Abstract
Nuclear transcription is repressed when eukaryotic cells enter mitosis. Mitotic repression of transcription of various cellular and viral gene promoters by RNA polymerase II can be reproduced in vitro either with extracts prepared from cells arrested at mitosis with the microtubule polymerization inhibitor nocodazole or with nuclear extracts prepared from asynchronous cells and the mitotic protein kinase cdc2/cyclin B. Purified cdc2/cyclin B kinase is also sufficient to inhibit transcription in reconstituted transcription reactions with biochemically purified and recombinant basal transcription factors and RNA polymerase II. The cyclin-dependent kinase inhibitor p21Waf1/Cip1/Sdi1 can reverse the effect of cdc2/cyclin B kinase, indicating that repression of transcription is due to protein phosphorylation. Transcription rescue and inhibition experiments with each of the basal factors and the polymerase suggest that multiple components of the transcription machinery are inactivated by cdc2/cyclin B kinase. For an activated promoter, targets of repression are TFIID and TFIIH, while for a basal promoter, TFIIH is the major target for mitotic inactivation of transcription. Protein labeling experiments indicate that the p62 and p36 subunits of TFIIH are in vitro substrates for mitotic phosphorylation. Using the carboxy-terminal domain of the large subunit of RNA polymerase II as a test substrate for phosphorylation, the TFIIH-associated kinase, cdk7/cyclin H, is inhibited concomitant with inhibition of transcription activity. Our results suggest that there exist multiple phosphorylation targets for the global shutdown of transcription at mitosis.
Many aspects of transcriptional regulation in eukaryotic cells involve reversible phosphorylation events; these include phosphorylation of both general and gene-specific transcription factors (20, 21). Examples of both positive and negative effects of protein phosphorylation have been reported (for reviews, see references 20 and 21). One such example of transcriptional control mediated by phosphorylation is the global repression of nuclear transcription that occurs when cells enter mitosis (22, 36). Over the years, numerous hypotheses have been put forward to explain mitotic repression of transcription (for reviews, see references 12, 16, and 32), including condensation of interphase chromatin into mitotic chromosomes, dissociation of transcription factors or RNA polymerase from the chromatin template, and inactivation of the basal transcription machinery by protein phosphorylation (13, 40, 50). For the genes transcribed by RNA polymerase III (pol III) (such as 5S rRNA and tRNA genes), previous studies have documented that the activity of the general class III transcription factor TFIIIB is greatly diminished in extracts from synchronized mitotic cells (49) or by the conversion of an interphase Xenopus egg extract to the mitotic state by the addition of recombinant cyclin B1 protein (13, 16, 50). In the latter experiments, the recombinant cyclin formed a complex with the p34cdc2 kinase subunit present in the extract and, after a series of specific phosphorylation and dephosphorylation events, the active form of the cdc2/cyclin B kinase (maturation-mitosis promoting factor) was generated (45, 46). Inhibition of transcription has been shown to be due to the enzymatic action of this kinase on a TFIIIB subunit (or a repressor protein that binds to and inactivates TFIIIB) (13, 16).
Similar to class III gene transcription, transcription of mRNA-coding genes by RNA polymerase II (pol II) is also repressed at mitosis. We have shown that purified cdc2/cyclin B kinase is sufficient to inhibit transcription by pol II in a reconstituted transcription system (27). Recently, Segil et al. (40) reported that the general pol II transcription factor TFIID isolated from mitotic cells is multiply phosphorylated and inactive in supporting activator-dependent transcription. TFIID is composed of the TATA-binding protein (TBP) and TBP-associated factors (TAFs), and the TAFs have been shown to be involved in activator-dependent transcription (for reviews, see references 17 and 33). The activity of mitotic TFIID can be restored by dephosphorylation, showing that a protein phosphorylation event regulates TFIID during mitosis. Thus, for both activated pol II transcription (40) and pol III transcription (13, 49), a TBP-associated factor is inactivated at mitosis. In the work of Segil et al. (40), only TFIID was purified from mitotic cells; thus, it is not clear whether other targets of mitotic regulation exist in the pol II transcription machinery. Indeed, mitotic TFIID was found to be defective in only activator-dependent transcription, suggesting that other general transcription factors might be targets for regulation of basal levels of transcription. Here we report that mitotic repression of pol II transcription can be reproduced in vitro with whole-cell and nuclear extracts or with a reconstituted transcription system and purified cdc2/cyclin B kinase isolated from the mitotic Xenopus egg extract. Our results show that in addition to TFIID, the general transcription factor TFIIH is also a target for mitotic repression of pol II transcription. We have found that, concomitant with inhibition of TFIIH transcriptional activity, the cdk7/cyclin H kinase activity associated with TFIIH is also inhibited by cdc2 phosphorylation. These results are consistent with models for the role of the TFIIH-associated cdk7 kinase in the transcription cycle (5, 33).
MATERIALS AND METHODS
Cell extracts and purification of activated cdc2/GST-cyclin B kinase.
The human lymphoid cell line H9 (ATCC HTB 176) was grown in suspension culture in RPMI medium (BioWhittaker) supplemented with 10% fetal calf serum (Tissue Culture Biological). To arrest cells in mitosis, nocodazole (Sigma) was added to log-phase cultures to a final concentration of 600 ng/ml and the cells were incubated for an additional 16 h (40). Whole-cell extracts were prepared from equivalent numbers of asynchronous and mitotic cells by hypotonic lysis (6). Protein concentrations were determined by the Bradford method, using the reagents available from Bio-Rad, and both of the whole-cell extracts contained 20 mg of protein per ml. When comparing asynchronous and mitotic cell extracts, equivalent amounts of extract protein were used. Xenopus mitotic cell extracts were generated by the addition of a recombinant sea urchin B1 cyclin–glutathione S-transferase (GST) fusion protein (46) to an interphase extract prepared from unfertilized eggs (16). Mitotic conversion was monitored by induction of histone H1 kinase activity (16, 50). The activated cdc2/cyclin B kinase was recovered from the extract with p13-agarose beads or with glutathione-Sepharose beads as described previously (16, 50). Equal volumes of extract and packed beads were incubated on a rotator at 4°C for 1 h. Unbound proteins were removed by washing the resin with buffer EB (16), and the immobilized kinase was used in phosphorylation reactions (see below). The cdc2/cyclin B kinase was eluted from the glutathione-Sepharose resin with 5 mM glutathione, yielding a final protein concentration of approximately 0.25 μg/μl. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed that the kinase preparation contained only p34cdc2 and GST-cyclin B (and the low-molecular-weight Escherichia coli glutathione-Sepharose binding proteins that are found in all preparations of GST fusion proteins) (50).
Transcription factors and in vitro transcription reactions.
HeLa cell nuclear extract was purchased from Promega and used as recommended by the manufacturer. For transcription reactions with the whole-cell extracts from H9 cells, 2 μl of extract was used in a final reaction volume of 25 μl. pol II and transcription factors were the generous gifts of Osvaldo Flores (Tularik, Inc., South San Francisco, Calif.). Reconstituted transcription reactions were performed as described elsewhere (10, 26). The following DNA templates (100 to 200 ng of DNA per 25-μl reaction volume) were used in this study: plasmid E1B-GL, containing the major immediate-early promoter (MIEP) element of cytomegalovirus (CMV) linked to a guanineless cassette (26); pγP4.0 γ-globin (37), digested with PstI to generate a 622-base-long runoff transcript; TdT, the terminal deoxynucleotidyl transferase Inr element (44) linked to pCAT (a gift of P. Ghazal, Scripps Research Institute), digested with EcoRI to generate a 400-base-long transcript; pCAT-HIV-1 (43), digested with EcoRI to generate a 400-base-long transcript; pHIV+TAR-G400 (56) containing a 400-base-long guanineless cassette; and the adenovirus (Ad) major late promoter (MLP) linked to a guanineless cassette (38). Reconstituted transcription reaction mixtures contained either TFIID (DE-52 fraction, 1 μg) or recombinant human TBP (50 ng) and a mixture of recombinant (TFIIB, 20 ng; TFIIE, 100 ng of each subunit) and partially purified (TFIIF, 200 ng of the TSK-phenyl fraction; TFIIH, 400 ng; TFIIA, 5 μg of DE-52 fraction; and pol II, 100 ng of the DEAE-5PW fraction) transcription factors in a final volume of 25 μl. Some experiments utilized a TFIIF-TFIIH fraction (10). Protein purification methods have been described elsewhere (10). Nuclear extract or transcription factors were incubated with empirically determined amounts (1 to 3 μl) of cdc2 kinase at ambient temperature as described below. Generally, 1 μl of affinity-purified kinase yields maximal inhibition of transcription in a 30-min reaction. Transcription complexes were allowed to form for 1 h at 30°C. For transcription experiments involving adding back immunopurified TFIIH, approximately 10 μl of packed TFIIH-coated protein A-Sepharose beads (prepared as described below) were included in the transcription complex assembly step. This was followed by a transcription step with 10 μCi of [α-32P]CTP or [α-32P]ATP, 600 μM remaining unlabeled nucleoside triphosphates (NTPs) 10 μM CTP or ATP (depending on which NTP radioisotope was used), and, for the templates linked to guanineless cassettes, RNase T1 (Boehringer Mannheim; 25 U per reaction) for 1 h at 30°C. RNA was purified by extraction with RNAzol (TelTest, Friendswood, Tex.) and analyzed by electrophoresis on a denaturing (8.3 M urea) 6% polyacrylamide gel. Autoradiograms were obtained by exposure of the dried gel to Kodak XAR-5 or BioMax film with DuPont Cronex Lightning Plus intensifying screens for 15 to 18 h at −80°C. Relative levels of transcription were estimated by storage phosphorimage analysis using Kodak storage phosphor screens (SO 230) and a Molecular Dynamics SF PhosphorImager. The data were analyzed by using ImageQuant software (Molecular Dynamics) in the volume integration mode after appropriate background subtraction.
Immunopurification of TFIIH.
Antibodies to the subunits of TFIIH (p62, p89 ERCC3, cdk7, and cyclin H) were the generous gifts of Osvaldo Flores (Tularik, Inc.) and Jean-Marc Egly (Strasbourg), and polyclonal antibodies to p62 and p89 were purchased from Santa Cruz Biotechnology. Antibodies to human TBP were purchased from Promega. Packed protein A-Sepharose beads (Pharmacia; 25 μl) were washed three times with cold phosphate-buffered saline (PBS). Antibodies (1 to 5 μl) were attached to these beads by incubating them for 1 to 2 h in 100 μl of PBS at 4°C with constant shaking. The antibody-coated beads were rinsed three times with cold PBS to remove unbound antibodies. HeLa cell nuclear extract (Promega; 2 μl at 12 mg of protein per ml), asynchronous or nocodazole-arrested H9 cell extract (2 μl at 20 mg of protein per ml), or 50 μl of cdc2-HeLa kinase solution (described below) plus PBS to a final volume of 100 μl was added to these beads. This mixture was incubated 1 to 2 h at 4°C with constant shaking. The beads were then washed four times with a cold solution of PBS containing 0.1% Triton X-100 to remove unbound proteins and then four times with cold PBS to remove residual Triton X-100. Prior to use, the protein-coated beads were equilibrated by rinsing them once in the appropriate reaction buffer.
Kinase reactions.
cdc2 phosphorylation of HeLa cell nuclear extract was performed in a solution containing kinase buffer (20 mM HEPES, 8 mM sodium β-glycerophosphate, 5 mM EGTA, 10 mM MgCl2; pH 7.3), 20 μM ATP, an empirically determined amount of affinity-purified cdc2/cyclin B kinase, 2 μl of HeLa cell nuclear extract, and deionized H2O to a final volume of 50 μl. This solution was incubated for 1 h at ambient temperature. The cdc/cdk kinase inhibitor p21 was purified as described in reference 25. For reactions involving p21 inhibition of cdc2, the standard kinase reaction mixture minus HeLa cell extract was preincubated with the appropriate concentration of affinity-purified p21 for 30 min at ambient temperature. HeLa cell extract was then added, and the reaction mixture was incubated for an additional 1 h at ambient temperature. A fusion protein consisting of GST and the carboxy-terminal domain of the largest subunit of pol II (GST-CTD) was expressed and affinity purified as previously described (35). Phosphorylation of CTD by immunopurified TFIIH was performed in a solution consisting of kinase buffer, 100 μM ATP, 80 μCi of [γ-32P]ATP, 0.5 μg of affinity-purified GST-CTD (35), and deionized H2O to a final volume of 50 μl. This solution was added to 25 μl (packed volume) of TFIIH-coated Sepharose beads, and the suspension was incubated for 1 h at ambient temperature with slow rotation. The beads were spun down, the supernatant was removed, and proteins were precipitated with 25% (vol/vol) trichloroacetic acid and separated by SDS-PAGE on a 10% polyacrylamide gel. Phosphorylation of histone H1 (2 μg) was performed as previously described (16), using 2 μl of H9 cell extract per 20-μl reaction volume. HeLa cell nuclear extract proteins were subjected to phosphorylation by p13-agarose-immobilized kinase (20 μl of packed resin), using [γ-32P]ATP (56 μCi) and 100 μM unlabeled ATP in a reaction volume of 20 to 40 μl. Incubations were for 1 h on a rotator at ambient temperature as described previously (16, 50). Biochemically isolated TFIIH was also subjected to phosphorylation by the immobilized kinases. The p13-agarose beads were removed by brief centrifugation, and immunoprecipitation of TFIIH from the supernatant was carried out as described above. Proteins were eluted from the protein A-Sepharose beads by heating the beads at 95 to 100°C in SDS sample buffer, and the labeled species were analyzed by SDS-PAGE and autoradiography. For Western blot analysis, the immunoreactive species were visualized by enhanced chemiluminescence and autoradiography, using the Amersham ECL system. Molecular masses of prestained markers (Amersham Rainbow markers) are indicated alongside the gels in the figures.
RESULTS
pol II transcription is repressed in extracts of cells arrested at mitosis.
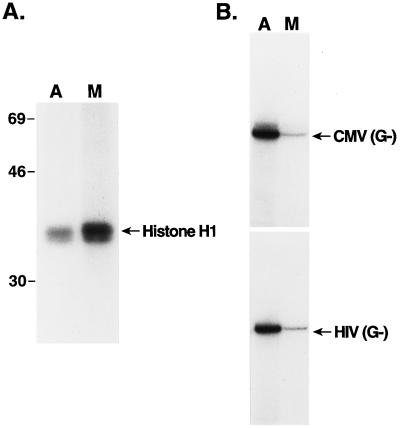
We prepared whole-cell extracts from asynchronous cultures of the lymphoid cell line H9 and similar extracts from cells arrested at mitosis with the microtubule polymerization inhibitor nocodazole (40). One hallmark of mitosis is the induction of potent mitotic kinases such as cdc2/cyclin B, (mitosis-maturation promoting factor) (46). To test for the induction of protein kinase activity in the mitotic cells, equivalent amounts of protein from the two extracts were used in phosphorylation experiments with [γ-32P]ATP and histone H1 as a test substrate (Fig. 1A). A three- to fourfold induction of H1 kinase activity was observed in the mitotic extract relative to the induction achieved in the asynchronous cell extract. These extracts were also used in transcription experiments with either the human CMV MIEP (26) or the human immunodeficiency virus type 1 (HIV-1) (56) promoter, both linked to a guanineless cassettes. The transcriptional activities of the HIV-1 and CMV promoters were reduced five- and eightfold, respectively, in the mitotic extracts compared with the activities in extracts from asynchronous cells (Fig. 1B; compare lanes A and M). Thus, mitotic repression of pol II transcription was reproduced with these extracts.
FIG. 1.
Transcription is repressed in whole-cell extracts prepared from mitotic cells in culture. (A) Histone H1 kinase activity of asynchronous (A) and mitotic (M) cell extracts. Equivalent amounts of extract protein were used as a source of kinase, along with histone H1 and [γ-32P]ATP, as described in Materials and Methods. Radiolabeled histone H1 was visualized by autoradiography of an SDS-polyacrylamide gel. The positions of molecular mass markers (denoted in kilodaltons) are indicated. (B) Transcription of the human CMV MIEP (CMV) and the HIV-1 promoter (HIV), both linked to guanineless cassettes (G−), was monitored, using equivalent amounts of extract protein from asynchronous (A) or mitotic (M) cells, as described in Materials and Methods. The gel autoradiogram is shown.
Repression of pol II gene transcription in vitro.
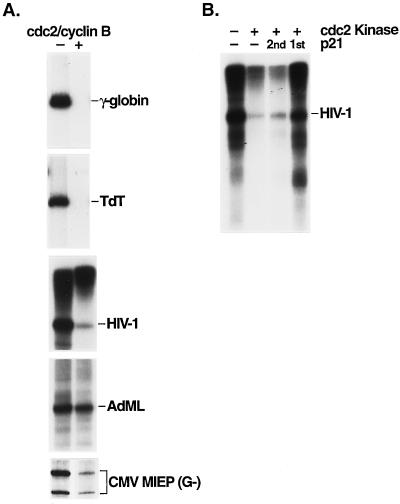
To identify the factor(s) (or polymerase) which is inactivated for transcription at mitosis, we asked whether biochemically purified cdc2/cyclin B kinase could repress pol II transcription in a nuclear extract prepared from asynchronous cultures of HeLa cells, in a manner similar to the pol III system (13, 16, 50). To isolate the kinase, a high-speed extract from unfertilized Xenopus eggs was converted to the mitotic state by the addition of a protease-resistant recombinant cyclin B1-GST fusion protein (16). Mitotic conversion was monitored by the induction of protein kinase activity, using histone H1 as a test substrate, and the active cdc2/cyclin B kinase was isolated from the extract by glutathione-Sepharose affinity chromatography, taking advantage of the GST fusion domain on the recombinant cyclin protein (16, 50). Protein kinase assays with histone H1 as the substrate confirmed the activity of the purified cdc2/cyclin B kinase (50) (see Fig. 7A). Transcription assays were performed with various cellular and viral gene promoters and with nuclear extract which had been incubated with both the cdc2/cyclin B kinase and ATP or with ATP alone prior to the addition of the DNA template (Fig. 2A). Significant repression of transcription (3 to 20-fold) was observed with each of the templates assayed, with the exception of the Ad MLP, which was not significantly repressed in this system (see Discussion). The extent of transcriptional repression varied with the particular template assayed, the amount of cdc2 kinase used in the reaction, and the length of the inhibition period. Conditions for maximal repression were determined empirically (see Materials and Methods). The templates used represent a variety of pol II promoter classes, including those containing TATA elements (γ-globin, HIV-1 promoter, Ad MLP, and CMV MIEP) and those lacking a TATA element (TdT). Some of these templates contain binding sites for various cellular activator proteins which are present in the HeLa cell nuclear extract; for example, the HIV-1 promoter contains binding sites for the factors Sp1 and USF. In contrast, the CMV promoter contains only a TATA box and an initiator element (Inr) and TdT contains only an Inr. Thus, with the exception of the Ad MLP, transcription repression by the cdc2/cyclin B kinase appears to be independent of promoter structure, indicating that general transcription factors or pol II is inactivated by the kinase.
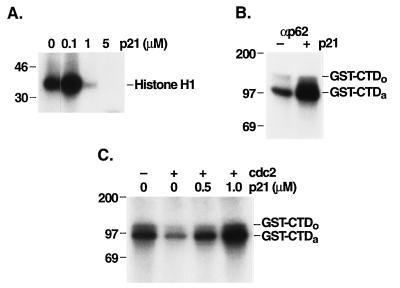
FIG. 7.
cdc2 inhibition of TFIIH kinase activity is reversed by the cyclin-dependent kinase inhibitor p21. (A) Affinity-purified cdc2/GST-cyclin B was incubated with the indicated concentrations of p21, and these mixtures were then used in kinase reactions with histone H1 as a substrate. The position of 32P-labeled histone H1 is indicated. The positions of molecular mass markers (in kilodaltons) are indicated to the left. (B) HeLa cell nuclear extract was incubated with (+) or without (−) 1 μM p21, and antibody to p62 (αp62) was used to immunopurify TFIIH from the treated or untreated extract. These immunoprecipitates were then used in kinase assays to phosphorylate GST-CTD. The positions of 32P-labeled GST-CTDo and GST-CTDa are shown. (C) Affinity-purified cdc2 was incubated without or with p21 at various concentrations as indicated. Next, HeLa cell extract was incubated with this p21-treated cdc2. Antibody to p62 was used to immunopurify TFIIH from the cdc2-treated HeLa cell extracts. The TFIIH immunoprecipitates were then used in kinase assays to phosphorylate GST-CTD. The positions of 32P-labeled GST-CTDo and GST-CTDa are shown.
FIG. 2.
Repression of pol II transcription with affinity-purified cdc2/GST-cyclin B kinase. (A) Products of transcription from the indicated templates were analyzed by denaturing gel electrophoresis and autoradiography. HeLa cell nuclear extract (2 μl) was incubated at ambient temperature for 30 min with 20 μM ATP in the absence (−) or presence (+) of 1 μl of kinase prior to the addition of each of the DNA templates (100 ng of PstI-digested human γ-globin, 100 ng of EcoRI-digested TdT, 200 ng of EcoRI-digested pCAT-HIV-1, 200 ng of HindIII-digested Ad MLP, and 100 ng of undigested CMV MIEP linked to a guanineless cassette [G−]). (B) The cyclin-dependent kinase inhibitor p21 reverses the transcription-inhibitory effect of the cdc2 kinase. p21 (2 μM final concentration) was incubated without (−) or with (+) 1 μl of cdc2 kinase for 15 min prior to addition of the HeLa cell nuclear extract (2 μl) (p21 1st) or after a 30-min incubation of the kinase and extract (p21 2nd). Runoff transcription was monitored with EcoRI-digested pCAT-HIV-1 DNA.
To show that protein phosphorylation is involved in the observed repression of pol II transcription, we monitored the effect of the cyclin-dependent kinase inhibitor p21Waf1/Cip1/Sdi1 (8, 14, 51) on cdc2-mediated repression (Fig. 2B). While cdc2/cyclin B kinase is not thought to be an important target for p21 cdk inhibitory activity in vivo, p21 has been shown to inhibit cdc2/cyclin B in vitro (51, 53), with a Ki of 0.4 μM (15). In agreement, we found that p21 inhibits histone H1 phosphorylation by cdc2/cyclin B at concentrations of 0.1 to 1 μM (see Fig. 7A). Inhibition of transcription could be prevented by incubating p21 and cdc2 kinase together prior to addition of the nuclear extract (Fig. 2B, lane p21 1st); however, p21 could not overcome the repressive effect of the kinase if it was added after the extract was incubated with the kinase (lane p21 2nd). Phosphorimage analysis indicated that the cdc2 kinase inhibited HIV-1 transcription by 87% in this assay, while prior incubation of the kinase with p21 fully restored the transcription activity of the extract to control levels. Similarly, cdc2-mediated repression was prevented if the nonhydrolyzable ATP analog 5′-adenylylimidodiphosphate (AMP-PNP) was included in the kinase reaction mixture (data not shown), indicating that the γ-phosphate of ATP is utilized in the repression mechanism.
Repression targets for basal pol II transcription.
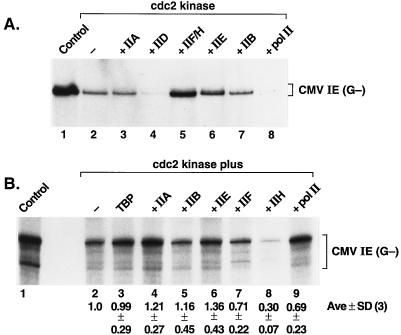
The human CMV MIEP was chosen as an example of a basal pol II promoter containing only a TATA box and an Inr. This promoter is equally active in reactions containing either TBP or TFIID, the other general transcription factors, and pol II, indicating that the TAFs are not required for CMV transcription (26). When a mixture of biochemically purified and recombinant general transcription factors and pol II (see Materials and Methods and reference 10) was incubated with the purified kinase, specific transcription was repressed (Fig. 3A; compare lanes 1 and 2). cdc2-mediated repression with this promoter was observed in reactions containing either purified TFIID (Fig. 3A) or recombinant TBP (27). This indicates that the TAFs are unlikely to be involved in repression of CMV transcription in this system. To identify the target of the kinase mediating repression, each of the highly purified transcription factors and pol II were added back to separate kinase-inhibited reactions (lanes 3 to 8). Only the fraction that contained factors TFIIF and TFIIH restored activity to a level similar to that of the control reaction. cdc2 treatment resulted in greater than 75% inhibition of transcription (compare lanes 1 and 2), and addition of the TFIIF-TFIIH fraction restored transcription activity to 65% of the control value (compare lanes 1 and 5). This suggests that inactivation of either TFIIF or TFIIH might be responsible for repression of CMV transcription by the cdc2 kinase. However, addition of TFIIE also partially restored transcription in this experiment (lane 6). Partial rescue with TFIIE might reflect an involvement of this factor in repression or the limitation of the quantity of this factor in the original transcription mixture. Curiously, addition of an excess of either TFIID or pol II reduced transcription to levels below that of the cdc2-inhibited reaction (compare lanes 4 and 8 with lane 2); this is likely due to the presence of an excess of these components in the reaction mixture, which might sequester factors required for the formation of transcription complexes.
FIG. 3.
cdc2/cyclin B kinase-mediated repression of basal transcription of the CMV MIEP. (A) The TFIIF-TFIIH fraction rescues kinase-mediated repression of CMV transcription. Reactions contained a mixture of transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, pol II, unlabeled NTPs, and [α-32P]CTP, as described in Materials and Methods, and (where indicated) 1 μl of the affinity-purified cdc2/GST-cyclin B kinase was added, followed by a 30-min incubation at ambient temperature, prior to the addition of the template DNA. Transcription factors or polymerase was added back to separate reactions as indicated above lanes 3 to 8. (B) cdc2 kinase inactivation of TFIIH. Each of the indicated factors was treated with 2 μl of cdc2/GST-cyclin B kinase and 0.2 mM ATP for 30 min at ambient temperature, and each of the nonphosphorylated factors or pol II was added back individually to separate reactions (except for the individual phosphorylated factor or polymerase in each reaction). Transcription with the CMV MIEP guanineless (G−) template was performed as described in Materials and Methods. Below each lane is shown the relative level of transcription (average value from three individual experiments ± standard deviation [SD]) as determined by phosphorimage analysis of the dried gel. Lanes 1, control reaction without added kinase; lanes 2, reaction products obtained when no protein was added back after the cdc2 kinase treatment.
To further establish which factor(s) is involved in mitotic repression, we incubated each of the factors (and pol II) individually with the purified cdc2/GST-cyclin B kinase and, after inhibition of kinase activity, added each of the treated factors to reaction mixtures containing the remaining untreated reaction components. Significant inhibition of transcription was observed in this experiment only with cdc2 treatment of TFIIH (Fig. 3B, lane 8). The relative levels of transcripts produced in each reaction were determined by storage phosphorimage analysis, and these values are shown below the autoradiogram in Fig. 3B. Transcription levels (and standard deviations for three independent experiments) are expressed relative to those of the control reaction, in which none of the reaction components was exposed to the kinase (lane 2). In this experiment, individual TFIIF and TFIIH fractions were used (10), and the results suggest that mitotic repression of basal pol II transcription is due, at least in part, to phosphorylation of a subunit(s) of TFIIH.
Factors involved in repression of an activated pol II promoter.
We chose HIV-1 as an example of a complex promoter containing both core promoter elements (a TATA box and an Inr or start site region) and upstream activator sites (three tandem Sp1 binding sites). To determine whether TFIIH is also involved in mitotic repression of HIV-1 transcription, an add-back experiment was performed in which the HeLa cell nuclear extract was first treated with cdc2 kinase in the presence of ATP to effect inhibition (Fig. 4A, lane 2) and the kinase was then inactivated by the addition of p21 (lanes 5 and 6). As before (Fig. 2B), p21 did not restore full activity to the kinase-treated reaction (lane 5). Addition of biochemically purified TFIIH restored HIV-1 transcription to ∼60% of the level of the control reaction (lane 6). In the absence of p21, TFIIH was unable to restore activity (lane 4), suggesting that the added TFIIH was rapidly inactivated by the kinase. As expected from the results reported by Segil et al. (40), addition of TFIID, but not recombinant TBP, to the kinase-inhibited nuclear extract also restored HIV-1 transcription to control levels (data not shown). We discuss below possible interpretations for this finding.
FIG. 4.
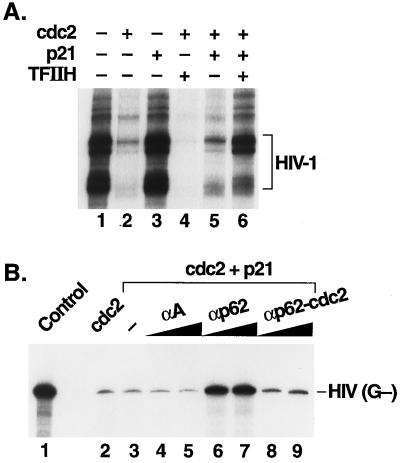
Rescue of cdc2-mediated repression of HIV-1 transcription with TFIIH. (A) Biochemically purified TFIIH rescues cdc2 repression in HeLa cell nuclear extract. Runoff transcription was monitored with EcoRI-digested pCAT-HIV-1 DNA. HeLa cell nuclear extract was treated with affinity-purified cdc2 kinase (1 μl) for 30 min in the reactions shown in lanes 2 and 4 to 6. p21 was added to a final concentration of 2.5 μM in the reactions shown in lanes 3, 5, and 6. TFIIH (200 ng) was added to the reactions shown in lanes 4 and 6. (B) Immunopurified TFIIH rescues cdc2-mediated repression. Transcription reactions with the HIV-1 promoter-enhancer linked to a guanineless (G−) cassette were performed with HeLa cell nuclear extract (lane 1, control) or with nuclear extract which had been treated with cdc2 kinase as described in the legend to Fig. 1 (lanes 2 to 9). After 30 min at ambient temperature, p21 was added to a final concentration of 2 μM (lanes 3 to 9), and after an additional 15-min incubation, the reaction mixtures were transferred to microcentrifuge tubes containing immunoprecipitates from the HeLa cell nuclear extract prepared with the indicated antibodies coupled to protein A-Sepharose beads (as described in Materials and Methods). The immunoprecipitates used in the reactions shown in lanes 8 and 9 were from HeLa cell extract pretreated with cdc2 kinase. The reactions shown in lanes 4, 6, and 8 contained 5 μl of packed beads, while the reactions shown in lanes 5, 7, and 9 contained 10 μl of packed beads. After an additional 15-min incubation, the DNA template was added and transcription was monitored as described in Materials and Methods. −, no beads; αA, anti-TFIIIA antibody and control extract; αp62, anti-p62 antibody and control extract; αp62-cdc2, anti-p62 antibody and cdc2-treated extract.
TFIIH was next isolated from both control and cdc2-treated HeLa cell nuclear extracts by immunopurification with an antibody to the p62 subunit of TFIIH (31). Immunopurified TFIIH was then tested for rescue of cdc2-mediated transcriptional repression as described above (Fig. 4B). As expected, we found that TFIIH isolated from the control extract (lanes 6 and 7), but not that from the cdc2-treated extract (lanes 8 and 9), was able to rescue cdc2-inhibited transcription (lanes 2 and 3). Similarly, mock-immunopurified TFIIH (prepared with an antibody to the 5S rRNA gene transcription factor TFIIIA) was unable to rescue transcriptional activity (lanes 4 and 5). These data support the finding that TFIIH transcriptional activity is repressed through the action of the cdc2 kinase (Fig. 3B).
Identification of TFIIH polypeptides phosphorylated by mitotic kinases.
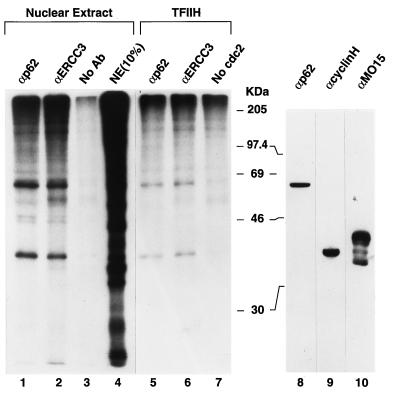
Protein labeling experiments were performed to identify the potential targets of mitotic phosphorylation in TFIIH. TFIIH is a multisubunit protein, with several subunits being involved in transcription as well as in DNA repair (ERCC2 and ERCC3) and protein phosphorylation (cdk7, cyclin H, and MAT1) (for reviews, see references 7 and 47). The HeLa cell nuclear extract was subjected to phosphorylation by [γ-32P]ATP, the cdc2/cyclin B kinase, and related mitotic kinases, immobilized on agarose beads (containing the yeast p13suc1 gene product [16]) (Fig. 5, lanes 1 to 4). After removal of the beads by a brief centrifugation, TFIIH was isolated from the nuclear extract by immunoprecipitation with antibodies to the subunits of TFIIH, and the labeled polypeptides were visualized by SDS-PAGE and autoradiography. Lane 4 of Fig. 5 contained an aliquot of the labeled nuclear extract prior to immunoprecipitation (denoted 10% NE), and the profiles of phosphoproteins present in immunoprecipitates with antibodies to the p62 and ERCC3 subunits of TFIIH are shown in lanes 1 and 2, respectively. As a control, a mock immunoprecipitation reaction was carried out with protein A-Sepharose beads but no antibody (lane 3). Similar patterns of labeled polypeptides were observed in both of the TFIIH immunoprecipitates; two major labeled species with apparent molecular masses of 36 and 62 kDa, respectively, were observed. A similar set of polypeptides were found to be substrates for in vitro phosphorylation when biochemically purified TFIIH (10) was incubated with the immobilized kinase (lanes 5 and 6). These labeled polypeptides were not observed for reactions lacking the kinase (lane 7), demonstrating that labeling of these polypeptides is not due to autophosphorylation of TFIIH. To identify the labeled polypeptides of TFIIH, a Western blot of the nuclear extract was probed with antibodies to TFIIH subunits (lanes 8 to 10). The labeled polypeptides migrated to the positions of the p62 and cyclin H subunits of TFIIH. Given the similar molecular masses of cdk7, cyclin H, and MAT1, peptide mapping will be required for an unambiguous identification of p36.
FIG. 5.
TFIIH polypeptides are phosphorylated in vitro by mitotic kinases. HeLa cell nuclear extract (10 μg of protein per 20-μl reaction volume) was treated with mitotic kinases bound to p13-agarose beads (20 μl per reaction) and [γ-32P]ATP for 1 h at ambient temperature. After a brief centrifugation to remove the agarose beads, the reaction volume was brought to 100 μl with PBS and TFIIH was immunoprecipitated with protein A-Sepharose beads (25 μl per reaction) prebound with 2.5 μl of rabbit polyclonal serum to the p62 (lane 1) and ERCC3 (lane 2) subunits of TFIIH (αp62 and αERCC3, respectively) or no antibody (No Ab) (lane 3). The immunoprecipitates were washed three times with 150 μl of PBS containing 0.1% (vol/vol) Triton X-100, and precipitated proteins were eluted from the Sepharose beads with SDS sample buffer. Lane 4 shows a 1/10-volume aliquot of the labeled nuclear extract proteins (NE) prior to immunoprecipitation. Biochemically purified TFIIH (600 ng of protein) was phosphorylated with p13-bound kinase, as described above, and after removal of the kinase, TFIIH was immunoprecipitated with anti-p62 antibody (lane 5) or anti-ERCC3 antibody (lane 6) and analyzed as described above. Autophosphorylation of TFIIH is shown in lane 7. The autoradiogram of an SDS-10% PAGE gel is shown. A Western blot of the HeLa cell nuclear extract was probed with the indicated antibodies to TFIIH (anti-p62, anti-cyclin H, and anti-MO15) subunits (lanes 8 to 10, respectively). The positions of molecular mass markers (in kilodaltons) are indicated.
pol II CTD kinase activity of TFIIH is inactivated by the cdc2 kinase.
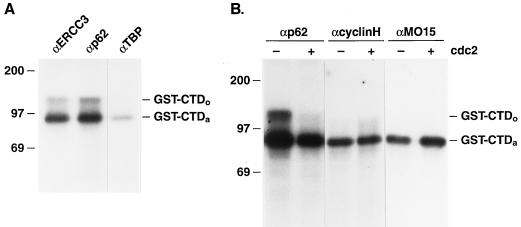
Several studies have documented that the cdk7/cyclin H kinase activity associated with TFIIH is able to phosphorylate the carboxy-terminal domain (CTD) of the largest subunit of pol II (4, 7, 54). We have used a GST-CTD fusion protein (35) as a test substrate for phosphorylation with immunopurified TFIIH (Fig. 6). Previous studies have demonstrated that the heptapeptide repeats within the CTD of this GST fusion protein (rather than the GST domain) are phosphorylated in vitro (35). We found that TFIIH, immunopurified with antibodies to either the ERCC3 or p62 subunit of TFIIH, was capable of phosphorylating this test substrate, giving rise to both the hypophosphorylated (CTDa) and hyperphosphorylated (CTDo) forms (Fig. 6A). As a control, immunoprecipitates from the HeLa cell nuclear extract, prepared with an antibody to TBP, showed only very low levels of CTD phosphorylation. We next asked what effect cdc2 kinase treatment of the nuclear extract might have on the CTD kinase activity of TFIIH. HeLa cell nuclear extract was treated as before with cdc2 kinase, and TFIIH was isolated by immunopurification with antibody to the p62 subunit. Figure 6B shows that cdc2 treatment reduced the total CTD kinase activity of TFIIH approximately threefold and reduced CTDo kinase activity approximately fivefold (ranging from four- to sevenfold in four independent experiments). Similar results were obtained with TFIIH immunopurified with an antibody to the p89 ERCC3 subunit (data not shown). Since cdk7/cyclin H has been shown to exist in TFIIH-associated and nonassociated forms (42), we wished to know whether the total nuclear cdk7 activity was affected by cdc2 treatment of the extract. To this end, we immunopurified total nuclear cdk7/cyclin H with antibodies to each of these subunits and tested these immunoprecipitates for CTD kinase activity (Fig. 6B). Immunopurified cdk7/cyclin H isolated with antibodies to either the cyclin H or cdk7 (MO15) subunit was able to phosphorylate the CTD test substrate, albeit at with lower CTD kinase activity than anti-p62- or anti-ERCC3-immunopurified TFIIH (42). Additionally, and in agreement with previous studies (52), only the hypophosphorylated (CTDa) form was observed with total cdk7/cyclin H immunoprecipitates. More significantly, kinase activity in the anti-cyclin H and anti-MO15 immunoprecipitates was not affected by cdc2 treatment of the nuclear extract prior to immunopurification.
FIG. 6.
CTD phosphorylation using immunopurified proteins with antibodies to TFIIH subunits. (A) Antibodies to TFIIH subunits ERCC3 (p89) (αERCC3) and p62 (αp62) or to TBP (αTBP) were used for immunoprecipitation from the HeLa cell nuclear extract. These immunoprecipitates were then used in kinase assays to phosphorylate GST-CTD, resulting in 32P-labeled GST-CTDo and GST-CTDa as indicated. Molecular mass markers (in kilodaltons) are indicated to the left. (B) Antibodies to TFIIH subunit p62, (αp62), cyclin H, (αcyclinH), or MO15 (cdk7) (αMO15) were used to immunopurify proteins from untreated (−) or cdc2-treated (+) HeLa cell nuclear extract. Kinase assays were performed with GST-CTD as a substrate.
To determine whether the observed repression of TFIIH-associated CTD kinase activity is due to the enzymatic action of cdc2/cyclin B, we asked whether the cdk inhibitor p21 would block cdc2-mediated inactivation of TFIIH CTD kinase (as it did for transcription [Fig. 2B]). As controls for this experiment, Fig. 7A shows that p21 is a potent inhibitor of cdc2 activity at concentrations of 1 μM or higher (using histone H1 as a substrate), and Fig. 7B shows that similar amounts of p21 do not inhibit (but rather actually stimulate) TFIIH-associated CTD kinase activity. This stimulation could be due to p21 inhibition of cdc/cdk kinases present in the HeLa cell nuclear extract. Thus, p21 inhibits cdc2/cyclin B activity but not cdk7/cyclin H activity, as previously described (15), making p21 an ideal reagent with which to assay the effects of cdc2 on transcription and cdk7 activity of TFIIH. Most significantly, the inhibition of TFIIH-associated CTD kinase by cdc2 treatment of the nuclear extract is reversed by p21 (Fig. 7C) at p21 concentrations that inhibit cdc2 activity (Fig. 7A). This experiment strongly suggests that it is the protein kinase activity of cdc2 that is responsible for inhibition of the TFIIH-associated CTD kinase.
TFIIH restores transcriptional activity to the mitotic extract.
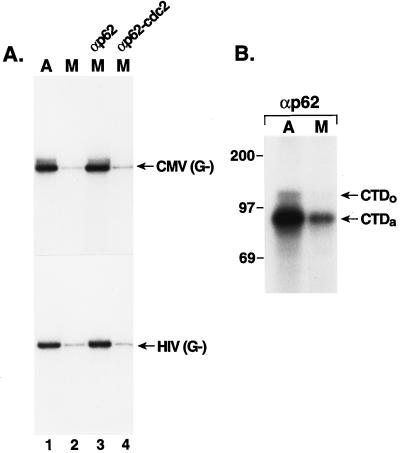
To establish whether the low level of transcription observed with the nocodazole-arrested mitotic cell extract, compared to that of the asynchronous cell extract (Fig. 1B), is due to inhibition of TFIIH activity, we performed an add-back experiment with TFIIH isolated by immunoprecipitation from the asynchronous cell nuclear extract (Fig. 8A). As before (Fig. 1B), the transcriptional activity of the mitotic extract is reduced, compared to that of the asynchronous cell extract, for both the CMV MIEP and the HIV-1 promoter (compare lanes 1 and 2 of Fig. 8A). Importantly, TFIIH, which was immunopurified from the asynchronous HeLa cell nuclear extract, restores transcriptional activity to the mitotic cell extract for both the CMV and HIV-1 promoters (lane 3). We found that the cyclin-dependent kinase inhibitor p21 had to be added to the nocodazole-arrested mitotic extract for rescue of transcription by TFIIH to be observed; if p21 was omitted from these reactions, TFIIH did not restore transcriptional activity (data not shown). This suggests that in the absence of p21, TFIIH is inactivated by a p21-sensitive cyclin-dependent kinase in the mitotic extract. When TFIIH was immunopurified from the cdc2/cyclin B-treated HeLa cell extract, rescue of the transcriptional activity of the mitotic extract was not observed (lane 4), again demonstrating inactivation of TFIIH by cdc2/cyclin B. To ensure that these results did not reflect TFIIH being a limiting component in the cell extracts, we added back immunopurified TFIIH to the asynchronous cell extract; no significant differences in the activities of several promoters (CMV, HIV-1, and Ad ML) were observed upon this addition, suggesting that TFIIH is not a limiting component in the cell extracts (data not shown).
FIG. 8.
TFIIH transcriptional activity and TFIIH-associated CTD kinase activity are repressed in the mitotic extract. (A) Transcription of the CMV MIEP or the HIV-1 promoter was monitored (as guanineless cassettes) with equivalent amounts of asynchronous (A) (lane 1) or mitotic (M) (lane 2) cell extract. p21 was added to these reaction mixtures to a final concentration of 1 μM. In lane 3, the mitotic extract was supplemented with TFIIH isolated from asynchronous HeLa cell nuclear extracts by immunoprecipitation with antibody to the p62 subunit of TFIIH. In lane 4, TFIIH was isolated from the cdc2/cyclin B kinase-treated HeLa cell extract and added to the mitotic extract. (B) The CTD kinase activity of TFIIH from either the asynchronous (A) or mitotic (M) H9 cell extract, isolated by immunoprecipitation with antibody to the p62 (αp62), was monitored with [γ-32P]ATP and GST-CTD as the test substrate. The positions of 32P-labeled GST-CTDo and GST-CTDa are shown.
TFIIH-associated CTD kinase activity is repressed in the mitotic extract.
TFIIH was immunopurified from both the nocodazole-arrested mitotic extract and the asynchronous cell extract for phosphorylation experiments with GST-CTD as the test substrate. Similar to the transcription results (Fig. 8A), the TFIIH-associated CTD kinase activity was repressed in the mitotic cell extract, compared to the activity in the asynchronous cell extract (Fig. 8B; compare lanes A and M). This result is similar to the repression of TFIIH-associated kinase activity found in the cdc2/cyclin B-treated HeLa cell nuclear extract (Fig. 6B).
DISCUSSION
We have shown that mitotic repression of pol II transcription can be reproduced in vitro with extracts prepared from cells arrested at mitosis with the microtubule polymerization inhibitor nocodazole or with nuclear extracts prepared from asynchronous cells converted to the mitotic state with the cdc2/cyclin B mitotic kinase. We find that several cellular and viral promoters are inhibited in these systems, including both TATA-containing and TATA-less promoters (Fig. 2A). Strikingly, however, the Ad MLP is not inhibited in our in vitro systems; we discuss the implications of this observation below. We showed that inhibition of transcription by the cdc2 kinase is due to protein phosphorylation, since the cyclin-dependent kinase inhibitor p21 will block transcription inhibition by the kinase (Fig. 2B). It is well established that cdc2/cyclin B can activate other protein kinases (reviewed in reference 16); thus, we cannot exclude the possibility that inhibition in our in vitro systems was due to the action of a secondary kinase(s) present in both the nuclear extract (Fig. 2) and the reconstituted transcription system (Fig. 3), which is activated by the cdc2/GST-cyclin B kinase. This possibility seems unlikely, however, since direct phosphorylation of TFIIH by the cdc2/GST-cyclin B kinase is sufficient to repress transcription (Fig. 3B).
Studies in several systems have shown that many gene-specific and basal transcription factors are displaced from the chromatin template during mitosis (18, 19, 30, 32), and a recent study found that the majority of the basal factor TFIID is relocalized from the interphase nucleus to the cytoplasm in late mitotic prophase (40). Further, Segil et al. (40) reported that the TFIID TAFs isolated from mitotic cells are highly phosphorylated and that mitotic TFIID is inactive in supporting activator-dependent transcription. Indeed, the results of our current in vitro studies with the HIV-1 promoter are entirely consistent with the findings of Segil et al. We found that TFIID, but not TBP, could restore HIV-1 transcription after cdc2 treatment of a nuclear extract, suggesting that a TAF(s) is the likely target for cdc2-mediated repression of this viral promoter. On the basis of previous findings indicating that a TBP-associated component of the pol III transcription factor TFIIIB is inactivated at mitosis (13, 49), it is not surprising that TFIID activity is regulated in a similar fashion. Segil et al. (40) reported that basal transcription, unlike activator-dependent transcription, was not affected by mitotic phosphorylation of TFIID by more than 50%. Thus, our present results, identifying TFIIH as a potential target for mitotic repression of transcription at mitosis, are not in conflict with the results of Segil et al. and suggest that multiple targets for mitotic repression exist in the basal transcription machinery. It is curious, however, that addition of either TFIID or TFIIH can restore activity to the HIV-1 promoter. One plausible explanation for this finding is that both TFIID and TFIIH are at limiting concentrations in the original HeLa cell nuclear extract and that addition of either factor to the kinase-inhibited extract will stimulate transcription. Careful titration studies are needed to assess this possibility.
We have used a simplified pol II transcription system consisting of only the basal transcription factors, pol II, and the CMV MIEP as an example of a basal promoter (10, 26). This template contains only a TATA box and an Inr and will support only basal transcription. Indeed, we found equivalent template activity with this promoter element whether we used the unfractionated HeLa cell nuclear extract (Fig. 2A) or reconstituted transcription reactions containing either recombinant TBP or biochemically isolated TFIID (Fig. 3; see also references 26 and 27). Additionally, transcription reactions containing either TBP or TFIID were inhibited to the same extent with the cdc2/cyclin B kinase (Fig. 3 and reference 27), providing evidence that the TAFs of TFIID are not involved in mitotic repression of this promoter. Transcription rescue and direct phosphorylation experiments suggest that the basal transcription factor IIH is the target of the cdc2/cyclin B kinase mediating repression in this system. We established that TFIIH is inactivated at mitosis in vivo by adding back TFIIH (isolated from an asynchronous cell extract) to the mitotic extract, with the result that the transcriptional activity of the mitotic extract was restored to the level of the asynchronous cell extract (Fig. 8A).
Now that TFIIH has been identified as a target of mitotic repression by the cdc2/cyclin B kinase, several questions must be addressed: namely, which subunits of TFIIH are phosphorylated at mitosis in vivo and which phosphorylation event(s) leads to inactivation of TFIIH activity. Our current protein labeling experiments demonstrate that two subunits of TFIIH, p36 and p62, are in vitro substrates for phosphorylation by mitotic kinases. Western blotting experiments suggest that these phosphoproteins correspond to the p62 and cyclin H subunits of TFIIH; however, given the similar molecular masses of cdk7, cyclin H, and MAT1, it is conceivable that p36 corresponds to MAT1 or cdk7 rather than to cyclin H. Future peptide mapping experiments with recombinant proteins are needed for an unambiguous assignment of the identity of p36. The role of the p62 subunit of TFIIH in transcription has not been established; however, the deduced sequence of p62 has one potential site for cdc2 phosphorylation (SPEGK, at amino acid positions 56 to 60 [9]). The observation that free nuclear cdk7/cyclin H activity is not affected by cdc2 phosphorylation, whereas the TFIIH-associated cdk7 activity is repressed by cdc2 treatment (Fig. 6B), suggests that phosphorylation of p62 may be responsible for repression of both the cdk7 kinase activity and the transcriptional activity of TFIIH. Future studies will determine the phosphorylation site(s) on p62 and will indicate whether this phosphorylation leads to inactivation of TFIIH at mitosis.
Our results clearly show that the cdk7 kinase activity associated with TFIIH is down-regulated in nocodazole-arrested mitotic H9 cell extracts and in cdc2-treated HeLa cell nuclear extracts. In contrast, a previous report from Egly and coworkers (1) showed that the CTD kinase activity of TFIIH did not change during the cell cycle in JEG-3 cells in culture. There are numerous technical differences between our work and the methods used by Adamczewski et al. (1). Most notably, these authors used a ctd4 peptide (containing only four heptapeptide repeats) as a test substrate, whereas in our experiments we used the entire pol II CTD linked to GST. Thus, hyperphosphorylation was not assayed by these authors. When using antibodies which immunoprecipitate cdk7 as part of intact TFIIH, such as anti-p89 or anti-p62, we have found that cdk7 hyperphosphorylation of the CTD is most significantly repressed in our mitotic extracts (Fig. 6B). When antibodies such as anti-cyclin H or anti-cdk7/MO15, which immunoprecipitate primarily free cdk7/cyclin H, are used, no CTD hyperphosphorylation is observed. This is consistent with previous studies showing that free cdk7/cyclin H (with or without MAT1) cannot hyperphosphorylate the CTD (52). Taken together, these results indicate that the CTD kinase activity of TFIIH-associated cdk7 is reduced in the mitotic extracts we prepare from the H9 lymphoid cell line.
Since TFIIH also possesses DNA helicase activity (39, 47), we investigated whether this activity of TFIIH is affected at mitosis. We attempted to use in this experiment immunopurified TFIIH isolated from the asynchronous and mitotic H9 cell extracts; however, in our hands, TFIIH isolated with any one of several polyclonal antibodies to the TFIIH subunits (anti-p62, anti-p34, anti-cyclin H, anti-cdk7/MO15, and anti-p89) exhibited extremely low levels of helicase activity in a standard assay (39), making any results virtually impossible to interpret or quantitate. However, these same preparations of immunopurified TFIIH exhibited high levels of transcriptional activity and cdk7 kinase activity. These findings suggest that the helicase activity of TFIIH is not involved in the mechanism of transcriptional repression at mitosis.
A current view of the pol II transcription cycle involves cycling of the pol II CTD between the nonphosphorylated and phosphorylated forms (5, 54). The nonphosphorylated form of pol II enters the preinitiation complex, and it is the cdk7/cyclin H kinase activity of TFIIH that is proposed to phosphorylate the CTD. Phosphorylation of the CTD then releases pol II from the initiation complex by disruption of specific protein-protein interactions (perhaps the CTD-TBP interaction [29, 48]), leading to RNA chain elongation. After termination, the CTD must be dephosphorylated to allow pol II to reenter the transcription cycle. One attractive model for repression of basal transcription by TFIIH phosphorylation is one in which this phosphorylation event inactivates the TFIIH-associated cdk7 kinase, thus trapping pol II in the nonphosphorylated form. Transcription complexes would be frozen in an inactive state until mitosis is completed and TFIIH is dephosphorylated as the cell enters the G1 phase of the cell cycle. Other studies have implicated CTD phosphorylation in mitotic repression of transcription: the pol II CTD can be directly phosphorylated in vitro by the cdc2/cyclin B kinase or related mitotic kinases (3, 54), and Reinberg and collaborators (54) have shown that phosphorylation of the CTD by the cdc2/cyclin B kinase disrupts the association of the polymerase with other components of the basal transcription machinery, namely TFIIB, TFIID, and TFIIF. These authors speculate that this phosphorylation and complex dissociation might be involved in mitotic repression of transcription. Recently, Corden and colleagues (11) reported that direct phosphorylation of the CTD by either cdc2 kinase or the TFIIH-associated cdk7 kinase inhibits transcription by pol II. These findings are consistent with the model mentioned above in which only the hypophosphorylated (CTDa) form of pol II can enter the preinitiation complex. We have found, however, that direct phosphorylation of pol II with the cdc2 kinase does not lead to repression in our reconstituted transcription system (Fig. 3B). An explanation for this discrepancy was provided by Corden and colleagues (11); these authors found that pol IIo was rapidly dephosphorylated in their extracts if ATP was used as the phosphate donor and that the polymerase could only be maintained in the hyperphosphorylated state (pol IIo) by using adenosine 5′-O-(3-thiotriphosphate) in place of ATP. Since we used ATP in our phosphorylation reactions (Fig. 3B), pol II might have been dephosphorylated and reactivated by phosphatases in our reconstituted transcription system. In another recent study (23), the level of total cellular hyperphosphorylated pol IIo was found not to vary during the cell cycle; however, the phosphorylation state of template-bound pol II was not determined in that study.
With regard to the lack of repression of Ad MLP transcription by the cdc2 kinase, numerous explanations for this observation can be suggested. First, several lines of evidence indicate that CTD phosphorylation may not be an obligatory step in transcription of the Ad MLP (41), and therefore lack of inhibition could be related to the mitotic inhibition of the CTD kinase activity of TFIIH. In general, it has been shown that transcription is not dependent on an intact CTD in purified systems (2, 24, 55), while the CTD is required in crude systems such as cell extracts (28). However, this requirement for CTD can be promoter dependent, and some promoters, such as the Ad MLP, do not require the CTD or its phosphorylation (31, 47). Since the Ad MLP does not appear to require CTD phosphorylation for its activity, a repression mechanism which involves CTD phosphorylation would be predicted not to affect this promoter. Conversely, other studies have suggested that the Ad MLP does require the CTD (28). Therefore, other explanations, such as promoter strength and/or template conformation and factor requirements for Ad MLP transcription (34), might also account for the lack of inhibition of the Ad MLP in our in vitro system. Supercoiled Ad MLP templates, such as those used in our experiments, have been shown not to require TFIIH in purified transcription assays (34). Future studies are needed to elucidate the molecular mechanisms by which cdc2 phosphorylation of components of the basal transcription machinery and/or polymerase leads to repression of transcription in vivo and in reconstituted in vitro systems.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health (GM26453) to J.M.G. R.W.K. is a fellow of the Leukemia Society of America.
We thank Osvaldo Flores, Peter Ghazal, Jean-Marc Egly, Phil Sharp, Michael Dahmus, and William Dynan for their gifts of plasmids and reagents, Rick Gulizia for the H9 cell line and technical assistance, and Peter Ghazal for numerous discussions.
REFERENCES
- 1.Adamczewski J P, Rossignol M, Tassan J-P, Nigg E A, Moncollin V, Egly J-M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Buermeyer A B, Thompson N E, Strasheim L A, Burgess R R, Farnham P J. The HIP1 initiator element plays a role in determining the in vitro requirement of the dihydrofolate reductase gene promoter for the C-terminal domain of RNA polymerase II. Mol Cell Biol. 1992;12:2250–2259. doi: 10.1128/mcb.12.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cisek L J, Corden J L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- 4.Dahmus M E. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- 5.Dahmus M E. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 6.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eucaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 7.Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 8.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 9.Fischer L, Gerard M, Chalut C, Lutz Y, Humbert S, Kanno M, Chambon P, Egly J-M. Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science. 1992;257:1392–1395. doi: 10.1126/science.1529339. [DOI] [PubMed] [Google Scholar]
- 10.Flores O, Lu H, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 11.Gebara M M, Sayre M H, Corden J L. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J Cell Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- 12.Gottesfeld J M, Forbes D J. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 13.Gottesfeld J M, Wolf V J, Dang T, Forbes D J, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- 14.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 cdk-interacting protein cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 15.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl P, Gottesfeld J M, Forbes D J. Mitotic repression of transcription in vitro. J Cell Biol. 1993;120:613–624. doi: 10.1083/jcb.120.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 18.Hershkovitz M, Riggs A D. Metaphase chromosome analysis by ligation-mediated PCR: heritable chromatin structure and a comparison of active and inactive X chromosomes. Proc Natl Acad Sci USA. 1995;92:2379–2383. doi: 10.1073/pnas.92.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu S C, Qi M, DeFranco D B. Cell cycle regulation of glucocorticoid receptor function. EMBO J. 1992;11:3457–3468. doi: 10.1002/j.1460-2075.1992.tb05425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 21.Jackson S P. Regulating transcription factor activity by phosphorylation. Trends Cell Biol. 1992;2:104–108. doi: 10.1016/0962-8924(92)90014-e. [DOI] [PubMed] [Google Scholar]
- 22.Johnson T C, Holland J J. Ribonucleic acid and protein synthesis in mitotic HeLa cells. J Cell Biol. 1965;27:565–574. doi: 10.1083/jcb.27.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E, Du L, Bregman D B, Warren S L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W Y, Dahmus M. The major late promoter of adenovirus-2 is accurately transcribed by RNA polymerases IIO, IIA, and IIB. J Biol Chem. 1989;264:3169–3176. [PubMed] [Google Scholar]
- 25.Kriwacki R W, Hengst L, Tennant L, Reed S I, Wright P E. Structural studies of p21waf1/cip1/sdi1 in the free and cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci USA. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee G, Wu J, Luu P, Ghazal P, Flores O. Inhibition of the association of RNA polymerase II with the preinitiation complex by a viral transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:2570–2574. doi: 10.1073/pnas.93.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leresche A, Wolf V J, Gottesfeld J M. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Kornberg R D. Interplay of positive and negative effectors in function of the C-terminal repeat domain of RNA polymerase II. Proc Natl Acad Sci USA. 1994;91:2362–2366. doi: 10.1073/pnas.91.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luscher B, Eisenman R N. Mitosis-specific phosphorylation of the nuclear oncoproteins Myc and Myb. J Cell Biol. 1992;118:775–784. doi: 10.1083/jcb.118.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mäkelä T P, Parvin J D, Kim J, Huber L J, Sharp P A, Weinberg R A. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez-Balbás M A, Dey A, Rabindran S K, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 34.Parvin J D, Timmers H T, Sharp P A. Promoter specificity of basal transcription factors. Cell. 1992;68:1135–1144. doi: 10.1016/0092-8674(92)90084-p. [DOI] [PubMed] [Google Scholar]
- 35.Peterson S R, Dvir A, Anderson C W, Dynan W S. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 36.Prescott D M, Bender M A. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- 37.Proudfoot N J, Shander M H, Manley J L, Gefter M L, Maniatis T. Structure and in vitro transcription of human globin genes. Science. 1980;209:1329–1336. doi: 10.1126/science.6158093. [DOI] [PubMed] [Google Scholar]
- 38.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H J, Chambon P, Egly J-M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 40.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 41.Serizawa H, Conaway R C, Conaway J W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor δ from rat liver. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheikhatter R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 43.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 44.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 45.Smythe C, Newport J W. Coupling of mitosis to the completion of S phase in Xenopus occurs via modulation of the tyrosine kinase that phosphorylates p34cdc2. Cell. 1992;63:787–797. doi: 10.1016/0092-8674(92)90153-4. [DOI] [PubMed] [Google Scholar]
- 46.Solomon M J, Glotzer M, Lee T H, Philippe M, Kirschner M W. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- 47.Svejstrup J Q, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 48.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 49.White R J, Gottlieb T M, Downes C S, Jackson S P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf V J, Dang T, Hartl P, Gottesfeld J M. Role of maturation-promoting factor (p34cdc2-cyclin B) in differential expression of the Xenopus oocyte and somatic-type 5S RNA genes. Mol Cell Biol. 1994;14:4704–4711. doi: 10.1128/mcb.14.7.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 52.Yankulov K Y, Bentley D L. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zang H, Hannon G J, Casso D, Beach D. p21 is a component of active cell cycle kinases. Cold Spring Harbor Symp Quant Biol. 1994;59:21–29. doi: 10.1101/sqb.1994.059.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Zawel L, Lu H, Cisek L J, Corden J L, Reinberg D. The cycling of RNA polymerase II during transcription. Cold Spring Harbor Symp Quant Biol. 1993;58:187–198. doi: 10.1101/sqb.1993.058.01.023. [DOI] [PubMed] [Google Scholar]
- 55.Zehring W A, Lee J M, Weeks J R, Jokerst R S, Greenleaf A L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Q, Sharp P A. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]