Abstract
Advances in a scientific discipline are often measured by small incremental steps. Here we report on two intertwined disciplines in the protein structure prediction field, the modeling of single chains and complexes, that have over decades emulated this pattern, as monitored by the community-wide blind prediction experiments CASP and CAPRI. However, over the last few years, dramatic advances were observed for the accurate prediction of single protein chains, driven by an upsurge of deep learning methodologies entering the prediction field. We review the main scientific developments that enabled these recent breakthroughs and feature the important role of blind prediction experiments in building up and nurturing the structure prediction field. We discuss how the new wave of AI-based methods is impacting the field of computational and experimental structural biology and highlight areas of future developments that deep learning methods are likely to thrust forward, provided major challenges are overcome.
Keywords: Protein-structure-prediction, CASP, CAPRI, artificial-intelligence, protein-interactions
Introduction
The problem of predicting the native 3D structure of a protein from its amino acid sequence has occupied the center stage in protein modeling research for over 5 decades owing to its inherent scientific interest and to the many potential applications that robust structure prediction algorithms would offer in areas such as the prediction of function from genome sequence, or designing new drugs to treat disease (104). In comparison, although the important functional role of protein-protein interactions and complexes was recognized in the sixties, methods for predicting the structure of complexes became a booming research area only since the turn of the century(136), fueled by the realization of the ubiquitous involvement of protein complexes in nearly all cellular processes.
The last decade has seen major advances in both types of prediction methodologies, due to a variety of factors. Notable has been the application of artificial intelligence (AI) methods, culminating with the recent phenomenal success of the AI-based algorithm AlphaFold2 by DeepMind in predicting the structures of single protein chains to accuracy levels rivaling with experimental methods (53). Important in nurturing and catalyzing these developments have been the blind prediction experiments of CASP (Critical Assessment of Structure predictions and CAPRI (Critical Assessment of PRedicted Interactions), focusing respectively, on the critical assessment of methods for the predicting the structure of proteins and protein complexes.
In this review we outline the progression in the methods developed for these two prediction tasks. We describe how the performance of prediction methods is evaluated by CASP and CAPRI and how progress is assessed. We highlight the role of blind predictions in building up the communities of methods developers and shaping the field. We end by offering our view on the impact the new wave of AI-based methods is having on the field of computational and experimental structural biology, and where the remaining challenges lie.
Prediction of protein 3D structure from sequence
Computational analysis of protein structures has been initiated by Shneior Lifson and his group in the 1960s, who extended the molecular mechanics approach developed for modeling small organic molecules to large molecular systems (42, 43). They introduced the “Consistent Force Field” (CFF) energy function, which led to the development of some of the most important all-atom potentials used today in protein modeling, including CHARMM (11), Amber (133) and ECEPP(50). All three potentials include covalent, non-covalent, and electrostatic energy terms as in the original CFF, with some additional terms, specific to each force field. These classical potentials have served well whenever various intrinsic properties of the protein needed to be investigated in vacuum; however, they were proven inadequate for a thermodynamic description of stable compact protein folds in solution, and unable to discriminate between native proteins and incorrectly folded models (97). The main reason was the failure to account for solvation effects, an important determinant of protein stability. These effects were usually incorporated by using these potentials in molecular dynamics (MD) simulations of the protein immersed in a box of explicit solvent molecules, an exercise, which remained prohibitive for protein structure prediction due to its computational burden, leading to problems of convergence and inadequate conformational sampling.
The next step forward was the addition of implicit solvation terms to the classical potentials. These included surface area-dependent empirical models used in conjunction with atomic solvation parameters (30) and continuum electrostatic terms evaluating the electrostatic contribution to the solvation free energy, using the finite difference Poisson–Boltzmann (PB) method (38) and various approximations to the Generalized Born (GB) treatment (99). Augmented with a surface area-based term to represent the nonpolar contribution to solvation and integrated into the classical potential functions, the resulting force-fields could identify the native states of peptides or proteins, albeit with limited accuracy (45, 147).
Such problems led to interest in extracting effective potentials from experimentally determined protein structures. A frequently used approach to deriving such potentials consists of computing frequencies of structural features (‘structural frequencies’) and converting these frequencies into free energy contributions (123). Following this approach many statistical (or knowledge-based) potentials were proposed (52, 117). Most of these potentials used simplified residue-based representations of the protein, reminiscent of the coarse-grained potentials used decades earlier in protein folding calculations (85). These relatively simple, computationally efficient potentials helped score and rank predicted protein models. When combined with various energy optimization methods they were also able to model the structure of very small proteins from their amino acid sequence, the so-called ab-initio protein modeling approach. But sampling the vast conformational space of average size proteins remained a problem. To address it, data on proteins sequences and known protein structures were increasingly relied upon.
Since evolutionarily related proteins adopt similar 3D structures(22), with the increasing number of experimentally determined protein structures this property gave rise to the method of homology modeling, also known as comparative- or template-based- modeling (29). The atomic-resolution structure of the "target" protein is modeled from its amino acid sequence and an experimental three-dimensional structure of an evolutionary related protein (the template). Aligned regions of the template backbone are simply copied into the target, whereas special prediction methods are used for adding loops in the non-aligned regions (32) and also for placing the side chains of non-conserved residues (32), and the resulting models are refined using molecular mechanics or molecular dynamics methods.
A notable development in conformational sampling, which in some ways bridges template-based and ab-initio methods has been the fragment-based assembly approaches, whereby models are built from short contiguous backbone fragments (typically 3–15 residues in length) taken from proteins of known structure, and assembled into full length models using Monte-Carlo simulated annealing or equivalent techniques (67, 119).
The next major advance in protein modeling has been the effective use of co-evolutionary information, enabled by the growing number of related sequences (39). The underlying hypothesis was that if mutations occurring at two positions in the aligned sequences are correlated, then these positions are likely to form a contact in 3D space (103). To find true evolutionary covariation between residues is difficult because one must minimize the effect of transitive correlations: indirect correlations that are observed, for example, when two residues contact the same third residue but do not actually contact each other. Transitive correlation can be removed by global statistical approaches involving either direct coupling analysis (90), pseudo-likelihood optimization (56), or machine learning (134). The approach has first been used to identify residue pairs that are in contact, and further extended to derive residue distance and dihedral angle distributions, all used as restraints in ab-initio modeling (103, 143). The more recent neural network-based learning methods further extend the use of multiple sequence alignment to end-to-end protein structure prediction, achieving previously unimaginable accuracy for a significant fraction of proteins (53), as will be discussed in the last section of this review.
Prediction of the 3D structure of protein complexes
Efforts to model protein-protein interactions started in the nineteen seventies, driven by the desire to explain aberrant protein-protein interactions caused by a single point mutant in sickle-cell hemogolobin (Hb-S)(84). The first protein docking algorithm, formulated as the task of modeling the atomic structure of a native protein complex from the structures of its components, was developed a few years later. This early incarnation of docking treated the interacting proteins as rigid bodies, used a coarse-grained representation of the protein developed for protein folding calculations (85) and searched for large surface patches with complementary shapes. Shape complementary was evaluated by the interface area formed by the contacting proteins (21), a geometric quantity representing the loss of solvent accessible surface area upon binding, itself related to the hydrophobic contribution to the binding free energy (20).
Ab-initio docking methods
Over the following two decade a variety of docking procedures were proposed (136, 137), including most notably the Fast Fourier Transform (FFT)-based methods (57) that currently dominate the field of protein docking. FFT-based methods enable speedy coarse-grained rigid-body searches capable of detecting shape complementary, as well as the evaluation of different properties of protein interfaces, such as hydrophobicity (127), or electrostatic and van der Waals interactions (9). Following these advances strategies were proposed to speed up high-resolution searches, required for accurately defining the molecular positions and orientations. These include the use of spherical polar Fourier expansion coefficients, shown to significantly accelerate the search for solutions that optimize properties of generated interfaces (112).
A series of rigid docking algorithms with variations on these fundamental principles (19, 35, 60, 112) underpin most of the docking procedures used today. One in particular forms the basis of a well frequented automatic docking server, ClusPro (62), which enables the reduction of the search space from 6 to 5 degrees of freedom by employing a Fourier transform in polar coordinate space, resulting in a 10-fold speed up over classical FFT approaches without compromising accuracy (100). A few alternative sampling algorithms such as enhanced sampling Monte-Carlo procedures (142), and algorithms incorporating heuristic methods based on Particle Swarm Optimization (PSO) (89) also hold their own.
Also worth mentioning, is the so-called data-driven docking methods, which involve the incorporation of distance restraints obtained from biophysical or biochemical data into the modeling protocol, thereby reducing the search space for the location of the native complex. One of the first approaches using these principles, now operating as a publicly available server (24), is the program HADDOCK (28). Similar restraints-based methodologies have been added to other protein docking methods (102, 132, 139). More recently, inter molecular contacts derived from data on residue co-evolution were also used as restraints in docking calculations with however, modest success (108).
Scoring docking poses
To further prioritize the large number of solutions (often in the thousands) produced by the docking calculations, these solutions are re-ranked using more sophisticated scoring functions. An important requirement for such functions has been that they be able to reliably percolate the most native-like binding modes to the top of the list. Aware of the challenge, the development of scoring schemes has been a major focus over the past two decades. These schemes span a wide range and are often combined with model optimization. Use is being made of atom or residue pair potentials, sometimes in combination with classical potential energy terms, but increasingly implementing different flavours of knowledge-based potentials. The latter are adapted from those developed for the structure prediction of single protein chains (46, 48, 145, 146). Among the most effective, are methods combining knowledge-based potentials with the evaluation of interatomic contact areas using Voronoi tessellation(98), methods that enrich knowledge-based potentials with evolutionary relationships (96), and methods augmented with deep learning models (86) or replaced with such models (111). A notable example is the Rosetta all-atom multi-component energy function (2), for its broad use in various molecular modeling applications including the evaluation of docking models. For many scoring schemes the rank of native-like solutions can be bolstered by clustering the top ranking docking poses based on the similarity of their interfaces and using cluster size and stability to perturbation(61) to rank models alone or as part of more complex ranking procedures (62).
Handling protein flexibility
With rigid-body search algorithms making up the core component of most docking procedures, it not surprising that these procedures do poorly when the interacting proteins exhibit moderate to high levels of flexibility (26). Nevertheless, with modifications to some rigid-body docking algorithms for so called ‘soft docking’ - allowing for some atomic clashes to be alleviated subsequently by standard molecular dynamics - this problem can be alleviated to a point. Another strategy coined ‘ensemble docking’ involves generating ensembles of conformers for individual components of a complex by molecular dynamics(121) or normal mode analysis (27), and systematically docking conformers from both ensembles to one another, with however mitigated results (71).
The inability to adequately address flexibility led to the design of algorithms, which introduced protein backbone movements and sidechain repacking of putative interface residues during the sampling process, with some success recorded for small to medium conformational changes upon complex formation (4, 10). However, to this day, it remains debatable as to whether modelling protein flexibility using available methods improves the quality of docked models sufficiently to justify the typical higher computational cost entailed (107). Nevertheless, new methodologies are continuing to address the problems associated with significant conformational change upon complex formation, some of which include a new wave machine learning approaches (44).
Template-based docking
Template-based docking was borne out by the increasing success of the homology modeling techniques for single protein chains, described above. With the growing number of available experimentally determined protein structures, it was soon realized that homology- or template-based modeling may be extended to pairs of homologous complexes, if at least some of their component parts show a degree of sequence similarity (3). This then led to the concept that the 3D structure of a complex can be modelled directly from the experimentally determined structures of other complexes. The accuracy of the method hinges on sensitive sequence searches and alignment to the appropriate complexed proteins (120). Interestingly, bearing in mind the constraints on accuracy described above, enough experimentally determined protein complex templates are available to model most native protein-protein interactions for any organism (70) - a concept underlined by the successful employment of the methodology in a number of recent CAPRI blind trials (74). Rapid searching for homologous complexes is now supported by annotated databases of such predicted relationships (69).
Scoring and ranking models derived from template-based docking conforms to the same principles as for classical docking, but with the potential advantage of having to score and rank fewer models. However, there is an obvious caveat, if the modelled complexes do not comply with the principle of conservation of homologous interfaces, the native complex will not be sampled. This contrasts with the ab-initio methods, where there is always a chance, provided flexibility does not dominate, of at least having a near-native model in the complete list of models generated. Moreover, there are clearly certain categories of interactions that are not conducive to this form of docking, the classic examples being antibody antigen complexes, where evolutionary relationships between the binding partners is not expected to be prevalent, thereby enforcing ab-initio docking methods (40).
Ab-initio and templates-based docking methods are clearly not mutually exclusive, and procedures are actively being developed to employ both in order to model the widest range of interactions possible, and to the highest level of accuracy, should the appropriate levels of sequence homology prevail (140); such pipelines have already been encoded into some automatic docking servers (109, 141). With the recent advances in deep learning approaches (see final section), both for components and full complex modelling, these two principal methodologies are likely to become seamlessly merged.
The blind prediction challenges
CASP
Critical Assessment of protein Structure Prediction (CASP) is a community-wide double-blind experiment for testing and comparing protein structure prediction (94). Every two years sequences of soon-to-be experimentally determined protein structures are collected and passed on to registered predictors. Predictors fall into two categories: teams of participants who usually have a period of three weeks to complete their work; and automatic servers, which must return a model within 72 hours, in principle without human intervention. Predictions are evaluated by independent assessors using well developed criteria. CASP provides research groups with an opportunity to test their protein structure prediction methods and delivers an independent assessment of the state of the art in protein structure modeling to the research community and software users. The results show what progress has been made during the previous two years and expose where future approaches should focus to improve the methodology.
The CASP experiment had a fairly modest start in 1994 with 35 participating research groups (95). Targets were provided in three prediction categories: comparative modeling, fold recognition or threading, and ab initio folding (95). The results of CASP1 demonstrated a sobering failure of prediction methods using physics-based potentials, shocking the protein folding community. The only meaningful predictions have been obtained using comparative modeling, and only for easy targets with closely related known structures. Such negative results made it difficult for protein scientist to continue publishing theoretical papers without participating in CASP. The pre-eminence of template-based approaches was further emphasized by CASP2 in 1996. By that time CASP has become more recognized as a much needed “blind” experiment that had the potential to introduce a new area of reproducibility and openness in protein structure prediction. The number of participating groups grew to 70, and there was some improvement in the predictions of more difficult targets (Figure 1). Improved sequence alignment tools, the use of multiple templates, and fragment assembly approaches further improved results at CASP3 and CASP4, but after that, improvements remained moderate, essentially until CASP13 in 2018. Although contacts maps based on co-evolutionary information were already present in several methods at CASP10 to CASP12, their effective use with deep learning led to a jump in prediction quality only at CASP13 (2018, about 100 participating groups), particularly for difficulty targets (Figure 1). While this improvement was already very significant, CASP14 in 2020 lead to a revolution by AlphaFold2, a neural network based end-to-end prediction method that will be further discussed in this paper. However, predictions by other predictor groups and servers have also become much better (Figure 1).
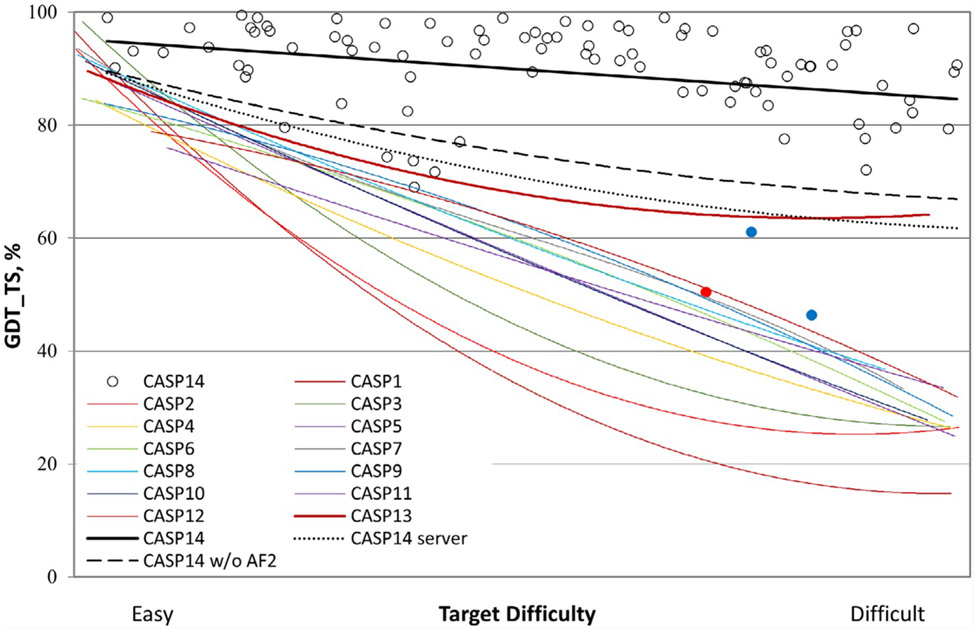
Figure 1: Backbone accuracy of the best models in each of the 14 CASP rounds.
Individual target points are shown for CASP14. The two targets with the lowest agreement with experiment are colored blue are NMR structures and red point represent the model of a a subunit of a cryo-EM-derived large heteromeric structure. The agreement metric, GDT_TS, is a multi-scale indicator of the closeness of the Cα atoms in a model to those in the corresponding experimental structure, and is reported as a percentage, ranging from 0 to 100. Because of experimental errors and artifacts, models with GDT-TS >90 are considered compatible with experiment in backbone accuracy (65). Target difficulty is based on sequence and structure similarity to other proteins with known experimental structures. (Reproduced from reference (66) with permission).
The CASP predictions are evaluated using a variety of quality measures (63) that are listed on the Protein Structure Prediction Center website (https://predictioncenter.org/). The most important measure is GDT_TS (Global DistanceTest Total Score), shown in Figure 1. Another important measure is the Local Distance Difference Test (lDDT), a superposition-free score that evaluates local distance differences of all atoms in a model, including validation of stereochemical plausibility. However, the ranking of CASP predictions is generally based on GDT_TS, computed over the alpha carbon atoms and reported as a percentage, ranging from 0 to 100, with higher values indicating a closer fit of a model to a given reference structure (see Figure 1 for details).
At CASP14 the targets were assigned to one of four classes of modeling difficulty, based on sequence and structure similarity to already experimentally determined structures: ‘TBM-Easy’ for straightforward template modeling targets, ‘TBM-Hard’ for more difficult homology modeling targets, ‘FM/TBM’ for those with only remote structural homologies and ‘FM’ (Free modeling) for the most difficult set with no detectable homology to known structures (65). However, with the significant improvement in prediction quality, for the ongoing CASP15 the distinction between template-based and template-free modeling is eliminated. As shown in Figure 1, until CASP13 the predictions were becoming substantially less accurate as the level of difficulty increased. However, this changed in 2018 with the introduction of deep learning methods at CASP13 that were able to yield predictions with GDT_TS over 60 for the most difficult targets. This trend was further strengthened at CASP14 (the black straight-line), as prediction quality starts at a GDT_TS of about 95, and rarely goes below 80. Although the outstanding performance at CASP14 is dominated by AlphaFold2, the dashed black line in Figure 1 shows that other groups also made substantial advances.
As shown in Figure 1, at CASP14 automated servers had similar performance to human groups (without the results for Alphafold2). Lastly, we note that the quality of many protein structure prediction servers is continuously evaluated by CAMEO (Continuous Automated Model EvaluatiOn), a fully automated assessment platform, which is a complement to the bi-annual CASP experiment (41, 113).
CAPRI
CAPRI (Critical Assessment of Predicted Interactions) is a community-wide experiment inspired by CASP. It was established in 2001 (51) to offer computational biologists the opportunity to test their algorithms in blind predictions of experimentally determined 3D structures of protein complexes, the ‘targets’, provided to CAPRI prior to publication. Experiments focusing on this prediction task were attempted only twice before, including once in 1996 by CASP (51), attracting limited interest. Since its inception, CAPRI prediction rounds have been managed in collaboration of the PDBe (Protein Databank Europe), at the European Bioinformatics Institute (EBI) ( https://www.ebi.ac.uk/pdbe/complex-pred/capri/ )
Due to the slower rate at which structures of protein complexes are being determined and offered as targets for prediction, CAPRI runs prediction rounds on a rolling basis, as targets become available (51). Like in CASP, participants include automatic servers, which must return models within 72 hours, and human predictors who are given 6-8 weeks to complete their predictions. Initially limited to homo- and hetero- protein-protein complexes, the panel of targets diversified to include protein-peptide interactions, complexes of proteins with RNA, DNA and oligosaccharides(78). With time, target size and complexity also increased, especially with the availability of large multimeric complexes solved to high resolution by cryo-EM. Recognizing the essential role scoring functions play in identifying native-like association modes, CAPRI also offers a scoring challenge upon the completion of each prediction round. In this challenge a larger set of anonymized models predicted by different groups and comprising both correct and incorrect binding modes, is made available to all participants to test new scoring functions independently from docking calculations (78, 82). The data comprising the consolidated ensemble of predicted complexes made available in the CAPRI scoring experiments have been compiled in a freely available benchmark dataset (the ScoreSet(83)), recently extended to include 19000 models of diverse complexes predicted by different methods for all CAPRI targets whose structure has been published.
The way the prediction problem is formulated for a given target has likewise evolved over the years to ensure the best use of the available data on the known structures of single protein chains and complexes. In early CAPRI rounds, predictor groups were offered the coordinates of the unbound structures for the components of the complex to predict. Occasionally the bound conformation of one of the components was provided as input in random orientation with sidechain coordinates stripped away (51). As the repertoire of 3D structures of single protein chains was progressive filled thanks in particular to the structural genomics initiatives (18), participants were invited to predict the structure of the target assembly starting from sequence information alone. This requires the integration of homology-based modeling of individual subunits with docking calculations or relying entirely on template-based modeling when adequate templates for the entire complex are available (see Methods). Identifying adequate templates is not trivial and has been a task members of the CAPRI community, who specialize in ab-initio docking calculations, had to learn to master using available resources such as HHPRED(36), PPI3D(23) or GalaxyWEB(59). The increased reliance on homology and template-based modeling was further catalysed starting in 2014, when CASP included the prediction of protein assemblies in their biannual prediction season in collaboration with CAPRI (80); a collaboration that has been continuing since.
Model quality measures
Objective criteria for independently evaluating the quality of predicted models by comparing them against the target structure, are a key component of blind prediction challenges such as CAPRI and CASP. When dealing with models representing protein complexes comprising two or more interacting partners, the evaluation criteria need to account for both local and global parameters of the molecular assembly. In CAPRI, the standard evaluation protocol involves the evaluation of three parameters for a given pair of interacting subunits (76): Two are based on residue-residue contacts and local backbone similarity respectively, and quantify different aspects of the fit between the model and the target binding interfaces. The third parameter quantifies the relative rigid-body global displacement between the binding partners in the model versus the target. Based on ranges in the values of these 3 parameters, defined by expert evaluation and validated by the community, models are assigned to 4 discrete categories: high-, medium-, and acceptable quality and incorrect (76). For targets representing higher order assemblies with multiple distinct interfaces, submitted models are evaluated by comparing each pair of interacting subunits in the model to each of the relevant pairs of interacting subunits in the target(80). The quality scores of the individual interfaces are then used to derive a global quality score for the full assembly. Several different formulations were tested over time, with the latest being a weighted sum of the quality scores of individual interfaces of the assembly (74).
This interface-centric evaluation reflects the essential role of the binding interface in defining the 3D structure of the complex. Even models that reproduce the structure of the native interface to lower accuracy (acceptable or medium accuracy) provide useful information that can be further exploited, whereas incorrect models that totally miss the binding interface are in general of little utility. The performance of individual groups is therefore ranked based only on the best model of respectively, acceptable- medium- or high- quality each group produces for a given target. This results in a coarse-grained ranking, which has its advantages (79), but may also be deemed cumbersome in comparison to continuous model quality metrics, which can be used to evaluate performance at different graininess levels and are more amenable to statistical analyses. The continuous DockQ metric, formulated as a weighted sum of the CAPRI model quality criteria(7) (see also Figure 2a) is an attractive alternative that CAPRI and other studies(13) have already been using to analyze prediction results. Still missing however, are evaluation criteria that seamlessly integrate quality measures of the interface region with those of the remainder of the protein structure.
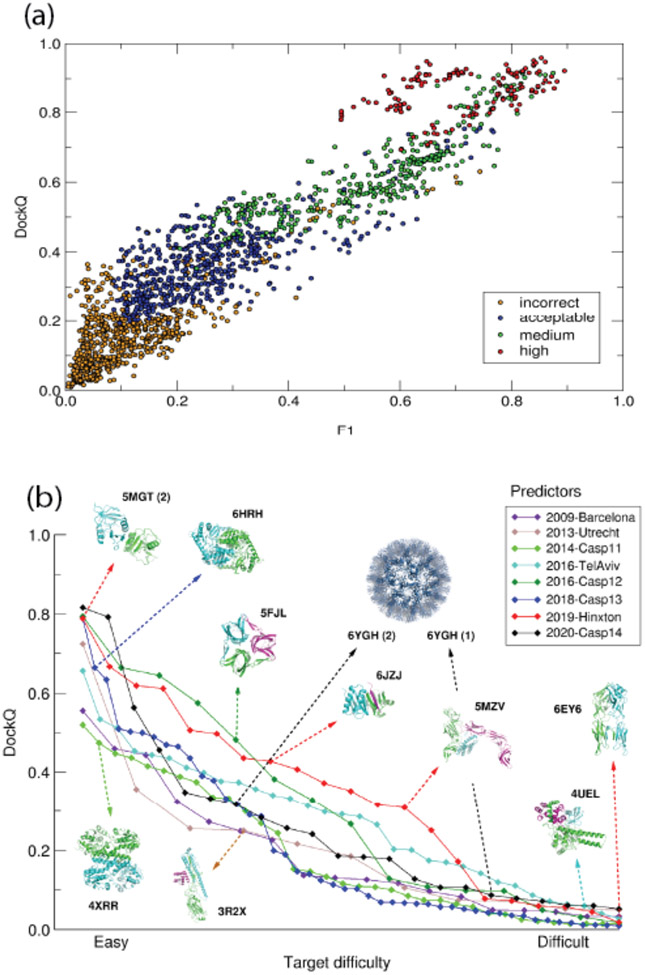
Figure 2: Accuracy levels of the best models of protein complexes in CAPRI and CASP-CAPRI prediction rounds, and the relation of the DockQ score to the CAPRI model quality categories.
(a) Scatter plots of DockQ values for models submitted by predictors for individual targets evaluated in ref (78) (vertical axis) as a function of f1 (f1= 2fnat (1-fnon_nat)/ (fnat + (1-fnon_nat)), where fnat is the fraction of native contacts recalled in the model, and (1-fnon_nat) is the fraction of predicted contacts that are native (79). Individual points are color-coded according to the CAPRI model quality category: incorrect (yellow), acceptable (blue), medium (green), high(red).
(b) DockQ values for the best models as a function of target difficulty. Individual colour coded plots refer to best models evaluated in individual CAPRI assessment periods and CASP-CAPRI prediction rounds between 2009-2020, as indicated in the legend. Examples of targets of different difficulty levels are shown together with their PDB-RCSB codes. Arrows indicate the rounds in which they were offered; numbers shown in parentheses following the PDB codes refer to the models of individual interfaces of the targets in question. Target difficulty is based on sequence and structure similarity to other proteins with known experimental structures.
Evaluating progress
CAPRI has been evaluating progress in intervals of about 3 years, with each evaluation performed on results for 10 – 30 targets achieved during the prediction rounds of the intervening period. In addition, the CAPRI team, which has been evaluating CAPRI prediction results since its inception, independently evaluated the results obtained by participants in the assembly prediction challenges of the CASP11-CASP14 prediction seasons. All the evaluations were performed on results achieved by human predictor groups (~40 on average), by automatic servers (increasing from about 3 to as many as 12 over the years), and by participants in the scoring challenges (15-20 groups on average). Detailed evaluations of these results and assessments of the progress achieved by the community have been reported in the associated publications, amply cited in this review. Here we provide a bird’s eye overview of the main trends.
Figure 2b plots the quality of predicted binding modes measured by the average DockQ score across human predictor groups as a function of the level of modeling difficulty of the corresponding targets. These binding modes were those evaluated in the successive dated periods since 2009, setting the chronological order of the results. The plots clearly illustrate the substantial variability in target difficulty levels during individual evaluation periods that persists over time, highlighting the challenge of evaluating differences in performance across time periods and challenges. Nonetheless, one observes that model quality improves with time for easy and medium difficulty targets but remains low for difficult targets. Examples of targets in different modeling categories and typical characteristics of these targets detailed in Figure 2b, clearly indicate that the modeling challenge differs substantially depending on the system at hand. Some large multi-component assemblies solved to high resolution by cryo-EM (not shown) are particularly challenging to model when they combine several characteristics of difficult to model complexes.
An important contribution of CAPRI community has been the development of automatic servers, the performance of which has steadily improved and diversified to the point of often rivaling with those of human predictors(75). Several of the best performing servers such as ClusPro(62), GalaxyPPDock (73), MDockPP (47) integrate docking procedures with template-based modeling and offer a panoply of handy tools for various modelling tasks, therewith gaining popularity with the wider scientific community.
Another area where CAPRI helped breaking new grounds is the prediction of protein-peptide complexes. This is an important category of complexes for which interest is rapidly growing given the important role recognition of short peptide motifs by protein domains plays in many regulatory processes (128). Recent methods, also implemented in several automatic servers, rose to the task of mastering the problem of modeling this challenging category of transient complexes, often to medium and high accuracy, despite their small binding interfaces, and the significant degree of flexibility of the bound peptide ligands(78).
For the other modeling problems CAPRI occasionally offers, such as the prediction of protein-nucleic acid complexes (81) of interface sidechain conformations and positions interfacial water molecules (77), or for protein oligosaccharide complexes, encouraging results were obtained (78). But the number of targets for these complexes was too low to draw any conclusions.
Considering the state of protein science before these blind prediction experiments, it is difficult to imagine that the current level of prediction technology could have been reached without CASP leading the way. With CAPRI following suit a few years later, both experiments created higher level of transparency not only in protein structure prediction but in computational biology in general, requiring the source code for most publications. They also built competitive and yet collaborative communities, promoting the increased exchange of ideas, and thus speeding up method development.
The breakthrough of DL-based prediction method: The current state of play
The last few years have witnessed a breakthrough in modelling the 3D structure of proteins. This breakthrough can be attributed to two primary factors. One is the extraordinary growth in protein sequence databases (126) coupled with a less prolific, yet notable, growth in the database of experimentally determined structures (131), both freely available in public depositories. The second is the progressive introduction of cutting-edge methods in deep learning to a maturing protein modelling field (5, 122, 124). A key role was also played by the community-wide initiatives that enabled the critical evaluation of the recent breakthrough methods for predicting the structure of single protein chains, recorded in the CASP13 and CASP14 prediction seasons(16). Without these three components the extraordinary achievement of the company Google DeepMind (118) would not have been possible. In the following we examine the impact of this remarkable achievement on charting the structural landscape of native proteins and their complexes using computations and experiments.
Predicting the structure of single chains
For the reasons invoked above, deep learning, a subfield of machine learning that utilises multi-layered artificial neural networks to extract patterns within large datasets, without the need to explicitly define their features (72, 114), was uniquely primed to make substantial contributions to protein structure prediction field. Taking advantage of this, the DeepMind scientists designed and employed their prediction engine AlphaFold2 (53) in the CASP 2020 season (54). This engine was trained on approximately 170,000 experimentally determined protein structures in the PDB (8) and massive data on multiply aligned protein sequences of related proteins, many of unknown structure (126), to produce models that rival in accuracy with experimentally determined protein structures, surpassing the results of their first program, AlphFold1 (116) shown to performed well 2 years earlier (115). The power of AlphaFold2 lies with its novel multi-component architecture that jointly embeds features from multiple sequence alignments (MSAs) and a residue pair representation, encoding spatial relationships between residues, and integrates graph-based components with attention learning (58). The pipeline also involves iterative refinements of predicted residue interactions based upon their predicted interactions to other residues, enabling it to encapsulate structural features to a higher level of accuracy in a fully differentiable end-to-end deep learning method(53).
To enable the community benefit from their monumental advance, DeepMind made their source code for the trained model of AlphaFold2 freely available to anyone wishing to make new predictions [https://github. com/deepmind/alphafold]. Following suit, the group of David Baker released their deep learning protocols for protein prediction and design, RoseTTAfold an algorithm exploring similar ideas to those of AlphaFold (6). In a further move to accelerate scientific research, DeepMind partnered with the European Bioinformatics Institute (EBI), to create AlphaFold-DB(130) providing access to predicted structures of single protein chains for the human proteome and other key organisms, as well as to the majority of the manually curated Uniport entries (SwissProt), with plans to further extend the coverage to over 100 million catalogued proteins, on 2022.
This new treasure trove of structural data and the associated software tools has had a watershed effect on the field of computational and experimental structural biology, generating a flurry of studies (16). Examples include the optimization of multiple sequence alignments fed into AlphaFold2 (12), and a community-wide study evaluating various aspects of the structural information that AlphaFold produced and the applications it enables (1). Evaluating these applications, has been greatly aided by two confidence measures Alphafold2 assigns to its predicted structures: a per-residue measure of confidence assigned to the local backbone structure, and another measuring the confidence associated with residue-pairwise distances (53). The first is usually high for structures domains, but low for linker regions, regions that may be flexible, intrinsically disordered or structured only in the context of a larger complex. The second is useful for assessing more global features, such as domain packing.
Results obtained from these first analyses suggest that AlphaFold2 can be used to substantially extend the structural information for model proteomes beyond what is enabled by homology modeling, provided its confidence metrics are critically interpreted. For example, while the atomic coordinates of regions modelled with low confidence may not be trustworthy, they can nevertheless be used to predict disordered regions more accurately than state-of-the-art methods (1) . Similar analyses, currently underway to characterize the structures of AlphaFold-DB whose number is orders of magnitude larger, should shed further light on the information that may be safely extracted from these predicted structures. Clearly missing however, is information on the dynamic properties of proteins, many of which adopt multiple conformational states that are essential for their function (i.e binding other proteins, nucleic acids, small molecule ligands, or switching between functionally active and inactive states) (93, 138). This is currently a serious limitation of deep learning approaches such as AlphaFold2 as recently discussed (33). Tackling this limitation is an important goal, which is receiving increased attention, as further noted below.
Notwithstanding these limitations, ready access to the predicted protein structures in AlpfaFold-DB and to the freely available code and resources such as ColabFold(88) for predicting new structures with AlphaFold2 or RoseTTAFold efficiently with more modest computational resources, is having a resounding impact on experimental determination of protein structures. Rather than putting experimental structural biology out of business it is offering new opportunities like never before. Combining these opportunities with the recent spectacular advances in cryo-EM techniques, is propelling the field to new levels. In several instances, hard to solve X-ray or cryo-EM structures have been elucidated by using AlphaFold models in molecular replacement protocols, (64, 87). AlphaFold and RoseTTAfold models have been used successfully to fit residual electron density of cryo-EM maps, most notably in a recent assembly of the human nuclear pore complex (91). This is clearly an area that should soon see major advances from closer integration of deep learning-based and other structure modeling approaches with emerging DL-based and experimental cryo-EM techniques (125, 144).
Prediction of protein complexes and assemblies
An obvious next frontier for DL-based protein structure prediction methods is the accurate prediction of complexes and larger protein assemblies, as witnessed by a flurry of recent studies reporting forays towards this goal. Several benchmarking studies suggest that extensions of DL-based methods to the prediction of protein complexes will provide a major advance over traditional docking methods. In these studies AlphaFold2, was ‘tricked’ into successfully modeling the structure of a set of protein complexes of known stoichiometry, albeit not consistently to high accuracy, by feeding it the concatenated sequences of the interacting component proteins. (13). Better performance, was reported for AlphaFold-Multimer the inference engine of AlphaFold, directly trained on protein complexes from the PDB (31). For the same benchmark dataset of heteromeric interfaces AlphaFold-Multimer produced acceptable prediction (DockQ ≥ 0.23 ) for ~67% of the interfaces, but high accuracy predictions(DockQ ≥0.80) for only 23%, an improvement of 25% and 11% respectively, over the modified alphafold2 version. More modest improvement was achieved for homomeric interfaces generally associated with higher binding affinity, for which larger fractions could be predicted to acceptable (69%) and high accuracy (34%), respectively. At the same time ways have been proposed to integrate AlphaFold2 predictions of complexes with classical docking calculations and using the predicted complexes as templates for AlphaFold2 to significantly improve the performance of either method used independently(37).
Furthermore, several studies have suggested that AlphaFold and RoseTTafold, can be used to extend the structural coverage of model interactomes beyond what is enabled by homology modeling (of complexes, or of single chains followed by docking (92)). For example, for the human interactome AlphaFold predicted ~1,400 high confidence models of complexes displaying no homology to a known structure (15). Both prediction methods were used of identify interacting proteins and model their complexes in Baker’s yeast (49) and the human mitochondrion (105), deriving in each case new structural information for functionally important complexes.
While these early results are very promising, they also indicate that significant room remains for improvement. Further optimizing these methods to tackle complexes spanning a wide range of binding partners, binding affinities, and functional states, has the potential to lead to significant breakthroughs for these prediction problems. However, fulfilling this potential will not be effortless. The structural coverage of the protein complexes that form in living cells – the body of data AI methods need to ‘learn’ from – is orders of magnitude smaller than the current structural coverage of single protein chains. Furthermore, the formation of many of the more transient complexes featuring lower binding affinities, such as those associated with signal transaction processes, is highly context dependent. Currently however, the ability of experimental structural biology to adequately sample the physiologically relevant contexts of complexes is limited. Third, modeling the dynamic properties of the component proteins, which govern the conformational changes associated with binding, will be an important bottleneck to overcome.
What next?
Although this new wave of applied deep learning methods in the protein structure prediction field have rocked the biological sciences, with numerous applications in view (55), much work will be needed to mitigate important current limitations. Top of the list is the problem of accounting for dynamic flexibility, within single chains, the association process and within the complex (106). This is important for understanding and modelling the functional states of proteins, including those of intrinsically disordered proteins of which there is a natural abundance (68). Next, and related to the first, DL-based methods are still incapable to interpret the effects of single point mutations; backbone movements are simply not replicated when one amino acid is substituted for another, as benchmarked in several recent studies (14, 101).
Here too deep learning methodologies are offering a way forward by first ‘understanding’ the conformational states a protein samples (f.e. in known structures) and the likely transition paths between them (110). This ‘understanding’ is then used to further extend the sampled conformational space, by generating experimentally unobserved but native-like protein conformations, as recently described with an Autoencoder method (25). These descriptions of multiple conformations of a given protein will have to be integrated with the data on multiple sequence alignments to model structures corresponding to specific functional states. Crucial for training and testing this type of methods will be the development of benchmark datasets of physiologically pertinent structures of single chain proteins and complexes that incorporate information on the sampled conformational landscape.
Addressing these challenges will impact all areas of protein structure predictions including that of protein assemblies. Here progress will also depend on the ability of deep learning algorithms to restrain the sampling of the vast number of potential binding modes closer to the basin of native-like solutions, even in cases where co-evolution signals derived from the multiple sequence alignments are weak (such as for complexes with antibodies, or host pathogen interactions): a problem that AlphaFold seems to struggles with (37). Improving the ability to recognize native-like binding modes – the model ranking problem will likewise be important. Promising results towards the latter goal have recently been reported by standalone deep learning models implementing convolutional neural networks (CNNs) and other methods (17, 111, 135). Ultimately ranking models of protein complexes needs to display some correlation with binding affinities, a goal that scoring methods have pursued with modest success(34, 129).
As end-to-end machine learning methodologies are improved and mastered by the wider structural biology community, it will become routine to model a significant fraction of proteins and the complexes they form just from their amino acid sequences, ultimately negating the need for intermediate steps, such as searching for and utilising closest structural templates. One likewise expects these new methodologies to be extended to modelling nucleic acids, more particularly RNA but also DNA, and the complexes they form with proteins within the cell. Here too a major challenge will be to collate enough experimental data to train and validate machine learning methods. The way forward would be a closer integration of computational and experimental approaches. This would involve combining emerging methods for extracting information on structural heterogeneity in macromolecular complexes from the cryo-EM data obtained from endogenous material with AI-based structure prediction algorithms and molecular simulation techniques. All these are important developing areas where blind prediction initiatives will continue to play a major role.
Funding information
Sandor Vajda and Dima Kozakov acknowledge support from grants R35GM118078, R01 GM140098, and RM1135136 from the National Institute of General Medical Sciences, and DMS 2054251 from the National Science Foundation. Paul Bates acknowledges support by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001003), the UK Medical Research Council (FC001003), and the Wellcome Trust (FC001003).
References
- 1.Akdel M, Pires D, Porta Pardo E, Jänes J, Zalevsky A, et al. 2021. A structural biology community assessment of AlphaFold 2 applications. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford RF, Leaver-Fay A, Jeliazkov JR, O'Meara MJ, DiMaio FP, et al. 2017. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J Chem Theory Comput 13: 3031–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloy P, Ceulemans H, Stark A, Russell RB. 2003. The relationship between sequence and interaction divergence in proteins. J Mol Biol 332: 989–98 [DOI] [PubMed] [Google Scholar]
- 4.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. 2008. Principles of flexible protein-protein docking. Proteins 73: 271–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek M, Baker D. 2022. Deep learning and protein structure modeling. Nat Methods 19: 13–14 [DOI] [PubMed] [Google Scholar]
- 6.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, et al. 2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373: 871–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu S, Wallner B. 2016. DockQ: A Quality Measure for Protein-Protein Docking Models. PLoS One 11: e0161879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. 2000. The Protein Data Bank. Nucleic acids research 28: 235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blom NS, Sygusch J. 1997. High resolution fast quantitative docking using Fourier domain correlation techniques. Proteins 27: 493–506 [PubMed] [Google Scholar]
- 10.Bonvin AM. 2006. Flexible protein-protein docking. Curr Opin Struct Biol 16: 194–200 [DOI] [PubMed] [Google Scholar]
- 11.Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, et al. 2009. CHARMM: the biomolecular simulation program. J Comput Chem 30: 1545–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant AS, Goddard CA, Huguenard JR, Knudsen EI. 2015. Cholinergic control of gamma power in the midbrain spatial attention network. J Neurosci 35: 761–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant P, Pozzati G, Elofsson A. 2022. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 13: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buel GR, Walters KJ. 2022. Can AlphaFold2 predict the impact of missense mutations on structure? Nature Structural & Molecular Biology 29: 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke DF, Bryant P, Barrio-Hernandez I, Memon D, Pozzati G, et al. 2021. Towards a structurally resolved human protein interaction network. bioRxiv: 2021.11.08.467664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callaway E 2020. 'It will change everything': DeepMind's AI makes gigantic leap in solving protein structures. Nature 588: 203–04 [DOI] [PubMed] [Google Scholar]
- 17.Cao Y, Shen Y. 2020. Energy-based graph convolutional networks for scoring protein docking models. Proteins 88: 1091–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandonia JM, Brenner SE. 2006. The impact of structural genomics: expectations and outcomes. Science 311: 347–51 [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Li L, Weng Z. 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52: 80–7 [DOI] [PubMed] [Google Scholar]
- 20.Chothia C 1974. Hydrophobic bonding and accessible surface area in proteins. Nature 248: 338–9 [DOI] [PubMed] [Google Scholar]
- 21.Chothia C, Janin J. 1975. Principles of protein-protein recognition. Nature 256: 705–8 [DOI] [PubMed] [Google Scholar]
- 22.Chothia C, Lesk AM. 1986. The relation between the divergence of sequence and structure in proteins. EMBO J 5: 823–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dapkunas J, Timinskas A, Olechnovic K, Margelevicius M, Diciunas R, Venclovas C. 2017. The PPI3D web server for searching, analyzing and modeling protein-protein interactions in the context of 3D structures. Bioinformatics 33: 935–37 [DOI] [PubMed] [Google Scholar]
- 24.de Vries SJ, van Dijk M, Bonvin AM. 2010. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 5: 883–97 [DOI] [PubMed] [Google Scholar]
- 25.Degiacomi MT. 2019. Coupling Molecular Dynamics and Deep Learning to Mine Protein Conformational Space. Structure 27: 1034–40 e3 [DOI] [PubMed] [Google Scholar]
- 26.Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. 2020. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 28: 1071–81 e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobbins SE, Lesk VI, Sternberg MJ. 2008. Insights into protein flexibility: The relationship between normal modes and conformational change upon protein-protein docking. Proc Natl Acad Sci U S A 105: 10390–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez C, Boelens R, Bonvin AM. 2003. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–7 [DOI] [PubMed] [Google Scholar]
- 29.Dunbrack RL Jr., Gerloff DL, Bower M, Chen X, Lichtarge O, Cohen FE. 1997. Meeting review: the Second meeting on the Critical Assessment of Techniques for Protein Structure Prediction (CASP2), Asilomar, California, December 13-16, 1996. Fold Des 2: R27–42 [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg D, McLachlan AD. 1986. Solvation energy in protein folding and binding. Nature 319: 199–203 [DOI] [PubMed] [Google Scholar]
- 31.Evans R, O'Neill M, Pritzel A, Antropova N, Senior AW, et al. 2021. Protein complex prediction with AlphaFold-Multimer. BioRxiv [Google Scholar]
- 32.Fiser A, Sali A. 2003. ModLoop: automated modeling of loops in protein structures. Bioinformatics 19: 2500–1 [DOI] [PubMed] [Google Scholar]
- 33.Fleishman SJ, Horovitz A. 2021. Extending the New Generation of Structure Predictors to Account for Dynamics and Allostery. J Mol Biol 433: 167007. [DOI] [PubMed] [Google Scholar]
- 34.Fleishman SJ, Whitehead TA, Strauch EM, Corn JE, Qin S, et al. 2011. Community-wide assessment of protein-interface modeling suggests improvements to design methodology. J Mol Biol 414: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabb HA, Jackson RM, Sternberg MJ. 1997. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol 272: 106–20 [DOI] [PubMed] [Google Scholar]
- 36.Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, et al. 2020. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 72: e108. [DOI] [PubMed] [Google Scholar]
- 37.Ghani U, Desta I, Jindal A, Khan O, Jones G, et al. 2021. Improved Docking of Protein Models by a Combination of Alphafold2 and ClusPro. bioRxiv: 2021.09.07.459290 [Google Scholar]
- 38.Gilson MK AS, Honig BH. 1988. Calculating the electrostatic potential of molecules in solution: Method and error assessment. Journal of computational chemistry 9: 337–35 [Google Scholar]
- 39.Gobel U, Sander C, Schneider R, Valencia A. 1994. Correlated mutations and residue contacts in proteins. Proteins 18: 309–17 [DOI] [PubMed] [Google Scholar]
- 40.Guest JD, Vreven T, Zhou J, Moal I, Jeliazkov JR, et al. 2021. An expanded benchmark for antibody-antigen docking and affinity prediction reveals insights into antibody recognition determinants. Structure 29: 606–21 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas J, Roth S, Arnold K, Kiefer F, Schmidt T, et al. 2013. The Protein Model Portal--a comprehensive resource for protein structure and model information. Database (Oxford) 2013: bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagler AT, Huler E, Lifson S. 1974. Energy functions for peptides and proteins. I. Derivation of a consistent force field including the hydrogen bond from amide crystals. J Am Chem Soc 96: 5319–27 [DOI] [PubMed] [Google Scholar]
- 43.Hagler AT, Lifson S. 1974. Energy functions for peptides and proteins. II. The amide hydrogen bond and calculation of amide crystal properties. J Am Chem Soc 96: 5327–35 [DOI] [PubMed] [Google Scholar]
- 44.Harmalkar A, Gray JJ. 2021. Advances to tackle backbone flexibility in protein docking. Curr Opin Struct Biol 67: 178–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho BK, Dill KA. 2006. Folding very short peptides using molecular dynamics. PLoS Comput Biol 2: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang SY, Zou X. 2008. An iterative knowledge-based scoring function for protein-protein recognition. Proteins 72: 557–79 [DOI] [PubMed] [Google Scholar]
- 47.Huang SY, Zou X. 2010. MDockPP: A hierarchical approach for protein-protein docking and its application to CAPRI rounds 15-19. Proteins 78: 3096–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang SY, Zou X. 2011. Statistical mechanics-based method to extract atomic distance-dependent potentials from protein structures. Proteins 79: 2648–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humphreys IR, Pei J, Baek M, Krishnakumar A, Anishchenko I, et al. 2021. Computed structures of core eukaryotic protein complexes. Science 374: eabm4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isogai Y, Nemethy G, Scheraga HA. 1977. Enkephalin: conformational analysis by means of empirical energy calculations. Proc Natl Acad Sci U S A 74: 414–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, et al. 2003. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins 52: 2–9 [DOI] [PubMed] [Google Scholar]
- 52.Jernigan RL, Bahar I. 1996. Structure-derived potentials and protein simulations. Curr Opin Struct Biol 6: 195–209 [DOI] [PubMed] [Google Scholar]
- 53.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, et al. 2021. Applying and improving AlphaFold at CASP14. Proteins 89: 1711–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jumper J, Hassabis D. 2022. Protein structure predictions to atomic accuracy with AlphaFold. Nature Methods 19: 11–12 [DOI] [PubMed] [Google Scholar]
- 56.Kamisetty H, Ovchinnikov S, Baker D. 2013. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A 110: 15674–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. 1992. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proceedings of the National Academy of Sciences of the United States of America 89: 2195–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knyazev B, Taylor GW, Amer M. 2019. Understanding attention and generalization in graph neural networks. Advances in neural information processing systems 32 [Google Scholar]
- 59.Ko J, Park H, Heo L, Seok C. 2012. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40: W294–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozakov D, Brenke R, Comeau SR, Vajda S. 2006. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 65: 392–406 [DOI] [PubMed] [Google Scholar]
- 61.Kozakov D, Hall DR, Beglov D, Brenke R, Comeau SR, et al. 2010. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19. Proteins 78: 3124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, et al. 2017. The ClusPro web server for protein-protein docking. Nat Protoc 12: 255–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kryshtafovych A, Monastyrskyy B, Fidelis K. 2016. CASP11 statistics and the prediction center evaluation system. Proteins 84 Suppl 1: 15–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kryshtafovych A, Moult J, Albrecht R, Chang GA, Chao K, et al. 2021. Computational models in the service of X-ray and cryo-electron microscopy structure determination. Proteins 89: 1633–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. 2021. Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins 89: 1607–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. 2021. Critical assessment of methods of protein structure prediction (CASP)—Round XIV. Proteins: Structure, Function, and Bioinformatics 89: 1607–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuhlman B, Bradley P. 2019. Advances in protein structure prediction and design. Nat Rev Mol Cell Biol 20: 681–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulkarni P, Uversky VN. 2018. Intrinsically Disordered Proteins: The Dark Horse of the Dark Proteome. Proteomics 18: e1800061. [DOI] [PubMed] [Google Scholar]
- 69.Kundrotas PJ, Anishchenko I, Dauzhenka T, Kotthoff I, Mnevets D, et al. 2018. Dockground: A comprehensive data resource for modeling of protein complexes. Protein Sci 27: 172–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. 2012. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci U S A 109: 9438–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuroda D, Gray JJ. 2016. Pushing the Backbone in Protein-Protein Docking. Structure 24: 1821–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521: 436–44 [DOI] [PubMed] [Google Scholar]
- 73.Lee H, Seok C. 2017. Template-Based Prediction of Protein-Peptide Interactions by Using GalaxyPepDock. Methods Mol Biol 1561: 37–47 [DOI] [PubMed] [Google Scholar]
- 74.Lensink MF, Brysbaert G, Mauri T, Nadzirin N, Velankar S, et al. 2021. Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment. Proteins 89: 1800–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lensink MF, Brysbaert G, Nadzirin N, Velankar S, Chaleil RAG, et al. 2019. Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment. Proteins 87: 1200–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lensink MF, Mendez R, Wodak SJ. 2007. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins 69: 704–18 [DOI] [PubMed] [Google Scholar]
- 77.Lensink MF, Moal IH, Bates PA, Kastritis PL, Melquiond AS, et al. 2014. Blind prediction of interfacial water positions in CAPRI. Proteins 82: 620–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lensink MF, Nadzirin N, Velankar S, Wodak SJ. 2020. Modeling protein-protein, protein-peptide, and protein-oligosaccharide complexes: CAPRI 7th edition. Proteins 88: 916–38 [DOI] [PubMed] [Google Scholar]
- 79.Lensink MF, Velankar S, Baek M, Heo L, Seok C, Wodak SJ. 2018. The challenge of modeling protein assemblies: the CASP12-CAPRI experiment. Proteins 86 Suppl 1: 257–73 [DOI] [PubMed] [Google Scholar]
- 80.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, et al. 2016. Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment. Proteins 84 Suppl 1: 323–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lensink MF, Velankar S, Wodak SJ. 2017. Modeling protein-protein and protein-peptide complexes: CAPRI 6th edition. Proteins 85: 359–77 [DOI] [PubMed] [Google Scholar]
- 82.Lensink MF, Wodak SJ. 2013. Docking, scoring, and affinity prediction in CAPRI. Proteins 81: 2082–95 [DOI] [PubMed] [Google Scholar]
- 83.Lensink MF, Wodak SJ. 2014. Score_set: a CAPRI benchmark for scoring protein complexes. Proteins 82: 3163–9 [DOI] [PubMed] [Google Scholar]
- 84.Levinthal C, Wodak SJ, Kahn P, Dadivanian AK. 1975. Hemoglobin interaction in sickle cell fibers. I: Theoretical approaches to the molecular contacts. Proc Natl Acad Sci U S A 72: 1330–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levitt M 1976. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol 104: 59–107 [DOI] [PubMed] [Google Scholar]
- 86.Liu J, Wu T, Guo Z, Hou J, Cheng J. 2022. Improving protein tertiary structure prediction by deep learning and distance prediction in CASP14. Proteins 90: 58–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCoy AJ, Sammito MD, Read RJ. 2022. Implications of AlphaFold2 for crystallographic phasing by molecular replacement. Acta Crystallogr D Struct Biol 78: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mirdita M, Ovchinnikov S, Steinegger M. 2021. ColabFold - Making protein folding accessible to all. bioRxiv: 2021.08.15.456425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moal IH, Chaleil RAG, Bates PA. 2018. Flexible Protein-Protein Docking with SwarmDock. Methods Mol Biol 1764: 413–28 [DOI] [PubMed] [Google Scholar]
- 90.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, et al. 2011. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A 108: E1293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, et al. 2022. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376: eabm9506. [DOI] [PubMed] [Google Scholar]
- 92.Mosca R, Ceol A, Aloy P. 2013. Interactome3D: adding structural details to protein networks. Nat Methods 10: 47–53 [DOI] [PubMed] [Google Scholar]
- 93.Motlagh HN, Wrabl JO, Li J, Hilser VJ. 2014. The ensemble nature of allostery. Nature 508: 331–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moult J 2005. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol 15: 285–9 [DOI] [PubMed] [Google Scholar]
- 95.Moult J, Pedersen JT, Judson R, Fidelis K. 1995. A large-scale experiment to assess protein structure prediction methods. Proteins 23: ii–v [DOI] [PubMed] [Google Scholar]
- 96.Nadaradjane AA, Guerois R, Andreani J. 2018. Protein-Protein Docking Using Evolutionary Information. Methods Mol Biol 1764: 429–47 [DOI] [PubMed] [Google Scholar]
- 97.Novotny J, Bruccoleri R, Karplus M. 1984. An analysis of incorrectly folded protein models. Implications for structure predictions. J Mol Biol 177: 787–818 [DOI] [PubMed] [Google Scholar]
- 98.Olechnovic K, Venclovas C. 2017. VoroMQA: Assessment of protein structure quality using interatomic contact areas. Proteins 85: 1131–45 [DOI] [PubMed] [Google Scholar]
- 99.Onufriev A, Case DA, Bashford D. 2002. Effective Born radii in the generalized Born approximation: the importance of being perfect. J Comput Chem 23: 1297–304 [DOI] [PubMed] [Google Scholar]
- 100.Padhorny D, Kazennov A, Zerbe BS, Porter KA, Xia B, et al. 2016. Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc Natl Acad Sci U S A 113: E4286–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pak MA, Markhieva KA, Novikova MS, Petrov DS, Vorobyev IS, et al. 2021. Using AlphaFold to predict the impact of single mutations on protein stability and function. bioRxiv: 2021.09.19.460937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pallara C, Jimenez-Garcia B, Romero M, Moal IH, Fernandez-Recio J. 2017. pyDock scoring for the new modeling challenges in docking: Protein-peptide, homo-multimers, and domain-domain interactions. Proteins 85: 487–96 [DOI] [PubMed] [Google Scholar]
- 103.Pearce R, Zhang Y. 2021. Deep learning techniques have significantly impacted protein structure prediction and protein design. Curr Opin Struct Biol 68: 194–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pearce R, Zhang Y. 2021. Toward the solution of the protein structure prediction problem. J Biol Chem 297: 100870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pei J, Zhang J, Cong Q. 2021. Human mitochondrial protein complexes revealed by large-scale coevolution analysis and deep learning-based structure modeling. bioRxiv: 2021.09.14.460228 [DOI] [PubMed] [Google Scholar]
- 106.Perrakis A, Sixma TK. 2021. AI revolutions in biology: The joys and perils of AlphaFold. EMBO Rep 22: e54046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Porter KA, Desta I, Kozakov D, Vajda S. 2019. What method to use for protein-protein docking? Curr Opin Struct Biol 55: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Porter KA, Padhorny D, Desta I, Ignatov M, Beglov D, et al. 2019. Template-based modeling by ClusPro in CASP13 and the potential for using co-evolutionary information in docking. Proteins 87: 1241–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quignot C, Rey J, Yu J, Tuffery P, Guerois R, Andreani J. 2018. InterEvDock2: an expanded server for protein docking using evolutionary and biological information from homology models and multimeric inputs. Nucleic Acids Res 46: W408–W16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramaswamy VK, Musson SC, Willcocks CG, Degiacomi MT. 2021. Deep Learning Protein Conformational Space with Convolutions and Latent Interpolations. Physical Review X 11: 011052 [Google Scholar]
- 111.Renaud N, Geng C, Georgievska S, Ambrosetti F, Ridder L, et al. 2021. DeepRank: a deep learning framework for data mining 3D protein-protein interfaces. Nat Commun 12: 7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ritchie DW, Kemp GJ. 2000. Protein docking using spherical polar Fourier correlations. Proteins 39: 178–94 [PubMed] [Google Scholar]
- 113.Robin X, Haas J, Gumienny R, Smolinski A, Tauriello G, Schwede T. 2021. Continuous Automated Model EvaluatiOn (CAMEO)-Perspectives on the future of fully automated evaluation of structure prediction methods. Proteins 89: 1977–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schmidhuber J 2015. Deep learning in neural networks: an overview. Neural Netw 61: 85–117 [DOI] [PubMed] [Google Scholar]
- 115.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, et al. 2019. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins 87: 1141–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, et al. 2020. Improved protein structure prediction using potentials from deep learning. Nature 577: 706–10 [DOI] [PubMed] [Google Scholar]
- 117.Shen MY, Sali A. 2006. Statistical potential for assessment and prediction of protein structures. Protein Sci 15: 2507–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Silver D, Huang A, Maddison CJ, Guez A, Sifre L, et al. 2016. Mastering the game of Go with deep neural networks and tree search. Nature 529: 484–9 [DOI] [PubMed] [Google Scholar]
- 119.Simons KT, Kooperberg C, Huang E, Baker D. 1997. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol 268: 209–25 [DOI] [PubMed] [Google Scholar]
- 120.Singh A, Dauzhenka T, Kundrotas PJ, Sternberg MJE, Vakser IA. 2020. Application of docking methodologies to modeled proteins. Proteins 88: 1180–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smith GR, Sternberg MJ, Bates PA. 2005. The relationship between the flexibility of proteins and their conformational states on forming protein-protein complexes with an application to protein-protein docking. J Mol Biol 347: 1077–101 [DOI] [PubMed] [Google Scholar]
- 122.Suh D, Lee JW, Choi S, Lee Y. 2021. Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction. Int J Mol Sci 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tanaka S, Scheraga HA. 1976. Medium- and long-range interaction parameters between amino acids for predicting three-dimensional structures of proteins. Macromolecules 9: 945–50 [DOI] [PubMed] [Google Scholar]
- 124.Torrisi M, Pollastri G, Le Q. 2020. Deep learning methods in protein structure prediction. Comput Struct Biotechnol J 18: 1301–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tuting C, Kyrilis FL, Muller J, Sorokina M, Skalidis I, et al. 2021. Cryo-EM snapshots of a native lysate provide structural insights into a metabolon-embedded transacetylase reaction. Nat Commun 12: 6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.UniProt C. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49: D480–D89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vakser IA, Aflalo C. 1994. Hydrophobic docking: a proposed enhancement to molecular recognition techniques. Proteins 20: 320–9 [DOI] [PubMed] [Google Scholar]
- 128.Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, et al. 2014. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev 114: 6733–78 [DOI] [PubMed] [Google Scholar]
- 129.Vangone A, Bonvin AM. 2015. Contacts-based prediction of binding affinity in protein-protein complexes. Elife 4: e07454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, et al. 2022. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic acids research 50: D439–D44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Velankar S, Burley SK, Kurisu G, Hoch JC, Markley JL. 2021. The Protein Data Bank Archive. Methods Mol Biol 2305: 3–21 [DOI] [PubMed] [Google Scholar]
- 132.Vreven T, Schweppe DK, Chavez JD, Weisbrod CR, Shibata S, et al. 2018. Integrating Cross-Linking Experiments with Ab Initio Protein-Protein Docking. J Mol Biol 430: 1814–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. 2004. Development and testing of a general amber force field. J Comput Chem 25: 1157–74 [DOI] [PubMed] [Google Scholar]
- 134.Wang S, Sun S, Li Z, Zhang R, Xu J. 2017. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput Biol 13: e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang X, Flannery ST, Kihara D. 2021. Protein Docking Model Evaluation by Graph Neural Networks. Frontiers in Molecular Biosciences 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wodak SJ, Janin J. 2002. Structural basis of macromolecular recognition. Adv Protein Chem 61: 9–73 [DOI] [PubMed] [Google Scholar]
- 137.Wodak SJ, Mendez R. 2004. Prediction of protein-protein interactions: the CAPRI experiment, its evaluation and implications. Curr Opin Struct Biol 14: 242–9 [DOI] [PubMed] [Google Scholar]
- 138.Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, et al. 2019. Allostery in Its Many Disguises: From Theory to Applications. Structure 27: 566–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xia B, Vajda S, Kozakov D. 2016. Accounting for pairwise distance restraints in FFT-based protein-protein docking. Bioinformatics 32: 3342–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yan Y, Wen Z, Wang X, Huang SY. 2017. Addressing recent docking challenges: A hybrid strategy to integrate template-based and free protein-protein docking. Proteins 85: 497–512 [DOI] [PubMed] [Google Scholar]
- 141.Yan Y, Zhang D, Zhou P, Li B, Huang SY. 2017. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 45: W365–W73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang Z, Schindler CE, Lange OF, Zacharias M. 2015. Application of Enhanced Sampling Monte Carlo Methods for High-Resolution Protein-Protein Docking in Rosetta. PLoS One 10: e0125941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zheng W, Li Y, Zhang C, Zhou X, Pearce R, et al. 2021. Protein structure prediction using deep learning distance and hydrogen-bonding restraints in CASP14. Proteins 89: 1734–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhong ED, Bepler T, Berger B, Davis JH. 2021. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods 18: 176–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou H, Skolnick J. 2011. GOAP: a generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys J 101: 2043–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou H, Zhou Y. 2002. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci 11: 2714–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou R 2003. Free energy landscape of protein folding in water: explicit vs. implicit solvent. Proteins 53: 148–61 [DOI] [PubMed] [Google Scholar]