Abstract
Candida albicans, a prominent opportunistic pathogenic fungus in the human population, possesses the capacity to induce life-threatening invasive candidiasis in individuals with compromised immune systems despite the existence of antifungal medications. When faced with macrophages or neutrophils, C. albicans demonstrates its capability to endure oxidative stress through the utilization of antioxidant enzymes. Therefore, the enhancement of oxidative stress in innate immune cells against C. albicans presents a promising therapeutic approach for the treatment of invasive candidiasis. In this study, we conducted a comprehensive analysis of a library of drugs approved by the Food and Drug Administration (FDA). We discovered that halofantrine hydrochloride (HAL) can augment the antifungal properties of oxidative damage agents (plumbagin, menadione, and H2O2) by suppressing the response of C. albicans to reactive oxygen species (ROS). Furthermore, our investigation revealed that the inhibitory mechanism of HAL on the oxidative response is dependent on Cap1. In addition, the antifungal activity of HAL has been observed in the Galleria mellonella infection model. These findings provide evidence that targeting the oxidative stress response of C. albicans and augmenting the fungicidal capacity of oxidative damage agents hold promise as effective antifungal strategies.
Keywords: halofantrine hydrochloride, oxidative response inhibitor, oxidative damage agents, Candida albicans
1. Introduction
Candida albicans, a significant opportunistic pathogenic fungus in humans, has the potential to cause fatal invasive candidiasis in immunocompromised patients [1,2], despite the availability of antifungal drugs such as polyenes, azoles, and echinocandins [3]. The primary defense mechanism against C. albicans involves phagocytosis mediated by innate immune cells, including macrophages and neutrophils [4]. Upon engulfment of C. albicans, innate immune cells employ a major antifungal strategy by generating toxic reactive oxygen species (ROS), resulting in oxidative damage to C. albicans [5,6]. Indeed, ROS have the capability to initiate the oxidation of proteins, lipids, and nucleic acids, leading to impaired functionality of these vital biological macromolecules, ultimately triggering programmed cell death in the fungal pathogen [7]. C. albicans exhibits the ability to withstand oxidative stress by means of antioxidant enzymes, namely catalase, glutathione peroxidase, and superoxide dismutase when confronted with macrophages or neutrophils [5]. In individuals with profound innate immunodeficiency, such as those suffering from cancer, organ transplantation, and acquired immune deficiency syndrome (AIDS), C. albicans can persist in the bloodstream and establish colonies within internal organs, leading to the development of invasive candidiasis [8]. Thus, augmenting oxidative stress in innate immune cells against C. albicans represents a potential therapeutic strategy for managing invasive candidiasis.
Augmenting intracellular ROS levels in C. albicans represents a viable fungicidal approach akin to using amphotericin B and miconazole, which results in cell death [9,10,11]. Our prior investigations have demonstrated that increasing ROS levels in C. albicans using baicalein, berberine, and osthole enhances the antifungal efficacy of antifungal agents [12,13,14]. However, the generation of ROS, which is cytotoxic to C. albicans, is contingent upon dysfunctions in the tricarboxylic acid (TCA) cycle and the respiratory chain and may also prove toxic to mammalian cells. An alternative antifungal strategy involves weakening C. albicans’ resistance to oxidative damage. Cationic stress, resulting from an increased influx of K+ into the phagosome of innate immune cells [15], has been shown to inhibit C. albicans’ response to oxidative stress and render it highly susceptible [16,17]. The regulation of C. albicans’ response to ROS primarily depends on the transcription factors Cap1 and Hog1 [5]. The deletion of CAP1 and HOG1 genes in C. albicans rendered the organism susceptible to oxidative stress [18] and reduced its resistance to host cell-mediated killing, leading to diminished virulence in Caenorhabditis elegans, Galleria mellonella, and mouse infection models [17,19,20]. Consequently, targeting the response of C. albicans to ROS and augmenting the fungicidal efficacy of innate immune cells represents a promising therapeutic approach. Nevertheless, the number of compounds that function as antifungal agents through this mechanism remains limited.
Here, we performed a high throughput screening of a Food and Drug Administration (FDA)-approved drug library and identified that halofantrine hydrochloride (HAL) could enhance the antifungal activities of oxidative damage agents (plumbagin, menadione, and H2O2) by suppressing C. albicans’ response to ROS. We further found that the mechanism of inhibiting the oxidative response action of HAL depends on Cap1. In addition, the antifungal activity of HAL has been observed in the G. mellonella infection model. These findings demonstrate that inhibiting C. albicans’ response to oxidative stress and enhancing the fungicidal ability of innate immunity cells is a promising antifungal strategy.
2. Materials and Methods
2.1. Strains, Primers, Agents, and Cultural Conditions
The present study utilized C. albicans’ strains, which are documented in Table 1, and primers, which are documented in Table 2. C. albicans’ strains were cultured in a YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) at 30 °C. Deletion and green fluorescence protein (GFP)-tagged mutants were constructed using a synthetic medium (0.67% yeast nitrogen base without amino acids, 2% dextrose, 2% agar). The FDA-approved drug library (MedChemExpress, Shanghai, China) consisted of 2372 drugs that were dissolved in dimethylsulfoxide (DMSO) at 10 mM. Plumbagin (Topscience, Shanghai, China) and menadione (Topscience, Shanghai, China) were dissolved in DMSO to prepare the stock solutions at 6.4 mg/mL and 102.4 mM, respectively.
Table 1.
Strains used in this study.
| Gene Accession No. | Strain | Genotype | Source or Reference |
|---|---|---|---|
| SC5314 | Wild type | [21] | |
| SN152 | his1/his1 arg4/arg4 leu2/leu2 | [21] | |
| 3636640 | cap1Δ/cap1Δ | cap1::HIS1/cap1::ARG4 leu2/leu2 | This study |
| 3637393 | hog1Δ/hog1Δ | hog1::HIS1/hog1::ARG4 leu2/leu2 | This study |
| 3645633 | rad53Δ/rad53Δ | rad53::HIS1/rad53::ARG4 leu2/leu2 | This study |
| 3636187 | ybp1Δ/ybp1Δ | ybp1::HIS1/ybp1::ARG4 leu2/leu2 | This study |
| 3644471 | gpx3Δ/gpx3Δ | gpx3::HIS1/gpx3::ARG4 leu2/leu2 | This study |
| 3636640 | CAP1-GFP | his1/his1 arg4/arg4 CAP1/cap1::CAP1-GFP-LEU2 | This study |
Table 2.
Primers used in this study.
| Gene Accession No. | Gene Name | Primer Name a | Primer Sequence (5′ to 3′) b |
|---|---|---|---|
| Primers for genes deletion | |||
| 3636640 | CAP1 | CAP1 P1 | GATTACTAATTATTCTTTGAC |
| CAP1 P3 | cacggcgcgcctagcagcggGAATAAGGATAGTTGAAAATG | ||
| CAP1 P4 | gtcagcggccgcatccctgcCATTAATCAAGTTAGTGGTGG | ||
| CAP1 P6 | CTAGTTGAATCAAAGAAAGCC | ||
| 3637393 | HOG1 | HOG1 P1 | GCCTTGCTTATGTTCACAAAC |
| HOG1 P3 | cacggcgcgcctagcagcggCATTTTCTTATATGCTTTATC | ||
| HOG1 P4 | gtcagcggccgcatccctgcTCTTCAAAAATACAAGCTAGC | ||
| HOG1 P6 | TCGTAAGGACGGTATTACAGC | ||
| 3645633 | RAD53 | RAD53 P1 | CTAGTTTTCATCTTGATCTTG |
| RAD53 P3 | cacggcgcgcctagcagcggTGTAGTTTGGTAAATTAAGGG | ||
| RAD53 P4 | gtcagcggccgcatccctgcATTTAGCATATATACAAGCAT | ||
| RAD53 P6 | GAACGGAGATGGCAACATGTG | ||
| 3636187 | YBP1 | YBP1 P1 | GCTAGTTTATCCCCTCTTATG |
| YBP1 P3 | cacggcgcgcctagcagcggAAATTGAAATGGCTCAATGGT | ||
| YBP1 P4 | gtcagcggccgcatccctgcTGTATATGTATGTAACTACGT | ||
| YBP1 P6 | CTTCACCATTACCATCTCATC | ||
| 3644471 | GPX3 | GPX3 P1 | GCTGTCAAACCATTGGAGCTC |
| GPX3 P3 | cacggcgcgcctagcagcggTGATGATTGTTGATAATTGTA | ||
| GPX3 P4 | gtcagcggccgcatccctgcAAATACAGTAGTATTATACAT | ||
| GPX3 P6 | CGGGCAGGTCAATGCCAAACC | ||
| Universal primer 2 | ccgctgctaggcgcgccgtgACCAGTGTGATGGATATCTGC | ||
| Universal primer 5 | gcagggatgcggccgctgacAGCTCGGATCCACTAGTAACG | ||
| Primers for diagnosing the genes null mutants | |||
| 3636640 | CAP1 | CAP1 Ucheck | CTGGCTGGCTTATACTCTAAC |
| CAP1 Dcheck | ATCTGGATCGATCTCTGCAAG | ||
| 3637393 | HOG1 | HOG1 Ucheck | AGGTAGTGTTGGTGTTATCAC |
| HOG1 Dcheck | GAAGCATTTGGATAAATTGGG | ||
| 3645633 | RAD53 | RAD53 Ucheck | CGAAATACGATACGTTAGACG |
| RAD53 Dcheck | TTGGACATTGAGCATGTTCGG | ||
| 3636187 | YBP1 | YBP1 Ucheck | GGTATTTTGGTTGGGATTGGG |
| YBP1 Dcheck | TGAATGTTCTTAAACTTGCCG | ||
| 3644471 | GPX3 | GPX3 Ucheck | TGTGTCATGTCACGTGATAAC |
| GPX3 Dcheck | CATAGCCATCAATCTCTTGGT | ||
| 8048008 | HIS1 | HIS1 Left | ATTAGATACGTTGGTGGTTC |
| HIS1 Right | AACACAACTGCACAATCTGG | ||
| 8049225 | ARG4 | ARG4 Left | ACACAGAGATACCTTGTACT |
| ARG4 Right | ACGGAGTACCACATACGATG | ||
| Primers for GFP-tagged C-terminals of Cap1 | |||
| 3636640 | CAP1 | Cap1gfp-F1 | GCTGATGTGAATCAATTACTAGAGCGAAGTATAAAACATCCCCAGGTCGACTCTAGATC |
| Cap1gfp-R1 | GAAATACCGTAAAATAAATTAAACCCACCACTAACTTGATTCTTTCCTGCGTTATCCTG | ||
| Cap1gfp-F2 | AAAGCTAAATGTTCTGAAAAGGGAGTAGTGATAAATACTGCTGATGTGAATCAATTACTA | ||
| Cap1gfp-R2 | ATATAAATACAAAAAAATAAAGCCAAATAGATGTCAATTGAAATACCGTAAAATAAATTA | ||
| Cap1check-F | GAAGTTGTGCCGGCACCTCC | ||
| Cap1check-R | AGATGATGTTGATTATGGTG | ||
| VP8 | GAATAATTCTTCACCTTTAGAGATGGT | ||
| VP19 | TGCAGATATCCATCACACTGG | ||
| Primers for qRT-PCR | |||
| 3636640 | CAP1 | CAP1-rtF | TGGGTTCATCTTCATCGT |
| CAP1-rtR | TTGGGCACTGGGTTACTT | ||
| 3639495 | CAT1 | CAT1-rtF | AAGAGTTGTCCACGCTAA |
| CAT1-rtR | GAACCTAATTCACCACCA | ||
| 3639313 | TTR1 | TTR1-rtF | ATTGCCTCCAAATCCTAT |
| TTR1-rtR | TGTTGACCACCAATAAAG | ||
| 3636195 | ACT1 | ACT1-rtF | TTGATTTGGCTGGTAGAG |
| ACT1-rtR | ATGGCAGAAGATTGAGAA | ||
Annotation: a Ucheck, upstream check primer for target genes; Dcheck, downstream check primer for target genes; Left, upstream check primer for the HIS1 and ARG4 genes; Right, downstream check primer for the HIS1 and ARG4 genes; rtF, forward primer for qRT-PCR; rtR, reverse primer for qRT-PCR. b Lowercase sequences correspond to exogenous, complementary sequences that were added to primers 2, 3, 4, and 5 to facilitate mutually primed synthesis during the second round of fusion PCR.
2.2. High-Throughput Screening
To identify drugs that potentiate oxidative stress damage to C. albicans, a logarithmic long-term culture of C. albicans was diluted to a concentration of 1 × 103 cells/mL in a YPD medium containing 1 μg/mL plumbagin. Subsequently, 1 μL of each FDA-approved drug was added to 199 μL of the cell suspension, resulting in a concentration of 50 μM in each well of a 96-well plate, which was then incubated at 30 °C for 48 h. The criterion for drug inclusion was a growth reduction of >50%, as determined by OD600 values, in a combination of an FDA-approved drug and plumbagin compared to plumbagin monotherapy.
2.3. Minimum Inhibition Concentration (MIC) Assay
The MIC assay was conducted in accordance with the guidelines outlined in the Clinical and Laboratory Standards Institute (CLSI) M27-A3. YPD medium was utilized, with strains inoculated at a concentration of approximately 1 × 103 cells/mL and 100 μL per well in 96-well plates. Serial dilutions of compounds ranging from 50 μM to 0.1 μM were added to each well. These plates were then incubated at 30 °C for 24 h. Following incubation, the optical densities were measured at an absorbance of 600 nm, using a Multiskan Sky (Thermo Fisher Scientific, Waltham, MA, USA). The MIC was determined as the concentration of the compound that suppressed 50% or more of cellular growth, as evidenced by the OD600 measurement, in comparison to the control.
2.4. Dose–Matrix Titration Assay
The dose–matrix titration assay, as outlined in the reference, was employed to evaluate the synergistic effects of drugs [22]. In summary, drug A was diluted in a two-fold serial manner across columns of a 96-well plate, with each well containing 50 μL of drug A at a concentration four times higher than the final drug concentration. Similarly, drug B was dispensed in a two-fold serial dilution manner across rows of the plate, with each well containing 50 μL of drug B at a concentration four times higher than the final drug concentration. Following this, a volume of 50 μL of the diluted drug B was subsequently transferred to the plate containing drug A. Subsequently, a volume of 100 μL of overnight diluted C. albicans cultures (1 × 103 cells/mL) was added to all wells containing the drugs and incubated at 30 °C for 24 h. The synergy between drugs A and B was assessed using the Loewe additivity model, employing the fractional inhibitory concentration index (FICI). Synergism is indicated by an FICI value of ≤0.5, additive effects by an FICI value of 0.5 < FICI ≤ 1, indifference by an FICI value of 1 < FICI ≤ 4, and antagonism by an FICI value greater than 4 [23].
2.5. Disruption of Target Genes
Fusion PCR methodology was utilized to generate null mutants of genes of interest from the C. albicans’ strain SN152, as previously described in reference [21]. In brief, the flanking sequences of the target genes were amplified using primers P1 and P3 or P4 and P6, with the genomic DNA of SN152 serving as the template. The selectable HIS1 or ARG4 marker was then amplified using universal primers 2 and 5, with plasmids pSN52 or pSN69 as the template. Subsequently, a fusion product was generated using primers P1 and P6, with three PCR products serving as the template. The transformation was selected on synthetic media supplemented with the necessary auxotrophic supplements.
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
The methodology for total RNA extraction and qRT-PCR was conducted according to the procedures described in a previous study [24]. To ensure the absence of genomic DNA contamination, the isolated RNA was subjected to DNase I treatment (Takara, Beijing, China). First-strand cDNAs were synthesized using a reverse transcription PCR cDNA synthesis kit (Takara, China). Triplicate independent qRT-PCR analyses were performed using the Roche Lightcycler 96 Fluorescence Quantitative PCR Instrument and TB Green Premix Ex TaqTM II (Takara, China). The ACT1 gene served as the internal control.
2.7. C-Terminal of Proteins Tagging GFP
A PCR-based approach was employed to amplify the desired DNA cassettes within plasmid pCPC64 for tagging the C-terminal of Cap1 with GFP [25]. The generation of DNA cassettes with 78 bp homology regions to the CAP1 gene was achieved through two rounds of PCR utilizing primers F1 plus R1 and F2 plus R2, respectively. The resulting product was subsequently transformed into C. albicans cells to generate a mutant strain with the C-terminal of Cap1 tagged with GFP.
2.8. Western Blot Analysis
C. albicans’ cells were washed with sterile water once and then were subjected to cell lysis using the Bead Ruptor12 System (OMNI International, Kennesaw, GA, USA) in lysis buffer (PBS containing 5 mM EDTA (pH 8.0), 1 mM PMSF, 1.0% Protease Inhibitor Cocktail). Proteins were separated by 4–20% SDS–PAGE and transferred to a PVDF membrane. After blocking, anti-GFP antibodies (Santa Cruz, Dallas, TX, USA) or anti-tubulin antibodies (Abbkine, Atlanta, GA, USA) were used for probing GFP-tagged proteins or the tubulin, which were then detected using the secondary antibody goat anti-mouse IgG-HRP (Santa Cruz) and the Pierce™ ECL system (Thermo Fisher Scientific, Waltham, MA, USA) [26].
2.9. G. mellonella Infection Model
G. mellonella larvae, obtained from the Tianjin Huiyude Biotech Company, were selected based on an average weight of 300 mg and randomly assigned to four groups (n = 10 per group), with any larvae displaying signs of melanization being excluded. The larvae were infected with 5 μL of an SN152 suspension (7.0 × 105 cells/larvae) using a Hamilton syringe and subsequently treated with a single injection of HAL (0, 0.5, 1, 2 mg/kg). All G. mellonella larvae were incubated at 30 °C for eight days. The mortality of G. mellonella was evaluated daily and subjected to statistical analysis using the Kaplan–Meier method, specifically employing the log-rank test [22,27].
3. Results
3.1. HAL Enhances the Antifungal Activities of Oxidative Damage Agents
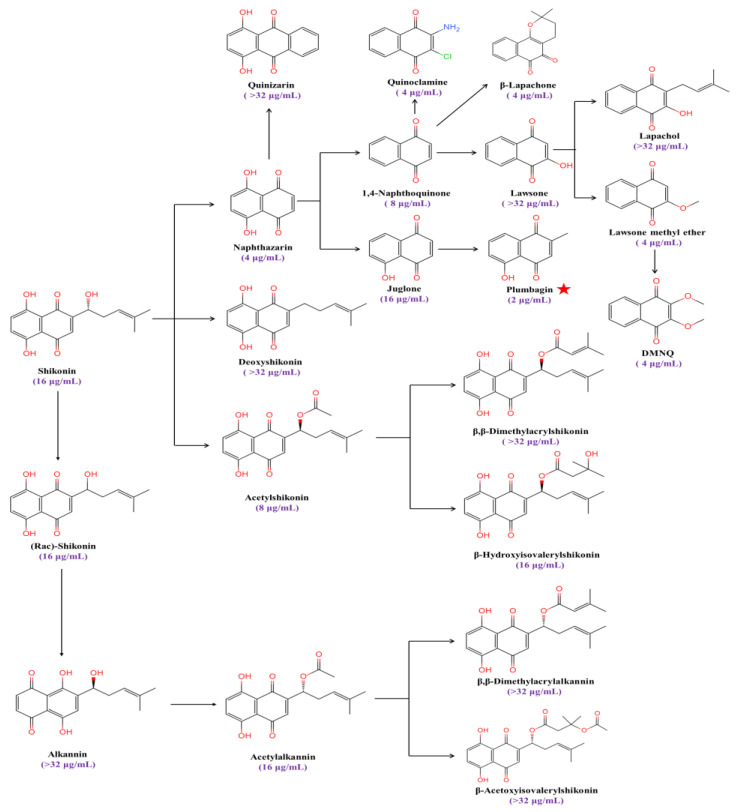
Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) is a strong inducer of ROS and causes oxidative stress in fungi [28,29]. We found that the MIC value of plumbagin against C. albicans’ SC5314 is 2 μg/mL, with the strongest antifungal activity among a series of anthraquinone analogs (Figure 1).
Figure 1.
The MIC values of anthraquinone analogs against C. albicans. The red star symbolizes the compound exhibiting the lowest MIC value against C. albicans.
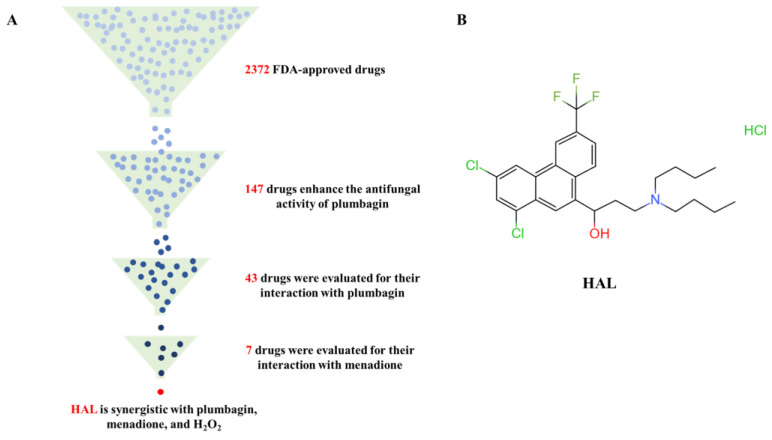
In this study, we performed a high-throughput screening of the FDA-approved drug library containing 2372 drugs to determine candidate agents that are synergistic with the ROS inducer plumbagin. Previous studies demonstrated that loss of Cap1 and Hog1 increased the susceptibility of C. albicans to oxidizing agents [10,30,31] but had no function in the growth of C. albicans [32]. Therefore, we utilize three criteria to screen candidate inhibitors of the oxidative stress response of C. albicans: a candidate inhibitor should (1) have weak antifungal activity but (2) enhance the antifungal activity of ROS inducers, and (3) ROS inducers can also enhance its antifungal activity. We tested the antifungal activity of 50 μM of each of the 2372 drugs on C. albicans’ SN152, grown in YPD medium with 1 μg/mL plumbagin in 96-well plates. We found that 147 compounds enhanced the antifungal activity of plumbagin, and the relative growth of C. albicans treated with the combination of candidate compounds and plumbagin is less than 50% of that of plumbagin alone after incubation at 30 °C for 24 h (Figure 2A, Table 3). We further investigated the ability of these compounds to enhance the antifungal activity of plumbagin (1 μg/mL), using the MIC assay by diluting these candidate compounds from 50 μM to 0.1 μM in a 2-fold ratio. We excluded 30 drugs for subsequent study as these drugs have potent antifungal activities (MIC values are less than 0.1 μM). We precluded 10 drugs for which plumbagin antagonized their antifungal activity, and 64 for which plumbagin slightly enhanced their antifungal activities (the ratio of MIC alone to the MIC in the presence of plumbagin is equal or less than 2) (Figure 2A, Table 3). Subsequently, we used a dose–matrix titration assay to determine the interaction between the remaining 43 drugs and plumbagin by the FICI. We included seven drugs as candidate oxidative response inhibitors as they synergize with plumbagin. Among them, the combination of HAL and plumbagin had the lowest FICI value (FICI = 0.0938) (Figure 2B, Table 4). We further tested the interaction of each of the seven drugs and menadione (2-methyl-1,4-naphthoquinone), another ROS inducer [29]. Only HAL was synergistic with menadione, with an FICI value of 0.1563 (Figure 2A, Table 5). We then gauged the combination of HAL and H2O2; as expected, they are synergistic (FICI = 0.2526) (Table 6). These findings suggest that HAL is a promising synergistic agent for ROS inducers.
Figure 2.
(A) Identification of HAL for enhancing the antifungal activity of ROS inducers from an FDA-approved compound library. (B) HAL chemical structure.
Table 3.
Compounds enhance the antifungal activity of plumbagin.
| No. | Drugs | MIC (μM) | Fold Change of MIC (MICalone/MICcombined) | |
|---|---|---|---|---|
| Alone | Combination with Plumbagin | |||
| 1 | Almonertinib hydrochloride | >50 | 0.78 | 64 |
| 2 | Bosutinib | >50 | 12.5 | 4 |
| 3 | Ceritinib dihydrochloride | 50 | 6.25 | 8 |
| 4 | Cetylpyridinium chloride | 6.25 | 1.56 | 4 |
| 5 | Cinacalcet | >50 | 12.5 | 4 |
| 6 | Clomiphene citrate | 25 | 6.25 | 4 |
| 7 | Dacomitinib | >50 | 12.5 | 4 |
| 8 | Halofantrine hydrochloride | >50 | 6.25 | 8 |
| 9 | Ilaprazole | >50 | 6.25 | 8 |
| 10 | Nilotinib monohydrochloride monohydrate | >50 | 12.5 | 4 |
| 11 | Pimavanserin tartrate | 50 | 12.5 | 4 |
| 12 | Tafenoquine Succinate | 25 | 3.13 | 8 |
| 13 | Triflupromazine hydrochloride | >50 | 6.25 | 8 |
| 14 | Vilanterol trifenatate | >50 | 1.56 | 32 |
| 15 | Vortioxetine | 25 | 6.25 | 4 |
| 16 | Vortioxetine hydrobromide | 50 | 12.5 | 4 |
| 17 | Alectinib | >50 | 0.78 | 64 |
| 18 | Amiodarone hydrochloride | >50 | 12.5 | 4 |
| 19 | Amphotericin B | 0.78 | <0.1 | 8 |
| 20 | Benzethonium chloride | 12.5 | 3.13 | 4 |
| 21 | Bleomycin hydrochloride | 50 | 6.25 | 8 |
| 22 | Ceritinib | 50 | 6.25 | 8 |
| 23 | Chlorhexidine | 25 | 3.13 | 8 |
| 24 | Clioquinol | 25 | 6.25 | 4 |
| 25 | Disulfiram | 12.5 | 3.13 | 4 |
| 26 | Domiphen bromide | 25 | 3.13 | 8 |
| 27 | Ebastine | 25 | 6.25 | 4 |
| 28 | Fingolimod | 12.5 | 1.56 | 8 |
| 29 | Ibudilast | >50 | 12.5 | 4 |
| 30 | Magnolol | 50 | 6.25 | 8 |
| 31 | Menadione | 50 | 12.5 | 4 |
| 32 | Olmutinib | >50 | 6.25 | 8 |
| 33 | Pinaverium bromide | 50 | 3.13 | 16 |
| 34 | Ponatinib | >50 | 6.25 | 8 |
| 35 | Rolapitant | 50 | 12.5 | 4 |
| 36 | Sertindole | >50 | 12.5 | 4 |
| 37 | Sonidegib | >50 | 0.78 | 64 |
| 38 | Sultiame | 50 | 6.25 | 8 |
| 39 | Tegaserod maleate | 50 | 12.5 | 4 |
| 40 | Telotristat ethyl | 12.5 | 3.13 | 4 |
| 41 | Telotristat etiprate | 25 | 3.13 | 8 |
| 42 | Thonzonium bromide | 12.5 | 0.78 | 16 |
| 43 | Triclosan | 12.5 | 1.56 | 8 |
| 44 | Butoconazole nitrate | <0.1 | NA | NA |
| 45 | Cinacalcet hydrochloride | <0.1 | NA | NA |
| 46 | Clotrimazole | <0.1 | NA | NA |
| 47 | Econazole nitrate | <0.1 | NA | NA |
| 48 | Efinaconazole | <0.1 | NA | NA |
| 49 | Everolimus | <0.1 | NA | NA |
| 50 | Fenticonazole Nitrate | <0.1 | NA | NA |
| 51 | Isavuconazole | <0.1 | NA | NA |
| 52 | Isoconazole nitrate | <0.1 | NA | NA |
| 53 | Itraconazole | <0.1 | NA | NA |
| 54 | Ketoconazole | <0.1 | NA | NA |
| 55 | Luliconazole | <0.1 | NA | NA |
| 56 | Micafungin sodium | <0.1 | NA | NA |
| 57 | Miconazole nitrate | <0.1 | NA | NA |
| 58 | Neticonazole hydrochloride | <0.1 | NA | NA |
| 59 | Oxiconazole nitrate | <0.1 | NA | NA |
| 60 | Posaconazole | <0.1 | NA | NA |
| 61 | Rapamycin | <0.1 | NA | NA |
| 62 | Sertaconazole nitrate | <0.1 | NA | NA |
| 63 | Sulconazole mononitrate | <0.1 | NA | NA |
| 64 | Temsirolimus | <0.1 | NA | NA |
| 65 | (+)-Ketoconazole | <0.1 | NA | NA |
| 66 | Amorolfine hydrochloride | <0.1 | NA | NA |
| 67 | Dasatinib | <0.1 | NA | NA |
| 68 | Econazole | <0.1 | NA | NA |
| 69 | Isavuconazonium sulfate | <0.1 | NA | NA |
| 70 | Lapatinib | <0.1 | NA | NA |
| 71 | Neticonazole | <0.1 | NA | NA |
| 72 | Tioconazole | <0.1 | NA | NA |
| 73 | Voriconazole | <0.1 | NA | NA |
| 74 | Atracurium besylate | 25 | 50 | 0.50 |
| 75 | Broxyquinoline | 1.56 | 3.13 | 0.50 |
| 76 | Cetrorelix Acetate | 6.25 | 50 | 0.13 |
| 77 | Revefenacin | 25 | 50 | 0.50 |
| 78 | Adapalene | 25 | 50 | 0.50 |
| 79 | Bifonazole | 3.13 | 6.25 | 0.50 |
| 80 | Dimetridazole | 12.5 | 25 | 0.50 |
| 81 | Dioscin | 0.2 | 3.13 | 0.06 |
| 82 | Terbinafine | 1.56 | 3.13 | 0.50 |
| 83 | Terbinafine hydrochloride | 1.56 | 3.13 | 0.50 |
| 84 | 10-Undecenoic acid | >50 | 50 | 1 |
| 85 | 10-Undecenoic acid zinc salt | >50 | 50 | 1 |
| 86 | Aclacinomycin A hydrochloride | 25 | 12.5 | 2 |
| 87 | Atorvastatin | 12.5 | 12.5 | 1 |
| 88 | Aviptadil acetate | >50 | >50 | 1 |
| 89 | Bleomycin sulfate | 25 | 12.5 | 2 |
| 90 | Blonanserin | >50 | 50 | 1 |
| 91 | Chlorprothixene | >50 | 25 | 2 |
| 92 | Chlorquinaldol | 3.13 | 1.56 | 2 |
| 93 | Ciclopirox | 50 | 50 | 1 |
| 94 | Dronedarone hydrochloride | 25 | 12.5 | 2 |
| 95 | Fluconazole | 6.25 | 6.25 | 1 |
| 96 | Fluvastatin sodium | 3.13 | 3.13 | 1 |
| 97 | Fosravuconazole lysine ethanolate | 12.5 | 12.5 | 1 |
| 98 | Josamycin | >50 | 50 | 1 |
| 99 | Lonafarnib | >50 | >50 | 1 |
| 100 | L-Thyroxine sodium | >50 | 50 | 1 |
| 101 | Miltefosine | 50 | 50 | 1 |
| 102 | Mycophenolic acid | 3.13 | 3.13 | 1 |
| 103 | Natamycin | 25 | 25 | 1 |
| 104 | Nintedanib esylate | >50 | 50 | 1 |
| 105 | Nitroxoline | 6.25 | 6.25 | 1 |
| 106 | Otilonium bromide | 3.13 | 1.56 | 2 |
| 107 | Penfluridol | >50 | 25 | 2 |
| 108 | Piroctone olamine | 50 | 50 | 1 |
| 109 | Pitavastatin Calcium | 1.56 | 0.78 | 2 |
| 110 | Pramocaine hydrochloride | 50 | 50 | 1 |
| 111 | Tamoxifen Citrate | 50 | 25 | 2 |
| 112 | Teprenone | 50 | 50 | 1 |
| 113 | Vilazodone | >50 | 25 | 2 |
| 114 | Visomitin | 25 | 12.5 | 2 |
| 115 | Abiraterone | >50 | 50 | 1 |
| 116 | Armillarisin A | >50 | >50 | 1 |
| 117 | Atorvastatin hemicalcium salt | 12.5 | 6.25 | 2 |
| 118 | Auranofin | >50 | 50 | 1 |
| 119 | Bepridil hydrochloride hydrate | 50 | 50 | 1 |
| 120 | Bithionol | 50 | 50 | 1 |
| 121 | Boceprevir | 50 | 50 | 1 |
| 122 | Bromperidol | 25 | 25 | 1 |
| 123 | Calcium lactate | 25 | 25 | 1 |
| 124 | Carmofur | >50 | 50 | 1 |
| 125 | Cerivastatin sodium | 1.56 | 1.56 | 1 |
| 126 | Cetylpyridinium chloride monohydrate | 6.25 | 3.13 | 2 |
| 127 | Chloroxine | 3.13 | 3.13 | 1 |
| 128 | Ciclopirox olamine | >50 | 50 | 1 |
| 129 | Degarelix | >50 | 50 | 1 |
| 130 | Dichlorisone acetate | >50 | 50 | 1 |
| 131 | Dronedarone | 25 | 12.5 | 2 |
| 132 | Eltrombopag | >50 | 50 | 1 |
| 133 | Fingolimod hydrochloride | 6.25 | 3.13 | 2 |
| 134 | Flucytosine | >50 | 25 | 2 |
| 135 | Fluspirilene | >50 | 50 | 1 |
| 136 | Hexylresorcinol | 50 | 50 | 1 |
| 137 | Levamlodipine besylate | >50 | >50 | 1 |
| 138 | Nintedanib | >50 | 50 | 1 |
| 139 | Octenidine dihydrochloride | 1.56 | 1.56 | 1 |
| 140 | Osimertinib mesylate | >50 | 25 | 2 |
| 141 | Propofol | >50 | 50 | 1 |
| 142 | Rosuvastatin calcium | 25 | 25 | 1 |
| 143 | Tamoxifen | 12.5 | 12.5 | 1 |
| 144 | Tavaborole | 0.78 | 0.78 | 1 |
| 145 | Terconazole | 0.2 | 0.2 | 1 |
| 146 | Ticagrelor | >50 | 25 | 2 |
| 147 | Zinc Pyrithione | 0.78 | 0.39 | 2 |
Annotation: NA: Not applicable. ■: 30 drugs are excluded as these drugs have potent antifungal activities. ■: 10 drugs were excluded as plumbagin antagonizes them. ■: 64 drugs were excluded as plumbagin slightly affected their antifungal activities.
Table 4.
Drugs were evaluated for their interaction with plumbagin.
| No. | Antifungal Agents | MIC (μM) | FIC | FICI | Outcome | |
|---|---|---|---|---|---|---|
| Alone | Combination | |||||
| 1 | HAL | 50 | 1.56 | 0.03125 | 0.09375 | Synergy |
| Plumbagin | 4 | 0.25 | 0.0625 | |||
| 2 | Ceritinib dihydrochloride | 50 | 12.5 | 0.25 | 0.3125 | Synergy |
| Plumbagin | 4 | 0.25 | 0.0625 | |||
| 3 | Vortioxetine hydrobromide | 50 | 12.5 | 0.25 | 0.3125 | Synergy |
| Plumbagin | 4 | 0.25 | 0.0625 | |||
| 4 | Disulfiram | 25 | 3.13 | 0.125 | 0.375 | Synergy |
| Plumbagin | 4 | 1 | 0.25 | |||
| 5 | Ponatinib | 50 | 12.5 | 0.25 | 0.5 | Synergy |
| Plumbagin | 4 | 1 | 0.25 | |||
| 6 | Tafenoquine succinate | 25 | 6.25 | 0.25 | 0.5 | Synergy |
| Plumbagin | 4 | 1 | 0.25 | |||
| 7 | Thonzonium bromide | 12.5 | 3.13 | 0.25 | 0.5 | Synergy |
| Plumbagin | 4 | 1 | 0.25 | |||
| 8 | Almonertinib hydrochloride | 50 | 3.13 | 0.0625 | 0.5625 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 9 | Ebastine | 25 | 3.13 | 0.125 | 0.625 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 10 | Menadione | 50 | 6.25 | 0.125 | 0.625 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 11 | Sultiame | 50 | 6.25 | 0.125 | 0.625 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 12 | Vilanterol trifenatate | 50 | 6.25 | 0.125 | 0.625 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 13 | Amiodarone hydrochloride | 50 | 12.5 | 0.25 | 0.75 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 14 | Cinacalcet | 50 | 12.5 | 0.25 | 0.75 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 15 | Domiphen bromide | 12.5 | 3.13 | 0.25 | 0.75 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 16 | Ilaprazole | 50 | 12.5 | 0.25 | 0.75 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 17 | Rolapitant | 50 | 12.5 | 0.25 | 0.75 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 18 | Amphotericin B | 0.78 | 0.39 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 19 | Bleomycin hydrochloride | 25 | 12.5 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 20 | Cetylpyridinium chloride | 6.25 | 3.13 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 21 | Clioquinol | 25 | 12.5 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 22 | Clomiphene (citrate) | 25 | 12.5 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 23 | Fingolimod | 6.25 | 3.13 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 24 | Pinaverium bromide | 0.78 | 0.39 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 25 | Tegaserod (maleate) | 50 | 25 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 26 | Telotristat ethyl | 12.5 | 6.25 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 27 | Telotristat etiprate | 12.5 | 6.25 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 28 | Triclosan | 25 | 12.5 | 0.5 | 1 | Addition |
| Plumbagin | 4 | 2 | 0.5 | |||
| 29 | Alectinib | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 30 | Benzethonium chloride | 12.5 | 12.5 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 31 | Bosutinib | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 32 | Ceritinib | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 33 | Chlorhexidine | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 34 | Dacomitinib | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 35 | Ibudilast | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 36 | Magnolol | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 37 | Nilotinib monohydrochloride monohydrate | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 38 | Olmutinib | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 39 | Pimavanserin tartrate | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 40 | Sertindole | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 41 | Sonidegib | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 42 | Triflupromazine hydrochloride | 50 | 50 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
| 43 | Vortioxetine | 25 | 25 | 1 | 2 | Indifferent |
| Plumbagin | 4 | 4 | 1 | |||
Annotation: ■: 15 drugs are excluded because their relationship with plumbagin is indifferent. ■: 21 drugs were excluded because their relationship with plumbagin is additive.
Table 5.
Drugs were evaluated for their interaction with menadione.
| No. | Antifungal Agents | MIC (μM) | FIC | FICI | Outcome | |
|---|---|---|---|---|---|---|
| Alone | Combination | |||||
| 1 | HAL | 50 | 1.56 | 0.03125 | 0.1563 | Synergy |
| Menadione | 32 | 4 | 0.125 | |||
| 2 | Tafenoquine Succinate | 25 | 12.5 | 0.5 | 0.625 | Addition |
| Menadione | 32 | 4 | 0.125 | |||
| 3 | Ceritinib dihydrochloride | 50 | 25 | 0.5 | 0.75 | Addition |
| Menadione | 32 | 8 | 0.25 | |||
| 4 | Disulfiram | 6.25 | 3.13 | 0.5 | 1 | Addition |
| Menadione | 32 | 16 | 0.5 | |||
| 5 | Ponatinib | 50 | 25 | 0.5 | 1 | Addition |
| Menadione | 32 | 16 | 0.5 | |||
| 6 | Thonzonium bromide | 12.5 | 6.25 | 0.5 | 1 | Addition |
| Menadione | 32 | 16 | 0.5 | |||
| 7 | Vortioxetine hydrobromide | 50 | 25 | 0.5 | 1 | Addition |
| Menadione | 32 | 16 | 0.5 | |||
Table 6.
HAL combined with H2O2 against C. albicans.
| Strain | Antifungal Agents | MIC | FIC | FICI | Outcome | |
|---|---|---|---|---|---|---|
| Alone | Combination | |||||
| SN152 | HAL | >25 μM | 0.78 μM | 0.0156 | 0.2526 | Synergy |
| H2O2 | 3.13 mM | 0.78 mM | 0.25 | |||
| hog1Δ/Δ | HAL | 1.56 μM | 0.39 μM | 0.25 | 0.5 | Synergy |
| H2O2 | 1.56 mM | 0.39 mM | 0.25 | |||
| rad53Δ/Δ | HAL | >25 μM | 1.56 μM | 0.03125 | 0.2813 | Synergy |
| H2O2 | 3.13 mM | 0.78 mM | 0.25 | |||
| cap1Δ/Δ | HAL | >25 μM | >25 μM | 1 | 2 | Indiferent |
| H2O2 | 1.56 mM | 1.56 mM | 1 | |||
| ybp1Δ/Δ | HAL | >25 μM | >25 μM | 1 | 2 | Indiferent |
| H2O2 | 0.78 mM | 0.78 mM | 1 | |||
| gpx3Δ/Δ | HAL | >25 μM | 3.13 μM | 0.0625 | 0.5625 | addition |
| H2O2 | 1.56 mM | 0.78 mM | 0.5 | |||
3.2. HAL Inhibits the Oxidative Stress Response in C. albicans
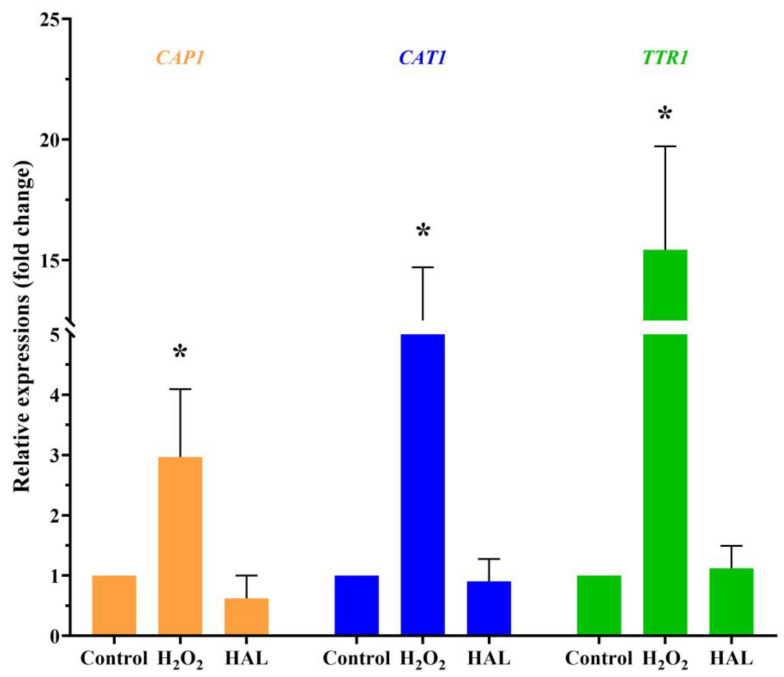
Is it possible that HAL acts as an oxidant that enhances the antifungal activity of ROS inducers? Exposure to cationic stress, such as sodium chloride (NaCl), inhibits the oxidative stress response of C. albicans [16]. If HAL is indeed an oxidant, it would be expected that HAL and NaCl would mutually enhance their respective antifungal activities. However, the interaction between HAL and NaCl appears indifferent (Table 7). On the other hand, in the presence of plumbagin, HAL and NaCl exhibit additive interactions (Table 7), indicating that HAL, like NaCl, can inhibit C. albicans’ oxidative stress response. One of the mechanisms employed by C. albicans to withstand oxidative stress involves active transcriptional responses to oxidative stress. This includes the activation of specific genes, such as the CAP1 gene (C3_02220W_A), encoding an AP-1 bZIP transcription factor, the CAT1 gene (C1_06810W_A), encoding catalase, and the TTR1 gene (C1_00490C_A), encoding glutaredoxin [5]. When C. albicans are exposed to a 10 mM concentration of H2O2 for 1 h, an upregulation in the expression levels of the three genes is observed. Conversely, a high concentration of HAL (20 μM) has minimal impact on the expression of these three genes (Figure 3). These findings indicate that HAL acts as an inhibitor of the oxidative stress response in C. albicans, thereby enhancing the antifungal activities of oxidative damage agents.
Table 7.
The interaction between HAL and NaCl.
| Conditions | Agents | MIC | FIC | FICI | Outcome | |
|---|---|---|---|---|---|---|
| Alone | Combination | |||||
| in the absence of plumbagin | HAL | >25 μM | >25 μM | 1 | 2 | Indifferent |
| NaCl | 1000 mM | 1000 mM | 1 | |||
| in the presence of plumbagin (2 μg/mL) | HAL | 0.78 μM | 0.39 μM | 0.5 | 1 | Addition |
| NaCl | 62.5 mM | 31.25 mM | 0.5 | |||
Figure 3.
The transcription levels of the CAP1, CAT1, and TTR1 genes in response to 10 mM H2O2 and 20 μM HAL for 1 h were measured by qRT-PCR. The significance of differences was determined by one-way ANOVA analysis, followed by Dunnett’s multiple comparisons test (* p < 0.05).
3.3. The Inhibitory Effect of HAL on Oxidative Stress Response Depends on the Cap1–Ybp1 Signaling Pathway
Three signaling pathways play a crucial role in response to ROS in C. albicans, including the transcriptional factor Cap1, the stress-activated protein kinase Hog1, and the DNA damage checkpoint kinase Rad53 [5]. Since HAL can inhibit the response to oxidative stress in C. albicans, HAL may target one of the signaling pathways that mediate C. albicans’ responses to ROS. To verify this conjecture, we constructed the CAP1 (C3_02220W_A), HOG1 (C2_03330C_A), and RAD53 (C3_03810W_A) genes’ null mutants (cap1Δ/cap1Δ, hog1Δ/hog1Δ, and rad53Δ/rad53Δ) in C. albicans. We found that the synergistic effect between HAL and H2O2 became indifferent in the cap1Δ/cap1Δ mutant, while their interaction remained synergistic in the hog1Δ/hog1Δ and rad53Δ/rad53Δ mutants (Table 6), suggesting that the inhibitory effect of HAL on oxidative stress response depends on the transcriptional factor Cap1.
Ybp1 is encoded by the YBP1 (C1_13960W_A) gene in C. albicans, binds to cytoplasmic pools of Cap1, and forms a complex with Cap1 [19], protecting Cap1 from ubiquitin-mediated degradation [33]. Like in the cap1Δ/cap1Δ mutant, HAL can no longer enhance the antifungal activity of H2O2 against the ybp1Δ/ybp1Δ mutant (Table 6). These findings indicate that the inhibitory effect of HAL on the antioxidant ability of C. albicans depends on the presence of Cap1 and Ybp1.
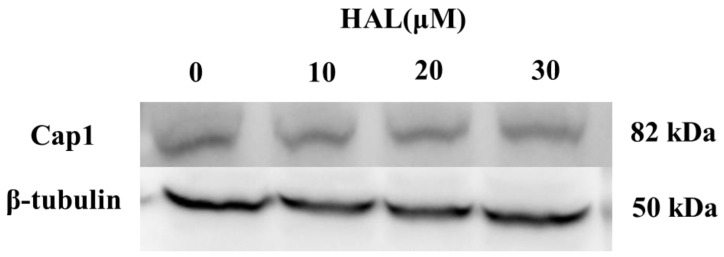
Cap1 will degrade when it cannot form a complex with Ybp1 [19]. Therefore, we conjecture that HAL inhibits the antioxidant ability of C. albicans by disrupting the interaction between Cap1 and Ybp1 and improving the degradation of Cap1. To verify this hypothesis, we tagged the C-termini of Cap1 with a GFP tag [25]. We then cultured the Cap1–GFP mutant in the presence (10 μM, 20 μM, 30 μM) or absence of HAL, respectively, and then treated it with 10 mM H2O2 for 1 h. We tested the expression of Cap1 using an anti-GFP antibody and found that the HAL did not affect the Cap1 levels in the presence of H2O2 (Figure 4), which refuted our conjecture that HAL should disrupt the interaction of Cap1 and Ybp1.
Figure 4.
The expression levels of Cap1 in the presence (10 μM, 20 μM, and 30 μM) or absence of HAL were detected by immunoblotting.
Following exposure of C. albicans to H2O2, Cap1 is activated by oxidation through a glutathione peroxidase Gpx3 [19]. Oxidized Cap1 (Cap1ox) can detach from the nuclear export factor Crm1, resulting in the accumulation of Cap1 in the nucleus and inducing the expression of genes with antioxidant functions [5]. When HAL and H2O2 are used together, they exhibit additive interactions against the gpx3Δ/gpx3Δ mutant (Table 6), suggesting that, like Gpx3, HAL may inhibit the activation and nucleation of Cap1 by inhibiting its oxidation or phosphorylation, thereby inhibiting the antioxidant activity of Cap1.
3.4. HAL Exhibits Antifungal Activity in the G. mellonella Infection Model
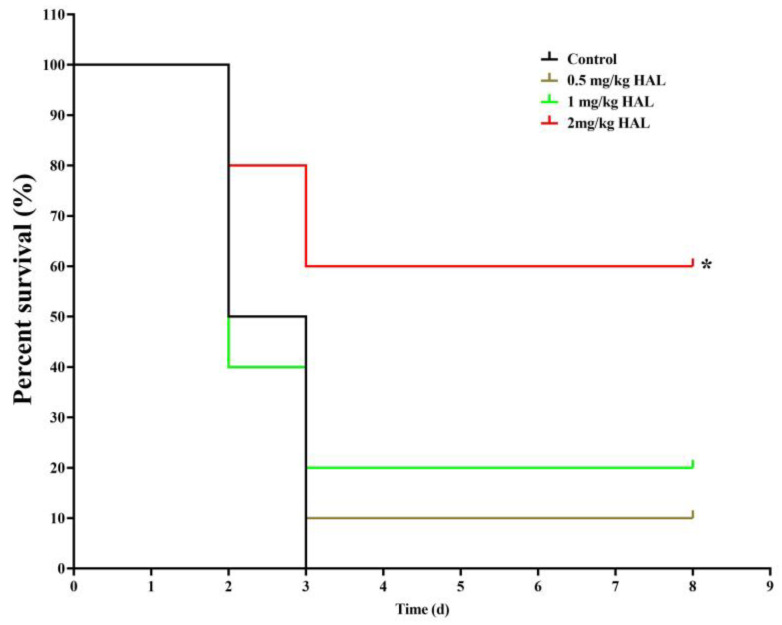
We used the G. mellonella infection model to test the antifungal efficacy of HAL in vivo, as G. mellonella utilizes phagocytic cells (hemocytes) as part of their host defense [34,35]. We divided the G. mellonella larvae into four groups, with ten larvae in each group: (1) a control group receiving no drug treatment, (2) a group treated with 0.5 mg/kg HAL, (3) a group treated with 1 mg/kg HAL, and (4) a group treated with 2 mg/kg HAL. Our observations revealed a mortality rate of 100% in the control group throughout the 8-day observation period. However, upon administration of 2 mg/kg HAL, the mortality rate of infected mice decreased to 40% (Figure 5). These in vivo experiments prove that HAL has antifungal activity against C. albicans in the G. mellonella infection model.
Figure 5.
HAL exhibits antifungal efficacy in the G. mellonella infection model. Survival curves of larvae infected with SN152 (7.0 × 105 cells/larvae) and injected with 0 mg/kg, 0.5 mg/kg, 1 mg/kg, 2 mg/kg doses of HAL. Each curve represents a group of 10 larvae (n = 10), monitored daily for survival for up to 8 days after infection. The significance of differences was determined by the Kaplan–Meier method, followed by the log-rank test (* p < 0.05).
4. Discussion
In this study, we performed a high-throughput screening of an FDA-approved drug library and identified that HAL could enhance the antifungal activities of oxidative damage agents by suppressing C. albicans’ response to ROS. We further found that HAL inhibits the oxidative stress response of C. albicans, depending on Cap1. In addition, the antifungal activity of HAL has been observed in the G. mellonella infection model. These findings demonstrated that inhibiting the oxidative stress response of C. albicans, thereby enhancing the antifungal activity of oxidative damage agents and innate immunity cells, is a promising antifungal strategy.
Candidiasis, primarily attributed to C. albicans, presents a significant risk to human health, and the availability of effective drugs for its treatment remains limited [26]. Consequently, there is an imperative to explore the development of novel antifungal medications. However, the comprehensive creation of new antifungal compounds with potent antifungal properties and optimal safety profiles, rendering them clinically valuable, necessitates substantial investments of time and resources. Considering this, repurposing FDA-approved drugs to treat candidiasis can circumvent the need for extensive safety assessments and reduce the associated time and financial burdens [36]. Numerous studies have demonstrated that FDA-approved non-antifungal drugs, such as statins and sertraline, can exhibit antifungal properties [37,38]. Following this drug repurposing strategy, our study conducted a high-throughput screening of a library comprising 2372 FDA-approved compounds and found that HAL effectively enhanced the antifungal activities of oxidative damage agents and exhibited antifungal activity in the G. mellonella infection model.
Inhibiting the oxidative stress response of C. albicans presents a promising strategy for enhancing the efficacy of antifungal treatment. Phagocytic cells are known to exert antifungal effects by generating ROS to eliminate C. albicans. Therefore, it is plausible to increase the intracellular ROS levels of C. albicans, intensify their oxidative damage, and synergistically enhance the antifungal activity of phagocytic cells or oxidative damage agents. However, there is a significant degree of conservation in the proteins comprising the mitochondrial respiratory chain between C. albicans and mammals. Consequently, the dysfunction of the mitochondrial respiratory chain induced by these compounds not only enhances the generation of ROS in C. albicans’ cells but also elevates ROS levels in mammalian cells. This ultimately leads to oxidative damage and cytotoxicity in mammalian cells, thereby restricting the potential clinical utility of these compounds. Consequently, it becomes imperative to investigate novel approaches to intensify the oxidative damage inflicted upon C. albicans. Loss of Cap1 increases susceptibility to menadione, H2O2, and host phagocytes [17,19,30], suggesting that the transcriptional factor Cap1 is important for the virulence of C. albicans [20] and is a valuable target for antifungal treatment. In the present study, we found that HAL can be repurposed as an inhibitor of the oxidative stress response of C. albicans to enhance the antifungal activity of oxidative damage agents in vitro. We further found that the mechanism of inhibiting the oxidative response action of HAL depends on inhibiting the transcriptional activity of C. albicans’ Cap1. In this study, we provide a novel antifungal strategy that inhibits the transcriptional activity of Cap1 and enhances the antifungal activity of oxidative damage agents.
Several mini-host models, including Drosophila melanogaster, Caenorhabditis elegans, and G. mellonella, have been utilized to investigate the pathophysiology of various fungal species [39]. Of interest is the G. mellonella infection model, which offers advantages such as cost effectiveness, ease of use, and independence from specialized infrastructures. The small size of G. mellonella larvae facilitates convenient manipulation, and their ability to withstand temperatures of 37 °C adds to their suitability for experimentation. The observation of melanization of larvae, decreased mobility, and mortality allows for easy detection of experimental outcomes [40]. The immune system of G. mellonella consists of phagocytic cells, which play a crucial role in the host defense by neutralizing and eliminating pathogens. This organism has been widely utilized in medical mycology research to investigate the virulence of pathogens and assess the effectiveness of antifungal treatments [41]. Thus, in this study, we employed the G. mellonella infection model to evaluate the antifungal properties of HAL, and our findings demonstrate that HAL effectively safeguards G. mellonella against C. albicans’ infection.
As an antimalarial drug, HAL has recently been reported to enhance the antifungal activity of amphotericin B against Cryptococcus and Candida species [42], suggesting that HAL may affect the fungal oxidative stress system since amphotericin B culminates C. albicans in death through the production of cytotoxic ROS [9]. In this study, we further uncovered the mechanism of HAL and its actions. The toxicity of HAL is low in G. mellonella, suggesting a promising clinical application. However, HAL is a blocker of delayed rectifier potassium current via the inhibition of the human-ether-a-go-go-related gene (hERG) channel [43,44]. Therefore, further research should develop a series of HAL analogs without inhibiting the hERG channel.
5. Conclusions
In conclusion, our study has identified that HAL has the potential to enhance the antifungal activities of oxidative damage agents (plumbagin, menadione, and H2O2) by suppressing the response of C. albicans to ROS. Furthermore, we discovered that the mechanism behind HAL’s inhibition of the oxidative response involves the inhibition of Cap1’s transcriptional activity. Additionally, the antifungal activity of HAL has been observed in the G. mellonella infection model. These findings provide evidence that targeting the oxidative stress response of C. albicans and enhancing the fungicidal ability of innate immunity cells could serve as a promising antifungal strategy.
Author Contributions
Conceptualization, H.L. and Y.J.; methodology, J.X., L.W. and Z.F.; software, J.X.; validation, S.H., J.Y. and Y.F.; formal analysis, J.X., L.W. and Z.F.; investigation, J.X., L.W. and Z.F.; data curation, S.H., J.Y. and Y.F.; writing—original draft preparation, J.X. and H.L.; writing—review and editing, H.L. and Y.J.; visualization, J.X.; supervision, H.L. and Y.J.; project administration, Y.J.; funding acquisition, Y.J. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This study received financial support from the National Key Research and Development Program of China (No. 2022YFC2303004 and No. 2021YFC2300404), the National Natural Science Foundation of China (No. 82020108032), and the Innovation Program of Shanghai Municipal Education Commission (202101070007-E00094).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Denning D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024 doi: 10.1016/S1473-3099(23)00692-8. [DOI] [PubMed] [Google Scholar]
- 2.Lu H., Hong T., Jiang Y., Whiteway M., Zhang S. Candidiasis: From cutaneous to systemic, new perspectives of potential targets and therapeutic strategies. Adv. Drug Deliv. Rev. 2023;199:114960. doi: 10.1016/j.addr.2023.114960. [DOI] [PubMed] [Google Scholar]
- 3.Zhen C., Lu H., Jiang Y. Novel Promising Antifungal Target Proteins for Conquering Invasive Fungal Infections. Front. Microbiol. 2022;13:911322. doi: 10.3389/fmicb.2022.911322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Zou Y., Chen X., Li H., Yin Z., Zhang B., Xu Y., Zhang Y., Zhang R., Huang X., et al. Innate immune responses against the fungal pathogen Candida auris. Nat. Commun. 2022;13:3553. doi: 10.1038/s41467-022-31201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dantas A.D.S., Day A., Ikeh M., Kos I., Achan B., Quinn J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules. 2015;5:142–165. doi: 10.3390/biom5010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda J.E.A., Baronetti J.L., Sotomayor C.E., Paraje M.G. Oxidative and nitrosative stress responses during macrophage–Candida albicans biofilm interaction. Med. Mycol. 2019;57:101–113. doi: 10.1093/mmy/myx143. [DOI] [PubMed] [Google Scholar]
- 7.Atriwal T., Chawla M., Hussain A., Alajmi M.F., Abid M. Reactive oxygen mediated apoptosis as a therapeutic approach against opportunistic Candida albicans. Adv. Protein Chem. Struct. Biol. 2021;125:25–49. doi: 10.1016/bs.apcsb.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Erwig L.P., Gow N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016;14:163–176. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- 9.Belenky P., Camacho D., Collins J.J. Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep. 2013;3:350–358. doi: 10.1016/j.celrep.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guirao-Abad J.P., Sánchez-Fresneda R., Román E., Pla J., Argüelles J.C., Alonso-Monge R. The MAPK Hog1 mediates the response to amphotericin B in Candida albicans. Fungal Genet. Biol. 2020;136:103302. doi: 10.1016/j.fgb.2019.103302. [DOI] [PubMed] [Google Scholar]
- 11.Regidor P.A., Thamkhantho M., Chayachinda C., Palacios S. Miconazole for the treatment of vulvovaginal candidiasis. In vitro, in vivo and clinical results. Review of the literature. J. Obstet. Gynaecol. 2023;43:2195001. doi: 10.1080/01443615.2023.2195001. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y., Wang Y., Yan L., Liang R.-M., Dai B.-D., Tang R.-J., Gao P.-H., Jiang Y.-Y. Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: Endogenous ROS augmentation. J. Proteome Res. 2009;8:5296–5304. doi: 10.1021/pr9005074. [DOI] [PubMed] [Google Scholar]
- 13.Fu Z., Lu H., Zhu Z., Yan L., Jiang Y., Cao Y. Combination of Baicalein and Amphotericin B Accelerates Candida albicans Apoptosis. Biol. Pharm. Bull. 2011;34:214–218. doi: 10.1248/bpb.34.214. [DOI] [PubMed] [Google Scholar]
- 14.Li D.-D., Chai D., Huang X.-W., Guan S.-X., Du J., Zhang H.-Y., Sun Y., Jiang Y.-Y. Potent In Vitro Synergism of Fluconazole and Osthole against Fluconazole-Resistant Candida albicans. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00436-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves E.P., Lu H., Jacobs H.L., Messina C.G.M., Bolsover S., Gabella G., Potma E.O., Warley A., Roes J., Segal A.W. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 16.Kaloriti D., Jacobsen M., Yin Z., Patterson M., Tillmann A., Smith D.A., Cook E., You T., Grimm M.J., Bohovych I., et al. Mechanisms underlying the exquisite sensitivity of candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. mBio. 2014;5:e01334-14. doi: 10.1128/mBio.01334-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kos I., Patterson M.J., Znaidi S., Kaloriti D., Dantas A.d.S., Herrero-De-Dios C.M., D’enfert C., Brown A.J.P., Quinn J. Mechanisms Underlying the Delayed Activation of the Cap1 Transcription Factor in Candida albicans following Combinatorial Oxidative and Cationic Stress Important for Phagocytic Potency. mBio. 2016;7:e00331. doi: 10.1128/mBio.00331-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien C.-T., Chen Y.-C., Liu Y.-C., Liang S.-H., Lin H.-H., Lin C.-H. The antimicrobial photodynamic inactivation resistance of Candida albicans is modulated by the Hog1 pathway and the Cap1 transcription factor. Med. Mycol. 2019;57:618–627. doi: 10.1093/mmy/myy079. [DOI] [PubMed] [Google Scholar]
- 19.Patterson M.J., McKenzie C.G., Smith D.A., Dantas A.d.S., Sherston S., Veal E.A., Morgan B.A., MacCallum D.M., Erwig L.-P., Quinn J. Ybp1 and Gpx3 Signaling in Candida albicans Govern hydrogen peroxide-induced oxidation of the Cap1 transcription factor and macrophage escape. Antioxidants Redox Signal. 2013;19:2244–2260. doi: 10.1089/ars.2013.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain C., Pastor K., Gonzalez A.Y., Lorenz M.C., Rao R.P. The role of Candida albicans AP-1 protein against host derived ROS in in vivo models of infection. Virulence. 2013;4:67–76. doi: 10.4161/viru.22700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noble S.M., Johnson A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang T., Xiong J., Wang L., Feng Z., Hang S., Yu J., Li W., Feng Y., Lu H., Jiang Y. Unexpected Inhibitory Effect of Octenidine Dihydrochloride on Candida albicans Filamentation by Impairing Ergosterol Biosynthesis and Disrupting Cell Membrane Integrity. Antibiotics. 2023;12:1675. doi: 10.3390/antibiotics12121675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson M.D., MacDougall C., Ostrosky-Zeichner L., Perfect J.R., Rex J.H. Combination antifungal therapy. Antimicrob. Agents Chemother. 2004;48:693–715. doi: 10.1128/AAC.48.3.693-715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Ji Z., Feng Y., Yan T., Cao Y., Lu H., Jiang Y. Myriocin enhances the antifungal activity of fluconazole by blocking the membrane localization of the efflux pump Cdr1. Front. Pharmacol. 2022;13:1101553. doi: 10.3389/fphar.2022.1101553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang P., Wang W., Igarashi Y., Luo F., Chen J. Efficient vector systems for economical and rapid epitope-tagging and overexpression in Candida albicans. J. Microbiol. Methods. 2018;149:14–19. doi: 10.1016/j.mimet.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Lu H., Li W., Whiteway M., Wang H., Zhu S., Ji Z., Feng Y., Yan L., Fang T., Li L., et al. A Small Molecule Inhibitor of Erg251 Makes Fluconazole Fungicidal by Inhibiting the Synthesis of the 14α-Methylsterols. mBio. 2023;14:e0263922. doi: 10.1128/mbio.02639-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Lu H., Zhang X., Whiteway M., Wu H., Tan S., Zang J., Tian S., Zhen C., Meng X., et al. Baicalein Acts against Candida albicans by Targeting Eno1 and Inhibiting Glycolysis. Microbiol. Spectr. 2022;10:e0208522. doi: 10.1128/spectrum.02085-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padhye S., Dandawate P., Yusufi M., Ahmad A., Sarkar F.H. Perspectives on medicinal properties of plumbagin and its analogs. Med. Res. Rev. 2012;32:1131–1158. doi: 10.1002/med.20235. [DOI] [PubMed] [Google Scholar]
- 29.Castro F.A.V., Mariani D., Panek A.D., Eleutherio E.C.A., Pereira M.D. Cytotoxicity mechanism of two naphthoquinones (menadione and plumbagin) in Saccharomyces cerevisiae. PLoS ONE. 2008;3:e3999. doi: 10.1371/journal.pone.0003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y., Wang Y., Dai B., Wang B., Zhang H., Zhu Z., Xu Y., Cao Y., Jiang Y., Zhang G. Trehalose is an important mediator of Cap1p oxidative stress response in Candida albicans. Biol. Pharm. Bull. 2008;31:421–425. doi: 10.1248/bpb.31.421. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y., Cao Y.-Y., Jia X.-M., Cao Y.-B., Gao P.-H., Fu X.-P., Ying K., Chen W.-S., Jiang Y.-Y. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free. Radic. Biol. Med. 2006;40:1201–1209. doi: 10.1016/j.freeradbiomed.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Noble S.M., French S., Kohn L.A., Chen V., Johnson A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gulshan K., Thommandru B., Moye-Rowley W.S. Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J. Biol. Chem. 2012;287:26796–26805. doi: 10.1074/jbc.M112.384719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durieux M.-F., Melloul S., Jemel S., Roisin L., Dardé M.-L., Guillot J., Dannaoui S., Botterel F. Galleria mellonella as a screening tool to study virulence factors of Aspergillus fumigatus. Virulence. 2021;12:818–834. doi: 10.1080/21505594.2021.1893945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fallon J., Kelly J., Kavanagh K. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol. Biol. 2012;845:469–485. doi: 10.1007/978-1-61779-539-8_33. [DOI] [PubMed] [Google Scholar]
- 36.Rossi S.A., de Oliveira H.C., Agreda-Mellon D., Lucio J., Mendes-Giannini M.J.S., García-Cambero J.P., Zaragoza O. Identification of Off-Patent Drugs That Show Synergism with Amphotericin B or That Present Antifungal Action against Cryptococcus neoformans and Candida spp. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01921-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavakkoli A., Johnston T.P., Sahebkar A. Antifungal effects of statins. Pharmacol. Ther. 2020;208:107483. doi: 10.1016/j.pharmthera.2020.107483. [DOI] [PubMed] [Google Scholar]
- 38.Spitzer M., Griffiths E., Blakely K.M., Wildenhain J., Ejim L., Rossi L., De Pascale G., Curak J., Brown E., Tyers M., et al. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol. Syst. Biol. 2011;7:499. doi: 10.1038/msb.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arvanitis M., Glavis-Bloom J., Mylonakis E. Invertebrate models of fungal infection. Biochim. Biophys. Acta. 2013;1832:1378–1383. doi: 10.1016/j.bbadis.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Andrea A., Krogfelt K.A., Jenssen H. Methods and Challenges of Using the Greater Wax Moth (Galleria mellonella) as a Model Organism in Antimicrobial Compound Discovery. Microorganisms. 2019;7:85. doi: 10.3390/microorganisms7030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jemel S., Guillot J., Kallel K., Botterel F., Dannaoui E. Galleria mellonella for the Evaluation of Antifungal Efficacy against Medically Important Fungi, a Narrative Review. Microorganisms. 2020;8:390. doi: 10.3390/microorganisms8030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freitas G.J., Ribeiro N.Q., Gouveia-Eufrasio L., Emidio E.C., Guimarães G.M., César I.C., Paixão T.A., Oliveira J.B., Caza M., Kronstad J.W., et al. Antimalarials and amphotericin B interact synergistically and are new options to treat cryptococcosis. Int. J. Antimicrob. Agents. 2023;62:106807. doi: 10.1016/j.ijantimicag.2023.106807. [DOI] [PubMed] [Google Scholar]
- 43.Tie H., Walker B.D., Singleton C.B., Valenzuela S.M., Bursill J.A., Wyse K.R., Breit S.N., Campbell T.J. Inhibition of HERG potassium channels by the antimalarial agent halofantrine. Br. J. Pharmacol. 2000;130:1967–1975. doi: 10.1038/sj.bjp.0703470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traebert M., Dumotier B., Meister L., Hoffmann P., Dominguez-Estevez M., Suter W. Inhibition of hERG K+ currents by antimalarial drugs in stably transfected HEK293 cells. Eur. J. Pharmacol. 2004;484:41–48. doi: 10.1016/j.ejphar.2003.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.