Summary
Background
Crimean-Congo Haemorrhagic Fever Virus is a tick-borne bunyavirus prevalent across Asia, Africa, the Middle East, and Europe. The virus causes a non-specific febrile illness which may develop into severe haemorrhagic disease. To date, there are no widely approved therapeutics. Recently, we reported an alphavirus-based replicon RNA vaccine which expresses the CCHFV nucleoprotein (repNP) or glycoprotein precursor (repGPC) and is protective against lethal disease in mice.
Methods
Here, we evaluated engineered GPC constructs to find the minimal enhancing epitope of repGPC and test two RNA vaccine approaches to express multiple antigens in vivo to optimize protective efficacy of our repRNA.
Findings
Vaccination with repNP and a construct expressing just the Gc antigen (repGc-FL) resulted in equivalent immunogenicity and protective efficacy compared to original repNP + repGPC vaccination. This vaccine was protective when prepared in either of two vaccine approaches, a mixed synthesis reaction producing two RNAs in a single tube and a single RNA expressing two antigens.
Interpretation
Overall, our data illustrate two vaccine approaches to deliver two antigens in a single immunization. Both approaches induced protective immune responses against CCHFV in this model. These approaches support their continued development for this and future vaccine candidates for CCHFV and other vaccines where inclusion of multiple antigens would be optimal.
Funding
This work was supported by the Intramural Research Program, NIAID/NIH, HDT Bio and MCDC Grant #MCDC2204-011.
Keywords: Crimean-Congo haemorrhagic fever virus, Vaccine, Replicon, Dual-antigen
Research in context.
Evidence before this study
Crimean-Congo haemorrhagic fever virus (CCHFV) is a tick-borne bunyavirus which can cause severe haemorrhagic disease in humans. Currently there are no widely approved vaccines for CCHFV. However multiple platforms have been evaluated and protection has been seen in NP-only, GPC-only, and NP/GPC expressing vaccines. While NP-only vaccines have been protective, GPC is the target of neutralizing antibodies. However, there is no consensus on the immune correlates of protection, and it is unclear which viral antigens are optimal for vaccine development.
Added value of this study
Here we build upon our recent report on a protective CCHFV vaccine expressing the viral nucleoprotein (repNP) and glycoprotein precursor (repGPC). This vaccine protects primarily via non-neutralizing anti-NP antibodies and GPC-specific T-cells. Here, we sought to develop a bivalent, dual-antigen vaccine by identifying the minimal enhancing epitope in repGPC. We compared constructs expressing the CCHFV-Gn (Gn-sol) and CCHFV-Gc in soluble (Gc-sol) and full-length (Gc-FL) forms and found that although both Gc constructs elicited robust immune responses, only the full-length Gc was as protective as the full glycoprotein precursor when administered alongside repNP RNA. This data indicates that modifications of the glycoproteins are important considerations in developing vaccines and eliciting protective immune responses. In addition, we developed two dual-antigen vaccine approaches including a mixed synthesis reaction which produces two individual RNAs in a single synthesis reaction, simplifying manufacturing, and a bivalent single-RNA which expresses both the CCHFV-NP and CCHFV-GcFL. This bivalent construct (repGcFL-NP) was optimized by inclusion of the minimal enhancing GPC epitope, Gc-FL, and two sub genomic promoters. This construct was as efficacious as original repNP + repGPC vaccination.
Implications of all the available evidence
Overall, our data indicates that protection from CCHFV disease can be optimized by identifying minimal epitopes within GPC which induce protective immune responses. This is important in future vaccine development since the CCHFV-GPC is large and complex and presents difficulty in manufacturing expression plasmids or proteins. In addition, our data presents two bivalent vaccination approaches which show how two antigens may be efficiently combined in a single vaccination to simplify manufacturing without compromising immunogenicity and efficacy.
Introduction
Crimean-Congo haemorrhagic fever virus (CCHFV) is a negative-sense, RNA virus in the Bunyavirales order with a wide geographic distribution.1,2 CCHFV outbreaks, as well as seroprevalence, have been reported across Africa, Europe, the Middle East, India, and Asia.2,3 This distribution is closely linked to the prevalence of Hyalomma genus ticks, the main reservoir of CCHFV.1,2 CCHFV is transmitted either directly via tick bite or indirectly, by transmission to agricultural livestock and subsequent infection of humans during activities such as butchering, where the possibility of contact with infected blood is high.1,2 With climate change, ticks have the potential to expand their range,4 and they can be introduced as invasive species,5 further increasing the number of people at risk for CCHFV infection. There have also been several reports of nosocomial human-to-human transmission, and this primarily occurs amongst healthcare workers.6 Disease itself begins as a non-specific febrile illness characterized by fever, headache, and myalgia before progressing into a severe haemorrhagic stage characterized by extensive internal and external hemorrhaging.1,2 Generally, progression into severe disease takes ∼5–6 days post-exposure and case fatality is over 30%.1,2 Due to these factors and the lack of therapeutics and vaccines, CCHFV is on the World Health Organization's list of high priority pathogens.
However, mechanisms of CCHFV pathogenesis are poorly understood, and this severely limits development of interventions. The genomic organization of CCHFV is complex and vaccines based on the CCHFV nucleoprotein (NP) and glycoprotein precursor (GPC) have been explored.7,8 Interestingly, multiple vaccine platforms using NP and/or GPC report varying efficacy from complete to no protection and there is no consensus on what immune responses are required for protection.8 Across vaccine candidates, GPC can be the target of neutralizing and non-neutralizing antibodies and may stimulate T-cell responses; NP can also induce antibodies and T-cell responses and is highly conserved, making it attractive for pan-CCHFV vaccines.8
Recently, we have characterized an alphavirus-based replicating RNA (repRNA) vaccine expressing either the CCHFV NP (repNP) or the glycoprotein precursor (repGPC), the combination of which was protective after a single, low-dose immunization against lethal CCHFV challenge in mice.9 RepRNA is efficiently delivered into cells by a cationic nanocarrier, termed LION™, optimized for the intramuscular delivery of repRNA.10 LIONs can be stockpiled for years at refrigerator temperatures and combined with repRNA immediately prior to vaccine administration which is beneficial for pandemic preparedness.10 Once inside the cell, repRNA mimics a natural alphavirus infection and stimulates the innate immune response while driving expression of heterologous antigens for a robust, antigen-specific adaptive response.11 LION restricts most of that innate immune response to the local injection site, mitigating the dose-dependent systemic reactogenicity observed when repRNA is delivered by lipid nanoparticles.10 Previously, we used this platform to protect against infections by mycobacterium tuberculosis, Zika virus, enterovirus D68, and SARS-CoV-2 in vivo; the latter achieving emergency use authorization in India with ongoing clinical trials in Brazil, South Korea, and the United States.10,12, 13, 14, 15, 16 In our previous report on our CCHFV vaccine, we found that repNP primarily stimulates B-cell responses and induces high titers of non-neutralizing, anti-NP antibodies and, remarkably, this vaccine is protective on its own.9 Although repGPC was only partially protective on its own, inclusion with repNP conferred maximum protection9 likely through eliciting CCHFV-specific T-cells.
To further optimize our vaccine for pre-clinical and clinical development, in this report we identified the minimal enhancing epitope within the GPC and we combined this epitope with the CCHFV-NP onto a single-bivalent RNA. This approach simplifies manufacturing, testing, and deployment of the vaccine and we demonstrate that bivalent repRNAs can elicit immune responses to both antigens. In addition, we describe a mixed synthesis repRNA reaction which, along with our single-bivalent RNA, provides two feasible and highly efficacious approaches for clinical development and manufacturing of this and future vaccines.
Methods
Ethics
All work involving infectious CCHFV was done in a biocontainment level 4 at the Rocky Mountain Laboratories, NIAID, NIH, Hamilton, MT in accordance with guidelines put forth by the Institutional Biosafety Committee (IBC). All animal work was approved by the Rocky Mountain Laboratories Institutional Animal Care and Use Committee (protocol #2020-76) in accordance with guidelines from the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Office of Animal Welfare, the United States Department of Agriculture in an association for Assessment and Accreditation of Laboratory Animal Care-Accredited Facility. Mice were housed in HEPA-filter cage systems enriched with nesting material and commercial food and water available ad libitum.
Vaccine preparation
repRNAs expressing CCHFV strain Hoti antigens were constructed and vaccine prepared with complexation of repRNAs to LION as previously described.13
Mice vaccinations and infections
Equal numbers of male and female wild-type C57BL6/J mice (stock# 000664) were purchased from Jackson Laboratories at approximately 8-weeks of age at time of initial vaccination. Mice were vaccinated with a single intramuscular injection of 50 μL to the hind limb. Total RNA delivered was 1 μg for all vaccinations and for mice receiving two RNAs, mice received 0.5 μg of each RNA. Vaccination appeared well tolerated and no vaccine-mediated adverse effects were observed. For challenge, mice were given an intraperitoneal injection of 2.5 mg MAR1-5A3 antibody (Leinco) followed by intraperitoneal injection of 100 TCID50 CCHFV strain UG3010. All procedures were done under isoflurane anesthesia. End-point euthanasia was performed under deep anesthesia via terminal cardiac puncture followed by cervical dislocation.
Viral stock
CCHFV strain UG3010 was originally provided by Eric Bergeron, Centers for Disease Control and Prevention. UG3010 was grown, titered, and sequence confirmed as previously described.9
Enzyme-linked immunoassay (ELISA)
An in-house ELISA using whole CCHFV Hoti antigen was used to quantify CCHFV-specific antibody responses from vaccinations previously described.17 Limits of detection are based on background absorbance of negative samples. In addition, we developed an in-house recombinant antigen ELISA (rELISA) using CCHFV rNP, rGn, and rGc based on the prototype CCHFV strain 10200 (The Native Antigen Company) protein. rELISA was developed the same as the aforementioned ELISA except that 1 μg/mL rNP, rGn, or rGc protein was diluted in PBS and adsorbed to Nunc Maxisorp plates overnight instead of whole CCHFV Hoti antigen. Internal positive controls for NP, Gn and Gc consisting of monoclonal antibodies specific for these antigens were included. Anti-NP (Clone 9D5) and anti-Gc (clone 11E7) from BEI resources and anti-Gn (clone JE12) from Native Antigen were used. Endpoint titers are reported as the reciprocal of the last dilution to provide signal 2X above background.
Enzyme-linked immunosorbent spot assay (ELISpot)
Evaluation of CCHFV-specific T-cell responses from vaccination was performed as previously described using CCHFV strain Hoti peptides (Genscript) and mouse single-color IFNγ kit (Immunospot).9
Immunofluorescence assay (IFA)
IFA was done to visualize CCHFV-NP and CCHFV-GPC protein expression from repRNAs as previously described.9
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA was extracted from blood and tissue samples using commercially available RNA isolation kits (Qiagen) and was quantified as previously described.9
Media tissue culture infectious dose 50 assay (TCID50)
TCID50 assay was done to quantify infectious virus in the blood, liver, and spleen as previously described.9
Neutralization assay
Antibody neutralization capacity was evaluated with neutralization assay using infectious CCHFV strain Hoti as previously described.9 Titers are reported as the reciprocal of the last dilution to show complete neutralization of CCHFV in vitro.
Histology
Tissues were fixed in 10% Neutral Buffered Formalin x2 changes, for a minimum of 7 days. Tissues were placed in cassettes and processed with a Sakura VIP-6 Tissue Tek, on a 12-h automated schedule, using a graded series of ethanol, xylene, and PureAffin. Embedded tissues are sectioned at 5 μm and dried overnight at 42 degrees C prior to staining. Specific anti-CCHFV immunoreactivity was detected using Rabbit anti-CCHFV N IBT (Bioservices, cat#04-0011) at a 1:2000 dilution. The secondary antibody is the Immpress-VR anti-rabbit IgG polymer kit Vector Laboratories cat#MP-6401. The tissues were then processed for immunohistochemistry using the Discovery Ultra automated stainer (Ventana Medical Systems) with a ChromoMap DAB kit Roche Tissue Diagnostics cat#760-159.
Statistics
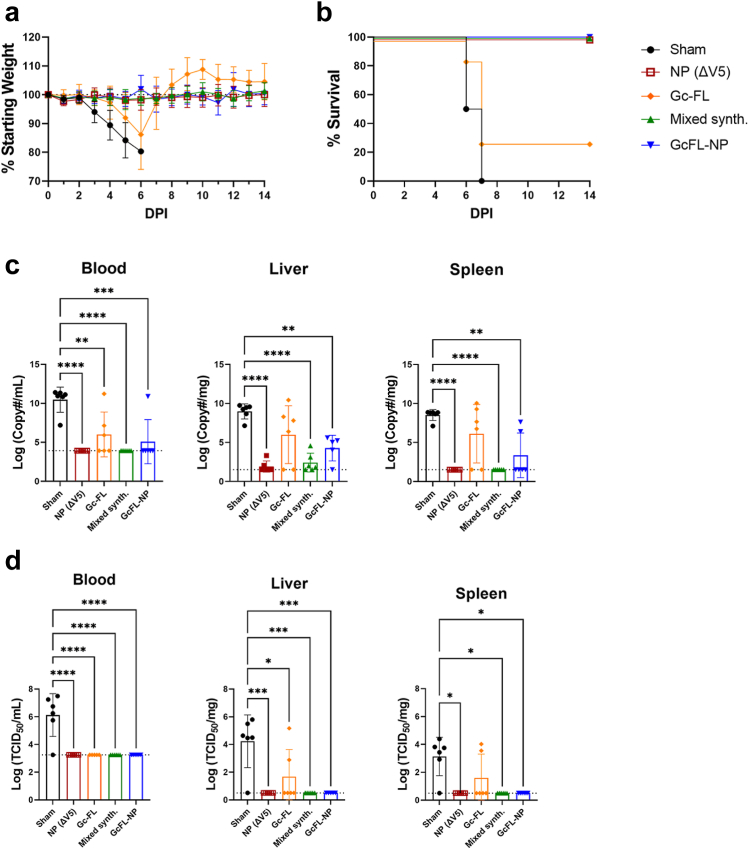
Statistical tests indicated in figure legends were chosen as recommended by and performed using GraphPad Prism 9. Mice were assigned to groups randomly and sample size determined based on previous experience with CCHFV mouse models. Pathologists were blinded towards study groups. One animal in the repGc-FL group was euthanized early due to dermatitis resulting in N = 7 for this survival group (Fig. 6a and b).
Fig. 6.
Bivalent and mixed synthesis repRNA protect against lethal CCHFV challenge. Mice prime-only vaccinated with sham, repNP(ΔV5), repGc-FL, mixed synthesis, or bivalent RNA on day −28 were treated with a MAR1-5A3 antibody and infected with a lethal dose of CCHFV strain UG3010 on D0. Mice (N = 8) were monitored daily for (a) weight loss and until day 14 p.i. for (b) survival. On D5 p.i., groups of mice (N = 6) were evaluated for control of (c) viral genome copies via qRT-PCR and (d) infectious virus via TCID50 in indicated tissues. Sham and repNP(ΔV5) group data are duplicated from Supplementary Figure S2 for comparison. Dashed lines indicate limit of detection. Significance was calculated using one-way ANOVA; ns P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
Role of funding
Funders had no input on study design, data collection, interpretation, data analysis, writing of report, or decision to publish.
Results
repGPC-T2A-NP vaccination elicits robust T-cell but impaired B-cell responses
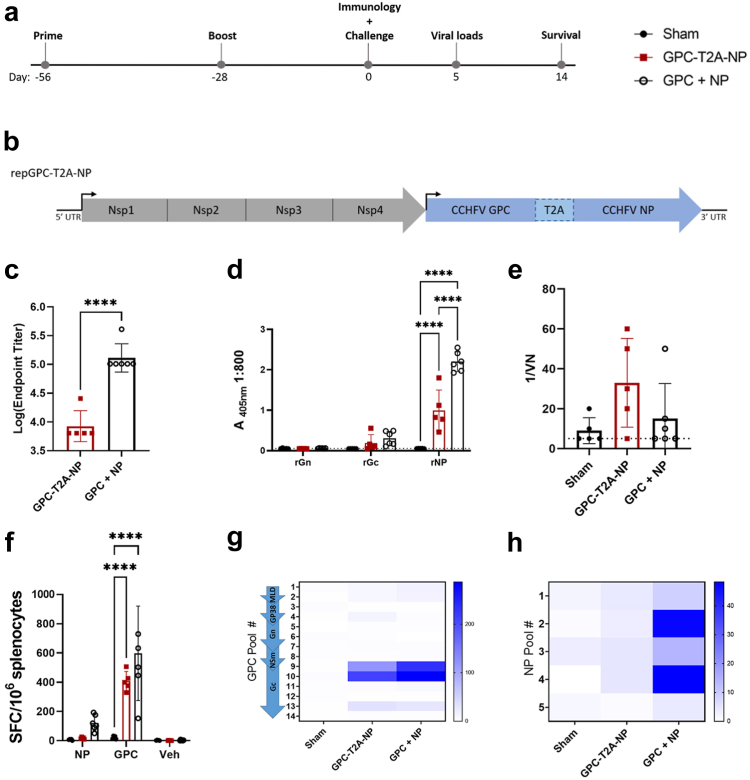
Based on our previous data showing that vaccination with repNP and repGPC provided optimal protection, we sought to determine if a single, bivalent repRNA could be protective.15 To evaluate immunogenicity of this single RNA in vivo, we vaccinated wild-type C57BL6/J mice with 1 μg of sham (expressing an irrelevant green fluorescent protein (GFP) antigen), repGPC-T2A-NP, or repNP + repGPC RNA complexed to LION administered intramuscularly (IM) with a prime-boost regimen (Fig. 1a).9 Our single, bivalent RNA, repGPC-T2A-NP, expresses both NP and GPC separated by a self-cleaving peptide motif derived from Thosea asigna (Fig. 1b). Surprisingly, compared to repNP + repGPC, repGPC-T2A-NP had a significantly diminished antibody response to whole virus and, specifically, the CCHFV NP (Fig. 1c and d). Similar to our previous studies, vaccine elicited antibody responses had low titers of anti-Gc antibodies and little-to-no neutralizing activity (Fig. 1d and e). Further, neither vaccination induced anti-Gn antibodies (Fig. 1d). Consistent with diminished immunogenicity, immunofluorescence assay (IFA) showed that cells transfected with repGPC-T2A-NP had diminished NP and GPC expression compared to cells transfected with individual repNP or repGPC (Supplementary Figure S1). We also evaluated T-cell responses via IFNγ ELISpot and both vaccinations induced significantly increased T-cell responses against the CCHFV GPC, but not the NP, compared to sham vaccination (Fig. 1f–h). These responses were directed against peptide pools 9 and 10 which span the end of the Nsm and N-terminal domain of the Gc (Fig. 1g). Overall, although both vaccinations elicited similar T-cell responses towards the CCHFV GPC, repGPC-T2A-NP had a diminished anti-NP antibody response compared to repNP + repGPC.
Fig. 1.
repGPC-T2A-NP elicits equivalent cellular but not humoral immune response compared to repNP + repGPC. WT C57BL6/J mice were (a) vaccinated with 1ug of Sham, repGPC-T2A-NP, or repGPC + repNP RNA on days −56 and −28 relative to lethal CCHFV challenge. (b) repGPC-T2A-NP is a bivalent, single RNA expressing the CCHFV GPC and NP under a single promoter, separated by a T2A self-cleaving site. On D0, groups of mice (N = 6) were evaluated for immune response to CCHFV. Antibody response was evaluated via (c) endpoint titers measured using a whole virion IgG ELISA, (d) recombinant antigen (rAg) ELISA to the CCHFV nucleoprotein (NP) and mature glycoproteins (Gn and Gc), and (e) neutralization assay using infectious virus. Dashed lines indicate limit of detection. Cellular immune response was evaluated via IFNу ELISpot with cumulative responses against either NP or GPC (f) and (g and h) heat maps showing the distribution of cellular responses to peptide pools spanning the entire CCHFV GPC and NP. DMSO vehicle (Veh) is also shown. Significance was calculated using one-way ANOVA; ns P > 0.05, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
repGPC-T2A-NP confers partial protection against lethal CCHFV challenge
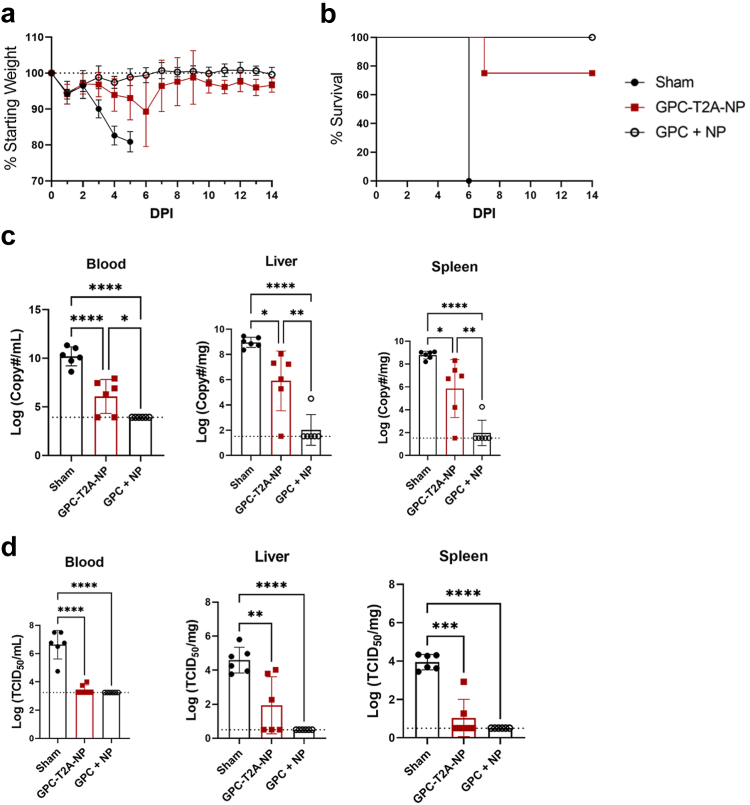
Next, we compared the efficacy of repGPC-T2A-NP vs. repGPC + repNP vaccinations against a lethal heterologous CCHFV challenge. 4 weeks post-boost, groups of mice were treated with 2.5 mg of MAR1-5A3 antibody to block the type I IFN response only at time of challenge and to render mice susceptible to CCHFV infection and disease.18,19 In addition, to stringently assess vaccine efficacy, mice were infected with a heterologous CCHFV strain, UG3010, which differs from vaccine antigens in amino acid sequence by 4.5% in the NP, 25.6% across the whole GPC and 14% specifically in the Gn and Gc glycoproteins. In the sham-vaccinated group, weight loss began on D3 p.i. and continued until mice had succumbed to disease by D6 p.i. (Fig. 2a and b). This was associated with high viral genome copies and infectious virus in the blood, liver, and spleen at day 5, shortly before the mice succumbed (Fig. 2c and d). Like our previous studies, repNP + repGPC vaccination conferred 100% protection against lethal disease with no infectious virus and little-to-no viral RNA in the blood, liver and spleen (Fig. 2a–d). In contrast, repGPC-T2A-NP vaccination only partially protected from CCHFV disease with 3 of 8 mice exhibiting weight loss and 2 of 8 mice succumbing (Fig. 2a and b). Consistently, these mice had significantly higher viral genome copies in the blood, liver and spleen compared to repNP + repGPC vaccination, although viral genome copies were still significantly decreased compared to sham vaccination (Fig. 2c). Consistent with breakthrough clinical disease, three repGPC-T2A-NP mice had detectable infectious virus in these tissues as well (Fig. 2d). Cumulatively, our immunology and viral challenge data indicate that repGPC-T2A-NP confers only partial protection against lethal CCHFV infection, likely due to the diminished anti-NP antibody response.
Fig. 2.
Vaccination with repGPC-T2A-NP partially protects against lethal CCHFV challenge. Mice vaccinated with Sham, repGPC-T2A-NP, or repGPC + repNP RNA were treated with MAR1-5A3 antibody and infected with a lethal dose of 100 TCID50 CCHFV strain UG3010. Mice (N = 8) were (a) weighed daily and monitored for (b) survival until day 14 post-infection (p.i.). On D5 p.i., groups of mice (N = 6) were evaluated for (c) viral genome copies via qRT-PCR and (d) infectious virus via tissue culture infectious dose 50 (TCID50) assay in the blood, liver, and spleen. Dashed lines indicate limit of detection. Significance was calculated using one-way ANOVA; ns P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
repNP + engineered GPC variants, repGc-sol and repGc-FL, elicit robust B and T cell responses
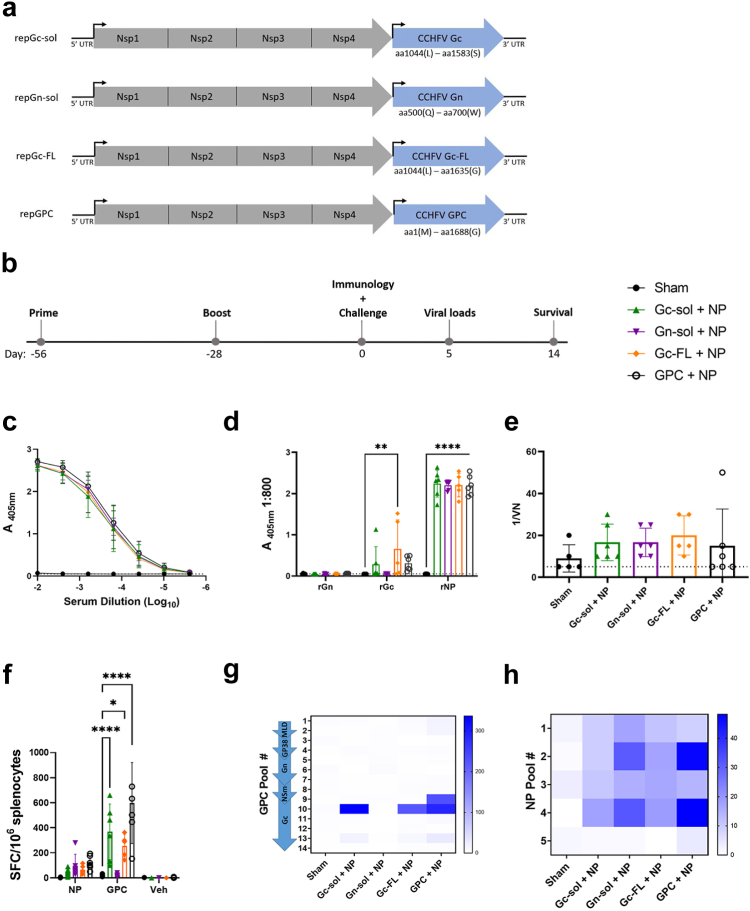
We hypothesized that the diminished antibody response and efficacy of repGPC-T2A-NP may be due to size and complexity of the CCHFV GPC, particularly when translated in the same open reading frame as NP via a ribosomal skipping mechanism. In addition, the plasmid template used to make repGPC RNA was difficult to produce (data not shown), complicating further pre-clinical and clinical development. Thus, we sought to refine the enhancing epitope within GPC that confers the enhanced protection observed in repNP + repGPC vaccination. Three engineered GPC variants were tested including soluble versions of both the Gn and Gc (Gc-sol, Gn-sol) and a full-length version of Gc (Gc-FL) that retained the native transmembrane domain (Fig. 3a). Mice were vaccinated with 1 μg of repNP + repGc-sol, repNP + repGn-sol, and repNP + repGc-FL as above (Fig. 3b) and compared with the responses in sham and repNP + repGPC vaccinated mice (Fig. 1, Fig. 2). Compared to sham vaccinated animals, all groups induced significant and robust IgG antibody titers, primarily directed against the NP with similar titers in all groups (Fig. 3c and d). Interestingly, the repGc-FL vaccination, but not repGc-sol and repGPC, induced significantly higher anti-Gc antibody titers compared to sham (Fig. 3d). Neither the repGn-sol nor repGPC groups induced detectable anti-Gn antibody (Fig. 3d). Consistent with our previous studies, antibodies in all vaccine groups had little to no neutralizing activity (Fig. 3e). In addition, CCHFV-specific T-cell responses were measured in repGc-sol, repGc-FL, and repGPC vaccinated groups while the repGn-sol vaccination did not elicit a measurable T-cell response (Fig. 3f and g). As before, none of the vaccines stimulated a strong NP T-cell response (Fig. 3f–h). Overall, both the repNP + repGc-sol and repNP + repGc-FL vaccinations elicited immune responses equivalent to the original repNP + repGPC vaccination.
Fig. 3.
repNP + repGc-sol or repGc-FL elicits robust humoral and cellular immunity. (a) Three engineered GPC variants, repGc-sol (aa1044-1583), repGn-sol (aa500-700), and repGc-FL (1044-1635), were constructed to compare to the full length repGPC (aa1-1688). WT C57BL6/J mice were (b) vaccinated with 1ug of Sham RNA or repNP RNA plus repGc-sol, repGn-sol, repGc-FL, or repGPC RNA prime-boost on days −56 and −28 relative to lethal CCHFV challenge. On D0, groups of mice (N = 6) were evaluated for immune response to CCHFV. Antibody response was evaluated via (c) whole virion IgG ELISA, (d) rNP, rGn, and rGc ELISA and (e) neutralization assay using infectious virus. Dashed lines indicate limit of detection. Cellular immune response was evaluated via IFNу ELISpot shown as (f) cumulative SFCs or (g and h) heat maps of cellular responses to peptide pools spanning the entire CCHFV GPC and NP. DMSO vehicle (Veh) is also shown. Sham and repGPC + repNP group data is duplicated from Figs. 1 and 2 for comparison. Significance was calculated using one-way ANOVA; ns P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
repGc-FL is the minimal enhancing epitope of repGPC when administered with repNP (repNP + repGc-FL)
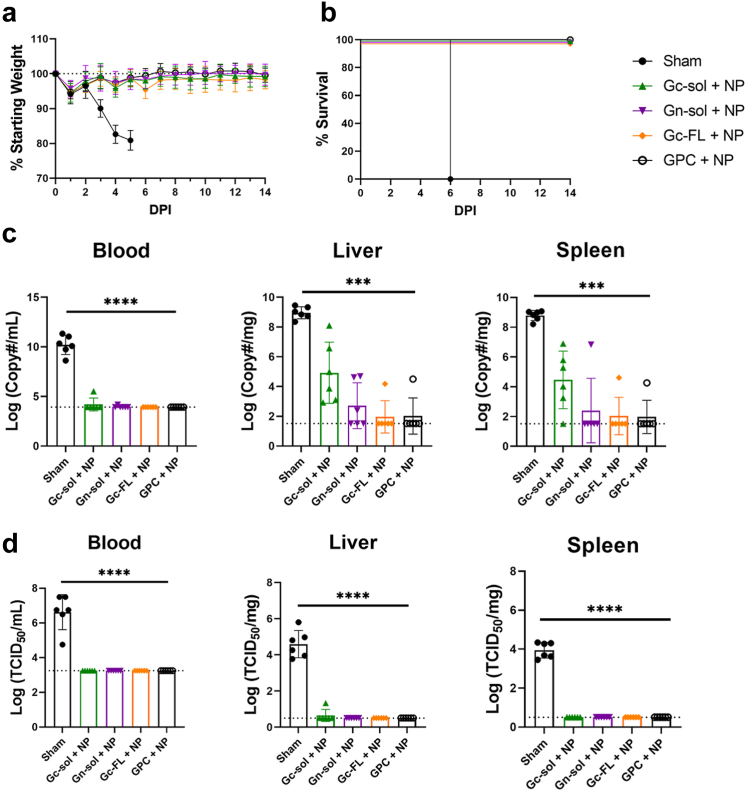
Next, we evaluated how immune responses to the repNP + repGPC engineered variants protected against lethal CCHFV challenge. Sham mice began to experience significant weight loss on D3 p.i. and all mice had succumbed to disease by D6 p.i. (Fig. 4a and b). In contrast, all other groups of mice were protected from weight loss, 100% survived (Fig. 4a and b), had little to no viremia and no infectious virus in either the liver or spleen (Fig. 4c and d). This is consistent with our previous data showing that repNP vaccination alone can confer protection from disease, death, and control infectious virus burden in these key tissues.9 However, viral RNA loads in the liver and spleen illustrated larger differences in efficacy with nearly all mice in the repNP + repGc-sol group exhibiting detectable viral RNA loads (Fig. 4c). The repGn-sol group also had diminished control of viral replication with 3 of 6 mice having detectable viral RNA in the liver compared to only 1 of 6 mice vaccinated with repGPC (Fig. 4c). On the other hand, our data demonstrate that vaccination with repNP + repGc-FL performed equivalently to the repNP + repGPC vaccine with only 1 mouse in each group positive by qRT-PCR in the liver and spleen (Fig. 4c). Overall, while all vaccinations prevented weight loss, conferred 100% survival, and significantly reduced viral genome copies and infectious virus, only the repNP + repGc-FL vaccination conferred protection equivalent to repNP + repGPC.
Fig. 4.
repNP + repGc-FL confers equivalent protection to repNP + repGPC vaccination. Mice vaccinated with sham RNA or repNP RNA plus repGc-sol, repGn-sol, repGc-FL, or repGPC RNA were treated with MAR1-5A3 antibody on D0 and infected with a lethal dose of 100 TCID50 CCHFV strain UG3010. Mice (N = 8) were monitored daily for (a) weight loss and for (b) survival until D14 p.i. On D5 p.i., groups of mice (N = 6) were evaluated for control of (c) viral genome copies via qRT-PCR and (d) infectious virus via TCID50. Dashed lines indicate limit of detection. Sham and repGPC + repNP group data is duplicated from Figs. 1 and 2 for comparison. Significance was calculated using one-way ANOVA; ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
repNP lacking the V5-epitope tag confers equivalent protection to epitope tagged repNP
Our repNP vaccine encodes a CCHFV NP with a C-terminal V5 epitope tag for in vitro expression characterization purposes. For eventual clinical development, we removed the V5 epitope tag from repNP, resulting in the repNP(ΔV5) construct and evaluated whether this would provide similar protection. RepNP(ΔV5) vaccination resulted in similar non-neutralizing, anti-NP antibody titers and T-cell responses to repNP after a single dose (Supplementary Figure S2a–e) and, during lethal CCHFV challenge, protected against weight loss while conferring 100% survival and significant control of viral genome copies with no infectious virus in evaluated tissues (Supplementary Figure S2f–i). Remarkably, repNP(ΔV5) vaccination conferred trending lower viral RNA loads compared to repNP in the blood and liver with significantly lower viral genome copies in the spleen, suggesting that removal of the V5 epitope tag may have increased efficacy of repNP vaccination and supports use of this construct for further development (Supplementary Figure S2h).
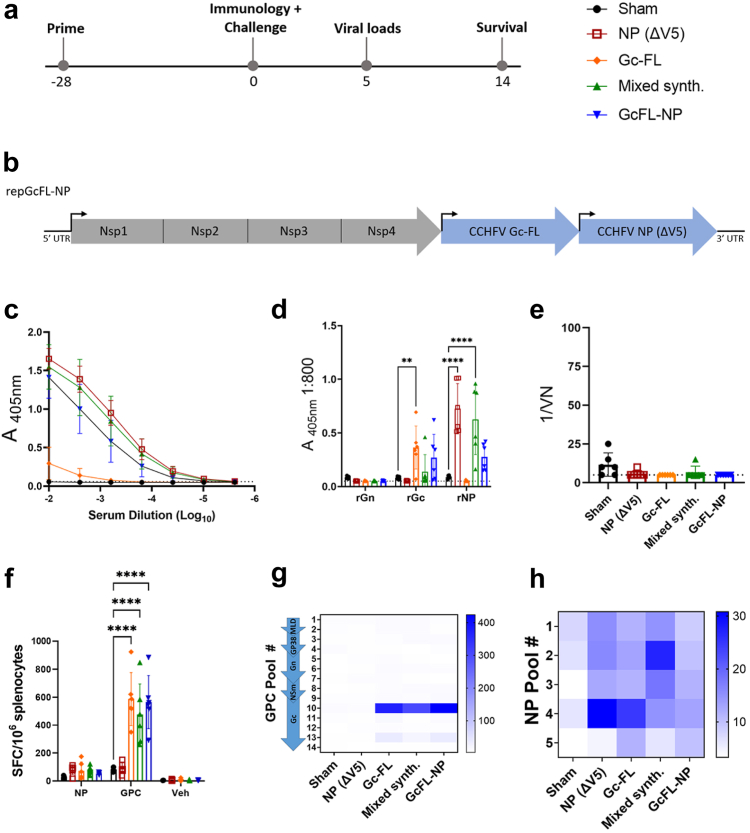
Bivalent repGcFL-NP and mixed synthesis repNP + repGc-FL RNA elicit robust B and T cell responses
With the protective subunit of repGPC narrowed to repGc-FL and confirmation that removal of the epitope tag on repNP did not negatively impact efficacy, we again tested a bivalent RNA approach to vaccination using a prime-only approach (Fig. 5a) as we have previously shown prime-only is sufficient to confer protection.9 In addition to using repNP(ΔV5) and the Gc-FL epitope to reduce the size of the expressed antigen we also utilized an alternative approach to generate a bivalent RNA. Our redesigned bivalent repGcFL-NP RNA (bivalent) contained two alphavirus subgenomic promoters driving synthesis of two independent subgenomic mRNAs, mediating translation of two separate open reading frames each encoding Gc-FL or NP(ΔV5) (Fig. 5b). This contrasts with repGPC-T2A-NP which produces a single subgenomic mRNA encoding one open reading frame that is translated into two protein products via a ribosomal skipping mechanism. We also evaluated a vaccine produced by a mixed synthesis reaction (mixed synth.) that produces individual repGc-FL and repNP(ΔV5) RNAs in one synthesis reaction thus reducing the number of in vitro transcription reactions, a costly step in manufacturing of clinical material. To evaluate these new constructs produced with different manufacturing protocols, we vaccinated wild-type mice with a single immunization of 1 μg of sham RNA, repNP(ΔV5), mixed synthesis, or bivalent RNA. Since our repGc-FL vaccine successfully induced significant antibodies against the Gc protein (Fig. 3d), we also vaccinated mice with repGc-FL alone, to determine if this response was sufficient for protection. Four weeks post-prime, groups of mice were evaluated for CCHFV-specific immune responses (Fig. 5a). Mice vaccinated with repNP(ΔV5), the mixed synthesis and bivalent RNAs had CCHFV-specific antibody responses (Fig. 5c) driven mostly by NP-specific antibody (Fig. 5d). In addition, these vaccinations also induced trending higher anti-Gc antibodies (Fig. 5d). Consistent with our previous data, repGc-FL vaccination did induce a low yet significant increase in anti-Gc antibodies though, as before, these were non-neutralizing (Fig. 5d and e). Further, repGc-FL, mixed synthesis, and bivalent RNA vaccinations induced significant T-cell responses against the GPC peptide pool 10 (Fig. 5f and g). As before, none of the vaccinations induced a significant T-cell response against the NP (Fig. 5f–h). Overall, both the mixed synthesis and bivalent RNA approaches induced significant anti-NP antibodies, like repNP(ΔV5) vaccination, and CCHFV-specific T-cell responses, like repGc-FL and previous repNP + repGPC vaccination. Furthermore, our data also demonstrate that our single, bivalent RNA approach can elicit immune responses against two distinct antigens.
Fig. 5.
Bivalent repGcFL-NP RNA and mixed synthesis repGc-FL + repNP RNA elicit robust humoral and cellular immunity. WT C57BL6/J mice were vaccinated with 1 μg of sham, repNP (ΔV5), repGc-FL, mixed synthesis (repGc-FL and repNP RNA produced in a single reaction), or bivalent (repGcFL-NP) RNA (a) prime-only on day-28 relative to CCHFV challenge. (b) repGcFL-NP is a bivalent, single RNA with two promoters driving expression of individual CCHFV Gc-FL and NP RNAs. On D0, groups of mice (N = 6) were evaluated for immune response to CCHFV via (c) whole virion IgG ELISA, (d) rAg ELISA to the CCHFV Gn, Gc, and NP and (e) neutralization assay using infectious virus. Dashed lines indicate limit of detection. Cellular immune response was assessed via IFNу ELISpot shown as (f) cumulative SFCs or (g and h) heat maps of cellular responses to peptide pools spanning the entire CCHFV GPC and NP. DMSO vehicle (Veh) is also shown. Sham and repNP(ΔV5) group data are duplicated from Supplementary Figure S2 for comparison. Significance was calculated using one-way ANOVA; ns P > 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
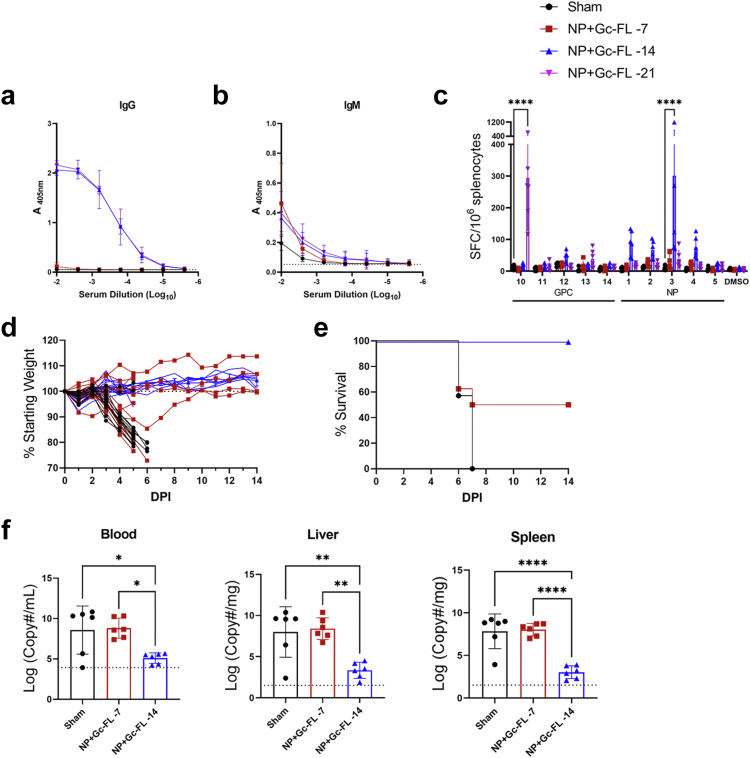
Bivalent repGcFL-NP and mixed synthesis RNA vaccinations protect against lethal CCHFV challenge
To test the efficacy of our new bivalent construct and mixed synthesis RNA, we challenged mice vaccinated with sham, repNP(ΔV5), repGc-FL, mixed synthesis, and bivalent RNA with a lethal dose of CCHFV as before. All mice in the repNP(ΔV5), mixed synthesis, and bivalent RNA groups were protected from weight loss and 100% survived (Fig. 6a and b). In contrast, despite significant anti-Gc antibodies, mice vaccinated with repGc-FL were significantly but, only partially protected with 6/7 mice experiencing weight loss and only 2 of 7 mice surviving challenge (Fig. 6a and b). At D5 p.i., all groups had significantly lower viremia but, only the repNP(ΔV5), mixed synthesis, and bivalent groups had significantly reduced viral genome copies in the liver and spleen (Fig. 6c). Remarkably, both the repNP(ΔV5) and mixed synthesis groups had no detectable viral RNA in the blood and spleen (Fig. 6c). In addition, no infectious virus was detected in the blood, liver, or spleen for repNP(ΔV5), mixed synthesis, or bivalent vaccinations (Fig. 6d). For mice vaccinated with repGc-FL alone, on the other hand, control of viral loads was incomplete (Fig. 6c) with no significant reductions of viral RNA in the liver or spleen and two mice were positive for infectious virus in the liver and spleen (Fig. 6d).
We also evaluated pathology and presence of viral antigen in formalin-fixed sections of liver and spleen from vaccinated mice. Liver samples from the sham vaccinated mice demonstrated random, multifocal to nearly diffuse hepatic necrosis with very little inflammation (Supplementary Figure S3a, Supplementary Figure S4a). Anti-CCHFV immunoreactivity was nearly diffuse in these samples except for a few cords of normal appearing hepatocytes (Supplementary Figure S3b, Supplementary Figure S4b). The liver samples from the other groups, except for Gc-FL, were essentially normal and did not express anti-CCHF immunoreactivity (Supplementary Figure S3c–l). An exception was the Gc-FL group which demonstrated hepatic necrosis ranging from mild to marked multifocal and coalescing necrosis (Supplementary Figure S3i) and CCHFV immunoreactivity (Supplementary Figure S3j). However, the anti-CCHFV immunoreactivity in repGc-FL alone mice was notably reduced from sham vaccinated animals indicating mice vaccinated with repGc-FL alone exerted some control against CCHFV. This is consistent with the significantly reduced infectious virus in this tissue (Fig. 6d). Spleen sections showed similar patterns (Supplementary Figure S5).
Cumulatively, our data suggest that non-neutralizing antibodies to NP are the primary correlate of protection. However, to evaluate the hypothesis that repRNA vaccination may prime low levels of humoral or cellular immunity that are rapidly boosted upon infection which in turn control the viral challenge, we measured antibody responses to NP, Gn and Gc, neutralizing activity and T-cell responses at day 14 PI. No anamnestic humoral response to any antigen nor any neutralizing activity was measured in any group (Supplementary Figure S6a and b). Instead, we measured significant declines in NP-specific antibodies after challenge (Supplementary Figure S6a) although it is unclear the explanation as we did not measure similar drops in titers measured using whole virus lysate in previous studies.9 Further, while the repGc-FL group did have a significant increase in T-cell responses after challenge, this increase was driven by one survivor and only two mice survived the challenge, suggesting that T-cells are not the primary determinants of protection (Supplementary Figure S6c). These data suggest that development of antibodies against the viral glycoproteins, including neutralizing antibodies, and cellular immunity to NP or the glycoproteins are not required prior to or after challenge to control the infection.
repRNA vaccination confers rapid protection against lethal challenge
Lastly, to determine how rapidly vaccination with our repRNA vaccine could confer protection, we vaccinated mice with repNP (ΔV5) + repGc-FL and evaluated immune responses 7-, 14- and 21-days post-vaccination (PV). One-week PV, IgG and IgM antibody responses against whole virion CCHFV antigen were low and T-cell responses had not yet developed (Fig. 7a–c). By two-weeks PV, mice had developed significant IgG antibody responses equivalent to those measured at 3 weeks PV (Fig. 7a). These data indicate repRNA vaccination results in robust anti-CCHFV humoral responses within two-weeks of vaccination. Interestingly, while we previously failed to measure an NP-specific T-cell response at 4 weeks PV (Fig. 1, Fig. 3, Fig. 5), we measured a significant but transient T-cell response against the CCHFV NP peptides two weeks post vaccination that was largely absent by three-weeks PV (Fig. 7c). Further, significant Gc-specific T-cell responses were not measured until 3 weeks PV (Fig. 7c) suggesting cellular immunity to Gc develops with slower kinetics than that to NP.
Fig. 7.
repRNA vaccines confer rapid protection. Mice vaccinated prime-only with sham or repNP(ΔV5) + repGc-FL RNA were euthanized seven days (T7), 14 days (T14) or 21 days (T21) after vaccination for evaluation of CCHFV-specific IgG (a) or IgM (b) by whole virion ELISA. CCHFV-specific T-cell responses were measured by IFNγ ELISpot (c). Seven or 14 days after vaccination, groups of mice were treated with MAR1-5A3 and lethally infected with CCHFV. Mice (N = 8) were monitored daily for (d) weight loss and until day 14 p.i. for (e) survival. On D5 p.i., groups of mice (N = 6) were evaluated for control of (f) viral genome copies via qRT-PCR in indicated tissues. Dashed lines indicate limit of detection. Significance was calculated using one-way ANOVA; ns P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. Data shown as mean plus standard deviation.
Upon lethal challenge, mice vaccinated one-week prior to challenge were partially protected against lethal disease (50% survival, 4/8), and three of those animals showed no weight loss (Fig. 7d and e). However, similar viral loads at day 5 PI were seen in these mice compared to sham-vaccinated mice (Fig. 7f) suggesting poor control of the viral challenge. Mice vaccinated two-weeks prior to CCHFV challenge were completely protected from clinical disease and had significantly reduced viral RNA loads in all tissues evaluated (Fig. 7d–f) demonstrating repRNA confers complete protection against CCHFV within two-weeks of vaccination.
Discussion
With the continued spread of Hyalomma ticks and growing number of people at risk for CCHFV, there is great need to develop a highly effective and safe vaccine. In this report we present continued pre-clinical development of the repRNA vaccine platform for CCHFV and identify key optimizations for eventual clinical trials. Importantly for public health, we showed that the repRNA could confer partial protection within one-week of vaccination and complete protection within two-weeks of vaccination suggesting the repRNA platform confers rapid immunity against CCHFV. Prompt protection against disease would be ideal for vaccines used to contain outbreaks. A viral replicon particle vaccine was able to confer partial protection against lethal CCHFV within three days of vaccination and complete protection within 7 days of vaccination20 and our data further demonstrate that vaccines against CCHFV can confer rapid protective immunity after a single immunization. Ongoing studies are evaluating the durability of this vaccine. This is another important consideration for deployment of vaccines in CCHFV endemic areas where the threat of infection with CCHFV is a persistent threat and limited health care resources make repeated vaccinations difficult to achieve.
In our previous CCHFV study, we found that protection conferred by repNP + repGPC is primarily mediated by non-neutralizing anti-NP antibodies and enhanced by GPC-specific T-cell responses.9 Our current study continues to support this hypothesis as protection against CCHFV challenge correlated with anti-NP antibody titers. Our repGPC-T2A-NP vaccine exhibited only partial protection that was associated with diminished anti-NP antibodies but robust Gc-specific cellular immunity. Partial protection against challenge at one-week post-vaccination and complete protection at 2 weeks post-vaccination also correlated with the levels of anti-CCHFV antibody responses. Evaluation of anamnestic antibody responses after viral challenge also indicate that rapid de novo responses after infection likely do not contribute to protection. Furthermore, our repGc-FL vaccine elicited low but significant amounts of anti-Gc antibodies along with robust CCHFV-specific T-cell responses. Nevertheless, mice vaccinated with repGc-FL alone exhibited >70% mortality indicating poor protection conferred by these responses. Our results here and previous results with the replicating RNA platform9 are in contrast to several other vaccine platforms that have shown efficacy with the CCHFV GPC alone21, 22, 23 and no efficacy with vaccine-expressed NP alone24 suggesting the vaccine platform is a key consideration for protective efficacy of CCHFV antigens. The repRNA backbone is based on Venezuelan Equine Encephalitis Virus (VEEV) and the non-structural proteins of VEEV modify the host cell environment to support viral replication and antigen expression.25 The CCHFV GPC is complex requiring multiple proteolytic processing events at various stages of the secretory pathway to form mature viral glycoproteins. It is possible that the non-structural proteins of VEEV negatively impact the proper expression or processing of CCHFV GPC leading to GPC being degraded. This in turn may lead to efficient presentation of GPC peptides to T-cells but poor B-cell responses. These effects would be absent from other vaccine platforms that report GPC-specific antibody responses. Similarly, our data indicate that non-neutralizing antibodies against the NP are required for protection. We have shown that the repNP platform induces antibodies of isotypes with high Fc-effector functions9 and it is possible that platforms in which NP failed to confer protection elicited antibodies of wrong isotype and poor Fc-effector function. Nevertheless, for most platforms demonstrating efficacy against CCHFV the correlates of protection have not been mechanistically investigated.
While our data consistently support the role of NP-specific antibodies in protection, the contribution of cellular immunity to vaccine-mediated protection for CCHFV remains largely unclear. We observed a significant but transient response to NP at 14 days post-vaccination while responses against Gc took several weeks to become detectable demonstrating cellular immunity to CCHFV antigens can occur with distinct kinetics. CCHFV-specific T-cell responses have been observed in other GPC-only or Gn/Gc including vaccines with high efficacy.26,27 Notably, a DNA-based vaccine expressing just the GPC, despite eliciting significant humoral immunity, required CD8 T-cells for protection with antibodies being dispensable for protection.28 However, our repGc-FL vaccine alone was poorly protective and a virus-like particle vaccine using ubiquitin-linked Gn/Gc antigens was only 40% protective despite robust T-cell responses,27 indicating that platform and modifications of the GPC can contribute to qualitatively different T-cell responses. Further, we have consistently found that repRNA vaccination induces a robust cellular immune response directed against the viral Gc protein and did not observe T-cell responses directed towards the Gn in mice vaccinated with the repGn-sol vaccine. IFA showed expression of the CCHFV-Gc from repGPC-T2A-NP, repGPC, repGc-sol, repGcFL, and repGcFL-NP however, Gn expression from repGn-sol could not be confirmed and this may account for the lack of Gn-specific immune responses (Supplementary Figure S1). This may also be an artifact of using inbred C57BL6/J mice throughout the studies. The narrow focus of cellular immunity against the viral Gc protein in vaccinated mice was not seen in cynomolgus macaques vaccinated with a DNA-based vaccine with CCHFV-specific T-cell responses instead directed across the viral GPC.29,30 Similarly, BALB/c and A129 mice vaccinated with a chimp-adenovirus vectored vaccine showed CCHFV-specific T-cell responses against the viral GP38, Gn, and central region of Gc.31 Ongoing studies are exploring vaccine responses in outbred mice. Additionally, we did not evaluate whether immune responses against the accessory protein GP38 contributed to repGPC protection. Immune responses against GP38 can confer significant protection32 and may influence GPC-based vaccine efficacy.23
Our data demonstrate that our bivalent repRNA platform efficiently expresses two antigens from a single RNA, Gc-FL and NP(ΔV5), to drive robust immune responses against both antigens after a single immunization. Further, we were able to measure robust anti-NP antibody responses and anti-Gc cellular immunity in mice vaccinated with our bivalent RNA demonstrating that this approach can elicit both humoral and cellular immunity to multiple antigens expressed by a single RNA (Fig. 5). Although the multivalent approach for CCHFV led to modest increases in viral control, this demonstrates a proof-of-concept for developing bivalent vaccines using our platform. Further, our mixed synthesis reaction simplifies manufacturing of multiple RNAs by producing two RNAs in a single reaction and we found this approach to confer similar efficacy as vaccination with independently produced RNAs. These approaches may be beneficial for vaccines against pathogens requiring continual updating to address emerging variants (e.g. SARS-CoV-2), vaccines that must cover distinct strains (e.g. influenza) or, pathogens with extensive geographic overlap, such as CCHFV and Rift Valley fever virus where vaccines conferring protection against multiple pathogens may be of public health benefit.3,33
In summary, we have optimized our original vaccination approach, identifying the minimal enhancing epitope in the CCHFV GPC and developing a single bivalent repGcFL-NP RNA and a mixed synthesis reaction which were highly immunogenic and efficacious as soon as two weeks post-vaccination against a lethal, heterologous CCHFV challenge in mice. These vaccine approaches induce high titers of non-neutralizing anti-NP antibodies and stimulate Gc-specific T cell responses. Our data continue to support the hypothesis that non-neutralizing antibodies directed against the CCHFV NP can confer robust protection against lethal infection and ongoing work seeks to mechanistically understand how these antibodies confer protection. These vaccines are well-tolerated, easily produced and administered, confer rapid protective immunity and are prime candidates for further development towards clinical trials. Ongoing studies are evaluating the durability of these immune responses and efficacy in non-human primate models of CCHF.
Contributors
D.W.H., J.H.E., S.S.L., and H.F. conceived and designed animal studies. D.W.H., K.M.-W., S.S.L., T.T., M.L., and E.M. performed experiments. D.W.H., C.S., J.H.E., S.S.L., and H.F. performed data analysis. D.W.H., S.S.L., and H.F. wrote the manuscript. D.W.H. and S.S.L. verified the underlying data. K.M.-W., C.S., T.T., M.L., E.M., and J.H.E. critically reviewed the manuscript. H.F. obtained funding. All authors have read and approved final version of the manuscript.
Data sharing statement
All data presented are available upon request.
Declaration of interests
J.E. has equity interest in HDT Bio. J.E. and A.K. are a co-inventor on U.S. patent application no. 62/993,307 “Compositions and methods for delivery of RNA” pertaining to formulations for RNA delivery. DWH, JE and HF are inventors on U.S. patent application number 63/365,015 “Replicating RNA vaccine for Crimean-Congo haemorrhagic fever virus” regarding the repRNA for use against CCHFV.
Acknowledgements
We thank the Rocky Mountain Laboratories Veterinary Branch (clinical veterinary services, histology core, veterinary animal care), Research Technology Branch Genomics and the Visual Medical Arts for their support of these studies. This study was supported by the Intramural Research Program of the NIAID/NIH and the Medical CBRN Defense Consortium grant #MCDC2204-011. Funders had no role in study design, data interpretation, or decision to publish.
Footnotes
Work completed in Hamilton, MT, USA.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105017.
Contributor Information
Heinz Feldmann, Email: feldmannh@niaid.nih.gov.
David W. Hawman, Email: David.hawman@nih.gov.
Appendix A. Supplementary data
References
- 1.Hawman D.W., Feldmann H. Recent advances in understanding Crimean-Congo hemorrhagic fever virus. F1000Res. 2018;7 doi: 10.12688/f1000research.16189.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawman D.W., Feldmann H. Crimean-Congo haemorrhagic fever virus. Nat Rev Microbiol. 2023;21:1–15. doi: 10.1038/s41579-023-00871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Organization WH Crimean-Congo hemorrhagic fever world health organization. 2023. https://www.who.int/health-topics/crimean-congo-haemorrhagic-fever#tab=tab_1 Available from:
- 4.Grandi G., Chitimia-Dobler L., Choklikitumnuey P., et al. First records of adult hyalomma marginatum and H. rufipes ticks (Acari: Ixodidae) in Sweden. Ticks Tick Borne Dis. 2020;11(3) doi: 10.1016/j.ttbdis.2020.101403. [DOI] [PubMed] [Google Scholar]
- 5.Egizi A., Bulaga-Seraphin L., Alt E., et al. First glimpse into the origin and spread of the Asian longhorned tick, Haemaphysalis longicornis, in the United States. Zoonoses Public Health. 2020;67(6):637–650. doi: 10.1111/zph.12743. [DOI] [PubMed] [Google Scholar]
- 6.Tsergouli K., Karampatakis T., Haidich A.B., Metallidis S., Papa A. Nosocomial infections caused by Crimean-Congo haemorrhagic fever virus. J Hosp Infect. 2020;105(1):43–52. doi: 10.1016/j.jhin.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Hawman D.W., Feldmann H. Crimean–Congo haemorrhagic fever virus. Nat Rev Microbiol. 2023;21(7):463–477. doi: 10.1038/s41579-023-00871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tipih T., Burt F.J. Crimean-Congo hemorrhagic fever virus: advances in vaccine development. Biores Open Access. 2020;9(1):137–150. doi: 10.1089/biores.2019.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leventhal S.S., Meade-White K., Rao D., et al. Replicating RNA vaccination elicits an unexpected immune response that efficiently protects mice against lethal Crimean-Congo hemorrhagic fever virus challenge. eBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erasmus J.H., Khandhar A.P., Guderian J., et al. A nanostructured lipid carrier for delivery of a replicating viral RNA provides single, low-dose protection against Zika. Mol Ther. 2018;26(10):2507–2522. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljungberg K., Liljestrom P. Self-replicating alphavirus RNA vaccines. Expert Rev Vaccines. 2015;14(2):177–194. doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- 12.ClinicalTrials.gov. 2023. https://clinicaltrials.gov/ct2/results?cond=&term=HDT-301&cntry=&state=&city=&dist= Available from:
- 13.Erasmus J.H., Khandhar A.P., O'Connor M.A., et al. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci Transl Med. 2020;12(555) doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawman D.W., Meade-White K., Clancy C., et al. Replicating RNA platform enables rapid response to the SARS-CoV-2 Omicron variant and elicits enhanced protection in naive hamsters compared to ancestral vaccine. eBioMedicine. 2022;83 doi: 10.1016/j.ebiom.2022.104196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erasmus J.H., Archer J., Fuerte-Stone J., et al. Intramuscular delivery of replicon RNA encoding ZIKV-117 human monoclonal antibody protects against Zika virus infection. Mol Ther Methods Clin Dev. 2020;18:402–414. doi: 10.1016/j.omtm.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen S.E., Erasmus J.H., Reese V.A., et al. An RNA-based vaccine platform for use against Mycobacterium tuberculosis. Vaccines (Basel) 2023;11(1):130. doi: 10.3390/vaccines11010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haddock E., Feldmann F., Hawman D.W., et al. A cynomolgus macaque model for Crimean-Congo haemorrhagic fever. Nat Microbiol. 2018;3(5):556–562. doi: 10.1038/s41564-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawman D.W., Meade-White K., Haddock E., et al. Crimean-Congo hemorrhagic fever mouse model recapitulating human convalescence. J Virol. 2019;93(18):e00554. doi: 10.1128/JVI.00554-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindquist M.E., Zeng X., Altamura L.A., et al. Exploring crimean-Congo hemorrhagic fever virus-induced hepatic injury using antibody-mediated type I interferon blockade in mice. J Virol. 2018;92(21):e01083. doi: 10.1128/JVI.01083-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spengler J.R., Welch S.R., Scholte F.E.M., et al. Viral replicon particles protect IFNAR(-/)(-) mice against lethal Crimean-Congo hemorrhagic fever virus challenge three days after vaccination. Antiviral Res. 2021;191 doi: 10.1016/j.antiviral.2021.105090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Appelberg S., John L., Pardi N., et al. Nucleoside-modified mRNA vaccines protect IFNAR(-/-) mice against crimean-Congo hemorrhagic fever virus infection. J Virol. 2022;96(3) doi: 10.1128/jvi.01568-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrison A.R., Shoemaker C.J., Golden J.W., et al. A DNA vaccine for Crimean-Congo hemorrhagic fever protects against disease and death in two lethal mouse models. PLoS Neglected Trop Dis. 2017;11(9) doi: 10.1371/journal.pntd.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suschak J.J., Golden J.W., Fitzpatrick C.J., et al. A CCHFV DNA vaccine protects against heterologous challenge and establishes GP38 as immunorelevant in mice. NPJ Vaccines. 2021;6(1):31. doi: 10.1038/s41541-021-00293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowall S.D., Buttigieg K.R., Findlay-Wilson S.J.D., et al. A Crimean-Congo hemorrhagic fever (CCHF) viral vaccine expressing nucleoprotein is immunogenic but fails to confer protection against lethal disease. Hum Vaccines Immunother. 2016;12(2):519–527. doi: 10.1080/21645515.2015.1078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhalla N., Sun C., Metthew Lam L.K., Gardner C.L., Ryman K.D., Klimstra W.B. Host translation shutoff mediated by non-structural protein 2 is a critical factor in the antiviral state resistance of Venezuelan equine encephalitis virus. Virology. 2016;496:147–165. doi: 10.1016/j.virol.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buttigieg K.R., Dowall S.D., Findlay-Wilson S., et al. A novel vaccine against Crimean-Congo haemorrhagic fever protects 100% of animals against lethal challenge in a mouse model. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinkula J., Devignot S., Akerstrom S., et al. Immunization with DNA plasmids coding for Crimean-Congo hemorrhagic fever virus capsid and envelope proteins and/or virus-like particles induces protection and survival in challenged mice. J Virol. 2017;91(10):e02076. doi: 10.1128/JVI.02076-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden J.W., Fitzpatrick C.J., Suschak J.J., et al. Induced protection from a CCHFV-M DNA vaccine requires CD8(+) T cells. Virus Res. 2023;334 doi: 10.1016/j.virusres.2023.199173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawman D.W., Ahlén G., Appelberg K.S., et al. A DNA-based vaccine protects against Crimean-Congo haemorrhagic fever virus disease in a cynomolgus macaque model. Nat Microbiol. 2021;6(2):187–195. doi: 10.1038/s41564-020-00815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawman D.W., Meade-White K., Leventhal S., et al. Accelerated DNA vaccine regimen provides protection against Crimean-Congo hemorrhagic fever virus challenge in a macaque model. Mol Ther. 2023;31(2):387–397. doi: 10.1016/j.ymthe.2022.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders J.E., Gilbride C., Dowall S., et al. Adenoviral vectored vaccination protects against Crimean-Congo haemorrhagic Fever disease in a lethal challenge model. eBioMedicine. 2023;90 doi: 10.1016/j.ebiom.2023.104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golden J.W., Shoemaker C.J., Lindquist M.E., et al. GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection. Sci Adv. 2019;5(7) doi: 10.1126/sciadv.aaw9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samy A.M., Peterson A.T., Hall M. Phylogeography of Rift Valley fever virus in Africa and the arabian peninsula. PLoS Negl Trop Dis. 2017;11(1) doi: 10.1371/journal.pntd.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.