Abstract
The meiosis-specific HOP1 gene is important both for crossing over between homologs and for production of viable spores. hop1 diploids fail to assemble synaptonemal complex (SC), which normally provides the framework for meiotic synapsis. Immunochemical methods have shown that the 70-kDa HOP1 product is a component of the SC. To assess its molecular function, we have purified Hop1 protein to homogeneity and shown that it forms dimers and higher oligomers in solution. Consistent with the zinc-finger motif in its sequence, the purified protein contained about 1 mol equivalent of zinc whereas mutant protein lacking a conserved cysteine within this motif did not. Electrophoretic gel mobility shift assays with different forms of M13 DNA showed that Hop1 binds more readily to linear duplex DNA and negatively superhelical DNA than to nicked circular duplex DNA and even more weakly to single-stranded DNA. Linear duplex DNA binding was enhanced by the addition of Zn2+, was stronger for longer DNA fragments, and was saturable to about 55 bp/protein monomer. Competitive inhibition of this binding by added oligonucleotides suggests preferential affinity for G-rich sequences and weaker binding to poly(dA-dT). Nuclear extracts of meiotic cells caused exonucleolytic degradation of linear duplex DNA if the extracts were prepared from hop1 mutants; addition of purified Hop1 conferred protection against this degradation. These findings suggest that Hop1 acts in meiotic synapsis by binding to sites of double-strand break formation and helping to mediate their processing in the pathway to meiotic recombination.
Meiosis is a crucial step in the cycle of sexual reproduction, since it reduces the chromosome complement to haploidy in preparation for fertilization. A single round of DNA replication is followed by two successive rounds of chromosome segregation to produce four haploid products. During the first division, the centromeres of homologous chromosomes are translocated to opposite poles of the meiotic spindle while sister centromeres remain associated with one another. The fidelity of this division depends on crossing over between the homologs, because the chiasmata thus formed provide for cohesion between the homologs as they become aligned on the metaphase plate. In many organisms, crossing over depends in turn on the elaboration of the synaptonemal complex (SC), which joins the homologs along their length during the period when meiotic recombination takes place (23, 32, 36). Cytologically, SC is seen as a tripartite structure, consisting of a central element flanked by two lateral elements that lie about 100 nm apart and are interconnected by transverse elements (43). It has been argued persuasively that only those recombination events that occur within the context of the SC generate stable chiasmata that are capable of facilitating proper disjunction (2, 12, 23).
The yeast Saccharomyces cerevisiae has served as an instructive model for genetic dissection of key mechanisms in meiosis. Combined genetic and cytological analyses have identified several genes that can broadly be classified on the basis of their meiotic phenotypes as providing exchange, pairing, and regulatory functions (2). Mutations in these genes generally cause a reduction in reciprocal exchange between homologs, leading to the production of spores that are inviable due to aneuploidy. Mutations in genes of the exchange category show defects in a broad range of specific functions, including the initiation of recombination, formation of double-strand breaks (DSBs), processing of recombination intermediates, and assembly of mature SCs. The pairing group is distinguished by mutants that show decreased recombination between homologs but may retain high levels of intrachromosomal recombination. These mutants—including hop1 (15), red1 (34), mek1 (35), and zip1 (44)—fail to assemble mature tripartite SCs during meiotic prophase I, leading to varied defects in meiotic disjunction. Epistasis analysis has indicated that HOP1 and RED1 function in the same pathway (29, 34), and detailed genetic and molecular dissection has provided evidence that their gene products interact directly as components of the SC (13, 17, 18, 34). Distinctive phenotypes of zip1 mutations reveal an important role for this gene in later stages of SC morphogenesis, and it has been shown that ZIP1 encodes a rod-like protein that is required for the organization of the central element of SC and for crossover interference (43, 44). Finally, regulatory genes, such as MER1 (11), are required for normal levels of recombination, with mutant alleles being characterized cytologically by their failure to form SCs to a normal extent.
Although the SC has been analyzed in great detail in numerous organisms (47), its molecular organization has only recently become accessible to investigation (23, 36). Immunochemical localization of the HOP1 product has shown that this protein is associated with the SC (16) and that it colocalizes with the RED1 product along the length of the chromosomes at early stages of synapsis (41). The deduced amino acid sequence reveals the presence of a putative zinc finger motif, which may serve as a crucial structural constituent involved in nucleic acid binding (16). To explore how HOP1 exerts its vital role in meiotic synapsis and recombination, we have devised a method to purify the gene product and have analyzed its binding to DNA in vitro. We find that Hop1 binds cooperatively and preferentially to duplex DNA and that G-rich oligonucleotides effectively compete for this binding. Furthermore, our experiments reveal that Hop1 protects free ends of added linear duplex DNA against an exonucleolytic DNase activity that is present in extracts of meiotic nuclei, suggesting a role for Hop1 in the processing of DSBs.
MATERIALS AND METHODS
Anti-TrpE-Hop1 antibodies were prepared as described previously (16). Restriction endonucleases were purchased from New England Biolabs. Unless otherwise stated, other reagents were purchased from Sigma. Circular single-stranded DNA and negatively superhelical DNA were prepared from bacteriophages M13 and M13 Gori1 as described previously (7). Negatively superhelical DNA was cleaved with appropriate restriction endonucleases to generate linear fragments of the desired length, separated by electrophoresis on polyacrylamide gels, and eluted. The DNA was labeled at either end with [γ-32P]ATP or [α-32P]dATP (3,000 Ci/mmol; Bhabha Atomic Research Center, Bombay, India) as described previously (37) and purified by gel electrophoresis. Single-stranded DNA substrates were prepared by heating linear DNA fragments at 85°C for 10 min and immediately cooling them to 4°C. Synthetic nucleic acids were purchased from Pharmacia LKB Biotechnology. The concentration of DNA is expressed as moles of nucleotide residues.
Strains, media, and genetic methods.
To prepare Hop1 protein from yeast, a HOP1 overexpression plasmid was created in two steps. First, a 4-kb PstI fragment from pNH33-2 (which contains the entire HOP1 coding sequence but lacks the upstream meiotic regulatory signals [16]) was cloned into the polylinker of the vector, pVZ1. This fragment was then removed by digestion with SphI and SalI and was ligated into the polylinker located downstream of the GAL10 promoter in plasmid pSJ101 to give pNH54-9. The (Cys-Ser) hop1 allele was cloned into pSJ101 in an identical construction starting with plasmid pNH54-9 (16) to give plasmid pNH91-9. Both pNH54-9 and pNH91-9 contain the 2μm plasmid origin of replication and the selectable marker LEU2.
Yeast media were prepared as described by Sherman et al. (40). YEPL contains 1% yeast extract, 2% peptone, and 2% lactic acid. Plasmids pSJ101, pNH54-9, and pNH91-9 were transformed into S. cerevisiae 334 (MATa pep4-3 prb1-1123 ura3-52 leu2-3112 reg1-501 gal1) (20) by the lithium acetate transformation procedure (21). For purification of Hop1, these transformants were grown in minimal medium lacking leucine.
Expression and purification of Hop1 from yeast.
YEPL (10 liters) was inoculated with 334/pNH54-9, and growth at 30°C was continued until the cell density reached 1.8 × 107 cells/ml. The production of Hop1 was then induced by the addition of galactose to a final concentration of 1%. After 10 h at 30°C, the cells were harvested by centrifugation, resuspended in 50% glycerol, and stored frozen at −80°C. All the steps of the following purification procedure were performed at 4°C. The cells were centrifuged from the thawed suspension and resuspended in 100 ml of NLB buffer (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 10% glycerol, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 1 mM benzamidine-HCl supplemented with 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 1 μg of pepstatin A per ml) for lysis. Extract was prepared from 50 g of frozen yeast cells by the use of a French press at 1,000 lb/in2 and clarified by centrifugation at 30,000 rpm for 45 min in a Beckman Ti-45 rotor. Solid ammonium sulfate (0.390 g/ml of extract) was added with continuous stirring over a period of 30 min at 4°C; after 1 h, the precipitated protein was pelleted at 12,000 rpm in an SS34 rotor. The precipitate was resuspended in 25 ml of NLB buffer and dialyzed against three changes of NLB buffer. The protein solution was then loaded by gravity onto a 100-ml DEAE-cellulose column (2 by 40 cm) which had been equilibrated with NLB buffer. The column was washed with 500 ml of equilibration buffer, during which the absorbance of the eluate at 280 nm fell to 0.02. The protein was eluted with a 500-ml linear gradient of 0.05 to 0.5 M NaCl in NLB buffer. Fractions containing Hop1, identified by Coomassie blue staining and by immunoblot assays, were combined and dialyzed against 2 liters of NLB buffer for 6 h. The dialysate was loaded onto a 10-ml heparin-agarose column (1.8 by 8 cm) which had been equilibrated with 50 ml of NLB buffer. The column was washed with 100 ml of NLB buffer, and the bound proteins were eluted with a 100-ml linear gradient of 0.05 to 0.4 M NaCl in NLB buffer. Fractions containing Hop1 were identified as described above, combined, and dialyzed against 1 liter of NLB buffer for 6 h. This highly enriched fraction was applied to a 10-ml double-stranded-DNA–cellulose column (1.8 by 8 cm) which had been equilibrated with NLB buffer. Hop1 was eluted with a 50-ml linear gradient of 0.05 to 0.5 M NaCl in NLB buffer. Column fractions were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting as described above. Fractions containing Hop1 were pooled and concentrated by using Centricon 30 microconcentrators (Amicon) and stored in NLB buffer containing 50% glycerol, instead of 10% glycerol, at −20°C. The final yield from 50 g of cells was 2.5 mg of Hop1.
Mutant (Cys-Ser) hop1 protein (16) was purified by a similar protocol with the following modifications. High-speed supernatant prepared from 25 g of induced 334/pNH91-9 cells, as described above, was chromatographed over a 40-ml SP-Sephadex column. Fractions containing mutant hop1 were pooled, dialyzed against NLB buffer, and adsorbed to a heparin-agarose column. The dialyzed hop1 protein was subsequently chromatographed on a fast protein liquid chromatography SP-5PW column. The protein-containing fractions were pooled and stored at −80°C. The yield of hop1 from 25 g of cells was 140 μg.
Protein analysis.
Protein concentrations were determined by the method of Bradford (5) with bovine serum albumin as the standard. Protein samples were analyzed by SDS-PAGE as described previously (25) and stained with Coomassie brilliant blue R-250. The amino-terminal sequence was determined by use of a gas-phase Sequenator (model 470A; Applied Biosystems). Atomic absorption spectroscopic analysis was done by a standard method at SmithKline Beecham Clinical Laboratories (Los Angeles, Calif.). Gel filtration was performed by high-pressure liquid chromatography with a Waters 300-SW sizing column run at a flow rate of 0.5 m/min. The elution profile was followed by a continuous assay of the optical density at 280 nm on a Kipp and Zonnen chart recorder. The average elution position, Kav, was computed by the equation Kav = (Ve − V0)/(Vt − V0), where Ve and Vt represent elution volumes for the sample and smallest standards, respectively, and V0 is the void volume. For sucrose gradient sedimentation, 5 ml of 5 to 20% sucrose gradients was run in an SW50.1 rotor for 5 hr at 200,000 × g and fractions collected from the bottom were assayed by polyacrylamide gel electrophoresis after being boiled in SDS.
Preparation of nuclear extracts.
Meiotic cells were brought to pachytene arrest by incubation at 36.5°C for 16 h, and nuclei were isolated as described previously (8). Briefly, cells from a 100-ml culture were harvested and resuspended in 10 ml of pretreatment solution (0.2 M Tris-HCl [pH 7.5], 2 mM EDTA, 1 M NaCl, 100 mM 2-mercaptoethanol) and incubated for 10 min at 24°C. The cells were pelleted by centrifugation, washed once with 10 ml of 50 mM KH2PO4-citrate (pH 5.8) containing 10% glycerol and 0.8 M sorbitol, and resuspended in 1 ml of spheroplast buffer (0.8 M sorbitol, 50 mM KH2PO4-citrate [pH 5.8]) containing 0.1 ml of glusulase (Dupont Corp.). After incubation at 37°C for 45 min, 20 ml of a solution (100 mM KH2PO4-citrate [pH 6.5], 10% glycerol, 1 M sorbitol, 0.5 mM MgCl2) was added and spheroplasts were collected by centrifugation. The pellet was resuspended in 5 ml of a solution (18% Ficoll [Sigma type 400], 40 mM KH2PO4-K2HPO4 [pH 6.5], 1 mM MgCl2), vortexed for 30 s, and incubated at 4°C. After 5 min, 5 ml of chilled HM buffer (0.4% Nonidet P-40, 2 M sorbitol in 20 mM KH2PO4-K2HPO4 [pH 6.5], 0.5 mM MgCl2, 24% glycerol, 8.4% Ficoll) was added and the spheroplasts were vortexed. The suspension was centrifuged at 12,000 rpm for 30 min in an SS34 rotor to remove the cell debris. The supernatant was centrifuged in a Beckman Ti-60 rotor at 30,000 rpm for 30 min. The nuclear pellet was resuspended in 500 ml of a solution consisting of 50 mM Tris-HCl (pH 7.5), 5 mM dithiothreitol, 100 mM NaCl, and 10% glycerol containing 0.1 mM phenylmethylsulfonyl fluoride supplemented with 1 μg each of pepstatin A, aprotinin, leupeptin, and benzamidine-HCl per ml and were lysed by sonication at 4°C. The suspension was centrifuged at 12,000 rpm in an SS34 rotor for 20 min. The supernatant was designated the nuclear extract.
Mobility shift assays.
Standard reaction mixtures (10 or 20 μl) for the mobility shift assays contained 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 0.1 mM ZnCl2, and the indicated concentrations of 32P-labeled DNA (10,000 cpm) and Hop1. After incubation at 24°C for 10 min, the reactions were terminated by adding 1 ml of 10× loading buffer (0.2% [wt/vol] bromphenol blue and 0.2% xylene cyanol containing 10% [vol/vol] glycerol) to each reaction mixture, and the individual samples were loaded onto either 5% polyacrylamide or 0.5% agarose gels and electrophoresed in 45 mM Tris–borate (pH 8.3) buffer for 3 h at 10°C. The gels were then dried and Hop1-DNA complexes were visualized by autoradiography. In the experiments in Fig. 4 and 9B to D, the DNA in the gel was transferred to a Nytran membrane, probed with 32P-labeled M13 DNA, and visualized by autoradiography.
FIG. 4.
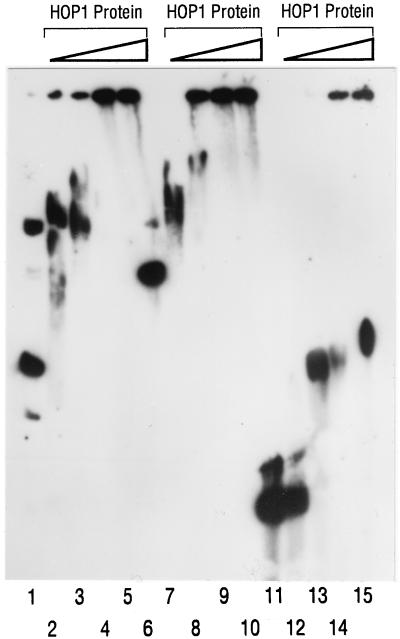
Effect of increasing the concentration of Hop1 on the formation of DNA-protein complexes with topologically different DNA substrates. The assay and reaction conditions were as described for Fig. 3, except that the concentration of Hop1 was varied. Lanes: 1 to 5, a mixture of negatively superhelical (form I; lower major band in lane 1) and nicked circular (form II; upper major band) DNA (70:30) plus Hop1 added to concentrations of 0, 0.2, 0.3, 0.4, and 0.5 μM, respectively; 6 to 10, linear duplex (form III) DNA plus Hop1 added to concentrations of 0, 0.2, 0.3, 0.4, and 0.5 μM, respectively; 11 to 15, single-stranded DNA plus Hop1 0, 0.2, 0.3, 0.4, and 0.5 μM, respectively.
FIG. 9.
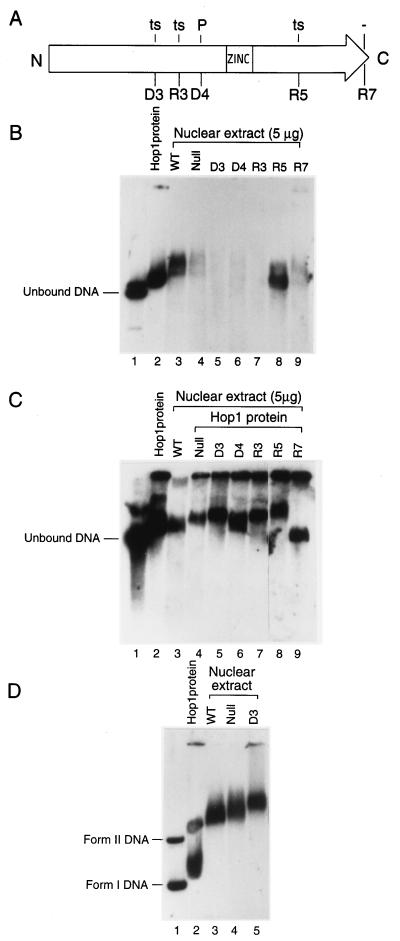
DNA binding and protection against exonuclease activity in nuclear extracts from HOP1 and various hop1 strains. (A) The open arrow represents the 605-amino-acid Hop1 polypeptide with amino-terminal (N) and carboxyl-terminal (C) ends. The effects on spore viability are indicated above the positions of in-frame linker insertion mutations that were analyzed: −, phenotypically null; ts, temperature sensitive; P, partial defect. The alleles are in-frame linker insertions from the studies of Friedman et al. (13). (B) Mobility shift and protection against DNase digestion occurring in the nuclear extracts of some mutants. Linear duplex DNA was incubated in a standard assay buffer as described for Fig. 3 with nuclear extracts from strains homozygous for the indicated alleles. Reaction mixtures containing 16 μM M13 linear duplex DNA, 5 mM MgCl2, and 0.1 μM Hop1 (lane 2) or 5 μg of protein of nuclear extracts from pachytene-arrested HOP1 (lane 3), hop1-null (lane 4), and hop1-linker insertion mutants (lanes 5 to 9) were incubated at 24°C for 30 min. The reactions were stopped by the addition of loading buffer. Samples were loaded onto a 0.5% agarose gel and analyzed by Southern blotting and autoradiography as described in Materials and Methods. (C) In vitro complementation by purified Hop1 of protection against DNase digestion in hop1 nuclear extracts. The assay and reaction conditions were as in panel B, except that the indicated mutant extracts were supplemented with Hop1 (0.1 μM). (D) Evidence that the DNase digestion by mutant nuclear extracts depends on a DSB-specific exonuclease activity. The binding-reaction mixtures contained a mixture (60:40) of negatively superhelical (form I) and nicked circular (form II) DNA with either purified Hop1 or the indicated nuclear extracts (5 μg of protein); the reactions were carried out as described for panel B.
Nitrocellulose filter binding assay.
Quantitative determination of Hop1-DNA complexes was done as described previously (24). Nitrocellulose filters (pore size, 0.22 μm; Sartorius) were pretreated with 0.5 M NaOH at 4°C for 30 min and then extensively washed with buffer (20 mM Tris-HCl [pH 7.5] containing 50 mM NaCl) prior to use. Reaction mixtures (10 μl) contained 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 0.1 mM ZnCl2, and the indicated amounts of labeled DNA and Hop1. After incubation at 24°C for 10 min, the samples were applied directly to the filter under suction and as soon as the liquid passed through the filter, the sample was washed with 4.5 ml of buffer (20 mM Tris-HCl [pH 7.5] containing 50 mM NaCl). The filters were dried, and the bound radioactivity was quantitated by liquid scintillation counting.
In competition experiments, Hop1 was incubated with 32P-labeled M13 linear duplex DNA for 10 min at 24°C before the addition of excess unlabeled competitor DNA. After incubation for 10 min, samples were passed through alkali-treated nitrocellulose filters, which were washed with buffer and dried, and the bound radioactivity was measured as indicated above.
RESULTS
Overexpression and purification of Hop1.
HOP1 is normally expressed in S. cerevisiae only under sporulation conditions, but our initial attempts to isolate sufficient Hop1 from meiotic cells for its characterization were hindered by its low abundance and limited solubility. We therefore chose to overproduce the protein in vegetative cells. To this end, we replaced the upstream meiotic regulatory sequences of the HOP1 gene with the galactose-inducible GAL10 promoter, creating plasmid pNH54-9 (Fig. 1A). In a similar construction with the (Cys-Ser)hop1 allele, we created plasmid pNH91-9 (see Materials and Methods). This allele, which is defective for function in vivo (16), contains a 1-bp mutation that results in a cysteine-to-serine change within the conserved zinc finger motif. The vector (pSJ101), as well as pNH54-9 and pNH91-9, was transformed into strain 334 (20), which is mutated for the major yeast proteases, pep4 and prb1. This strain also bears the reg1-501 mutation, which relieves the glucose repression of the galactose promoter and thereby facilitates induced expression. Hop1 was induced by the galactose shift method described in Materials and Methods, and maximal induction of the protein, which was detected as a band corresponding to its deduced molecular mass (70 kDa), was observed approximately 6 h after galactose addition (data not shown). This band, which was absent in extracts from an identically treated strain harboring the vector alone, cross-reacted with anti-trpE-Hop1 antibodies in Western blots (16).
FIG. 1.
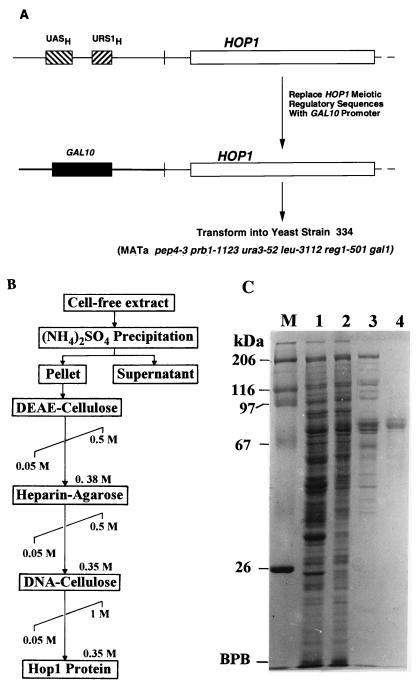
(A) Schematic representation of the relevant part of the GAL10-HOP1 construct used for overexpression of Hop1. URS1H exerts repression of the HOP1 gene in mitotic cells, while both UASH and URS1H function as activators during meiosis (46). (B) Scheme of purification of Hop1 (see Materials and Methods for details). (C) Samples containing 20 μg (lanes 1 to 3) or 5 μg (lane 4) of protein at various stages of purification were analyzed on an SDS–10% polyacrylamide gel and stained with Coomassie blue (25). Lanes: 1, cell-free lysate; 2, DEAE-cellulose; 3, heparin-agarose; 4, double-stranded-DNA–cellulose eluates; M, molecular mass markers; BPB, bromphenol blue.
We then devised a rapid method for purifying Hop1 by monitoring the enrichment of the 70-kDa band (Fig. 1B). Cell extracts were subjected to precipitation with ammonium sulfate at 60% saturation followed by successive rounds of chromatography on DEAE-cellulose, heparin-agarose, and DNA-cellulose columns. Hop1 eluted from all these matrices at moderate ionic strength. The final preparation was >95% pure for the 70-kDa protein when analyzed by SDS-PAGE (Fig. 1C). Identification of this protein as the HOP1 gene product was confirmed both by its staining with anti-trpE-Hop1 antibodies in Western blots (see above) and by direct amino acid sequencing. The amino-terminal sequence of the first 10 residues corresponded precisely to the residues predicted from the nucleotide sequence (data not shown). Purified Hop1 was devoid of either ATP-dependent or ATP-independent exonuclease or endonuclease activities in the presence of single- or double-stranded DNA, as indicated by standard assays (data not shown).
Oligomerization of Hop1.
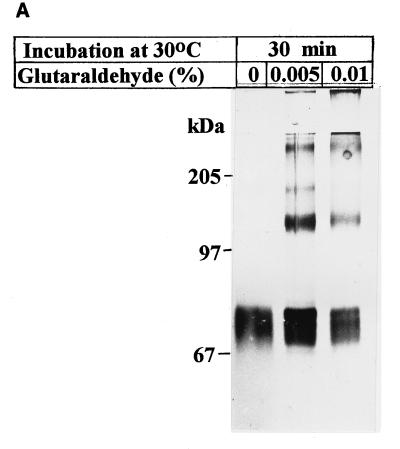
Immunolocalization of Hop1 with anti-Hop1 antibodies by both electron microscopy (16) and fluorescence microscopy (23a, 41) had shown that Hop1 is closely associated with the SC. The apparent high degree of its condensation within this region was suggestive of intermolecular interactions, and the complex pattern of intragenic complementation among hop1 alleles has also provided genetic evidence that Hop1 functions within a multimeric complex (1, 13). To establish whether purified Hop1 forms oligomers in vitro, we subjected solutions of the purified protein to glutaraldehyde cross-linking. The chemically modified products were separated by SDS-PAGE and visualized by silver staining (Fig. 2A). Although a substantial fraction of the protein remained monomeric, the presence of a discrete band at the position expected for a 130-kDa protein indicated the stabilization of a homodimeric form by cross-linking. Additionally, two weaker bands that corresponded in sizes to those expected for trimeric and tetrameric forms could be detected, suggesting that Hop1 forms oligomers of at least four subunits. The maximal number of Hop1 molecules capable of entering into oligomer formation was not evident because cross-linking at higher glutaraldehyde concentrations led to the formation of large aggregates that remained in the well (data not shown).
FIG. 2.
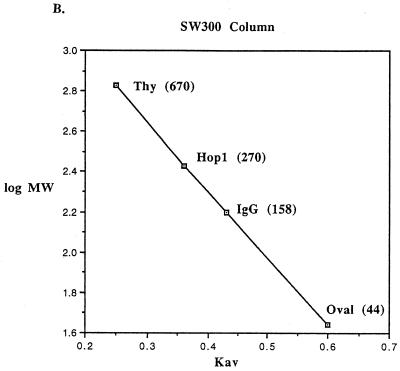
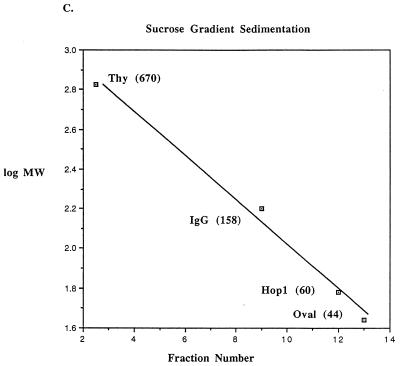
Hop1 exists in solution in the form of oligomers. (A) Cross-linking. The self-association of Hop1 in solution was examined by glutaraldehyde cross-linking. Reaction mixtures (50 μl) contained 20 mM Tris-HCl (pH 7.5), 1 mM ZnCl2, 0.35 μM Hop1, and glutaraldehyde in the indicated amounts. After incubation at 24°C for 30 min, the reaction was stopped by the addition of loading buffer. The samples were maintained at 95°C for 5 min and then electrophoresed on an SDS–7.5% polyacrylamide gel. The protein bands were visualized by silver staining. (B) Gel filtration. Hop1 (30 μg) was mixed with the indicated commercially supplied molecular mass standards (Thy, thyroglobulin; IgG, gamma globulin; Oval, ovalbumin), layered onto a sizing column (Waters SW300), and eluted at 0.5 m/min. The elution volume was computed as indicated in Materials and Methods. Values shown here and in panel C are the established molecular masses in kilodaltons of the standards and the computed apparent molecular mass of Hop1 derived from the plot. (C) Sucrose gradient sedimentation. Hop1 and the same standards as in panel B were subjected to sedimentation analysis in 5 to 20% sucrose gradients as indicated in Materials and Methods.
Gel filtration and sucrose gradient sedimentation experiments were also performed to establish the oligomeric state of purified Hop1. By comparison with globular protein standards (thyroglobulin, gamma globulin, and ovalbumin), native Hop1 was observed to elute from a sizing column at an elution volume expected for a globular protein with a molecular weight of 270,000 (Fig. 2B), which is nearly four times the actual molecular weight of the monomeric form as predicted from the sequence (16) and as assayed on SDS-containing gels (Fig. 1C). Since the migration rate on a gel exclusion (sizing) column is a function of both the size and shape of the protein, this result is consistent with Hop1 existing either as an oligomer or as an extremely extended monomer (with an estimated axial ratio of >20:1). To distinguish between these possibilities, we also subjected similar samples to sedimentation through 5 to 20% sucrose gradients along with the same molecular weight markers that were used for gel exclusion (Fig. 2C). The apparent molecular weight of Hop1 in this test was 60,000, ruling out the possibility that the native protein is an extremely extended monomer (which would have sedimented much more slowly). These findings are consistent only with the possibility that Hop1 is oligomeric, thus confirming the glutaraldehyde cross-linking evidence for its existence in a dimeric and/or larger oligomeric state.
Requirements for binding of Hop1 to linear duplex DNA.
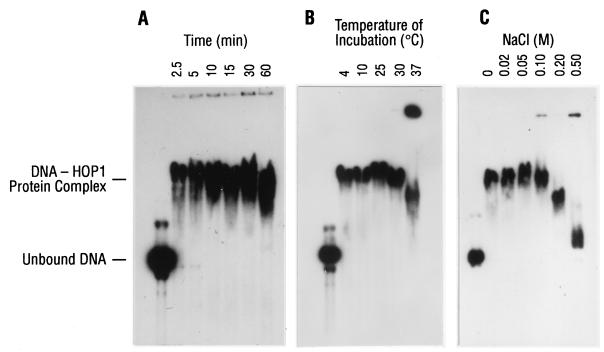
The localization of Hop1 along the SC, in conjunction with the presence of a putative zinc finger motif in the derived amino acid sequence, has raised the possibility that this protein plays its important role in synapsis by directly associating with DNA along the length of the chromatid. We originally attempted to isolate specific Hop1-binding sequences in yeast chromosomal DNA and obtained no evidence for specificity of binding to cloned DNA segments (data not shown). Accordingly, we then explored nonspecific binding to M13 DNA under a variety of conditions, using electrophoretic mobility shift assays to assess binding. In the presence of 0.1 mM Zn2+, Hop1 readily bound 32P-labeled linear duplex DNA, thereby changing its mobility in agarose gel electrophoresis (Fig. 3A). Varying the period of incubation prior to electrophoresis revealed that binding occurred maximally with the briefest incubation times tested (Fig. 3A) and was independent of temperature over a broad range (Fig. 3B), although a substantial fraction of the DNA formed aggregates with Hop1 that became trapped in the well after incubation at 37°C. Variation of ionic strength showed that binding was efficient in the presence of 0.1 M NaCl but was progressively decreased at higher concentrations, with a substantial loss of binding occurring at the highest concentration tested, 0.5 M NaCl (Fig. 3C). Collectively, these findings suggested that the reaction was optimal at 24°C in buffers containing physiological salt concentrations. The DNA-binding activity of Hop1 was unaffected by the addition of nucleotide triphosphates (data not shown).
FIG. 3.
Requirements for the binding of Hop1 to linear duplex DNA. Reactions were performed in a standard assay buffer (10 μl) containing 30 μM M13 Gori1 linear duplex DNA and 0.25 μM Hop1 as described in Materials and Methods. The reaction was terminated by the addition of loading buffer. Individual samples were loaded onto a 0.5% agarose gel and electrophoresed in 89 mM Tris borate buffer (pH 8.3) at 24°C for 16 h. The gel was stained with ethidium bromide (0.5 μg/ml), visualized under UV light, and transferred to a nylon membrane. The blot was probed with 32P-labeled denatured M13 DNA and visualized by autoradiography. (A) Time course of Hop1 binding to linear duplex DNA at 24°C. (B) Effect of incubation temperature on the binding of Hop1 to linear duplex DNA. (C) Effect of varying the concentration of NaCl on the binding of Hop1 to linear DNA. The assay was performed as described in Materials and Methods, except that the reaction mixtures contained NaCl in the amounts indicated.
Hop1 displays a hierarchy of binding to topologically different DNA substrates.
In light of evidence that the chromosomes of meiotic yeast cells contain DSBs and other forms of interruptions in the DNA during the formation of tripartite SC (2, 23), we wished to establish the topology of DNA substrates that are most favorable for binding Hop1. The formation of protein-DNA complexes upon titration of various DNA substrates with increasing concentrations of Hop1 was assayed by monitoring mobility shifts in agarose gels (Fig. 4). At the lowest protein concentration tested (0.2 μM), Hop1 bound readily to linear duplex (form III) DNA and to negatively superhelical (form I) DNA, while binding to nicked circular (form II) DNA was considerably lower (also see Fig. 7). At successively higher concentrations of Hop1, the entire populations of both form I and form III DNA showed decreasing mobility (increased binding) and about half of the molecules in each case were fully displaced to the origin at a Hop1 concentration of 0.3 μM. By contrast, most form II molecules showed no significant binding at this protein concentration. This phenomenon is clearly seen in the analogous experiments shown in Fig. 7B, where ZnCl2 is replaced with MgCl2 and the band of unbound form II DNA clearly persists at 0.3 μM, indicating that the increased accumulation of label at the origin consists almost entirely of form I DNA. Little binding to circular single-stranded DNA was seen for lower concentrations of Hop1. These findings indicate a strong affinity of Hop1 for duplex DNA and suggest that this binding is enhanced by the presence of either negative supercoiling or free double-stranded ends.
FIG. 7.
Role of divalent cations in the binding of Hop1 to different DNA substrates. (A) Effect of ZnCl2 on the binding of Hop1 to linear duplex (form III) DNA. The assays were performed as described in the legend to Fig. 3. Reaction mixtures contained 30 μM linear duplex DNA, 0.2 μM Hop1, and ZnCl2 or EDTA at the concentrations indicated. (B) Binding of Hop1 to topologically different DNA substrates in the presence of 1 mM MgCl2. Assays were performed as in Fig. 3, except that 1 mM MgCl2 was substituted for 0.1 mM ZnCl2. Lanes: 1 to 3, 30 μM total DNA as a mixture (80:20) of negatively superhelical (form I) DNA and nicked circular duplex (form II) DNA with 0, 0.2, and 0.4 μM Hop1, respectively (note that the migration of nicked circular [form II] DNA was unaffected); 4 to 6, linear duplex DNA with 0, 0.2, and 0.4 μM Hop1, respectively; 7 to 9, single-stranded DNA with 0, 0.2, and 0.4 μM Hop1, respectively.
Binding modes of Hop1 to single- and double-stranded DNA.
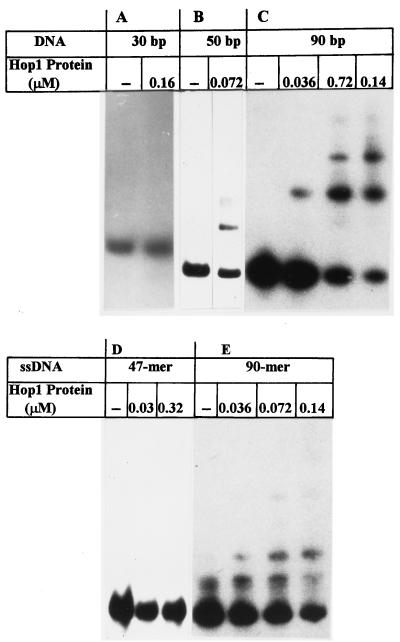
To determine the minimal length of the DNA substrate for Hop1 binding, we tested Hop1 binding to a series of linear single- and double-stranded DNA fragments of various lengths. Figure 5A to C illustrates the mobilities of DNA-protein complexes formed with various double-stranded DNA fragments upon incubation with increasing amounts of Hop1. Whereas no complex was detected for the 30-bp fragment, a small amount of DNA-protein complex was formed with the 50-bp fragment, suggesting that the formation of a DNA-bound Hop1 complex that is sufficiently stable to persist during electrophoresis requires a fragment larger than 30 bp. Consistent with this interpretation, the 90-bp fragment produced two distinct complexes, with the more abundant band (the higher-mobility species) appearing to contain a single unit of bound protein while the lower-mobility band presumably represents a doubly bound species. Formation of the latter species increased in abundance, as expected, with increasing protein concentration. For comparison, the patterns obtained with single-stranded DNA under identical conditions are also shown (Fig. 5D and E); a 90-mer single-stranded DNA fragment formed very little complex with Hop1, and the 47-mer single-stranded DNA formed none at all.
FIG. 5.
Site size determination of Hop1 binding to linear duplex DNA (A, B, and C) and single-stranded deoxyribonucleotides (ssDNA [D and E]). The assay and reaction conditions were as described for Fig. 3, except that the reaction mixtures contained 1.5 μM DNA fragments of the sizes indicated.
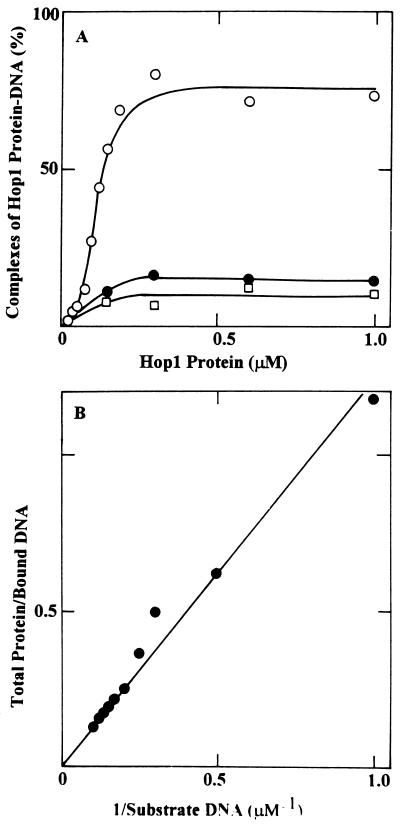
The results of these band shift studies were confirmed by a series of nitrocellulose filter-binding assays. Figure 6A shows an analysis in which a fixed amount of 32P-labeled linear duplex DNA was incubated with increasing concentrations of Hop1. The extent of Hop1 binding to 8.6-kb linear DNA (upper curve) rose with increasing protein concentrations in a sigmoidal manner, reaching a plateau at a stoichiometry of about 55 bp/monomer. We interpret the shape of this curve to mean that binding to a DNA molecule of this size is cooperative. Consistent with this interpretation, less than 20% as much Hop1 protein bound to an equivalent mass of the shorter DNA molecules, indicating considerably weaker binding. Although we cannot accurately assess the extent to which the nonoptimal conditions of the filter-binding assay may affect the affinity of DNA for Hop1, the reduced binding to smaller DNA molecules confirmed the cooperativity of binding. The nature of any multimeric complexes that might contribute to cooperativity remains to be explored. Since multimers of Hop1 protein exist in solution (as revealed by the cross-linking studies described above), these oligomers might serve to initiate binding. Alternatively, Hop1 protein may initiate binding as a monomer and undergo oligomerization along the DNA substrate.
FIG. 6.
Analysis of increasing concentrations of Hop1 on the formation of the DNA-protein complex by the nitrocellulose filter-binding assay. (A) Binding of Hop1 to linear duplex DNA of different lengths. The reaction mixtures contained 10 μM [3H]DNA with the indicated concentrations of Hop1 protein, and the assay was performed as described in Materials and Methods. ○, M13 Gori1 linear duplex DNA (8.6 kb); •, 100 bp; □, 50 bp. (B) Determination of the dissociation constant of binding of Hop1 to M13 Gori1 linear duplex DNA. The formation of Hop1-DNA complexes with different concentrations of [3H]DNA was achieved as described in Materials and Methods. The data is represented as a double reciprocal plot of total protein/bound DNA plotted against 1/substrate DNA concentration. The slope of the line yields a Kd of 1.4 × 10−7 M.
We then used these nitrocellulose filter-binding assays to determine the dissociation constant of the Hop1-duplex DNA complexes. The data obtained by titration of a fixed amount of Hop1 protein with increasing amounts of 32P-labeled linear DNA (8.6 kb) are presented as a Scatchard plot in Fig. 6B. The equilibrium dissociation constant (Kd) was found by this method to be on the order of 107 M−1.
Role of divalent metal ions in binding of Hop1 to DNA.
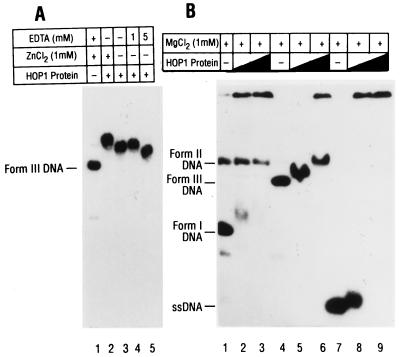
Site-specific mutagenesis has previously been used to assess the role of the putative zinc finger motif in this gene, and a Cys-to-Ser mutation was shown to result in the loss of HOP1 function in vivo (16). Accordingly, we wished to test the influence of added zinc ions on the affinity of Hop1 for DNA. Although electrophoretic mobility shift assays showed that purified Hop1 formed complexes with linear duplex DNA in the absence of added cations, the mobility of the labeled DNA (as complexed with Hop1) was further decreased by the addition of zinc ions (Fig. 7A, compare lanes 2 and 3), suggesting that this addition caused either a higher density of protein binding along the DNA or a conformational change in the complexes.
The absence of a more striking effect when Zn2+ was added led us to question the significance of the apparent zinc finger motif. A variety of different amino acid motifs containing cysteines and histidines—including Cys2/His2, Cys2/Cys2, and Cys6 forms—have been identified as zinc fingers (see reference 6 for a review), but none are identical in spacing to the pattern of Cys residues found in Hop1. We reasoned that if the novel pattern of residues present in Hop1 protein actually forms a zinc-binding structure that is essential for function, the DNA-binding activity that occurred without addition of ZnCl2 must reflect the presence of endogenous cation in the isolated protein. Atomic absorption spectroscopy was performed on our isolated wild-type Hop1, and it was found to contain 1 mol of Zn/mol of Hop1, while protein isolated from the Cys-to-Ser mutant extract lacked detectable zinc (Table 1). Dilution of wild-type Hop1 to the same concentration as in the hop1 protein (Cys→Ser) sample established that the assay was fully capable of detecting zinc at the level expected if the mutant protein had bound it as strongly as the wild type did (data not shown). Despite the apparent high affinity of the wild-type Hop1 for zinc ion, it appears likely that the ion can be removed by chelation, as indicated by the altered mobility of Hop1-DNA complexes upon incubation with EDTA (Fig. 7A, lanes 4 and 5).
TABLE 1.
Atomic absorption spectroscopic analysis of bound zinc in Hop1
| Sample | Amt of Hop1 (mol) | Amt of zinc (μg/100 ml) | Amt of zinc (mol) | Mol of zinc/mol of Hop1 |
|---|---|---|---|---|
| Buffer | 0 | <1 | <1.5 × 10−10 | |
| Hop1 | 1.2 × 10−8 | 99 | 1.4 × 10−8 | 1.2 |
| hop1 (Cys-to-Ser) | 5 × 10−10 | <1 | <1.5 × 10−10 | <0.3 |
We also tested the affinity of Hop1 for various topological forms of DNA in the presence of 1 mM MgCl2 and no added ZnCl2, perhaps better approximating the conditions for its function in vivo. As shown in Fig. 7B, these conditions led to decreased binding in comparison with the results in Fig. 4. In the presence of 1 mM MgCl2, Hop1 selectively retarded the mobility of negatively superhelical DNA and linear duplex DNA while failing to affect the mobility of nicked circular duplex DNA. The basis for this discrimination among topologically different forms of DNA by Hop1 is unknown, but we speculate that binding may be enhanced by partial unwinding of the duplex due either to negative supercoiling or to the thermal motion of double-strand cut ends.
Hop1 displays preferential binding to certain classes of DNA sequences.
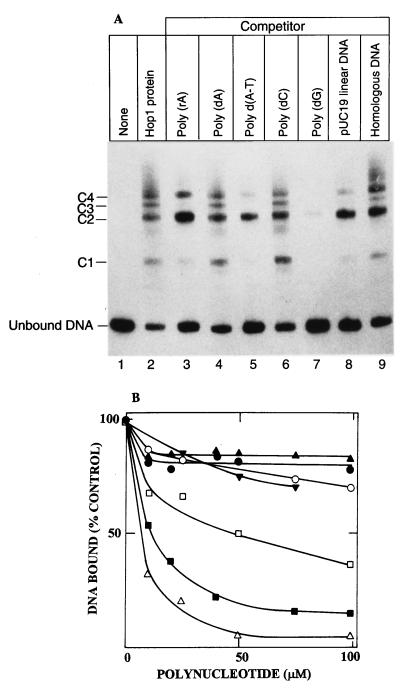
The fact that Hop1 readily bound strongly to duplex M13 DNA, as well as to every smaller restriction fragment derived from it that was tested, indicated the relative absence of specific sequence requirements for its binding. However, a general affinity for very short sequences that might be redundant in chromosomal DNA would have escaped detection by these assays. We therefore sought to determine whether various polynucleotides of simple redundant sequence might compete for binding with linear duplex M13 DNA. To this end, Hop1 was first incubated with stoichiometric amounts of a 32P-labeled 190-bp restriction fragment of M13 DNA and then challenged by the addition of various unlabeled synthetic oligonucleotides. Mobility shift assays (Fig. 8A) showed that the majority of the labeled DNA entered into the formation of four distinct Hop1-containing complexes, designated C1 to C4, when no competitor was added. When excess unlabeled competitor DNA was added and incubation was continued before analysis by gel electrophoresis and autoradiography, some of these complexes were lost. The greater relative abundance of C2 and C4 than other complexes both before and after competitors were added suggests that Hop1 protein may bind more stably in a pairwise manner (forming either dimers or tetramers of binding units) than when it binds singly. Regardless of the specific number of binding units, overall binding was entirely competed out by poly(dG) whereas incubation with poly(dA-dT) and pUC19 linear duplex DNA led to the loss of complexes C1, C3, and C4 but not C2. All of the nucleoprotein complexes resisted challenge by homologous DNA, as well as by poly(dC), poly(rA), and poly(dA).
FIG. 8.
Competitive inhibition of Hop1 binding to 32P-labeled M13 DNA by unlabeled DNA. (A) Electrophoretic gel mobility shift assay. A 190-bp M13 DNA fragment was generated by restriction endonuclease digestion. [32P]DNA (1.5 μM) was mixed with Hop1 (64 nM) in standard assay buffer as described in the legend to Fig. 3. After a 10-min incubation at 24°C, 100 μM of the indicated nucleic acid competitor was added and the incubation was continued for an additional 10 min. The nucleoprotein complexes were analyzed by nondenaturing PAGE (5% polyacrylamide) as described in Materials and Methods. (B) Quantitation of the extent of competition by a nitrocellulose filter binding assay. Hop1 (64 nM) was incubated with 1.5 μM [32P]DNA (190 bp) and challenged with different nucleic acid polymers at the concentrations indicated. Reaction mixtures were diluted with buffer and immediately filtered on nitrocellulose filters, which were assayed for bound radioactivity as described in Materials and Methods. The competitors were poly(dG) (▪), poly(dG-dC) · poly(dC-dG) (▵), poly(dA-dT) (□), poly(rA) (▾), poly(dC) (○), poly(dA) (•), and poly(rU) (▴).
Quantitative assessment of these effects under equilibrium conditions was performed by challenging the preformed 32P-labeled DNA-Hop1 complexes with different concentrations of the competitors and measuring the amounts of resistant complexes by a nitrocellulose filter-binding assay. As shown in Fig. 8B, poly(dG-dC) · poly(dC-dG) and poly(dG) competed most efficiently at low concentrations. Poly(dA-dT) was a less effective competitor, and several other homopolymers including poly(dA), poly(dC), poly(rA), and poly(rU) failed to compete detectably. We also note that heterologous (pUC19) DNA competed more effectively than homologous (M13) DNA. The reason for this difference is unknown, but we speculate that it reflects the specific molecular mechanism by which Hop1 complexes are translocated from the original bound substrate to a competing DNA that is added later.
Hop1 protects double-stranded DNA ends from exonuclease digestion that occurs in nuclear extracts of meiotic cells.
Recent genetic and biochemical studies have revealed a crucial role for DSBs in meiotic recombination (for reviews, see references 2 and 23). The 5′ ends of DSBs undergo resection to generate long single-stranded 3′ tails that are believed to act as intermediates in the formation of stable joint molecules. Molecular assays of hop1 mutants relative to the wild type have shown that DSB formation is drastically reduced (29) and that very few joint molecules are formed between the homologs (39). To gain further insight into the possible role of Hop1 in DSB repair, we have sought to determine how Hop1 may interact in vitro with duplex DNA that had been subjected to a double-strand cut. To this end, we prepared nuclear extracts from pachytene-arrested cells that are wild type for HOP1 or contain hop1 mutations including various small insertions that affect function in vivo (Fig. 9A) (13). Upon incubation of these extracts with linear duplex DNA, we found that extracts prepared from either wild-type or hop1-R5ts cells formed nucleoprotein complexes that migrated more slowly than free DNA (Fig. 9B). On the other hand, incubation of the same linear duplex DNA with nuclear extracts from the hop1 deletion strain or from the hop1-D3ts, hop1-D4p, hop1-R3ts, and hop1-R7 insertion mutants resulted in extensive digestion of the linearized DNA. Since no intermediate-size fragments of DNA were detected, it seems likely that the DNA had undergone digestion to fragments too small to be retained on the gel.
These findings suggested that an unspecified DNase activity is present in meiotic extracts of all these strains and that normal meiotic levels of functional Hop1 confer protection against this activity. Alternatively, it might have been the case that functional Hop1 had performed a function in vivo that altered the cells in such a manner that the nuclear extracts later prepared from them were devoid of the nuclease activity. To distinguish between these alternatives, in another set of assays we added purified Hop1 to an aliquot of each extract before incubation with linear duplex DNA. As shown in Fig. 9C, this addition of functional Hop1 prevented degradation of the linear DNA, consistent with the interpretation that the wild-type protein interferes directly with the action of a nuclease(s) present in the extracts, presumably by virtue of its demonstrable binding to the DNA. We also note that the probed DNA that was protected against degradation by supplementation with purified Hop1 was retarded in its gel migration to a greater extent than occurred for the unsupplemented extract from HOP1 cells. This indicates that the Hop1-DNA complexes formed in unsupplemented HOP1 extracts, although protected against degradation, were not fully saturated for Hop1 binding. In turn, this suggests that nonsaturating amounts of Hop1 preferentially bind to a subset of potential binding sites that more effectively confer protection against degradation.
Given the demonstrable ability of Hop1 to confer protection against nuclease digestion of linear duplex DNA, we wished to establish whether the digestion in question resulted principally from exonuclease and/or endonuclease activity. We therefore carried out similar experiments in which we incubated negatively superhelical DNA and nicked circular duplex DNA with nuclear extracts. As shown in Fig. 9D, neither of these circular DNA substrates was digested detectably in nuclear extracts from hop1 mutants. We conclude that the digestion of linear DNA must have resulted from the action of one or more exonuclease(s) and that endonuclease activity was insignificant to the reaction. Accordingly, it seems clear that Hop1 protects linear duplex DNA against degradation in these meiotic extracts by preventing an exonuclease from acting. The relevant exonuclease in these nuclear extracts remains to be identified. Other workers have demonstrated that the DNase encoded by NUC1 resides inside the mitochondria and plays a role in the extent of gene conversion tracts within the mitochondrial genome (51). Electron microscopic examination of fractionated nuclear isolates used for the preparation of these extracts has revealed few, if any, contaminating mitochondria (data not shown), but it remains possible that the NUC1 nuclease represents the predominant activity detected in the hop1 extracts. Regardless of whether the observed exonucleolytic activity corresponds to one that mediates resection of chromosomal DSBs in vivo, the present findings clearly demonstrate that Hop1 can confer protection against exonucleolytic attack and suggest that it may play a similar role in vivo.
We note that this Hop1-mediated protection against exonuclease activity appears largely independent of the sequence at the free end of the duplex DNA, since several linear duplexes generated by cleaving circular M13 DNA with different restriction endonucleases all yielded the same result (data not shown). We also note that negatively superhelical M13 DNA was retarded in every lane (Fig. 9D), including one from the extract that completely lacked Hop1 protein (lane Null). This result demonstrates that the mobility shifts seen for these complete nuclear extracts represent DNA-binding activities that can occur in the absence of HOP1, although Hop1 probably enters into the complexes when it is present. Therefore, DNA-binding proteins besides Hop1 must form complexes with linear duplex DNA, but these other proteins cannot confer protection against exonucleolytic degradation unless functional Hop1 is present as well.
DISCUSSION
In this study, we have isolated and characterized the product of the HOP1 gene to explore how it helps confer on meiotic yeast cells their capacity for homologous synapsis and recombination. The large-scale production of Hop1 for this purpose was facilitated by use of a galactose-regulatable HOP1 construct expressed in vegetative culture. Confirmation that the purified protein is the HOP1 gene product was provided by demonstrating its affinity in immunoblots for antibody raised against a trpE-Hop1 fusion protein. Furthermore, we have shown that the N-terminal sequence of 10 amino acids corresponded exactly to that predicted from the nucleotide sequence. Interestingly, gel electrophoretic analysis of purified Hop1 that had been subjected to chemical cross-linking has provided evidence that Hop1 exists in solution in oligomeric forms, including dimers, trimers, and tetramers. These findings corroborate evidence from interallelic complementation of insertional hop1 mutations that the protein functions as a multimer in vivo (1, 13).
How does Hop1 perform its key role in meiosis? The HOP1 gene had been identified in a screen for mutations that cause reduced recombination between homologs while retaining high levels of meiotic recombination between tandem sequences (15). hop1 mutants fail to assemble mature tripartite SCs, and most of the spores produced are inviable because of extensive meiotic nondisjunction. Analysis of the cloned HOP1 gene revealed that the derived gene product contains an essential zinc finger motif and that the expression of the gene is restricted to meiosis (16). Furthermore, antibody localization demonstrated that Hop1 is a constituent of the meiotic chromosomes (16, 41). These findings clearly suggest that Hop1 mediates its function in meiotic synapsis by a direct interaction with chromosomal DNA. In the present work, we have shown that purified Hop1 binds strongly and rather nonspecifically to double-stranded DNA in an ATP-independent manner while its binding to single-stranded DNA is substantially weaker. We have also shown that the binding to duplex DNA occurs rapidly at physiological ionic conditions, thereby suggesting the presence of Hop1-DNA complexes within meiotic cells throughout the period when the protein is present. Mobility shift and nitrocellulose filter-binding assays revealed that its binding to linear duplex DNA saturates at a stoichiometric ratio of approximately 1 monomer/55 bp. When short restriction fragments of duplex DNA were tested in mobility shift assays, Hop1 was seen to form nucleoprotein complexes with a fragment of 50 bp but not with one of 30 bp. Strikingly, the binding to DNA fragments of minimal length required severalfold-larger amounts of Hop1 to achieve saturation of binding, corroborating evidence from filter-binding assays with larger DNA substrates that the binding is cooperative.
These studies have revealed a striking hierarchy in the pattern of Hop1 binding to topologically different DNA substrates, with the strongest binding occurring on linear and supercoiled duplex DNA, less affinity for nicked circular duplexes, and yet weaker binding to single-stranded DNA. It is clearly implicit in these findings that the association of Hop1 previously shown by antibody staining (16) reflects a strong affinity of the protein for double-stranded DNA. In light of the substantial evidence that DSBs play a key role in the initiation of meiotic recombination (23), it seems reasonable to suppose that duplex DNA adjacent to DSBs is a preferred substrate for the assembly of Hop1. What, then, is the state of the DNA at the time of Hop1 binding? The greater binding to negatively supercoiled DNA than to single-stranded DNA suggests that the bound state entails partial (but not complete) unwinding of the DNA duplex. We speculate that the formation of such partially unwound complexes in relaxed DNA would entail a concerted interaction by several Hop1 molecules aligned along the DNA duplex and that the stochastic occurrence of this concerted reaction might be favored by an underwinding of the DNA duplex that occurs transiently next to cut ends. It seems likely that in the meiotic cell, Hop1 initially binds to negatively supercoiled DNA prior to DSB formation and then retains a high affinity for the ends once cutting has occurred. Whatever the pathway of functions, it is evident that Hop1 can play a decisive role at free ends in vitro, since it effectively protects against the exonuclease activity that is intrinsic to our meiotic nuclear extracts (Fig. 9).
In considering the binding of Hop1 to DNA, it must also be recognized that most of the assays reported here were performed in the absence of any other protein(s). Clearly, it may be the case that the interaction of Hop1 with chromosomal DNA both in vivo and in the meiotic nuclear extracts is altered by the presence of other constituents of the meiotic nucleus. The protein is large enough that only a portion of the molecule may be required for interaction with DNA while other portions could remain free to bind other components of the SC. Specifically, a complex pattern of genetic interactions suggests that Hop1 interacts not only with DNA but also with the RED1 gene product in performing its essential function in synapsis (13, 17, 18, 41).
The presence of a putative zinc finger motif in the derived Hop1 sequence had suggested a role for zinc ion in the function of this protein, and site-directed mutagenesis to create a cysteine-to-serine mutation confirmed the importance of this motif for HOP1 function in vivo (16). Although the present assays have shown that purified Hop1 binds DNA well without the addition of exogenous Zn2+, we have also shown that the purified protein contains 1 mol of Zn/mol of protein whereas no bound zinc was detectable in the protein isolated from a hop1 mutant defective in the putative zinc finger motif. We therefore believe it likely that the bound Zn2+ already present in the purified preparation fulfills a requirement for this cation in DNA binding. It may be relevant in this regard that the addition of MgCl2, rather than ZnCl2, resulted in an overall decline in the extent of Hop1 binding to all topological DNA forms that were tested. In particular, we detected a substantial reduction in the formation of nucleoprotein complexes with nicked circular duplex DNA when only Mg2+ was added. Since this form of DNA binds Hop1 less strongly than does linear duplex DNA even when Zn2+ is present, we posit that the marginal affinity of Hop1 for nicked circular DNA may be further compromised by substituting Mg2+ for Zn2+ in the potential binding sites within the protein.
The results presented in this report substantiate the concept that Hop1 exerts its influence on meiotic synapsis through a direct interaction with duplex DNA. Although we have found little sequence specificity in the binding of Hop1 to many DNA sequences that we have tested (data not shown), we found that G-rich oligonucleotides competed effectively for binding with M13 DNA while poly(dA-dT) sequences showed the same effect but to a lesser extent (Fig. 8). What role might the apparent affinity of Hop1 protein for G-rich DNA, and perhaps for poly(dA-dT), play in meiotic synapsis or recombination? We entertain the possibility that these sequences interact with Hop1 to form structural intermediates for the synapsis of homologous chromatids during meiotic prophase. It is perhaps relevant that Hotta and Stern (19) observed during zygotene in the Lilium anther that about 0.3% of the DNA, which is GC rich, undergoes delayed completion of its replication in a manner that is essential for full synapsis to occur. In addition, there have been several studies implicating G-rich sequences in yeast as hot spots for recombination (27, 33, 48), and, intriguingly, GT-rich sequences are preferentially found at sites of recA-mediated recombination in bacterial cells (45). On the other hand, we note that the frequencies of DSB formation and gene conversion in the ARG4 hot spot for meiotic recombination in yeast were reduced by a deletion that removed a poly(dA-dT) tract (38). A similar tract is also seen in the recombinational hot spot near THR4 (14, 52), perhaps consistent with a role for Hop1 in interactions with these sequences.
Molecular analysis of several recombination hot spots in yeast has provided compelling evidence that DSBs formed in these regions serve to initiate meiotic recombination (9, 26, 31, 49, 50). The enzyme that catalyzes DSB formation has recently been identified as the Spo11 protein, which has considerable sequence identity to the A subunit of archaeal topoisomerase VI; Spo11 becomes covalently attached to the 5′ end of the DNA molecule that has experienced DSB formation (3, 22). Mutational analysis of yeast meiosis has identified a number of other genes—including the RAD51-57 series, DMC1, MRE2, and MRE11—that are then crucial for the recombinational repair of the DSBs thus formed. Recent genetic studies have also implicated additional proteins that must interact with Rad51 and Rad52 proteins for efficient DSB repair (10). These findings, in conjunction with the results reported in our present study, lead us to propose that Hop1 plays a prominent role in meiotic DSB repair and recombination in vivo. Of the many genes essential for meiotic recombination in yeast, several specify proteins that are homologous to Escherichia coli RecA; these genes include RAD51, RAD55, RAD57, and DMC1 (2, 4, 23, 28). Like RecA protein, Rad51 protein polymerizes on single-stranded DNA in the presence of ATP to form helical nucleoprotein filaments that catalyze the formation of heteroduplex DNA in vitro (30, 42). By virtue of its homology to RecA, it has been proposed that Dmc1 also assembles on DNA, alone or together with Rad51, to produce nucleoprotein filaments (4, 30). The view that Dmc1 and Rad51 act in parallel in meiotic recombination is consistent with their colocalization in a punctate staining pattern during meiotic prophase (4). In hop1 mutants, such foci of Dmc1 localization arose at a later stage than usual and were only faintly detected in comparison with controls (4), indicating that Hop1 may act in some manner to modulate the formation or processing of DSBs.
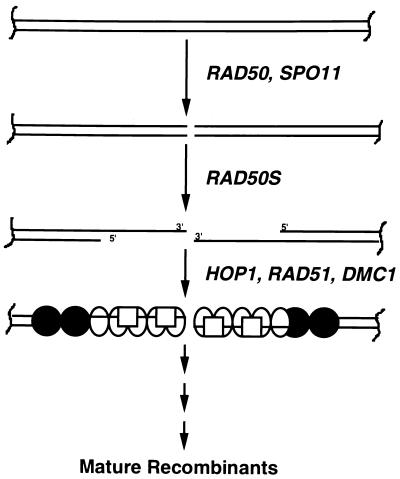
In light of these observations, we suggest that Hop1 binds at or near sites of DSB formation, serving both to modulate the initial cleavage and to delimit the extent of resection of the 5′ strands at the broken ends (Fig. 10). Single-stranded DNA, to which Hop1 binds much less strongly, might remain free for interaction with single-stranded-DNA-binding proteins, such as Rad51 and Dmc1, which in turn would target strand invasion within the homolog. It may also be the case, of course, that Hop1 directly recruits to the DSB other proteins that are required for enzymatic and/or structural roles in DNA repair during meiotic synapsis and recombination.
FIG. 10.
Hypothetical mechanism for the involvement of Hop1 in DSB repair. The cleavage of duplex DNA, presumably at a hot spot by the meiosis-specific Spo11 endonuclease, and resection of 5′ ends generate long 3′ single-stranded tails. Rad51 (ovals), Dmc1 (squares), and possibly other proteins that participate in recombinational repair may polymerize on the adjacent single-stranded DNA either individually or in combination. By binding more strongly to duplex DNA, Hop1 (circles) may regulate the extent of resection and/or other aspects of DSB formation and processing.
ACKNOWLEDGMENTS
This work was supported by fellowships from the Rockefeller Foundation, a grant from the Department of Science and Technology to K.M., and NIH research grants GM18541 to B.B. and GM44532 to Alexander D. Johnson. K.M.K. was the recipient of a fellowship from the Council of Scientific and Industrial Research, New Delhi, India. N.M.H. was supported by Damon Runyon-Walter Winchell Cancer Research Fund Fellowship DRG-965. K.M. thanks G. Padmanaban for his generous help.
REFERENCES
- 1.Ajimura M, Leem S-H, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atcheson C L, Esposito R E. Meiotic recombination in yeast. Curr Opin Genet Dev. 1993;3:736–744. doi: 10.1016/s0959-437x(05)80092-9. [DOI] [PubMed] [Google Scholar]
- 3.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:329–331. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 4.Bishop D K. RecA homologs of Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Coleman J E. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham R P, DasGupta C, Shibata T, Radding C M. Homologous pairing in genetic recombination: RecA protein makes joint molecules of gapped and closed circular DNA. Cell. 1980;20:223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- 8.Davidow L, Byers B. Enhanced gene conversion and postmeiotic segregation in pachytene-arrested Saccharomyces cerevisiae. Genetics. 1984;106:165–183. doi: 10.1093/genetics/106.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Massy B, Rocco V, Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donovan J W, Milne T D, Weaver D T. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 11.Engebrecht J, Roeder G S. Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics. 1989;121:237–247. doi: 10.1093/genetics/121.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engebrecht J, Hirsch J, Roeder G S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell. 1990;62:927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- 13.Friedman D B, Hollingsworth N M, Byers B. Insertional mutations in the yeast HOP1 gene: evidence for multimeric assembly in meiosis. Genetics. 1994;136:449–464. doi: 10.1093/genetics/136.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldway M, Sherman A, Zenvirth D, Arbel T, Simchen G. A short chromosomal region with major roles in yeast chromosome III meiotic disjunction, recombination and double-strand breaks. Genetics. 1993;133:159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingsworth N M, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollingsworth N M, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- 17.Hollingsworth N M, Johnson A D. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. Genetics. 1993;133:785–797. doi: 10.1093/genetics/133.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollingsworth N M, Ponte L. Genetic interactions between HOP1, RED1, and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast, Saccharomyces cerevisiae. Genetics. 1997;147:33–42. doi: 10.1093/genetics/147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotta Y, Stern H. Analysis of DNA synthesis during meiotic prophase in Lilium. J Mol Biol. 1971;55:337–355. doi: 10.1016/0022-2836(71)90322-6. [DOI] [PubMed] [Google Scholar]
- 20.Hovland P, Flick J, Johnston M, Sclafani R A. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 21.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 23.Kleckner N, Padmore R, Bishop D K. Meiotic chromosome metabolism: one view. Cold Spring Harbor Symp Quant Biol. 1991;56:729–743. doi: 10.1101/sqb.1991.056.01.082. [DOI] [PubMed] [Google Scholar]
- 23a.Klein, F., and B. Byers. Unpublished data.
- 24.Kumar A K, Mahalakshmi S, Muniyappa K. DNA-induced conformational changes in RecA protein: evidence for structural heterogeneity among nucleoprotein filaments of RecA protein-single stranded DNA. J Biol Chem. 1993;268:26162–26170. [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Wu T-C, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 28.Lovett S T. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene. 1994;142:103–106. doi: 10.1016/0378-1119(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 29.Mao-Draayer Y, Galbraith A M, Pittman D L, Cool M, Malone R E. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa T, Yu X, Shinohara A, Egelman E H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 31.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J, Jones J, Pringle J, editors. Molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 396–401. [Google Scholar]
- 33.Pluta A F, Zakian V A. Recombination occurs during telomere formation in yeast. Nature (London) 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- 34.Rockmill B, Roeder G S. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci USA. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rockmill B, Roeder G S. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- 36.Roeder G S. Sex and the single cell: meiosis in yeast. Proc Natl Acad Sci USA. 1995;92:10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schultes N P, Szostak J. A poly(dA/dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 40.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 41.Smith A V, Roeder G S. The yeast Red1 protein localizes to cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 43.Sym M, Engebrecht J, Roeder G S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 44.Sym M, Roeder G S. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 45.Tracy R B, Chedin F, Kowalczykowski S C. The recombination hot spot chi is embedded within islands of preferred DNA pairing sequences in the E. coli genome. Cell. 1997;90:205–206. doi: 10.1016/s0092-8674(00)80328-1. [DOI] [PubMed] [Google Scholar]
- 46.Vershon A K, Hollingsworth N M, Johnson A D. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol Cell Biol. 1992;12:3706–3714. doi: 10.1128/mcb.12.9.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Wettstein D, Rasmussen S W, Holm P B. The synaptonemal complex in genetic recombination. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 48.White M A, Dominska M, Petes T D. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu T-C, Lichten M. Meiosis-induced double-strand breaks determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 50.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zassenhaus H P, Denniger G. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr Genet. 1994;25:142–149. doi: 10.1007/BF00309540. [DOI] [PubMed] [Google Scholar]
- 52.Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]