Abstract
Although it is essential for protein synthesis to be highly accurate, a number of cases of directed ribosomal frameshifting have been reported in RNA viruses, as well as in procaryotic and eucaryotic genes. Changes in the efficiency of ribosomal frameshifting can have major effects on the ability of cells to propagate viruses which use this mechanism. Furthermore, studies of this process can illuminate the mechanisms involved in the maintenance of the normal translation reading frame. The yeast Saccharomyces cerevisiae killer virus system uses programmed −1 ribosomal frameshifting to synthesize its gene products. Strains harboring the mof2-1 allele demonstrated a fivefold increase in frameshifting and prevented killer virus propagation. In this report, we present the results of the cloning and characterization of the wild-type MOF2 gene. mof2-1 is a novel allele of SUI1, a gene previously shown to play a role in translation initiation start site selection. Strains harboring the mof2-1 allele demonstrated a mutant start site selection phenotype and increased efficiency of programmed −1 ribosomal frameshifting and conferred paromomycin sensitivity. The increased frameshifting observed in vivo was reproduced in extracts prepared from mof2-1 cells. Addition of purified wild-type Mof2p/Sui1p reduced frameshifting efficiencies to wild-type levels. Expression of the human SUI1 homolog in yeast corrects all of the mof2-1 phenotypes, demonstrating that the function of this protein is conserved throughout evolution. Taken together, these results suggest that Mof2p/Sui1p functions as a general modulator of accuracy at both the initiation and elongation phases of translation.
The ability of ribosomes to maintain the correct translational reading frame is fundamental to the integrity of translation and, ultimately, to cell growth and viability. The protein translational machinery has evolved to ensure that the intrinsic error rate of reading frame maintenance is extremely low, with error rates on the order of 1 misreading in 5,000 translational events (14, 30, 41). Although it is essential for protein synthesis to be highly accurate, in the last 10 years, a number of cases of directed ribosomal frameshifting have been reported in viruses, including retroviruses, coronaviruses, the L-A double-stranded RNA (dsRNA) virus and the Ty family of viruses in the yeast Saccharomyces cerevisiae, the dsRNA virus of Giardia lamblia, positive-single-stranded RNA viruses of plants, and bacteriophage T7. The efficiency of ribosomal frameshifting determines the relative ratios of Gag to Gag-Pol fusion protein available for viral particle morphogenesis, and changes in ribosomal frameshift efficiencies have major effects on the ability of cells to propagate viruses which use ribosomal frameshifting (reviewed in reference 13). In addition, programmed frameshifting has been utilized by a number of bacterial transposons, as well as in some bacterial cellular genes and the ornithine decarboxylase antizyme gene in mammals (for reviews, see references 9, 13, 20, and 22).
Frameshifting events in viruses typically produce fusion proteins in which the N- and C-terminal domains are encoded by two distinct, overlapping open reading frames. The two cis-acting mRNA elements that are required to promote −1 ribosomal frameshifting have been well defined. A slippery site, X XXY YYZ (the 0 frame is indicated by the spaces) (26), allows the simultaneous slippage of ribosome-bound A- and P-site tRNAs by one base in the 5′ direction (24, 25). A structural element, usually an RNA pseudoknot, is located immediately 3′ to the slippery site (6, 14, 39). The mRNA pseudoknot structure makes the ribosome pause over the slippery site and is thought to increase the probability of 5′ ribosomal movement (37, 42). These signals can direct elongating ribosomes to shift the reading frame with a frequency of 2 to 10% (reviewed in references 13 and 20). Thus, this naturally occurring molecular mechanism provides a powerful experimental system for probing how the maintenance of a translational reading frame is governed.
The dsRNA L-A virus in the yeast S. cerevisiae has two open reading frames. The 5′ gag gene encodes the Gag protein, and the 3′ pol gene encodes a multifunctional protein domain required for viral RNA packaging and replication. A −1 ribosomal frameshift event is responsible for the production of the Gag-Pol fusion protein. M1, a satellite dsRNA virus of L-A which encodes a secreted killer toxin, is encapsidated and replicated by using the gene products synthesized by the L-A virus (reviewed in reference 13). The combination of L-A and M1 constitutes the yeast “killer” virus system.
We are interested in identifying the trans-acting factors that govern programmed −1 ribosomal frameshifting (16). Screens for mutations that increased the programmed −1 ribosomal frameshift efficiencies in yeast cells identified chromosomal mutations that are called mof (for maintenance of frame; 13, 17, 18) and ifs (increased frameshifting; 29). The screen originally used to identify the mof mutants utilized a construct in which the lacZ gene was inserted downstream of the L-A −1 ribosomal frameshift signal and in the −1 reading frame relative to a translational start site. Recent results demonstrated that mof4-1 is a unique allele of the UPF1 gene, which affects both the nonsense-mediated mRNA decay pathway and programmed −1 frameshifting (11).
Among the mof mutants, strains harboring the mof2-1 allele demonstrated the greatest increase in programmed −1 ribosomal frameshifting efficiency and a temperature-sensitive (ts) cell cycle arrest phenotype (18). In this report, we present the results of the cloning of the wild-type MOF2 gene by complementation of the ts phenotype of mof2-1 strains. mof2-1 is a novel allele of SUI1, a gene previously shown to play a role in translation initiation start site selection (43). The results presented here demonstrate that, in addition to possessing a mutant start site selection phenotype, cells harboring the mof2-1 allele also promote increased efficiency of programmed −1 ribosomal frameshifting and paromomycin sensitivity in yeast cells, indicative of a reduction in translation fidelity. Addition of purified wild-type Mof2p/Sui1p to translationally competent extracts of mof2-1 cells is able to reduce −1 ribosomal frameshifting efficiencies back to wild-type levels, demonstrating that the Sui1/Mof2 protein is actively involved in both the initiation and elongation phases of protein synthesis. Expression of the human homolog of this protein in yeast was able to correct all of the mof2-1 phenotypes, demonstrating that the function of this protein is conserved throughout evolution. These results suggest that Mof2p/Sui1p may function as a general component necessary for translational accuracy.
MATERIALS AND METHODS
Strains and media.
The strains of S. cerevisiae used in this study are listed in Table 1. YPAD, YPG, SD, synthetic complete medium (35), and 4.7 MB plates for testing the killer phenotype were prepared as previously reported (18). Strains Y218 to Y221 were constructed by plasmid shuffling (35). Briefly, strain JD272 was transformed with plasmids carrying different alleles of the MOF2/SUI1 gene, including wild-type MOF2, mof2-1, sui1-1, and huISOSUI1. The chromosomal copy of the MOF2/SUI1 gene was then deleted and replaced with the hisG-URA3-hisG cassette (1). These strains were subsequently plated on media containing 5-fluoro-orotic acid, and ura mutant strains (mof2::hisG) were isolated.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YGC106 | MATa ade2,3 ura3 leu2 his7 can1 sap3 upf1::hisG | 11 |
| 742-9B | MATa leu2-1::pJD85 his3 and/or his4 ura3 mof2-1 | 18 |
| JD272 | MATα leu2 lys11 ura3-52 trp1ΔK− | This study |
| JD758 | MATa kar1-1 leu1 [L-AHN M1] K+ | 17 |
| 5X47 | MATa/MATα his1/+ trp1/+ ura3/+ K− R− | 17 |
| Y218 | MATα leu2 lys11 ura3-52 trp1Δ mof2::hisG pYcp22MOF2 K− | JD272 derivative; this study |
| Y219 | MATα leu2 lys11 ura3-52 trp1Δ mof2::hisG pYcp22mof2-1 K− | JD272 derivative; this study |
| Y220 | MATα leu2 lys11 ura3-52 trp1Δ mof2::hisG pYcp22sui1-1 K− | JD272 derivative; this study |
| Y221 | MATα leu2 lys11 ura3-52 trp1Δ mof2::hisG pGhuISOSUI1 K− | JD272 derivative; this study |
| sui1 | MATα leu2 his4-303(AUU) ura3-52 sui1-1 | 43 |
| sui2 | MATa leu2 his4-303(AUU) ura3-52 sui2-1 | 43 |
| SUI3 | MATα leu2 his4-303(AUU) ura3-52 ino1-13 SUI3-3 | 43 |
Molecular and genetic methods.
Transformations of yeast and Escherichia coli were performed as described previously (12). Cytoductions and the killer test were performed as previously described (17). Genetic crosses, sporulation tetrad analysis, and β-galactosidase activity assays were performed as previously described (11, 18). Testing for drug sensitivity of the various strains was performed as previously described (11). Northern blots for monitoring of the killer viruses were performed as previously described (15).
Plasmid constructions.
Plasmids p314-JD85-ter, p315-JD85-ter, p314-JD86-ter, and p315-JD86-ter, used for frameshifting β-galactosidase assays, were constructed as described before (11). p314-JD85-ter and p315-JD85-ter are the lacZ gene in the −1 reading frame relative to the initiation AUG and preceded by a −1 ribosomal frameshifting sequence from the L-A virus. p314-JD86-ter and p315-JD86-ter are the lacZ gene in the 0 reading frame relative to the initiation AUG and lacking the −1 ribosomal frameshifting sequence. Plasmids pTI24 and pJD115a, (see Fig. 1B) were described previously (16a, 18a).
FIG. 1.
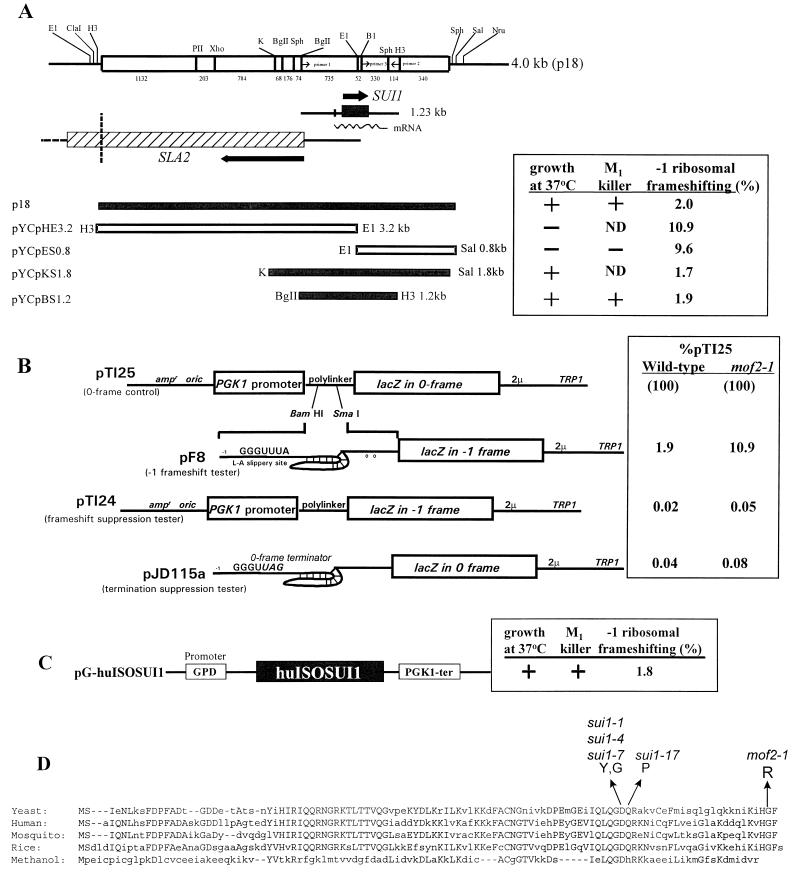
Molecular cloning of the MOF2 gene, characterization of the human homolog, and identification of the mof2-1 lesion. (A) At the top is a schematic representation of the 4.0-kb insert isolated from the YCp50 plasmid library (p18). Below are the locations of the SUI1 and SLA2 coding sequences. The open reading frame of SUI1 is indicated as an arrow and a shaded box within the 1.23-kb fragment. The subclones of plasmid p18 (left) and their effects on growth, −1 ribosomal frameshifting efficiency, and the M1 killer phenotype are shown on the right. ND, not done. (B) The elevated β-galactosidase activities observed in a mof2-1 strain were a result of elevated programmed −1 frameshifting efficiencies. The 0-frame reporter plasmid (pTI25), the programmed −1 frameshift reporter plasmid (pF8), the frameshift suppression reporter plasmid (pTI24), and the nonsense suppression plasmid (pJD115a) were transformed into either wild-type mof2-1 strains, and the β-galactosidase activities were monitored. The ratio of the tester plasmids (pTI24 and pJD115a) to the 0-frame control (pTI125) was determined, and the results are presented as percent pTI25). (C) The human homolog of the MOF2/SUI1 gene was obtained from a human cDNA library and cloned into the pG-1 vector harboring the yeast glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter and phosphatidylglycerol kinase 1 (PGK1) terminator sequences. Functional complementation tests were performed in a mof2-1 strain. (D) Sequence alignment of human, mosquito, rice, yeast, and Methanococcus (methanol) Sui1p/Mof2p homologs. The conserved amino acids are in uppercase. The mutation site of the mof2-1 allele was determined by a PCR strategy described in Materials and Methods. It is localized in the C terminus of the protein coding region and changes Gly105 to Arg. The mutation sites of the sui1 alleles are also shown (43).
The plasmids harboring HIS4AUG-lacZ or his4UUG-lacZ that were used for the his4UUG suppression experiments were kindly provided by T. F. Donahue (43). The two constructs are identical, except that a single base change in the his4UUG-lacZ allele alters the start codon from AUG to UUG.
Plasmids pYCp33sui1-1 and pYCp22sui1-1 were constructed as follows. The 1.2-kb BglII-HindIII fragment containing the sui1-1 allele was obtained from PCRs using primers 1 (5′-[BglII]-GACAGATCTGAATCT ATTCTGG-3′) and 2 (5′-[HindIII]-GACAAGCTTGGGATTCCATGAT-3′) (the underlined sequences are BglII and HindIII sites, respectively). Genomic DNA from a sui1-1 strain was used as the template for the PCR. The 1.2-kb BglII-HindIII-digested PCR products were cloned into vector pYCplac33 or pYCplac22. The presence of the sui1-1 mutation was confirmed by sequencing.
The FLAG-MOF2/SUI1 allele was constructed as follows. Primer a, containing the sequence which encodes the FLAG epitope, was linked in frame with the N terminus of Mof2p/Sui1p. Primer b corresponds to the 3′ end of the MOF2/SUI1 gene and contains a SalI site. The DNA fragment containing the FLAG epitope at the N terminus of Mof2p/Sui1p was generated from plasmid pYCp22MOF2 by PCR and cloned into E. coli expression vector pET-14b.
To delete the MOF2/SUI1 gene from the yeast chromosome, plasmid pKOM2 was prepared by first inserting the 1.2-kb BamHI-HindIII DNA fragment harboring the entire MOF2/SUI1 gene from YCpBH1.2 into a pUC19 vector and then replacing a 0.75-kb fragment of the MOF2 gene between the PstI and BamHI sites (containing the MOF2 transcription initiation site and part of the MOF2 coding region; see Fig. 3A) with a 3.0-kb DNA fragment harboring the hisG-URA3-hisG cassette. To make the mof2Δ strain, the PvuII-PvuII fragment from pKOM2 was used for transformation.
FIG. 3.
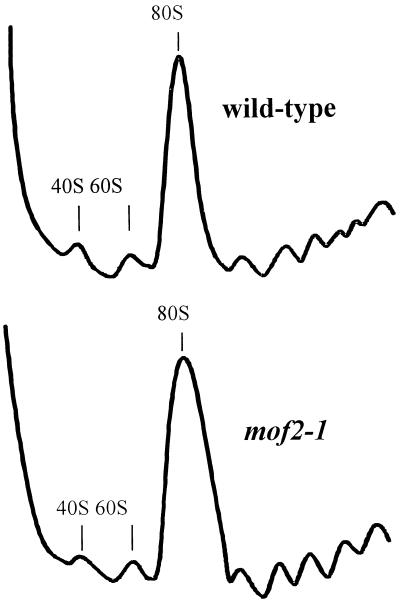
Analysis of polysome profiles from wild-type and mof2-1 cells. Wild-type (MOF2+) and mof2-1 cells were grown and harvested, and cytoplasmic extracts were prepared and fractionated on sucrose gradients as described in Materials and Methods. The A254s indicating the polysome profiles of wild-type and mof2-1 cells are shown.
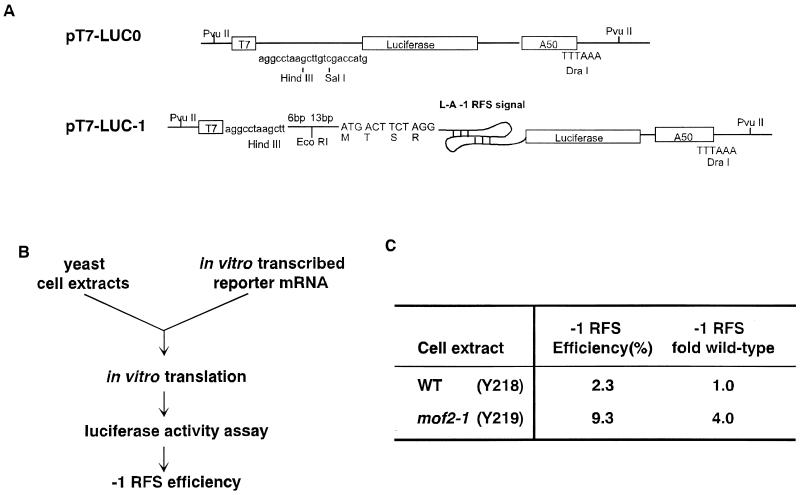
Plasmid pT7-LUC minus 3′ untranslated region A50 (referred to here as pT7-LUC0; see Fig. 3) was used to produce synthetic 0-frame luciferase-encoding mRNAs. The vector for production of the −1 ribosomal frameshift luciferase reporter mRNA was constructed as previously described (15).
Isolation and characterization of the MOF2 gene.
The MOF2 gene was cloned from a pYCp50 yeast genomic library. Strain JD742-9B (mof2-1) was transformed with this library, and transformants were screened by replica plating cells and monitoring their growth at either 24 or 37°C for 5 to 7 days. Colonies that grew at both temperatures were retested, and one strain harboring plasmid p18 was isolated. To confirm that the growth phenotype of the mof2-1 strain harboring the plasmid was a consequence of the plasmid, strains that lost the p18 plasmid were identified on 5-fluoro-orotic acid plates and reverted to a ts phenotype. The p18 plasmid was isolated and retransformed into a mof2-1 strain (35). The p18 plasmid rescued the ts growth phenotype of the mof2-1 strain. Subsequent subcloning and sequencing of the yeast genomic DNA fragment in plasmid p18 were used to identify the sequences from the yeast genome bank (see Fig. 1A). The following subclones, containing various DNA fragments, were based on yeast centromere plasmid pYCplac33 (see restriction map of the yeast genomic DNA fragment in Fig. 1): pYCpSLA2(HB) (2.4-kb HindIII-BglII DNA fragment), pYCpHE3.2 (3.2-kb HindIII-EcoRI DNA fragment), pYCpES0.8 (0.8-kb EcoRI-SalI DNA fragment), pYCpKS1.8 (1.8-kb KpnI-SalI DNA fragment), and pYCpBH1.2 (1.2-kb BglII-HindIII DNA fragment).
Identification of the mof2-1 mutation.
A PCR strategy was used to identify the mof2-1 allele by using primers 1 and 2 described above and primer 3 (5′-[EcoRI]-GATTTAAAGAGAATTCTTAAGG-3′ [the underlined sequence is an EcoRI site]). Genomic DNA (50 to 100 ng) was prepared (35) from the mof2-1 strain and used as the template in a PCR. Primers 1 and 2 were used to synthesize the 1.2-kb DNA fragment to construct pmof2BH, and primers 2 and 3 were used to synthesize the 0.4-kb DNA fragment to construct pmof2EH. Two PCR products from two different PCRs were used in the cloning reaction to minimize artifacts from the PCR. The PCR conditions used were as follows: 95°C for 5 min, 94°C for 1 min, 45 or 50°C for 1 min, and 72°C for 1.5 min for 25 cycles. The DNA fragments from the PCR were purified from a 1.5% agarose gel and used to replace the corresponding DNA fragment of the wild-type MOF2 gene which was on a YCplac33 vector as described above. Plasmids pmof2BH and pmof2EH were transformed into a mof2-1 strain (JD742-9B, Table 1), and the ts phenotype of the transformants was tested. The sequence of the plasmid that failed to rescue the ts phenotype of the mof2-1 allele was obtained to identify the mutation site. A single point mutation was identified from pmof2BH and pmof2EH. Hence, these two plasmids were both named pYCpmof2-1.
Isolation of the human homolog of the MOF2/SUI1 gene.
The human homolog of the MOF2/SUI1 gene (huISOSUI1) was obtained by a PCR approach. The primers used in the PCR were 5′-(BamHI)-AGCCGGATCCACCGAGGAAAAGGAACG-3′ and 5′-(SalI)-GGGAAAGTCGACTCATTGCAAGGAAATCC-3′ (the underlined sequences are BamHI and SalI sites, respectively). Four different sources of human cDNA libraries were used as templates for the PCRs. The conditions for the PCRs were the same as those described above. The products of the PCRs were purified, restriction enzyme digested, and cloned into low-copy yeast expression vector pG-1. All of the four cDNA library templates produced the same size of DNA fragment, and all were cloned and subjected to the functional tests.
Polysome analysis.
Cytoplasmic extracts, prepared as described by Baim et al. (3a), were fractionated on 15 to 50% sucrose gradients buffered with 50 mM Tris-acetate (pH 7.4)–50 mM NH4Cl–12 mM MgCl2–1 mM dithiothreitol. Gradients were centrifuged in an SW41 rotor at 40,000 rpm for 135 min at 4°C, fractionated, and analyzed by continuous monitoring of A254 (33a).
Purification of the Sui1p/Mof2p.
The MOF2/SUI1 allele was FLAG tagged at its N terminus and expressed in E. coli cells. E. coli cells harboring FLAG-tagged MOF2/SUI1 were grown at 20°C, and FLAG-Mof2p/Sui1p expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). A cytoplasmic extract was prepared and loaded onto a FLAG monoclonal antibody column. Fractions eluted from the column by either the FLAG peptide or glycine were analyzed by sodium dodecyl sulfate (SDS)–14% polyacrylamide gel electrophoresis (PAGE) and either stained with Coomassie blue or immunoblotted with an anti-FLAG monoclonal antibody. The function of purified wild-type Mof2p was tested in an in vitro translation system as described below.
In vitro frameshifting assay.
Plasmids pT7-LUC0 and pT7-LUC-1 were linearized with DraI, and a synthetic transcript containing a 7-Methyl-Gppp cap and a poly(A) tail was made by using T7 RNA polymerase and a MessageMachine kit (Ambion). The translation-competent yeast cell extracts were prepared from strains Y218 (wild type) and Y219 (mof2-1), as described by Tarun and Sachs (38), as follows. A yeast S30 cell extract was subjected to Sephadex G-25 superfine chromatography to remove low-molecular-weight translation inhibitors, and the peak void volume fractions were collected and pooled. The endogenous mRNAs were removed from the fractions by treatment with micrococcal nuclease. The in vitro frameshifting reaction mixture contained translation buffer (22 mM HEPES-KOH [pH 7.4], 120 mM potassium acetate, 2 mM magnesium acetate, 0.75 mM ATP, 0.1 mM GTP, 25 mM creatine phosphate, 0.04 mM amino acids, 1.7 mM dithiothreitol), 0.27-mg/ml creatine phosphate kinase, 0.07 mM methionine, 0.07-U/ml RNasin, 1.3-ng/ml mRNA (LUC0 or LUC-1 mRNA), and 70 μg of cell extract. The reactions were performed at 26°C for 1 h, stopped on dry ice, and thawed on ice, and luciferase activities were determined with a Turner 20/20 luminometer. The activity observed after addition of the frameshifting signal-containing LUC mRNA (LUC-1) was normalized to that of the 0-frame control (LUC0) and used to gauge frameshifting efficiencies in these cell extracts. We added 0, 40, 76, or 112 ng of purified Mof2 protein or an equal amount of protein storage buffer or 100 ng of bovine serum albumin (BSA) to the in vitro translation reaction mixtures, and the frameshifting efficiencies were calculated as described above. The functional half-lives of the transcripts (LUC-1 and LUC0) in the in vitro translation system in an individual cell extract, determined by measuring the luciferase activities at different time points after the reaction started, were approximately 40 min.
RESULTS
Isolation of the MOF2 gene.
A YCp50 yeast genomic library was used to clone the wild-type MOF2 gene by complementation of the ts growth phenotype of a mof2-1 strain. Plasmid p18 enabled a mof2-1 strain to grow at 37°C (Fig. 1A). Sequence data obtained from the insert of plasmid p18 confirmed that it harbors the complete SUI1 gene, whose product was shown to be involved in translation start codon selection (43), and all but the 3′-terminal portion of the SLA2 gene, which encodes a membrane cytoskeleton assembly protein (23; Fig. 1A). Further subcloning of this DNA fragment demonstrated that only plasmids containing the wild-type SUI1 gene complemented the ts growth phenotype of mof2-1 cells (Fig. 1A). Following sporulation and dissection of diploids resulting from a cross of ts mof2-1 and sui1-1 strains, analysis of 20 four-spore tetrads demonstrated that all four spores were ts, confirming that mof2-1 and sui1-1 map to the same genetic locus (data not shown).
Characterization of the MOF2/SUI1 gene.
The effects of expressing the MOF2/SUI1 gene in mof2-1 cells on programmed −1 frameshifting and M1 viral maintenance were monitored. Programmed −1 ribosomal frameshifting efficiency was determined in the mof2-1 strains harboring subclones of plasmid p18. Cells were transformed with either a plasmid that contains the lacZ coding region inserted downstream of a programmed −1 ribosomal frameshifting signal from the yeast L-A virus and in the −1 reading frame relative to a translational start site (−1 frameshift reporter) or a control plasmid containing the lacZ gene that lacks the frameshifting signal insertion and is in the 0 reading frame relative to the translational start codon (0-frame control; 14). The efficiencies of −1 ribosomal frameshifting were calculated by determining the ratio of the β-galactosidase activities in cells harboring the −1 frameshift reporter plasmid to activities from cells harboring the 0-frame control plasmid (11, 14). The programmed −1 ribosomal frameshifting efficiency was approximately 10% in a mof2-1 strain harboring only the YCplac33 vector or subclones lacking the entire MOF2/SUI1 gene (Fig. 1A, plasmids pYCpES0.8 and pYCpHE3.2). Programmed −1 ribosomal frameshifting efficiencies, however, decreased to approximately 2% in a mof2-1 strain harboring the wild-type MOF2/SUI1 gene. These levels are equivalent to those observed in wild-type cells (Fig. 1A, plasmids p18, pYCpBH1.2, and pYCpKS1.8).
Although programmed −1 ribosomal frameshifting appears to be elevated in mof2-1 cells, it is possible that the increased β-galactosidase activity was due to reasons other than changing programmed frameshift efficiencies. For example, increased β-galactosidase activity could be observed as a consequence of an increased translation frameshift and/or termination suppression or initiation of translation at a non-AUG or UUG codon. To rule out these possibilities, we monitored β-galactosidase activity by using reporter plasmids that did not contain the L-A −1 frameshift signal. Plasmid pTI24 (16a) is identical to 0-frame control plasmid pTI25, except that the lacZ gene is in the −1 frame with regard to the translational start site (Fig. 1B). Thus, the β-galactosidase activity generated from this plasmid is a consequence of an unprogrammed −1 ribosomal frameshift event. In addition, pJD115a (Fig. 1B) (18a) was used to test termination suppression. Plasmid pJD115a is derived from −1 frameshift plasmid pF8, except that a 0-frame termination codon disrupts the ability of the slippery site to promote efficient frameshifting. In addition, the lacZ gene is in the 0 frame with respect to the translation start site (Fig. 1B). Thus, the β-galactosidase activity generated from this plasmid should be a consequence of suppression of the nonsense codon. Isogenic wild-type and mof2-1 cells were transformed with these plasmids, β-galactosidase activities were measured, and the ratios of the two test plasmids to the 0-frame control (pTI25) were determined. The results demonstrated that the ratios of nonsense or frameshift suppression to the 0-frame control in wild-type and mof2 strains were 50- to 200-fold lower than the levels observed when the L-A programmed −1 frameshift site was present (Fig. 1B, compare pF8 to pTI24 and pJD115a). Taken together, these results demonstrate that the increase in β-galactosidase activity from the programmed −1 reporter plasmid observed in a mof2-1 strain (Fig. 1A) was most likely not a consequence of promiscuous translation initiation or enhanced nonsense or nondirected frameshift suppression (Fig. 1B).
The ability of cells to maintain the M1 virus was determined in a mof2-1 strain harboring subclones of the MOF2/SUI1 gene on a centromere plasmid. L-A and M1 were introduced into cells by cytoplasmic mixing (cytoduction), and these cells were replica plated onto a lawn of cells sensitive to the killer toxin. Cells maintaining the M1 virus secrete the killer toxin, creating a ring of growth inhibition (17). The results from these experiments indicate that although a mof2-1 strain harboring nonfunctional MOF2/SUI1 subclones cannot maintain M1 (Fig. 1A, plasmids pYCpES0.8 and pYCpHE3.2), a mof2-1 strain containing the wild-type MOF2/SUI1 gene is able to propagate M1 (Fig. 1A, plasmids p18, pYCpBH1.2, and pYCpKS1.8). The presence of the M1 virus was confirmed by determining that the M1 dsRNA was present in extracts of these cells (data not shown). Taken together, these results demonstrated that expression of the MOF2/SUI1 gene in a mof2-1 strain restored growth at 37°C, lowered programmed −1 ribosomal frameshifting efficiency to levels observed in wild-type cells, and restored the ability of cells to propagate the M1 virus.
The human homolog of the MOF2/SUI1 gene functions in yeast.
A human homolog of the MOF2/SUI1 gene product (huISOSUI1) has recently been identified by assembling expressed sequence tags (21). Several polypeptide fragments sequenced from rabbit reticulocyte eucaryotic initiation factor 1 (eIF-1) were shown to be similar to yeast Sui1p and huISOSUI1 (27). Sequence alignment of the yeast, rice, mosquito, Methanococcus, and human Sui1 proteins shows high levels of amino acid sequence similarity, suggesting that the gene for Sui1 has been well conserved throughout a wide range of organisms and during evolution (21; Fig. 1D).
Yeast Sui1p and huISOSUI1 share 60% identity and 80% similarity (21; Fig. 1D). To test whether mammalian eIF-1 can substitute for its yeast counterpart, Sui1p/Mof2p, in vivo, we cloned the human homolog of the MOF2/SUI1 gene from a human cDNA library by PCR and expressed it from a single-copy plasmid containing the constitutive yeast glyceraldehyde-3-phosphate dehydrogenase promoter and phosphatidylglycerol kinase 1 terminator (see Materials and Methods). This plasmid was transformed into a mof2-1 strain and introduced into a mof2Δ/sui1Δ strain by plasmid shuffling (see Materials and Methods). The results from these experiments are summarized in Fig. 1C and Table 2. The plasmid expressing huISOSUI1 supported the growth of mof2Δ/sui1Δ cells at both permissive and nonpermissive temperatures, allowed cells to exhibit wild-type phenotypes of −1 ribosomal frameshifting efficiency, and enabled these cells to propagate the M1 virus (Table 2). These results indicate that mammalian eIF-1 can function in yeast cells.
TABLE 2.
Characterization of frameshifting efficiencies in isogenic MOF2/SUI1 cells
| Strain (genotype) | β-Galactosidase activitya
|
Frameshifting efficiency (%)
|
Fold wild-type frameshifting
|
Maintenance or loss of M1 dsRNAb | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | −1 | +1 | −1 | +1 | −1 | +1 | ||
| Y218 (wild type) | 43.18 | 0.91 | 1.39 | 2.1 | 3.2 | 1.0 | 1.0 | + |
| Y219 (mof2-1) | 45.46 | 4.86 | 1.64 | 10.7 | 3.6 | 5.1 | 1.2 | − |
| Y220 (sui1-1) | 44.70 | 2.41 | 1.38 | 5.4 | 3.1 | 2.6 | 1.0 | + |
| Y221 (huISOSUI1) | 32.38 | 0.65 | 0.97 | 2.0 | 3.0 | 1.0 | 1.0 | + |
These experiments were conducted with isogenic SUI1/MOF2 strains. The values shown are average measurements of at least three individual colonies. β-Galactosidase activity is presented as units of optical density at 420 nm per hour per unit of optical density at 600 nm, with variations of no more than 15%. The −1 or +1 frameshifting efficiency was determined by measuring the ratio of β-galactosidase activity in a strain harboring either the −1 or +1 frameshifting reporter plasmid to the activity in the same strain harboring the 0-frame control plasmid. The frameshifting increases (fold wild type) are mutant/wild-type frameshifting efficiency ratios. In the last column, the M1 killer phenotype was determined by cytoduction in which the donor strain was JD758.
The maintenance (+) or loss (−) of M1 dsRNA was analyzed by 1.5% agarose gel electrophoresis from nucleic acids extracted from the cytoductants.
Identification and characterization of the mof2-1 lesion in the SUI1 gene.
The mutation responsible for the mof2-1 allele was determined by PCR cloning as described previously (11). DNA fragments containing different regions of the MOF2/SUI1 gene were isolated by PCR from the genomic DNA of mof2-1 cells, cloned into a yeast shuttle vector, and transformed into mof2-1 cells (11; see Materials and Methods). The DNA fragment containing the mutation was identified by determining which fragment of the MOF2/SUI1 gene failed to complement the mof2-1 ts growth phenotype. The mof2-1 mutation is near the carboxyl terminus of the protein, converting highly conserved Gly105 to Arg (Fig. 1D). As described below, this mutation was confirmed to be responsible for all of the mof2-1 defects. Interestingly, both the mof2-1 and sui alleles were identified in screens that monitored translational fidelity (18, 43). The mutations that affect either translation initiation (sui1 alleles) or programmed −1 frameshifting (mof2-1 allele) are both located very near the carboxyl terminus of the protein in highly conserved amino acids Asp81 and Gln82 within the context LQGDQR (sui1 alleles) (21, 43) or Gly105 (mof2-1 allele) within the conserved HGF motif, indicating a potential functional domain in this region.
Strains harboring the mof2-1 allele allow translation initiation at a non-AUG codon.
Strains harboring the previously identified sui1 alleles allow translation to begin at a UUG codon in the absence of an AUG start codon (8, 43). We examined whether a strain harboring the mof2-1 allele was also able to initiate translation at a UUG codon by using a set of isogenic strains constructed to eliminate possible differences in the genetic background for the sui1-1 and mof2-1 alleles. Since the MOF2/SUI1 gene is essential for cell survival, the isogenic mof2-1, sui1-1, and wild-type (MOF2+/SUI1+) strains were constructed by plasmid shuffling (35). Wild-type, sui1-1 or mof2-1 alleles on centromere-based plasmids were introduced into a wild-type strain (JD272) to support cell viability when the MOF2/SUI1 gene was deleted from the chromosome (see Materials and Methods). mof2Δ/sui1Δ strains carrying either the plasmid-borne sui1-1 or mof2-1 allele were ts for growth, indicating that the plasmid-borne mutant alleles function analogously to their chromosomal counterparts. The strain containing the wild-type MOF2/SUI1 gene was able to grow at both 24 and 37°C.
The isogenic wild-type (MOF2+/SUI1+), mof2-1, and sui1-1 cells were transformed with constructs containing either a normal AUG start codon in the HIS4 gene or one in which the AUG codon was changed to a UUG codon and fused in frame with the lacZ gene (HIS4-AUG-lacZ and his4-UUG-lacZ; 43). The β-galactosidase activities were determined, and the ratios of the activities from his4-UUG-lacZ and HIS4-AUG-lacZ were calculated to represent the level of translation initiation at a UUG codon. The results of these experiments are summarized in Table 3. Similar to a sui1-1 strain, mof2-1 cells were able to initiate translation at a UUG codon at efficiencies equivalent to that observed in sui1-1 cells. The basal levels of the β-galactosidase expression from HIS4-AUG-lacZ were similar in wild-type and mof2-1 cells (Table 3), indicating that there were no significant translation initiation defects in the mof2-1 strain. The basal levels of β-galactosidase expression from HIS4-AUG-lacZ in a sui1-1 strain was reduced less than twofold (Table 3). Further, strains harboring the huISOSUI1 allele did not utilize UUG efficiently as a translation start codon, consistent with the phenotypes observed in cells harboring the yeast wild-type MOF2/SUI1 gene (Table 3).
TABLE 3.
his4UUG suppression in MOF2/SUI1 strains
| Strain (genotype)a | HIS4AUG-lacZ β-galactosidase activity | his4UUG-lacZ β-galactosidase activity | UUG/AUG ratio (%) | UUG suppression (fold wild type) |
|---|---|---|---|---|
| Y218 (wild type) | 8.6 | 0.22 | 2.5 | 1.0 |
| Y219 (mof2-1) | 7.1 | 1.14 | 16.0 | 6.4 |
| Y220 (sui1-1) | 4.6 | 0.68 | 15.6 | 6.2 |
| Y221 (huISOSUI1) | 6.1 | 0.19 | 3.0 | 1.2 |
These experiments were performed with isogenic SUI1/MOF2 strains in which the SUI1/MOF2 gene was deleted from the chromosome and the specific alleles were carried on centromere-based plasmids. Low-copy-number plasmids containing either HIS4AUG-lacZ or his4UUG-lacZ were transformed into each strain. The two constructs are identical, except that the start codon in his4UUG-lacZ is altered from AUG to UUG. The values shown are averages of two independent experiments using three individual colonies from each strain for the β-galactosidase assay. β-Galactosidase activity is presented as units of optical density at 420 nm per hour per unit of optical density at 600 nm. The UUG/AUG ratio was calculated by dividing the β-galactosidase activity in a strain harboring his4UUG-lacZ by the β-galactosidase activity in the same strain harboring HIS4AUG-lacZ. UUG suppression is expressed as the UUG/AUG ratio of the mutant divided by the UUG/AUG ratio of the wild type.
The mof2-1 allele of SUI1, but not the sui2-1 or SUI3-3 allele, affects programmed ribosomal frameshifting.
Since mutations in the MOF2/SUI1 gene affect programmed −1 ribosomal frameshifting, we investigated the possibility that these effects are solely attributable to translation initiation defects. Strains harboring sui2 and SUI3 alleles have the same levels of translation initiation at a UUG codon as those with sui1 alleles, as monitored by using the his4-UUG-lacZ reporter construct (8). Therefore, we asked whether the strains harboring the sui alleles affect programmed −1 ribosomal frameshifting as well. The −1 ribosomal frameshifting reporter constructs were introduced into wild-type, mof2-1, sui1-1, sui2-1, and SUI3-3 strains to determine the effect of these mutations on −1 ribosomal frameshifting efficiency. The results of these experiments indicated that the −1 programmed frameshifting efficiencies in wild-type or mof2-1 cells were 2.3 and 8.5%, respectively, consistent with the results observed previously (Fig. 1A). The sui1-1 allele increased programmed −1 ribosomal frameshifting approximately twofold (Table 4). Although sui2-1 and SUI3-3 have the same or even higher levels of translation initiation at a UUG codon than sui1-1 or mof2-1 (8), these alleles did not affect the efficiency of programmed −1 ribosomal frameshifting significantly (Table 4).
TABLE 4.
Assay of ribosomal frameshifting in sui suppressors
| Straina | β-Galactosidase activity
|
Frameshifting efficiency (%) | Fold wild-type frameshifting | |
|---|---|---|---|---|
| 0 | −1 | |||
| Wild type | 24.8 | 0.57 | 2.3 | 1.0 |
| mof2-1 | 29.7 | 2.54 | 8.5 | 3.7 |
| sui1-1 | 32.8 | 1.47 | 4.5 | 2.0 |
| sui2-1 | 6.9 | 0.19 | 2.7 | 1.2 |
| SUI3-3 | 46.8 | 1.30 | 2.8 | 1.2 |
The strains used in these experiments were either from the MOF2 isogenic strain (the wild type and a mof2-1 mutant) or from isogenic sui strains (sui1-1, sui2-1, and SUI3-3 mutants; 43). The −1 frameshifting reporter or 0-frame control plasmid was transformed into each strain, and three individual colonies from each strain were tested for β-galactosidase activity. The results had variations of no more than 11%. The −1 frameshifting efficiency was determined by the ratio of β-galactosidase activity in a strain harboring the −1 ribosomal frameshifting reporter plasmid to the activity in the same strain harboring the 0-frame control plasmid. The frameshifting increase values (fold wild type) are ratios of the frameshifting efficiency of mutant cells to that of wild-type cells.
The differences in −1 ribosomal frameshifting in mof2-1 and sui1-1 cells were further characterized in the isogenic mof2-1 and sui1-1 strains (Y219 and Y220) to eliminate possible difference in the genetic background. The results of these experiments demonstrated that the efficiencies of −1 ribosomal frameshifting in mof2-1 and sui1-1 strains were 10.7 and 5.4%, equivalent to 5.1- and 2.6-fold increases, respectively, compared to wild-type cells (Table 2). The steady-state levels of the −1 frameshift reporter lacZ transcripts in all of these strains were equivalent (data not shown), suggesting that the different frameshifting efficiencies observed in these cells were not due to differences in the stability of this mRNA. Further, the ability of these strains to maintain the M1 killer virus was monitored as described above. The results demonstrated that cells harboring the wild-type (MOF2+/SUI1+) or sui1-1 allele were able to maintain the M1 dsRNA virus, while strains harboring the mof2-1 allele could not maintain the virus (Table 2, last column). These data are consistent with the large change in −1 ribosomal frameshifting efficiency observed in a mof2-1 strain, an effect that promotes loss of the M1 virus (17, 18). The higher −1 frameshifting efficiency in mof2-1 cells compared to sui1-1 cells suggests that the frameshifting defects in mof2-1 cells are more severe. Given that the levels of translation initiation at a UUG codon are equivalent for the mof2-1, sui1-1, sui2-1, and SUI3-3 mutants (8), their different effects on −1 ribosomal frameshifting cannot be attributed to a general defect in translation initiation. Thus, the MOF2/SUI1 allele-specific effects suggest the involvement of Mof2p/Sui1p in both the initiation and elongation phases of protein synthesis.
The mof2-1 and sui1-1 mutants do not affect +1 ribosomal frameshifting.
The results described above indicate that programmed −1 ribosomal frameshifting is affected in mof2-1 cells. We next determined whether +1 programmed ribosomal frameshifting is also affected by this mutation. Isogenic wild-type (MOF2+/SUI1+), sui1-1, and mof2-1 strains were transformed with a +1 frameshifting reporter construct which harbors the lacZ reporter gene inserted downstream of a programmed +1 frameshifting signal from the yeast Ty1 retrotransposable element in the +1 reading frame relative to the translational start site (4). The efficiencies of +1 ribosomal frameshifting were measured by determining the ratio of the β-galactosidase activities in cells harboring the +1 frameshift reporter plasmid to the activities in cells harboring the 0-frame control plasmid. The results demonstrated that the +1 frameshifting efficiencies in mof2-1, sui1-1, and wild-type cells were all the same (Table 2), indicating that the increased frameshifting efficiency in mof2-1 cells is specific for −1 programmed ribosomal frameshifting rather than a consequence of a general defect in translational fidelity.
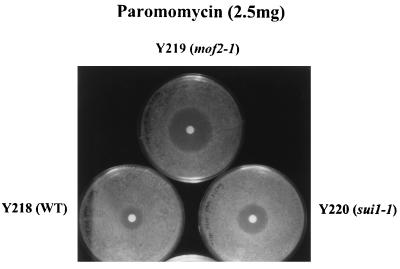
The mof2-1 strain is sensitive to paromomycin.
Strains harboring mutations that lower translational fidelity have been shown to be hypersensitive to the aminoglycoside antibiotic paromomycin, a drug that is thought to increase the frequency of misreading in yeast (33, 36). The paromomycin sensitivities of the isogenic mof2-1, sui1-1, and wild-type strains were tested. As shown in Fig. 2, drug sensitivity can be monitored by determining the size of the zone of growth inhibition around a drug-containing disc on a lawn of cells. The results demonstrated that mof2-1 cells were more sensitive to paromomycin than were sui1-1 cells. Interestingly, sui1-1 cells had an intermediate level of sensitivity to the drug. This result mirrors our observation that although a mof2-1 strain shows the highest increases in −1 ribosomal frameshifting efficiency, programmed −1 ribosomal frameshifting efficiencies were slightly increased in a sui1-1 strain compared to wild-type cells. Increased paromomycin sensitivity is also consistent with our previous findings which showed cells with the mof4-1 allele of the UPF1 gene to be paromomycin sensitive (11). Thus, the paromomycin sensitivities of mof2-1 and sui1-1 cells further support the possible role of Mof2p/Sui1p in controlling translational fidelity.
FIG. 2.
Drug sensitivity tests of isogenic MOF2/SUI1 strains. Cells were grown in C-Trp medium to mid-log phase and diluted to an optical density at 600 nm of 0.4 to 0.5, and a 300-μl volume of cells was spread on the C-Trp plates. A disc containing 2.5 mg of paromomycin was placed on the plate, and the ring of cell growth inhibition around the disc was measured after incubation at 24°C for 4 to 5 days. These experiments were repeated with at least two independent colonies and performed with different concentrations of drugs. WT, wild type.
Strains harboring the mof2-1 allele do not demonstrate a polysome defect.
The results described above indicated that the basal levels of β-galactosidase expression from HIS4-AUG-lacZ were similar in wild-type and mof2-1 cells, indicating that there were no significant translation initiation defects in the mof2-1 strain (Table 3). To confirm and extend this result, we analyzed the polysomes found in wild-type and mof2-1 cells. Extracts from wild-type and mof2-1 cells were prepared, and polysomes were analyzed on sucrose gradients (see Materials and Methods). The results are shown in Fig. 3. Comparison of the polysome profiles of wild-type and mof2-1 cells demonstrated no dramatic polysome defect (Fig. 3).
The defect in −1 ribosomal frameshifting in mof2-1 cells can be recapitulated in an in vitro translation system.
To investigate whether Mof2p is directly involved in modulating programmed −1 ribosomal frameshifting, translationally competent yeast cell extracts from wild-type and mof2-1 strains lacking the killer virus were prepared (Materials and Methods; 38). Capped and polyadenylated transcripts containing the luciferase protein coding region either lacking (0-frame control, pT7-LUC0; Fig. 4A) or containing the L-A −1 ribosomal frameshift signal (−1 frameshift reporter, pT7-LUC-1; Fig. 4A) were synthesized in vitro (see Materials and Methods). These mRNAs were added to the translation extract, and the amount of luciferase synthesized was determined by a luciferase activity assay (Fig. 4B). Programmed −1 ribosomal frameshifting efficiencies were monitored in vitro by determining the ratio of luciferase activity from the −1 frameshift reporter transcript (pT7-LUC-1) to that from the 0-frame control mRNA (pT7-LUC0). Consistent with the results observed in vivo, extracts prepared from wild-type cells promoted a −1 ribosomal frameshifting efficiency of 2.3% and extracts prepared from a mof2-1 strain had a −1 ribosomal frameshifting efficiency of 9.3% (Fig. 4B). Thus, the increased −1 ribosomal frameshifting observed in strains harboring the mof2-1 allele can be recapitulated in an in vitro translation system. The increase in programmed ribosomal frameshifting was not a consequence of stabilizing the reporter transcripts in these extracts, since the functional half-lives of the pT7-LUC0 and pT7-LUC-1 transcripts were the same in reaction mixtures containing either wild-type or mof2-1 cell extracts (data not shown).
FIG. 4.
In vitro frameshifting assay with yeast cell extracts. (A) The capped and polyadenylated in vitro-synthesized transcripts, used for in vitro translation reactions, shown contain the luciferase protein coding region either lacking (0-frame control [LUC0]) or containing (LUC-1) a −1 ribosomal frameshift (RFS) site. (B) Equal amounts (20 ng) of RNA were added to the in vitro translation system with an individual cell extract. The level of protein expression was measured by a luciferase assay, and the efficiency of −1 frameshifting was determined by the ratio of luciferase activity from LUC-1 to that from LUC0. The values shown are averages of at least three independent experiments done in triplicate. WT, wild type.
Purified Mof2p restores wild-type levels of ribosomal frameshifting in extracts prepared from mof2-1 cells.
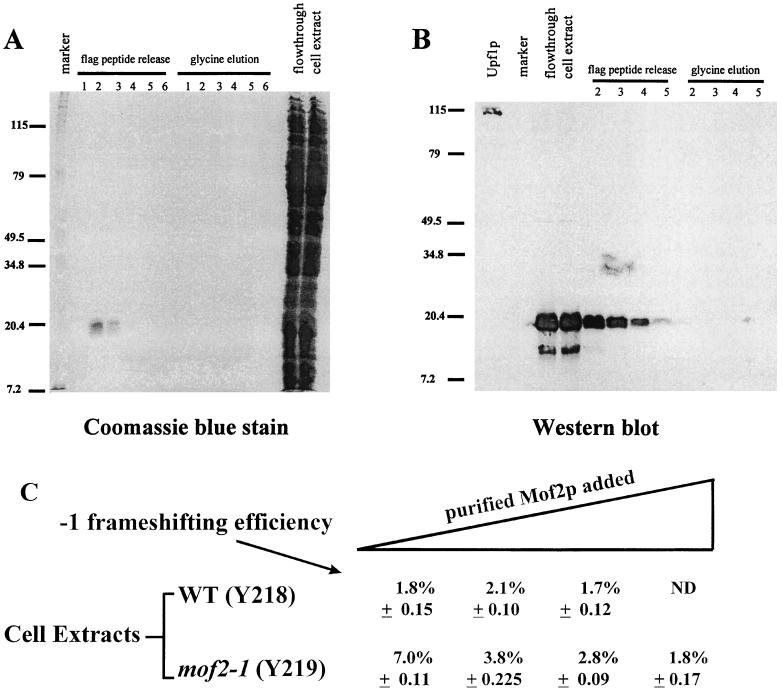
We next examined whether purified Mof2p/Sui1p can correct the programmed −1 ribosomal frameshifting defect in an extract prepared from a mof2-1 strain. The FLAG-MOF2/SUI1 allele, which harbors an epitope tag at the amino terminus of its protein coding region, was constructed and utilized to detect and purify Mof2p/Sui1p. Cells harboring the FLAG-MOF2/SUI1 allele had the same growth and frameshifting phenotypes as cells harboring the wild-type MOF2/SUI1 gene (data not shown). FLAG-MOF2/SUI1 was expressed in E. coli, and cytoplasmic extracts were prepared and applied to an anti-FLAG immunoaffinity chromatography column. Mof2p/Sui1p was subsequently eluted from the antibody column with the FLAG peptide. As shown in the Coomassie blue-stained SDS-PAGE gel in Fig. 5A, fractions eluted from the antibody column by the FLAG peptide contained a single band with an apparent molecular mass of approximately 17 kDa, consistent with previous reports that Sui1p is 16 kDa (32). Furthermore, proteins in the FLAG peptide-eluted fractions were able to react with the anti-FLAG antibody, as detected by immunoblotting (Fig. 5B), indicating that the purified protein was encoded by the FLAG-MOF2/SUI1 allele. Increasing amounts of purified Mof2p/Sui1p were added to the in vitro translation reaction mixtures and programmed −1 ribosomal frameshifting efficiency was determined as described above (Fig. 5C). The results demonstrate that exogenously added Mof2p/Sui1p can reduce the efficiency of programmed −1 ribosomal frameshifting in extracts of mof2-1 cells to the level observed in extracts of wild-type cells (Fig. 5C). As a control, experiments were performed in which 200 ng of BSA was added as a nonspecific control. The results demonstrated that addition of BSA to the reactions did not reduce programmed −1 frameshift efficiencies in a mof2 extract (data not shown). The added Sui1p/Mof2p had no apparent effect on the efficiency of frameshifting in a wild-type cell extract (Fig. 5C). These results strongly suggest that Mof2p/Sui1p is directly involved in regulating the efficiency of programmed −1 ribosomal frameshifting.
FIG. 5.
Purification of Mof2p and in vitro frameshifting assay. (A and B) A cytoplasmic extract of E. coli cells expressing FLAG-MOF2/SUI1 was prepared and loaded onto a FLAG monoclonal antibody column. Fractions eluted from the column were separated by SDS-PAGE and monitored by staining with Coomassie blue (A) or immunoblotting with an anti-FLAG monoclonal antibody (B). (C) In vitro frameshifting assay using yeast cell extracts and purified FLAG-Mof2/Sui1 protein. In vitro translation extracts and reactions were prepared as described by Tarun and Sachs (38), with strains Y218 (wild type [WT]) and Y219 (mof2-1 in Y218). Capped and polyadenylated mRNA (LUC-1 and LUC0) were used to program the in vitro translation reactions by using either the wild-type or mof2-1 extract. Zero, 40, 76, or 112 ng of purified Mof2 protein was added to the in vitro translation reaction mixtures (from left to right), and the frameshifting efficiency is defined as the ratio (percentage) of luciferase activity produced from the −1 ribosomal frameshift mRNA (LUC-1) to the luciferase activity produced from the 0-frame control (LUC0). The luciferase activity from the 0-frame control from extracts prepared from either wild-type or mof2 cells was varied between 20 and 30 U. Reactions were prepared in triplicate, and the values shown are averages of two sets of independent experiments. The standard deviations from these experiments are shown. The values to the left of A and B are molecular sizes in kilodaltons.
DISCUSSION
We previously identified a set of yeast chromosomal mutants having increased efficiencies of programmed −1 ribosomal frameshifting. We have been characterizing these trans-acting factors to elucidate the role of the protein synthetic machinery in this posttranscriptional regulatory mechanism. In this study, we isolated the MOF2 gene and showed that it is isogenic with SUI1. Sui1p was previously shown to be important in start site selection in translation initiation (43). Our studies on the mof2-1 allele of the SUI1 gene indicate that it is also required to help maintain the appropriate reading frame during the elongation phase of translation. Based on these observations, we hypothesize that Mof2p/Sui1p is a pivotal protein involved in the accuracy of both the initiation and elongation phases of the translation program. The role of the MOF2/SUI1 gene product is probably not unique to yeast, since a human homolog of this gene has been identified and can function in yeast cells. Each of these points will be discussed below.
The human Sui1p homolog, eIF-1, functions in yeast cells to modulate programmed −1 ribosomal frameshifting.
The wild-type MOF2 gene was shown to be allelic to SUI1 (Fig. 1). A human homolog of Sui1p/Mof2p, called huISOSUI1 (eIF-1), has recently been identified. huISOSUI1 shares 60% identity and 80% similarity with Mof2p/Sui1p (Fig. 1D) (21). eIF-1 is one of the smallest translation initiation factors defined with an in vitro translation system (40). In vitro studies have shown that eIF-1 stimulates tRNA or mRNA binding to the 40S ribosomal subunit and the 80S ribosomal complex (40). We have shown that the huISOSUI1-encoding gene suppressed the effects of the mof2-1 mutation. A mof2-1 strain harboring the huISOSUI1-encoding gene was viable at 37°C, showed an efficiency of programmed −1 frameshifting equivalent to that of wild-type MOF2/SUI1 cells, and was able to maintain the M1 killer virus (Fig. 1B and Table 2). Taken together, these results demonstrate that the SUI1/MOF2 gene is conserved between mammalian and yeast cells. Thus, understanding the mechanism of how this conserved protein functions to monitor the translational process in yeast will aid our understanding of the underlying mechanisms governing translation in mammalian systems.
Mof2p/Sui1p is a general monitor of translational fidelity.
The mof2-1 allele resulted in elevated −1 ribosomal frameshifting efficiency (Table 2), loss of the M1 killer virus (Table 2), greater paromomycin sensitivity (Fig. 2), and altered translation initiation codon selection (Table 3). Isolation of the wild-type MOF2 gene demonstrated that it is identical to the previously isolated SUI1 gene. The sui mutants (i.e., sui1, sui2, and SUI3) were originally identified as mutations that allowed an initiator Met-tRNAiMet to mismatch base pair with a UUG codon in the absence of an AUG initiator codon (43). The SUI2 and SUI3 genes were shown to encode yeast translation initiation factors eIF-2α and eIF-2β, respectively (10, 19). The SUI1 gene product is not part of the eIF-2 complex (43). Recent results have demonstrated that 30% of Sui1p is associated with the eIF-3 complex (32). eIF-3 is thought to stabilize the eIF-2–GTP–Met–tRNAiMet complex on the 40S ribosomal subunit and prevent 60S ribosomal subunit joining (31). However, 70% of Sui1p is not associated with eIF-3 and thus may function independently (3).
Our results suggest direct involvement of Sui1p/Mof2p in the regulation of programmed −1 ribosomal frameshifting efficiencies. The experiments using the sui2-1 and SUI3-3 mutants demonstrate that general initiation defects cannot account for the extent of the observed increases in programmed −1 ribosomal frameshifting and inability to propagate M1 observed in mof2-1 strains. The possibility that the increased β-galactosidase activity observed in mof2-1 strains might be a consequence of utilizing an internal UUG translation initiation site can be eliminated since the ribosomal frameshift transcript sequence does not contain any UUG codons in frame with the lacZ reporter gene. Although both the mof2-1 and sui1-1 alleles are capable of promoting translation initiation at a UUG codon, only mof2-1 strains lose the M1 killer virus (Table 2), demonstrating that the effect on programmed ribosomal frameshifting is most pronounced with the mof2-1 allele. The elevated programmed −1 ribosomal frameshifting efficiency observed in mof2-1 cells can be recapitulated in an in vitro frameshifting assay (Fig. 4B), and addition of purified wild-type Mof2p can repair the defect in programmed −1 ribosomal frameshifting in extracts prepared from mof2-1 cells (Fig. 5C). This observation is particularly significant since it demonstrates a direct effect of Mof2p/Sui1p on translation elongation. Taken together, these results strongly indicate that Sui1p/Mof2p is directly involved in the monitoring of −1 ribosomal frameshifting.
Interestingly, the effects of the mof2-1 allele were specific to programmed −1 frameshifting but did not affect Ty1-programmed +1 ribosomal frameshifting (Table 2). Yeast cells harbor two different viral systems that utilize programmed ribosomal frameshifting. The frameshift site found in the Ty1 retrotransposons induces a +1 frameshift (5), while the L-A site induces a −1 frameshift event (13). Both frameshift processes require specific signals (or slippery sites) and a ribosomal pausing event. They differ, however, in their requirements for A- and P-site occupancy of the ribosome. The +1 frameshift event is favored by an empty A site occurring after peptide bond formation and translocation (20). The −1 frameshift event, however, requires occupancy of both the A and P sites (13). Thus, the specific effects observed in strains harboring a mof2-1 mutation on −1, but not +1, programmed frameshifting are consistent with a role of this protein in maintaining fidelity at early ribosome-dependent proofreading steps such as those modulated by EF-1α (15).
How is Sui1p/Mof2p involved in monitoring the fidelity of multiple steps in translation?
Appropriate recognition of RNA signals is necessary for proper regulation of RNA-mediated posttranscriptional events. Altered recognition of RNA signals can lead to inappropriate regulation of gene expression. Mutations in the MOF2/SUI1 gene allow initiation of translation at non-AUG codons and also reduce a very specific aspect of fidelity during translation elongation. We hypothesize that Mof2p/Sui1p is a key component of a complex that is required to monitor translational fidelity and that mutations in the MOF2/SUI1 gene alter this process by reducing translational fidelity at the level of codon-anticodon recognition.
If this hypothesis is correct, then how does this small protein monitor translational fidelity? Although the final answer still has to be determined, the paradigm for fidelity in translation initiation and elongation may give us a clue to this problem. Initiation and elongation both utilize GTP hydrolysis as a key component employed to regulate the accuracy of these processes (7, 28, 31, 34). We hypothesize that Mof2p/Sui1p is a factor that regulates the proofreading activity and that mutations in its gene can reduce fidelity by altering the kinetics of ribosome-associated events. We hypothesize several models by which the mof2-1 allele reduces the ability of ribosomes to recognize near-cognate tRNA-mRNA interactions that are normally rejected by both the 43S preinitiation complex and the ribosome during the elongation phase of protein synthesis.
Mof2p/Sui1p can be potentially involved in a number of steps in the proofreading process. First, it is possible that Mof2p/Sui1p affects the GTP hydrolysis rate, so that the fidelity of RNA recognition is reduced. Mutations in the MOF2/SUI1 gene may alter the rate of GTP hydrolysis for eIF-2 or elongation factor 1 (EF-1), leading to reduced fidelity during the initiation and elongation phases of translation. The MOF2/SUI1 gene does not demonstrate any similarity to other known genes harboring nucleoside triphosphate-binding motifs, suggesting that this protein is not a GTP-hydrolyzing protein. It is possible, however, that Sui1p/Mof2p can alter GTP hydrolysis by functioning as a GTPase-activating protein to enhance GTP hydrolysis by other proteins. Potential targets of Mof2p/Sui1p include eIF-2, EF-1α, and eEF-2. These are all known GTP-hydrolyzing proteins involved in translation initiation and elongation. We are currently examining whether purified Sui1p/Mof2p has GTPase activity or can modify the GTPase activities of other proteins. If this is the case, regulation of GTP hydrolysis as a sensor of the accuracy of the cognate codon-anticodon interactions would serve as a general modulator in translation (34).
Second, the Mof2p/Sui1p mutation may affect the affinity of the aa-tRNA or the aa-tRNA binding factor (eIF-2 or EF-1α) for the ribosome P or A site, respectively. An enhanced affinity would increase the residence time at the ribosome, even in the absence of a noncognate interaction, such as between the UUG codon and Met-tRNAi. This would increase the probability that the intrinsic GTPase activity would occur, depositing an incorrect but near-cognate aa-tRNA. Similarly during elongation, while the intrinsic rate of GTP hydrolysis is low, a sufficient increase in residence time would allow hydrolysis to occur spontaneously. This would provide the increased A-site residency required by −1 programmed frameshifting (13).
Third, errors in translation may be sensed at the P site (2). Reduced P-site editing may allow the ribosome to tolerate a noncognate codon-anticodon pair. The initial binding of Met-tRNAi at the initiating codon occurs at the P site. This mechanism would allow the ribosome to better retain the near cognate tRNA both for the initial binding of Met-tRNAi at a UUG codon and consequent to the ribosomal slippage after programmed −1 frameshifting.
Mof2p/Sui1p appears to be a general monitor of the fidelity of RNA recognition that is actively involved in both the initiation and elongation phases of protein synthesis. Thus, these results suggest that Mof2p/Sui1p is a key regulator that is involved in the monitoring of at least two very important aspects of gene expression. Consequently, understanding how Mof2p/Sui1p functions will greatly contribute to our understanding of how fidelity of RNA recognition is regulated within the cell.
ACKNOWLEDGMENTS
This work was supported by a grant (GM48631) from the National Institutes of Health and an American Heart Association Established Investigator Award to S.W.P. and by grant 8-97 to J.D.D. from the UMDNJ Foundation.
We thank Carlos Gonzalez, Maria Ruiz-Echevarria, Shuang Zhang, Kevin Czaplinski, and Philip Farabaugh for discussions of the work and critical reading of the manuscript. We thank Thomas Donahue for sending us strains and plasmids and Alan Hinnebusch for strains harboring mutations in the GCD10 gene.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson R P, Menninger J R. Tests of the ribosome editor hypothesis. III. A mutant Escherichia coli with a defective ribosome editor. Mol Gen Genet. 1987;209:313–318. doi: 10.1007/BF00329659. [DOI] [PubMed] [Google Scholar]
- 3.Asano K, Kinzy T G, Merrick W C, Hershey J W B. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Sci. 1997;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- 3a.Baim S B, Pietras D F, Eustice D C, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985;5:1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasundaram D, Dinman J D, Wickner R B, Tabor C W, Tabor H. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belcourt M F, Farabaugh P J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley I A, Dingard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess S M, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- 8.Castilho-Valavicius B, Yoon H, Donahue T F. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics. 1990;124:483–495. doi: 10.1093/genetics/124.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 10.Cigan A M, Pabich E K, Feng L, Donahue T F. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares sequence identity with the human α subunit. Proc Natl Acad Sci USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Dinman J D, Peltz S W. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 13.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinman J D, Icho T, Wickner R B. A −1 ribosomal frameshifting in a double-stranded RNA virus of yeast forms a Gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinman J D, Kinzy T G. Translational misreading: mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 16.Dinman J D, Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Peptidyl-transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc Natl Acad Sci USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Dinman J D, Icho T, Wickner R B. A −1 ribosomal frameshifting in a double-stranded RNA virus of yeast forms a Gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinman J D, Wickner R B. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Dinman J D, Wickner R B. 5S rRNA is involved in fidelity of translational reading frame. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue T F, Cigan A M, Pabich E K, Castilho-Valavicius B. Mutations at a Zn(II) finger motif in the yeast eIF2β gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 20.Farabaugh P J. Post-transcriptional regulation by Ty retrotransposon of Saccharomyces cerevisiae. J Biol Chem. 1995;270:10361–10364. doi: 10.1074/jbc.270.18.10361. [DOI] [PubMed] [Google Scholar]
- 21.Fields C, Adams M D. Expressed sequence tags identify a human isolog of the SUI1 translation initiation factor. Biochem Biophys Res Commun. 1994;198:288–291. doi: 10.1006/bbrc.1994.1040. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi S-I, Murakami Y. Rapid and regulated degradation of ornithine decarboxylase. Biochem J. 1995;306:1–10. doi: 10.1042/bj3060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtzman D A, Yang S, Drubin D G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacks T, Madhani H D, Masiraz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacks T, Power M D, Masiarz F R, Luciw P, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 26.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 27.Kasperaitis M A M, Voorma H O, Thomas A M. The amino acid sequence of eukaryotic translation initiation factor 1 and its similarity to yeast initiation factor SUI1. FEBS Lett. 1995;365:47–50. doi: 10.1016/0014-5793(95)00427-b. [DOI] [PubMed] [Google Scholar]
- 28.Kozak M. Regulation of translation in eukaryotic systems. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 29.Lee S I, Umen J G, Varmus H E. A genetic screen identifies cellular factors involved in retroviral −1 frameshifting. Proc Natl Acad Sci USA. 1995;92:6587–6591. doi: 10.1073/pnas.92.14.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menninger J R. Peptidyl transfer RNA dissociates during protein synthesis from ribosomes of Escherichia coli. J Biol Chem. 1975;251:3392–3398. [PubMed] [Google Scholar]
- 31.Merrick W. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naranda T, Macmillan S E, Donahue T F, Hershey J W. SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2307–2313. doi: 10.1128/mcb.16.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer E, Wilhelm J, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 33a.Peltz S W, Donahue J L, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodnina M V, Pape T, Fricke R, Kuhn L, Wintermeyer W. Initial binding of the elongation factor Tu·GTP·aminoacyl-tRNA complex preceding codon recognition on the ribosome. J Biol Chem. 1996;271:646–652. doi: 10.1074/jbc.271.2.646. [DOI] [PubMed] [Google Scholar]
- 35.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 36.Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 37.Somogyi P, Jenner A J, Brierley I A, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarun S Z, Sachs A B. A common function for mRNA 3′ and 5′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 39.TenDam E, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- 40.Thomas A, Spaan W, Van Steeg H, Voorma H O, Benne R. Model of action of protein synthesis factor eIF-1 from rabbit reticulocytes. FEBS Lett. 1980;116:67–71. doi: 10.1016/0014-5793(80)80530-8. [DOI] [PubMed] [Google Scholar]
- 41.Thompson R C, Dix D B. Accuracy of protein biosynthesis. J Biol Chem. 1982;257:6677–6682. [PubMed] [Google Scholar]
- 42.Tu C, Tzeng T-H, Bruenn J A. Ribosomal movement impeded at a pseudoknot required for ribosomal frameshifting. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon H, Donahue T F. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]