SUMMARY
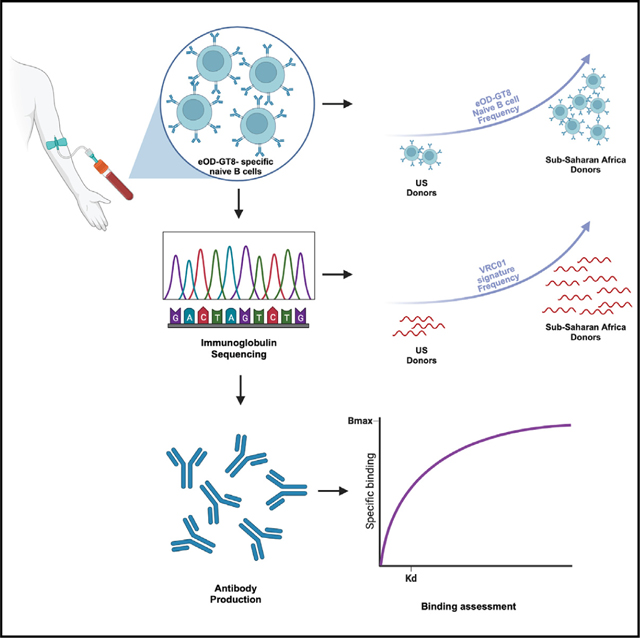
HIV gp120 engineered outer domain germline-targeting version 8 (eOD-GT8) was designed specifically to engage naive B cell precursors of VRC01-class antibodies. However, the frequency and affinity of naive B cell precursors able to recognize eOD-GT8 have been evaluated only in U.S. populations. HIV infection is disproportionally concentrated in sub-Saharan Africa, so we seek to characterize naive B cells able to recognize eOD-GT8 in sub-Saharan cohorts. We demonstrate that people from sub-Saharan Africa have a higher or equivalent frequency of naive B cells able to engage eOD-GT8 compared with people from the U.S. Genetically, the higher frequency of eOD-GT8-positive cells is accompanied by a higher level of naive B cells with gene signatures characteristic of the VRC01 class, as well as other CD4bs-directed antibodies. Our study demonstrates that vaccination with eOD-GT8 in sub-Saharan Africa could be successful at expanding and establishing a pool of CD4bs-directed memory B cells from naive precursors.
In brief
Germline-targeting HIV vaccine strategies depend on the frequency of vaccine-specific naive B cell precursors. Matassoli et al. show that the frequency and immunoglobulin signature of B cells binding the immunogen eOD-GT8 in sub-Saharan African cohorts are higher or equivalent to those of a U.S. cohort, suggesting that it could prove effective in Africa.
Graphical Abstract
INTRODUCTION
To date, VRC01-class antibodies are some of the broadest and most potent anti-HIV broad neutralizing antibodies (bnAbs) isolated from chronically infected HIV patients.1–3 All VRC01-class antibodies use the variable heavy chain (HC) gene VH1–2*02 paired with a light chain (LC) harboring an unusually short complementarity determining region 3 (CDRL3) of 5 amino acids (aa).1–4 Unlike most antibodies that use mainly the CDRH3 region to bind to the cognate epitope, VRC01-class antibodies primarily use CDRH2 regions to bind the HIV Env CD4 binding site (CD4bs) epitope, mimicking how the CD4 receptor binds to this region.3–5 To achieve high neutralization breadth of HIV isolates, VRC01-class antibodies typically have high levels of somatic hypermutation (SHM). It has been estimated that it took more than a decade for VRC01 to mature into a potent bnAb from its unmutated common ancestor (UCA).6 Several full-length or truncated HIV glycoprotein (gp) monomers and stabilized trimers have been engineered to specifically elicit this class of CD4bs antibodies as potential vaccine immunogens.7,8 However, although most of these HIV proteins can be recognized by mature bnAbs, they fail to bind their UCAs or inferred germline (iGL) versions,9,10 questioning their function as an immunogenic antigen capable of efficiently engaging and activating naive B cells precursors to bnAbs in HIV-uninfected individuals.
Evolution of several bnAbs has been successfully tracked from HIV-infected people.11–13 This comprehensive understanding of their ontology from UCA to potent bnAb has aided the design of new immunogens modified to bind naive B cell precursors with the intent of driving antibody lineages with successive immunogens to become broadly neutralizing.14 The design of immunogens that can engage the UCA/iGLs and initiate the process has focused on the removal of key glycans that partially interfere with antibody accessibility to a potentially neutralizing epitope.7,15,16 HIV gp120 engineered outer domain germline-targeting version 8 (eOD-GT8) has been successfully designed to specifically engage the iGL of VRC01.17 Immunization with eOD-GT8 in VH1–2 transgenic mice led to activation and development of functional VRC01-like antibodies.18,19 Studies in HIV-uninfected humans demonstrated that eOD-GT8 binds naive B cells with the canonical VRC01 immunoglobulin (Ig) gene signature,17,20 along with other members of the VRC01 class (VRC23, N6, and PCIN63) and other CD4bs non-VRC01-class antibodies.21 Furthermore, a phase I clinical trial conducted in the U.S. using eOD-GT8 as immunogen demonstrated that it is highly capable of engaging and expanding VRC01-class naïve precursors in the human population22 (NCT03547245). This suggests that eliciting VRC01-like bnAbs from naive precursors with sequential immunization may be feasible. However, both precursor frequency and immunization studies with eOD-GT8 were conducted in a U.S. population. The burden of the HIV pandemic is much more concentrated in sub-Saharan Africa, accounting for approximately 70% of all people living with HIV and 65% of new HIV infections worldwide.23 Therefore, evaluating the naive B cell compartment in this population is important to understand whether a similar vaccine strategy would be equally successful. Here, we compared the frequency and Ig repertoire of eOD-GT8 naive B cells in HIV-negative human volunteers from the U.S. and different countries of sub-Saharan Africa (South African, Uganda, Rwanda, Tanzania, and Kenya), as well as Thailand, which has medium HIV incidence globally but has one of the highest HIV incidence rates in Asia.24 We found that individuals from Thailand and sub-Saharan Africa have equal or higher frequency of circulating eOD-GT8-specific naive B cells than U.S. donors. In addition, repertoire analysis showed a higher frequency of known CD4bs subclass antibody signatures in individuals from sub-Saharan African countries, including non-VH1–2 CD4bs antibodies. This suggests that vaccine strategies designed to elicit VRC01-like bnAbs could be successful in regions with the highest HIV burden.
RESULTS
Frequency of HIV CD4bs-specific naive B cells is high in donors living in sub-Saharan Africa
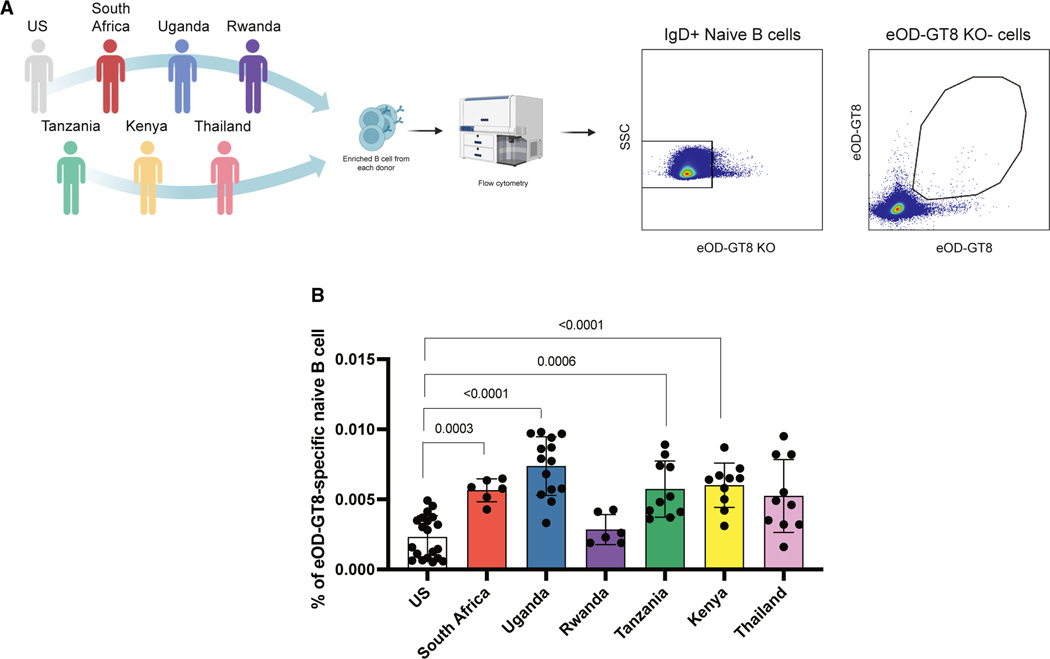
To detect eOD-GT8-specific naive B cells binding the HIV Env CD4bs, we enriched for total B cells from about 1 × 108 peripheral blood mononuclear cells (PBMCs) from each individual donor separately and stained them with fluorescently labeled eOD-GT8 and a mutated version of eOD-GT8 with the CD4bs epitope knocked out (KO11). Using flow cytometry, we measured the frequency of live CD14— CD56— CD3— CD19+ CD20+ IgD+ KO11— eOD-GT8++ naive B cells able to specifically recognize the CD4bs epitope (CD4bs specific) on eOD-GT8 (Figures 1A and S1). Interestingly, apart from Rwanda, individuals from sub-Saharan African countries had significantly higher frequencies of CD4bs-specific naive B cells compared with U.S. donors (Figure 1B). Samples from Thailand had similar frequencies as the U.S. cohort (Figure 1B).
Figure 1. African cohorts harbor higher frequencies of eOD-GT8+ cells among naive IgD+ B cells.
(A) Schematic representation of the cohorts used and gate strategy.
(B) Frequencies of circulating eOD-GT8+ IgD+ naive B cells among different cohorts. Statistical significance was determined using the two-tailed Mann-Whitney test. Each dot represent a individual donor (for the number of donors analyzed from each location, see Table S1).
VRC01-class signatures are enriched in sub-Saharan African cohorts
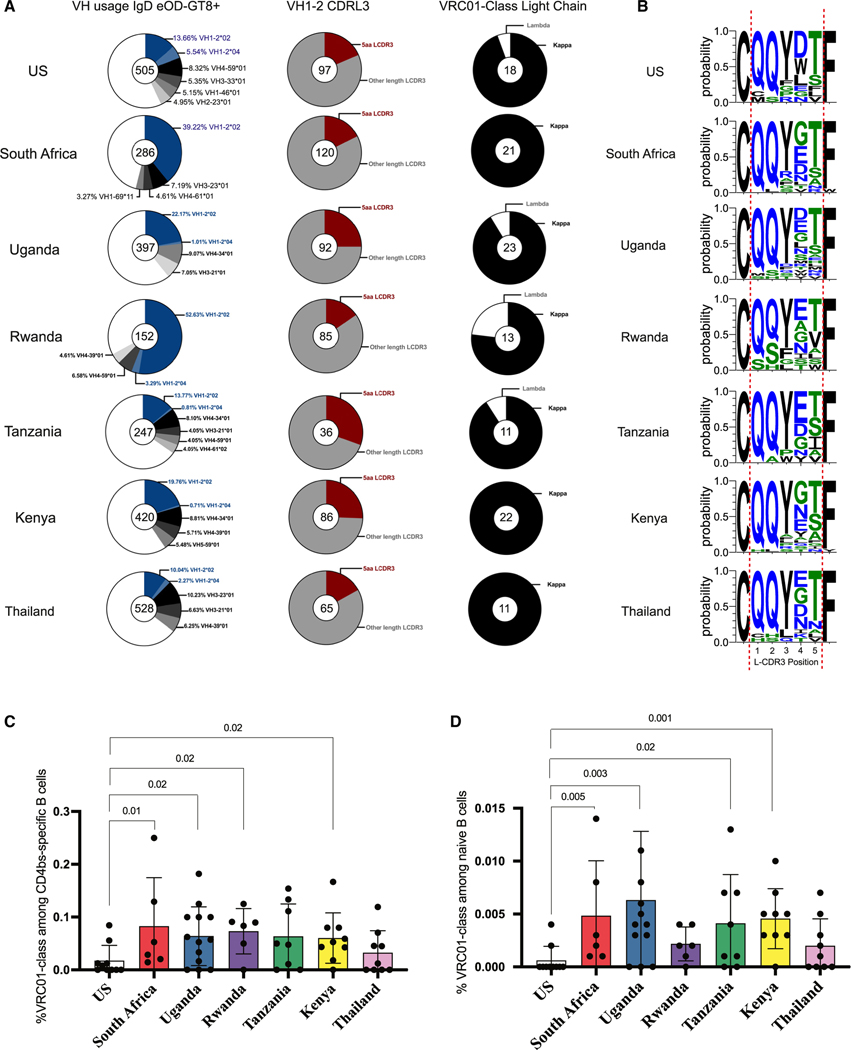
To better understand the differences among CD4bs-specific naive B cells from different cohorts, we single-cell-sorted all eOD-GT8 double-positive naive B cells from each individual donor separately (Figure S1), and amplified and sequenced the Ig genes to analyze the Ig repertoire. VRC01-class antibodies are characterized by a VH1–2*02 or VH1–2*04 allele paired with an LC containing a CDRL3 with 5 aa. The distribution of the VH gene use among CD4bs-specific naive B cells showed VH1–2 as a predominant gene family in all groups, with increased frequencies in the South African and Rwanda cohorts (Figure 2A, left panel). However, a variety of other Ig HC variable genes could also be observed (Figure 2A, left panel). Fifteen percent to 30% of all VH1–2 HCs were paired with a CDRL3 5 aa long kappa chain (Figure 2A, middle and right panels). An LC with a CDRL3 of 5 aa is rare in the human repertoire, and almost all 5 aa CDRL3 chains recovered from all cohorts were paired with a VH1–2 HC gene (Figure S2). Most non-VH1–2 HCs were paired with the usual 9 aa CDRL3 (Figure S2). Interestingly, many of the kappa LC 5 aa CDRL3s paired with VH1–2 HCs had the amino acid sequence “QQYET,” especially in the Rwanda and Tanzania cohorts (Figure 2B). Mature VRC01-class antibodies harbor a consensus “QQYEF” in the CDRL3,4 indicating that these cells are close to the mature bnAb CDRL3. The frequency of VRC01-class naive B cells was estimated by multiplying two frequencies: (1) the frequency of CD4bs-specific (KO—/eOD-GT8++) naive B cells among all naive B cells sorted by fluorescence-activated cell sorting (FACS) and (2) the frequency of VRC01-class B cell receptors (BCRs) among all CD4bs-specific naive B cells with sequenced BCRs. Both frequencies were significantly higher in South Africa, Uganda, and Kenya cohorts compared with the U.S. cohort (Figures 2C and 2D). Together, these data indicate that CD4bs-specific eOD-GT8+ naive B cells from sub-Saharan cohorts are not only in higher frequency in the periphery, but also include many B cells expressing Igs with genetic characteristics of VRC01-class germline precursors.
Figure 2. Increased VRC01-class gene signature among African cohort.
(A) Pie chart of VH use among eOD-GT8+ naive B cells from different cohorts; blue slices indicate the VH1–2 gene, and gray shades indicate the other most frequent VH genes (left panel). VH1–2+ sequences paired with a 5 aa L-CDR3 (red, middle panel) and VRC01-class naive B cells expressing kappa (κ) or lambda (λ) LCs among total VRC01-class naive B cells. Total B cell sequences are indicated at the center of each pie chart.
(B) Weblogo comparing L-CDR3 sequences of VRC01-class naive B cells from all different cohorts.
(C) Frequency of VRC01-class signatures among eOD-GT8+ naive B cells for each cohort.
(D) Frequency of VRC01-class signatures among total IgD+ naive B cells for each cohort. Statistical significance was determined using the two-tailed Mann-Whitney test.
VH1–2 HC genes make up 3%–4% of the circulating B cell repertoire in the U.S. population.25,26 To confirm that the enrichment of VH1–2 gene use among eOD-GT8+ naive B cells from sub-Saharan African cohorts was not due to higher overall VH1–2 use, we bulk-sorted total IgD+ naive B cells (Figure S1) from 5 individuals from African countries (Uganda, Kenya, and Tanzania) and 5 from U.S. donors and used 10X Genomics to sequence the Ig repertoire. We recovered 2,981 productive paired HC and LC from the U.S. cohort and 1,983 from the sub-Saharan African cohort (Figure S3A). Overall, we observed similar VH gene use between the two cohorts, with about 3% VH1–2 use in both populations (Figure S3A). The CDRL3 length of LCs paired with a VH1–2 HC was also similar in both cohorts, with a 9-aa-long CDRL3 accounting for about 50% of all LCs in both cohorts (Figure S3B). Thus, the higher frequency of VH1–2 HC with a 5 aa CDRL3 in eOD-GT8+ naive B cells in the sub-Saharan cohort was specific to the CD4bs-specific naive B cells and not a general characteristic of the naive B cell repertoire.
Naive B cells with other CD4bs bnAb signatures are engaged by eOD-GT8 in sub-Saharan African cohorts
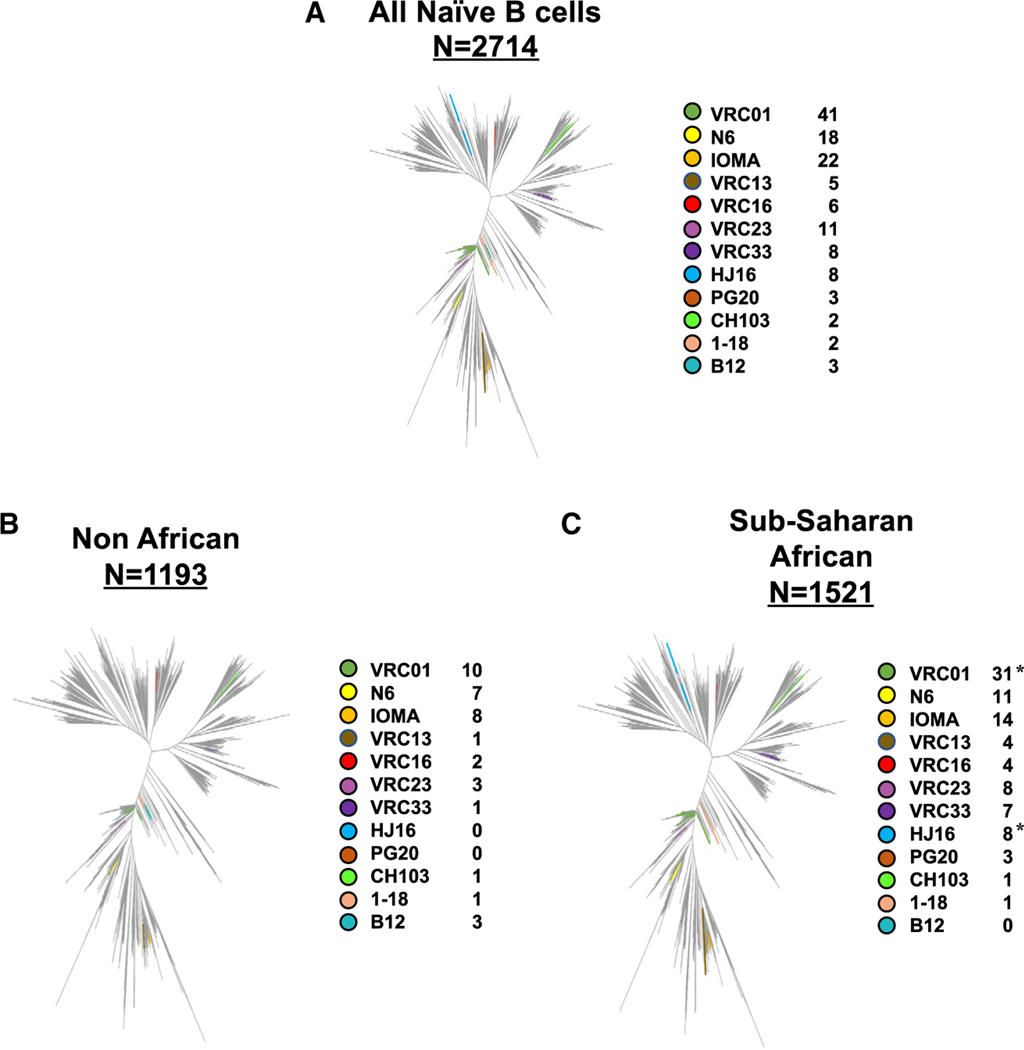
Although eOD-GT8 was specifically designed to bind VRC01-class naive B cells, it also engages other CD4bs Ig class signatures in the U.S. population.21 To more comprehensively investigate the CD4bs Ig gene signatures in our dataset, we concatenated Ig HC and LC sequences from naive epitope-specific eOD-GT8-positive B cells and performed phylogenetic analyses with respect to known CD4bs antibody gene signatures, including dissecting the VRC01 class into sub-classes.3 We were able to identify several paired Ig sequences classified as IOMA and VRC23 in addition to a few additional signatures linked to VRC16 (Figure 3A). Importantly, within the VRC01 class, we also identified N6, which, as a mature antibody, has a neutralization breadth near 100%.1 We next divided the Ig sequences into two groups, those from sub-Saharan African or non-African cohorts and assessed whether the different signatures identified were differentially represented in naive B cells from the two groups. We observed in the CD4bs-specific eOD-GT8+ Ig sequences from sub-Saharan Africans a significantly higher frequency of VRC01 and HJ16 Ig classes compared with Ig sequences from non-Africans (Figures 3B and 3C). There was no appreciable difference in the distribution of the other Ig gene signatures across the two groups (Figures 3B and 3C). Thus, although there were numerically more Igs with known CD4bs signatures isolated from sub-Saharan Africans, the overall CD4bs-specific antibodies classes identified from eOD-GT8+ naive B cells was similar in the two groups.
Figure 3. CD4bs antibody lineages pulled out by eOD-GT8 in different cohorts.
Dendrograms based on the HC and LC paired sequence of all Ig sequences isolated in the study combined (A) or separately by Non-African (B) and Sub-Saharan African cohorts (C). Number indicates total number of sequences analyzed, and colors further delineate the CD4bs antibody lineage as indicated in the legend. Statistical significance was determined using the two-sided Fisher’s exact test.
Precursor VRC01-class antibodies from sub-Saharan African and U.S. cohorts bind to eOD-GT8 with similar affinity
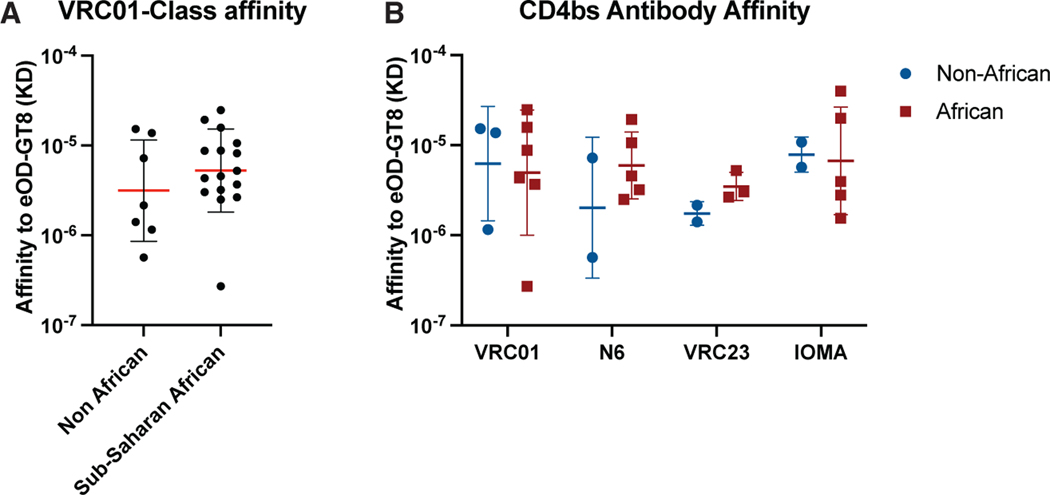
Finally, we expressed a few monoclonal antibodies (mAbs) from VRC01-class signature (VH1–2 paired with an LC with 5 aa CDRL3) (Figure 4A) and other CD4bs epitope-specific classes isolated from naive B cells in both U.S. and sub-Saharan African individuals (Figure 4B) and tested their affinity for eOD-GT8. All mAbs produced and tested were able to bind eOD-GT8 with a wide range of affinities for eOD-GT8 (Figure 4). In addition, there was no overall difference in the binding affinity of genetically diverse CD4bs epitope-specific mAbs isolated from the U.S. or African individuals (Figure 4). Thus, CD4bs epitope-specific naive precursors from the two cohorts appeared to engage eOD-GT8 with similar affinity.
Figure 4. Affinity of CD4bs antibodies for eOD-GT8.
(A) Monovalent affinities of VRC01-class Abs (VH1–2 paired with 5 aa CDRL3) derived from eOD-GT8+ naive B cells isolated from non-African(7 antibodies) and sub-Saharan African (15 antibodies) individuals.
(B) Monovalent affinities of different CD4bs antibody classes derived from eOD-GT8+ naive B cells isolated from non-African and sub-Saharan African individuals.
DISCUSSION
Germline-targeting and sequential immunization strategies are designed to target and expand rare naive B cells and lead to development of bnAbs by gradually educating them to recognize epitopes as displayed on wild-type HIV.16,27,28 This complex approach is required for the development of HIV bnAbs because conserved epitopes targeted by neutralizing antibodies are generally not accessible to germline Igs expressed by naive B cells.29,30 Promising results in mice18,28,31 and human B cell repertoire analyses17,21 led to a phase I human vaccine trial in healthy volunteers with a eOD-GT8 60-mer nanoparticle vaccine designed to bind the inferred UCA of the CD4bs bnAb VRC01.22 Recent results from this trial conducted in the U.S. showed that eOD-GT8 vaccination successfully induces a pool of CD4bs-specific memory B cells with VRC01 and other HIV bnAb signatures.22
The success of the eOD-GT8 immunogen is due in part to a relatively large naive precursor frequency of VRC01-class CD4bs-specific naive B cells in most immunized participants from the U.S. However, little is known about precursor frequencies in genetically diverse populations from other parts of the world. Here, we investigated the frequency and Ig repertoire of CD4bs-specific naive B cells engaged by eOD-GT8 in individuals living in low-risk (U.S.), medium-risk (Thailand), and high-risk (sub-Saharan Africa countries) areas for HIV infection. Results showed that VRC01, IOMA, and other classes of CD4bs gene signatures can be found in donors from all groups, with even higher frequencies in donors living in sub-Saharan Africa compared with donors living in the U.S. and Thailand. This suggests that vaccination with eOD-GT8 of populations living in areas with a much higher burden of HIV infection could prove successful at expanding bnAb precursors.
Frequencies of naive B cells able to bind eOD-GT8 from donors living in the U.S. in this study are overall in line with previous work.17 Furthermore, the increase in CD4bs Ig signatures in the sub-Saharan African individuals was not due to overall populational repertoire difference, as the HC gene use among total naive B cells from African individuals aligned with gene use in the U.S. population.25,26 Moreover, many of the VRC01-class precursor naive B cells that we identified presented with a 5 aa CDRL3 sequence motif of “QQYET,” which is only 1 aa “away” from the “QQYEF” motif present in the same region of the mature VRC01. This may facilitate progression of these cells toward broad neutralization. A considerable portion of the VRC01-class precursor signatures that we identified showed relation to N6, which is by far one of the broadest CD4bs-specific bnAbs, with neutralization breadth near 100%.1 In addition, analyses on mAbs expressed from naive B cells showed that eOD-GT8+ B cells from the sub-Saharan African cohort bound the immunogen with the same affinity as B cells from the U.S. population. This affinity level has been shown to be sufficient to induce a pool of memory B cells upon eOD-GT8 nanoparticle vaccination.22
It is unclear why CD4bs-specific naive B cells are more frequent in donors living in sub-Saharan Africa. One hypothesis is that living in an environment with overall higher antigenic burden32–34 might lead the naive cell pool to evolve toward broader antigenic coverage. If this is the case, a naive B cell pool shaped to cover a greater variety of antigens might result in a circumstantial increase of rare combinations of Ig HC and LC genes, such as the VRC01 class. Future studies analyzing naive B cell compartment repertoire will add insight into populations living in different parts of the world.
In conclusion, our study indicates that targeting and developing VRC01-class CD4bs-specific naive B cell precursors into B cells expressing broadly neutralization antibodies is a possible strategy in sub-Saharan African countries with high incidence of HIV.
Limitations of the study
This study describes increased frequency of eOD-GT8-specific naive B cells in cohorts from sub-Saharan Africa countries compared with the U.S. However, we acknowledge that sub-Saharan Africa comprises 46 countries with several ethnicities, and our study is limited to only 6 countries in that region. In addition, we did not investigate deeply the genetic differences that could explain the reason for this specificity difference among cohorts. Future studies are needed to understand the molecular basis behind these observations.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sarah F. Andrews (sarah.andrews2@nih.gov).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Antibody sequence data have been deposited at GenBank and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Study design and cohorts
The study was designed to assess whether human volunteers of different regions of the world living in both low and high-risk areas for HIV infection (USA, Thailand and sub-Saharan Africa) (Table S1) would be equally suitable for vaccination with eOD-GT8 and to collect useful information for possible future trial design with eOD-GT8 to be conducted in HIV-endemic areas. For those purposes, a large number of cryopreserved PBMC were obtained from HIV-negative individuals living in USA either from the National Institutes of Health (NIH) blood bank or the PBMC repository at the Duke University; South Africa samples were obtained from FRESH cohort in Durban, South Africa26; Rwanda samples from International AIDS Vaccine Initiative (IAVI); Uganda samples from either IAVI or Military HIV Research Program (MHRP); Tanzania, Kenya and Thailand samples obtained from MHRP (Table S1). All individuals tested negative for HIV at the time of sampling and none of them was previously enrolled in any HIV vaccine trial or had an HIV-positive partner.
METHOD DETAILS
Fluorophore-conjugation of eOD-GT8 and KO11
Biotinylated eOD-GT8 and eOD-GT8-KO11 were produced as previously described.17 Conjugation with a fluorophore was done by mixing 2.5μg of eOD-GT8 with 1μg of either Streptavidin-PE (Life Technologies) or Streptavidin-BV711 (BD Biosciences) in a final volume of 15μg diluted in PBS for at least 45 min at 4◦C protected from light. The KO11 was conjugated with Streptavidin-APC (Life Technologies) following the same protocol.
B cell enrichment, flow cytometry and sorting of single eOD-GT8+ naive B cells
Cryopreserved PBMCs were thawed, treated with Benzonase nuclease (Millipore Corp.), washed with PBS, and enriched for B cells by magnetic negative selection using the Human Pan-B cell Enrichment Kit (Miltenyi) according to manufacturer’s instruction. Enriched B cells were then washed with PBS and first stained with viability dye Aqua Fluorescent Reactive (ThermoFisher) for 2 min followed by staining with the following human antibodies: IgM BB700 (customized from BD Bioscience), CD21-PE594 (BD Bioscience), CD22-PECY5 (Biolegend), CD20-APCH7 (BD Bioscience), IgG-BUV395 (BD Bioscience), CD38-BUV661 (BD Bioscience), CD19-BUV805 (BD Bioscience), IgD-BV570 (BD Bioscience), CD27-BV605 (Biolegend), CD56-BV510 (Biolegend), CD14-BV510 (Biolegend) and CD3-BV510 (Biolegend) in addition to the conjugated eOD-GT8 and KO-11, all diluted in Brilliant stain Buffer (BD Bioscience) for 30 min at 4◦C protected from light. Cells were washed twice with PBS containing 0.1% BSA and analyzed on FACS Symphony A5 (BD Biosciences) using Diva software. Cells were gated on live singlets CD3− CD14− CD56− CD19+ CD38lo CD27+ CD20+ IgG-IgD+ KO- and all double-positive eOD-GT8 cells were single cell sorted (Figure S1) into 96-well plates coated with 5μL of TCL buffer (Qiagen) containing 1% of 2-Mercaptoethanol (Sigma-Aldrich). After sorting, plates were sealed and immediately frozen at −80◦C until further processing. Flow cytometry analyses were performed by using the Flow-Jo software (Three Star Inc).
Sequencing and analysis of immunoglobulin genes of eOD-GT8+ naive B cells
Single sorted eOD-GT8-specific naive B cell Ig sequencing was done fallowing the adapted version of the SMARTSeq-V4 protocol by 5′ RACE published before.35 Briefly, single cell RNA was purified using magnetic beads (RNAclean beads – Beckman Coulter), followed by incubation with TSoligo2_polydT and subsequent cDNA synthesis with a template switching oligo (TSO) was performed using SMARTseq reagents. cDNA was then amplified using TSO primers. Excess oligos and dNTPs were removed from amplified cDNA with EXO-CIP cleanup kit (New England BioLabs). HC and LC were then enriched by amplifying cDNA with TSO forward and IgG/IgM revere or IgK/IgL reverse primer pools. Ig amplicons were used to prepare Illumina-ready libraries using Nextera XT reagents (doi: https://doi.org/10.1038/s41467–022-35456–2). We obtained paired heavy and light chain Ig sequences from an average of 70% of the single cells. Ig sequences were assembled using BALDR (https://doi.org/10.1186/s13073–018-0528–3) and filtered (https://github.com/scharch/filterBALDR). V(D)J were annotated using SONAR v4.3 (https://doi.org/10.3389/fimmu.2016.00372) in single cell mode.
Bulk sort and Ig sequencing of total naive B cells
Cryopreserved PBMC were thawed, treated with Benzonase nuclease (Millipore Corp.), washed with PBS and each donor was individually stained with a different TotalSeq anti-human Hashtag (Biolegend) for 15min. Cells were washed with PBS containing 0.1% BSA and then stained with viability dye Aqua Fluorescent Reactive (Invitrogen) for 2 min followed by staining with the following human antibodies: IgM BB700 (customized from BD Bioscience), CD21-PE594 (BD Bioscience), CD22-PECY5 (Biolegend), CD20-APCH7 (BD Bioscience), IgG-BUV395 (BD Bioscience), CD38-BUV661 (BD Bioscience), CD19-BUV805 (BD Bioscience), IgD-BV570 (BD Bioscience), CD27-BV605 (Biolegend), CD56-BV510 (Biolegend), CD14-BV510 (Biolegend) and CD3-BV510 (Biolegend) for 30 min at 4◦C protected from light. Cells were washed twice with PBS containing 0.1% BSA and analyzed on FACS Symphony A5 (BD Biosciences) using Diva software. Cells were gated on live singlets CD3− CD14− CD56− CD19+ CD38lo CD27+ CD20+ IgG-IgD+ and all IgD+ naive B cell were bulk sorted in a 1.5mL tube containing RPMI with 10% FBS.
After sorting, cDNA from bulk-sorted total naive B cells was immediately made following 10X Genomics Chromium Next GEM Single Cell 5′ v1 Kit according to manufacturer’s instructions. VDJ genes were amplified from cDNA and libraries were made following manufacturer’s instructions. VDJ libraries were sequenced on Novaseq (Illumina) targeting at least 10,000 reads per cell.
Phylogenetic analysis of CD4bs sequences
Stand alone IgBlast was used to assign antibody V(D)J germline gene.36 Paired heavy and light chain germline gene and CDRL 3 length were used as criteria to assign different CD4bs antibody classes (Table S2). A neighbor joining tree was built based on protein sequences of isolated antibodies with ClustalW.37 Dendroscope was used to visualize assigned CD4bs antibodies on neighbor joining trees.38
Monoclonal antibody expression from eOD-GT8-specific naive B cells
Paired HC and LC sequences from eOD-GT8-specific naive B cells were cloned and synthesized by Genscript. Expi293 cells (Thermo Fisher) were co-transfected with plasmids encoding both HC and LC with ExpiFectamine (Thermo Fisher). After 6 days of expression, monoclonal antibodies were purified from the cell supernatant using protein A Sepharose (Pierce).
eOD-GT8 binding assay
Kinetics and affinity of antibody-antigen interactions were measured on Carterra LSA using HC30M Sensor Chip (Carterra) and 1x HBS-EP+ pH 7.4 running buffer (20x stock from Teknova, Cat. No H8022) supplemented with BSA at 1 mg/mL. The chip surface was prepared for ligand capture following Carterra software instructions. Approximately 2000–3000 RU of capture antibody (SouthernBiotech Cat no 2047–01) in 10 mM Sodium Acetate pH 4.5 was amine coupled to the chip. Phosphoric Acid 1.7% was used as a regeneration solution with 30 s contact time and injected three times per each cycle. Solution concentration of ligands were at 1 μg/mL and contact time was 5 min. Raw sensograms were analyzed using Kinetics software (Carterra) using interspot and blank double referencing, Langmuir modeling. Analyte concentrations were quantified on NanoDrop 2000c Spectrophotometer using absorption signal at 280 nm.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyzes was performed using Prism8 software. Specific statistical tests used for datasets is indicated in the figure legends. p values equal or less than 0.05 was considered significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-Human IgM | BD Biosciences | Custom, Clone G20–127 |
| Anti-Human CD21 | BD Biosciences | RRID: AB_2738231 |
| Anti-Human CD20 | BD Biosciences | RRID: AB_1727449 |
| Anti-Human IgG | BD Biosciences | RRID: AB_2738683 |
| Anti-Human CD38 | BD Biosciences | Cat# 612969 |
| Anti-Human CD19 | BD Biosciences | RRID: AB_2873553 |
| Anti-Human IgD | BD Biosciences | Custom, Clone IA6–2 |
| Anti-Human CD22 | Biolegend | RRID: AB_2074592 |
| Anti-Human CD27 | Biolegend | RRID: AB_2561450 |
| Anti-Human CD56 | Biolegend | RRID: AB_2561944 |
| Anti-Human CD14 | Biolegend | RRID: AB_2561946 |
| Anti-Human CD3 | Biolegend | RRID: AB_2561943 |
| Anti-Human Hashtag 1 | Biolegend | RRID: AB_2801031 |
| Anti-Human Hashtag 2 | Biolegend | RRID: AB_2801032 |
| Anti-Human Hashtag 3 | Biolegend | RRID: AB_2801033 |
| Anti-Human Hashtag 4 | Biolegend | RRID: AB_2801034 |
| Anti-Human Hashtag 5 | Biolegend | RRID: AB_2801035 |
| Anti-Human Hashtag 6 | Biolegend | RRID: AB_2820042 |
| Anti-Human Hashtag 7 | Biolegend | RRID: AB_2820043 |
| Anti-Human Hashtag 8 | Biolegend | RRID: AB_2820044 |
| Anti-Human Hashtag 9 | Biolegend | RRID: AB_2820045 |
| Anti-Human Hashtag 10 | Biolegend | RRID: AB_2820046 |
|
| ||
| Biological samples | ||
|
| ||
| Human PBMC from US | NIH Blood bank and Duke University | N/A |
| Human PBMC from South Africa | FRESH Cohort | N/A |
| Human PBMC from Rwanda | IAVI | N/A |
| Human PBMC from Uganda | IAVI and MHRP | N/A |
| Human PBMC from Tanzania | MHRP | N/A |
| Human PBMC from Kenya | MHRP | N/A |
| Human PBMC from Thailand | MHRP | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Streptavidin PE | ThermoFisher | Cat# S21388 |
| Streptavidin APC | ThermoFisher | Cat# S32362 |
| Streptavidin BV711 | BD Biosciences | Cat# 563262 |
| Fixable live/dead stain | ThermoFisher | Cat# L34957 |
| Brilliant Stain Buffer | BD Biosciences | Cat# 563794 |
| Protein A Sepharose | GE LifeSciences | Cat# 17–1279-03 |
| 2-Mercaptoethanol | ThermoFisher | Cat# 21985023 |
| dNTPs | ThermoFisher | Cat# 18427088 |
| TCL Buffer | Qiagen | Cat# 1031576 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Expi293 Expression System Kit | ThermoFisher | Cat# A14635 |
| RNA Clean XP Beads | Beckman Coulter | Cat# A63987 |
| 10x Lysis Buffer | Clontech | Cat# 635013 |
| SMARTScribe RT | Clontech | Cat# 639538 |
| RNAse Inhibitor | Clontech | Cat# 2313B |
| SeqAmp PCR Buffer | Clontech | Cat# 638509 |
| SeqAmp DNA Polymerase | Clontech | Cat# 638509 |
| AMPure XP Beads | Beckman Coulter | Cat# A63881 |
| EXO-CIP Cleanup kit | New England Biolabs | Cat# E1050L |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human Expi293F | ThermoFisher | Cat# A14527 |
|
| ||
| Software and algorithms | ||
|
| ||
| Flowjo 10 | FlowJo | https://www.flowjo.com |
| Prism 7/8 | Graphpad | https://www.graphpad.com/ |
| Seaview | RRID: SCR_015059 | |
| Standalone IgBlast | NCBI | https://www.ncbi.nlm.nih.gov/igblast/ |
| Kinetics software | Carterra | https://carterra-bio.com/resources/kinetics-software/ |
Highlights.
Sub-Saharan African cohorts have high frequencies of eOD-GT8-specific naive B cells
eOD-GT8-specific naive B cells in this cohort express VRC01-class antibody precursor genes
IOMA, VRC23, and VRC16 HIV CD4bs-specific antibody signatures are also detected
ACKNOWLEDGMENTS
We thank David Ambrozak and Richard Nguyen for help with FACS, David Brown, and Garcia Ambrosia for help with the acquisition of demographics information. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests. This work was supported by a cooperative agreement (W81XWH-18–2-0040) between the Henry M. Jackson Foundation (HJF) for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD). This research was funded, in part, by the National Institute of Allergy and Infectious Diseases. The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army, the DOD, or the HJF. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70–25.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113450.
REFERENCES
- 1.Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. (2015). Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 161, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. (2013). Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. (2010). Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. (2015). Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell 161, 470–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. (2013). Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medina-Ramirez M, Garces F, Escolano A, Skog P, de Taeye SW, Del Moral-Sanchez I, McGuire AT, Yasmeen A, Behrens AJ, Ozorowski G, et al. (2017). Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 214, 2573–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, and Dimitrov DS (2009). Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, and Stamatatos L (2013). Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 9, e1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. (2013). Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. (2011). Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. (2014). Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamatatos L, Pancera M, and McGuire AT (2017). Germline-targeting immunogens. Immunol. Rev. 275, 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. (2017). Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep. 19, 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. (2013). Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereñ o-Orbea J, Kalyuzhniy O, Deresa I, et al. (2016). HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. (2015). Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell 161, 1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sok D, Briney B, Jardine JG, Kulp DW, Menis S, Pauthner M, Wood A, Lee EC, Le KM, Jones M, et al. (2016). Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Toy L, Kos JT, Safonova Y, Schief WR, Havenar-Daughton C, Watson CT, and Crotty S (2021). Vaccine genetics of IGHV1–2 VRC01-class broadly neutralizing antibody precursor naive human B cells. NPJ Vaccines 6, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havenar-Daughton C, Sarkar A, Kulp DW, Toy L, Hu X, Deresa I, Kalyuzhniy O, Kaushik K, Upadhyay AA, Menis S, et al. (2018). The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 10, eaat0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leggat DJ, Cohen KW, Willis JR, Fulp WJ, deCamp AC, Kalyuzhniy O, Cottrell CA, Menis S, Finak G, Ballweber-Fleming L, et al. (2022). Vaccination induces HIV broadly neutralizing antibody precursors in humans. Science 378, eadd6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, et al. (2018). Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1736–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vannakit R, Andreeva V, Mills S, Cassell MM, Jones MA, Murphy E, Ishikawa N, Boyd MA, and Phanuphak N (2020). Fast-tracking the end of HIV in the Asia Pacific region: domestic funding of key populationled and civil society organisations. Lancet HIV 7, e366–e372. [DOI] [PubMed] [Google Scholar]
- 25.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, and Georgiou G (2015). In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 21, 86–91. [DOI] [PubMed] [Google Scholar]
- 26.Briney B, Inderbitzin A, Joyce C, and Burton DR (2019). Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medina-Ramírez M, Sanders RW, and Klasse PJ (2014). Targeting B-cell germlines and focusing affinity maturation: the next hurdles in HIV-1-vaccine development? Expert Rev. Vaccines 13, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. (2015). HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton DR, and Mascola JR (2015). Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sliepen K, and Sanders RW (2016). HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev. Vaccines 15, 349–365. [DOI] [PubMed] [Google Scholar]
- 31.Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. (2016). Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell 166, 1445–1458.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clerici M, Butto S, Lukwiya M, Saresella M, Declich S, Trabattoni D, Pastori C, Piconi S, Fracasso C, Fabiani M, et al. (2000). Immune activation in africa is environmentally-driven and is associated with upregulation of CCR5. Italian-Ugandan AIDS Project. Aids 14, 2083–2092. [DOI] [PubMed] [Google Scholar]
- 33.Cohen CR, Moscicki AB, Scott ME, Ma Y, Shiboski S, Bukusi E, Daud I, Rebbapragada A, Brown J, and Kaul R (2010). Increased levels of immune activation in the genital tract of healthy young women from sub-Saharan Africa. Aids 24, 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chachage M, Podola L, Clowes P, Nsojo A, Bauer A, Mgaya O, Kowour D, Froeschl G, Maboko L, Hoelscher M, et al. (2014). Helminth-associated systemic immune activation and HIV co-receptor expression: response to albendazole/praziquantel treatment. PLoS Negl. Trop. Dis. 8, e2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lima NS, Musayev M, Johnston TS, Wagner DA, Henry AR, Wang L, Yang ES, Zhang Y, Birungi K, Black WP, et al. (2022). Primary Exposure to SARS-CoV-2 Variants elicits convergent epitope specificities, immunoglobulin V gene usage and public B cell clones. Preprint at bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye J, Ma N, Madden TL, and Ostell JM (2013). IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, and Thompson JD (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, and Rupp R (2007). Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinf. 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Antibody sequence data have been deposited at GenBank and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.