Abstract
Polymyxins are the last line of defense against multidrug-resistant (MDR) Gram-negative bacterial infections. However, this last resort has been threatened by the emergence of superbugs carrying the mobile colistin resistance gene-1 (mcr-1). Given the high concentration of matrix metalloproteinase 3 (MMP-3) in bacterial pneumonia, limited plasma accumulation of colistin (CST) in the lung, and potential toxicity of ionic silver (Ag+), we designed a feasible clinical transformation platform, an MMP-3 high-performance lung-targeted bio-responsive delivery system, which we named “CST&Ag@CNMS”. This system exhibited excellent lung-targeting ability (>80% in lungs), MMP-3 bio-responsive release property (95% release on demand), and synergistic bactericidal activity in vitro (2–4-fold minimum inhibitory concentration reduction). In the mcr-1+ CST-resistant murine pneumonia model, treatment with CST&Ag@CNMS improved survival rates (70% vs. 20%), reduced bacteria burden (2–3 log colony-forming unit [CFU]/g tissue), and considerably mitigated inflammatory response. In this study, CST&Ag@CNMS performed better than the combination of free CST and AgNO3. We also demonstrated the superior biosafety and biodegradability of CST&Ag@CNMS both in vitro and in vivo. These findings indicate the clinical translational potential of CST&Ag@CNMS for the treatment of lung infections caused by CST-resistant bacteria carrying mcr-1.
Keywords: Colistin-resistant bacteria, MCR-1, Lung targeting, Matrix metalloproteinase 3, Synergistic action, Bacterial severe pneumonia
Graphical abstract
Development of a lung-targeted bio-responsive platform for treatment of severe pneumonia caused by mcr-1+ bacteria.
Highlights
-
•
High-performance lung-targeted bio-responsive platform was fabricated via emulsion crosslinking method.
-
•
The platform possessed excellent lung-targeted capability with targeting efficiency >80%.
-
•
The platform improved the drug release performance with the help of NFF-3.
-
•
The platform could combat MCR-1+ bacterial infections in two mouse pneumonia models.
Nomenclature
- CFU
Colony-forming unit
- CRE
Carbapenem-resistant G- Enterobacteriaceae
- DL
Drug loading
- EE
Encapsulation efficiency
- ELISA
Enzyme-linked immunosorbent assay
- FBS
Fetal bovine serum
- FDA
Food and Drug Administration
- FIC
Fractional inhibitory concentration
- FICI
Fractional inhibitory concentration index
- GO
Gene Ontology
- H&E
Hematoxylin and eosin
- IVIS
In Vivo Imaging System
- LB
Luria-Bertani
- MFI
Mean fluorescence intensity
- MIC
Minimum inhibitory concentration
- OD
Optical density
- PBS
Phosphate-buffered saline
- PI
Propidium iodide
- RBC
Red blood cells
- SD
Standard deviation
- SEM
Scanning electron microscopy
- SLF
Simulated lung fluids
- TCS
Two-component system
- XRD
X-ray diffraction
- ZraP
Zinc resistance-associated protein
1. Introduction
Mortality associated with severe community-acquired pneumonia remains a major concern [1]. Gram-negative (G−) bacteria, including Klebsiella pneumoniae (Kp) and Escherichia coli (Ec), have emerged as significant causes of pneumonia in recent years [[2], [3], [4]]. The rising prevalence of colistin-resistant Kp and Ec exacerbates the threat posed by these pathogens because colistin (CST) is the last-line therapy available to overcome multidrug-resistant (MDR) pathogens [5]. Accordingly, concerns regarding resistance to CST have grown significantly since the first plasmid-borne transmissible CST-resistant gene-mobile colistin-resistant gene-1 (mcr-1) was discovered in 2015 in China [6]. This situation is complicated by the ability of mcr-1 to coexist with other antimicrobial resistance genes, such as carbapenemases [[7], [8], [9], [10], [11]] and extended-spectrum β-lactamases (ESBL) [[12], [13], [14]], and co-transfer with mcr-3 or mcr-5 [[15], [16], [17]].
The mechanism by which CST combats G− bacteria involves cell membrane lysis. This lysis process is caused by electrostatic attraction between the cationic positively charged polymyxin and the anionic negatively charged phosphate group of lipid A, which is anchored on the outer leaflet of the bacterial membrane [18,19]. The transmembrane protein, MCR-1, which is responsible for CST resistance, contains an essential Zn (II) cofactor in its active site. More importantly, MCR-1 functions as a phosphoethanolamine (pEA) transferase, catalyzing the addition of phosphorylethanolamine (pEtN) to lipid A head groups of lipopolysaccharide (LPS), thereby neutralizing the bacteria membrane charge [20]. This modification makes it challenging for positively charged CST to attach cell membranes, leading to drug resistance.
In order to address the rising concerns around antibiotic resistance, combination therapy using an accessible antibiotic and a resistance inhibitor has been deemed a safer and more cost-efficient alternative than developing new antimicrobial leads [[21], [22], [23], [24], [25]]. However, only three studies have so far attempted to find a suitable MCR-1 breaker to collaboratively combat CST-resistant pathogens carrying mcr-1 [[26], [27], [28]]. Scientists have developed a growing interest in metal-oriented agents since metal ions can readily replace the metal cofactors of enzymes mediating resistance to antibiotics [23,[29], [30], [31], [32], [33]]. Recently, Li et al. revealed that the aseptic ulcer bismuth (Bi) drug can restore metallo-β-lactamase (MBL)-positive bacteria to β-lactam antibiotics by substituting the zinc ion [Zn (II)] cofactors with Bi (III) at the inactive active site of MBLs, hampering their ability to decompose the β-lactam ring in carbapenem [29]. The authors also demonstrated that auranofin, an antirheumatic drug, can be used as a double inhibitor of MBL and MCR [28]. Here, based on previous discovery [27], we confirmed that AgNO3 could significantly synergize with CST. Although silver ion (Ag+) is approved by the U.S. Food and Drug Administration (FDA) as a topical antimicrobial agent [34], we explored the possibility of administering Ag+ intravenously (i.v.) as an adjuvant in combination therapies.
When administered via an i. v. Route, microspheres of a suitable size (7.0–15 μm) exhibit excellent passive lung-targeting effects, improve drug concentrations in damaged tissues, and reduce the distribution of drugs in physiological tissues [35]. Meanwhile, chitosan is widely used in biological biomaterials owing to its excellent properties, including biodegradability, biocompatibility, and non-toxicity, as well as anti-inflammatory, antioxidant, and antibacterial activity [36]. We, therefore, selected chitosan as a drug carrier. Additionally, MMP-3 has been demonstrated to have a higher concentration in bacterial infection microenvironments (IMEs) [22,37]. Hence, we anticipated that our drug delivery system could be responsive to upregulated MMP-3 in IMEs.
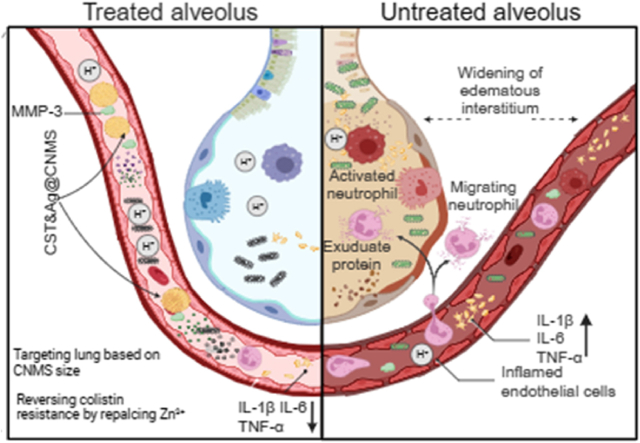
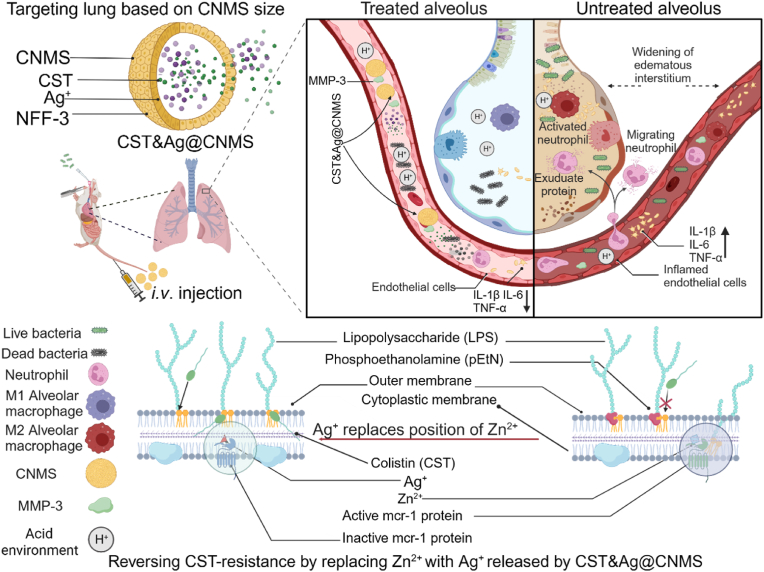
In the present study, we developed a feasible clinical transformation platform, MMP-3 bio-responsive release chitosan microsphere (CNMS), which we name “CST&Ag@CNMS,” in which CST and Ag+ are co-loaded to improve target densities and antibacterial and anti-inflammatory effects in the lungs. Scheme 1 shows the concept of a lung-targeting platform reversing CST resistance via metal replacement against CST-resistant bacterial pneumonia. This lung-targeted delivery system sensitizes G− bacteria to CST by substituting Zn2+ with Ag+ in the active-site pocket of MCR-1. For mice exposed to this polymer via an i. v. Route, optimal CNMS with appropriate diameters can deliver an adequate dose of drugs to the lung without introducing any noticeable side effects. Furthermore, in the CST-resistant bacterial pneumonia mouse model, treatment with CST&Ag@CNMS improved mice survival rates, reduced bacterial burden, and mitigated inflammatory response by forwarding the conversion from classical activated macrophages (M1-type macrophages) to selectively activated macrophages (M2-type macrophages). These findings indicate the potential clinical translation of CST&Ag@CNMS to treatment of severe bacterial pneumonia caused by mcr-1+ bacteria.
Scheme 1.
High-performance lung-targeted bio-responsive platform for severe bacterial pneumonia therapy via reversing colistin-resistant bacteria carrying mcr-1. Created using BioRender.com..
2. Material and methods
Materials: CST, chitosan, and IR783 were purchased from Sigma-Aldrich (USA). Enzyme-linked immunosorbent assay (ELISA) kits for interleukin (IL)-1β, tumor necrosis factor (TNF)-α, and IL-6 were purchased from Shanghai Dakewe Biotech Co., Ltd (China). Phosphate-buffered saline (PBS), Luria-Bertani (LB) broth powder, and hydroxyethyl piperazine ethanesulfonic acid (HEPES) were purchased from Shanghai Saiwei Biotechnology Co., Ltd (China). Silver nitrate (AgNO3) was purchased from Merck (USA). Metal compounds mentioned in this study, except AgNO3, were kindly provided as a gift by the Ruijin Hospital affiliated to Shanghai Jiao Tong University. NFF-3 peptide was purchased from Shanghai Apeptide Co., Ltd (China).
Bacteria: All test strains used in this study are exhibited in Table S1.
Synthesis of chitosan NFF-3 microsphere (CNMS): First, CNMS was synthesized by emulsion crosslinking, as in previous studies, with some modifications [38]. Briefly, aqueous phase (1% w/w) was prepared by dissolving 100 mg chitosan in 10 mL distilled water containing 300 μL acetic acid at 37 °C. CST, AgNO3, and MMP-3 substrate (NFF-3 peptide, Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys (DNP)-NH2) with or without IR783 were mixed into this dispersed phase. The oil phase was maintained at 47 mL paraffin oil containing 2880 μL Span80 and 120 μL Twain20. The aqueous phase was added drop-by-drop into the oil phase and continuously mixed at 600 revolutions per min (rpm) for 3 h to form a stable water-in-oil emulsion. Subsequently, 1.5 mL glutaraldehyde solution (25% concentration) was added to the mixture and crosslinked further for 6 h. To completely remove the free CST, AgNO3, and unreacted organic reagents, the obtained products were then separated by centrifugation at 3000 rpm. The sediment was washed with petroleum ether twice and then with water and anhydrous ethanol 3–4 times each, before a final wash with water. The product was refrigerated at −20 °C overnight and then freeze-dried to obtain 10 μm CNMS. CNMS-S and CNMS-L were prepared in the same manner as CNMS-M, except that the total volumes of oil phase used were 50 (800 rpm) and 45 mL (500 rpm), respectively.
Characterization analysis: The zeta-potential of microspheres was determined using Master-sizer (Omni, Brookhaven, US). In brief, 5 mg of each sample was suspended in distilled water (pH = 7.4), and the zeta-potential was measured. The particle size of CNMS was measured using a laser particle analyzer (S3500, China). All measurements were taken at 25 °C in triplicate (n = 3).
Morphology and structure: An electron optical microscope (Olympus) linking with an online charge-coupled device (Pixlink) was used to observe the morphology and structure of droplets and microspheres. More detailed structures and topography were examined using scanning electron microscopy (SEM; RISE-MAGNA, TESCAN). An ultrathin layer of gold (5 nm) was coated with CNMS before the SEM measurement. The SEM was coupled with energy-dispersive X‐ray spectroscopy (EDX) to confirm the composition of the metallic particles. Fluorescence was observed using a fluorescence microscope (Olympus).
X-ray diffraction (XRD): XRD tests were conducted on a table-type XRD (Aeris, China). Copper (Cu)-Kα radiation (λ = 1.5406 Å) was employed at a scanning rate of 0.5 °C min−1 over 10–60 °C. The XRD test voltage and current were 40 kV and 150 mA, respectively.
Fourier-transform infrared (FT-IR) spectroscopy: The spectra of chitosan, CST, AgNO3, chitosan microsphere (CMS), CNMS, and CST&Ag@CNMS were captured using FT-IR spectroscopy (Nicolet 6700, THERMO FISHER, USA). Each sample was scanned three times between 4000 and 400 cm−1 at a resolution of 2 cm−1 and averaged for that sample.
Lung target ability, biodistribution, and degradation of IR783@CNMS in vivo: Healthy mice were administered IR783@CNMS (50 mg kg−1) via tail vein injection to investigate the change in morphology of IR783@CNMS in vivo. Mice were monitored using the In Vivo Imaging System (IVIS imaging, PerkinElmer, USA) at 2, 12, 24, and 36 h, and then sacrificed. The main organs, including the heart, liver, spleen, lung, and kidney, were harvested and rinsed in phosphate-buffered saline (PBS), and then fluorescence signals of IR783 were measured using the IVIS system (PerkinElmer, USA). Quantitative analysis of the fluorescence of the main organs was also performed by the IVIS system. The lung was fixed in 4.0% paraformaldehyde, embedded in paraffin, and stained with hematoxylin and eosin (H&E) for observation with light microscopy. To identify the degradation of CNMS, dried CNMS was incubated in simulative body fluid at 37 °C with slight shaking. We extracted 200 μL CNMS at fixed time points, and representative time-dependent morphology changes of CNMS were observed using an SEM.
Evaluation of the drug loading and encapsulation efficiency: The encapsulation efficiency (EE) of CST was fixed by dissolving 1 mg dried CST&Ag@CNMS in 5 mL 3% acetic acid solution and shaking vigorously for 30 s. The mixed solvent was filtered, and the filtered products were added to 5 mL 3% acetic acid solution to a set volume of 10 mL. This phase was then measured using ultraviolet (UV) spectroscopy (UV-3600i Plus, Shimadzu, Japan) to calculate the CST content based on the previously derived calibration curve. Absorbance was monitored at λ = 192 nm. Drug loading efficiency (DL) and EE were set using the following formulas:
| DL (%) = Ma/Mm |
| EE (%) = Ma/Mt |
where Ma and Mt are the actual and theoretical CST contents of CNMS, respectively, and Mm is the weight of the CST-loaded CNMS. We used inductively coupled plasma mass spectrometry (ICP-MS) to confirm the content of AgNO3. Finally, large CNMS (CNMS-L; EE of CST at 56.15 ± 3.17%, Ag+ at 30.14 ± 2.13%), medium (CNMS-M; CST EE at 58.23 ± 4.12%, Ag+ at 30.27 ± 4.13%), and small CNMS (CNMS-S; EE of CST at 40.25 ± 2.10%, Ag+ at 16.25 ± 5.21%)) were prepared. Calculation revealed that drug loading of Ag+ and colistin was approximately 10% and 50%, respectively.
Drug release: The in vitro drug release of CNMS loaded with CST was determined using the dialysis method in simulated lung fluids [SLFs; potential of hydrogen (pH) = 7.4 and pH = 5.5] at a temperature of 37 °C. We enveloped 1 mg mL−1 of 2 mL CST-loaded CNMS in the dialysis bag (MWCO 100 kDa), immersed it in 48 mL PBS solution, and stirred horizontally at 100 rpm. Aliquots of 2 mL were periodically (0, 0.5, 1, 2, 3, 4, 5, 6, 9, 11, 21, 23, and 24 h) removed from the conical flask to quantity CST release with UV–vis at the wavelength of 193 nm, and 2 mL fresh PBS was supplied to maintain an equal volume of 50 mL. Drug aggregate release was determined as the percentage of the quantity of CST at a certain time, accounting for the initial CST amount.
Initial screening of antimicrobial activity on antibiotics and different metals: Metal salts used for screening were hafnium boride (HfB2), cobalt chloride (CoCl2), zinc chloride (ZnCl2), cupric chloride (CuCl2), zirconium tetrachloride (ZrCl4), aluminum nitrate nonahydrate [Al (NO3)3], ferrous chloride (FeCl2), bismuth trichloride (BiCl3), potassium chloride (KCl), nickel chloride (NiCl2), and AgNO3. Briefly, overnight cultures of MCR-1 Ec34 and KPN18 were created at 1:1000 dilution and allowed to regrow to the mid-log phase; they were then diluted to an optical density (OD)600 of 0.6 in each well of a 96-well plate. Bacterial suspensions were exposed to CST therapy (2 μg mL−1), in both the absence and presence of different metal ions at a set concentration (50 μg mL−1). Bacterial growth inhibition was examined by monitoring OD600 for 0, 3, 6, 18, and 24 h. Wells without antibiotics or metallic compounds were used for growth controls, and wells with only LB were used as negative controls.
Measurement of minimum inhibitory concentration (MIC) and fractional inhibitory concentration (FIC): MICs were measured using a standard broth microdilution assay. Briefly, in a typical assay, the bacteria (Ec34, Ec50, Ec62, Ec99, Ec5505, Kp4, Kp18, Kp31, Kp38, and Kp5445) were grown overnight and diluted 1:100 in LB broth at 37 °C for 3 h to achieve the logarithmic phase. The original number of bacteria was adjusted to approximately 5 × 105 CFU mL−1, and the final volume was 100 μL in each well (96-well plate). Subsequently, the bacterial suspensions were exposed to 0, 1, 2, 4, 8, 16, 32, and 64 μg mL−1 CST and 0, 0.5, 1, 2, 4, 8, 16, 32, and 64 μg mL−1 AgNO3, either individually or in combination. LB broth without bacteria was used as a negative control; bacteria suspension without any drug was used as a positive control. The 96-well pad was placed on a 37 °C stirrer for 24 h. Aliquots of bacterial suspension were measured using UV analysis at 24 h. The OD value of each well was measured with a spectra microplate reader (BioTek) at 600 nm. MIC is the minimum agent concentration that causes 95% bacterial death. After the MIC experiment, 10 μL of the content of each well was spread evenly onto an LB agar. The dry liquid was incubated at 37 °C for 18 h, and the colonies were counted. The fractional inhibitory concentration (FIC) for each drug is determined by dividing each drug's MIC when used in combination by each drug's MIC when used alone. The fractional inhibitory concentration index (FICI) is the sum of the FICs of two substances, and synergy is set at an FIC index ≤0.5.
| FIC = MIC (Combination) /MIC(CST) + MIC(Combination)/MIC(AgNO3) |
SEM: A medium-logarithm growth phase culture of Kp4 or Ec34 was prepared at 5 × 105 CFU mL−1 using LB-modified media. The suspension was exposed to ¼; MIC AgNO3, ¼; MIC CST, AgNO3, and CST combination, or PBS, and then they were fixed overnight with paraformaldehyde (4 wt%) at 4 °C. The specimens were then washed thrice with PBS, followed by dehydration with an ascending ethanol series (25%, 50%, 75%, and 100%) and another series of dehydration with tert-butanol for 10 min repeated three times. The dried specimens were sprayed with gold for SEM examination and were pictured under a high vacuum with 2.0 kV acceleration voltage.
Live/dead staining bacteria: Ec34 and Kp4 were cultured overnight, diluted in fresh LB, and incubated at 37 °C until the OD600 value reached 0.6 nm. The original number of bacteria was adjusted to approximately 105–106 CFU mL−1. The suspensions were treated with CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS, with no treatment as a negative control. After incubation for around 6 h, the bacterial pellets were collected through centrifugation, resuspended in saline solution, and treated with 1 μL propidium iodide (PI) and SYTO 9 fluorescent dyes. The bacteria were then incubated in the dye for a total of 30 min in a dark environment. The fluorescent images of stained bacteria were obtained using a fluorescent microscope. Green fluorescence indicated living bacteria, while red fluorescence signified dead bacteria.
Bacteria 24-h time-growth assay: In a typical assay, the bacteria (Ec34, Ec50, Ec62, Ec99, Ec5505, Kp4, Kp18, Kp31, Kp38, and Kp5445) were grown overnight and diluted in 1:100 LB broth at 37 °C for 3 h to achieve the logarithmic phase. The original number of bacteria was adjusted to approximately 105–106 CFU mL−1, and the final volume was 100 μL in each well (96-well plate). The bacterial suspensions were then exposed to ¼ MIC CST and ¼ MIC AgNO3, either individually or in combination. LB broth without bacteria was used as a negative control, while bacteria suspension without any drug served as a positive control. The 96-well pad was placed on a 37 °C stirrer for 24 h. Aliquots of bacterial suspension were analyzed by UV at different time intervals (0, 1, 2, 4, 6, 8, and 24 h). Data from three independent experiments were averaged over 24 h. All assays were triplicated and performed three times on different days.
Zn2+displacement based on equilibrium dialysis: Equilibrium dialysis assay was generated as described previously to monitor the process of Zn2+ displacement [29]. Briefly, various concentrations of CST&Ag@CNMS was incubated in metal-free ICP-MS buffer, containing Zn3- MCR-1-S, at 25 °C with slight shaking for about 6 h. Subsequently, the samples were dialyzed in HEPES buffer to remove free metal ions and analyzed using an ICP-MS spectrometer. Three replicate assays (n = 3) were performed in all tests, and results were expressed as mean ± standard deviation (SD). The graphs were filled using the Hill-plot equation.
RNA-seq and transcriptomic analysis: The Kp4 and Ec34 were grown in an LB medium to the logarithmic phase and treated with CST&Ag@CNMS for 6 h, with three replicates for each sample. The bacterial cells were then washed thrice with PBS. The cells were collected by centrifugation and transferred into liquid nitrogen for 30 min and a −80 °C refrigerator for 24 h. Then, the cell pellet was sent to Shanghai OY Biomedical Technology Company for RNA-seq for further transcriptomic analysis.
Molecular docking and molecular dynamic: This analysis was conducted in three stages, including protein pretreatment, identification of active sites, and analysis of Molecular Mechanics–Generalized Born Surface Area (MMGBSA). For protein pretreatment, the MCR-1 protein corresponding crystal structure (PDB ID: 5YLF) was obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database. The Protein Preparation Wizard module of the Schrödinger software was used for obtaining protein crystals, regenerating the states of native ligands, optimizing H-bond assignment, minimizing protein energy, and removing waters. To identify active sites, we first used the SiteMap module in Schrödinger to predict the best binding sites. We next used the Receptor Grid Generation module in the Schrödinger software to set the most suitable Enclosing box that would predict the perfect package of combined sites. Then, the active site of the MCR-1 protein was obtained based on this prediction. Finally, we analyzed the MMGBSA of Ag+, Zn2 +, and MCR-1 protein. MMGBSA dG Bind can approximate the free energy of ligand-protein binding, such that the lower the binding free energy, the higher the binding stability of ligands and proteins.
Cell lines and culture conditions: Lung carcinoma (A549) cells were cultured with F-12 K Medium supplemented with 10% heat-inactivated fetal bovine serum (FBS); endothelial cells (EA.hy926) were cultured in Dulbecco's modified Eagle's medium (Adamas) supplemented with 10% heat-inactivated FBS and alveolar macrophage cells (MHS) for Roswell Park Memorial Institute medium (1640 RPIM, Adamas) supplemented with 10% heat-inactivated FBS at 37 °C in a humidified atmosphere of 5% CO2.
Cellular toxicity assay: Alveolar lung epithelial cells (A549), endothelial cells (EA.hy926), and alveolar macrophage cells (MHS) were seeded into a 96-well plate (104 cells/well) and cultured for 12 h. Subsequently, the cells were treated with AgNO3, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS at various concentrations for 24 h. Cell viability was measured using a standard Cell Counting Kit-8 (CCK8) assay and calculated using a Microplate Reader at OD450 nm (Biotek).
Cell morphology staining: Approximately 5 × 105 cells were planted per pore in a 12-well plate. Overnight, cell cultures were washed with PBS thrice, supplemented with 4% formaldehyde, and incubated at room temperature for 15–20 min for cell fixation. Then, cells were permeabilized with 0.5% Triton X-100 at room temperature for 10 min, blocked with blocking solution (3% bovine serum albumin in PBS), and incubated with phalloidin in the dark overnight at 4 °C. Subsequently, the cells were treated with 4’,6-diamidino-2-phenylindole (DAPI) solution for 10 min, and fluorescence images were captured using a fluorescent inverted microscope (Olympus).
Hemolysis test: The hemolytic activity of the CMS was performed according to methods previously reported. We harvested 1 mL of fresh murine blood from laboratory-bred strain (BALB/c) mice and mixed it with ethylenediamine tetraacetic acid (EDTA) for stability. After centrifugation at 1000 rpm for 10 min at room temperature, serum was removed, and red blood cells (RBCs) were washed five times with isotonic saline solution. The RBCs were resuspended in fresh saline solution to achieve a 2% RBC suspension. In the positive group, 500 μL 2% RBCs suspension and 500 μL of deionized water were added in a 1.5-mL centrifuge tube, while in the negative group, 500 μL deionized water and 500 μL 2% RBCs suspension were added. Subsequently, 500 μL of RBCs and 500 μL of material were mixed, and the final concentration of material was 5 mg mL−1. All groups were incubated at room temperature for 4 h, centrifuged and pictured, and the supernatant was obtained to measure the absorbance values at 541 nm. The hemolysis rate was determined using the following equation:
| Hemolysis (%) = (As−An)/(Ap−An) × 100% |
where As refers to the absorbance of samples, A is the absorbance of the negative control, and Ap represents the absorbance of the positive group.
Mice: Animal welfare and all animal experiments were conducted according to the requirements of the Animal Care and Experimental Protocol Committee of Ruijin Hospital, Affiliated to Shanghai Jiao Tong University Medicine, and established guidelines for laboratory animals. Male BALB/c mice aged 6–8 weeks (25–30 g) were used throughout. These mice were housed in decent laboratory conditions in accordance with the standards of microbiology and neurology. The feeding environment and exercise quality also met standards for experimental animals. All the efforts were made to alleviate the suffering of animals.
Creation of acute lung infection model and samples harvest: The mice were anesthetized by intraperitoneal (i.p.) injection of 100 mg kg−1 sodium pentobarbital, intubated by tracheotomy, and ventilated with room air using a small animal ventilator. Their tracheas were exposed by placing them supine, head-up on a board. Next, 106 CFU of Kp or 107 CFU of Ec was dripped onto each mouse, and the mouse was kept supine for 5 min after the intervention. After 2 h, the mice were randomly divided into six groups (n = 15 for each group), including: PBS (100 μL), CNMS (20 mg kg−1, 100 μL), CST (10 mg kg−1, 100 μL), CST + AgNO3 (10 mg kg−1 for CST and 2 mg kg−1 for AgNO3, 100 μL), CST&Ag@CNMS (20 mg kg−1, 100 μL) and healthy categories based on various treatments of drug formulations. Five mice in each group were sacrificed by cervical dislocation approximately 24 h after treatment, and orbital blood was collected and placed in a tube containing blood in ice. A small incision was made on the trachea, and 500 μL PBS was perfused using a needle of a 20-gauge catheter into the trachea to obtain lung bronchoalveolar lavage fluid (BALF). This step was performed by perfusing 0.5 mL PBS three times. Then the main organs, including the heart, liver, spleen, lung, and kidney, were removed from the sacrificed mice.
The BALF was centrifuged at 2000 rpm for 10 min and blood was centrifuged at 3000 rpm for 20 min. ELISA was used to measure levels of TNF-α, IL-1β, and IL-6 in the supernatant of the BALF. The sediment was resuspended with 1 mL PBS and supplemented with RBC lysate. Then, the suspension was collected to measure the cell number using a cell counting instrument. The wet weight of the left lung tissue in the five mice from each group was recorded; the tissue was then placed in a drying oven and reweighed when dry. Another part of the lung tissue of each mouse was divided into two parts; one was subjected to standard H&E staining and immunofluorescence staining procedures, while the other was subjected to issue homogenization for bacteria colony counting. The final 10 mice were observed for survival analysis.
Measurement of mean fluorescence intensity (MFI). The MFI of each image was acquired and measured using Image J software. A single channel picture was chosen and transformed into a 16-bit image. A drawing pen was then used to circle the area of the tissue section. Once the desired tissue area is confirmed, click on the ‘Analyze’ button to select the ‘Measure’ option. A window will then pop up named ‘Results’ which includes a number of measurements that the software made on the selected area including Area, Mean, etc.
Statistical analysis: Each independent experiment was repeated three times for all statistical analyses using Prism 9.0 software (GraphPad Prism Software Inc.). Data were presented as mean ± standard error of the mean. One‐way analysis of variance followed by Dunnett's post hoc test was used to evaluate multiple comparisons when there were more than two groups. P ≤ 0.05 was considered to be significant. Ns represented no significance; * represents p < 0.05; ** represents p < 0.01; *** represents p < 0.001; and **** represents p < 0.0001.
3. Results and discussion
3.1. Primary screening confirmed Ag + as an adjuvant to reverse CST resistance
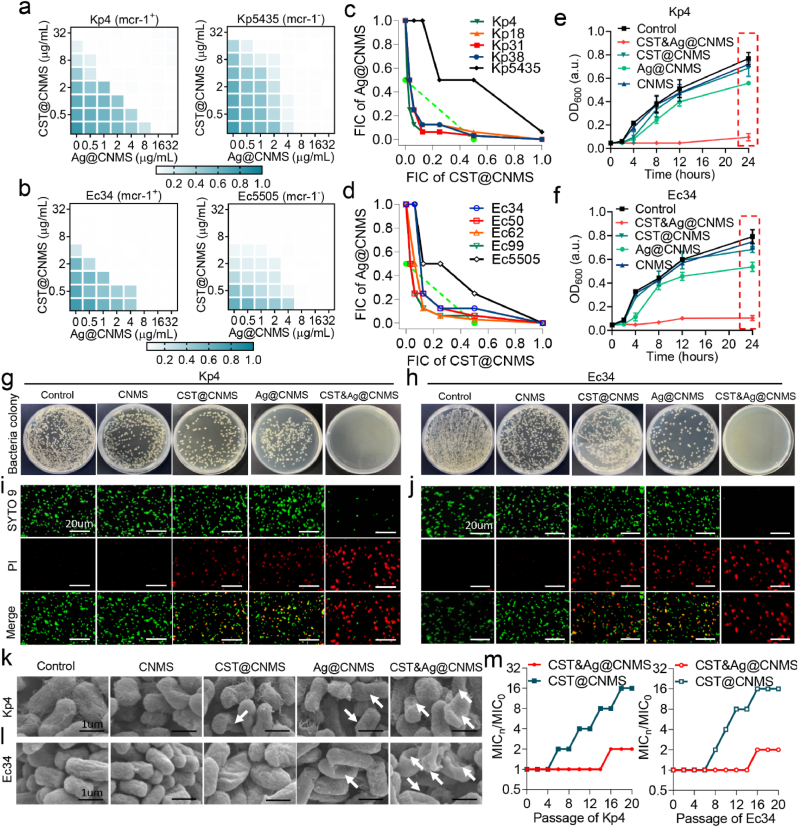
A number of metal compounds were screened to identify a suitable booster for CST against Kp and Ec strains carrying the mcr-1 gene. According to the Clinical and Laboratory Standards Institute (CLSI), a CST concentration of 4 μg mL−1 is defined as the CST-resistant breakpoint for Enterobacteriaceae [39]. In this study, 10 CST-resistant strains including four mcr-1+ Kps (Kp4, Kp18, Kp31, and Kp38), four mcr-1+ Ecs (Ec34, Ec50, Ec62, and Ec99), one mcr-1- Kp (Kp5435), and one mcr-1- Ec (Ec5505), were selected. We confirmed the presence of mcr-1 using polymerase chain reaction and electrophoresis experiments (Fig. S1, Table S1 and Table S2). The growth curve assay was first used to evaluate the synergistic effects between CST (2 μg mL−1) and 11 metal compounds (5 μg mL−1) against two representative mcr-1+ carriers (Kp4 and Ec34) (Fig. S2). In agreement with findings of Sun's studies [27], we further confirmed CST and AgNO3 exhibited synergistic bactericidal activity, while the other metallic compounds showed no noticeable effects. We, therefore, selected AgNO3 as a potential inhibitor of MCR-1 in developing a drug delivery platform.
Ag+ compound has been shown to irreversibly inactivate MCR-1 [27], although whether it can potentiate antibiotic activity as an inhibitor of MCR-1 in selected bacteria remains unclear. Hence, we explored the possibility of using Ag+ as a CST adjuvant by testing the MIC of CST and Ag+ against selected eight MCR-1+ strains and two MCR-1− strains. We found that the MIC of CST toward these strains ranged from 4 to 32 μg mL−1, while the MIC of AgNO3 ranged from 4 to 16 μg mL−1 (Table S1). Subsequently, the standard checkerboard microdilution method was used to observe the interrelation between CST and Ag+ against CST-resistant Kp and Ec strains (Figs. S3 and S4). CST monotherapy at the concentration of 16 μg mL−1 and 8 μg mL−1 showed no growth toward either MCR-1+ Kp4 or MCR-1+ Ec4. However, after adding Ag + to CST for Kp4 and Ec34 bearing the mcr-1 gene, the MIC of CST decreased to 2 μg mL−1 and 1 μg mL−1, respectively, at sublethal concentrations of Ag+. Bacterial growth curves further demonstrated the time-dependent growth inhibition effects of the CST and AgNO3 combination at ¼ MIC against MCR-1+ Kp and Ec, but the use of each agent individually had limited impact on restraining bacterial growth over time (Figs. S3 and S4). Minor or zero growth was not observed when CST was combined with AgNO3 at ¼ MIC concentration for MCR-1− Kp5435 and Ec5505 (Figs. S3 and S4).
Additionally, to identify the generality of Ag+ as a CST sensitizer, we added sublethal concentrations of Ag + to CST treatments in other strains carrying mcr-1, including Kp18, Kp31, Kp38, Ec50, Ec62, and Ec99. Surprisingly, we also noticed significantly enhanced bactericidal activity through the standard checkerboard microdilution method and bacterial growth curves (Tables S1 and S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6). We then illustrated the synergistic effect using isobolograms for all selected strains in this paper. For Kp4, we observed a significant synergistic effect between CST and Ag+, with a FICI of 0.375 (synergism was determined as FICI ≤0.5, below the dashed green line) (Fig. S7a) [40]. In contrast, for Kp5435 carrying no mcr-1 genes, no such synergism was observed, but an additive effect was noticed with a FICI of 0.75 (an additive effect was determined as 0.5 < FICI ≤1). (Fig. S7a). A similar phenomenon was observed for Ec (Fig. S7b).
Serial passage of Kp4 and Ec34 showed that resistance levels toward CST were greatly elevated upon exposure to CST at subinhibitory concentrations, as the MIC value increased by 8- and 16-fold over 20 consecutive generations. However, the increase in resistance was less pronounced with the combined use of CST and Ag+, with only a 2-fold increase (Fig. S8). These findings indicate that Ag+ sensitizes CST against G− Enterobacteriaceae and attenuates the potential emergence of resistance.
3.2. Synthesis and characteristics of CST&Ag@CNMS
To develop a biofriendly drug delivery system, CMS were prepared based on a previously reported method with some alterations [41]. Earlier studies have demonstrated that MMP-3 is highly expressed in IMEs [42]. As an MMP-3 substrate, NFF-3 peptide [Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys (Dnp)-NH2] can be degraded quickly when encountered with MMP-3. Detailed information regarding NFF-3 is provided in Fig. S9. To enhance rapid drug release in the targeted site, we modified CMS by adding 1 mg NFF-3 peptide [(Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys (Dnp)-NH2)] into 1 mL 2% acetic acid solution, followed subsequent steps as previously described [43,44]. We then used SEM–energy-dispersive spectrometer (SEM–EDS) images to identify the morphology and stable size of drug-loaded CMS after modification (Fig. 1a and b). The modified NFF-3 CNMS had an approximate spherical shape with a diameter of about 12 μm. When administered i. v., microspheres with a suitable size (∼10 μm) exhibit excellent passive lung-targeting effects, which have been proven and utilized in bacteria-infected mouse models in two previous studies [45,46]. A passive lung-targeting microsphere usually has a diameter slightly larger than that of capillaries (∼7.5 μm) in healthy adults. It can thus be mechanically filtrated by alveolar capillaries after being pumped out by the heart. This theoretically explains why this kind of microsphere displays decent lung-targeting ability and a good safety profile [47]. Lin et al. designed lung-targeting lysostaphin poly (dl-lactide-co-glycolide) (PLGA) microspheres for the treatment and prevention of methicillin-resistant Staphylococcus aureus pneumonia. The diameter of microspheres is approximately 11.36 ± 5.19 μm m, with good biocompatibility and biodegradability at this size [45]. A similar study conducted in 2020 also proved the excellent safety of this size of PLGA microsphere [46]. Elemental mapping showed that Ag+ was successfully loaded inside CNMS (Fig. S10). As previously reported, chitosan in acetic acid 2% solution presents a positive zeta-potential (60.92 ± 0.92 mV) due to protonation of the amino group [48]. The average potential of final NFF-3 CMS loaded with AgNO3 and CST was 5.61 ± 0.44 mV (Fig. S11).
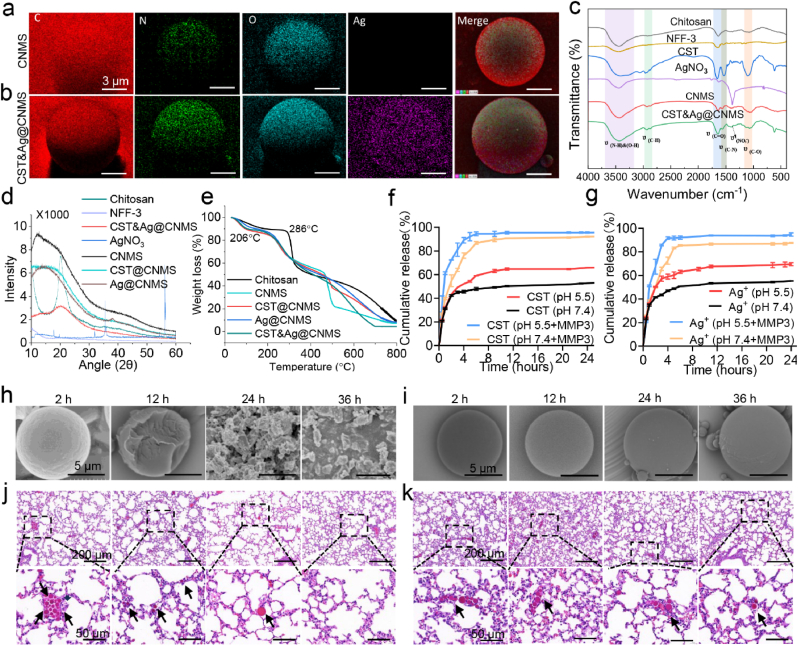
Fig. 1.
Design and characterization of CNMS. a & b SEM–EDS analysis of metallic elements C, N, O, and Ag of (a) CNMS and (b) CST&Ag@CNMS. c Comparison of the FT-IR spectra of chitosan, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS. d X-ray diffractogram of chitosan, CST and AgNO3, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS. e Thermogravimetric analysis (TGA) of chitosan, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS. f Cumulative in vitro release behavior of CST and AgNO3 from CST&Ag@CNMS at 37 °C in simulated lung fluids (SLFs) without MMP-3 (pH, ∼7.4–5.5) (n = 3). g Cumulative in vitro release behavior of CST and AgNO3 from CST&Ag@CNMS at 37 °C in SLFs with MMP-3 (pH, ∼7.4–5.5). Results are expressed as mean ± SD (n = 3). h & i Representative SEM images of CMS-M during the process of degradation; incubated at 37 °C in (h) PBS (pH, 5.5) in the presence of MMP-3 enzyme and (i) normal PBS (pH, 7.4) without MMP-3 enzyme. Scale bars: 5 μm. j & k Representative histology images over time of lungs exposed to microspheres after i. v. Injection. (j) LPS-treated lungs (2, 12, 24, and 36 h after injection of CNMS). Upper Scale bar 200 μm, lower scale bar 50 μm. (k) Healthy lungs (2, 12, 24, and 36 h after injection of CNMS). Upper Scale bar 200 μm, lower scale bar 50 μm. The images represent at least three independent experiments.
The structure and combination of CNMS were further analyzed using FT-IR. As indicated in Fig. 1c, compared with CNMS alone, the peaks were enhanced in the range 3600–3200 cm−1 due to stretching vibrations from the O–H and N–H groups. The polymer also showed characteristic bands of amide and alcohol corresponding to CST at 1637 cm−1 and 1200–1000 cm−1, respectively, suggesting the successful coating of CST. The anti-symmetric telescopic vibration characteristic absorption peak of nitrate appeared at 1381 cm−1, indicating the successful encapsulation of nitrate. The XRD diffractogram of pure chitosan (Fig. 1d), showed a crystalline nature with intense peaks at 2θ = 20.08°, which is consistent with previous studies [49]. Due to the strong intra- and intermolecular interactions of chitosan molecules, chitosan microspheres exhibit a semi-crystalline structure, characterized by the formation of hydrogen bonds between the amino-, hydroxyl-, acetylamine-, and other functional groups within the chitosan molecules. These strong interactions provide a certain degree of organization for the crystal structure of chitosan [43,50]. The crystallinity of CNMS decreases due to the polypeptide and silver ion effect, which is evidenced by diminution of the peak at 2θ = 20.08°, which is partly consistent with a previous report [50]. According to Fig. 1e, the thermal stability of microspheres formed by crosslinking was poorer than that of chitosan alone, as evidenced by the decline of maximum decomposition temperature from 286 °C to 206 °C. This finding may be attributed to reduced hydrogen bonds between molecules [51]. These findings combine to demonstrate that CNMS was successfully prepared.
The degradation property of the bio-responsive CST&Ag@CNMS was performed in SLFs, pH = 7.4 or pH = 5.5, with or without the MMP-3 enzyme. CST&Ag@CNMS released 52.89% CST at pH = 7.4 in the absence of MMP-3, significantly lower than the 92.39% released at pH = 5.5 in the presence of MMP-3 in 24 h (Fig. 1f). Similarly, in vitro drug release of AgNO3 from CST&Ag@CNMS reached 94.68% at pH = 5.5 with MMP-3, but only reached 55.33% at pH = 7.4 without MMP3. Furthermore, the AgNO3 release was 87.62% at pH = 7.4 with MMP-3 and 69.55% at pH = 5.5 without MMP3 (Fig. 1g). In general, microspheres with a larger size (>100 μm) have greater structural stability than smaller ones (<10 μm) [35,52]. In this study, we constructed chitosan microspheres with a size of ∼10 μm, which demonstrated excellent properties in terms of rapid material degradation and drug release. Moreover, we further improved the release and degradation properties of the microspheres, including MMP-3-response release and degradation, and protonated and swollen -NH2 of chitosan according to bacterial IMEs [53]. Additionally, degradation of chitosan is known to occur in vertebrates by lysosomes and several bacterial enzymes [54]. In short, the fabricated CNMS should be degraded in a timely and adequate manner in the human complex internal environment.
To better understand particle degradation and its relationship with the release mechanism, the CNMS surface morphology was observed in PBS at pH = 7.4 or pH = 5.5, with and without the MMP-3 enzyme. In PBS at pH = 5.5 with MMP-3 (Fig. 1h), the number of broken fragments increased over time. After immersion in this PBS solution for 36 h, the CNMS degraded substantially, with the appearance of CNMS resembling debris. However, after immersing in PBS at pH = 7.4 without MMP-3 (Fig. 1i), the microsphere remained unchanged. To assess whether CNMS biodegradation occurred using a similar method in vivo after i. v. Administration, CNMS was injected into LPS-treated mice, and the status of their lungs in situ was evaluated (Fig. 1j). CNMS appeared to degrade equally rapidly in vivo compared with in vitro, with all CNMS having dissolved within 36 h, indicating ideal biodegradability. Furthermore, this faster degradation was not observed in healthy mice (Fig. 1k). Release processes were also curved with equations, including first-order equations, Higuchi equations, and zero-order equations [55,56]. Results showed that the drug release from CNMS was consistent with first-order dynamics, indicating that drug release predominantly occurs via the diffusion mechanism [57]. According to the first-level release model, the initial release is most rapid, which is possibly because the compound is responsive to low pH and higher concentrations of MMP-3 (Figs. S12a and b). Taken together, these results demonstrate that CST&Ag@CNMS achieves rapid release in IMEs. This rapid silver and CST release in acidic environments enhances the local drug concentration at the infection site and decreases off-target drug effects.
3.3. Lung-targeting ability of CNMS with different sizes in vivo
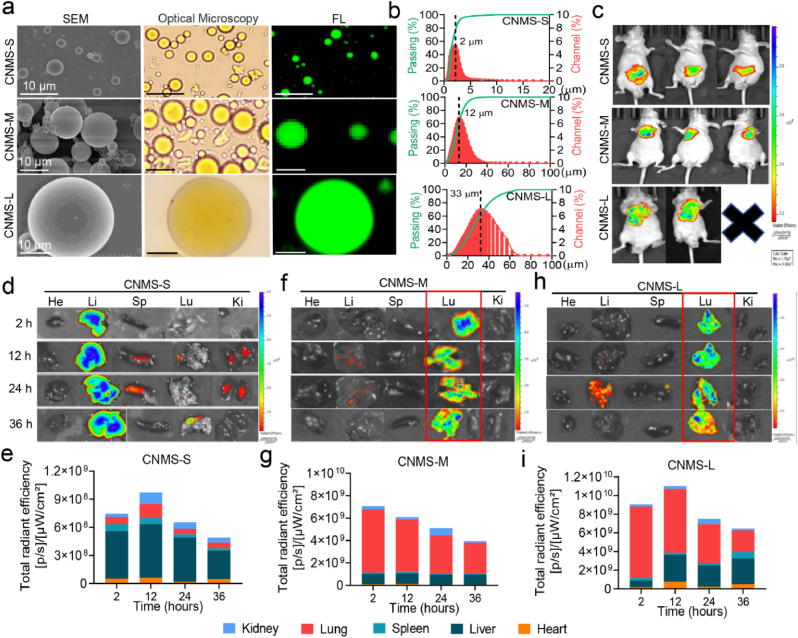
Given the potential toxicity of Ag+ and low blood concentration of CST in the lung, a targeted drug delivery system is mandatory to increase drug content in key sites while reducing aggregation and injury in non-targeted organs. In order to observe drug distribution visually in vivo, the near-infrared fluorescent dye IR783 was encapsulated into chitosan, forming a stable IR783@MS microsphere. As the size of the microsphere is vital in determining the efficiency of passive lung targeting, CNMS with large (CNMS-L), medium (CNMS-M), and small (CNMS-S) diameters were prepared by adjusting the ratio of oil and water and the emulsification rate. The microstructures of CNMS were observed using SEM, optical microscopy, and fluorescence microscopy (Fig. 2a). A Malvern Laser Particle Size Analyzer revealed that CNMS-L consisted of round droplets with an average size of 28.36 ± 5.28 μm, while the sizes of CNMS-M and CNMS-S were 12.74 ± 3.46 μm and 1.56 ± 0.62 μm, respectively (Fig. 2b).
Fig. 2.
Lung-targeting ability and biosafety of fluorescent CNMS with large (CNMS-L), medium (CNMS-M), and small (CNMS-S) diameters. A Representative image of SEM, bright-field, and fluorescent images of the three types of CNMS. b Size distribution of three types of CNMS. c–i (c) In vivo and ex vivo representative fluorescence imaging and corresponding fluorescence quantitative in the heart (He), liver (Li), spleen (Sp), lung (Lu), and kidney (Ki) of healthy mice after tail vein injection (i.v.) with (d & e) IR783@CMS-S, (f & g) IR783@CMS-M, and (h & i) IR783@CMS- L for 24 h, respectively. Images are representative of at least three independent experiments.
We selected the CNMS with the greatest lung-targeting ability through i. v. Injection of the surviving mice with the three CNMS types, all of which were labeled with IR783. Fluorescence imaging in vivo and ex vivo showed that both IR783@CNMS-M and IR783@CNMS-L accumulated predominantly in the lungs, with fluorescence accounting for nearly 80% of the total. In contrast, IR783@CNMS-S was mainly distributed in the liver (Fig. 2c–i; Fig. S13). One of the mice in the CNMS-L group died within 24 h of the experiment, probably due to pulmonary embolism and subsequent artery infarction owing to the large diameter of CNMS-L. Based on biosafety, lung-targeting ability, and drug-entrapment efficiency, CNMS-M was selected for the remaining experiments described in this article.
3.4. Antibacterial activity of CST&Ag@CNMS in vitro
We examined whether the antibacterial efficiency of Ag+ and CST is still observed after encapsulation in CNMS by exploring the antibacterial activity based of these polymers. We tested the MICs of CNMS loaded with CST, AgNO3, or both. The results indicated that encapsulating CST or AgNO3 in CNMS particles led to a 2- to 4-fold decrease in the antimicrobial effect of some bacteria (Tables S1 and S2). Notably, a 4-fold increase in MIC for netilmicin sulfate toward P. aeruginosa after formulation into poly (lactic-co-glycolic acid) nanoparticles has previously been reported, which was attributed to the gradual release of the drug from the particles [58]. A 2-fold concentration range is considered acceptable, and as a result, the precision of the broth dilution method is usually taken to be within a 2-fold concentration range [59]. Our research showed that the MIC of CST&Ag@CNMS was reduced by 2–4-fold compared with single drug loaded particles. More importantly, the MIC of CST&Ag@CNMS was not greater than that of free drugs. A standard checkerboard microdilution assay revealed that Ag+ encapsulated in CNMS remained a strong adjuvant of CST-loaded particles for the MCR-1+ bacteria, Kp4, Ec34, and other MCR-1+ strains (Fig. 3a and b; Fig. S14). We also observed a similar synergistic effect with FICIs of 0.1875 and 0.25, respectively (Fig. 3c and d). By contrast, no such synergism was observed for Kp5435 or Ec5505, neither of which carry the mcr-1 gene. Bacterial growth curves further demonstrated the time-dependent growth inhibition effects of CST&Ag@CNMS against MCR-1+ Kp and Ec, while the use of each drug individually rarely inhibited bacteria growth over time (Fig. 3e and f; Fig. S15). By contrast, substantial growth was observed at the same concentration of CST&Ag@CNMS for MCR-1− Kp5435 and Ec5505 (Fig. 3e and f). The antibacterial properties were also analyzed using the dilution plating experiment. At the same material concentration, the results of Kp4 and Ec34 CFUs also aligned with the standard checkerboard microdilution method. No colony growth was observed in the CST&Ag@CNMS group, in contrast to the other groups (Fig. 3g and h).
Fig. 3.
CST&Ag@CNMS exhibits strong bactericidal activity in vitro. a and b Representative heat plots of microdilution checkerboard assays for the combination of CST@CNMS and Ag@CNMS against (a) MCR-1+ Kp4 and MCR-1− Kp5435, (b) MCR-1+ Ec34 and MCR-1− Ec5505. c and d Isobolograms of the combination of CST@CNMS and Ag@CNMS against (c) different MCR-1+ Kp strains (Kp4, Kp18, Kp31, and Kp38), MCR-1− Kp strains (Kp5435), and (d) MCR-1+ Ec strains (Ec34, Ec50, Ec62, and Ec99) and MCR-1− Ec strains (Ec5505). The green dashed line indicates the ideal isobole, where drugs act additively and independently. Data points below this line indicate synergism. e and f Bacteria time-growth curves for CST@CNMS or Ag@CNMS monotherapy or their combination therapy against (e) MCR-1+ Kp4 and MCR-1− Kp5435 and (f) MCR-1+ Ec34 and MCR-1− Ec5505 during 24-h incubation. The concentrations of CST@CNMS and AgNO3@CNMS are used at ¼ MIC for each bacterium. g and h Representative agar plate images of different treatments killing (g) Kp4 and (h) Ec34. i and j Representative fluorescence images of (i) Kp4 and (j) Ec34 treated with control, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS. Live cells were stained positive for SYTO 9 (green), while dead cells were visualized by PI staining (red). Scale bars: 20 μm. k and l Representative standard error of the mean of (k) Kp4 and (l) Ec34 at 8 h after treatment with control, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS, respectively. White arrows indicate damaged cell membrane. m Resistance acquisition curves during serial passage with the subinhibitory concentration of CST@CNMS or the combination of CST@CNMS and Ag@CNMS against MCR-1+ strains at ¼ MIC concentrations for Kp4 and Ec34. This test was performed after every two passages. All the experiments were conducted at least three times at different time points.
To visualize the state of bacteria exposed to different types at fixed amounts of CNMS and to explore the underlying mechanism of the antipathogens, we stained Kp4 and Ec34 with live/dead dye. In this staining method, PI stains cells with an injured membrane in a red color, while SYTO9 dyes all cells green; however, the green color of SYTO9 is affected or diminished if PI is also present. Despite a percentage of red cells appearing in CST@CNMS and Ag@CNMS, only large patches of red fluorescence were observed in the final CNMS, suggesting an enhanced ability to disrupt bacterial cell membrane after combining Ag+ and CST (Fig. 3i and j; Fig. S16). To further observe the alteration of bacterial structure and explore the underlying mechanisms, we performed SEM to capture high-definition images of the pathogens. The SEM images revealed that the bacterial morphologies were nearly all wrinkled and deformed in the final group. In contrast, the morphological changes in the CST@CNMS and Ag@CNMS groups were less severe, and the bacteria in the control group remained smooth and intact (Fig. 3k and l).
Similar to previous findings in free drugs, resistance to CST@CNMS was significantly elevated upon exposure to CST@CNMS at subinhibitory concentrations, as its MIC value increased by 16-fold over a period of 20 serial passages. By contrast, only a slight increase was observed in the CST&AG@CNMS group (Fig. 3m). Our combined data demonstrated that Ag+, even when loaded in CNMS, still restores the antibacterial efficiency of CST against MCR-1+ bacteria. Additionally, CST&Ag@CNMS combines the advantages of Ag+ and CST to kill pathogens effectively in vitro by disrupting the bacteria membrane.
3.5. Mechanisms by which Ag + restores colistin efficacy
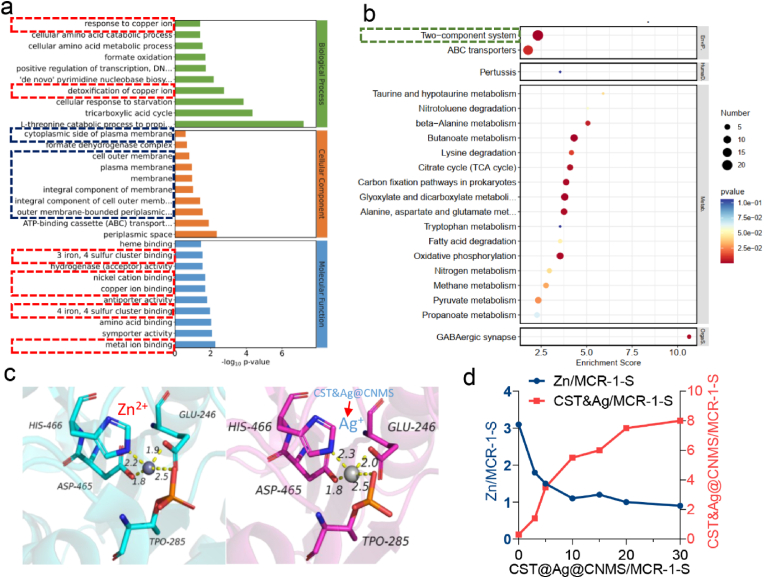
RNA sequencing before and after CST&Ag@CNMS treatment was carried out to explore the potential mechanism for the observed decreased resistance of CST to MCR-1+ CST-resistant bacteria. Genes differentially expressed at significant levels between cases and controls were analyzed using the Gene Ontology (GO) database to identify related biological processes. As shown in Fig. 4a, the metal ion (red dotted line) and components of both the outer and integral cellular membrane (blue dotted line) were the most enriched GO terms, indicative of the significant effect on the cell membrane of Ec34 and associated metal ion pathways. We demonstrated bacterial cell membrane damage via bacteria live/dead staining assay (Fig. 3i and j) and SEM images (Fig. 3k and l). However, the exact mechanism of alteration in the metal ion process remains to be investigated. The Kyoto Encyclopedia of Genes and Genomes enrichment analysis (Fig. 4b) revealed that a two-component system (TCS) has the highest number of differential expression analyses among the pathways. In G− bacteria, TCS acts as a typical signal transduction system that plays a significant role in regulating cellular communication [3,60]. TCS consists of histidine kinase protein GacS that detects a specific signal and the response regulatory protein GacA that modulates the response by regulating gene expression [60]. Metal ions are among the external signals sensed by TCSs. Zn2+- zinc resistance-associated protein (ZraP) was previously reported to sense Zn2+ and to modulate colistin sensitivity through LPS modifications [61,62]. Taken together, it is plausible that ZraP responded to Zn2+ dysregulation and induced further LPS redecoration.
Fig. 4.
Ag + restores colistin efficacy by displacing the Zn2+ cofactor. A GO terms enrichment of Differentially Expressed Genes (DEGs) in bacteria treated with CST&Ag@CNMS. b Pathway enrichment of DEGs in bacteria treated by CST&Ag@CNMS. c Cartoon of tetra-nuclear Zn center and Ag center in the active-site pocket of the MCR-1-S enzyme. d The substitution of Zn2+ of Zn MCR-1 by CST&Ag@CNMS via equilibrium dialysis. The metal content was determined by ICP-MS. All the experiments were conducted at least three times at different time points.
One possible mechanism by which Ag+ restores colistin activity against MCR-positive bacteria has been proposed in previous research, which stated that Ag+ inactivates MCR-1 by displacement of the Zn2+ cofactor [27]. To compare our results with findings of the previous study, and to further characterize the mechanism of CST&Ag@CNMS against MCR-1 function, the MCR-1 protein crystal structure obtained from the RCSB PDB database (PDBID: 5YLF) was subjected to molecular dynamics simulation analysis and processing using Schrödinger software. The native Zn-bound MCR-1 results demonstrated that Zn2+ could bind to the cavity on the surface of the MCR-1 protein, forming a salt bridge with residues TPO285, HIS466, GlU246, and ASP465, with bond lengths of 2.50 Å, 2.20 Å, 1.90 Å, and 1.80 Å, respectively (Fig. 4c; Fig. S17). Similarly, computational insights from molecular docking analysis revealed that the Ag+ forms salt bridges with residues TPO285, HIS466, GlU246, and ASP465 located within the active-site pocket of the MCR-1 protein, with bond lengths of 2.50 Å, 2.30 Å, 2.00 Å, and 1.80 Å, respectively (Fig. 4c; Fig. S17). MMGBSA analysis showed the MMGBSA dG binding value for Zn2+ and MCR-1 was 3.13 kcal mol−1, indicating a high binding free energy and unstable binding between Zn2+ and MCR-1 protein. By contrast, the MMGBSA dG binding for Ag+ and MCR-1 was −20.71 kcal mol−1, suggesting a low binding free energy and relatively stable binding between Ag+ and MCR-1. These results further validate the substituting ability of Ag + for Zn2+ in the MCR-1 enzyme, elucidating the excellent inhibitory ability of Ag+ on MCR-1 enzyme activity.
Based on the findings of transcriptomics analysis and molecular simulation docking, we observed the binding ability of CST&Ag@CNMS to the enzyme of MCR-1 and the corresponding replacement of Zn2+ by ICP-MS. As previously reported, intact MCR-1-S can bind to 3 M equivalents of Zn2+ [27]. We then set out to explore the characteristics of the CST&Ag@CNMS binding enzyme and the subsequent process of releasing Zn2+. With CST&Ag@CNMS supplementation, the binding stoichiometry of Zn2+ to MCR-1-S gradually reduced, while the chemical metering of CST&Ag@CNMS to MCR-1-S progressively increased. As a result, almost all Zn2+ ions were replaced with CST&Ag@CNMS. However, after adding excessive Zn2+ to the former solution, we did not observe any noticeable changes in the binding of metals to MCR-1-S, indicating irreversible Zn2+ release (Fig. S18). Therefore, we can see that CST&Ag@CNMS improves the sensitivity of MCR-1+ bacteria to CST by replacing Zn2+ in the MCR-1 protein pocket.
3.6. In vitro biosafety of CST&Ag@CNMS
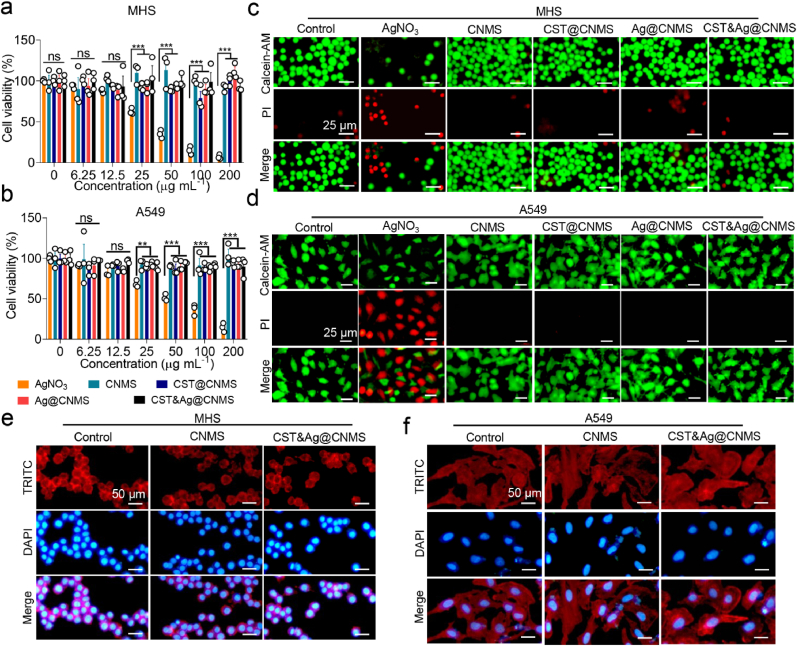
We selected alveolar lung epithelial cell A549, endothelial cell EA. hy926, and alveolar macrophage cell MHS to evaluate the cytotoxicity of CNMS using Cell Counting Kit-8 (CCK8 assay), as these particles are intended for venous injection and lung targeting. CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS showed excellent biological compatibility since cell viabilities were >80% after 24 h incubation with different treatments at the indicated concentrations (Fig. 5a and b; Fig. S19a). We also investigated whether the final concentrations of Ag + selected in the current study exerted a toxic effect in vitro. Although unwrapped Ag + began to show a slight cytotoxicity at a concentration of 25 μg mL−1, Ag+ loaded in CNMS still presented a higher degree of cell safety, even at 200 μg mL−1, probably due to the sustained release of CNMS.
Fig. 5.
Biosafety evaluation of CNMS in vitro shows CNMS alleviates toxicity of Ag+. a & b Survival of (a) MHS cells and (c) A549 cells after 24 h exposure to an increasing concentration of AgNO3, CNMS, CST@CNMS, AgNO3@CNMS, and CST&Ag@CNMS. c & d Representative fluorescent images of (b) MHS cells and (d) A549 cells after treatment with AgNO3, CNMS, CST@CNMS, Ag@CNMS, and CST&Ag@CNMS (25 μg mL−1) for 24 h. The dead and viable cells were visualized by PI staining (red) and calcein AM (green), respectively. Scale bar, 25 μm. e & f TRITC/DAPI staining of (e) MHS cells and (f) A549 cells after exposure to medium, CNMS, and CST&Ag@CNMS for 1 day, respectively. Scale bar, 50 μm. Data are shown as mean ± SEM, n = 5. ns, no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Images are representative of at least three independent experiments.
The result of the in vitro safety study was further confirmed by the cell live/dead staining assay. Incubation with 25 μg mL−1 CNMS, Ag@CNMS, CST@CNMS, and CST&Ag@CNMS for 24 h did not increase PI positive cells (red fluorescence) compared with the untreated group (Fig. 5c and d; Fig. S19b). The morphology of the cocultured cells was also investigated using TRITC/DAPI staining (Fig. 5e and f; Fig. S20). When different lines cells were treated with CNMS or drug-loaded CNMS, the cell structure remained as unchanged and healthy as the control group (Fig. 5e and f; Fig. S20). Collectively, when incubated with CNMS loaded with or without drugs, A549, MHS, and EA. hy926 cells thrived with normal structure, cell adhesion, proliferation, and viability.
3.7. In vivo therapeutics of CST&Ag@CNMS against Kp4-induced severe pneumonia
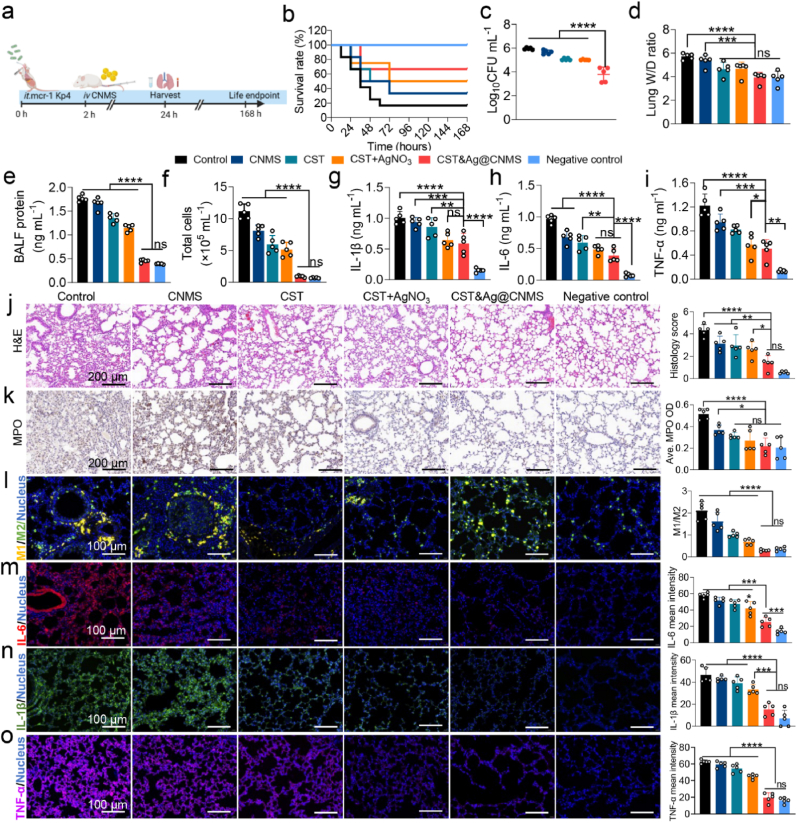
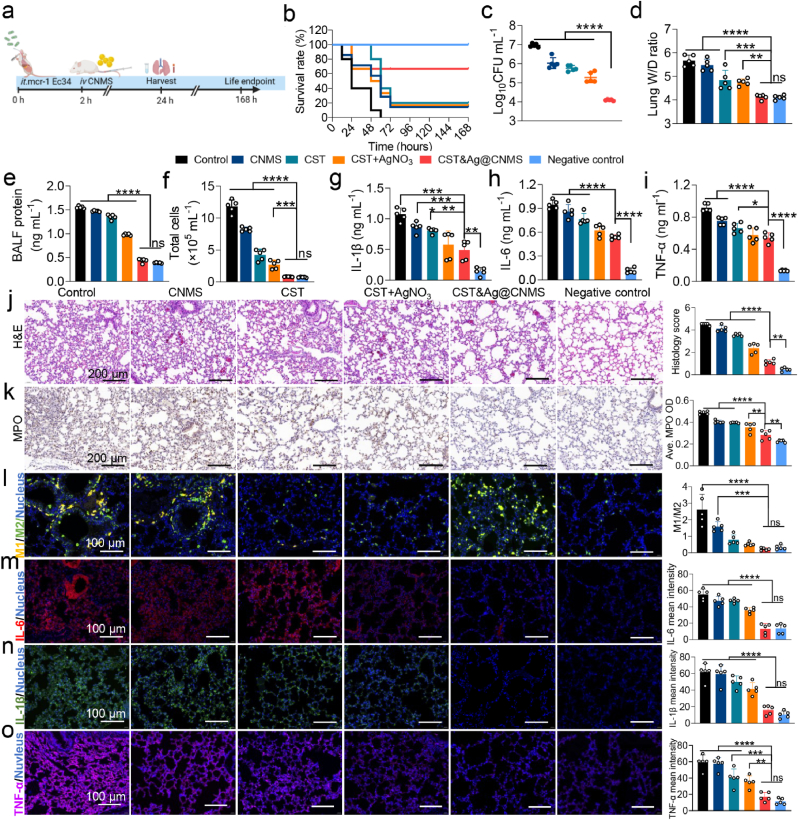
We used a mouse model of acute bacterial pneumonia to test whether the synergistic effects of Ag+ and CST observed in vitro are also be observed in vivo after treatment using drugs loaded with CNMS. 2 h after being intratracheally (i.t.) inoculated with Kp4 (106 CFU per mouse), the mice with acute pneumonia were injected i. v. With PBS, CNMS, CST, CST&AgNO3, and CST&Ag@CNMS, respectively (Fig. 6a). After 24 h, the CST&Ag@CNMS group had a higher survival rate (60%) compared with those treated with PBS (20%), CNMS (30%), CST (30%), or CST + AgNO3 (50%) (Fig. 6b). Similarly, the CST&Ag@CNMS combination significantly mitigated bacteria burden, while the bacterial colony in the lung homogenate was not noticeably reduced in the other groups (Fig. 6c; Fig. S21). The reason for the superior bactericidal activity in vivo may be due to the higher concentrations of CST and Ag+ in the alveoli released from the bio-responsive CST&Ag@CNMS compared with free drugs, indicating the specific lung-targeting properties of CST&Ag@CNMS (Fig. 6c; Fig. S21). To compare the degree of pulmonary interstitial edema with the different treatment regimens, the lung wet/dry weight (W/D) ratio and levels of protein in BALF were calculated. Infected mice without any treatment displayed the highest ratio and the largest protein level, while the final group exhibited values close to the negative control, indicating the alleviation of edema and interstitial fluid leakage in mouse lungs (Fig. 6d and e). In summary, treatments with CST&Ag@CNMS protected against CST-resistant MCR-1+ Kp4-induced pneumonia, decreased the formation of bacteria colony, and alleviated pulmonary edema.
Fig. 6.
Treatment with CST&Ag@CNMS effectively attenuated MCR-1+ Kp4 pneumonia. an Experimental protocol of post-infection treatment studies in Kp4 pneumonia mouse models. Mice were infected with Kp4 via intratracheal inoculation (i.t.). 2 h after infection, mice were treated with PBS, CNMS, CST, AgNO3+CST, and CST&Ag@CNMS via i. v. Injection, respectively. Samples were harvested 24 h after infection, and the survival endpoint was calculated at 168 h b Percentage survival of pneumonia mice treated with PBS, CNMS, CST, AgNO3+CST, CST&Ag@CNMS, and control. c Bacterial CFUs in lung homogenates at 24 h (left) after infection in different treated groups. Data are shown as mean ± SEM, n = 5. d Lung (Right) wet/dry weight ratio of mice after infection exposed to different treatments. Data are shown as mean ± SEM, n = 5. e –i (e) BALF protein, (f) total cell amount, (g) IL-1β, (h) IL-6, and (i) TNF-α in BALF collected at 24 h post-infection in different treated groups. Data are shown as mean ± SEM, n = 5. j Representative images of H&E stained and corresponding quantification of H&E score. k immunohistochemically stained (MPO) lung sections and average MPO OD at 24 h post-infection (m). Scale bar, 200 μm. l –o Representative images of immunofluorescence staining with (l) M1/M2, (m) IL-1β (n), IL-6, and (o) TNF-α of lung sections and quantification of immunofluorescence intensity of themselves. Data are shown as mean ± standard error of the mean. ns, no significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Since eliminating pathogens and maintaining a balance of immune stationary are the principal objectives of pneumonia therapy, we further evaluated lung inflammation and cytokine levels in mice exposed to different treatments. Hence, the total number of cells collected in BALF were counted. As presented in Fig. 6f, there was an increase in the number of cells in infected mice treated with PBS, indicating the most severe infection, while the CST&Ag@CNMS-treated group showed a decrease in the number of cells. The neutrophil cell count in the model group (77.24%) was considerably higher compared with the final group (33.65%) (Fig. S22a). We hypothesized that the lower neutrophil count might be associated with a lower bacterial load in the lungs of mice treated with CST&Ag@CNMS. As expected, the proinflammatory cytokines IL-1β (Fig. 6g; Fig. S22b), IL-6 (Fig. 6h; Fig. S22c), and TNF-α (Fig. 7i; Fig. S22d) both in BALF and serum showed a greater diminution in the CST&Ag@CNMS category compared with others.
Fig. 7.
Treatment with CST&Ag@CNMS effectively attenuated MCR-1+ Ec34 pneumonia. an Experimental protocol of post-infection treatment studies in Kp4 pneumonia mouse models. Mice were infected with Ec34 via intratracheal inoculation (i.t.). 2 h after infection, mice were treated with PBS, CNMS, CST, AgNO3+CST, and CST&Ag@CNMS via i. v. Injection, respectively. Samples were harvested 24 h after infection, and survival endpoint was calculated at 168 h b Percentage survival of pneumonia mice treated with PBS, CNMS, CST, AgNO3+CST, CST&Ag@CNMS, and control. c Bacterial CFUs in lung homogenates at 24 h (left) after infection in different treated groups. Data are shown as mean ± SEM, n = 5. d Lung (Right) wet/dry weight ratio of mice after infection exposed to different treatments. Data are shown as mean ± SEM, n = 5. e –i Levels of (e) BALF protein, (f) total cell amount, (g) IL-1β (h), IL-6, and (i) TNF-α in BALF collected at 24 h post-infection in different treated groups. Data are shown as mean ± SEM, n = 5. j Representative images of H&E stained and corresponding quantification of H&E score. k Immunohistochemically stained (MPO) lung sections and average MPO OD at 24 h post-infection (m). Scale bar, 200 μm. l–o Representative images of immunofluorescence staining with (l) M1/M2, (m) IL-1β, (n) IL-6, and (o) TNF-α of lung sections and quantification of immunofluorescence intensity of themselves. Data are shown as mean ± standard error of the mean. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
H&E staining of the lungs revealed that CST&Ag@CNMS notably alleviated bacteria-induced lung pneumonia, as characterized by inflammatory cell infiltration, broadening alveolar walls, necrosis of the bronchiole mucosa, and inflammatory exudate fluid of the alveolar cavity. These conditions were also more or less prevalent in the other groups (Fig. 6j). Myeloperoxidase (MPO) is an enzyme found in neutrophils, which acts as a critical inflammatory mediator to provoke enhanced inflammatory states [63]. In the model group, MPO accounted for a significantly large proportion in the whole lung section tissue, compared with its much smaller influence in both the negative control group and the CST&Ag@CNMS group, as demonstrated by immunohistochemical staining and the mean OD values of MPO (Fig. 6k). In the early stages of inflammation, LPS binds to the toll-like receptor 4 (TLR4) on macrophages, transforming them into M1-type macrophages and provoking an inflammatory cytokine storm [64]. Hence, we assessed the anti-inflammatory effects of inflamed macrophages on lung tissue using immunofluorescence staining. The proportion of M1-like macrophages (CD86+) in the CST&Ag@CNMS group was significantly lower than for other groups, while M2-like macrophages (CD163+) exhibited the opposite trend (Fig. 6l; Fig. S23). The ideal anti-inflammatory properties exhibited by the final product may be attributed to its robust antibacterial characteristics and the ability of chitosan to regulate macrophage polarization [65,66]. Consistently, immunofluorescence staining of the proinflammatory cytokines, IL-6, IL-1β, and TNF-α, in the lung corresponded with levels in the BALF and serum (Fig. 6m–o; Figs. S24–S26). Taken together, these results show that CST&Ag@CNMS treatment protects mice from pneumonia-related lung injury arising from an excessive inflammatory response by relieving inflammation and reducing cytokine levels.
3.8. In vivo therapeutics of CST&Ag@CNMS against Ec34-infected pneumonia
Due to the presence of mcr-1 in carbapenem-resistant G− Enterobacteriaceae (CRE), we selected Ec34 as a molding strain, which is resistant to both carbapenem and CST. A schematic of our in vivo animal experiments is presented in Fig. 7a. The CST&Ag@CNMS group exhibited an elevated survival rate (Fig. 7b) and excellent antibacterial efficiency against MCR-1+ MDR Ec34 (Fig. 7c; Fig. S27). After CST&Ag@CNMS treatment, bacteria colonies showed a ∼2–3 log reduction. Therefore, the final product maintained strong sterilization properties in CRE carrying mcr-1.
Besides the suppression of bacterial growth, prevention or inhibition of a cytokine storm could be crucial in saving the lives of individuals suffering from severe pneumonia, as previously reported [37]. Indeed, CST&Ag@CNMS significantly reduced the degree of lung tissue edema, as shown by a lower lung W/D ratio and reduced BALF protein levels in the final group (Fig. 7d and e). CST&Ag@CNMS also reduced the levels of proinflammatory cytokines, including IL-1β, IL-6, and TNF-α, protein concentration, and total cell count in BALF compared with free CST, CST + AgNO3, and CNMS (Fig. 7f–i; Fig. S28a). The suppression was also manifest systemically, with corresponding reductions in serum levels of IL-1β, IL-6, and TNF-α in the presence of CST&Ag@CNMS (Fig. S28b).
Histological analysis, including H&E staining and immunofluorescent (IF) staining, revealed excessive pulmonary edema, alveolar inflammatory cell exudation/infiltration, and alveolar wall thickening in lungs infected with Ec34 (Fig. 7j–o). However, these pathological features were substantially reduced in the CST&Ag@CNMS group, resulting in nearly normal lung histology after CST&Ag@CNMS treatment (Fig. 7j). A similar phenomenon was observed in the evaluation of MPO and the M1/M2 macrophage ratio (Fig. 7k and i; Fig. S29). Levels of proinflammatory cytokines, including IL-6, L-1β, and TNF-α, in the serum of treated and untreated mice yielded similar results to those obtained by IF observation in the lungs (Fig. 7m–o; Figs. S30–S32). Collectively, our results demonstrate the ability of CST&Ag@CNMS to kill MCR-1+ CRE and effectively relieve inflammation in vivo.
3.9. Biosafety of CST&Ag@CNMS in vivo
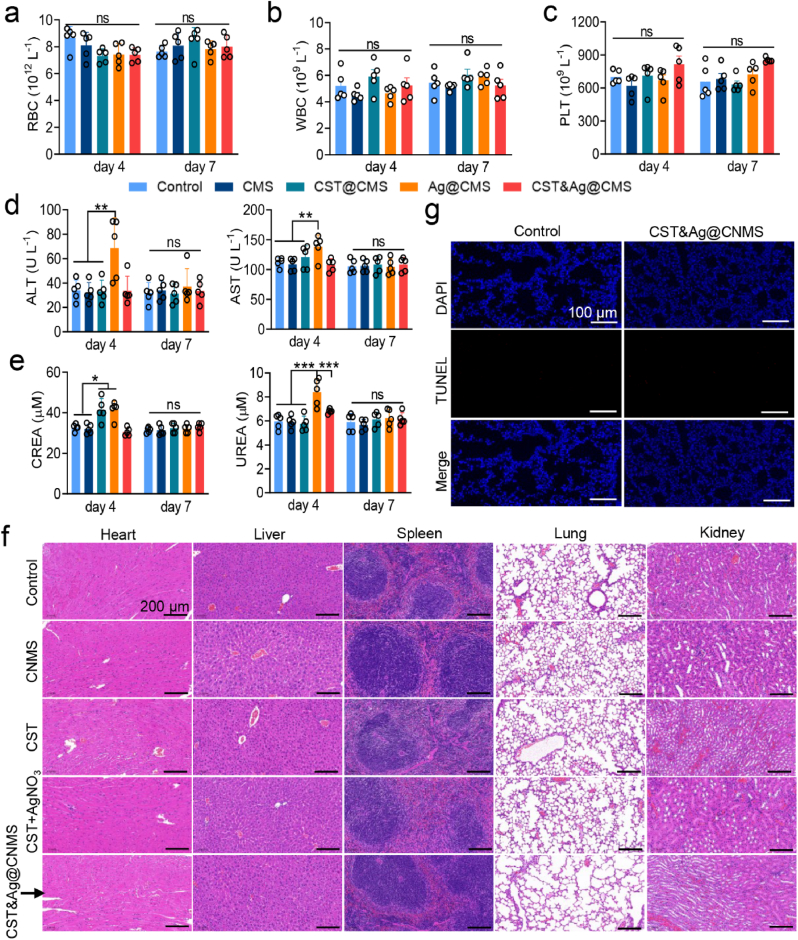
Given the short- and long-term potential safety hazards of CST&Ag@CNMS, we evaluated their effects on the blood chemistry and function of major organs using healthy mice by measuring vital enzyme content and performing H&E staining. After treatment with the different agents for 3 days, we collected the blood and main organs of the treated mice on Days 4 and 7. There was no difference in hemocyte parameters between the groups (Fig. 8a–c), although we noticed slightly elevated concentrations of the hepatic enzymes alanine aminotransferase (ALT) and aminotransferase aspartate (AST) in the group treated with a combination of free drugs, as previously reported (Fig. 8d) [67]. Nevertheless, in the CST&Ag@CNMS group, the concentrations of hepatic enzymes were stable within the normal range throughout the study duration, indicating that the toxic effects of Ag+ were mitigated by CNMS. Likewise, indicators of kidney function, including creatinine and urea, exhibited similar trends to the hepatic enzymes, with the final polymers showing no visible kidney injury (Fig. 8e).
Fig. 8.
Safety evaluation in vivo evidenced biosafety of CST&Ag@CNMS. a–c Routine blood indices of (a) RBCs, (b) white blood cells, and (c) platelets on Days 4 and 7 after 3 days of consecutive injections of CNMS, CST, CST + AgNO3, and CST&Ag@CNMS (n = 5). d–e Blood biochemical values of (d) ALT and AST, (e) creatinine and urea on Days 4 and 7 in same above groups (n = 5). f H&E staining of heart, liver, spleen, lung, and kidney on Day 7 after 3 days of injection with different agents. Scale bar, 200 μm g TUNEL staining of the lung, on Day 7 post-injection, when the nuclei were counterstained with DAPI (blue) and apoptotic cells stained with TUNEL (red). Scale bar, 100 μm. Error bars represent means ± standard error of the mean from at least three biological replicates.
As further illustrated by H&E staining 72 h after injection, the heart, liver, spleen, lung, and kidney tissues showed no obvious pathological damage, suggesting the biocompatibility of CST&Ag@CNMS, and the absence of side effects relating to Ag+ and CST (Fig. 8f). Consistently, the cell apoptosis levels for lung slices reflected showed the same effect, as revealed by the deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay (Fig. 8g). Meanwhile, a hemolytic test of CNMS loaded with CST, Ag+, or their combination, at a concentration of 100 μg mL−1 was performed to further evaluate blood biosafety. As shown in Fig. S33, the treated RBCs did not show obvious damage. Based on the above results, mice treated with CST&Ag@CNMS did not experience any acute or chronic toxicity, either topically or systemically. In contrast, CST&Ag@CNMS exhibited excellent biosafety, and may even act as an antidote.
4. Conclusions
The present study developed a feasible potentially clinical translational MMP-3 bio-responsive release chitosan microsphere (CNMS) by co-loading CST and Ag+, named as CST&Ag@CNMS, to improve the target densities and antibacterial and anti-inflammatory effects in the lungs. This lung-targeted delivery system sensitizes G− bacteria to CST by substituting Zn2+ with Ag+ in the active-site pocket of MCR-1, lower the toxicity and increase the bioavailability of both CST and Ag+. We showed that in both in vitro and in vivo mouse models of lung infection mediated MCR-1+ CST-resistant Kp and Ec, the micron particles exhibited enhanced antibacterial and anti-inflammatory activities compared with free drugs, without introducing any visible side effects. Moreover, this polymer exhibited an intriguing lung-targeting ability and biosafety. Compared with the time- and cost-consuming nature of developing novel antibiotics, these findings support an alternative approach to strengthening the antibiotic arsenal against bacterial infections.
Data availability
All data that support the findings of this study are presented in the main text and the supplementary information. Additional data are available from the corresponding author upon reasonable request.
Funding
This study was supported by the Natural Science Foundation of Shanghai [grant number 23ZR1456800], the Interdisciplinary Program of Shanghai Jiao Tong University [grant number YG2021ZD07], the Science and Technology Commission of Shanghai Municipality [grant number 20Y11901100], the Clinical Science and Technology Innovation Project of SHCD [grant number SHDC22021212], the National Natural Science Foundation of China [grant number 82002188], the Scientific Research Project Plan of Shanghai Municipal Health Commission [grant number 20204Y0145], and the Guangci Discipline Group Construction of Public Health and Disaster Emergency Center [grant number XKQ-09].
Ethics
All animal experiments were approved by Animal Investigation Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine.
CRediT authorship contribution statement
Decui Cheng: Writing – original draft, Software, Resources, Methodology, Investigation, Data curation. Rui Tian: Software, Methodology, Investigation, Formal analysis. Tingting Pan: Project administration. Qiang Yu: Methodology, Formal analysis. Li Wei: Data curation. Jiaozhi Liyin: Software, Resources, Methodology. Yunqi Dai: Validation, Methodology, Investigation. Xiaoli Wang: Writing – review & editing, Visualization, Funding acquisition, Conceptualization. Ruoming Tan: Writing – review & editing, Validation, Supervision. Hongping Qu: Writing – review & editing, Funding acquisition, Conceptualization. Min Lu: Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
None.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.02.017.
Contributor Information
Xiaoli Wang, Email: wxl12028@rjh.com.cn.
Ruoming Tan, Email: trm11714@rjh.com.cn.
Hongping Qu, Email: qhp10516@rjh.com.cn.
Min Lu, Email: lumin111@sjtu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Liu Y.N., et al. Infection and co-infection patterns of community-acquired pneumonia in patients of different ages in China from 2009 to 2020: a national surveillance study. Lancet Microbe. 2023;4(5):e330–e339. doi: 10.1016/S2666-5247(23)00031-9. [DOI] [PubMed] [Google Scholar]
- 2.Nair G.B., Niederman M.S. Updates on community acquired pneumonia management in the ICU. Pharmacol. Ther. 2021;217:107663. doi: 10.1016/j.pharmthera.2020.107663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., et al. DeepSecE: a deep-learning-based Framework for multiclass Prediction of secreted Proteins in gram-negative bacteria. Research. 2023;6:258. doi: 10.34133/research.0258. (Wash D C) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attallah N.G.M., et al. Anti-Biofilm and antibacterial Activities of Cycas media R. Br secondary metabolites: in silico, in vitro, and in vivo approaches. Antibiotics. 2022;11(8) doi: 10.3390/antibiotics11080993. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taglialegna A. Reviving colistin. Nat. Rev. Microbiol. 2023;21(7) doi: 10.1038/s41579-023-00907-0. 411–411. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y.Y., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Poirel L., et al. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect. Dis. 2016;16(3):281. doi: 10.1016/S1473-3099(16)00006-2. [DOI] [PubMed] [Google Scholar]
- 8.Du H., et al. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect. Dis. 2016;16(3):287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang X., et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infect. 2018;7(1):122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., et al. Rapid increase in prevalence of carbapenem-resistant Enterobacteriaceae (CRE) and emergence of colistin resistance gene mcr-1 in CRE in a hospital in henan, China. J. Clin. Microbiol. 2018;56(4) doi: 10.1128/JCM.01932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojas L.J., et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin. Infect. Dis. 2017;64(6):711–718. doi: 10.1093/cid/ciw805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X.F., et al. Possible transmission of mcr-1-harboring Escherichia coli between companion animals and human. Emerg. Infect. Dis. 2016;22(9):1679–1681. doi: 10.3201/eid2209.160464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falgenhauer L., et al. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect. Dis. 2016;16(3):282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 14.Moawad A.A., et al. Antimicrobial resistance in Enterobacteriaceae from healthy broilers in Egypt: emergence of colistin-resistant and extended-spectrum β-lactamase-producing Escherichia coli. Gut Pathog. 2018;10:39. doi: 10.1186/s13099-018-0266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernández M., et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 2017;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda A., et al. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int. J. Antimicrob. Agents. 2018;51(1):163–164. doi: 10.1016/j.ijantimicag.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Belaynehe K.M., et al. Emergence of mcr-1 and mcr-3 variants coding for plasmid-mediated colistin resistance in Escherichia coli isolates from food- producing animals in South Korea. Int. J. Infect. Dis. 2018;72:22–24. doi: 10.1016/j.ijid.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Poirel L., Jayol A., Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms Encoded by Plasmids or chromosomes. Clin. Microbiol. Rev. 2017;30(2):557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon R.A., Chopra I. Polymyxin B and polymyxin B nonapeptide alter cytoplasmic membrane permeability in Escherichia coli. J. Antimicrob. Chemother. 1986;18(5):557–563. doi: 10.1093/jac/18.5.557. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y., et al. Mechanistic insights into transferable polymyxin resistance among gut bacteria. J. Biol. Chem. 2018;293(12):4350–4365. doi: 10.1074/jbc.RA117.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyers M., Wright G.D. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nature Reviews. Microbiology. 2019;17(3):141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y., Ma Q.J.S.M. 2022. Bacterial Infection Microenvironment-Responsive Porous Microspheres by Microfluidics for Promoting Anti-infective Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., et al. Advances in the delivery systems for oral antibiotics. Biomedical Technology. 2023;2:49–57. [Google Scholar]
- 24.Torres A., et al. Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect. Dis. 2018;18(3):285–295. doi: 10.1016/S1473-3099(17)30747-8. [DOI] [PubMed] [Google Scholar]
- 25.Kaye K.S., et al. Effect of meropenem-Vaborbactam vs piperacillin-Tazobactam on clinical Cure or Improvement and microbial Eradication in complicated urinary tract infection: the TANGO I randomized clinical trial. JAMA. 2018;319(8):788–799. doi: 10.1001/jama.2018.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., et al. Gold drugs as colistin adjuvants in the fight against MCR-1 producing bacteria. J. Biol. Inorg. Chem. 2023;28(2):225–234. doi: 10.1007/s00775-022-01983-y. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q., et al. vol. 119. 2022. Re-sensitization of mcr carrying multidrug resistant bacteria to colistin by silver. (Proceedings of the National Academy of Sciences of the United States of America). 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H., et al. Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin. Nat. Commun. 2020;11(1):5263. doi: 10.1038/s41467-020-18939-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R., et al. Bismuth antimicrobial drugs serve as broad-spectrum metallo-β-lactamase inhibitors. Nat. Commun. 2018;9(1):439. doi: 10.1038/s41467-018-02828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Barajas N., de Jesús Martín-Camacho U., Pérez-Larios A. Mechanisms of metallic nanomaterials to induce an antibacterial effect. Curr. Top. Med. Chem. 2022;22(30):2506–2526. doi: 10.2174/1568026622666220919124104. [DOI] [PubMed] [Google Scholar]
- 31.Wong K.I., et al. Combating drug-resistant helicobacter pylori infection with zinc peroxide-based nanoparticles: a ROS reservoir via photochemical reaction. Chem. Eng. J. 2024;483:149287. [Google Scholar]
- 32.Lu G., et al. Injectable nano-micro Composites with anti-bacterial and osteogenic Capabilities for minimally invasive Treatment of osteomyelitis. Adv. Sci. 2024 doi: 10.1002/advs.202306964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., et al. Bio-responsive sliver peroxide-nanocarrier Serves as broad-spectrum metallo-β-lactamase Inhibitor for combating severe pneumonia. Adv. Mater. 2023 doi: 10.1002/adma.202310532. [DOI] [PubMed] [Google Scholar]
- 34.FDA,. Drugs@FDA: FDA-Approved Drugs. Available from:: https://www.accessdata.fda.gov/scripts/cder/daf/.
- 35.Wang L., et al. ROS-sensitive Crocin-loaded chitosan microspheres for lung targeting and attenuation of radiation-induced lung injury. Carbohydr. Polym. 2023;307:120628. doi: 10.1016/j.carbpol.2023.120628. [DOI] [PubMed] [Google Scholar]
- 36.Dinarello C.A. Anti-inflammatory agents: present and future. Cell. 2010;140(6):935–950. doi: 10.1016/j.cell.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C.Y., Gao J., Wang Z. Bioresponsive nanoparticles targeted to infectious microenvironments for sepsis management. Adv. Mater. 2018;30(43) doi: 10.1002/adma.201803618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo J., et al. Chitosan microsphere used as an effective system to deliver a linked antigenic peptides vaccine protect mice against acute and chronic toxoplasmosis. Front. Cell. Infect. Microbiol. 2018;8:163. doi: 10.3389/fcimb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.CLSI . 31st ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2021. Performance Standards for Antimicrobial Susceptibility Testing, M100. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyers M., Wright G.D. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019;17(3):141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 41.Guo J., et al. Chitosan microsphere used as an effective system to deliver a linked antigenic peptides vaccine protect mice against acute and chronic toxoplasmosis. Front. Cell. Infect. Microbiol. 2018;8:163. doi: 10.3389/fcimb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi S., et al. Matrix metalloproteinase 3 as a valuable marker for patients with COVID-19. J. Med. Virol. 2021;93(1):528–532. doi: 10.1002/jmv.26235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jyoti K., et al. Inhalable bioresponsive chitosan microspheres of doxorubicin and soluble curcumin augmented drug delivery in lung cancer cells. Int. J. Biol. Macromol. 2017;98:50–58. doi: 10.1016/j.ijbiomac.2017.01.109. [DOI] [PubMed] [Google Scholar]
- 44.Zhou T., et al. Hypoxia and matrix metalloproteinase 13-responsive hydrogel microspheres alleviate osteoarthritis progression in vivo. Small. 2023 doi: 10.1002/smll.202308599. [DOI] [PubMed] [Google Scholar]
- 45.Lin X., et al. Lung-targeting lysostaphin Microspheres for methicillin-resistant Staphylococcus aureus pneumonia Treatment and prevention. ACS Nano. 2021;15(10):16625–16641. doi: 10.1021/acsnano.1c06460. [DOI] [PubMed] [Google Scholar]
- 46.Agnoletti M., et al. Monosized polymeric microspheres designed for passive lung targeting: biodistribution and pharmacokinetics after intravenous administration. ACS Nano. 2020;14(6):6693–6706. doi: 10.1021/acsnano.9b09773. [DOI] [PubMed] [Google Scholar]
- 47.Loira-Pastoriza C., Todoroff J., Vanbever R. Delivery strategies for sustained drug release in the lungs. Adv. Drug Deliv. Rev. 2014;75:81–91. doi: 10.1016/j.addr.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 48.Aghbashlo M., et al. Tuning chitosan's chemical structure for enhanced biological functions. Trends Biotechnol. 2023;41(6):785–797. doi: 10.1016/j.tibtech.2022.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Bezerra M.C., et al. Microstructure and properties of thermomechanically processed chitosan citrate-based materials. Carbohydr. Polym. 2022;278:118984. doi: 10.1016/j.carbpol.2021.118984. [DOI] [PubMed] [Google Scholar]
- 50.Cerón A.A., et al. Synthesis of chitosan-lysozyme microspheres, physicochemical characterization, enzymatic and antimicrobial activity. Int. J. Biol. Macromol. 2021;185:572–581. doi: 10.1016/j.ijbiomac.2021.06.178. [DOI] [PubMed] [Google Scholar]
- 51.Yu S., et al. Facile fabrication of functional chitosan microspheres and study on their effective cationic/anionic dyes removal from aqueous solution. Int. J. Biol. Macromol. 2019;134:830–837. doi: 10.1016/j.ijbiomac.2019.04.208. [DOI] [PubMed] [Google Scholar]
- 52.Yang H., et al. Extracellular matrix-mimicking nanofibrous chitosan microspheres as cell micro-ark for tissue engineering. Carbohydr. Polym. 2022;292:119693. doi: 10.1016/j.carbpol.2022.119693. [DOI] [PubMed] [Google Scholar]
- 53.Mahdavinia G.R., Etemadi H., Soleymani F. Magnetic/pH-responsive beads based on caboxymethyl chitosan and κ-carrageenan and controlled drug release. Carbohydr. Polym. 2015;128:112–121. doi: 10.1016/j.carbpol.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 54.Kean T., Thanou M. Biodegradation, biodistribution and toxicity of chitosan. Adv. Drug Deliv. Rev. 2010;62(1):3–11. doi: 10.1016/j.addr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Sopade P.A. Modelling multiphasic starch digestograms with multiterm exponential and non-exponential equations. Carbohydr. Polym. 2022;275:118698. doi: 10.1016/j.carbpol.2021.118698. [DOI] [PubMed] [Google Scholar]
- 56.Gao G.F., et al. Predicting drug release and degradation kinetics of long-acting microsphere formulations of tacrolimus for subcutaneous injection. J. Contr. Release. 2021;329:372–384. doi: 10.1016/j.jconrel.2020.11.055. [DOI] [PubMed] [Google Scholar]
- 57.Martín-Camacho U.J., et al. Drug delivery system of green synthesized Ti-Cu nanocomposite. Mater. Lett. 2022;321:132437. [Google Scholar]
- 58.Kolate A., et al. Polymer assisted entrapment of netilmicin in PLGA nanoparticles for sustained antibacterial activity. J. Microencapsul. 2015;32(1):61–74. doi: 10.3109/02652048.2014.944951. [DOI] [PubMed] [Google Scholar]
- 59.Jorgensen J.H., Ferraro M.J. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 2009;49(11):1749–1755. doi: 10.1086/647952. an Official Publication of the Infectious Diseases Society of America. [DOI] [PubMed] [Google Scholar]
- 60.Song H., Li Y., Wang Y. Two-component system GacS/GacA, a global response regulator of bacterial physiological behaviors. Engineering Microbiology. 2023;3(1):100051. [Google Scholar]
- 61.Rome K., et al. The two-component system ZraPSR Is a novel ESR that Contributes to intrinsic antibiotic Tolerance in Escherichia coli. J. Mol. Biol. 2018;430(24):4971–4985. doi: 10.1016/j.jmb.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 62.Appia-Ayme C., et al. ZraP is a periplasmic molecular chaperone and a repressor of the zinc-responsive two-component regulator ZraSR. Biochem. J. 2012;442(1):85–93. doi: 10.1042/BJ20111639. [DOI] [PubMed] [Google Scholar]
- 63.Hickey M.J. MPO and neutrophils: a magnetic attraction. Blood. 2011;117(4):1103–1104. doi: 10.1182/blood-2010-11-317479. [DOI] [PubMed] [Google Scholar]
- 64.Locati M., Curtale G., Mantovani A. Diversity, mechanisms, and Significance of macrophage plasticity. Annual Review of Pathology. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu P., et al. Chitooligosaccharides alleviate hepatic fibrosis by regulating the polarization of M1 and M2 macrophages. Food Funct. 2022;13(2):753–768. doi: 10.1039/d1fo03768d. [DOI] [PubMed] [Google Scholar]
- 66.Panahi, H Kazemi Shariat, et al. Current and emerging applications of saccharide-modified chitosan: a critical review. Biotechnol. Adv. 2023;66:108172. doi: 10.1016/j.biotechadv.2023.108172. [DOI] [PubMed] [Google Scholar]
- 67.Morones-Ramirez J.R., et al. Silver enhances antibiotic activity against gram-negative bacteria. Sci. Transl. Med. 2013;5(190):190ra81. doi: 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are presented in the main text and the supplementary information. Additional data are available from the corresponding author upon reasonable request.