Abstract
A strictly aerobic, Gram-stain-negative, rod-shaped, and motile bacterium, designated strain KMM 296, isolated from the coelomic fluid of the mussel Crenomytilus grayanus, was investigated in detail due to its ability to produce a highly active alkaline phosphatase CmAP of the structural family PhoA. A previous taxonomic study allocated the strain to the species Cobetia marina, a member of the family Halomonadaceae of the class Gammaproteobacteria. However, 16S rRNA gene sequencing showed KMM 296’s relatedness to Cobetia amphilecti NRIC 0815T. The isolate grew with 0.5–19% NaCl at 4–42 °C and hydrolyzed Tweens 20 and 40 and L-tyrosine. The DNA G+C content was 62.5 mol%. The prevalent fatty acids were C18:1 ω7c, C12:0 3-OH, C18:1 ω7c, C12:0, and C17:0 cyclo. The polar lipid profile was characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, phosphatidic acid, and also an unidentified aminolipid, phospholipid, and a few unidentified lipids. The major respiratory quinone was Q-8. According to phylogenomic and chemotaxonomic evidence, and the nearest neighbors, the strain KMM 296 represents a member of the species C. amphilecti. The genome-based analysis of C. amphilecti NRIC 0815T and C. litoralis NRIC 0814T showed their belonging to a single species. In addition, the high similarity between the C. pacifica NRIC 0813T and C. marina LMG 2217T genomes suggests their affiliation to one species. Based on the rules of priority, C. litoralis should be reclassified as a later heterotypic synonym of C. amphilecti, and C. pacifica is a later heterotypic synonym of C. marina. The emended descriptions of the species C. amphilecti and C. marina are also proposed.
Keywords: marine bacteria, Cobetia amphilecti, Cobetia marina, Cobetia crustatorum, phylogenomics, core genome, pan-genome analysis, taxonomy
1. Introduction
The genus Cobetia was created by Arahal et al. [1] to reclassify the species Halomonas marina. At the time of writing, the genus Cobetia comprises five validly published species names, including C. marina as the type species, C. amphilecti, C. crustatorum, C. litoralis, and C. pacifica, all isolated from different marine environments [1,2,3]. The genus accommodates Gram-stain-negative, aerobic, heterotrophic, halophilic, and rod-shaped bacteria, which can move by means of a single polar flagellum and/or two to seven lateral flagella [1,2,3]. Many strains of the genus Cobetia have been reported as a source of molecules and activities of biotechnological interest, and the genomic analysis of these strains has revealed their potential for the biosynthesis of biosurfactants, aromatic hydrocarbon degradation, inorganic carbon fixation, the synthesis and production of polyhydroxy-alkanoates, surface colonization, and alginate degradation [4,5,6,7,8]. Earlier, in the course of a survey of Halomonas-like bacteria inhabiting different areas of the Northwest Pacific, the strain KMM 296 was isolated from the coelomic fluid of the mussel Crenomytilus grayanus, collected from the Sea of Japan, and initially identified as a representative of the species C. marina (formerly Deleya marina) [9,10]. Later, the results of phylogenetic analysis based on the 16S rRNA gene sequence revealed the closest relationship of the strain KMM 296 to the strain C. amphilecti NRIC 0815T, with 100% sequence similarity. It should be noted that the genome of the strain KMM 296 (GenBank accession no. NZ_JQJA00000000.1) was sequenced [11] due to its ability to produce the highly active periplasmic alkaline phosphatase CmAP belonging to the PhoA alkaline phosphatase family [9,10,11,12,13,14]. Although the structural, biochemical, and catalytic properties of CmAP have been thoroughly studied, its exact physiological role still remains unknown due to the presence of several genes encoding for alkaline phosphatases with different structures in the KMM 296 genome [11]. In addition, CmAP was found to exhibit dephosphorylating activity towards bacterial lipopolysaccharides (LPS, endotoxins) similarly to other PhoA alkaline phosphatases from invertebrates and mammals, including humans [15,16,17,18,19]. Human intestinal alkaline phosphatase (IAP) provides an innate defense against endotoxins by altering the molecules to eliminate their pyrogenicity, resulting in an overall decrease in inflammatory processes. Thus, new data on the LPS-detoxifying activity of CmAP might lead to the development of a novel therapeutic approach for neutralizing the effects of bacterial endotoxins, such as Crohn’s disease, endotoxic shock, aging, etc. [17,18,19].
In the present study, we clarify the taxonomic position of the strain KMM 296 as a member of the C. amphilecti species and specify the description of the species C. amphilecti based on the results of phylogenomic analysis, phenotypic characterization, and experiments on the DNA-DNA hybridization between the strains KMM 296 and C. amphilecti NRIC 0815T. Since the 16S rRNA gene does not provide sufficient resolution to delineate Cobetia species [3], a huge number of published genomes of Cobetia strains cannot be affiliated to the existing species or have been wrongly designated. As a result, the precise identification of Cobetia strains other than C. marina and C. crustatorum strains remains a challenge, largely due to the lack of genome sequences for the type strains of C. litoralis, C. pacifica, and C. amphilecti. Therefore, in this work, we have presented data on the sequencing and genome analysis of the type strains NRIC 0814T, NRIC 0815T, and NRIC 0813T. A comparison of the genomic sequences and phenotypic characteristics suggests that the species C. amphilecti and C. litoralis belong to a single species. Moreover, based on the data obtained in this study, we propose the species C. marina and C. pacifica to be considered as the representatives of a single species. In accordance with the rules of priority, C. litoralis should be reclassified as a later heterotypic synonym of C. amphilecti, and C. pacifica is a later heterotypic synonym of C. marina. The emended descriptions of the species C. amphilecti and C. marina are also proposed.
2. Materials and Method
2.1. Strain Cultivation
Strain KMM 296 was obtained from the collection of marine microorganisms (KMM) at the G.B. Elyakov Pacific Institute of Bioorganic Chemistry FEB RAS (Vladivostok, Russia) and cultivated at 28 °C on marine agar (MA, Difco) and stored at −80 °C in marine broth (Difco) supplemented with 20% (v/v) glycerol. The type strains C. amphilecti NRIC 0815T (KMM 1561T), C. litoralis NRIC 0814T (KMM 3880T), C. pacifica NRIC 0813T (KMM 3879T), and C. marina LMG 2217T were kindly provided to us by NODAI Culture Collection Center (NRIC, Tokyo University of Agriculture, Tokyo, Japan) and the Belgian Coordinated Collection of Microorganisms (BCCM, Ghent University, Ghent, Belgium), respectively, and used as the reference strains for comparative taxonomic analysis.
2.2. Morphological, Biochemical, and Physiological Characterization
The physiological, morphological, and biochemical properties of strain KMM 296 were studied using the standard methods. The isolate was also examined in the API 20E, API 20NE, API 50 CH, API 32 ID GN, and API ZYM galleries (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions, except that the galleries were incubated at 28 °C. Gram staining was performed as recommended in [20]. Oxidative or fermentative utilization of glucose was determined on Hugh and Leifson’s medium modified for marine bacteria [21]. Catalase activity was tested via addition of 3% (v/v) H2O2 solution to a bacterial colony and observation for the appearance of gas. Oxidase activity was determined by using tetramethyl-p-phenylenediamine. Degradation of agar, starch, casein, gelatin, chitin, DNA, and urea and production of acid from carbohydrates, hydrolysis of Tween 80, nitrate reduction, production of hydrogen sulfide, acetoin (Voges-Proskauer reaction), and indole were tested according to standard methods [20]. The temperature range for growth was assessed on marine agar (MA). Tolerance to NaCl was assessed in medium containing 5 g Bacto Peptone (Difco), 2 g Bacto Yeast Extract (Difco), 1 g glucose, 0.02 g KH2PO4, and 0.05 g MgSO4·7H2O per liter of distilled water with 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3, 4, 5, 6, 8, 10, 12, 15, 17, 19, and 20% (w/v) of NaCl. Susceptibility to antibiotics was examined via the routine disc diffusion plate method. Discs were impregnated with the following antibiotics: ampicillin (10 μg), benzylpenicillin (10 U), carbenicillin (100 μg), cefalexin (30 μg), cefazolin (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), kanamycin (30 μg), lincomycin (15 μg), nalidixic acid (30 μg), neomycin (30 μg), ofloxacin (5 μg), oleandomycin (15 μg), oxacillin (10 μg), polymyxin B (300 U), rifampicin (5 μg), streptomycin (30 μg), tetracycline (5 μg), and vancomycin (30 μg).
2.3. Whole-Cell Fatty Acid, Polar Lipid, and Respiratory Quinone Composition
For comparative whole-cell fatty acid and polar lipid analysis, the strains KMM 296 and C. amphilecti NRIC 0815T were grown under optimal physiological conditions for both strains at 30 °C for 24 h on MA. Cellular fatty acid methyl esters (FAMEs) were prepared according to the methods described by Sasser [22], using the standard protocol of Sherlock Microbial Identification System (version 6.0, MIDI), and analyzed using a GC-21A chromatograph (Shimadzu) equipped with a fused-silica capillary column (30 m × 0.25 mm) coated with Supercowax-10 and SPB-5 phases (Supelco) at 210 °C. FAMEs were identified by using equivalent chain-length measurements and comparing the retention times to those of authentic standards. The polar lipids of the strains studied were extracted using the chloroform/methanol extraction method of Bligh and Dyer [23]. Two-dimensional TLC of polar lipids was carried out on silica gel 60 F254 (10 × 10 cm; Merck) using chloroform/methanol/water (65:25:4, v/v) in the first dimension and chloroform/methanol/acetic acid/water (80:12:15:4, v/v) in the second dimension [24]. The spray reagent used to reveal the spots was molybdophosphoric acid. Isoprenoid quinones were extracted with chloroform/methanol (2:1, v/v) and purified via TLC using a mixture of n-hexane and diethyl ether (85:15, v/v) as the solvent. The isoprenoid quinone composition of the strain KMM 296 was characterized via HPLC (Shimadzu LC-10A) using a reversed-phase type Supelcosil LC-18 column (15 cm × 4.6 mm) and acetonitrile/2-propanol (65:35, v/v) as a mobile phase at a flow rate of 0.5 mL min−1, as described previously [25].
2.4. The 16S rRNA Gene Sequencing and DNA–DNA Hybridization
DNA was extracted from 0.1–0.2 g of the bacterial cells (wet weight), using an extraction protocol by Sambrook and Russell [26]. PCR was carried out using the universal oligonucleotide primers 11F (5′-GTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′), as described by Weisburg [27], and the GeneAmp PCR System 2720 (Applied Biosystems, Singapore, Singapore). PCR amplicons were used as templates for sequencing amplification using a BigDye Terminator version 3.1 Cycle sequencing kit (Applied Biosystems). The purified sequencing products were analyzed via electrophoresis using a 50 cm capillary array with an ABI Prism 3130xL DNA sequencer (Applied Biosystems, Hitachi, Japan), and the sequence was assembled with SeqScape version 2.6 (Applied Biosystems). The sequences obtained were deposited in NCBI GenBank under the accession numbers presented by Noskova et al. [28] and analyzed against the referent phylotypes, based on the type strain 16S rRNA gene sequences and whole-genome assemblies in the EzBioCloud database [29].
The DNA–DNA hybridization between the strain KMM296 and C. amphilecti NRIC 0815T was performed spectrophotometrically, and initial renaturation rates were recorded as described by De Ley et al. [30].
2.5. Whole-Genome Shotgun Sequencing and Phylogenetic Analysis
The genomic DNA was obtained from the bacterial cultures of eight Cobetia strains, namely, C. amphilecti NRIC 0815T, C. litoralis NRIC 0814T, C. pacifica NRIC 0813T, Cobetia sp. 1AS1, Cobetia sp. 2AS, Cobetia sp. 3AK, Cobetia sp. 10Alg 146, and Cobetia sp. 29-18-1, using NucleoSpin Microbial DNA Mini kit (Macherey-Nagel, Düren, Germany), following the manufacturer’s instructions. Whole-genome shotgun sequencing was carried out on an Illumina MiSeq platform using Nextera DNA Flex kits, with a 150 bp paired-end sequencing kit (Illumina, San Diego, CA, USA). The sequence quality was assessed via FastQC version 0.11.8 [FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 28 December 2021)], and reads were trimmed using Trimmomatic version 0.38 [31]. Filtered reads were assembled de novo with SPAdes version 3.15.3 [32]. The draft genomes of the strains C. amphilecti NRIC 0815T, C. litoralis NRIC 0814T, C. pacifica NRIC 0813T, Cobetia sp. 1AS1, Cobetia sp. 2AS, Cobetia sp. 3AK, Cobetia sp. 10Alg 146, and Cobetia sp. 29-18-1 were annotated using NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [33] and deposited in GenBank/EMBL/DDBJ under the accession numbers JASCSA000000000, JARWKV000000000, JASCSB000000000, JARWKU000000000, JARWKQ000000000, JARWKR000000000, JARWKT000000000, and JARWKS000000000, respectively.
All publicly available Cobetia genomes were retrieved from the Reference sequence (RefSeq) database at NCBI using the NCBI-genome-download version 0.3.0 (https://github.com/kblin/ncbi-genome-download, accessed on 16 March 2023, n = 28) [34]. The accession numbers for the genomes used in this study are listed in Table 1.
Table 1.
The accession numbers and general attributes of 36 Cobetia spp. genomes used in this study.
| Strain | Accession ID | Genome Size, bp | G+C (mol%) | Contigs | Completeness (%) | Contamination (%) | Isolation Source |
|---|---|---|---|---|---|---|---|
| Cobetia sp. UCD-24C | GCF_001306765.1 | 4,229,986 | 62.5 | 51 | 98.9 | 1.76 | Seagrass sediment |
| C. amphilecti B2M13 | GCF_018860945.1 | 4,289,324 | 62.5 | 58 | 95.88 | 0.91 | Artificial alginate particle |
| Cobetia sp. 2AS1 | GCF_014876835.1 | 4,248,424 | 62.5 | 49 | 95.96 | 0.9 | Coastal sediment |
| Cobetia sp. 2AS | GCF_029846355.1 | 4,247,060 | 62.5 | 39 | 99.15 | 1.3 | |
| Cobetia sp. 1AS1 | GCF_029846435.1 | 4,235,090 | 62.5 | 43 | 99.27 | 1.71 | Coastal seawater |
| C. litoralis NRIC 0814T | GCF_029846315.1 | 4,621,254 | 62.5 | 51 | 99.15 | 2.93 | Sandy sediment |
| Cobetia sp. MC34 | GCF_018340035.1 | 4,022,416 | 62.5 | 175 | 95.47 | 0.87 | Fish-landing facility |
| Cobetia sp. 1CM21F | GCF_023161745.1 | 4,261,659 | 62.5 | 24 | 95.88 | 0.49 | Sea cave |
| Cobetia sp. 29-18-1 | GCF_029846405.1 | 4,117,019 | 62.5 | 70 | 99.35 | 1.48 | Esperiopsis digitata |
| C. amphilecti NRIC 0815T | GCA_030010415.1 | 4,171,304 | 62.5 | 112 | 98.54 | 1.34 | Internal tissue, Amphilectus digitatus |
| Cobetia sp. 4B | GCF_018831605.1 | 4,325,922 | 62.5 | 3 | 99.41 | 1.33 | Current Humbolt system, Heterostera chilensis |
| Cobetia sp. AM6 | GCF_009617955.1 | 4,229,996 | 62.5 | 1 | 98.74 | 1.36 | Japan: Tokyo |
| C. amphilecti N-80 | GCF_020217465.1 | 4,160,095 | 62.5 | 1 | 98.74 | 1.28 | Marine sediment |
| C. amphilecti KMM 296 | GCF_000754225.1 | 3,965,007 | 62.5 | 97 | 98.29 | 1.28 | Crenomytilus grayanus |
| Cobetia sp. Dlab-2-U | GCF_024124585.1 | 4,144,083 | 62.5 | 137 | 96.63 | 1.3 | Coral surface mucus layer and tissue Diploria labyrinthiformis |
| Cobetia sp. Dlab-2-AX | GCF_024124625.1 | 4,001,795 | 62.5 | 20 | 96.29 | 1.3 | |
| C. marina 402 | GCF_013350055.1 | 3,978,956 | 62.5 | 132 | 95.93 | 0.6 | Seawater, aquarium |
| Cobetia sp. cqz5-12 | GCF_016495405.1 | 4,209,007 | 62.5 | 1 | 99.45 | 1.71 | Brown algae Sargassum fusiforme |
| Cobetia sp. MB87 | GCF_011319755.1 | 3,101,384 | 62.5 | 12 | 81.26 | 1.58 | Sea cucumber gut |
| C. pacifica GPM2 | GCF_009931455.1 | 4,195,186 | 62.5 | 1 | 99.59 | 1.33 | Neopyropia tenera |
| Cobetia sp. 10Alg 146 | GCF_029846385.1 | 4,095,141 | 62.5 | 33 | 99.44 | 1.68 | Ahnfeltia tobuchiensis |
| Cobetia sp. 3AK | GCF_029846335.1 | 4,073,243 | 62.5 | 58 | 99.44 | 1.71 | Coastal seawater |
| C. pacifica NRIC 0813T | GCA_030010515.1 | 4,066,371 | 62.5 | 42 | 99.44 | 1.3 | Sandy sediment |
| Cobetia sp. MMG027 | GCF_027947415.1 | 4,168,882 | 62.5 | 47 | 99.24 | 1.71 | - |
| Cobetia sp. MM1IDA2H-1 | GCF_002916775.1 | 4,151,052 | 62 | 88 | 99.65 | 1.33 | Eulittoral intertidal pond at sea level |
| C. marina MM1IDA2H-1AD | GCF_900119965.1 | 4,155,178 | 62 | 105 | 99.65 | 1.33 | - |
| Cobetia sp. 5-11-6-3 | GCF_013374055.1 | 4,120,053 | 62.5 | 40 | 99.44 | 1.3 | Seaweed |
| C. marina T1 | GCF_005144735.1 | 4,177,239 | 62 | 21 | 99.44 | 1.28 | Saltwater |
| C. marina NBRC 15607 | GCF_006540105.1 | 4,184,377 | 62.5 | 113 | 99.24 | 1.68 | - |
| Cobetia sp. 5-25-4-2 | GCF_013374075.1 | 4,118,344 | 62.5 | 41 | 99.44 | 1.3 | Seaweed |
| Cobetia sp. ICG0124 | GCF_004006355.1 | 3,345,506 | 63 | 1 | 90.85 | 2.1 | Marine waters |
| C. marina JCM 21022T | GCF_001720485.1 | 4,176,400 | 62.5 | 1 | 99.51 | 1.51 | Littoral water |
| Cobetia sp. L2A1 | GCF_009796845.1 | 4,118,938 | 57.5 | 1 | 98.9 | 1.28 | Arctic Ocean beach |
| Cobetia sp. QF-1 | GCF_002213105.1 | 4,084,184 | 57.5 | 31 | 99.04 | 0.93 | Seawater |
| C. crustatorum SM1923 | GCF_007786215.1 | 4,215,468 | 57.5 | 163 | 98.83 | 0.82 | Surface seawater |
| C. crustatorum JO1T | GCF_000591415.1 | 4,049,952 | 57.5 | 138 | 90.05 | 0.52 | Fermented shrimp |
Type strains, C. litoralis NRIC 0814T, C. amphilecti NRIC 0815T, C. pacifica NRIC 0813T, C. marina JCM 21022T, C. crustatorum JO1T, are shown in bold.
The pan-genome for these 36 Cobetia strains (Table 1) was reconstructed using the microbial pan-genomics workflow in Anvi’o version 7.1 (minbit = 0.5; mcl-inflation = 2; min-occurrence = 1) [35]. The genomes were organized based on the distribution of gene clusters using the MCL algorithm (Distance: Euclidean; linkage: Ward). For average nucleotide identity (ANIm) calculation, we used the program ‘anvi-compute-genome-similarity’ with ‘-program pyANI’ flag. Amino acid identity (AAI) and in silico DNA–DNA hybridization (dDDH) values between the strains were calculated with the online server ANI/AAI-Matrix [36] and the TYGS platform (formula d4), respectively [37]. To produce the phylogenetic tree of the genus Cobetia, 1432 single-copy core gene sequences for each strain were extracted from the pan-genome and concatenated (composite length of 465,701 bp) using the program ‘anvi-get-sequences-for-gene-clusters’ with ‘-concatenate-gene-clusters’ flag. Resulting FASTA files were cleaned up by removing nucleotide positions that had gap characters in more than 50% of the sequences using the trimAl version 1.4.1 [38]. A core-genome phylogeny was reconstructed with IQ-TREE version 2.2.0.3 under the WAG model with non-parametric bootstrapping using 100 replicates [39]. The pan-genome and core-genome modelling were estimated with PanGP v.1.0.1 using a power-law regression model based on Heap’s law and exponential regression, respectively, as described by Tettelin et al. [40].
Fonts and sizes in all figures were edited manually in Adobe Photoshop CC 2018 for better visualization.
3. Results and Discussion
3.1. Morphological, Biochemical, and Physiological Characterization
The strain KMM 296 was shown to be a strictly aerobic, heterotrophic, Gram-stain-negative and motile bacterium that formed a slightly yellow-colored colony on MA and required NaCl or seawater for growth. It was positive for cytochrome oxidase and catalase and did not hydrolyze agar, casein, gelatin, starch, Tween 80, DNA, urea, or chitin (Table 2).
Table 2.
The different characteristics of the strain KMM 296 and its closest relative C. amphilecti NRIC 0815T.
| Characteristic | KMM 296 |
C. amphilecti NRIC 0815T |
|---|---|---|
| Source and site of isolation | Mollusk C. grayanus, the Sea of Japan, Pacific Ocean | Sponge A. digitatus, the Gulf of Alaska, Pacific Ocean |
| Temperature range for growth (°C) | 4–42 | 4–42 |
| Salinity range for growth (% NaCl) | 0.5–19 | 0–20 |
| Nitrate reduction | - | + |
| Hydrolysis of: | ||
| Tween 80 | - | + |
| DNA | - | + |
| Acid production from: | ||
| D-Fructose, D-lactose | - | + |
| L-Arabinose, D-melibiose, L-rhamnose | + | - |
| Assimilation of: | ||
| Amygdalin, maltose, sodium malonate, glycogen, capric acid, valeric acid, 3-hydroxybutiric acid, L-proline | + | - |
| N-acetylglucosamine, L-serine | - | + |
| Enzyme activities: | ||
| Valine arylamidase, cysteine arylamidase | - | + |
| Trypsin | + | - |
| DNA G+C content (mol%) | 62.5 | 62.5 |
Both strains were positive in the following tests: respiratory type of metabolism and motility; slightly yellowish colony color; hydrolysis of tyrosine and Tweens 20 and 40; catalase, oxidase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, and α-glucosidase activities via the PNPG test; acid production from D-galactose, D-glucose, and D-mannose; assimilation of L-arabinose, D-melibiose, L-rhamnose, sucrose, maltose, and D-mannitol; susceptibility to ampicillin, carbenicillin, cephalexin, cephazolin, chloramphenicol, erythromycin, gentamicin, kanamycin, nalidixic acid, neomycin, ofloxacin, polymyxin, rifampicin, streptomycin, tetracycline, and vancomycin; and resistance to benzylpenicillin, lincomycin, oleandomycin, and oxacillin. Both strains were negative for the following tests: arginine dihydrolase, lipase (C14), α-chymotrypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, α-mannosidase, and α-fucosidase activities; the hydrolysis of aesculin, agar, casein, chitin, gelatin, starch, and urea; acid production from raffinose, D-ribose, D-xylose, sucrose, trehalose, D-cellobiose, N-acetylglucosamine, glycerol, inositol, D-sorbitol, sodium acetate, trisodium citrate, and D-mannitol; and the production of H2S and indole.
The strains KMM 296 and C. amphilecti NRIC 0815T shared many common phenotypic features, such as the respiratory type of metabolism, motility by means of 1–2 polar and/or 2–3 lateral flagella, the ability to grow at 4–42 °C, the presence of catalase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, and α-glucosidase activities, and the assimilation of sucrose, maltose, sodium malonate, glycogen, D-mannitol, D-glucose, 3-hydroxybutyric acid, and L-proline (Table 2). They could not synthesize arginine dihydrolase, lipase (C14), cystine arylamidase, α-chymotrypsin, N-acetyl-β-glucosaminidase, β-glucosidase, α-galactosidase, β-glucuronidase, α-mannosidase, or α-fucosidase; hydrolyze agar, chitin, aesculin, gelatin, starch, urea, or Tween 80; produce acid from D-mannose, melibiose, raffinose, L-rhamnose, D-ribose, N-acetylglucosamine, inositol, D-sorbitol, glycerol, or D-mannitol; reduce nitrate to nitrite or assimilate L-arabinose, D-mannose, N-acetylglucosamine, adipate, phenylacetate, itaconic acid, sodium acetate, propionic acid, trisodium citrate, or 4-hydroxybenzoic acid. However, the strain KMM 296 can be distinguished from its closest phylogenetic relative by several phenotypic traits, including the presence of cytochrome oxidase and the ability to assimilate capric and valeric acids, the inability to produce acid from a set of carbohydrates and to assimilate D-glucose, D-mannitol, maltose, D-gluconate, L-malate, L-rhamnose, N-acetylglucosamine, D-ribose, inositol, suberic acid, lactic acid, L-alanine, potassium 5-ketogluconate, 3-hydroxybenzoic acid, L-serine, salicin, melibiose, L-fucose, L-arabinose, L-histidine, and potassium 2-ketogluconate (Table 2).
The above findings can extend the phenotypic characteristics that were reported for the species C. amphilecti by Romanenko et al. [3] after the justification of the placement of the strain KMM 296 in this species.
3.2. Whole-Cell Fatty Acid, Polar Lipid, and Respiratory Quinone Composition in the Strains KMM 296 and C. amphilecti NRIC 0815T
The predominant fatty acids (>5% of the total) in the strain KMM 296 were C18:1 ω7c, C12:0 3-OH, C18:1 ω7c, C12:0, and C17:0 cyclo (Table 3).
Table 3.
Fatty acid profiles (%) of the strains KMM 296 and C. amphilecti NRIC 0815T.
| Fatty Acid * | KMM 296 | C. amphilecti NRIC 0815T |
|---|---|---|
| C10:0 | 3.2 | 4.3 |
| C12:0 | 11.0 | 8.7 |
| C16:0 | 21.6 | 21.9 |
| C17:0 cyclo | 5.2 | 9.4 |
| C16:1 ω7c | 38.2 | 32.5 |
| C18:1 ω7c | 14.9 | 12.7 |
| C19:1 ω6c | 1.6 | tr |
| C12:0 3-OH | 15.8 | 20.3 |
* Data are from the present study. tr, trace amount (≤1%).
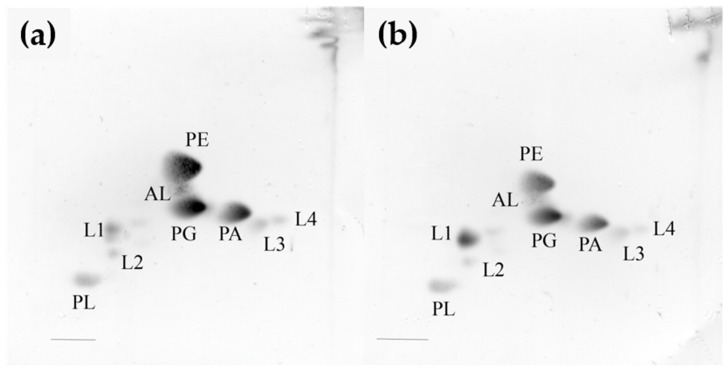
The polar lipid profile of the strain KMM 296 was characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, phosphatidic acid, an unidentified aminolipid, an unidentified phospholipid, and unidentified lipids, and it was found to be similar to that of C. amphilecti NRIC 0815T (Figure 1a,b). The major respiratory quinone was Q-8, which is common among members of the class Gammaproteobacteria.
Figure 1.
Two-dimensional thin-layer chromatogram of polar lipids extracted from the strains KMM 296 (a) and C. amphilecti NRIC 0815T (b). PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PC, phosphatidylcholine; PA, phosphatidic acid; AL, unidentified amino lipid; L, unidentified lipid; PL, unknown phospholipid.
3.3. The 16S rRNA Gene Phylogenetic Analysis of the Strains KMM 296, C. marina LMG 2217T, and C. amphilecti NRIC 0815T
Phylogenetic analysis based on 16S rRNA gene sequences revealed that the strain KMM 296 demonstrated only 99.5% sequence similarity to C. marina LMG 2217T (=JCM 21022T), whereas it was found to be identical to the type strain of another validly published Cobetia species, C. amphilecti, with 100% sequence similarity [3,28]. This suggests that the strain KMM 296 can be placed in this species instead of C. marina, the strains of which were predominantly isolated from degraded alga thallus [10]. In addition, the comparative genome analysis and phylogenomic analysis of the family Halomonadaceae, implemented by Tang et al. [41], indicated that the significant differences between C. marina JCM 21022T and the strain KMM 296 (formerly C. marina KMM 296) resulted from sequence insertions or deletions and chromosomal recombination [13,41].
3.4. GC Comparison between Strains KMM 296 and C. amphilecti NRIC 0815T
The genomic GC content of the strain KMM 296 was 62.5 mol%, as determined using the genome sequencing data [13]. This value was slightly lower than that obtained via the thermal denaturation method (62.7 mol%) [10]. The DNA–DNA relatedness between the isolate KMM 296 and the strain C. amphilecti NRIC 0815T, which was determined via the experimental DNA–DNA hybridization method, was 92%. This value was higher than the 70% threshold used for assigning bacterial strains to the same genomic species [42] and it strongly suggested that the two strains belong to the single species, C. amphilecti.
The closest evolutionary distances between the type strains of the species C. amphilecti and C. litoralis, on the one hand (99.93% 16S rRNA gene sequence similarity), and C. marina and C. pacifica, on the other hand (100% 16S rRNA gene sequence similarity), calculated using the EzBioCloud 16S RNA database tools and discussed earlier [28], also suggest that the species C. amphilecti and C. litoralis belong to one species and the species C. marina and C. pacifica could also be joined to a single species.
However, the comparison of the whole-genome sequences of all Cobetia spp. strains, which are currently deposited in the NCBI GenBank database (Table 1), revealed that each genome contains up to seven 16S rRNA genes, with different levels of similarity (99.86–100%) within one strain, as well as between the different species [28]. Therefore, comprehensive whole-genome-based analyses are required for Cobetia species demarcation.
3.5. Whole-Genome-Based Phylogeny and Analysis of Cobetia Strains
In total, 36 Cobetia strains were chosen for phylogenetic and comparative analyses, 8 of which have been sequenced in this study (3 type strains, NRIC 0814T, NRIC 0815T, and NRIC 0813T, and 5 isolates, 2AS1, 2AS, 1AS1, 29-18-1, and 10Alg 146). Twenty-eight genomes of the Cobetia spp. isolates were retrieved from the RefSeq database at NCBI. The genomic dataset included the genomes of the type strains of five Cobetia species according to the previous taxonomy classification [1,2,3], nine Cobetia spp. strains, and twenty-two unclassified Cobetia isolates. The overall features of the genomes are listed in Table 1. The genome size ranged from 3.1 to 4.6 Mbp, while the GC content varied slightly and was 62–63% except for four strains with 57–57.5%, including C. crustatorum JO1T (Table 1). Apparently, such values might be due to the difference in the genome completeness levels. According to the NCBI Quality analysis (CheckM) [43], the assemblies showed 81.26–99.65% completeness and 0.49–2.93% contamination (Table 1).
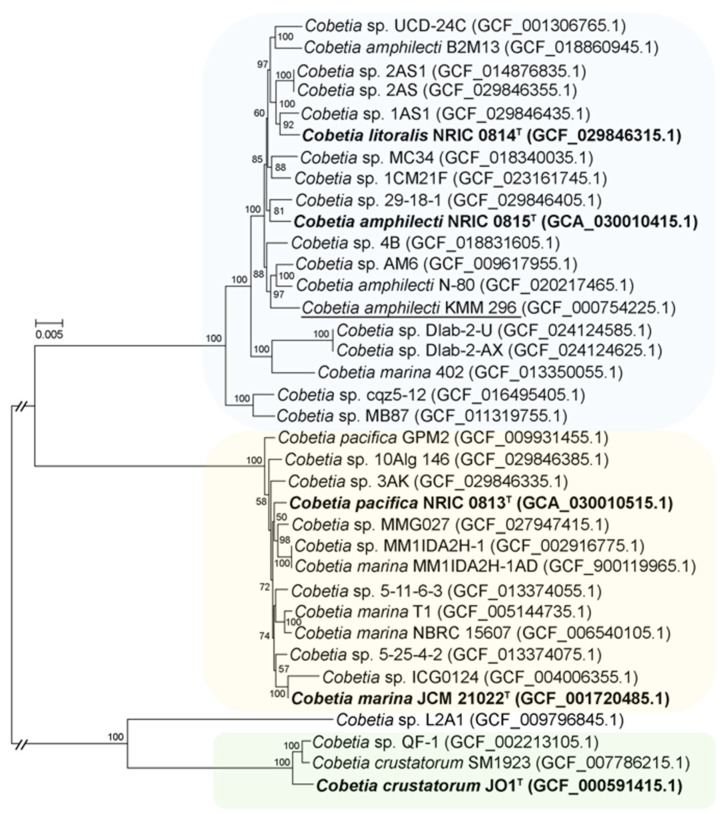
A core-genome phylogeny was used to estimate the phylogenetic relationships of the Cobetia strains (Figure 2). According to the phylogenetic tree, the strains fall into four clades with subclades (bootstrap values = 100).
Figure 2.
Maximum-likelihood phylogeny of the genus Cobetia based on concatenated 1432 single-copy core gene sequences and reconstructed with IQ-TREE with non-parametric bootstrapping using 100 replicates, including Bar, with 0.005 substitutions per amino acid position. The corresponding accession numbers for the genomes are given in parentheses. The type strains of the previously classified Cobetia species are shown in bold; strain KMM 296 is underlined.
The first one included three subclades and contained nineteen strains, among which two type strains, C. litoralis NRIC 0814T and C. amphilecti NRIC 0815T, clustered at the same subclade. The second clade consisted of 13 strains, including the type strains C. pacifica NRIC 0813T and C. marina JCM 21022T. The third clade branched distantly and consisted of two C. crustatorum and two Cobetia spp. strains. According to the obtained topology of the phylogenomic tree, it is clear that five described species may actually represent only three species (Figure 2).
The values of the phylogenomic metrics ANIb, AAI, and dDDH are further evidence redefining the species assignments within the genus Cobetia (Table S1). The obtained ANIb, AAI, and dDDH values for fourteen strains showing the same phylogenomic grouping, including the type strains C. litoralis NRIC 0814T and C. amphilecti NRIC 0815T (Figure 2), were found to be 96.43–96.95%, 97.64–99.99%, and 70.1–100%, respectively. The group of thirteen strains clustered together, including the type strains C. pacifica NRIC 0813T and C. marina JCM 21022T (Figure 2), showed the ranges of 97.14–98.19%, 98.2–99.99%, and 80.4–100% for the ANIb, AAI, and dDDH values, respectively. These values between C. crustatorum JO1T, C. crustatorum SM1923, and Cobetia sp. QF-1 (Figure 2) were 98.4–98.9%, 98.44–99.18%, and 89.5–91.7%, respectively. The strain Cobetia sp. L2A1 shared corresponding values of 80.8–85.7%, 86.27–90.89%, and 24.4–29.3% with the Cobetia spp. type strains. Considering the thresholds of 95–96% ANI, 95–96% AAI, and 70% dDDH defined for species demarcation, the type strains C. litoralis NRIC 0814T, C. amphilecti NRIC 0815T, C. pacifica NRIC 0813T, and C. marina JCM 21022T do not belong to the corresponding originally assigned species [44]. The high values confirm the phylogenetic grouping of those strains, which are likely to represent two instead of four separate species (Figure 2). The phylogenomic metrics of the strain Cobetia sp. L2A1 were below the cutoff scores, implying that it might belong to a novel species of the genus Cobetia (Figure 2, Table S1).
3.6. Pan-Genome-Based Phylogeny and Analysis of Cobetia Strains
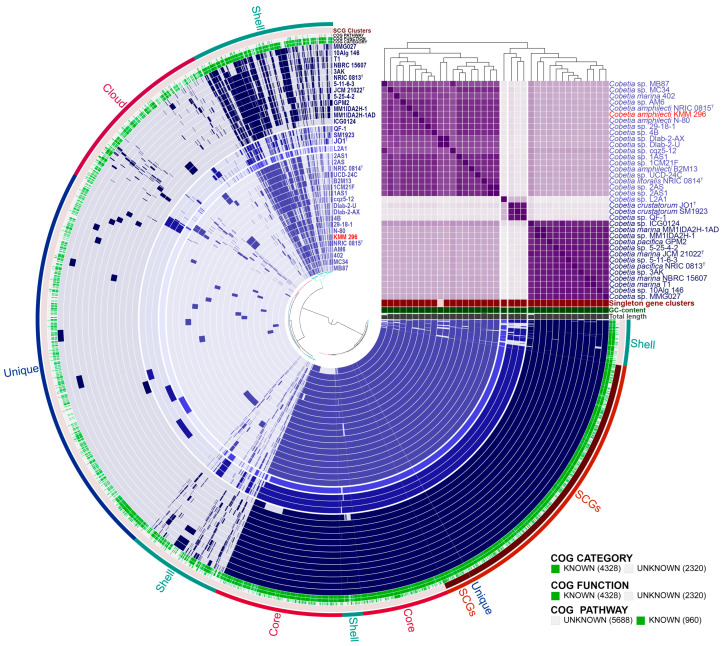
The pan-genome analysis of the genus Cobetia was performed to determine its genetic heterogeneity and phylogenetic relationships (Figure 3). The pan-genome is presented by a set of gene clusters (GCs), among which are the conserved core and the accessory shell and the cloud genes. The core genes are found in ≥95% of the genomes, the shell genes are found in more than 10% and less than 95% of the genomes, and the cloud genes are present in ≤10% of the genomes. Moreover, the single-copy genes (SCGs), as a part of the core, are found in all strains, while the unique genes (singletons) from the cloud are strain-specific. The pan-genome of 36 strains of the genus Cobetia comprised a total of 6648 gene clusters (distance: Euclidean; linkage: Ward) with 123,892 gene calls that include 2471 core gene clusters (93,289 genes in all 36 genomes), 1469 gene clusters in the shell (26,722 genes), and 2708 in the cloud (3881 genes), including 1902 gene clusters (1920 genes) of singletons. It is interesting that 62 GCs were found belonging exclusively to the strains grouped with the type strain C. crustatorum JO1T, while 20 GCs were found exclusively in the strains, clustered with the type strains C. pacifica NRIC 0813T and C. marina JCM 21022T (Figure 3). However, only one GC was found to be common for the 14 strains, grouped with C. litoralis NRIC 0814T and C. amphilecti NRIC 0815T, indicating a high rate of genomic intraspecies reorganization within their populations and the species diversification depending on their free or host-associated lifestyle [28].
Figure 3.
The pan-genome of 36 Cobetia spp. strains. Circle bars represent the presence/absence of 6648 pan-genomic clusters in each genome. Gene clusters are organized as core, soft-core, shell, and cloud gene clusters using Euclidian distance and Ward ordination. The heatmap in the upper right corner displays pairwise values of average nucleotide identity (ANIm) in percentages. The strain KMM 296 is colored in red.
The core and unique gene clusters were further annotated into COG classes. The core genes were related to the following classes: translation and ribosomal biogenesis (10.48%), amino acid transport and metabolism (9.58%), cell envelope synthesis (7.35%), energy production and conversion (6.95%), transcription (6.65%), carbohydrate metabolism and transport (6.35%), lipid metabolism (6.06%), inorganic ion transport and metabolism (5.91%), coenzyme metabolism (5.46%), post translational modifications (5.21%), replication and repair (4.42%), and signal transduction (3.82%). The classes for nucleotide metabolism and transport, secondary metabolite synthesis, defense mechanisms, cell cycle control, and intracellular trafficking and secretion were in a minority in the core (1.24–3.07%).
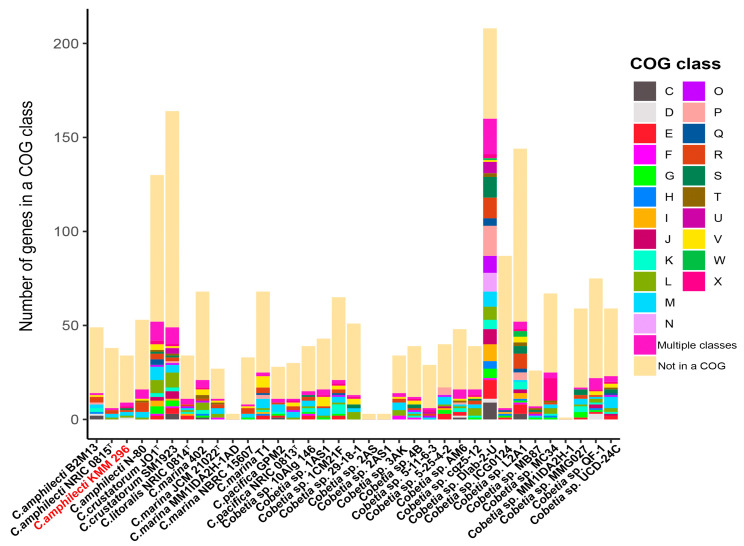
It is worth noting that each of the Cobetia genomes contains from one to two hundred and eight unique genes (Figure 4). The largest numbers of singletons were observed in the genomes of Cobetia sp. Dlab-2-U (208 genes), C. crustatorum SM1923 (164), Cobetia sp. L2A1 (144), and Cobetia sp. QF-1 (130). The genomes of C. marina MM1IDA2H-1AD, Cobetia sp. 2AS, Cobetia sp. 2AS1, and Cobetia sp. MM1IDA2H-1 account for the smallest number of the unique genes—3, 3, 3, and 1, respectively. The remaining genomes harbor from 26 to 87 unique genes. According to the COG class annotation of these unique genes, the most abundant functional classes were cell wall/membrane/envelope biogenesis (14.12% of total unique gene clusters), general functional prediction only (10.63%), replication and repair (9.8%), defense mechanisms (6.98%), amino acid metabolism and transport (6.48%), and transcription (6.15%).
Figure 4.
The number of unique genes assigned to a functional class (COG) among strains of Cobetia spp. Classes: C, energy production and conversion; D, cell cycle control and mitosis; E, amino acid metabolism and transport; F, nucleotide metabolism and transport; G, carbohydrate metabolism and transport; H, coenzyme metabolism, I, lipid metabolism; J, translation; K, transcription; L, replication and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover, chaperone functions; P, inorganic ion transport and metabolism; Q, secondary structure; R, general functional prediction only; S, function unknown; T, signal transduction; U, intracellular trafficking and secretion; V, defense mechanisms; W, extracellular structures; X, mobilome: prophages, transposons. Multiple classes—genes assigned to two or more COG categories. Not in a COG—COG not defined. The strain KMM 296 is colored in red.
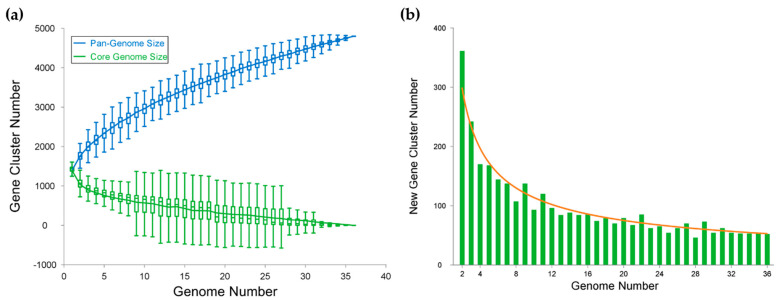
According to the modeling of the pan- and core genome sizes upon the addition of new genomes into the dataset, the pan-genome of Cobetia spp. is an open one with a γ value of 0.43 (Figure 5a). The best-fit regression curve for a pan-genome is rising upwards, implying an expanding pan-genome, while the core genome’s curve tends to reach a plateau [40]. Moreover, the fitting of the curve to a power law showed that the number of the new gene cluster discovery with the adding of the new genome would add 52 and 29 genes to the pan-genome, as predicted for the 37th and 100th sequenced genomes, respectively (Figure 5b).
Figure 5.
Pan-genome modelling. (a) Gene accumulation curves for the pan-genome and the core genome of 36 Cobetia genomes. Pan-genome curve: y = 889.06x0.43 + 535.51. Core genome curve: y = 1250.45e−0.07x − 29.77. (b) The new gene cluster number plot, curve: y = 452.537x−0.6.
4. Conclusions
The genus Cobetia currently includes five species with validly described names, C. marina, C. amphilecti, C. crustatorum, C. litoralis, and C. pacifica. However, the shared 16S rRNA gene sequence identity being almost 100% between the species C. amphilecti and C. litorali, as well as between the species C. marina and C. pacifica, does not allow us to delineate the Cobetia species. In this work, we have presented data on the sequencing and genome analysis of the type strains of C. amphilecti, C. litoralis, and C. pacifica. Our phylogenomic and pan-genomic analyses of the genus Cobetia, based on the 8 genome sequences presented in this study and 28 publicly available genome sequences, confirm the taxonomic status of only three species: C. marina, C. amphilecti, and C. crustatorum. The strain Cobetia sp. L2A1 was proven to be a member of a novel species. In addition, the taxonomic status of all Cobetia strains with available genomes has been clarified.
In summary, based on the results of genomic, phylogenetic, phenotypic, and chemotaxonomic studies, we suggested that the species C. litoralis should be placed in the species C. amphilecti, and the species C. pacifica should be included in the species C. marina. In accordance with the priority rules, C. litoralis should be reclassified as a later heterotypic synonym of C. amphilecti and C. pacifica is a later heterotypic synonym of C. marina. The emended descriptions of the species C. amphilecti and C. marina are proposed.
Emended description of the species Cobetia amphilecti (Romanenko et al., 2013)
The description of the species Cobetia amphilecti and Cobetia litoralis is as given by Romanenko et al. (2013), with the following amendments. Cells are oxidase-positive and motile by means of 1–2 polar and/or 2–5 lateral flagella. Some strains can request seawater or NaCl for growth. The predominant fatty acids (>5% of the total fatty acids) are C16:1 ω7c, C12:0 3-OH, C16:0, C18:1 ω7c, C17:0 cyclo, and C12:0. The polar lipid profile is characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, phosphatidic acid, an unidentified aminolipid, the two unidentified phospholipids, and the four unidentified lipids. The major respiratory quinone is Q-8. The genomic DNA G+C content is 62.5 mol%.
Emended description of the species Cobetia marina (Cobet et al., 1970, Arahal et al., 2002, Romanenko et al., 2013)
The description of the species Cobetia marina and Cobetia pacifica is as given by Arahal et al. (2002) and Romanenko et al. (2013), with the following amendments. Cells are oxidase-positive and motile by means of 1–2 polar and/or 2–5 lateral flagella. The predominant fatty acids (>5% of the total fatty acids) were C16:1 ω7c, C12:0 3-OH, C16:0, C17:0 cyclo, C18:1 ω7c, and C12:0. The polar lipid profile is characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, phosphatidic acid, an unidentified aminolipid, the two unidentified phospholipids, and the four unidentified lipids. The major respiratory quinone is Q-8. The genomic DNA G+C content is 62.2–62.4 mol%.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom14020196/s1, Table S1: The phylogenomic metrics ANIb, AAI, and dDDH calculated for the Cobetia spp. strains and isolates.
Author Contributions
Conceptualization, O.N. and M.I.; methodology, O.N. and N.O.; software, N.O.; validation, M.I., L.B. and L.T.; investigation, N.Z., E.D., A.S., E.B. and Y.N.; writing—original draft preparation, O.N., N.O. and L.B.; writing—review and editing, M.I.; visualization, N.O. and O.N.; project administration, M.I.; funding acquisition, L.T. and M.I. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole-genome shotgun sequences of the strains Cobetia amphilecti NRIC 0815T, Cobetia litoralis NRIC 0814T, Cobetia pacifica NRIC 0813T, Cobetia sp. 1AS1, Cobetia sp. 2AS, Cobetia sp. 3AK, Cobetia sp. 10Alg 146, and Cobetia sp. 29-18-1 were deposited in GenBank/EMBL/DDBJ under the accession numbers JASCSA000000000, JARWKV000000000, JASCSB000000000, JARWKU000000000, JARWKQ000000000, JARWKR000000000, JARWKT000000000, and JARWKS000000000, respectively.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by a grant from the Ministry of Science and Higher Education, Russian Federation 15.BRK.21.0004 (contract no. 075-15-2021-1052).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Arahal D.R., Castillo A.M., Ludwig W., Schleifer K.H., Ventosa A. Proposal of Cobetia marina gen. nov., comb. nov., within the family Halomonadaceae, to include the species Halomonas marina. Syst. Appl. Microbiol. 2002;25:207–211. doi: 10.1078/0723-2020-00113. [DOI] [PubMed] [Google Scholar]
- 2.Kim M.S., Roh S.W., Bae J.W. Cobetia crustatorum sp. nov., a novel slightly halophilic bacterium isolated from traditional fermented seafood in Korea. Int. J. Syst. Evol. Microbiol. 2010;60:620–626. doi: 10.1099/ijs.0.008847-0. [DOI] [PubMed] [Google Scholar]
- 3.Romanenko L.A., Tanaka N., Svetashev V.I., Falsen E. Description of Cobetia amphilecti sp. nov., Cobetia litoralis sp. nov. and Cobetia pacifica sp. nov., classification of Halomonas halodurans as a later heterotrophic synonym of Cobetia marina and emended descriptions of the genus Cobetia and Cobetia marina. Int. J. Syst. Evol. Microbiol. 2013;63:288–297. doi: 10.1099/ijs.0.036863-0. [DOI] [PubMed] [Google Scholar]
- 4.Ibacache-Quiroga C., Canales C., Charifeh M., Dinamarca M.A. Genome sequence of Cobetia sp. strain MM1IDA2H-1, a hydrocarbon-degrading and biosurfactant-producing marine bacterium. Genome Announc. 2017;5:e00132-17. doi: 10.1128/genomeA.00132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu W., Cong B., Lin J., Zhao L., Liu S. Complete genome sequencing and comparison of two nitrogen-metabolizing bacteria isolated from Antarctic deep-sea sediment. BMC Genom. 2022;23:713. doi: 10.1186/s12864-022-08942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng W., Yan X., Xiao J., Chen Y., Chen M., Jin J., Bai Y., Wang Q., Liao Z., Chen Q. Isolation, identification, and whole genome sequence analysis of the alginate-degrading bacterium Cobetia sp. cqz5-12. Sci. Rep. 2020;10:10920. doi: 10.1038/s41598-020-67921-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen M., Jablonski P., Altermark B., Irgum K., Hansen H. High natural PHA production from acetate in Cobetia sp. MC34 and Cobetia marina DSM 4741T and in silico analyses of the genus specific PhaC2 polymerase variant. Microb. Cell Fact. 2021;20:225. doi: 10.1186/s12934-021-01713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriya H., Takita Y., Matsumoto A., Yamahata Y., Nishimukai M., Miyazaki M., Shimoi H., Kawai S.J., Yamada M. Cobetia sp. bacteria, which are capable of utilizing alginate or waste Laminaria sp. for poly(3-hydroxybutyrate) synthesis, isolated from a marine environment. Front. Bioeng. Biotechnol. 2020;8:974. doi: 10.3389/fbioe.2020.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanova E.P., Mikhailov V.V., Plisova E.J., Balabanova L.A., Svetashev V.V., Vysockyi M.V., Stepanenko V.I., Rasskazov V.A. Characterization of the marine bacterium Deleya marina producing highly active alkaline phosphatase and associated with the mussel Crenomytilus grayanus. Russ. J. Mar. Biol. 1994;20:340–545. [Google Scholar]
- 10.Ivanova E.P., Christen R., Sawabe T., Alexeeva Y.V., Lysenko A.M., Chelomin V.P., Mikhailov V.V. Presence of ecophysiologically diverse populations within Cobetia marina strains isolated from marine invertebrate, algae and the environments. Microbes Environ. 2005;20:200–207. doi: 10.1264/jsme2.20.200. [DOI] [Google Scholar]
- 11.Balabanova L.A., Golotin V.A., Kovalchuk S.N., Babii A.V., Shevchenko L.S., Son O.M., Kosovsky G.Y., Rasskazov V.A. The Genome of the marine bacterium Cobetia marina KMM 296 isolated from the mussel Crenomytilus grayanus (Dunker, 1853) Russ. J. Mar. Biol. 2016;42:106–109. doi: 10.1134/S106307401601003X. [DOI] [Google Scholar]
- 12.Plisova E.Y., Balabanova L.A., Ivanova E.P., Kozhemyako V.B., Mikhailov V.V., Agafonova E.V., Rasskazov V.A. A highly active alkaline phosphatase from the marine bacterium Cobetia. Mar. Biotechnol. 2005;7:173–178. doi: 10.1007/s10126-004-3022-4. [DOI] [PubMed] [Google Scholar]
- 13.Golotin V., Balabanova L., Likhatskaya G., Rasskazov V. Recombinant production and characterization of a highly active alkaline phosphatase from marine bacterium Cobetia marina. Mar. Biotechnol. 2015;17:130. doi: 10.1007/s10126-014-9601-0. [DOI] [PubMed] [Google Scholar]
- 14.Balabanova L., Podvolotskaya A., Slepchenko L., Eliseikina M., Noskova Y., Nedashkovskaya O., Son O., Tekutyeva L., Rasskazov V. Nucleolytic enzymes from the marine bacterium Cobetia amphilecti KMM 296 with antibiofilm activity and biopreservative effect on meat products. Food Control. 2017;78:270–278. doi: 10.1016/j.foodcont.2017.02.029. [DOI] [Google Scholar]
- 15.Buinovskaya N.S., Bakholdina S.I., Balabanova L.A. Dephosphorylation of lipopolysaccharides by alkaline phosphatase from marine bacterium. Vestn. FEB RAS. 2018;6:80–81. [Google Scholar]
- 16.Yang Y., Wandler A., Postlethwait J., Guillemin K. Dynamic evolution of the LPS-detoxifying enzyme intestinal alkaline phosphatase in Zebrafish and other vertebrates. Front. Immunol. 2012;3:314. doi: 10.3389/fimmu.2012.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalles J.P. Intestinal alkaline phosphatase: Multiple biological roles in maintenance of intestinal homeostasis and modulation by diet. Nutr. Rev. 2010;68:323–332. doi: 10.1111/j.1753-4887.2010.00292.x. [DOI] [PubMed] [Google Scholar]
- 18.Hamarneh S.R., Mohamed M.M., Economopoulos K.P., Morrison S.A., Phupitakphol T., Tantillo T.J., Gul S.S., Gharedaghi M.H., Tao Q., Kaliannan K., et al. A novel approach to maintain gut mucosal integrity using an oral enzyme supplement. Ann. Surg. 2014;260:706–714. doi: 10.1097/SLA.0000000000000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kühn F., Adiliaghdam F., Cavallaro P.M., Hamarneh S.R., Tsurumi A., Hoda R.S., Munoz A.R., Dhole Y., Ramirez J.M., Liu E., et al. Intestinal alkaline phosphatase targets the gut barrier to prevent aging. JCI Insight. 2020;26:e134049. doi: 10.1172/jci.insight.134049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerhardt P., Murray R.G.E., Wood W.A., Krieg N.R. Methods for General and Molecular Bacteriology. American Society for Microbiology; Washington, DC, USA: 1994. p. 224. [Google Scholar]
- 21.Lemos M.L., Toranzo A.E., Barja J.L. Modified medium for oxidation-fermentation test in the identification of marine bacteria. Appl. Environ. Microbiol. 1985;40:1541–1543. doi: 10.1128/aem.49.6.1541-1543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasser M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. MIDI, Inc.; Newark, DE, USA: 1990. Tech. Note 101. [Google Scholar]
- 23.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 24.Collins M.D., Shah H.M. Fatty acid, menaquinone and polar lipid composition of Rothia dentosacariosa. Arch. Microbiol. 1984;137:247–249. doi: 10.1007/BF00414552. [DOI] [Google Scholar]
- 25.Komagata K., Suzuki K.I. Lipid and cell wall analysis in bacterial systematics. Methods Microbiol. 1987;19:161–207. [Google Scholar]
- 26.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd ed. Volume 1 Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 27.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noskova Y., Seitkalieva A., Nedashkovskaya O., Shevchenko L., Tekutyeva L., Son O., Balabanova L. Are the closely related Cobetia strains of different species? Molecules. 2021;26:690. doi: 10.3390/molecules26030690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Ley J., Cattoir H., Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 31.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eren A.M., Esen O.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L., Delmont T.O. Anvi’o: An advanced analysis and visualization platformfor ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-R L.M., Konstantinidis K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4:e1900v1. [Google Scholar]
- 37.Meier-Kolthoff J.P., Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tettelin H., Riley D., Cattuto C., Medini D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Tang X., Xu K., Han X., Mo Z., Mao Y. Complete genome of Cobetia marina JCM 21022T and phylogenomic analysis of the family Halomonadaceae. J. Ocean. Limnol. 2018;36:528–536. doi: 10.1007/s00343-017-6239-6. [DOI] [Google Scholar]
- 42.Wayne L.G., Brenner D.J., Colwell R.R., Grimont P.A.D., Kandler O., Krichevsky M.I., Moore L.H., Moore W.E.C., Murray R.G.E., Stackebrandt E., et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 43.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome shotgun sequences of the strains Cobetia amphilecti NRIC 0815T, Cobetia litoralis NRIC 0814T, Cobetia pacifica NRIC 0813T, Cobetia sp. 1AS1, Cobetia sp. 2AS, Cobetia sp. 3AK, Cobetia sp. 10Alg 146, and Cobetia sp. 29-18-1 were deposited in GenBank/EMBL/DDBJ under the accession numbers JASCSA000000000, JARWKV000000000, JASCSB000000000, JARWKU000000000, JARWKQ000000000, JARWKR000000000, JARWKT000000000, and JARWKS000000000, respectively.