Abstract
Most small nuclear RNAs (snRNAs) are synthesized by RNA polymerase II, but U6 and a few others are synthesized by RNA polymerase III. Transcription of snRNA genes by either polymerase is dependent on a proximal sequence element (PSE) located upstream of position −40 relative to the transcription start site. In contrast to findings in vertebrates, sea urchins, and plants, the RNA polymerase specificity of Drosophila snRNA genes is intrinsically encoded in the PSE sequence itself. We have investigated the differential interaction of the Drosophila melanogaster PSE-binding protein (DmPBP) with U1 and U6 gene PSEs. By using a site specific protein-DNA photo-cross-linking assay, we identified three polypeptide subunits of DmPBP with apparent molecular masses of 95, 49, and 45 kDa that are in close proximity to the DNA and two additional putative polypeptides of 230 and 52 kDa that may be integral to the complex. The 95-kDa subunit cross-linked at positions spanning the entire length of the PSE, but the 49- and 45-kDa subunits cross-linked only to the 3′ half of the PSE. The same polypeptides cross-linked to both the U1 and U6 PSE sequences. However, there were significant differences in the cross-linking patterns of these subunits at a subset of the phosphate positions, depending on whether binding was to a U1 or U6 gene PSE. These data suggest that RNA polymerase specificity is associated with distinct modes of interaction of DmPBP with the DNA at U1 and U6 promoters.
In higher eukaryotes, four of the major small nuclear RNAs (snRNAs) found in spliceosomes (U1, U2, U4, and U5) are synthesized by RNA polymerase II (RNAP II), but U6 snRNA is synthesized by RNA polymerase III (RNAP III) (2, 3, 9, 18, 22). The U6 gene promoter is representative of an unusual class of RNAP III promoters that contain a TATA box but lack internal promoter elements (5, 15, 19, 30). Promoters of snRNA genes transcribed by either RNAP II or RNAP III contain an essential proximal sequence element (PSE) at a conserved location approximately 40 to 65 bp upstream of the transcription start site that is required for the initiation of snRNA transcription (4, 9, 18, 26, 29, 30, 32, 36).
In Drosophila melanogaster, the PSE is more specifically named the PSEA to distinguish it from a second, even more proximal conserved element, PSEB, present in the promoters of Drosophila snRNA genes transcribed by RNAP II (36). The PSEB is located at the TATA box position but is a poor TATA box sequence (consensus CATGGAg/aA) (16). PSEB is separated from the upstream PSEA by 8 bp of nonconserved sequence (16, 36). Drosophila U6 genes, on the other hand, contain a canonical TATA box rather than a PSEB, and the TATA box is separated by 12 bp from the upstream PSEA (4).
In vertebrates and sea urchins, the PSEs of U1 and U2 genes are interchangeable with the PSEs of U6 genes (14, 17, 23). In these organisms, the PSEs are therefore not responsible for the determination of RNAP specificity. Surprisingly, recent results from our lab revealed that the U1 and U6 PSEAs are not interchangeable in the Drosophila system. In fact, the RNAP specificity of Drosophila snRNA genes is determined by the sequence of the 21-bp-long PSEA itself, not by the presence of a TATA box versus PSEB or by differences in the spacing of these elements relative to the PSEA (11). Our results indicated that the U1 and U6 PSEAs, even though they were identical at 16 of 21 base positions, exclusively recruited RNAP II and RNAP III, respectively (11). Further data indicated that all five base differences between the U1 and U6 PSEAs contributed to RNAP specificity, but the differences at base positions 19 and 20 played a major role in determining the RNAP specificity of the Drosophila snRNA promoter (11).
The PSE-binding protein (PBP) was first identified in the human system in HeLa cell extracts (31). It was further characterized by two groups and alternatively named proximal transcription factor (PTF) (20) or snRNA activator protein complex (SNAPc) (25). This factor was capable of activating U1 and U2 transcription in vitro by RNAP II and U6 and 7SK transcription by RNAP III (8, 25, 34). PTF and SNAPc each contain four subunits with approximate molecular masses of 180 to 200, 50 to 55, 44 to 45, and 43 to 45 kDa (8, 34). Molecular cloning and sequencing of the three smallest subunits indicated that they are identical in SNAPc and PTF (1, 7, 8, 24, 35). Because each of these subunits is required for snRNA transcription by either RNAP II or RNAP III, it seems that the same protein complex functions at both classes of vertebrate snRNA gene promoters. The fact that the PSEA plays a major role in determining RNA polymerase specificity in Drosophila poses the question of whether a similar complex functions at both RNAP II and RNAP III snRNA promoters in insects.
Our lab recently reported the partial purification and characterization of the D. melanogaster PSEA-binding protein (DmPBP) (27). Like SNAPc/PTF, DmPBP is a multisubunit protein, and it is capable of specifically activating transcription of Drosophila U1 and U6 genes in vitro in a soluble nuclear extract (27). By using a site-specific protein-DNA photo-cross-linking technique (13), we here show that DmPBP contains three subunits that are in close proximity to the DNA. The site-specific cross-linking assay has further allowed us to map the translational and rotational positions of these subunits relative to the PSEA. Importantly, the data also reveal that significant conformational differences exist in the DmPBP-DNA complexes depending on whether they are assembled with the U1 or U6 PSEA sequence. These differences may lead to the recruitment of polymerase-specific factors in subsequent steps of preinitiation complex assembly.
MATERIALS AND METHODS
Source of DmPBP.
The soluble nuclear fraction (SNF) was prepared by using purified nuclei from 0- to 12-h Drosophila embryos as described by Kamakaka et al. (12) and modified by Su et al. (27). A fraction enriched in DmPBP activity was prepared by fractionating the SNF on DEAE-cellulose and heparin-agarose chromatography columns as previously described (27). The fraction eluting from the heparin-agarose column in 300 mM KCl (HA300 fraction) was approximately 10-fold enriched relative to the SNF in the DmPBP DNA-binding activity as estimated by an electrophoretic mobility shift assay (EMSA). The HA300 fraction was dialyzed against 25 mM HEPES (adjusted to pH 7.6 with KOH)–12.5 mM MgCl2–0.1 mM EDTA–10% (by volume) glycerol–100 mM KCl and was concentrated by centrifugation in an Ultrafree-15 centrifugal filtration device (Millipore) to a final protein concentration of approximately 2.8 mg/ml. This fraction was used for all of the EMSA and photo-cross-linking experiments described in this report.
DNA probes and competitors for EMSA analysis.
The radiolabeled U1 PSEA probe used for Fig. 2 consisted of the 80-base-long nontemplate strand oligonucleotide (shown in the lower section of Fig. 1) annealed to its complement. The U6 PSEA probe consisted of a similar double-stranded oligonucleotide but with U6-specific changes at five positions (Fig. 1). For use as EMSA probes, they were radiolabeled with [γ-32P]ATP and polynucleotide kinase. Each of these probes also contains a PSEB sequence 8 bp downstream of the U1 or U6 PSEA. The flanking sequences (and those between the PSEA and PSEB) do not correspond to sequences found in the wild-type U1 or U6 promoters, but they are identical to those in constructs used previously to investigate the cis-acting determinants of RNAP specificity at Drosophila snRNA gene promoters (11). When used to promote transcription, the first sequence (with the U1 PSEA) specifically promoted RNAP II transcription in vitro, and the second sequence (with the U6 PSEA) specifically promoted RNAP III transcription (11).
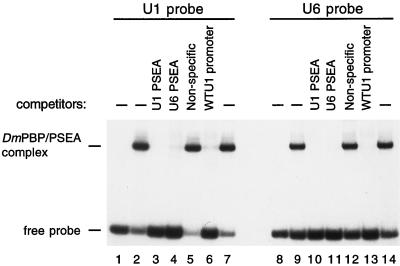
FIG. 2.
The U1 and U6 PSEA sequences interact with the same DNA-binding activity (i.e., DmPBP). EMSAs were performed with a radiolabeled probe that contained the U1 PSEA sequence (lanes 1 to 7) or a similar probe that contained the U6 PSEA sequence (lanes 8 to 14). Reactions also contained 4 μl of a heparin-agarose fraction partially enriched for the DmPBP DNA-binding activity (11), except those in lanes 1 and 8, which contained no added protein. Reactions in lanes 3, 4, 10, and 11 each contained 1 pmol (100 ng) of unlabeled competitor DNA oligonucleotides that were identical in sequence to the radiolabeled U1 or U6 PSEA probes as indicated above the lanes. Reactions shown in lanes 5 and 12 each contained 100 ng of a nonspecific oligonucleotide as a competitor. Those loaded in lanes 6 and 13 each contained 1 pmol (50 ng) of an oligonucleotide with the sequence of the wild-type U1 promoter. This latter oligonucleotide contains no sequences in common with the radiolabeled probes except for the 21-bp PSEA. Titration of the specific competitor oligonucleotides in other experiments (not shown) indicated that the inhibition of binding was dependent on the concentration of the competing oligonucleotides.
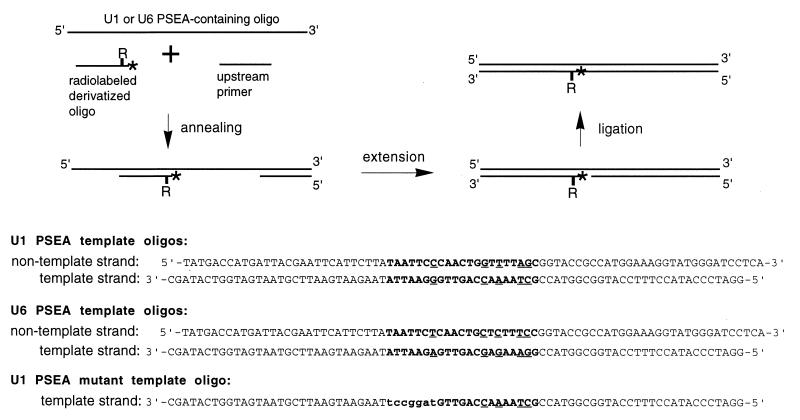
FIG. 1.
Preparation of 32P-labeled site-specific photo-cross-linking probes. The double-stranded probes were prepared by using long (∼80 nucleotides) synthetic template oligonucleotides (shown in the lower part), chemically derivatized oligonucleotides labeled at the 5′ end with 32P, and appropriate upstream primers. The “R” stands for an azidophenacyl moiety coupled to a phosphorothioate residue in the downstream primer, and the asterisk indicates a 32P radiolabel. For details, see Materials and Methods. The lower part shows the actual sequences of the long template oligonucleotides used to generate the probes. Note that the U1 and U6 PSEA template oligonucleotides have exactly the same sequence with the exception of five base changes (denoted by underlines) within the sequence of the PSEA (shown in boldface). The mutant template contains seven base changes at the upstream end of the U1 PSEA which are indicated by lowercase letters.
U1 and U6 PSEA competitor oligonucleotides (used for Fig. 2, lanes 3, 4, 10, and 11) were prepared by annealing the template and nontemplate DNA strand oligonucleotides shown in the lower part of Fig. 1. A second and distinct competitor oligonucleotide that contained the U1 PSEA (shown in boldface) was prepared by annealing the oligonucleotides 5′-CGCGTTCGTTGCAATTCCCAACTGGTTTTAGCTGCTCAGCC-3′ and 5′-ATGGCTGAGCAGCTAAAACCAGTTGGGAATTGCAACGAACG-3′. This double-stranded oligonucleotide, which contains the wild-type U1-95.1 gene promoter sequence from positions −72 to position −32, shares only the PSEA sequence with the competitor oligonucleotides described above. The sequence flanking the PSEA is unrelated, and there is no PSEB. This oligonucleotide was used as the competitor in lanes 6 and 13 of the EMSA shown in Fig. 2, as well as in the photo-cross-linking assays that contained specific competitor. The nonspecific competitor used in the same experiments was comprised of the annealed oligonucleotides 5′-GATCAAACCGCGCGCTGCATGCCGGGAGCACCAC-3′ and 5′-GATCGTGGTGCTCCCGGCATGCAGCGCGCGGTTT-3′.
EMSA reaction conditions.
Protein-DNA complexes were formed in a 10-μl reaction volume containing 4 μl of DmPBP HA300 fraction and a final buffer composition of 12.5 mM HEPES (pH 7.6)–50 mM KCl–6.25 mM MgCl2–0.05 mM EDTA–5% (by volume) glycerol. Samples also contained 1 μg of poly(dI-dC) · poly(dI-dC) and 1 μg of poly(dG-dC) · poly(dG-dC). When competitor oligonucleotides were added, they were incubated with HA300 fraction for 5 min before addition of the probe (20,000 cpm). Then the reaction mixtures were incubated at room temperature for 30 min. Samples were electrophoresed in 5% (29:1 acrylamide/bisacrylamide ratio) native gels in a running buffer consisting of 45 mM Tris base, 45 mM boric acid, and 1.25 mM Na2EDTA (pH 8.3) at 200 V for 1 h at room temperature. Gels were dried prior to autoradiography.
Site-specific protein-DNA photo-cross-linking probes.
Fifty different probes, each containing cross-linking reagent at a unique position, were prepared to scan through the U1 and U6 PSEA sequences on both the template and nontemplate strands. The cross-linking reagent and method have been described previously by Yang and Nash (33) and Lagrange et al. (13). Briefly, DNA oligonucleotides 25 bases long were synthesized on a 1-μmol scale, using solid-phase β-cyanoethylphosphoramidite chemistry on an ABI392 automated synthesizer. Phosphorothioate was incorporated 5′ of the third nucleotide from the 5′ end of the oligonucleotide during synthesis by using tetraethylthiuram disulfide (Applied Biosystems). The phosphorothiolate-substituted oligonucleotides were gel purified, and 50 nmol of each was derivatized with 1 mg of azidophenacyl bromide (Sigma) in 220 μl of methanol and 55 μl of water for 3 h at 37°C. The oligonucleotides were ethanol precipitated, and 10 pmol of each was phosphorylated by using [γ-32P]ATP (7,000 Ci/mmol) and polynucleotide kinase.
The double-stranded site-specific cross-linking probes were prepared as outlined in Fig. 1. A derivatized oligonucleotide and an upstream primer (10 pmol of each) were annealed to 2 pmol of a 79-mer or 80-mer template oligonucleotide that contained the template or nontemplate strand sequence of the U1 or U6 PSEA. The primers were extended by using T4 DNA polymerase and the four deoxynucleoside triphosphates. T4 DNA ligase was used to seal the nick at the 5′ end of the derivatized oligonucleotide, and the product was gel purified to eliminate excess primer, incomplete extension products, and any remaining unligated product.
To prepare probes with cross-linking reagent in the template strand, the 80-base-long U1 and U6 PSEA nontemplate strand oligonucleotide shown in the lower section of Fig. 1 were used as templates for the synthesis of the second strand. For incorporation of cross-linking reagent into the nontemplate strand, the U1 or U6 template strand oligonucleotides (Fig. 1) were used. To generate mutant cross-linking probes a U1 template strand oligonucleotide synthesized with seven base alterations at the upstream end of the PSEA was used (Fig. 1). Each of the 50 chemically derivatized probes that contained wild-type sequence retained its ability to bind to DmPBP by EMSA, but the mutant probes were not recognized (data not shown).
Photo-cross-linking reactions and analysis.
Samples of the derivatized DNA fragments (20,000 cpm) were mixed with HA300 fraction in a 10-μl reaction volume as described for EMSA. After 30 min at 25°C in the dark, reaction mixtures were irradiated with UV light (emission maximum at 312 nm) for 10 min, using a Spectrolinker XL-1500 UV cross-linker (Spectronic Corporation). Nuclease digestions were performed by adding 0.5 μl each of 200 mM CaCl2, 4 mM phenylmethylsulfonyl fluoride, leupeptin (100 μg/ml), aprotinin (200 μg/ml), and 2 μl (2 U) of DNase I (Promega). Following a 15-min incubation at 25°C, 0.7 μl of 10% sodium dodecyl sulfate (SDS) was added and the mixtures were heated at 65°C for 3 min. The following items were then added: 0.69 μl of 1.75 M acetic acid, 0.57 μl of 30 mM ZnSO4, 0.57 μl of 4 mM phenylmethylsulfonyl fluoride, and 0.4 μl (370 U) of S1 nuclease (Gibco BRL). Samples were incubated at 37°C for 20 min. The reactions were terminated by the addition of 1 μl of 1.5 M Tris-HCl (pH 8.8), 7.7 μl of 10 M urea, and 6 μl of 5× loading dye (0.3125 M Tris-HCl [pH 6.8], 50% glycerol, 10% SDS, 24% 2-mercaptoethanol, 0.125% bromphenol blue). The samples were heated for 3 min at 95°C and electrophoresed through SDS–10% polyacrylamide gels. Gels were dried, and the cross-linked peptides were detected by autoradiography.
For the experiments shown in Fig. 4C in which the DmPBP complex was purified by EMSA, the reactions were scaled up 40-fold: 160 μl of HA300 fraction was incubated with 800,000 cpm of probe in a 400-μl volume. Following UV exposure, the DmPBP-PSEA complex was separated on a 5% native gel. The shifted band was detected by autoradiography and was sliced from the gel. The protein-DNA complex was eluted in a Centrilutor (Amicon) and concentrated by using Centricon-10 microconcentrators. Bovine serum albumin (100 μg per sample) was added to reduce the nonspecific adhesion of the cross-linked protein. Samples were digested with nuclease and loaded onto an SDS gel.
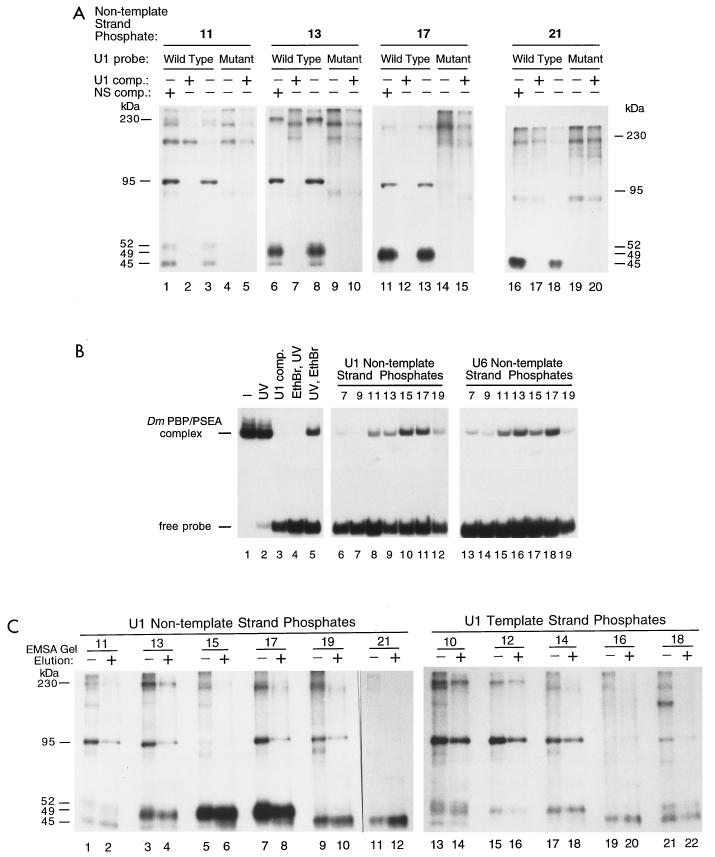
FIG. 4.
(A) Specificity of the photo-cross-linking reactions. Cross-linking reactions were carried out at selected phosphate positions on the nontemplate strand of the U1 PSEA as indicated above each panel. Lanes 3, 8, 13, and 18 show reactions performed identically to those shown in Fig. 3. The reactions loaded in lanes 4, 9, 14, and 19 were performed under the same conditions but using a mutant PSEA probe (bottom of Fig. 1) not recognized by DmPBP. Reactions loaded in the remaining lanes were performed similarly but in addition contained 250 ng of either a U1 PSEA-specific competitor (comp.) oligonucleotide or a nonspecific competitor oligonucleotide as indicated above the lanes. The sequences of the competitor oligonucleotides are shown in Materials and Methods. Panels shown represent different lengths of exposures to optimize the clarity of the bands in each individual panel. (B) Demonstration that the photo-cross-linking probes can be covalently cross-linked to the DmPBP activity originally identified by EMSA. Reactions shown in lanes 1 to 5 were performed with a U1 photo-cross-linking probe derivatized at position 17 in the nontemplate strand. Following incubation of the probe with the HA300 fraction which contained the DmPBP activity, all samples (except the one loaded in lane 1) were irradiated with UV light. A U1 PSEA competitor oligonucleotide was included in the incubation shown in lane 3. Ethidium bromide (final concentration, 0.11% [wt/vol]) was added to the sample loaded in lane 4 10 min prior to UV irradiation, but the order was reversed for the sample shown in lane 5 (i.e., UV irradiation followed by ethidium bromide). Reactions loaded in lanes 6 to 19 were performed as the one shown in lane 5 but with a variety of cross-linking probes derivatized at selected phosphate positions in either the U1 or U6 PSEA nontemplate strand as indicated above the corresponding lanes. (C) Identification of stably associated subunits of DmPBP by native gel purification of DmPBP-DNA complexes prior to SDS gel electrophoresis. Photo-cross-linking probes were derivatized at selected phosphate positions on the template or nontemplate strand of the U1 PSEA, incubated with the HA300 DmPBP fraction, and irradiated with UV light. Samples shown in the even-numbered lanes were then electrophoresed in EMSA gels, and the band corresponding to the DmPBP-DNA complex was sliced out. The complex was then eluted, concentrated, digested with nucleases, and run on SDS gels. Samples shown in the odd-numbered lanes were prepared using the standard protocol without EMSA purification.
RESULTS
PSEA sequences from Drosophila U1 and U6 genes compete for the same DNA binding activity.
A 32P-labeled synthetic double-stranded DNA oligonucleotide that contained the U1 PSEA sequence was prepared and used as a probe in EMSA analysis. Incubation of this probe with a protein fraction (HA300) enriched in DmPBP activity resulted in the appearance of a single band of retarded mobility following native gel electrophoresis (Fig. 2, lanes 2 and 7). Previous work indicated that this activity specifically and exclusively protected the region of the PSEA sequence from nuclease digestion in a DNase I footprinting assay (27). Furthermore, it was shown that the DmPBP activity detected by EMSA cofractionated over three subsequent chromatography columns with an activity that specifically stimulated U1 and U6 transcription in a PSEA-dependent manner (27). Consistent with its ability to activate both U1 and U6 transcription, the DmPBP activity detected by EMSA (Fig. 2) was competed by oligonucleotides that contained either the U1 or U6 PSEA sequences but not by a nonspecific oligonucleotide that lacked a PSEA sequence (lanes 3 to 5). Moreover, an oligonucleotide that contained the U1 PSEA, but no other sequence in common with the probe, also competed for this DNA-binding activity (lane 6).
In a reciprocal assay with a 32P-labeled probe that contained the U6 PSEA sequence, we detected a DmPBP-DNA complex that was similar in mobility to that formed with the fragment containing the U1 PSEA (Fig. 2, lanes 9 and 14). This complex was specifically competed by the homologous U6 sequence as well as by a similar oligonucleotide that contained the U1 PSEA sequence instead (Fig. 2, lanes 10 and 11). Importantly, the complex was also competed by an oligonucleotide that contained the U1 PSEA but no other sequences in common with the U6 probe (Fig. 2, lane 13). These results indicate that the same DmPBP activity is capable of binding specifically to both the U1 and U6 gene PSEA sequences.
Site-specific protein-DNA photo-cross-linking identifies DmPBP subunits and reveals their differential interaction with the U1 and U6 PSEA sequences.
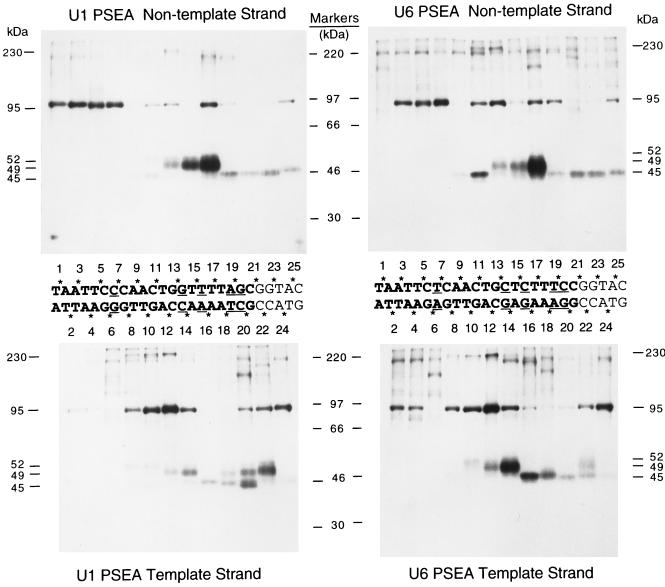
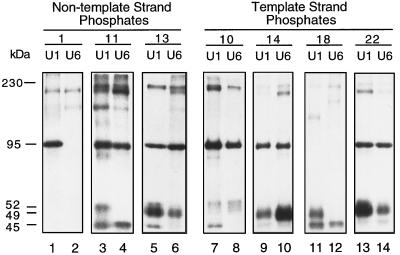
We used a site-specific protein-DNA photo-cross-linking technique (13) to investigate the interaction of DmPBP with the DNA. Our goals were threefold: (i) to gain knowledge about the subunit structure of DmPBP; (ii) to study the arrangement of these subunits relative to the DNA; and (iii) to determine whether DmPBP-DNA complexes formed with the U1 and U6 PSEAs exhibit distinctive features. To accomplish these goals, we prepared 50 different site-specific probes that scanned through the template and nontemplate strands of the U1 and U6 PSEAs at every other phosphate position. In the center of Fig. 3, the phosphate positions that were individually derivatized with cross-linking reagent are indicated by asterisks above and below the sequences of the U1 PSEA (left) and the U6 PSEA (right). Phosphate position 1 corresponds to the phosphate that links the first and second bases of the PSEA. Odd-numbered phosphates were derivatized on the nontemplate strand, and even-numbered phosphates were analyzed on the template strand. Following UV-mediated photo-cross-linking and extensive nuclease digestion, the cross-linked polypeptides were separated by SDS gel electrophoresis and detected by autoradiography.
FIG. 3.
Site-specific protein-DNA photo-cross-linking reactions that scan through the PSEA sequences of Drosophila U1 and U6 genes. Fifty different radiolabeled site-specific photo-cross-linking probes were incubated in separate reactions with the HA300 fraction that contains the DmPBP activity. Following UV irradiation, polypeptides that cross-linked to the DNA were detected by SDS gel electrophoresis and autoradiography. The sequences of the U1 (left) and U6 (right) PSEAs (and five bases downstream of the PSEAs) are shown between the upper and lower panels. The five-base-pair differences between the U1 and U6 sequences are indicated by underlining. The phosphate positions on the template and nontemplate strands that were derivatized with cross-linking reagent are indicated by asterisks. The numbers below the upper panels and above the lower panels indicate the individual lanes that contain the cross-linking results from the correspondingly numbered phosphate positions. (According to our numbering system, the phosphate designated 1 is the phosphate that links the first and second nucleotides in the PSEA. This differs from the standard International Union of Pure and Applied Chemistry convention, which would require that all the phosphate positions on the nontemplate strand be increased by a value of 1.) The upper panels show the results of cross-linking to the nontemplate strand of the U1 PSEA (left) and to the nontemplate strand of the U6 PSEA (right). The lower panels show the results of cross-linking to the complementary template strands. The positions migrated by molecular weight markers are shown in the center. The molecular masses of the specifically cross-linked polypeptides, identified in this and later experiments (Fig. 4 and 5), are indicated to the far left and far right.
Bands corresponding to three polypeptides with apparent molecular masses of 45, 49, and 95 kDa were readily visualized in the autoradiograms (Fig. 3). Each of these three polypeptides cross-linked in a distinctive manner that was dependent upon the site of incorporation of the cross-linking reagent (Fig. 3). When the nontemplate strand was analyzed, the pattern of cross-linking was generally similar whether DmPBP was bound to a U1 PSEA or a U6 PSEA (upper left and right panels, respectively). However, there were a few notable differences. For example, the 95-kDa polypeptide cross-linked strongly to phosphate position 1 in the U1 PSEA, but it failed to cross-link to the same position in the U6 PSEA. At positions 11, 13, and 19 in the nontemplate strand, there were differences in the relative cross-linking intensity of the 95-kDa subunit depending on whether a U1 or U6 PSEA sequence was used. Third, the 45-kDa subunit cross-linked with a greater intensity to position 11 in the nontemplate strand of the U6 PSEA than to the same position in the U1 PSEA.
Differences in the cross-linking patterns dependent on whether DmPBP was bound to a U1 or U6 PSEA were more pronounced when the template strand was analyzed (Fig. 3, lower left and lower right panels, respectively). The same three polypeptides (45, 49, and 95 kDa) that intensely cross-linked to the nontemplate strand PSEA sequences also cross-linked strongly to the template strand, and again each polypeptide cross-linked with a distinctive pattern that depended on the position of the cross-linking reagent. Although there were many similarities in the cross-linking of DmPBP to the U1 and U6 sequences, there were also several pronounced differences. For example, the 49-kDa polypeptide cross-linked most intensely to position 22 of the U1 PSEA but to position 14 of the U6 PSEA. The 45-kDa polypeptide cross-linked most intensely to position 20 of the U1 PSEA but to position 16 of the U6 PSEA. The 95-kDa polypeptide cross-linked strongly to positions 2 and 4 of the U6 PSEA, but it cross-linked weakly to the identical positions in the U1 PSEA sequence. From these data, we conclude that the 45-, 49-, and 95-kDa polypeptides approach the backbone of the DNA differently depending on whether DmPBP is bound to a U1 or U6 PSEA sequence.
Specificity of the cross-linking reactions.
Besides the three most intense bands discussed above, a number of weaker bands were visible in the autoradiograms shown in Fig. 3. Conceivably, these could be integral subunits of DmPBP that are not as closely associated with the DNA. Alternatively, since the HA300 fraction is still a relatively crude preparation of DmPBP, these bands could be due to nonspecific photo-cross-linking, or they could derive from other proteins that are recruited to the DNA by DmPBP. To differentiate between these possibilities, as well as to confirm that the previously identified three polypeptides were legitimate components of DmPBP, an extensive series of control experiments were performed.
First, a mutant PSEA sequence that had the first seven bases altered at the 5′ end of the PSEA was used to prepare cross-linking probes that were not effectively recognized by DmPBP. The seven-base mutation utilized in the PSEA sequence was previously shown to destroy U1 promoter activity (36) and to prevent complex formation with DmPBP in an EMSA (data not shown). As additional controls for specificity, reactions that contained specific and nonspecific competitor oligonucleotides were included with each set of incubations.
Figure 4A shows results using probes derivatized at several different positions on the nontemplate strand of the U1 PSEA. At phosphate position 11, four bands (mobilities corresponding to 45, 52, 95, and 230 kDa) cross-linked to the wild-type probe (Fig. 4A, lane 3) but not to the mutant probe (lane 4). Moreover, these four bands were competed by a competitor oligonucleotide containing the U1 PSEA sequence (lane 2) but not by a nonspecific oligonucleotide (lane 1). A band migrating at approximately 210 kDa also appeared to be competed by the specific oligonucleotide but not by the nonspecific oligonucleotide (lanes 1 to 3). However, this band, and another one running at approximately 150 kDa, cross-linked to the mutant probe as well (lane 4). Therefore, these two bands are not specific to the DmPBP-DNA complex.
When the cross-linker was located at position 13 of the nontemplate strand of the U1 PSEA (Fig. 4A, lane 8), five bands (45, 49, 52, 95, and 230 kDa) were cross-linked to the probe. (In other experiments [data not shown], a better resolution of the 52-kDa band was obtained.) None of these five bands cross-linked to the mutant PSEA sequence, and all were competed by the U1-specific oligonucleotide but not by the nonspecific oligonucleotide (lanes 6 to 9). With cross-linker at position 17, only the 49- and 95-kDa bands were cross-linked specifically, and at position 21, only the 45-kDa subunit was specifically cross-linked (lanes 11 to 20). In summary, the experiments shown in Fig. 4A confirmed the specificity of the three interactions previously discussed and provided an indication that two additional polypeptides (52 and 230 kDa) may be associated with the DmPBP activity as well.
As a second approach to examining the specificity of the reactions, photo-cross-linked samples were treated with ethidium bromide prior to being electrophoresed through EMSA gels (Fig. 4B). Ethidium bromide intercalates into DNA and can disrupt sequence-specific protein-DNA interactions but does not disrupt covalent links between protein and DNA. Figure 4B (lanes 1 to 5) shows results using a probe that was internally labeled with 32P and derivatized with cross-linking reagent at phosphate position 17 in the U1 nontemplate strand. Lane 1 shows a reaction in which essentially all of the probe was shifted to a position in the gel characteristic of the DmPBP-DNA complex. UV irradiation of the sample only marginally reduced the stability of the complex (lane 2), but a U1 competitor oligonucleotide completely disrupted complex formation (lane 3). When ethidium bromide was added to the mixture prior to UV irradiation, the DmPBP-DNA complex was similarly disrupted (lane 4). However, if DmPBP was covalently cross-linked to the DNA by UV irradiation prior to the addition of ethidium bromide, the signal still remained (lane 5). This indicates that the DmPBP-DNA complex identified by EMSA indeed contained polypeptide constituents that were covalently cross-linked to the DNA. It then follows that these cross-linked polypeptides from the DmPBP complex should be detectable in SDS gels (as demonstrated in Fig. 3).
Figure 4B (lanes 6 to 19) shows the results of similar experiments in which the cross-linking reagent was introduced at a number of other positions within the U1 or U6 probes. In each case, ethidium bromide was added to the samples following covalent cross-linking by UV irradiation. At each position, a covalently cross-linked DmPBP-DNA complex was visible, with the exception of position 9 of the U1 nontemplate strand, which correlates with the finding that at this position no cross-linked polypeptides were detected by SDS gel electrophoresis (Fig. 3, upper left panel). At each derivatized phosphate, there is a reasonable correlation between the intensity of the shifted EMSA band and the sum of the intensities of bands detected on the SDS gels in Fig. 3. In the absence of ethidium bromide, all of the probes were strongly shifted (data not shown).
A third and final method was used to further correlate the radiolabeled bands visualized on SDS gels with the DmPBP-DNA complex detected by EMSA. Cross-linking reactions were scaled up 40-fold, and the covalently cross-linked products were electrophoresed through a native EMSA gel. The specific band corresponding to the DmPBP-DNA complex was sliced from the gel and eluted, and the cross-linked polypeptides were analyzed by SDS gel electrophoresis (Fig. 4C). The even-numbered lanes show results obtained when the DmPBP complex was purified by EMSA gel electrophoresis prior to analysis on SDS gels, and the odd-numbered lanes show results obtained in a cross-linking reaction without this prior purification step (conditions similar to those used in the experiments shown in Fig. 3). The EMSA purification step simplified the electrophoretic profile by eliminating certain background bands that apparently were due to nonspecific cross-linking or due to proteins only weakly associated with the DmPBP-DNA complex prior to EMSA gel electrophoresis.
A careful examination of the even-numbered lanes in Fig. 4C revealed five bands in the EMSA-purified DmPBP-DNA complex that cross-linked specifically to the PSEA. Of the phosphate positions analyzed, the 45-kDa band cross-linked most intensely to positions 19 and 21 of the nontemplate strand (lanes 10 and 12) but was also visible in lanes 2, 4, 14, 20, and 22. The 49-kDa product cross-linked very intensely at positions 15 and 17 (lanes 6 and 8) but was also clearly visible in lanes 4, 16, 18, and 22. The 95-kDa polypeptide cross-linked at a number of positions on both the template and nontemplate strands. A weakly cross-linked 52-kDa band was present in lanes 2 and 14, and the 230-kDa band was clearly evident in lanes 4, 14, and 16. These results identify five distinct bands on the SDS gel associated with the DmPBP activity and confirm the data shown in Fig. 4A that was obtained using the mutant probe and competitors.
Side-by-side comparison of the cross-linking patterns of DmPBP to U1 and U6 PSEAs at selected phosphate positions.
The results presented in Fig. 3 revealed a number of differences in the cross-linking patterns depending on whether DmPBP was bound to a U1 or U6 PSEA sequence. To more closely examine these differences, reactions were performed with the U1 and U6 PSEA probes, and the cross-linking products were run side by side in SDS gels (Fig. 5). At position 1 in the nontemplate strand, the 95-kDa subunit cross-linked strongly to the U1 PSEA, but cross-linking to the U6 PSEA at this position was undetectable (compare lanes 1 and 2). At position 11, the 52-kDa polypeptide cross-linked only to the U1 PSEA, but the 45-, 95-, and 230-kDa bands cross-linked to both the U1 and U6 PSEAs (lanes 3 and 4). All five polypeptides cross-linked to position 13 of the U1 PSEA (lane 5), but only the 49-, 95-, and 230-kDa bands cross-linked to this position in the U6 PSEA (lane 6).
FIG. 5.
Differential cross-linking of DmPBP to the U1 and U6 PSEAs at selected phosphate positions. Photo-cross-linking probes derivatized at a variety of positions on the template or nontemplate strands of the U1 or U6 PSEAs (as indicated above the lanes) were cross-linked to the HA300 DmPBP fraction. Reactions performed with the U1 and U6 PSEAs were run side by side on SDS gels to facilitate the comparison of the cross-linking patterns.
On the template strand, an even larger number of differences could be noted. The results at several of these positions are shown in Fig. 5, lanes 7 to 14. It is particularly notable that at position 14, the 49-kDa subunit cross-linked more intensely to the U6 PSEA than to the U1 PSEA (lanes 9 and 10), yet at position 22, the opposite was true (lanes 13 and 14). At phosphate position 18 (lanes 11 and 12), the 45-kDa protein cross-linked to both PSEA sequences, yet the 49-kDa subunit cross-linked only to the U1 PSEA. In summary, the same polypeptides were cross-linked to the U1 and U6 PSEA sequences, but there was variation in how they approached the DNA phosphate backbone depending on the PSEA sequence bound.
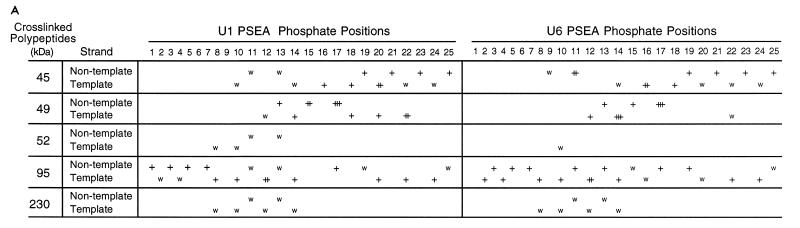
Summary of the cross-linking data and graphical visualization of the arrangement of DmPBP subunits relative to the DNA.
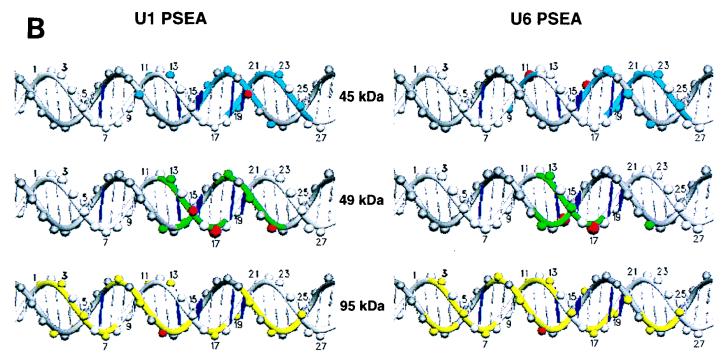
Figure 6A summarizes in tabular form the results of the cross-linking data at all 50 positions analyzed on the template and nontemplate strands of the U1 and U6 PSEA sequences. Four levels of cross-linking intensities, ranging from weak to very strong, are indicated. In Fig. 6B, the data for the three subunits of DmPBP (45, 49, and 95 kDa) that cross-linked strongly to the PSEA are projected onto B-form DNA. Although the DNA may be distorted from B form when bound to DmPBP, Fig. 6B nevertheless provides visual information about the surfaces of the DNA approached by the various subunits of DmPBP. (Since the 52- and 230-kDa bands were always weak, it is less likely that they represent polypeptides intimately associated with the DNA.)
FIG. 6.
Summary of the results of the site-specific photo-cross-linking of DmPBP subunits to the U1 and U6 PSEAs. (A) Tabular representation of the cross-linking results accumulated from the experiments shown in Fig. 3 to 5 and other experiments not shown. Four different relative intensities of cross-linking, ranging from weak (w) to very strong (+++), are indicated. (B) Representation of the cross-linking data on B-form DNA. The template strand is differentiated from the nontemplate strand by a darker gray shading. Phosphate positions are indicated by spheres on the DNA backbone strands. Phosphates on the nontemplate strand are numerically labeled. Phosphate positions that covalently cross-linked to the indicated subunits are shown in color. Those that cross-linked very strongly (++ or +++ in panel A) are indicated by red spheres. Furthermore, the DNA backbone strand is colored surrounding those phosphate positions that were cross-linked with a relative intensity of + or greater, whereas it is left uncolored near positions where there was weak or no detectable cross-linking. The five base pairs that are different in the U1 and U6 PSEAs are indicated by a thicker dark blue shading. This illustration is not meant to suggest that the DNA is necessarily in B form when bound to DmPBP. This illustration was generated by using the program SETOR (6).
(i) The 45-kDa subunit.
The smallest subunit detected in the photo-cross-linking assay interacted with the DNA near the 3′ end of the 21-bp PSEA sequence and beyond. With the DNA sequence oriented as shown in Fig. 6B, the 45-kDa subunit apparently approaches the DNA from above, spanning the minor groove between positions 16 and 25 (Fig. 6B, upper left graphic). The strongest cross-link between the 45-kDa subunit and the U1 PSEA occurred at position 20 of the template strand, which is on the front face of the DNA oriented as in Fig. 6B. In contrast, with the U6 PSEA (Fig. 6B, upper right graphic), the strongest cross-links occurred with position 11 of the nontemplate strand and position 16 of the template strand. This suggests that the 45-kDa subunit may be more intimately associated with the major groove between positions 11 and 16 when DmPBP is bound to the U6 PSEA versus its interaction with the U1 PSEA.
(ii) The 49-kDa subunit.
The cross-linking pattern to the U1 PSEA (Fig. 6B, left middle graphic) indicates that the 49-kDa subunit approaches the front surface of the DNA (in the orientation shown) and spans the major groove between positions 13 and 22 and the minor groove between positions 12 and 17. On the U6 PSEA, the contacts with positions 18 to 22 on the template strand are either missing or are greatly reduced (Fig. 6B, middle right graphic), suggesting that the 49-kDa subunit is less intimately associated with the far 3′ end of the U6 PSEA than it is with the corresponding region of the U1 PSEA. The strong cross-link to position 15 of the U1 nontemplate strand is replaced with a very strong cross-link to position 14 of the template strand of the U6 PSEA, indicating a more intimate approach to the minor groove of the U6 PSEA between phosphates 12 to 17. Together, these data suggest that the 49-kDa subunit contacts the PSEA primarily through the major groove on the U1 PSEA but through minor groove interactions on the U6 PSEA.
(iii) The 95-kDa subunit.
The cross-linking pattern of the 95-kDa subunit is very complex. This subunit interacts extensively with the DNA throughout both the U1 and U6 PSEAs and even beyond into the 3′-flanking sequence. Within the 5′ half of the PSEA, the 95-kDa subunit approaches from the front surface of the DNA (as oriented in Fig. 6B); however, toward the 3′ end of the PSEA, the interaction is primarily with the lower face. When all of the cross-linking interactions are taken into consideration, the 3′ end of the PSEA appears to be nearly completely buried within the DmPBP complex.
The 95-kDa polypeptide is the only subunit closely associated with the upstream third of the PSEA sequence. Since the first seven nucleotides of the PSEA are essential for activity and recognition by DmPBP, it follows that the 95-kDa subunit almost certainly plays a role in the binding of DmPBP sequence specifically to the PSEA. It is interesting that differences in the cross-linking pattern occur at the 5′ end of the U1 and U6 PSEAs even though the first 6 bp are identical (and there is only one difference in the first 13 bp). This suggests that the differential protein-DNA interactions that occur in the 3′ half of the PSEA can be transmitted, presumably through allosteric effects, to establish differences in the local protein-DNA environment at the 5′ end of the PSEA as well.
DISCUSSION
Identification of polypeptides associated with the DmPBP activity.
We have used site-specific protein-DNA photo-cross-linking (13) to investigate the interaction of the D. melanogaster PSEA-binding protein with the U1 and U6 gene PSEA sequences. By using this technique, we have identified three polypeptides (45, 49, and 95 kDa) that cross-linked strongly at a number of phosphate positions and therefore reside in relatively close proximity to the DNA. We identified by SDS gel electrophoresis two additional bands (52 and 230 kDa) that cross-linked specifically to the PSEA, yet weakly and at fewer locations. Among other possibilities, these latter two bands could represent subunits of DmPBP that do not reside in close proximity to the DNA. Alternatively, it is conceivable that these bands arise from position-specific incomplete nuclease digestion. For example, we cannot exclude the possibility that a fraction of the cross-links to the 95-kDa polypeptide at positions 8 to 14 result in exceptional stabilization of the complex and corresponding resistance to nuclease digestion, resulting in the appearance of a 230-kDa band. Further studies will be required to assess the relationship of the putative 52- and 230-kDa polypeptides to DmPBP structure and function. On the other hand, the data provide striking evidence for three DmPBP subunits (45, 49, and 95 kDa) that are intimately associated with the DNA. The length of the arm of the cross-linking reagent used in these studies conceivably permits covalent links to be formed with polypeptides that approach within about 11 Å of the phosphate phosphorus atom (13). The length and flexibility of this arm undoubtedly account at least in part for the fact that multiple subunits can often be cross-linked to the same derivatized phosphate position.
Human SNAPc/PTF contains at least four distinct subunits, three of which are in the size range of 43 to 55 kDa (8, 34). This raises the possibility that the Drosophila polypeptides found in the 45-, 49-, and 52-kDa bands may be homologous to the human subunits of similar size. The fourth subunit in human SNAPc/PTF (SNAP190 or PTFα) has a molecular mass of 180 to 200 kDa and can be photo-cross-linked to a bromodeoxyuridine (BrdU)-substituted probe, indicating that it is in close contact with the DNA (34). Since our photo-cross-linking data indicate that the Drosophila 95-kDa subunit makes extensive contacts with the DNA throughout the PSEA sequence, the 95-kDa polypeptide may be functionally homologous to the largest human subunit. The gene for human SNAP190 has recently been cloned and was found to code for a protein with 4.5 Myb repeats (10). Interestingly, the Drosophila U1 and U6 PSEA sequences used in the studies reported here each contain a consensus Myb recognition element at positions 9 to 13 (sequence AACNG [21, 28]). This sequence occurs in a region of the PSEA contacted primarily by the 95-kDa subunit (Fig. 6).
In earlier work from this lab, a probe that contained BrdU as a photo-cross-linking agent cross-linked strongly to DmPBP polypeptides that ran with apparent relative mobilities corresponding to 59 and 61 kDa on SDS gels (27). We have since investigated the relationship between those bands and the ones identified in the current study, and we have determined that the discrepancy in apparent molecular masses is due to the nuclease digestion protocols used following covalent cross-linking. When the cross-linking studies with the BrdU-substituted probe were repeated with the nuclease digestion protocol described in Materials and Methods, the cross-linked product comigrated with the 49-kDa subunit identified in the current study (data not shown), whereas digestion according to the previous procedure using DNase I and micrococcal nuclease resulted in bands migrating at 59 to 61 kDa (27). The less stringent nuclease digestion protocol used in the earlier study apparently left a larger number of nucleotides residually attached to the protein that slowed its migration through the SDS gel. Thus, the polypeptide that cross-linked strongly to the earlier probe (which had BrdU incorporated at positions 15, 16, 17, and 18 in the nontemplate strand) appears identical to the 49-kDa polypeptide identified in the present study. Since the 49-kDa polypeptide cross-linked very strongly to phosphate 17 in the nontemplate strand, experiments with both types of cross-linking reagent (BrdU or phosphorothioate based) suggest that the 49-kDa subunit is in very close contact with the DNA in this region of the PSEA. Henry et al. (7) demonstrated that the 50-kDa subunit of SNAPc could be cross-linked to DNA by using a BrdU-substituted probe. It is therefore possible that the 50-kDa subunit of SNAPc and the 49 kDa subunit of DmPBP may be homologous polypeptides.
RNAP specificity and the differential binding of DmPBP to the U1 and U6 PSEAs.
In vertebrates, sea urchins, and plants, the PSEs upstream of RNAP II-transcribed and RNAP III-transcribed snRNA genes are interchangeable. However, this is not true in Drosophila where the sequence of the PSEA itself plays a major role in determining RNAP specificity (11). The finding that the PSEA in Drosophila is the major determinant of RNAP specificity predicts that DmPBP should interact differently with the PSEA sequences from U1 and U6 genes. The photo-cross-linking data obtained in the present study provide evidence that this indeed is true. Although the PSEA sequences in the photo-cross-linking probes are identical at 16 of 21 base positions and the flanking sequences in the photo-cross-linking probes are completely identical, we detect significant differences in the cross-linking patterns to the two probes.
The four bases at positions 14, 16, 19, and 20 differ between the U1 and U6 PSEAs, and each contributes to the RNAP specificity of the PSEA (11). It is therefore of interest that many of the most significant differences in the photo-cross-linking patterns occur near to these bases. For example, at phosphate position 14 on the template strand, the 49-kDa subunit cross-linked much more intensely to the U6 PSEA than to the U1 PSEA (Fig. 3, 5, and 6). Conversely, the same subunit cross-linked strongly to positions 18 and 20 of the U1 PSEA but did not cross-link to these same positions of the U6 PSEA. (In the numbering scheme used, the phosphate is adjacent to but on the downstream side of the base with the same number.)
The 45-kDa subunit also exhibited significant differences in its cross-linking pattern that correlate with the base differences responsible for polymerase specificity. For example, the cross-linking intensities of this subunit are greatest at position 16 in the template strand when bound to the U6 PSEA but greatest at position 20 when bound to the U1 PSEA (Fig. 3 and 6). Nonetheless, differences in the cross-linking of the 45-kDa subunit also occurred at more distant sites where the U1 and U6 PSEAs are identical in sequence (e.g., position 11 on the nontemplate strand). Thus, differential interactions resulting from base differences at one site can affect the local protein-DNA environment at a distant site as well.
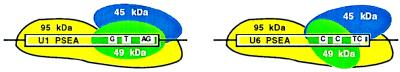
Relative position of the DmPBP subunits on the PSEA.
The data presented in Fig. 6 yielded information about the arrangement of the subunits of DmPBP relative to the DNA. Figure 7 presents a schematic model that is consistent with the photo-cross-linking data. The 95-kDa subunit is in close proximity to the DNA over the entire length of the PSEA. If the DNA is oriented such that this subunit approaches the front surface of the DNA in the 5′ half of the PSEA, then toward the 3′ half of the PSEA the 95-kDa subunit would primarily contact the DNA from below. The 49- and 45-kDa subunits also interact specifically with the 3′ half of the PSEA (Fig. 7). In the model, the 49-kDa subunit approaches from the front surface of the DNA, whereas the 45-kDa subunit approaches the PSEA from above. The 49-kDa subunit is intimately associated with the extreme 3′ end of the U1 PSEA by major groove contacts, but these are missing with the U6 PSEA (Fig. 6 and 7).
FIG. 7.
Schematic model of the subunits of DmPBP bound to the U1 and U6 PSEAs. DmPBP assumes a different conformation depending on whether it is bound to a U1 PSEA (left) or to a U6 PSEA (right). In comparison to the interaction with the U1 PSEA, the 49-kDa subunit lacks contacts with the extreme 3′ end of the U6 PSEA, and the 45-kDa subunit has stronger contacts with the central region of the U6 PSEA. The position of four of the five base differences between the U1 and U6 PSEAs are shown.
Our results are therefore consistent with a mechanism in which the same core set of DmPBP subunits bind to the U1 and U6 PSEAs, but they do so differently, resulting in conformational differences in the overall DmPBP-DNA complex (Fig. 7). Five base differences between the U1 and U6 PSEAs are sufficient to induce these two distinct modes of binding. We believe that these conformational differences in the DmPBP-DNA complex may lead during subsequent steps of preinitiation complex assembly to the exclusive recruitment of the correct polymerase-specific factors and the respective RNAP themselves. We have obtained no evidence to support the existence of polymerase-specific factors that interact directly with the PSEA sequences. If such factors were present in the HA300 fraction, we would expect to detect them in the photo-cross-linking assay. However, we cannot rule out that direct interactions between the PSEA and polymerase-specific factors may occur in a more completely assembled snRNA preinitiation complex.
ACKNOWLEDGMENTS
We are very grateful to Thierry Lagrange of Danny Reinberg’s lab, who gave us many helpful hints and suggestions for preparing and using the photo-cross-linking probes. Without his advice, this project would have been much more difficult. We thank Michael Sawaya of Joseph Kraut’s lab for assistance with molecular graphics. We thank George Kassavetis and Peter Geiduschek for helpful discussions, and we thank Anca Segall for critically reading the manuscript prior to submission.
This work was supported by grant GM-33512 from the National Institutes of Health, by the California Metabolic Research Foundation, and by the San Diego State University Department of Chemistry, College of Sciences, and Office of Faculty Affairs. During the course of this work, Y.W. was supported by an Arne N. Wick predoctoral research fellowship and a postdoctoral fellowship from the California Metabolic Research Foundation.
REFERENCES
- 1.Bai L, Wang Z X, Yoon J B, Roeder R G. Cloning and characterization of the β subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol Cell Biol. 1996;16:5419–5426. doi: 10.1128/mcb.16.10.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahlberg J E, Lund E. The genes and transcription of the major small nuclear RNAs. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Heidelberg, Germany: Springer-Verlag KG; 1988. pp. 38–70. [Google Scholar]
- 3.Dahlberg J E, Lund E. How does III × II make U6? Science. 1991;254:1462–1463. doi: 10.1126/science.1962205. [DOI] [PubMed] [Google Scholar]
- 4.Das G, Henning D, Reddy R. Structure, organization, and transcription of Drosophila U6 small nuclear RNA genes. J Biol Chem. 1987;262:1187–1193. [PubMed] [Google Scholar]
- 5.Das G, Henning D, Wright D, Reddy R. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J. 1988;7:503–512. doi: 10.1002/j.1460-2075.1988.tb02838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans S V. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 7.Henry R W, Ma B C, Sadowski C L, Kobayashi R, Hernandez N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 8.Henry R W, Sadowski C L, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 10.Hernandez, N. 1997. Personal communication.
- 11.Jensen R C, Wang Y, Hardin S B, Stumph W E. The proximal sequence element (PSE) plays a major role in establishing the RNA polymerase specificity of Drosophila U-snRNA genes. Nucleic Acids Res. 1998;26:616–622. doi: 10.1093/nar/26.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamakaka R T, Tyree C M, Kadonaga J T. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc Natl Acad Sci USA. 1991;88:1024–1028. doi: 10.1073/pnas.88.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagrange T, Kim T-K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J-M, Haberman R P, Marzluff W F. Common factors direct transcription through the proximal sequence elements (PSEs) of the embryonic sea urchin U1, U2, and U6 genes despite minimal sequence similarity among the PSEs. Mol Cell Biol. 1996;16:1275–1281. doi: 10.1128/mcb.16.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J-M, Parsons R A, Marzluff W F. Transcription of the sea urchin U6 gene in vitro requires a TATA-like box, a proximal sequence element, and sea urchin USF, which binds an essential E box. Mol Cell Biol. 1994;14:2191–2200. doi: 10.1128/mcb.14.3.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo P C H, Mount S M. Drosophila melanogaster genes for U1 snRNA variants and their expression during development. Nucleic Acids Res. 1990;18:6971–6979. doi: 10.1093/nar/18.23.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobo S M, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 18.Lobo S M, Hernandez N T. Transcription of snRNA genes by RNA polymerases II and III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 127–159. [Google Scholar]
- 19.Murphy S, Di Liegro C, Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–87. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- 20.Murphy S, Yoon J-B, Gerster T, Roeder R G. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 22.Parry H D, Scherly D, Mattaj I W. Snurpogenesis: the transcription and assembly of U snRNP components. Trends Biochem Sci. 1989;14:15–19. [Google Scholar]
- 23.Parry H D, Tebb G, Mattaj I W. The Xenopus U2 gene PSE is a single, compact, element required for transcription initiation and 3′ end formation. Nucleic Acids Res. 1989;17:3633–3644. doi: 10.1093/nar/17.10.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadowski C L, Henry R W, Kobayashi R, Hernandez N. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc Natl Acad Sci USA. 1996;93:4289–4293. doi: 10.1073/pnas.93.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadowski C L, Henry R W, Lobo S M, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 26.Stefanovic B, Marzluff W F. The proximal element is capable of determining proper temporal expression of the embryonic sea urchin U2 snRNA gene. Gene Expression. 1994;4:1–18. [PMC free article] [PubMed] [Google Scholar]
- 27.Su Y, Song Y, Wang Y, Jessop L, Zhan L C, Stumph W E. Characterization of a Drosophila proximal-sequence-element-binding protein involved in transcription of small nuclear RNA genes. Eur J Biochem. 1997;248:231–237. doi: 10.1111/j.1432-1033.1997.t01-1-00231.x. [DOI] [PubMed] [Google Scholar]
- 28.Tanikawa J, Yasukawa T, Enari M, Ogata K, Nishimura Y, Ishii S, Sarai A. Recognition of specific DNA sequences by the c-myb protooncogene product: Role of three repeat units in the DNA-binding domain. Proc Natl Acad Sci USA. 1993;90:9320–9324. doi: 10.1073/pnas.90.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vankan P, Filipowicz W. A U-snRNA gene-specific upstream element and a -30 ‘TATA box’ are required for transcription of the U2 snRNA gene of Arabidopsis thaliana. EMBO J. 1989;8:3875–3882. doi: 10.1002/j.1460-2075.1989.tb08566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waibel F, Filipowicz W. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res. 1990;18:3451–3458. doi: 10.1093/nar/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldschmidt R, Wanandi I, Seifart K H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;10:2595–2603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wendelburg B J, Marzluff W F. Two promoter elements are necessary and sufficient for expression of the sea urchin U1 snRNA gene. Nucleic Acids Res. 1992;20:3743–3751. doi: 10.1093/nar/20.14.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S-W, Nash H A. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc Natl Acad Sci USA. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon J-B, Murphy S, Bai L, Wang Z, Roeder R G. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon J-B, Roeder R G. Cloning of two proximal sequence element-binding transcription factor subunits (γ and δ) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol Cell Biol. 1996;16:1–9. doi: 10.1128/mcb.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamrod Z, Tyree C M, Song Y, Stumph W E. In vitro transcription of a Drosophila U1 small nuclear RNA gene requires TATA box-binding protein and two proximal cis-acting elements with stringent spacing requirements. Mol Cell Biol. 1993;13:5918–5927. doi: 10.1128/mcb.13.9.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]