, incubated with lysates from cells expressing N-myc-p110α, washed, and assayed for lipid kinase activity. Lipid kinase activity bound to wild-type p85 was defined as 100%. (C) Lysates from Sf-9 cells expressing N-myc-p110α were incubated in the absence or presence of wild-type p85 or p85-S608A and then assayed for lipid kinase activity (expressed as a percentage of lipid kinase activity in the absence of p85). (D) Lysates from Sf-9 cells expressing N-myc-p110α were incubated in the absence or presence of immunopurified wild-type p85 or p85-S608A and then assayed for lipid kinase activity (expressed as in panel C). (E) Sf-9 cells were infected with N-myc-p110α alone or were coinfected with wild-type p85 or p85-S608A. Lipid kinase activity in the cell lysates was determined (expressed as in panel C). Inset: p110α expression was determined by blotting with anti-p110α antibody. (F) Lysates containing N-myc-p110α and wild-type p85 or p85-S608A were incubated in the absence or presence of bisphosphopeptide (1 μM) for 1 h, and lipid kinase activity was determined. Activation was expressed as fold stimulation over activity in the absence of phosphopeptide. All determinations were made in duplicate or triplicate, and the data are the means ± standard errors of the means of four separate experiments.
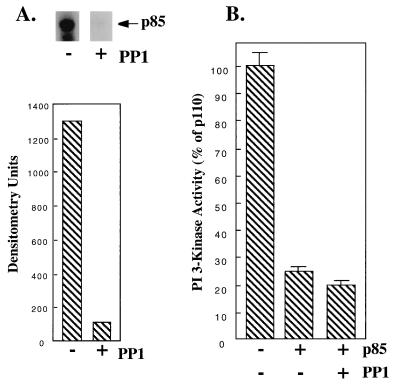
To determine whether other potential phosphorylation sites could account for the observed inhibition of p110α by p85, we labeled p85-infected Sf-9 cells with [32P]orthophosphate and immunoprecipitated them with anti-p85 antibodies. p85 was phosphorylated in Sf-9 cells but could be completely dephosphorylated by treatment with protein phosphatase 1 (Fig. 4A). Dephosphorylated p85 was fully capable of inhibiting p110α when mixed with the enzyme in vitro (Fig. 4B). Our data suggest that phosphorylation of p85 is not required for its inhibition of p110α.
FIG. 4.
Inhibition of p110α by dephosphorylated p85. (A) Forty-eight hours after infection with p85 baculovirus, Sf-9 cells were labeled with [32P]orthophosphate for 4 h. The cells were lysed, treated with recombinant protein phosphatase 1 (0.5 μg) or not treated, and immunoprecipitated with anti-p85–protein A–Sepharose beads. Proteins were eluted and separated by SDS-PAGE, and the dried gel was visualized by autoradiography and quantitated with a Molecular Dynamics PhosphorImager. (B) Lysates from Sf-9 cells expressing p85 were treated in the absence or presence of recombinant protein phosphatase 1 (0.5 μg). p85 was purified by absorption on anti-p85–protein A–Sepharose beads and mixed with p110α, and lipid kinase activity was determined. All determinations were made in triplicate, and the data are the means ± standard errors of the means of two separate experiments.
Activity of monomeric p110α in mammalian cells: requirement for p85 is supplanted by bulky N-terminal tags.
In contrast with the data from insect cells, several laboratories have suggested that p110α expressed in mammalian cells is inactive as a monomer and requires binding to p85 for activity (11, 18). However, p110β is highly active as a monomer in mammalian cells (13). Although the reason for these discrepancies has not been clear, we noted that the studies examining p110α used C-terminal epitope tags, whereas the study with p110β used an N-terminal tag, a Tris-HA cassette inserted between residues 30 and 31.
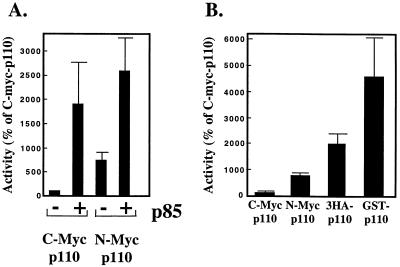
We reexamined this question with HEK 293T cells by transiently expressing bovine p110α containing an epitope tag at either its N or C terminus. In agreement with previous reports, C-terminally myc-tagged p110α had little activity as a monomer, but its specific activity was increased nearly 20-fold by coexpression with p85 (Fig. 5A). Interestingly, placement of the myc tag at the N terminus of p110α increased its specific activity eightfold relative to the activity of C-myc-110. The specific activity of N-myc-p110α was increased approximately threefold more by coexpression with p85, i.e., to a level similar to that seen in cells transfected with C-myc-p110α and p85.
FIG. 5.
Activity of epitope-tagged p110α: comparison of N-terminal versus C-terminal tags. (A) HEK 293T cells were transfected with expression vectors for C-myc- or N-myc-p110α (30 μg) plus control vector or an expression vector for p85 (30 μg). Forty-eight hours after transfection the cells were lysed, and anti-myc immunoprecipitates were prepared. The immune complexes were assayed for lipid kinase activity. Parallel samples were eluted, separated by SDS-PAGE, and blotted sequentially with anti-p110α and anti-p85 antibodies. PI 3′-kinase specific activity was calculated relative to p110α expression, as measured by blotting. Data from different experiments were normalized to the activity of C-myc-p110α in each experiment. All determinations were performed in triplicate, and the data are the means ± standard errors of the means from five separate experiments. (B) HEK 293T cells were transfected with 30 (C-myc-p110α, N-myc-p110α, and GST-p110α) or 45 μg (3HA-p110α) of DNA. The cells were lysed after 48 h, and anti-myc, anti-HA, and anti-GST immunoprecipitates were prepared. Lipid and protein determinations and data normalization were performed as described for panel A. All determinations were performed in triplicate, and the data are the means ± standard errors of the means from four separate experiments.
We next compared the activities of p110α isoforms containing various N-terminal tags. p110α was modified by the addition of myc, Tris-HA, or GST tags as indicated and expressed in HEK 293T cells. As seen before, placement of the myc tag at the N terminus of p110α increased its activity by approximately eightfold relative to C-myc-p110α (Fig. 5B). However, an N-terminal Tris-HA cassette (38 amino acids) increased activity by 20-fold relative to C-myc-p110α, whereas an N-terminal GST tag increased activity by 45-fold (Fig. 5B). It should be noted that the expressed proteins were immunoprecipitated with different antitag antibodies, making it difficult to directly compare their expression levels. Nonetheless, the immunoprecipitates were assayed under identical conditions and were subjected to blotting with the same anti-p110 antibody. Thus, the determination of p110 specific activity was unaffected by potential differences in recovery during immunoprecipitation.
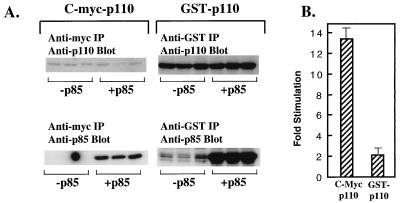
These data suggest that in mammalian cells, the attachment of a bulky N-terminal tag to the amino terminus of p110α can substitute for p85, which also binds to the N terminus of p110α. We therefore examined the effect of p85 on the C-myc-p110 and GST-p110 constructs. HEK 293T cells were transfected with C-myc-p110 and GST-p110 (30 and 5 μg of DNA, respectively) in the absence or presence of p85. Both constructs could associate with p85, as shown by anti-p85 immunoblotting of the anti-myc and anti-GST immunoprecipitates (Fig. 6A, bottom). When we measured the specific activities of C-myc-p110 and GST-p110, we found that the specific activity of C-myc-p110α in cells cotransfected with p85 was increased 13-fold relative to the activity of C-myc-p110α monomers (Fig. 6B). In contrast, the activity of GST-p110 was increased only twofold by coexpression with p85, relative to the activity of GST-p110 alone (Fig. 6B). Thus, p85 significantly increased the activity of C-myc-p110 but had only a small effect on GST-p110. These data suggest that both p85 and the GST N-terminal tag increase the specific activity of p110α by a similar mechanism. This mechanism is unlikely to involve a specific activating protein-protein interaction and is more likely to reflect a stabilization of the overall conformation of p110α. Although GST-p110 is already much more active than C-myc-p110, the ability of p85 to increase its activity by an additional two times suggests that it is still not maximally stabilized. The effect of p85 on an additionally stabilized GST-p110 is examined below.
FIG. 6.
Effect of p85 on C-myc-p110α versus GST-p110α. HEK 293T cells were transfected with expression vectors for C-myc p110α or GST-p110α in the presence of a control vector or the expression vector for p85. (A) Anti-myc or anti-GST immunoprecipitates (IP) were blotted with anti-p110 antibody (top lanes) or anti-p85 antibody (lower lanes). (B) Protein and lipid kinase activities in anti-myc or anti-GST immunoprecipitates were calculated as described in the legend for Fig. 5A. The data are expressed as the fold stimulation for each construct in the presence versus the absence of p85. All determinations were performed in triplicate, and the data are the means ± standard errors of the means from two separate experiments.
To directly test whether p85 stabilizes p110α in mammalian cells, we examined the effect of p85 on the half-life of p110α in metabolically labeled cells. HEK 293T cells were transfected with C-myc-p110α in the absence or presence of p85 and labeled with [35S]methionine for 16 h. The cells were then chased in medium containing cold methionine for the indicated times, and C-myc-p110α was immunoprecipitated with anti-myc antibodies (Fig. 7). In the absence of p85, C-myc-p110α turned over with a half-life of approximately 1 h and was completely gone by 5 h. In contrast, C-myc-p110α/p85 dimers were significantly more stable, with an approximate half-life of 5 h. We also found that the half-life of GST-p110 in [35S]methionine-labeled cells was significantly greater than that of C-myc-p110 (data not shown). Thus, p110α was stabilized by dimerization with p85 or linkage to a bulky N-terminal tag.
FIG. 7.
Stabilization of p110 by p85 in mammalian cells. HEK 293T cells were transfected with vectors for C-myc-p110 in the absence or presence of p85. Thirty-six hours after transfection the cells were labeled with [35S]methionine overnight and then chased for the indicated times in medium containing 10× methionine. The cells were lysed, and immunoprecipitated p110 was separated by SDS-PAGE and visualized by autoradiography. The data are representative of two separate experiments.
Monomeric p110α is temperature sensitive in mammalian cells.
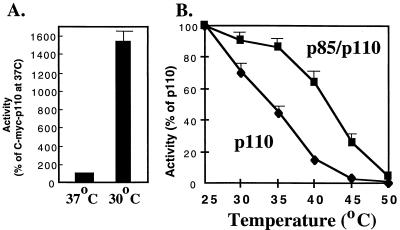
While these data suggest that the rescue of monomeric p110α activity by p85 in mammalian cells reflects a stabilization of the catalytic subunit, they do not explain why this is not observed when the proteins are expressed in insect cells. We noted that a major difference between mammalian and insect culture systems is the low temperature (27°C) used for Sf-9 cells and reasoned that this might affect the stability of a conformationally labile p110α monomer. We therefore compared the activities of C-myc-p110α monomers in HEK 293T cells grown at 37 versus 30°C. Strikingly, the specific activity of C-myc-p110α in HEK 293T cells was increased 15-fold by culture at 30°C (Fig. 8A), consistent with idea that monomeric p110α is conformationally unstable.
FIG. 8.
Effect of temperature on the activity of p110α. (A) HEK 293T cells were transfected with expression vectors for C-myc-p110α and then maintained for 48 h at 37 or 30°C. The cells were then lysed, and protein expression and lipid kinase activities were determined as described in the legend for Fig. 5A. All determinations were performed in triplicate, and the data are the means ± standard errors of the means (SEM) from two experiments. (B) Recombinant p110 or p85/p110 dimers were incubated at the indicated temperatures for 30 min. After being chilled on ice, the samples were assayed for lipid kinase activity at 22°C. The data are the means ± SEM from three separate experiments.
The effect of p85 on thermal stability was examined in vitro by producing p110α monomers and p85/p110α dimers in Sf-9 cells and then incubating them for 30 min at various temperatures prior to assay at 22°C (Fig. 8B). In the absence of p85, p110α was strikingly sensitive to elevated temperatures and lost half its activity after 30 min at 35°C. In contrast, the activity of p85/p110α was minimally affected until temperatures reached 40°C or higher. These data confirm the hypothesis that monomeric p110 is thermally unstable and suggest that p85 increases the activity of p110α in mammalian cells by stabilizing the catalytic subunit.
Inhibition of p110α by p85 in mammalian cells.
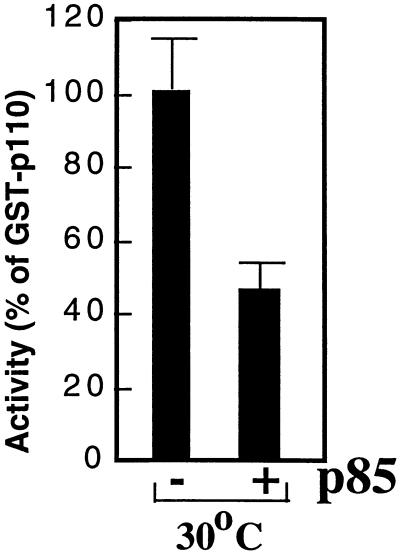
To further reconcile the data derived from the insect and mammalian systems, we sought to determine whether p85 acts as an inhibitor of p110α in mammalian cells. To do this, it was necessary to experimentally segregate the stabilizing effects of p85 from its potential inhibitory effects. We therefore measured the activities of p110α and p85/p110α under conditions designed to maximize the stability of the p110α monomer. We transfected HEK 293T cells with the GST-p110α cDNA in the presence of empty vector or p85 expression plasmid, and cultured the cells at 30°C. p85 immunoblots of anti-GST immunoprecipitates demonstrated that p85 associated with GST-p110α in cells grown at 30°C (data not shown). Interestingly, coexpression of p85 with GST-p110 in HEK 293T cells caused a 50% decrease in the specific activity of GST-p110, compared with the activity of GST-p110 alone (Fig. 9). This result is similar to the data obtained with recombinant baculovirus p110α (Fig. 1 and 2). These data show that under conditions that optimize the stability of monomeric p110α in mammalian cells, p85 inhibits p110α in a manner similar to that seen in insect cells.
FIG. 9.
Inhibition of p110 by p85 in mammalian cells. HEK 293T cells were transfected with GST-p110α in the presence of control vector or p85 expression vector as indicated. After 48 h at 30°C, the cells were lysed and protein and lipid kinase activities were determined. The activities were normalized to the activity of GST-p110α. All determinations were performed in triplicate, and the data are representative of four separate experiments.
DISCUSSION
Our data show that p85 has two distinct effects on the activity of p110α. On the one hand, p85 binds to the N terminus of p110α and inhibits the activity of the p110α monomer. This effect is clearly seen under experimental conditions in which monomeric p110 is stable. On the other hand, p85 stabilizes p110α. This effect is most clearly seen in mammalian cells at 37°C, a temperature at which monomeric p110α is unstable. The net activity of p110α in a given experimental protocol reflects a balance between the inhibitory and stabilizing effects of p85 on p110α.
With regard to the inhibitory effect, we show that the p85 regulatory subunit of PI 3′-kinase decreases the activity of the p110α catalytic subunit produced in Sf-9 cells. Given that the binding of p85 to p110α is extremely stable (45), the basal state of a p85/p110α dimer is inhibited relative to the activity of monomeric p110α. This inhibition appears to be a direct result of dimerization with p85, since it can be readily reconstituted in vitro and in fact can be seen with p85 expressed in bacteria (data not shown). Similarly, we find that inhibition of p110α by p85 is unaffected by mutation of the Ser-608 inhibitory site of p85 or by dephosphorylation of p85 with protein phosphatase 1. We have no information as to the level of Ser-608 phosphorylation in p85 produced in Sf-9 cells and can make no comment on potential additional roles for Ser-608 in the regulation of p110α activity. However, the inhibition we record here is clearly independent of p85 phosphorylation. Our results differ from those of Woscholski et al., who reported that coexpression of p85 with p110 induced a phosphorylation-dependent 17-fold increase in the Km for ATP (45). This increase was only seen by using PI[4,5]P2 as a lipid substrate and was not seen when p85 was reconstituted with p110α in vitro. These data may reflect a different phenomenon than the one described in the present study, since the inhibition we observe is clearly seen with PI, PI[4]P, or PI[4,5]P2 and does not require the coexpression of the two subunits.
We and others have previously shown that the activity of p85/p110α dimers is increased when the SH2 domains of p85 bind to phosphotyrosyl proteins containing appropriate sequence motifs (2, 5). This increase presumably reflects a conformational change induced by phosphopeptide binding, since p85 remains bound to p110 in the presence of phosphotyrosyl proteins (2). Our current data suggest that the increased activity of p85/p110α heterodimers when bound to phosphoproteins such as IRS-1 is not a true activation of p110α, but rather a disinhibition of the p85/p110α heterodimer. Thus, activation of p85/p110α dimers in mitogen-stimulated cells reflects a transition between inhibited and disinhibited states. Using recombinant enzyme and varying the concentration of either lipid or ATP, we find that relative Vmax/Km values for p85/p110α dimers are decreased by 60 to 85% compared to that seen with monomeric p110α. The relative Vmax/Km values for p85/p110α dimers are increased by incubation with bisphosphorylated peptide, to within 75 to 95% of that seen with monomeric p110α. However, the activity of “activated” p85/p110α dimers never exceeds that of an equivalent amount of monomeric p110α.
It is interesting to note that while inhibition of p110α by p85 was seen by using PI, PI[4]P, or PI[4,5]P2 as the substrate, the subsequent activation of the p85/p110α dimers by phosphopeptides was less effective with PI[4,5]P2 as a substrate than with the other lipids. In this regard, Rameh and coworkers have noted that PI[3,4,5]P3 binds to the p85 SH2 domains and can compete with tyrosyl-phosphorylated IRS-1 for p85 binding (30). Thus, it is possible that the in vitro-produced PI[3,4,5]P3 competes with phosphopeptide for SH2 domain occupancy, reducing the resultant activation of p85/p110α dimers.
In contrast to our data obtained with recombinant enzyme produced in Sf-9 cells, previous studies have shown that the activity of p110α is significantly increased when the isoform is coexpressed with p85 in mammalian cells (18), and attachment of the iSH2 domain to p110 has been used to produce a constitutively active enzyme (15). How can one reconcile this apparent activation of p110 by p85 in mammalian cells with the inhibition we observe in Sf-9 cells?
The explanation suggested by our data is that p110α is unstable as a monomer at 37°C but can be stabilized by the binding of p85 to its N terminus. Although the binding of p85 to p110α requires specific interactions between the iSH2 domain of p85 and the N terminus of p110α (12, 14, 19), the stabilization of p110α by p85 appears to be relatively nonspecific. It can be replaced by either synthesis at low temperature or the attachment of a bulky N-terminal tag. The ability of these nonspecific interventions to mimic the effects of p85 suggest that the regulatory subunit acts by stabilizing the overall conformation of the catalytic subunit, rather than by inducing a specific activated conformation in p110α. This is supported by the increased half-life of p85/p110α dimers as opposed to p110α monomers in mammalian cells and by the finding that p85 protects p110α from thermal inactivation. Thus, the apparent activation of p110α by p85 in mammalian cells at 37°C does not reflect the difference between an enzyme in its basal and activated states. Instead, it reflects the difference between a nonfunctional, destabilized enzyme and an enzyme in its basal state.
Of the two effects of p85 on the activity of p110α, the stabilizing effects are predominant when the activities of p110 monomers and p85/p110 heterodimers in mammalian cells at 37°C are compared (a 15- to 20-fold difference). However, the stabilizing effects of p85 are less impressive when the activity of monomeric GST-p110 is compared to that of p85/GST-p110 in mammalian cells (a twofold difference). When GST-p110 is further stabilized by growth of the cells at 30°C, we find that the p110α catalytic subunit is maximally active as a monomer and is inhibited 50% by dimerization with the p85 regulatory subunit. This degree of inhibition is somewhat less than the 80% inhibition we see with p110α from insect cells. The lesser degree of inhibition could be due to interactions between p85 and additional regulatory molecules present in HEK 293T cells. Alternatively, although the conditions were designed to experimentally segregate the stabilizing and inhibitory effects of p85, it is likely that p85 is still somewhat stabilizing, even in mammalian cells at 30°C. This is consistent with the finding that monomeric p110α loses activity after 30 min at 30°C in vitro. Attempts to culture HEK 293T cells at lower temperatures were unsuccessful. Nonetheless, our data show that p85 has qualitatively similar effects on p110α in mammalian and insect cells under conditions that augment the stability of the p110α monomer.
Our data are not inconsistent with the data presented by Klippel et al. and Hu et al., who showed that the activity of C-myc-tagged p110α was rescued by either coexpression with the iSH2 domain of p85 or direct linkage of the iSH2 domain to the N terminus of p110α (15, 18). The iSH2 domain of p85 presumably stabilizes monomeric p110α in much the same way as p85, leading to an apparent activation in mammalian cells at 37°C. The iSH2 domain does not, however, appear to be a p110α activator, since binding of the recombinant iSH2 domain to recombinant p110α in vitro has no effect on p110α activity (46a). Similarly, the effects of a tethered iSH2 domain are mimicked by a tethered GST moiety, suggesting that conformational stability rather than specific activating interactions are involved.
In intact cells, the activity of p85/p110α dimers may also be affected by their binding to GTP-bound ras and CDC42, SH3 domains from Src family kinases, or proline-rich proteins that bind to the p85 SH3 domain (10, 28, 32, 47). It is not yet clear whether these mechanisms of activation are distinct from activation via phosphotyrosine-SH2 domain binding and whether they would lead to levels of activity above that seen with monomeric p110α. Thus, it is possible that activation of p85/p110α above the level of monomeric p110α occurs in the more complex environment of the intact cell. Moreover, an additional contribution to intracellular 3-phosphoinositide production would come from the targeting of p85/p110α to cell membranes. The ability of the p85 regulatory subunit to modulate p110α activity in response to these disparate inputs will be of great mechanistic interest for further studies.
ACKNOWLEDGMENTS
We thank Zhong-Yin Zhang and Steve Almo for numerous helpful discussions. We thank Michael Waterfield for the p110α cDNA and Zhong-Yin Zhang for the recombinant PP1.
This work was supported by grants to J.M.B. from the American Diabetes Association and National Institutes of Health grant GM55692. J.M.B. is an Established Scientist of the American Heart Association, New York Affiliate, and is a recipient of a Scholar Award from the Irma T. Hirschl Trust. J.M. was supported by a fellowship from the Juvenile Diabetes Foundation.
REFERENCES
- 1.Antonetti D A, Algenstaedt P, Kahn C R. Insulin receptor substrate 1 binds two novel splice variants of the regulatory subunit of phosphatidylinositol 3-kinase in muscle and brain. Mol Cell Biol. 1996;16:2195–2203. doi: 10.1128/mcb.16.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backer J M, Jr, Myers M G, Shoelson S E, Chin D J, Sun X J, Miralpeix M, Hu P, Margolis B, Skolnik E Y, Schlessinger J, White M F. The phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booker G W, Gout I, Downing A K, Driscoll P C, Boyd J, Waterfield M D, Campbell I D. Solution structure and ligand-binding site of the SH3 domain of the p85α subunit of phosphatidylinositol 3-kinase. Cell. 1993;73:813–822. doi: 10.1016/0092-8674(93)90259-s. [DOI] [PubMed] [Google Scholar]
- 4.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:231–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter C L, Auger K R, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, Cantley L C. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J Biol Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 6.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhand R, Hara K, Hiles I, Bax B, Gout I, Panayotou G, Fry M J, Yonezawa K, Kasuga M, Waterfield M D. PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J. 1994;13:511–521. doi: 10.1002/j.1460-2075.1994.tb06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhand R, Hiles I, Panayotou G, Roche S, Fry M J, Gout I, Totty N F, Truong O, Vicendo P, Yonezawa K, Kasuga M, Courtneidge S A, Waterfield M D. PI 3-kinase is a dual specificity enzyme: autoregulation by an intrinsic protein-serine kinase activity. EMBO J. 1994;13:522–533. doi: 10.1002/j.1460-2075.1994.tb06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escobedo J A, Navankasattusas S, Kavanaugh W M, Milfay D, Fried V A, Williams L T. cDNA cloning of a novel 85 kD protein that has SH2 domains and regulates binding of PI3-kinase to the PDGF β-receptor. Cell. 1991;65:75–82. doi: 10.1016/0092-8674(91)90409-r. [DOI] [PubMed] [Google Scholar]
- 10.Guinebault C, Payrastre B, Racaud-Sultan C, Mazarguil H, Breton M, Mauco G, Plantavid M, Chap H. Integrin-dependent translocation of phosphoinositide 3-kinase to the cytoskeleton of thrombin-activated platelets involves specific interactions of p85-α with actin filaments and focal adhesion kinase. J Cell Biol. 1995;129:831–842. doi: 10.1083/jcb.129.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiles I D, Otsu M, Volinia S, Fry M J, Gout I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty N F, Hsuan J J, Courtneidge S A, Parker P J, Waterfield M D. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell. 1992;70:419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 12.Holt K H, Olson A L, Moye-Rowley W S, Pessin J E. Phosphatidylinositol 3-kinase activation is mediated by high-affinity interactions between distinct domains within the p110 and p85 subunits. Mol Cell Biol. 1994;14:42–49. doi: 10.1128/mcb.14.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu P, Mondino A, Skolnik E Y, Schlessinger J. Cloning of a novel, ubiquitously expressed human phosphatidylinositol 3-kinase and identification of its binding site on p85. Mol Cell Biol. 1993;13:7677–7688. doi: 10.1128/mcb.13.12.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu P, Schlessinger J. Direct association of p110β phosphatidylinositol 3-kinase with p85 is mediated by an N-terminal fragment of p110β. Mol Cell Biol. 1994;14:2577–2583. doi: 10.1128/mcb.14.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Q, Klippel A, Muslin A J, Fantl W J, Williams L T. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 16.Joly M, Kazlauskas A, Corvera S. Phosphatidylinositol 3-kinase activity is required at a postendocytic step in platelet-derived growth factor receptor trafficking. J Biol Chem. 1995;270:13225–13230. doi: 10.1074/jbc.270.22.13225. [DOI] [PubMed] [Google Scholar]
- 17.Kapeller R, Prasad K V S, Janssen O, Hou W, Schaffhausen B S, Rudd C E, Cantley L C. Identification of two SH3-binding motifs in the regulatory subunit of phosphatidylinositol 3-kinase. J Biol Chem. 1994;269:1927–1933. [PubMed] [Google Scholar]
- 18.Klippel A, Escobedo J A, Hirano M, Williams L T. The interaction of small domains between the subunits of phosphatidylinositol 3-kinase determines enzyme activity. Mol Cell Biol. 1994;14:2675–2685. doi: 10.1128/mcb.14.4.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klippel A, Escobedo J A, Hu Q, Williams L T. A region of the 85-kilodalton (kDa) subunit of phosphatidylinositol 3-kinase binds the 110-kDa catalytic subunit in vivo. Mol Cell Biol. 1993;13:5560–5566. doi: 10.1128/mcb.13.9.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotani K, Yonezawa K, Hara K, Ueda H, Kitamura Y, Sakaue H, Ando A, Chavanieu A, Calas B, Grigorescu F, Nishiyama M, Waterfield M D, Kasuga M. Involvement of phosphoinositide 3-kinase in insulin- or IGF-1-induced membrane ruffling. EMBO J. 1994;13:2313–2321. doi: 10.1002/j.1460-2075.1994.tb06515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Leevers S J, Weinkove D, MacDougall L K, Hafen E, Waterfield M D. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- 25.McIlroy J, Chen D, Wjasow C, Michaeli T, Backer J M. Specific activation of p85-p110 phosphatidylinositol 3′-kinase stimulates DNA synthesis by ras- and p70 S6 kinase-dependent pathways. Mol Cell Biol. 1997;17:248–255. doi: 10.1128/mcb.17.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molz L, Chen Y W, Hirano M, Williams L T. Cpk is a novel class of Drosophila PtdIns 3-kinase containing a C2 domain. J Biol Chem. 1996;271:13892–13899. doi: 10.1074/jbc.271.23.13892. [DOI] [PubMed] [Google Scholar]
- 27.Otsu M, Hiles I, Gout I, Fry M J, Ruiz-Larrea F, Panayotou G, Thompson A, Dhand R, Hsuan J, Totty N, Smith A D, Morgan S J, Courtneidge S A, Parker P J, Waterfield M D. Characterization of two 85 kD proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 28.Pleiman C M, Hertz W M, Cambier J C. Activation of phosphatidylinositol-3′ kinase by Src-family kinase SH3 binding to the p85 subunit. Science. 1994;263:1609–1612. doi: 10.1126/science.8128248. [DOI] [PubMed] [Google Scholar]
- 29.Pons S, Asano T, Glasheen E, Miralpeix M, Zhang Y, Fisher T L, Myers M G, Jr, Sun X J, White M F. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol Cell Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rameh L E, Chen C-S, Cantley L C. Phosphatidylinositol (3,4,5)P3 interacts with SH2 domains and modulates PI 3-kinase association with tyrosine-phosphorylated proteins. Cell. 1995;83:821–830. doi: 10.1016/0092-8674(95)90195-7. [DOI] [PubMed] [Google Scholar]
- 31.Roche S, Koegl M, Courtneidge S A. The phosphatidylinositol 3-kinase α is required for DNA synthesis induced by some, but not all, growth factors. Proc Natl Acad Sci USA. 1994;91:9185–9189. doi: 10.1073/pnas.91.19.9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Viciana P, Warne P H, Vanhaesebroeck B, Waterfield M D, Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 33.Rordorf-Nikolic T, Van Horn D J, Chen D, White M F, Backer J M. Regulation of phosphatidylinositol 3′-kinase by tyrosyl phosphoproteins. Full activation requires occupancy of both SH2 domains in the 85-kDa regulatory subunit. J Biol Chem. 1995;270:3662–3666. doi: 10.1074/jbc.270.8.3662. [DOI] [PubMed] [Google Scholar]
- 34.Ruderman N, Kapeller R, White M F, Cantley L C. Activation of phosphatidylinositol-3-kinase by insulin. Proc Natl Acad Sci USA. 1990;87:1411–1415. doi: 10.1073/pnas.87.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schu P V, Takegawa K, Fry M J, Stack J H, Waterfield M D, Emr S D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 36.Skolnik E Y, Margolis B, Mohammadi M, Lowenstein E, Fischer R, Drepps A, Ullrich A, Schlessinger J. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 37.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 38.Stephens L R, Eguinoa A, Erdjument-Bromage H, Lui M, Cooke F, Coadwell J, Smrcka A S, Thelen M, Cadwallader K, Tempst P, Hawkins P T. The Gβgamma sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell. 1997;89:105–114. doi: 10.1016/s0092-8674(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 39.Stoyanov B, Volinia S, Hanck T, Rubio I, Loubtchenkov M, Malek D, Stoyanova S, Vanhaesebroeck B, Dhand R, Nurnberg B, Gierschik P, Seedorf K, Hsuan J J, Waterfield M D, Wetzker R. Cloning and characterization of a G protein-activated human phosphoinositide-3 kinase. Science. 1995;269:690–693. doi: 10.1126/science.7624799. [DOI] [PubMed] [Google Scholar]
- 40.Sun X J, Rothenberg P, Kahn C R, Backer J M, Araki E, Wilden P A, Cahill D A, Goldstein B J, White M F. The structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 41.Tolias K F, Cantley L C, Carpenter C L. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 42.Vanhaesebroeck B, Welham M J, Kotani K, Stein R, Warne P H, Zvelebil M J, Higashi K, Volinia S, Downward J, Waterfield M D. p110-delta, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virbasius J V, Guilherme A, Czech M P. Mouse p170 is a novel phosphatidylinositol 3-kinase containing a C2 domain. J Biol Chem. 1996;271:13304–13307. doi: 10.1074/jbc.271.23.13304. [DOI] [PubMed] [Google Scholar]
- 44.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall L K, Stein R, Zvelebil M J, Domin J, Panaretou C, Waterfield M D. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woscholski R, Dhand R, Fry M J, Waterfield M D, Parker P J. Biochemical characterization of the free catalytic p110α and the complexed heterodimeric p110α · p85α forms of the mammalian phosphatidylinositol 3-kinase. J Biol Chem. 1994;269:25067–25072. [PubMed] [Google Scholar]
- 46.Yao R, Cooper G M. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 46a.Yu, J., and J. M. Backer. Unpublished data.
- 47.Zheng Y, Bagrodia S, Cerione R A. Activation of phosphoinositide 3-kinase by Cdc42Hs binding to p85. J Biol Chem. 1994;269:18727–18730. [PubMed] [Google Scholar]
- 48.Zvelebil M J, MacDougall L, Leevers S, Volinia S, Vanhaesebroeck B, Gout I, Panayotou G, Domin J, Stein R, Pages F, Hoga F, Salim K, Linacre J, Das P, Panaretou C, Wetzker R, Waterfield M. Structural and functional diversity of phosphoinositide 3-kinases. Philos Trans R Soc Lond. 1996;351:217–223. doi: 10.1098/rstb.1996.0019. [DOI] [PubMed] [Google Scholar]