Abstract
Gene targeting is a very powerful tool for studying mammalian development and physiology and for creating models of human diseases. In many instances, however, it is desirable to study different modifications of a target gene, but this is limited by the generally low frequency of homologous recombination in mammalian cells. We have developed a novel gene-targeting strategy in mouse embryonic stem cells that is based on the induction of endogenous gap repair processes at a defined location within the genome by induction of a double-strand break (DSB) in the gene to be mutated. This strategy was used to knock in an NH2-ezrin mutant in the villin gene, which encodes an actin-binding protein expressed in the brush border of the intestine and the kidney. To induce the DSB, an I-SceI yeast meganuclease restriction site was first introduced by gene targeting to the villin gene, followed by transient expression of I-SceI. The repair of the ensuing DSB was achieved with high efficiency (6 × 10−6) by a repair shuttle vector sharing only a 2.8-kb region of homology with the villin gene and no negative selection marker. Compared to conventional gene-targeting experiments at the villin locus, this represents a 100-fold stimulation of gene-targeting frequency, notwithstanding a much lower length of homology. This strategy will be very helpful in facilitating the targeted introduction of several types of mutations within a gene of interest.
The ability to introduce specific alterations of endogenous genes into the germ line of mice via targeted mutagenesis in embryonic stem (ES) cells has represented a major breakthrough in mouse genetics. Gene inactivation has been widely used to examine the effects of loss of function in various biological processes such as development, cellular biology, and physiology. This has already permitted the accumulation of new insights into gene function and also the creation of mouse models of human genetic diseases. Introduction of subtle mutations at specific locations of the mammalian genome is also useful to refine genetic analysis and to produce models of genetic diseases which do not necessarily result from null mutations. Several strategies have been developed, each aimed at generating subtle mutations in a given gene (6). One common limitation to all current gene-targeting procedures is the low frequency of correct targeting. This becomes a serious problem especially with use of two successive rounds of targeting, a method common to several strategies used for the generation of mutated genes devoid of foreign selection sequences. Therefore, attempts have been made to increase the efficiency of gene targeting by several means, such as increasing the size of the region of homologies with the target locus, using isogenic genomic DNA, or improving the selection procedures (6).
In this report, we present an alternative approach to overcome these limitations which relies on the observation that double-strand ends of broken chromosomes are highly recombinogenic (reviewed in reference 5). Double-strand breaks (DSB) are frequently associated with DNA alteration events in eukaryotes (4, 31); during meiosis in Saccharomyces cerevisiae, for example, transient DSB are induced at a number of positions known to be hot spots for recombination (23). It has recently been shown that a unique DSB can specifically be induced in the yeast (12), plant (24), and mammalian (8, 22, 26, 28) genomes by using the yeast I-SceI meganuclease. The I-SceI protein is an endonuclease responsible for intron homing in yeast mitochondria, a process that apparently proceeds by DSB repair (18); I-SceI endonuclease can induce recombination in yeast nuclei (12). In mammalian cells, the yeast meganuclease I-SceI has been shown to efficiently induce a DSB in a chromosomal target containing an I-SceI recognition sequence. This allows DNA break repair with high frequency by recombination with a donor molecule homologous to the regions flanking the break (7, 8, 22, 25, 26, 28).
We reasoned that the introduction of a DSB in an endogenous gene could increase targeting frequency at this natural locus through stimulation of the cellular recombination machinery. The gene encoding villin, a major component of the actin cytoskeleton of intestine and kidney cells (13), was chosen to develop this gene targeting strategy. We found that induction of a DSB in the target gene by using the meganuclease I-SceI resulted in greatly enhanced homologous replacement by the incoming DNA, even when the length of genomic DNA homology is reduced.
MATERIALS AND METHODS
Constructs and electroporation of ES cells.
The targeting construct was made as follows. An I-SceI restriction site was introduced in a unique XhoI site flanking the 5′ end of the neomycin resistance (neo) gene (pMC1neo; Stratagene), using an oligodimer (sense oligonucleotide, TCGAGTAGGGATAACAGGGTAAT; antisense oligonucleotide, TCGAGATTACCCTGTTATCCCTA). We derived from the PGK-hygromycin resistance (Hygror) gene (32) two nonfunctional Hygror cassettes, hygro A-B and hygro B-C. These two cassettes shared an 800-bp region of homology (region B), between the AatII and SacII restriction sites. Homologous recombination of the two genes will lead to a functional Hygror gene (hygro A-B-C). When electroporated into ES cells, neither of the hygro A-B and hygro B-C cassettes was able to confer hygromycin resistance. The hygro B-C fragment is 1.5 kb long, starting at the SacII restriction site of the PGK-Hygror gene and including part of the coding sequence, the stop codon, and the simian virus 40 poly(A) region. This fragment was ligated 3′ to the I-SceI/neo cassette. This I-SceI/neo hygro B-C cassette was then introduced in a unique KpnI site present in a 9.6-kb BglII-BamHI fragment isolated from a λDASHII phage containing 16 kb of the mouse villin gene (kindly furnished by G. Tremp, Rhone Poulenc Rorer) and subcloned in pBS/KS+ (Stratagene). Insertion of the I-SceI/neo hygro B-C cassette disrupted the second exon of the villin gene. A 2-kb thymidine kinase (TK) cassette (33) was subcloned in the unique ClaI site flanking the 5′ end of the construct. The resulting pvillin I-SceI/neo hygro B-C targeting construct contains 6.1 kb of 5′ and 3.5 kb of 3′ villin genomic sequence flanking a 2.5-kb I-SceI/neo hygro B-C cassette.
The replacement construct was made as follows. A 5′ 2-kb BamHI-NcoI villin gene fragment (located upstream of the initiation codon) was subcloned upstream of the 1-kb NH2-terminal domain of human ezrin cDNA fused to nucleotides encoding the 11-amino-acid carboxy terminus of the vesicular stomatitis virus glycoprotein G (1). The 1.5-kb-long hygro A-B fragment includes the PGK promoter and part of the Hygror gene coding sequence ending at the AatII restriction site of the PGK-Hygror gene. This hygro A-B cassette was subcloned downstream of the 3-kb villin-NH2-ezrin fragment, resulting in the pvillin NH2-ezrin hygro A-B replacement construct. A total of 2 × 107 CK35 ES cells (9) were electroporated with 20 μg of the NotI-linearized targeting construct. G418 (300 μg/ml) and gancyclovir (2 μM) were added 36 h after plating for 8 days. Cell culture was performed in Dulbecco modified Eagle medium (Gibco-BRL) supplemented with 1 mM sodium pyruvate, 5% fetal calf serum (Seromed, Berlin, Germany), 1,000 U/ml LIF (ESGRO; Gibco-BRL) per ml, and 50 mM β-mercaptoethanol (Gibco-BRL) as described previously (9). The G418-resistant, gancyclovir-resistant (Gangr) clones were isolated, and their genotypes were analyzed by Southern blotting. The I-SceI-targeted ES clone (ES 3.1) was chosen for further experiments.
The supercoiled pI-SceI expression plasmid (allowing expression of the yeast endonuclease I-SceI under the control of the cytomegalovirus promoter [7]) and the supercoiled pvillin NH2-ezrin hygro A-B replacement construct (20 μg of each) were coelectroporated into the 2 × 107 ES 3.1 cells obtained in the first targeting step. Hygromycin (150 μg/ml) was added 36 h after plating. Hygror ES clones were isolated after 10 to 12 days.
Southern blot analysis.
Genomic DNAs of ES clones obtained after selection with G418 and gancyclovir in the first targeting step were digested with the ScaI endonuclease. Correct gene targeting was analyzed with a 3′ external probe (0.4-kb BamHI-HincII) (data not shown). After amplification, the I-SceI-targeted ES 3.1 clone was further analyzed after BglII digestion using a 5′ internal probe (0.5-kb BglII-StuI). When required, DNA was digested with the commercially available I-SceI restriction enzyme (Boehringer, Mannheim, Germany). Hygror ES clones obtained in the second targeting step were characterized by Southern blotting after digestion of the genomic DNAs with the BglII endonuclease. A 5′ external probe (0.5-kb BglII-StuI) and an internal probe (2 kb; Hygror gene) were used to analyze the 5′ and 3′ homologous recombination events, respectively.
RESULTS
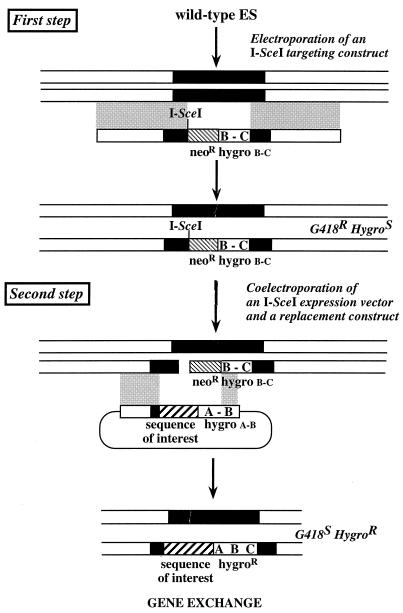
The use of DSB to enhance homologous recombination at a given locus is based on the two-step strategy depicted in Fig. 1. To test this experimental design, we chose to introduce an ezrin cDNA into the villin gene, a natural locus. In the first step, an I-SceI restriction site was introduced into the villin locus, using the pvillin I-SceI/neo hygro B-C targeting vector. I-SceI is a yeast rare-cutter endonuclease (18) that has been shown to initiate DSB in the mammalian genome, naturally devoid of endogenous I-SceI target sequences. In the second step, a unique DSB was induced at the villin locus and the effect of DSB on homologous integration of a pvillin NH2-ezrin hygro A-B replacement vector in the targeted ES cells was assessed (Fig. 1). To facilitate recovery of the targeted clones, we combined to this scheme the plug-and-socket strategy developed by Detloff et al. (11), which is based on the restoration by homologous recombination of a functional Hygror gene.
FIG. 1.
Strategy for the induction of gene replacement upon DSB repair in a natural locus. First step, gene targeting of the I-SceI restriction site in a natural locus by homologous recombination. Second step, cotransfection of expression plasmid pI-SceI and of the replacement construct. Expression of the meganuclease I-SceI leads to cleavage of the targeted gene at the I-SceI site. The ensuing DSB is repaired by gene exchange with the replacement construct. This allows the introduction in the locus of any sequence of interest (reporter gene, mutated allele, etc.). Selection of the recombined ES cells by hygromycin was possible due to the restoration of a functional Hygror gene (hygro A-B-C).
Introduction of the I-SceI recognition site into the endogenous villin locus.
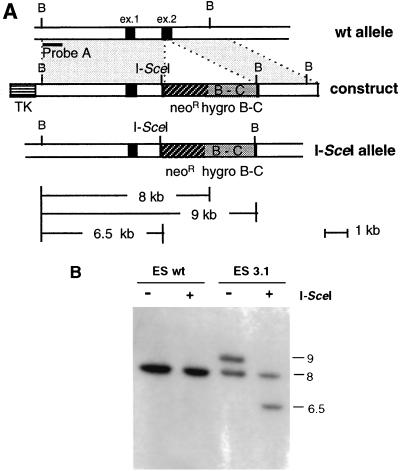
We prepared a targeting vector, pvillin I-SceI/neo hygro B-C (Fig. 2A), containing one I-SceI restriction site, a G418r cassette, and a 1.5-kb partial, nonfunctional Hygror cassette (hygroB-C) flanked with 6.1 and 3.5 kb of villin isogenic genomic DNA at the 5′ and 3′ ends, respectively. A negative selection step was possible due to the addition of a TK counterselection cassette at the 5′ end of the construct. The linearized vector was electroporated into 2 × 107 CK35 ES cells, and the cells were cultured in the presence of G418 and gancyclovir. Of 60 G418 Gancr clones recovered, one ES clone (ES 3.1) was correctly targeted with the pI-SceI/neo hygro B-C targeting construct, as demonstrated by Southern blot analysis. This clone displays a modified allele with the I-SceI/neo hygro B-C gene sequences in exon 2 of the villin gene (Fig. 2B). This analysis also showed that the meganuclease I-SceI is able to specifically cleave the targeted allele in vitro (Fig. 2B, lane 4).
FIG. 2.
Introduction of the I-SceI restriction site in the villin locus by gene targeting. (A) Diagram of the mouse villin locus (wild-type [wt] allele), the targeting construct, and the I-SceI-targeted allele (I-SceI allele). From top to bottom, the dark rectangles represent the two first exons of the villin gene, and the black bar represents the probe A (0.5-kb BglII-StuI, 5′ probe) used for hybridization. BglII (B) and I-SceI restriction sites are indicated. (B) Southern blot analysis of wild-type (wt) and targeted ES (clone 3.1) cells. Genomic DNAs of ES cells were digested with (lanes 2 and 4) or without (lanes 1 and 3) meganuclease I-SceI followed by BglII and then hybridized with probe A. The 9.0-kb band represents the targeted allele and is further cleaved into a 6.5-kb band when digested with I-SceI. Numbers on the right indicate sizes of the bands in kilobases.
I-SceI-induced recombination at the villin locus.
The ES 3.1 clone was then used in the second step. ES cells were electroporated with 20 μg of supercoiled plasmid pvillin NH2-ezrin hygro A-B with or without the I-SceI expression plasmid (pI-SceI). Transient expression of I-SceI induces a unique DSB by in vivo digestion at the target locus. The replacement construct is composed of a 1.0-kb 5′ ezrin mutant cDNA flanked 5′ with a 2-kb region of the murine villin promoter and 3′ with the 1.5-kb hygro A-B cassette.
No Hygror ES clones were recovered when plasmid pvillin NH2-ezrin hygro A-B alone was electroporated (Table 1, experiments 1 and 2). This finding suggests that under the conditions used, homologous recombination between the modified villin locus and the incoming replacement construct was not achieved. In contrast, when plasmid pI-SceI was electroporated together with the replacement construct, 105 and 139 clones, respectively, survived to the hygromycin selection step in two independent experiments (Table 1, experiments 3 and 4).
TABLE 1.
Targeting frequency data
| Plasmid | Expta | No. of Hygror clones | No. of targeted clones/no. analyzed |
|---|---|---|---|
| p-villin NH2 ezrin | 1 | 0 | |
| 2 | 0 | ||
| p-villin NH2 ezrin + pI-SceI | 3 | 105 | 15/15 |
| 4 | 139 | 9/9 |
In all experiments, 2 × 107 cells were electroporated.
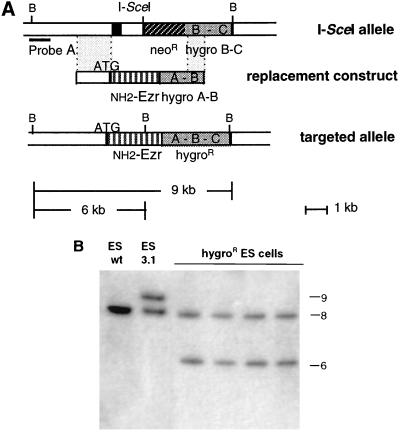
Hygror clones should result from the recombination between the modified villin allele of the 3.1 clone and the replacement construct, since either hygro A-B or hygro B-C alone is not able to confer hygromycin resistance to ES cells (data not shown). To analyze the molecular nature of the recombination event, we performed Southern blot analysis of 24 Hygror clones from experiments 3 and 4. Hybridization of BglII-digested genomic DNA with a 5′ external probe reveals that all Hygror clones have lost the 9-kb band characteristic of the I-SceI allele, while they exhibit a 6-kb band diagnostic of a correct 5′ homologous recombination event between the replacement construct and the I-SceI allele (Fig. 3). Accuracy of the 3′ homologous recombination event was confirmed by using a Hygror gene probe (data not shown). Thus, 100% of the analyzed clones have been correctly targeted, a result that strongly suggests that all of the Hygror clones contained the same allelic modification. The homologous recombination frequency could therefore be estimated at 6 × 10−6 (Table 1).
FIG. 3.
Gene replacement of the I-SceI-targeted villin locus after I-SceI expression. (A) Diagram of the I-SceI-targeted allele (I-SceI allele), the replacement construct, and the villin-NH2-ezrin recombinant locus (targeted allele). BglII (B) and I-SceI restriction sites are indicated. The black bar represents the probe A (0.5-kb BglII-StuI, 5′ external probe) used for hybridization. (B) Southern blot analysis of genomic DNAs of ES wild-type (wt), 3.1, and Hygror clones. DNA was digested with BglII and probed with probe A. The 9.0-kb band represents the I-SceI-targeted allele that resulted in a 6.0-kb band after I-SceI expression and homologous integration of the replacement construct. Sizes are indicated in kilobases.
DISCUSSION
Rare-cutting endonucleases provide a powerful tool for genome manipulation. Several studies have shown that expression of such endonucleases in mammalian cells stimulated homologous recombination between a transfected repair matrix and a randomly integrated DNA construct containing an I-SceI recognition site (7, 8, 22, 25, 26, 28). Here, the I-SceI recognition site was introduced into a natural locus by gene targeting, and we analyzed whether induction of a DSB in a natural locus affected the gene-targeting frequency. The strategy that we have developed is applicable when several rounds of gene targeting at a specific locus are needed. As a first step, we introduced an I-SceI recognition site at the villin locus by a standard gene-targeting procedure. Subsequently, we showed that highly efficient gene targeting could be obtained upon coelectroporation of the modified ES cells with an I-SceI-expressing vector and a villin replacement vector in a circular form. Thus, ES cells with a modified allele having a unique I-SceI recognition site can be used to efficiently introduce any desired modification at the locus. When needed, repetition of the second step with different replacement constructs would allow rapid and efficient recovery of the corresponding recombinant ES cells.
Our data indicate that introduction of a site-specific DSB in a natural locus allows gene targeting with high frequency. Indeed, we observed a gene-targeting frequency of 6 × 10−6 when a DSB was induced in the target locus, whereas no homologous recombination event could be observed when pI-SceI was omitted. The frequency for a conventional gene-targeting experiment is highly dependent on several parameters, including the nature of the locus and the size of homology between the targeting construct and the endogenous locus (10, 16). In several independent conventional gene-targeting experiments at the villin locus, we obtained a frequency of 5 × 10−8, using replacement constructs sharing 8 to 10 kb of homology with the endogenous locus (step 1 of this study and reference 24a). Strikingly, in the second step, which includes a DSB and involves the same genomic region, the targeting frequency was at least 100 times higher, notwithstanding a much lower length of homology (2.8 kb between the targeting construct and the modified villin locus). This high efficiency might rely on the DSB repair mechanism used for the integration of foreign DNA in the target locus, which requires less homology between the replacement construct and the target locus than classical gene-targeting procedures (3, 20, 21). Furthermore, the homologous recombination frequency that we have observed is probably underestimated because not every cell received both constructs. Moreover, the quantity of replacement construct that we have electroporated may be limiting. Thus, even higher efficiency might be obtained by including the cytomegalovirus–I-SceI cassette into the replacement vector and transfecting ES cells with a higher quantity of the replacement construct.
Previous studies of I-SceI-induced gene replacement into randomly integrated transgenes has disclosed a high rate of one-sided homologous recombination events in mammalian cells. This could account for 45 and 21% of drug-resistant clones in NIH 3T3 and ES cells, respectively (25, 28). One-sided homologous recombination events were not observed in the 24 Hygror clones analyzed here, which suggests that it is a relatively rare event at the endogenous villin locus under the conditions used. Whether this observation is specific to the villin locus and/or the targeting construct or rather depends on the route of introduction of the recombination constructs (electroporation versus calcium phosphate transfection) awaits further analysis.
To alleviate the screening of recombined clones during the second step, we used a strategy similar to the plug-and-socket strategy previously described by Detloff et al. (11). They reported that using linearized constructs, insertion (O-type) targeting events occurred in their experiment, noticeably reducing the proportion of desired replacement recombinational (Ω-type) events among the drug-resistant clones. Our strategy, based on DSB repair using a circular matrix, appears to be very efficient, as all Hygror ES clones elicited the expected replacement targeting event. The differences between the results of these two experiments might be due to the design of the targeting constructs or to differences between the recombinational processes involved. More experiments will be needed to resolve this issue.
Introduction of I-SceI in mammalian cells is apparently nontoxic. Due to the size of the mammalian genome, the probability that an endogenous 18-bp I-SceI restriction site exists in the genome of ES cells is very low. Moreover, even if I-SceI induces a DSB elsewhere in the genome, it would probably be repaired by interchromosomal gene conversion (12). After transient expression of I-SceI, no obvious effects were observed in ES cells; in particular, pI-SceI-transfected ES cells formed apparently normal embryoid bodies after in vitro differentiation (data not shown).
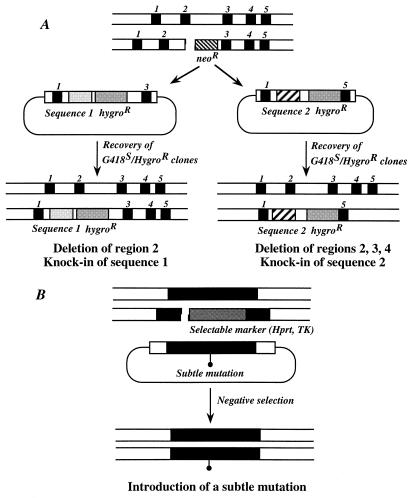
In contrast to other gene-targeting procedures (2, 14, 17, 29), the modified target locus is altered such that the endogenous repair machinery can be stimulated. This helps to overcome the major limitation of the gene targeting procedure, i.e., the low frequency of adequate gene targeting, especially when various mutations of the same locus are needed. Moreover, as short regions of homology are sufficient, replacement constructs may be smaller than previously required and therefore easier to handle. This gene-targeting procedure is very versatile, allowing the knock-in of any sequence of interest in a locus and/or creation of various deletions (Fig. 4A). In addition, it has been suggested from studies in yeast (15) and mammalian (28) cells that chromosomal DSB is preferentially repaired in regard to the unbroken matrix. This implies that any modification present in the replacement vector (unbroken) will be copied into the target locus (harboring the DSB). Therefore, this approach offers an efficient way to introduce subtle mutations at desired locations in the genome (Fig. 4B). Combination of the DSB-mediated gene-targeting procedure with site-specific recombinase-based strategies (19, 27) should increase the range of genetic manipulation of the mammalian genome.
FIG. 4.
Various applications of I-SceI-induced gene-targeting strategy. (A) Depending on the design of the replacement construct, the same I-SceI-targeted allele can be modified in different ways. Use of different replacement constructs allows the introduction of any sequence of interest in the locus or any deletion of genomic sequences (production of truncated protein, targeted deletion of cis-acting regulatory sequences, domain swapping, etc.). Two examples showing different deletions of variable length together with knock-in of a given sequence are illustrated. (B) Because of the mechanisms operating during the second step (i.e., DSB gap repair), any modification (point mutation, small deletion, etc.) present in the replacement vector would be introduced with high efficiency in the locus. This is an advantage compared to other gene-targeting strategies where a DNA mismatch repair mechanism, for example, may limit the cointroduction of other modifications (30).
In conclusion, we have constructed an ES cell line carrying the recognition site of the meganuclease I-SceI in a natural locus, the villin gene. Our data suggest that the yeast endonuclease I-SceI can specifically induce gene-targeting and homologous recombination events with high frequency, allowing specific and highly efficient gene replacement in ES cells. The use of I-SceI in gene-targeting experiments will greatly enhance the possibility of obtaining mutations needed for a comprehensive analysis of gene function.
ACKNOWLEDGMENTS
We specially thank M. Buchwald, head of the Research Department at the Sick Children Hospital in Toronto, Canada, who spent 6 months as a sabbatical fellow at the Curie Institute in Paris. He played a key role in the success of these experiments, and his continuous stimulating advice in the ES cell program is specially acknowledged. We are grateful to S. Memet for the gift of the PGK-Hygror cassette. We thank R. Golsteyn, S. Holmes, and E. Ferrary for critical reading of the manuscript. We also thank F. Apiou and B. Dutrillaux.
This work was supported by grants from the Centre National de la Recherche Scientifique, Institut Pasteur, Institut Curie, Ligue National contre le Cancer, Association pour la Recherche sur le Cancer, MENESR (ACC-SV1), and Rhone-Poulenc SA. F.J. was a recipient of a long-term EMBO fellowship.
REFERENCES
- 1.Algrain M, Turunen O, Vaheri A, Louvard D. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askew G R, Doetschman T, Lingrel J B. Site-directed point mutations in embryonic stem cells: a gene-targeting tag-and-exchange strategy. Mol Cell Biol. 1993;13:4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayares D, Chekuri L, Song K Y, Kucherlapati R. Sequence homology requirements for intermolecular recombination in mammalian cells. Proc Natl Acad Sci USA. 1986;83:5199–5203. doi: 10.1073/pnas.83.14.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein C, Bernstein H, editors. Aging, sex, and DNA repair. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 5.Bollag R J, Waldman A S, Liskay R M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- 6.Bronson S K, Smithies O. Altering mice by homologous recombination using embryonic stem cells. J Biol Chem. 1994;269:27155–27158. [PubMed] [Google Scholar]
- 7.Choulika A, Perrin A, Dujon B, Nicolas J F. The yeast I-SceI meganuclease induces site-directed chromosomal recombination in mammalian cells. C R Acad Sci (Paris) 1994;317:1013–1019. [PubMed] [Google Scholar]
- 8.Choulika A, Perrin A, Dujon B, Nicolas J F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen-Tannoudji M, Marchand P, Akli S, Sheardown S A, Puech J-P, Kress C, Gressens P, Nassogne M-C, Beccari T, Muggleton-Harris A L, Evrard P, Stirling J L, Poenaru L, Babinet C. Disruption of murine Hexa gene leads to enzymatic deficiency and to neuronal lysosomal storage, similar to that observed in Tay-Sachs disease. Mamm Genome. 1995;6:844–849. doi: 10.1007/BF00292433. [DOI] [PubMed] [Google Scholar]
- 10.Deng C, Capecchi M R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Detloff P J, Lewis J, John S W, Shehee W R, Langenbach R, Maeda N, Smithies O. Deletion and replacement of the mouse adult beta-globin genes by a “plug and socket” repeated targeting strategy. Mol Cell Biol. 1994;14:6936–6943. doi: 10.1128/mcb.14.10.6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairhead C, Dujon B. Consequences of unique double-stranded breaks in yeast chromosomes: death or homozygosis. Mol Gen Genet. 1993;240:170–180. doi: 10.1007/BF00277054. [DOI] [PubMed] [Google Scholar]
- 13.Friederich E, Pringault E, Arpin M, Louvard D. From the structure to the function of villin, an actin-binding protein of the brush border. Bioessays. 1990;12:403–408. doi: 10.1002/bies.950120902. [DOI] [PubMed] [Google Scholar]
- 14.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 15.Haber J E, Ray B L, Kolb J M, White C I. Rapid kinetics of mismatch repair of heteroduplex DNA that is formed during recombination in yeast. Proc Natl Acad Sci USA. 1993;90:3363–3367. doi: 10.1073/pnas.90.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasty P, Rivera P J, Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol. 1991;11:5586–5591. doi: 10.1128/mcb.11.11.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasty P, Ramirez S R, Krumlauf R, Bradley A. Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature. 1991;350:243–246. doi: 10.1038/350243a0. . (Erratum, 353:94.) [DOI] [PubMed] [Google Scholar]
- 18.Jacquier A, Dujon B. An intron encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 19.Kilby N J, Snaith M R, Murray J A H. Site-specific recombinases: tools for genome engineering. Trends Genet. 1993;9:413–421. doi: 10.1016/0168-9525(93)90104-p. [DOI] [PubMed] [Google Scholar]
- 20.Lin F L, Sperle K, Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990;10:113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin F L M, Sperle K, Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990;10:103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukacsovich T, Yang D, Waldman A S. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI. Nucleic Acids Res. 1994;22:5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. Molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 24.Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Robine, S., and F. Jaisser. Unpublished observations.
- 25.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer B. Site-Specific recombination: developments and applications. Curr Opin Biotechnol. 1994;5:521–527. doi: 10.1016/0958-1669(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 28.Smih F, Rouet P, Romanienko P J, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stacey A, Schnieke A, McWhir J, Cooper J, Colman A, Melton D W. Use of double-replacement gene targeting to replace the murine alpha-lactalbumin gene with its human counterpart in embryonic stem cells and mice. Mol Cell Biol. 1994;14:1009–1016. doi: 10.1128/mcb.14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steeg C M, Ellis J, Bernstein A. Introduction of specific point mutations into RNA polymerase by gene targeting in mouse embryonic stem cells: evidence for a DNA mismatch repair mechanism. Proc Natl Acad Sci USA. 1990;87:4680–4684. doi: 10.1073/pnas.87.12.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szostak J W, Orr-Weaver T L, Rothstein R J. The double-strand break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 32.te Riele H, Maandag E R, Clarke A, Hooper M, Berns A. Consecutive inactivation of both allele of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 33.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]