Abstract
The t(11;22) chromosomal translocation specifically linked to Ewing sarcoma and primitive neuroectodermal tumor results in a chimeric molecule fusing the amino-terminus-encoding region of the EWS gene to the carboxyl-terminal DNA-binding domain encoded by the FLI-1 gene. As the function of the protein encoded by the EWS gene remains unknown, we investigated the putative role of EWS in RNA polymerase II (Pol II) transcription by comparing its activity with that of its structural homolog, hTAFII68. We demonstrate that a portion of EWS is able to associate with the basal transcription factor TFIID, which is composed of the TATA-binding protein (TBP) and TBP-associated factors (TAFIIs). In vitro binding studies revealed that both EWS and hTAFII68 interact with the same TFIID subunits, suggesting that the presence of EWS and that of hTAFII68 in the same TFIID complex may be mutually exclusive. Moreover, EWS is not exclusively associated with TFIID but, similarly to hTAFII68, is also associated with the Pol II complex. The subunits of Pol II that interact with EWS and hTAFII68 have been identified, confirming the association with the polymerase. In contrast to EWS, the tumorigenic EWS–FLI-1 fusion protein is not associated with either TFIID or Pol II in Ewing cell nuclear extracts. These observations suggest that EWS and EWS–FLI-1 may play different roles in Pol II transcription.
Structural alteration or aberrant expression of transcription factors is often a critical event in tumorigenic transformation (13, 19, 22). Karyotypic analysis has revealed a tumor-specific t(11;22)(q24;q12) chromosomal translocation in 86% of both Ewing sarcoma and primitive neuroectodermal tumor, suggesting that the product of this rearrangement is involved in the formation of these malignancies (34). This chromosomal translocation fuses the EWS gene on chromosome 22 to the FLI-1 gene on chromosome 11 (8). EWS is a protein with unknown function containing an RNA-binding motif and an activation domain(s) (18, 24, 25). In the EWS–FLI-1 fusion protein, the RNA-binding motif containing the C-terminal half of EWS is replaced by the DNA-binding domain (DBD) of the FLI-1 protein. FLI-1 is a member of the ETS family of transcription factors which activate specific target genes by binding to their cognate DNA sequences through their DNA-binding regions, usually located at their carboxyl termini (2, 37). The replacement of the native transcription activation domain(s) of FLI-1 by the N-terminal region of EWS converts the nontransforming activator, FLI-1, into a transforming protein with new transcriptional activation potential. In the EWS–FLI-1 fusion protein, both the N-terminal domain of EWS and the DBD of FLI-1 are necessary for the transforming activity (20). Recently, the EWS gene was also shown to be involved in tumorigenesis by chromosomal translocation with other genes encoding either other members of the ETS family (Erg, ETV1, E1A-F, and FEV) or other transcription factors, including ATF-1, WT1, and the nuclear orphan receptor TEC1 (16, 17, 27, 35).
The gene encoding human TLS/FUS, a protein that is highly similar to EWS, has also been implicated in human sarcomas induced by chromosomal translocations (7, 30). In mixoid liposarcoma, the t(12;16) translocation fuses the TLS/FUS gene to that encoding the transcription factor CHOP. The function of the intact TLS/FUS protein is also unknown. Like EWS, it contains an RNA-binding motif and an activation domain. CHOP, a member of the C/EBP family of transcription factors, is expressed usually in response to various cellular stresses and can induce growth arrest. It has been demonstrated that the fusion of the N-terminal portion of either EWS or TLS/FUS to either the DBD (FLI-1) or the dimerization domain (CHOP) of a given transcription factor leads to tumorigenic transformation (39). These results suggest that the N-terminal domains of these sarcoma-associated proteins have an important and functionally similar function in the transformation of the oncogenic cells.
Recently, we have identified and characterized a novel transcription factor, hTAFII68, that shows extensive sequence similarity with the sarcoma-associated proteins EWS and TLS/FUS (3). Like EWS and TLS/FUS, hTAFII68 contains a consensus RNA-binding domain (RNP-CS) which allows it to bind not only RNA but also single-stranded DNA (ssDNA). hTAFII68 was identified on the basis of its substoichiometric association with a distinct TFIID subpopulation. TFIID is a multiprotein complex composed of the TATA-binding protein (TBP) and TBP-associated factors (TAFIIs) and is the factor that nucleates preinitiation complex formation on protein-coding genes (31). Antibodies raised against hTAFII68 coimmunoprecipitate a fraction of TFIID, and anti-TBP or anti-TAFII100 monoclonal antibodies (MAbs) coimmunopurify hTAFII68. Moreover, hTAFII68 is associated with another multiprotein complex, the human RNA polymerase II (Pol II) complex. Interestingly, hTAFII68 is able to enter into the preinitiation complex together with Pol II, suggesting that hTAFII68 has a role in transcription initiation and/or elongation. Like hTAFII68, TLS/FUS is associated with a subpopulation of TFIID complexes that are chromatographically distinct and functionally different from those containing hTAFII68 (3, 5, 15). These experiments strongly suggested that hTAFII68 and TLS/FUS play an important role in the cross talk between various components of the basal transcription machinery and that they may function by linking transcription initiation and elongation.
Recently, a Drosophila protein, termed Cabeza (33) or SARFH (14), that has high homology to TLS/FUS and EWS has been described. TLS/FUS, EWS, hTAFII68, and Cabeza all have particularly conserved RNA-binding motifs that deviate from the organization of such domains commonly found in most RNA-binding proteins. Thus, TLS/FUS, EWS, hTAFII68, and Cabeza all belong to a new subfamily of RNP-CS-containing proteins that we have called the TET family (3). TET family members all contain an acidic residue at the second position and a threonine in the fourth position of the RNP1 domain of their RNP-CS instead of hydrophobic residues found in most other RNA-binding proteins. In addition, the RNP-CS motifs of the TET family members contain an unusually long predicted loop immediately after the first α helix (3, 6, 23). The common structural features which are limited to the TET family members suggest that they bind RNA and/or ssDNA in a unique way. Moreover, Cabeza was found to be associated with the majority of active transcription units in preparations of polythene chromosomes from salivary gland nuclei (14), further indicating that the TET family members participate in a function common to the expression of most genes transcribed by Pol II.
The transformation of Ewing cells by EWS–FLI-1 is dependent on the activity of both the EWS N-terminal domain and the FLI-1 DBD (18, 39). To assess the contribution of the N-terminal domain of the EWS protein to the formation of human solid tumors, it is important to understand the normal function(s) of EWS. The structural homology between EWS and the transcription factor hTAFII68 (70% similarity among the full-length proteins) strongly suggested that there may be a functional homology between these proteins. Thus, we investigated whether EWS and the EWS–FLI-1 fusion protein are able to interact with the same multiprotein complexes as hTAFII68. We demonstrate that EWS, like hTAFII68, is able to associate with a portion of the basal transcription factor TFIID. Using an in vitro protein-protein interaction assay, we show that both EWS and hTAFII68 interact with several subunits (TAFIIs) of the TFIID complex. EWS, similarly to hTAFII68, copurifies with the endogenous Pol II. Moreover, the subunits of the Pol II complex that interact directly with either EWS or hTAFII68 were identified, further confirming the importance of EWS and hTAFII68 in Pol II transcription. Using Ewing cell nuclear extracts (NEs), we studied the association of EWS and the oncogenic fusion protein, EWS–FLI-1, with different multiprotein complexes. These experiments suggest that EWS and EWS–FLI-1 behave differently since EWS–FLI-1 cannot stably associate with any of the targets of EWS identified to date.
MATERIALS AND METHODS
Cell lines and NEs.
Two Ewing cell lines expressing different EWS–FLI-1 chimeric transcripts were used: COH (ICB104), which expresses a fusion transcript linking exon 10 of EWS to exon 6 of FLI-1 (EWS 10/FLI 6); and RD-ES, which expresses a type II fusion transcript linking exon 7 of EWS to exon 5 of FLI-1 (EWS 7/FLI 5) (12). NEs were prepared as previously described (5).
Immunization and antibody production.
To generate the anti-EWS polyclonal antibody (PAb) 677, a peptide corresponding to amino acids 136 to 152 of the EWS protein was synthesized, coupled to keyhole limpet hemacyanin carrier protein (Neosystem Laboratories), and used for immunization of rabbits. MAbs raised against hTAFII68 (2B10), hTAFII100 (2D2), hTBP (3G3 and 2C1), the C-terminal domain (CTD) of the largest subunit of Pol II (7G5), and FLI-1 (7.3) have been described previously (3–5, 9, 15, 21).
Immunoprecipitation and Western blot analysis.
Routinely, 100 to 500 μl (approximately 500 μg) of the indicated protein fractions was immunoprecipitated with 50 μl of protein G-Sepharose (Pharmacia) and approximately 2 μg of the different antibodies (as indicated in the figure legends). Antibody-protein G-Sepharose-bound protein complexes were washed three times with immunoprecipitation buffer (25 mM Tris-HCl [pH 7.9], 10% [vol/vol] glycerol, 0.1% Nonidet P-40, 0.5 mM dithiothreitol, 5 mM MgCl2) containing 0.5 M KCl and two times with immunoprecipitation buffer containing 100 mM KCl. After washing, 20 μl of the beads was boiled in sodium dodecyl sulfate (SDS) sample buffer, and protein was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Protein samples were then transferred to a nitrocellulose membrane and probed with the indicated primary antibodies. As secondary antibodies, either peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG)-IgM (heavy plus light chain)-specific (Jackson ImmunoResearch Laboratories, Inc.) or peroxidase-conjugated goat anti-mouse κ-type light-chain-specific (Southern Biotechnology Associates, Inc.) antibody was used. Detection with an enhanced chemiluminescence kit (Amersham) was performed by standard methods.
Construction of baculovirus expression vectors for EWS, hTAFIIs, and subunits of Pol II and protein expression.
The EWS cDNA (STA ET 19 [29]) was excised from the Bluescript vector (pBSK+) by EcoRI/DraI digestion and inserted in the EcoRI/SmaI sites of the pVL1392 vector. The hTAFII68 cDNA was excised from the pBSK+ vector by BamHI/XbaI digestion and inserted in the corresponding sites of the pVL1393 vector. The other constructions encoding the different hTAFIIs or hTBP have been described previously (9). Constructions of baculovirus expression vectors for the human Pol II subunits have previously been described (1). SF9 cell infection, plaque purification, and whole-cell extract (WCE) preparation were performed as previously described (1, 26).
Expression and purification of GST fusion proteins.
The cDNAs encoding the glutathione S-transferase (GST)–hTAFII68 or GST-EWS deletion mutants were amplified by PCR using the appropriate oligonucleotides with either BamHI/XhoI or EcoRI/XhoI sites. The PCR products were digested with the appropriate restriction enzymes and inserted in frame into the corresponding sites of the pGEX-4-T3 vector (Pharmacia). All constructions were sequenced. GST fusion protein overexpression and purification were performed as previously described (32).
Protein-protein interaction assay.
GST fusion proteins (1 to 2 μg) attached to 20 μl of glutathione-agarose (Pharmacia) were incubated with 200 to 500 μl of SF9 protein extracts containing the various TAFIIs or Pol II subunits in buffer G (25 mM Tris-HCl [pH 7.3], 10% [vol/vol] glycerol, 1% Triton X-100, 1 mM dithiothreitol, 5 mM MgCl2) containing 0.5 M NaCl for 2 h at room temperature. The beads were washed three times with 1 ml of buffer G containing 1 M NaCl and once with buffer G containing 100 mM NaCl. Beads were boiled in SDS sample buffer, and protein was analyzed by SDS-PAGE. The gel was either subjected to autoradiography or transferred to a nitrocellulose filter and probed with the appropriate antibodies.
Glycerol gradients.
HeLa and RD-ES cell NE (2 mg) and high-molecular-weight markers (Pharmacia) were separately centrifuged through a 20 to 40% glycerol gradient as described previously (10, 11). Each 4-ml gradient was then fractionated into 30 140-μl fractions; 25 μl from each fraction was analyzed by Western blotting using antibodies raised against the CTD of the largest subunit of Pol II (MAb 7G5), hTAFII100 (MAb 2D2), TBP (MAb 3G3), EWS (PAb 677), hTAFII68 (MAb 2B10), EWS (PAb 677), and EWS–FLI-1 (MAb 7.3). The glycerol gradient concentration in each fraction was determined to ensure that the gradients were linear. The Western blots were quantified with a Bio-Rad densitometer.
RESULTS
Similarly to TAFII68, EWS interacts with TFIID and copurifies with Pol II.
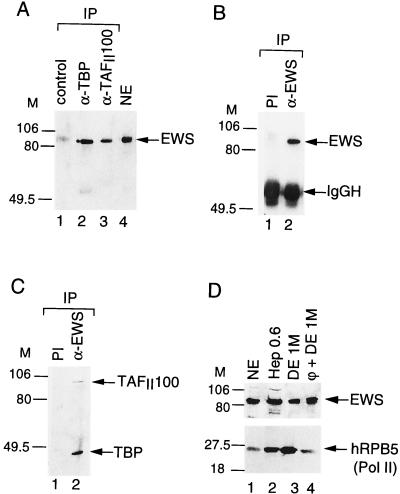
As two members of the TET family have previously been shown to be substoichiometric components of distinct TFIID complexes, we examined whether a third member of the TET family, EWS, can also associate with TFIID. TFIID complexes were immunopurified from HeLa cell NEs by using either an anti-TBP or an anti-TAFII100 MAb, and the presence of EWS in these complexes was verified by Western blot analysis using an anti-EWS PAb (Fig. 1A). While the anti-TBP and the anti-TAFII100 immunoprecipitations depleted all TBP and hTAFII100 from the NE (data not shown), about 10% of the input EWS was specifically retained in both the anti-TBP and the anti-TAFII100 immunoprecipitations but not in the control immunoprecipitation (carried out with an unrelated anti-GAL4 MAb; lane 1). Similar results were obtained when the immunoprecipitations were carried out in the presence of RNase (data not shown). These data indicate that a fraction of EWS can associate with TFIID. To further confirm the EWS-TFIID interaction and to analyze putative EWS-associated proteins, the anti-EWS PAb was tested for its ability to immunoprecipitate the nondenatured EWS protein. As shown in Fig. 1B, lane 2, the anti-EWS PAb recognized native EWS protein, since it immunoprecipitated EWS from the NE. Next we analyzed whether components of the TFIID complex would coimmunoprecipitate with EWS. Consistent with the anti-TBP and anti-TAFII100 immunoprecipitations, the anti-EWS PAb specifically coimmunoprecipitated about 5 to 10% of the input TBP and 15% of the input hTAFII100 (Fig. 1C, lane 2; see Materials and Methods). Analysis of either the EWS- or the hTAFII100-bound proteins by silver staining indicated that the association of EWS with TFIID is substoichiometric (data not shown). Together, these data demonstrate that, similarly to its structural homologs hTAFII68 and TLS/FUS, EWS can be found associated with a TFIID subpopulation in HeLa cell NEs.
FIG. 1.
EWS is associated with TFIID and copurifies with Pol II. (A) The anti-hTBP (α-TBP) and the anti-hTAFII100 (α-TAFII100) MAbs coimmunoprecipitate EWS from a HeLa cell NE. HeLa cell NE was immunoprecipitated (IP) with either an unrelated antibody (lane 1) or a MAb raised against TBP (3G3; lane 2) or hTAFII100 (2D2; lane 3). Beads were washed and boiled, and bound proteins were analyzed by Western blotting using an antibody raised against the N-terminal domain of EWS that recognizes the endogenous EWS protein in HeLa cell NE (lane 4). M, markers in kilodaltons. (B and C) The anti-EWS antibody coimmunoprecipitates components of the TFIID complex. HeLa cell NE was immunoprecipitated with either the anti-EWS PAb (lanes 2) or the preimmune serum (PI; lanes 1). Beads were washed and boiled, and bound proteins were analyzed by Western blotting using either the anti-EWS antibody (B) or the anti-TBP MAb 3G3 together with the anti-hTAFII100 MAb 2D2 (C). In panel B, the IgG heavy chain (IgGH) is indicated. (D) EWS copurifies with Pol II. The previously described chromatographic fractions obtained during the purification of Pol II (3) were tested by Western blotting using an antibody raised against either EWS (upper panel) or the fifth-largest subunit of Pol II (hRPB5; lower panel). Hep, Heparin-Ultrogel column; DE, DEAE 5PW HPLC column; φ, Phenyl-5PW HPLC column.
We have shown previously that only a fraction of the total cellular hTAFII68 is associated with TFIID and that hTAFII68 can also be found associated with Pol II (3). Thus, we analyzed whether EWS could copurify with Pol II and tested the fractions from our Pol II purification for the presence of EWS. As shown in Fig. 1D, EWS copurifies with Pol II over five chromatography columns, as determined by Western blotting using antibodies raised against EWS and the 25-kDa subunit of Pol II (hRPB5) (Fig. 1D) (3). The highly purified Pol II (lane 4) is free of other basal Pol II transcription factors and is active in transcription initiation and elongation (9). This result suggests that, like hTAFII68, EWS is tightly associated with Pol II (see also below).
Interaction of EWS and hTAFII68 with individual components of the TFIID complex.
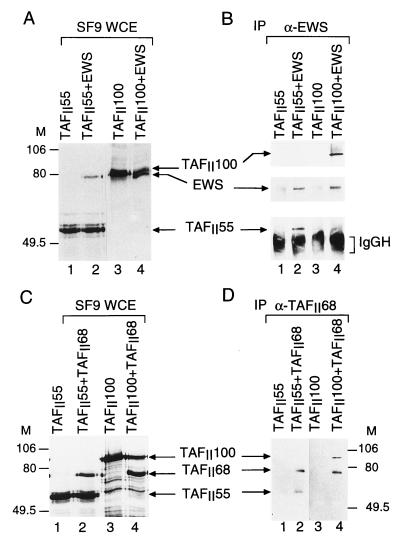
To identify the subunits of TFIID that interact directly with either EWS or hTAFII68, the TAFIIs and EWS or the TAFIIs and hTAFII68 were tested pairwise in a protein-protein interaction assay. To this end, cDNAs encoding most of the human TAFIIs and hTBP were inserted in baculovirus expression vectors (9), and each TFIID subunit was expressed either alone or together with EWS or hTAFII68 in SF9 cells. WCEs were made, and protein expression was tested (Fig. 2A and C). From these extracts, either EWS or hTAFII68 was immunoprecipitated, and bound proteins were analyzed by Western blotting (Fig. 2B and D; Table 1). Extracts in which EWS or hTAFII68 were either not expressed (Fig. 2B and D, lanes 1 and 3) or expressed alone (data not shown) served as negative controls for the immunoprecipitations. Since the overexpressed proteins in SF9 cell extracts greatly exceed (by at least 1,000-fold) the endogenous insect cell TAFII or EWS concentrations, these interaction studies indicate that both EWS and hTAFII68 bind directly to hTAFII100 (Fig. 2B and D, lanes 4), hTAFII55 (lanes 2), hTAFII28, and hTAFII18 (Table 1). The fact that EWS and hTAFII68 contact the same TAFIIs in this direct protein-protein interaction assay and that the anti-EWS PAb does not coimmunoprecipitate hTAFII68 from crude HeLa cell NE (data not shown) suggests that the presence of EWS and that of hTAFII68 in the same TFIID complex are mutually exclusive.
FIG. 2.
Interactions of EWS and hTAFII68 with other components of the human TFIID complex. SF9 cells were coinfected with recombinant baculoviruses expressing hTAFII100 and hTAFII55 either individually or pairwise with EWS (A) and hTAFII68 (C) as indicated. After 44 h of infection, proteins were radiolabeled with [α-35S]methionine and [α-35S]cysteine for 4 h. WCEs were made, proteins were separated by SDS-PAGE, and gels were dried and subjected to autoradiography. M, markers in kilodaltons. (B and D) From the protein extracts, EWS and TAFII68 were immunoprecipitated (IP) with either the anti-EWS (α-EWS) PAb (B) or the anti-hTAFII68 (α-TAFII68) MAb (D) as indicated. Resin-bound proteins were analyzed by Western blotting with antibodies raised against either EWS, hTAFII100, and hTAFII55 separately (B) or hTAFII68, hTAFII100, and hTAFII55 (D). In panel B, peroxidase-conjugated goat anti-mouse IgG-IgM-specific secondary antibodies were used; in panel D, peroxidase-conjugated goat anti-mouse κ-type light-chain-specific secondary antibody was used. IgGH, IgG heavy chain.
TABLE 1.
Comparison of the interactions of EWS and hTAFII68 with individual components of the TFIID complex in baculovirus-coinfected SF9 cells
| TFIID subunit | Interactiona
|
|
|---|---|---|
| TAFII68 | EWS | |
| TAFII250 | − | − |
| ΔNTAFII135 | − | − |
| TAFII100 | +++ | +++ |
| TAFII80 | ND | ND |
| TAFII55 | ++ | ++ |
| TBP | − | − |
| TAFII31 | ND | ND |
| TAFII30 | − | − |
| TAFII28 | + | + |
| TAFII20 | − | − |
| TAFII18 | + | + |
Interactions of EWS and hTAFII68 with each TFIID subunit were tested and the averages of at least three independent experiments similar to the one presented in Fig. 2 are shown. +++, ++, +, and −, strong, moderate, weak, and no interactions between EWS or hTAFII68 and a given hTAFII or TBP; ND, not determined.
Interaction of EWS and hTAFII68 with subunits of the RNA polymerase II.
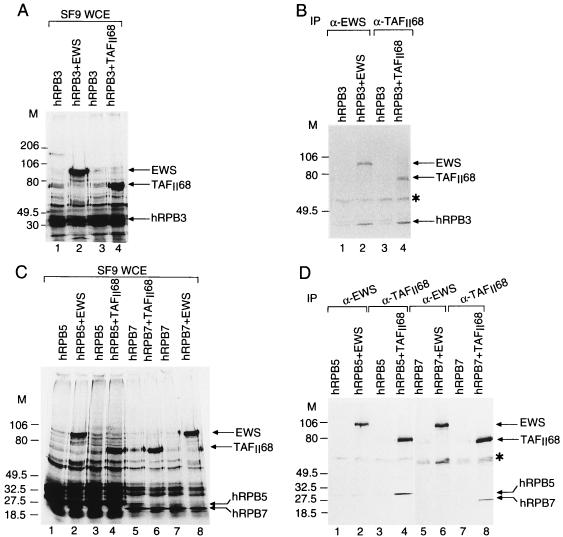
To confirm the tight association of EWS or hTAFII68 with Pol II, pairwise interactions of EWS or hTAFII68 with individual subunits of the Pol II complex were tested. cDNAs encoding almost all the human Pol II subunits were inserted in baculovirus expression vectors (1), and each subunit was expressed either alone or together with EWS or hTAFII68 in SF9 cells. Proteins were radiolabeled with [α-35S]methionine and [α-35S]cysteine, WCEs were made, and the protein expression was examined by autoradiography (Fig. 3A and C). From these extracts, either EWS or hTAFII68 was immunoprecipitated by using the appropriate antibodies, and EWS- or hTAFII68-bound proteins were analyzed (Fig. 3B and D and data not shown). Extracts in which EWS or hTAFII68 were not expressed served as negative controls for the immunoprecipitations. These interaction studies indicate that both EWS and hTAFII68 bind directly to hRPB3, a specific subunit of Pol II (Fig. 3B and Table 2). Moreover, hTAFII68 also interacts with two other Pol II subunits, hRPB5 and hRPB7 (Fig. 3D and Table 2). These results suggest that EWS and hTAFII68 may directly contact these Pol II subunits in the endogenous Pol II complex (see also Discussion).
FIG. 3.
Interactions between EWS or hTAFII68 and the different subunits of Pol II. (A and C) SF9 cells were coinfected with recombinant baculoviruses expressing subunits of Pol II either individually or pairwise with EWS and hTAFII68 as indicated. After 44 h of infection, proteins were radiolabeled with [α-35S]methionine and [α-35S]cysteine for 4 h. WCEs were made, proteins were separated by SDS-PAGE, and gels were dried and subjected to autoradiography. M, markers in kilodaltons. (B and D) From the WCEs, EWS or hTAFII68 was immunoprecipitated (IP) with either the anti-EWS (α-EWS) antibody or the anti-hTAFII68 (α-TAFII68) MAb as indicated. Resin-bound proteins were then analyzed by SDS-PAGE followed by autoradiography. The asterisk indicates a nonspecific protein species.
TABLE 2.
Comparison of the interactions of EWS and hTAFII68 with individual subunits of the Pol II in baculovirus-coinfected SF9 cells
| Pol II subunit(s) | Interactiona
|
|
|---|---|---|
| TAFII68 | EWS | |
| hRPB1 | ND | ND |
| hRPB2 | − | − |
| hRPB3 | ++ | ++ |
| hRPB4 | ND | ND |
| hRPB5 | ++ | − |
| hRPB6 | − | − |
| hRPB7 | ++ | − |
| hRPB8 | +/− | − |
| hRPB9 | − | − |
| hRPB10α, -10β, and -11 | +/− | − |
Interactions of EWS and hTAFII68 with the indicated Pol II subunits were tested, and the averages of at least three independent experiments similar to the one presented in Fig. 3 are shown. +++, ++, +, +/−, and −, strong, moderate, weak, very weak but detectable, and no interactions between EWS or hTAFII68 and a given Pol II subunit; ND, not determined.
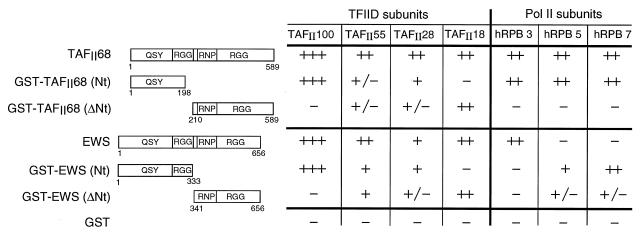
Mapping the domains of EWS and hTAFII68 which interact with the subunits of TFIID and Pol II.
Since the N-terminal domains of EWS and TLS/FUS play a specific role in tumorigenic processes (7, 8), it is important to characterize the interactions in which the N-terminal region of the TET proteins are involved. To this end, we generated GST fusion proteins which contain either the N-terminal (GST-EWSNt) or C-terminal (GST-EWSΔNt) halves of EWS and hTAFII68 (Fig. 4). Note that the GST-EWSNt fusion protein contains the N-terminal domain present in the type II EWS–FLI-1 oncogenic fusion protein. The GST fusion proteins (or GST alone) were expressed in Escherichia coli, bound on glutathione-agarose beads, and incubated with SF9 cell protein extracts in which the different human TAFIIs or Pol II subunits were overexpressed (see above and Fig. 4). The beads were then extensively washed, and bound proteins were analyzed by either Western blotting or autoradiography (Fig. 4). The immobilized N-terminal domain of either TAFII68 or EWS retained specifically TAFII100, while TAFII18 bound more specifically to the CTDs of hTAFII68 and EWS (Fig. 4). The binding of TAFII55 and TAFII28 to the truncated hTAFII68 or EWS fusion proteins was less specific, suggesting that they may interact with several regions of TAFII68 or EWS. These results indicate that the N-terminal regions of EWS and hTAFII68 clearly retain the interaction with hTAFII100 but do not retain, or retain only weakly, interactions with the other TAFIIs that were shown to interact with the full-length proteins.
FIG. 4.
Mapping the domains of EWS and hTAFII68 which interact with the subunits of TFIID and Pol II. Numbers in the diagrams refer to amino acid positions in either hTAFII68 or EWS. The results of the protein-protein interaction assay, using either baculovirus-overexpressed full-length proteins or E. coli-produced GST fusion proteins, are summarized as follows: +++, ++, +, +/−, and −, strong, moderate, weak, very weak but detectable, and no interactions between the indicated proteins.
Interestingly, all of the polymerase subunits which interacted with the full-length hTAFII68 bound specifically to the N-terminal region of TAFII68 (Fig. 4), indicating that the N-terminal region of hTAFII68 plays an important role in the tight association between the Pol II complex and hTAFII68. In contrast, the binding of the polymerase subunits to the different EWS fusion proteins was unexpected. The hRPB3 subunit, which bound to the full-length EWS, did not interact with the isolated regions of EWS, and hRPB5 and hRPB7, which did not interact with the full-length EWS, interacted with both halves of EWS (Fig. 4). These results, together with the observation that the full-length EWS is able to interact with its separated CTD (data not shown), suggest that either an intramolecular interaction(s) takes place within the full-length EWS or EWS is able to multimerize. Thus, it appears that the distinct domains of EWS are differently accessible in the full-length protein than they are in the separated GST fusion proteins. This finding further suggests that the full-length EWS and EWS–FLI-1 may interact differently with TFIID and Pol II.
EWS, but not EWS–FLI-1, interacts with TFIID and Pol II.
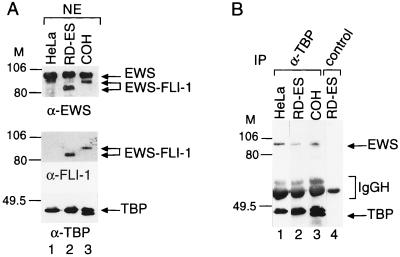
The fact that the N-terminal domain of EWS retained its ability to interact with TAFII100 prompted us to examine whether the oncogenic fusion protein EWS–FLI-1 is also associated with TFIID. To answer this question, we analyzed the TFIID composition from two Ewing sarcoma cell lines (RD-ES and COH); These cell lines express different fusion transcripts between EWS and FLI-1 which give rise to a 520-amino-acid fusion protein in the case of the RD-ES cells and a 582-amino-acid fusion protein in the case of the COH cells. NEs were made from these cell lines expressing the two different EWS–FLI-1 fusion proteins. Expression of the fusion oncoproteins was compared to that of the germ line EWS by Western blot analysis using the anti-EWS PAb (Fig. 5A, upper panel) and the anti-FLI-1 antibody (Fig. 4A, middle panel). The anti-EWS PAb was raised against a common region present in the N-terminal domains of both the EWS and EWS–FLI-1 fusion proteins, and the anti-FLI-1 antibody was raised against the C-terminal end of FLI-1 (21). In the NEs of the Ewing sarcoma cells, the anti-EWS antibody recognized both EWS and the EWS–FLI-1 proteins (Fig. 5A, upper panel, lanes 2 and 3). Moreover, it appears that both Ewing sarcoma cell lines tested express about three times less EWS–FLI-1 protein than germ line EWS. The same two EWS–FLI-1 fusion oncoproteins were also recognized by the anti-FLI-1 MAb in the NE of the Ewing sarcoma cell lines (Fig. 5A, middle panel, lanes 2 and 3); however, this antibody did not recognize any protein in the HeLa NE (lane 1). Next, we prepared TBP-containing complexes from the Ewing sarcoma cell and the HeLa NEs by an anti-TBP immunoprecipitation and tested the presence of EWS or EWS–FLI-1 in the immunoprecipitated TBP-containing complexes by Western blot analysis (Fig. 5B). Similar to the TFIID-EWS coimmunoprecipitation from HeLa cell NE (Fig. 1A and 5B, lane 1), EWS coimmunoprecipitated with TBP from the two Ewing sarcoma cell NEs (Fig. 5B, lanes 2 and 3). Moreover, the anti-TAFII100 MAb coimmunoprecipitated EWS from the RD-ES cell line (data not shown). In contrast, no EWS–FLI-1 was detected in the immunoprecipitated TFIID complexes by Western blot analysis using the anti-EWS PAb (lanes 2 and 3), even after very long exposures of the Western blots. Moreover, no EWS–FLI-1 fusion proteins were observed to be associated with the TBP-containing complexes when the antibody raised against the C terminus of FLI-1 was used (reference 21 and data not shown). This finding suggests that the oncogenic EWS–FLI-1 fusion proteins do not associate with TFIID in the two Ewing sarcoma cell lines tested.
FIG. 5.
The oncogenic fusion protein EWS–FLI-1 does not coimmunoprecipitate with the TFIID complex in Ewing sarcoma cell lines. (A) The PAb raised against the N-terminal domain of EWS recognizes both wild-type EWS and the two different EWS–FLI-1 fusion proteins in NEs from the two Ewing sarcoma cell lines, RD-ES (lane 2) and COH (lane 3). NEs from HeLa, RD-ES and COH cells were analyzed by Western blotting using the anti-EWS (α-EWS) antibody (upper panel), the anti-FLI-1 (α-FLI-1) MAb (middle panel), and the anti-TBP (α-TBP) MAb 3G3 (lower panel). M, markers in kilodaltons. (B) NEs from the various cell lines were immunoprecipitated (IP) with the anti-TBP MAb 3G3 (lane 1 to 3). Beads were washed and boiled, and bound proteins were analyzed by Western blotting with the anti-EWS antibody and the anti-TBP MAb. The control immunoprecipitation using an unrelated MAb is shown in lane 4.
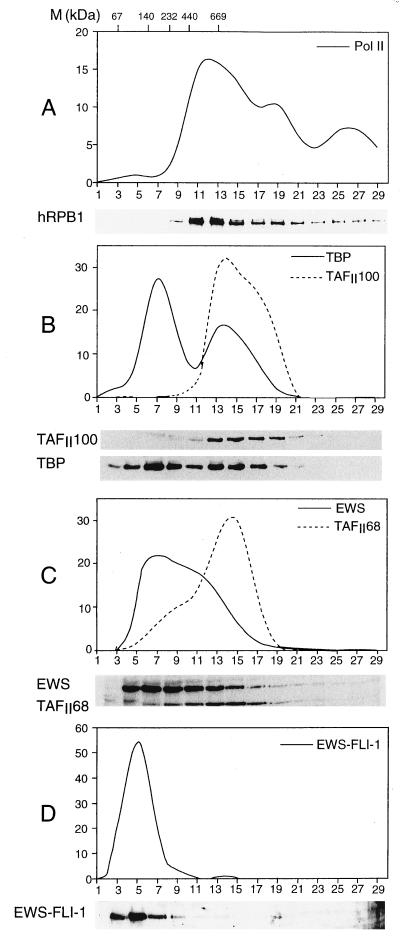
Using several different in vitro approaches, we have shown that portions of EWS and hTAFII68 are associated with either TFIID or Pol II. To further investigate the association of EWS and hTAFII68 with these multiprotein complexes under more physiological conditions, we determined the native molecular masses of hTAFII68 and EWS from the HeLa and RD-ES cell lines as well as the apparent native molecular mass of the oncogenic fusion protein EWS–FLI-1 from the RD-ES cell line. Human HeLa and RD-ES NEs were made and centrifuged through a 20 to 40% glycerol gradient, and no further manipulations were performed on the crude extracts to ensure that high-molecular-weight complexes remained intact. The sedimentation of hTAFII68, EWS, and EWS–FLI-1 was compared with that of components of TFIID (TBP and TAFII100) and Pol II (the largest subunit of the Pol II complex) multiprotein complexes, as well as markers of known molecular mass. With the exception of EWS–FLI-1, similar results were obtained in analyses of fractions from either HeLa or RD-ES NEs (Fig. 6 and data not shown). Most of hTAFII68 and about 40% of EWS cosedimented in fractions corresponding to high molecular masses (between 400 and 1,300 kDa [Fig. 6C, fractions 10 to 20]) that contained TFIID and a portion of Pol II (Fig. 6A and B). This cosedimentation further suggests that the previously found association of EWS and hTAFII68 with the TFIID and Pol II complexes may be physiologically relevant. In contrast, the majority of EWS–FLI-1 was detected by Western blot analysis using an anti-FLI-1 antibody in the low-molecular-mass-range fractions (between 67 and 160 kDa [Fig. 5D, fractions 2 to 9]). As both the TFIID and the Pol II complexes have native molecular masses greater than 600 kDa, this result, together with the immunoprecipitation data (see above), suggests that in contrast to EWS, EWS–FLI-1 is not stably associated with TFIID or Pol II.
FIG. 6.
Sedimentation of RD-ES cell NE through a 20 to 40% glycerol gradient indicates that the endogenous EWS–FLI-1 fusion protein is present in low-molecular-mass ranges. The relative sedimentations of the largest subunit of Pol II (A), TAFII100 and TBP (B), EWS and TAFII68 together (C), and EWS–FLI-1 (D) were determined by Western blotting using antibodies raised against either the CTD of the largest subunit of Pol II (A), TAFII100 and TBP (B), or EWS and TAFII68 (C). To better visualize EWS–FLI-1 that is only weakly detected in panel C by the EWS antibody, in panel D the anti-FLI-1 antibody was used. In each panel, the upper part shows a quantification of the Western blot. Values represent the percentage of a given protein present in each fraction compared to the total amount of this protein loaded on the glycerol gradient. Positions of markers (M) of known molecular mass standards are indicated at the top of panel A.
DISCUSSION
Possible functions of the TET proteins in transcription initiation and elongation.
Previously, two members of the TET family (hTAFII68 and TLS/FUS) were shown to interact with functionally different TFIID complexes (3). In this study, we show that the third human member of the TET family (EWS) can also associate with endogenous TFIID. These findings indicate that the TET family members have not only structural but also functional homology. hTAFII68 and TLS/FUS were described as specific TAFIIs since they were found to be associated with functionally distinct TFIID subpopulations (3, 5). Similarly, the association of EWS with TFIID is substoichiometric, suggesting that it associates only with a subpopulation of TFIID and thus EWS can also be considered a specific TAFII. The high homology among EWS, TLS/FUS, and hTAFII68 and common properties to associate with complexes involved in Pol II transcription suggest that they may play a common role in transcription initiation and/or elongation.
The fact that hTAFII68 and EWS interact with the same core TAFIIs and that hTAFII68 and TLS/FUS are not present in the same TFIID subpopulations strongly suggests that these proteins cannot both be present in the same TFIID complexes. In agreement with this conclusion, we could not coimmunoprecipitate EWS (or TLS/FUS) with hTAFII68 or vice versa. Moreover, the presence of one of these RNA- and/or ssDNA-binding proteins in a distinct TFIID complex may distinguish a particular TFIID complex, at least partly, from the other different TFIID complexes. Thus, the different TET proteins-containing TFIID complexes may have a specific role in the preinitiation complexes and/or may define the promoter selectivity of the distinct TFIID complexes.
Only a fraction of the total cellular amount of EWS binds to TFIID. Another fraction of endogenous EWS copurifies with the Pol II complex on five subsequent chromatographic columns, similarly to hTAFII68, suggesting an association between EWS and the Pol II complex. The association of EWS or hTAFII68 with Pol II was confirmed by mapping possible contact points between these two TET proteins and subunits of Pol II. This mapping indicated that while both EWS and hTAFII68 interacted with the third-largest subunit of the human Pol II (hRPB3), only hTAFII68 interacted with hRPB5 and hRPB7. Thus, despite the fact that the members of the TET family are functional homologs, they may differ in the capacity to interact with other proteins.
Involvement of the N-terminal domains of EWS and hTAFII68 in the interactions with TFIID and Pol II.
To understand more about the mechanisms by which the chimeric sarcoma-associated oncogenes induce tumor formation, it is important to study the involvement of their N-terminal domains in the above-described interactions (see also the introduction). TAFII100 was the only TFIID subunit that bound reasonably well to the N-terminal domain of EWS or hTAFII68. This finding suggests that the interactions between the endogenous TFIID complex and EWS–FLI-1 are considerably weaker than those between EWS and TFIID (see also below). Since the baculovirus-overexpressed EWS–FLI-1 is very insoluble, we could not investigate the interactions between the different TAFIIs and EWS–FLI-1. The other TAFII interactions with EWS and hTAFII68 either mapped in the CTDs of EWS and hTAFII68 or could not be clearly determined with the GST fusion proteins used. The interactions mapped between the CTD of EWS and the different TAFIIs suggest that this domain plays an important role in the stable association of full-length EWS with TFIID. These results suggest that the complex network of interactions occurring between EWS and the TFIID complex may be seriously impaired in the case of the EWS–FLI-1 oncogenic fusion protein that contains only the N-terminal domain of EWS (see also below).
The N-terminal domain of hTAFII68 retained the ability to interact with all of the Pol II subunits which were found to interact with the full-length protein, indicating that this domain of hTAFII68 plays an important role in the tight association between hTAFII68 and the Pol II complex. Unexpectedly, none of the isolated domains of EWS interact with the Pol II subunit which interacts with the full-length EWS. This finding suggests that within the EWS protein, an intramolecular interaction(s) occurs and that a particular conformation of EWS is involved in the interaction(s) with Pol II. Consistent with this hypothesis, an in vitro interaction between the full-length EWS and its C-terminal region can be detected (data not shown). Based on these observations, we propose a model where EWS may exist in the cells in a conformation in which the N-terminal domain of the protein is not accessible. This form of EWS may then bind to Pol II through hRPB3. However, following interaction with a certain cellular target(s) or after a posttranslational modification(s), EWS may change its conformation such that its N-terminal domain becomes accessible. This modified form of EWS would be able to interact with other Pol II subunits, hRPB5 and hRPB7. The interaction between the N-terminal domain of EWS and hRPB7 has also been identified independently in a yeast two-hybrid screen using the first 82 amino acids of EWS as a bait (28). Importantly, this short 82-amino-acid region of EWS has been previously shown to be sufficient for nearly full transforming activity of EWS (18). Thus, this interaction between the transcriptional activator EWS–FLI-1 and the Pol II complex, which may not occur between wild-type FLI-1 and Pol II, seems to play an important role in the deregulation of gene expression in the sarcoma cells.
EWS–FLI-1 cannot stably associate with Pol II and TFIID but may interact with these complexes as a transcriptional activator.
The aberrant transcription factor EWS–FLI-1 transforms the cells by either interfering in the function of germ line EWS, having a dominant-negative effect on the function of EWS, or modifying the FLI-1-regulated gene expression. The fact that EWS–FLI-1 does not coimmunoprecipitate with TFIID from RD-ES and COH cells (Fig. 5B) and that in the RD-ES cell NEs EWS–FLI-1 can be found predominantly in low-molecular-weight ranges indicates that EWS–FLI-1 is not stably associated with any multiprotein complexes in these cells. This suggests that the interactions mapped between the C-terminal half of EWS and the TFIID subunits are critical for the stable association of full-length EWS with the TFIID complex. Moreover, there is also a dramatic change in the Pol II subunits which interact either with the full-length EWS or with its N-terminal domain (Fig. 4). Thus, it is unlikely that EWS–FLI-1 can have a dominant-negative effect on the function of the portion of EWS which is associated with the TFIID and/or Pol II multiprotein complexes. Note that another portion of EWS may be involved in functions that are yet unknown. EWS–FLI-1 has been shown to function as a transcriptional activator on different FLI-1-binding sites containing test promoters (2). Our finding that EWS–FLI-1 is not associated with any multiprotein complexes is in agreement with this finding since to date none of the known transcriptional activators have been found tightly associated with TFIID or Pol II. However, transcriptional activators are known to interact with components of the basal transcription machinery, e.g., TAFIIs, TBP, and Pol II, to enhance transcription of the different target genes (36, 38). The efficiency and the stability of the interactions between the N-terminal domain of EWS and TFIID or Pol II may also be very different from those in which wild-type FLI-1 participates. These differences seems to be important for the transformation capability of EWS–FLI-1.
ACKNOWLEDGMENTS
We thank H. Kovar and R. Petermann for discussing unpublished results, I. Kolb-Cheynel for expression of hTAFIIs, Pol II subunits, and EWS in the baculovirus system; J. Ghysdael, V. Dubrovskaya, X. Jacq, A. C. Lavigne, G. Mengus, and I. Davidson for different reagents; F. J. Dilworth and C. Kedinger for critical reading of the manuscript; Y. Lutz for antibody preparations; the cell culture group for cells; and C. Werlé and J.-M. Lafontaine for illustrations and photography.
A.B. was supported by a fellowship from Association pour la Recherche contre le Cancer. This work was supported by grants from the CNRS, INSERM, Centre Hospitalier Universitaire Régional, Ministère de la Recherche et Technologie, and Fondation pour la Recherche Médicale to L.T. and the Association pour la Recherche contre le Cancer to C. Kedinger and L.T.
REFERENCES
- 1.Acker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M. Interactions between the human RNA polymerase II subunits. J Biol Chem. 1997;272:16815–16821. doi: 10.1074/jbc.272.27.16815. [DOI] [PubMed] [Google Scholar]
- 2.Bailly R A, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, Thomas G, Ghysdael J. DNA-binding and transcriptional activation properties of the EWS–FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertolotti A, Lutz Y, Heard D J, Chambon P, Tora L. hTAFII68 a novel RNA/SSDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 4.Besse S, Vigneron M, Pichard E, Puvlon-Dutilleul F. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 1995;4:143–161. [PMC free article] [PubMed] [Google Scholar]
- 5.Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly J M, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 8.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 9.Dubrovskaya V, Lavigne A-C, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIFb (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser R A, Rossignol M, Heard D J, Egly J M, Chambon P. SUG1, a putative transcriptional mediator and subunit of the PA700 proteasome regulatory complex, is a DNA helicase. J Biol Chem. 1997;272:7122–7126. doi: 10.1074/jbc.272.11.7122. [DOI] [PubMed] [Google Scholar]
- 11.Gerard M, Fischer L, Moncollin V, Chipoulet J M, Chambon P, Egly J M. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 12.Giovannini M, Biegel J A, Serra M, Wang J Y, Wei Y H, Nycum L, Emanuel B S, Evans G A. EWS-erg and EWS-Fli1 fusion transcripts in Ewing’s sarcoma and primitive neuroectodermal tumors with variant translocations. J Clin Invest. 1994;94:489–496. doi: 10.1172/JCI117360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst H C. Transcription factors as drug targets in cancer. Eur J Cancer. 1996;32A:1857–1863. doi: 10.1016/0959-8049(96)00214-6. [DOI] [PubMed] [Google Scholar]
- 14.Immanuel D, Zinszner H, Ron D. Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II. Mol Cell Biol. 1995;15:4562–4571. doi: 10.1128/mcb.15.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 16.Labelle Y, Zucman J, Stenman G, Kindblom L G, Knight J, Turc-Carel C, Dockhorn-Dworniczak B, Mandahl N, Desmaze C, Peter M, et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum Mol Genet. 1995;4:221–226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 17.Ladanyi M. The emerging molecular genetics of sarcoma translocations. Diagn Mol Pathol. 1995;4:162–173. doi: 10.1097/00019606-199509000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Lessnick S L, Braun B S, Denny C T, May W A. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 19.Lewin B. Oncogenic conversion by regulatory changes in transcription factors. Cell. 1991;64:303–312. doi: 10.1016/0092-8674(91)90640-k. [DOI] [PubMed] [Google Scholar]
- 20.May W A, Gishizky M L, Lessnick S L, Lunsford L B, Lewis B C, Delattre O, Zucman J, Thomas G, Denny C T. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci USA. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melot T, Gruel N, Doubeikovski A, Sevenet N, Teillaud J L, Delattre O. Production and characterization of mouse monoclonal antibodies to wild-type and oncogenic FLI-1 proteins. Hybridoma. 1997;16:457–464. doi: 10.1089/hyb.1997.16.457. [DOI] [PubMed] [Google Scholar]
- 22.Meyers S, Hiebert S W. Indirect and direct disruption of transcriptional regulation in cancer: E2F and AML-1. Crit Rev Eukaryotic Gene Expr. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 23.Nagai K, Oubridge C, Jessen T H, Li J, Evans P R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 24.Ohno T, Ouchida M, Lee L, Gatalica Z, Rao V N, Reddy E S. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- 25.Ohno T, Rao V N, Reddy E S. EWS/Fli-1 chimeric protein is a transcriptional activator. Cancer Res. 1993;53:5859–5863. [PubMed] [Google Scholar]
- 26.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors. W. H. New York, N.Y: Freeman; 1992. [Google Scholar]
- 27.Peter M, Couturler J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- 28.Petermann, R., B. M. Mossier, and H. Kovar. Personal communication.
- 29.Plougastel B, Zucman J, Peter M, Thomas G, Delattre O. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics. 1993;18:609–615. doi: 10.1016/s0888-7543(05)80363-5. [DOI] [PubMed] [Google Scholar]
- 30.Rabbitts T H, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 31.Roeder R G. Nuclear RNA polymerase: role of general initiation factors and cofactors in eukaryotic transcription. Methods Enzymol. 1996;273:165–171. doi: 10.1016/s0076-6879(96)73016-1. [DOI] [PubMed] [Google Scholar]
- 32.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 33.Stolow D T, Haynes S R. Cabeza, a Drosophila gene encoding a novel RNA binding protein, shares homology with EWS and TLS, two genes involved in human sarcoma formation. Nucleic Acids Res. 1995;23:835–843. doi: 10.1093/nar/23.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turc-Carel C, Aurias A, Mugneret F, Lizard S, Sidaner I, Volk C, Thiery J P, Olschwang S, Phillp I, Berger M P, et al. Chromosomes in Ewing’s sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12) Cancer Genet Cytogenet. 1988;32:229–238. doi: 10.1016/0165-4608(88)90285-3. [DOI] [PubMed] [Google Scholar]
- 35.Urano F, Umezawa A, Hong W, Kikuchi H, Hata J. A novel chimera gene between EWS and E1A-F, encoding the adenovirus E1A enhancer-binding protein, in extraosseous Ewing’s sarcoma. Biochem Biophys Res Commun. 1996;2:608–612. doi: 10.1006/bbrc.1996.0281. [DOI] [PubMed] [Google Scholar]
- 36.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 37.Wasylyk B, Hahn S L, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 38.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 39.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]