Abstract
A novel 761-amino-acid transcription factor, DMP1, contains a central DNA binding domain that includes three imperfect myb repeats flanked by acidic transactivating domains at the amino and carboxyl termini. D-type cyclins associate with a region of the DMP1 DNA binding domain immediately adjacent to the myb repeats to form heteromeric complexes which detectably interact neither with cyclin-dependent kinase 4 (CDK4) nor with DNA. The segment of D-type cyclins required for its interaction with DMP1 falls outside the “cyclin box,” which contains the residues predicted to contact CDK4. Hence, D-type cyclin point mutants that do not interact with CDK4 can still bind to DMP1. Enforced coexpression of either of three D-type cyclins (D1, D2, or D3) with DMP1 in mammalian cells canceled its ability to activate gene expression. This property was not shared by cyclins A, B, C, or H; did not depend upon CDK4 or CDK2 coexpression; was not subverted by a mutation in cyclin D1 that prevents its interaction with CDK4; and was unaffected by inhibitors of CDK4 catalytic activity. Introduction of DMP1 into mouse NIH 3T3 fibroblasts inhibited entry into S phase. Cell cycle arrest depended upon the ability of DMP1 to bind to DNA and to transactivate gene expression and was specifically antagonized by coexpression of D-type cyclins, including a D1 point mutant that does not bind to CDK4. Taken together, these findings suggest that DMP1 induces genes that inhibit S phase entry and that D-type cyclins can override DMP1-mediated growth arrest in a CDK-independent manner.
Entry into the cell division cycle from quiescence is stimulated by mitogens, which must be present throughout most of the first gap (G1) phase for cells to synthesize their chromosomal DNA (in S phase) (36). In mammalian cells, three D-type cyclins (D1, D2, and D3) are induced in a combinatorial, lineage-specific fashion in response to growth factor-mediated signaling and accumulate throughout the G1 interval (47). D-type cyclins enter into holoenzyme complexes with cyclin-dependent kinase 4 (CDK4) and CDK6 to trigger phosphorylation of the retinoblastoma protein (Rb) in mid- to late G1 phase (28, 29, 32). In its hypophosphorylated state, Rb prevents G1 exit by binding to transcription factors, such as the E2Fs, thereby inhibiting their activity (3, 9, 11, 18, 25, 57). Rb phosphorylation by the cyclin D- and E-dependent kinases (6, 15, 20, 48, 56), probably in a defined temporal sequence (4, 13, 43), releases E2Fs from Rb constraint and enables them to activate a series of genes that are required for S phase entry (8). Cells that overexpress cyclin D1 or that lack Rb function exhibit a decreased dependency on growth factors and a shortened G1 phase (14, 39, 42). Conversely, inhibition of cyclin D-dependent kinase prevents S phase entry in normal cells (1, 39) but does not affect G1 exit in cells that lack a functional Rb protein (10, 24, 26, 27, 31, 46). Therefore, it has been suggested that Rb, and perhaps related pocket proteins like p107 and p130, are the only physiologic substrates of cyclin D-dependent kinases whose phosphorylation is essential for S phase entry.
These observations do not preclude additional roles for D-type cyclins, and indeed, other functions for these proteins have been documented. For example, in proliferating cells, cyclin D-CDK complexes bind and sequester CDK inhibitors during G1 phase, enabling cyclin E-CDK2 complexes to exceed the inhibitory threshold (22, 37, 38, 49, 52). Mitogen withdrawal or treatment of cells with certain antiproliferative cytokines, such as transforming growth factor β, results in the loss of functional cyclin D-CDK complexes, thereby releasing latent pools of p27Kip1 and p21Cip1, which associate with cyclin E-CDK2 to extinguish its activity (12, 44, 45). This coordinated inhibition of both classes of G1 cyclin-dependent kinases leads to G1 phase arrest, usually within a single cell cycle. In this scenario, cyclin D-CDK complexes function stoichiometrically to titrate CDK inhibitors and CDK catalytic activity per se is not required (49).
Cyclin D-dependent kinases may also negatively regulate differentiation programs whose proper execution depends upon cell cycle arrest. For example, ectopic expression of cyclin D1 can cancel the differentiation-promoting effects of MyoD1 in mitogen-deprived myoblasts while, conversely, the inhibition of endogenous cyclin D-dependent kinase activity by CDK inhibitors can promote MyoD1-dependent differentiation, even in mitogen-stimulated cells (41, 51). Overexpression of cyclins D2 and D3 inhibits the ability of cultured interleukin-3-dependent myeloid blasts to undergo neutrophil differentiation in response to granulocyte colony-stimulating factor (21). Cyclin D1 also plays a crucial, lineage-specific role in breast lobular alveolar development (7, 50), and its overexpression in breast epithelium can trigger tumor formation (54). Surprisingly, D-type cyclins were found to bind directly to the estrogen receptor and were reported to enhance its transcriptional activity via a hormone- and CDK-independent mechanism (33, 58). The latter results raise the intriguing possibility that D-type cyclins might participate in physiologic processes that do not mechanistically depend upon CDKs at all.
Using a two-hybrid interactive screen with cyclin D2 as “bait,” a cyclin D-binding myb-like protein, designated DMP1, was isolated (17). DMP1 is a novel 761-amino-acid protein which, while not bearing overall homology to others in available databases, contains a central domain composed of three tandem, imperfect myb-like repeats flanked by acidic domains at both its N and C termini. In agreement with the idea that DMP1 might regulate transcription, the protein was shown to stimulate transcription at synthetic minimal promoters containing the nonameric consensus sequence CCCG(G/T)ATGT. That subset of consensus oligonucleotide sequences containing the GGA core can also interact with ETS-family transcription factors, whereas those containing the GTA core do not. When radiolabeled oligonucleotide probes containing the GTA core were used, DMP1 DNA binding activity was unambiguously detected in lysates of mammalian T cells, fibroblasts, and embryonic kidney cells and a single DMP1 mRNA was observed to be ubiquitously expressed at low levels in adult mouse tissues. DMP1 is a relatively poor substrate (compared to Rb) for cyclin D-dependent kinases. Although D-type cyclins bind to DMP1 both in vitro and in yeast and insect cells programmed to coexpress the recombinant proteins, DMP1 and CDK4 compete with one another for binding to D-type cyclins and ternary complexes are not formed. Preliminary data suggested that, in the presence or absence of CDK4, cyclin D2 interfered with DMP1-mediated transcriptional activity when vectors encoding both proteins were introduced together with a reporter gene into transformed 293T human embryonic kidney cells. As the latter cells lack functional Rb, the observed inhibition of DMP1-mediated transactivation did not depend upon Rb phosphorylation by cyclin D-dependent kinases. Although the physiologic role of DMP1 remains unclear, its properties nonetheless underscore a potential for Rb-independent control of gene expression by D-type cyclins.
We have now mapped the functional domains in DMP1 that are necessary for DNA binding, transactivation, and association with D-type cyclins. We show that DMP1 can induce cell cycle arrest in rodent fibroblasts which is overridden, in a CDK-independent manner, by D-type cyclins.
MATERIALS AND METHODS
Cells and culture conditions.
NIH 3T3 cells were cultured in a 5% CO2 incubator at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and penicillin and streptomycin (each at 100 U/ml; Gibco/BRL, Gaithersburg, Md.). Spodoptera frugiperda Sf9 cells were maintained at 27°C in Grace’s medium containing 10% FBS, Yeastolate, lactalbumin hydrolysate, and gentamicin (all from Gibco/BRL) in 100-ml spinner bottles. Baculoviruses were produced using a Bac-to-Bac Baculovirus expression system according to instructions of the manufacturer (Gibco/BRL, Gaithersburg, Md.) and propagated as previously described (20).
Construction of DMP1 mutants.
By PCR primer-based amplification, an EcoRI site was inserted at the expense of the ATG initiator codon of DMP1, enabling full-length DMP1 coding sequences to be excised with EcoRI from pBluescript-DMP1 (17) and reinserted downstream of the pSRα promoter and Flag epitope tag in the pFLEX1 mammalian expression vector (2). The resulting construct (pFLEX-DMP1) encodes Met-Asp-Tyr-Lys-Asp4-Lys instead of the original DMP1 initiator codon, enabling precipitation of the encoded product by the M2 monoclonal antibody directed to the Flag epitope (Kodak, New Haven, Conn.). C-terminal DMP1 truncation mutants M1 to M4 (see Fig. 1A) were created by using unique DMP1 restriction sites (EcoNI at codon 661, StuI at codon 520, NcoI at codon 458, and BstBI at codon 380). Plasmids codigested with either of these enzymes and with SmaI (at a unique site located in the 3′ polylinker) were treated with the Klenow fragment of DNA polymerase I and recircularized at the blunt ends; the resulting DMP1 fragments were excised with EcoRI and transferred into pFLEX1.
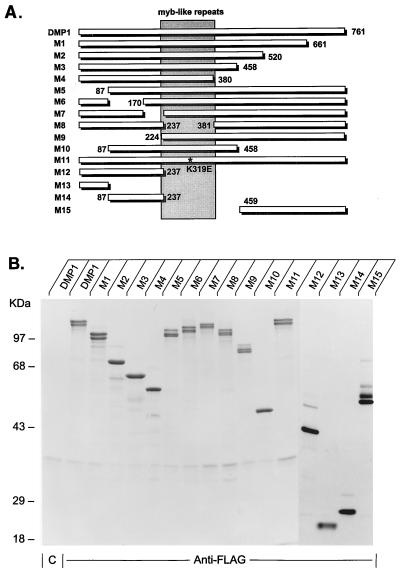
FIG. 1.
DMP1 mutants. (A) Schematic representation of wild-type DMP1 (top line) and various mutants (M1 to M15). All are deletion mutants except for M11, which contains a Glu-for-Lys substitution at codon 319 (K319E, marked with an asterisk) located within the second myb repeat. The numbers indicate the deletion boundaries, and the central region containing the three tandem myb repeats is shaded. (B) Metabolically labeled wild-type and mutant DMP1 proteins recovered from baculovirus vector-infected Sf9 cells and separated on denaturing polyacrylamide gels. The first lane on the left indicates the results of precipitation of wild-type DMP1 with an irrelevant control monoclonal antibody (designated C), as indicated at the bottom of the panel. All other lysates were precipitated with monoclonal antibody M2 directed to the Flag epitope at the DMP1 N terminus. The mobilities of markers of known molecular mass are indicated to the left of the panel.
The N-terminal truncation mutant M5 (see Fig. 1A) was created in the original pBluescript-DMP1 by digestion with XbaI (5′ DMP1 untranslated sequence) and BstEII (codon 86) followed by linker ligation to adjust the reading frame and create a 5′ EcoRI site. This was accomplished by preparing a partially overlapping, self-annealing double-stranded oligonucleotide (sense strand, 5′-CTAGAACTCGTGAATTC; antisense strand, 5′-GTCACGAATTCACGCGTT) containing the EcoRI site (underlined), thereby enabling subsequent transfer of the truncated DMP1 fragment into pFLEX1. Mutant M15 was created similarly, except that pBluescript-DMP1 was digested with XbaI and NcoI (codon 458) and religated with an analogous linker containing an EcoRI site. Mutant M10 was derived from M5 by truncation with NcoI (codon 458), codigestion with SmaI (3′ polylinker), and blunt-end ligation prior to transfer into pFLEX1.
Mutants M6 to M8 were created in pFLEX1-DMP1 by digestion at unique restriction sites and linker ligation, using a strategy analogous to that described above for mutant M5. Sites of restriction used were BstEII (codon 87), Eco47III (codon 170), SacI (codon 237), and BstBI (codon 380) (see Fig. 1A); for convenience, the linkers were designed to contain an internal MluI site, enabling confirmation of linker insertion after ligation.
Mutant M9 was prepared by amplifying DMP1 sequences, including codons 224 to 391, by using the following primers: sense, 5′-ACTTCTAGAGAATTCGTGGGAAAATACACTCCTGAA (EcoRI site and XbaI site underlined), and antisense, 5′-ACTGAATTCTCATGCAATTTGCCTTTTGATGGT. The amplified DNA was codigested with XbaI and BstBI (codon 380) and substituted for the unique XbaI-BstBI fragment in pBluescript-DMP1. The resulting plasmid was then digested with EcoRI, and the DMP1 fragment was transferred into pFLEX1.
The M11 point mutant (with a K-to-E mutation at position 319 [K319E]) was generated by two-step PCR with the following oligonucleotide primers: 5′-CAATGACTGGGCAACAATAGG (an upstream sense primer beginning 28 bp 5′ of the DMP1 AvrII site at codon 253), 5′-GATGGTCCACCATTTACTTCG (a downstream antisense primer beginning 19 bp 3′ of the BstBI site at codon 380), 5′-GCTCAGAAGAGCAATGCCGTT (K319E sense strand primer) (codon 319 is underlined), and 5′-AACGGCATTGCTCTTCTGAGC (K319E antisense strand primer) (codon 319 is underlined). PCRs were performed with either the upstream flanking oligonucleotide and the antisense oligonucleotide containing the mutation at codon 319 or the downstream oligonucleotide and the sense-strand oligonucleotide containing the codon 319 mutation. The two respective PCR products were gel purified, mixed, and joined by amplification using the upstream and downstream oligonucleotide primers. PCR products were redigested with AvrII and BstBI and subcloned at the expense of wild-type sequences into pBluescript-DMP1 before transfer to pFLEX1. All fragments synthesized by PCR were resequenced in their entirety to confirm their authenticity.
For expression in insect cells, the complete Flag-tagged DMP1 coding sequence and fragments encoding all of the above mutants were excised from pFLEX1 by digestion with BamHI and cloned into the pFastBac expression vector (Gibco/BRL). Mutant M12 was prepared in pFastBac-DMP1 by deleting the unique SacI fragment (DMP1 codon 237 to the 3′ polylinker), enabling direct in-frame ligation. Mutant M13 was obtained similarly following digestion of pFastBac-DMP1 with BstEII (codon 86) and XbaI (3′ polylinker), treatment with the Klenow fragment of polymerase I, and blunt-end ligation. Mutant M14 was generated from pFastBac-M5-DMP1 by deleting the unique SacI fragment as described above.
Other expression plasmids.
Deletion mutants of cyclin D1 were generated in pBluescript-D1 by restriction enzyme digestion of the plasmid DNA and removal of internal coding sequences (Δ1-99, Δ142-253, and Δ67-166 by using PstI, XmnI, and an MnlI partial digest, respectively followed by linker ligation to adjust the open reading frames and to provide an initiator codon in the case of D1 (Δ1-99). After confirmation by nucleotide sequencing, the cDNAs were recloned into the pFastBac baculovirus vector.
Based on studies with human cyclin D1 (58), we prepared an analagous mouse D1 mutant (K114E) which does not bind or activate CDK4. The entire D1 coding sequence was amplified by two-step PCR using a 5′ upstream primer (5′-GAATTCGGCCCGCGCCATGGAACACCAGCTCCTG [EcoRI and initiation codons are underlined]), a downstream antisense primer (5′-GGATCCTCAGATGTCCACATCTCG [BamHI and the termination codon, TGA in the sense strand, are both underlined]), a sense-strand primer containing mutated codon 114 (5′-TTGCCTCTAAGATGGAGGAGACCATTCCC [codon 114 is underlined]), and an antisense primer containing the mutated codon (5′-GGGAATGGTCTCCTCCATCTTAGAGGCCA [codon 114 is underlined]). The strategy was identical to that used for mutant M11 as described above. The resulting product was resequenced, excised with EcoRI and BamHI, and transferred into the pBJ5 expression vector (identical to pFLEX1, but lacking sequences encoding the Flag epitope). Coexpression in Sf9 cells was used to confirm that cyclin D1 (K114E) did not bind to CDK4 or activate its Rb kinase activity. Full-length cDNAs encoding wild-type and other mutant D cyclins, cyclins A, B, C, and H, and p27Kip1 were also cloned into the EcoRI site of pBJ5. Expression vectors encoding a wild-type or catalytically inactive CDK4 mutant (K35M) (28), CDK2 (39), p16INK4a (40), and p19INK4d (16) were prepared as previously described.
DMP1 and cyclin D association in insect cells.
Insect Sf9 cells infected with the indicated recombinant baculoviruses were metabolically labeled 24 to 48 h postinfection for an additional 16 h in methionine-free medium with 50 μCi of [35S]methionine (1,000 Ci/mmol; ICN, Irvine, Calif.) per ml. Infected cells (106) were lysed by repeated freezing and thawing in 300 μl of EBC buffer (50 mM Tris HCl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA) containing 2% aprotinin, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM sodium orthovanadate, and 0.1 mM sodium fluoride. For detection of Flag-tagged DMP1 or its complexes with D cyclins, 50 to 100 μl of lysate was diluted with 300 μl of EBC buffer; M2 beads (24 μl of a 1:1 suspension in phosphate-buffered saline [PBS] [Kodak]) were added and were recovered by centrifugation after incubation for 2 h at 4°C. With the exception of mutant M13, the quantity of DMP1 mutants was adjusted, based on their methionine content, to be approximately equimolar. The beads were washed 5 times in immunoprecipitation assay buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) containing protease and phosphatase inhibitors as described above, and the immunoprecipitated proteins were denatured and resolved on denaturing polyacrylamide gels containing SDS.
Purification of Flag-tagged DMP1 and Flag-tagged cyclin D1-DMP1 complexes.
Sf9 cells were infected for 24 h with a baculovirus vector encoding wild-type, Flag-tagged DMP1 or were coinfected with viruses encoding Flag-tagged cyclin D1 and untagged DMP1. Infected cells were metabolically labeled with [35S]methionine as described above and lysed in EBC buffer, and lysates cleared of debris were immunoprecipitated with M2 beads. The beads were washed five times with EBC buffer and suspended in 100 μl of EBC buffer. A 400-fold molar excess (20 μg) of FLAG-peptide was added to the suspended beads, which were incubated on a rotary shaker for 4 h at 4°C. Supernatants were collected after centrifugation, and portions were either subjected to electrophoresis on denaturing polyacrylamide gels or were used for electrophoretic mobility shift assays (EMSAs).
DMP1 and cyclin D association in NIH 3T3 fibroblasts.
In order to detect D-cyclins bound to DMP1, 8 × 105 NIH 3T3 cells were cotransfected with 5 μg of pFLEX-DMP1 or with DMP1 mutants together with 20 μg of the pBJ5-cyclin D2 expression vector. Cells were lysed in EBC buffer containing protease and phosphatase inhibitors, and cleared lysates were immunoprecipitated with M2 beads which were washed five times with EBC buffer, boiled in gel sample buffer, and separated by electrophoresis on denaturing 10% polyacrylamide gels. Proteins were transferred onto Immobilon membranes (Millipore, Bedford, Mass), which were probed with rat monoclonal antibodies to murine cyclin D2 (53). Sites of antibody binding were detected according to manufacturer’s instructions by using rabbit antiserum to rat immunoglobulin G, followed by protein A-conjugated horseradish peroxidase (EY Laboratories, San Mateo, Calif.). The filters were subjected to chemiluminescence detection (ECL detection kit; Amersham, Arlington Heights, Ill.).
EMSA.
EMSAs were performed as described previously (17), using lysates of normal or transfected NIH 3T3 cells as indicated. Washed cells were suspended in 10 mM HEPES (pH 7.9)–10 mM KCl–0.1 mM EDTA–0.1 mM EGTA–1 mM dithiothreitol (DTT)–0.5 mM PMSF and swollen on ice for 15 min. Nonidet P-40 was added to a final concentration of 0.5%, and after vigorous vortexing for 10 s, nuclei were collected by centrifugation and washed once in the same buffer. For all assays performed with nuclei from transfected cells, the nuclear pellets were routinely suspended in ice-cold high-salt buffer (20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF), and rocked at 4°C for 15 min, spun for 15 min in a microcentrifuge to clear debris, and the supernatant fluids were stored at −70°C until used. In experiments using untransfected cells, nuclei were suspended as indicated in low-salt extraction buffer (25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 1 mM PMSF, 1% Triton X-100), frozen on dry ice and thawed through two rapid cycles, and treated as described above. Double-stranded oligonucleotides (BS2 probe) containing binding site 2 (the nonameric DMP1 binding site included in BS2 is CCCGTATGT), which is recognized by DMP1 but not by ETS1 or ETS2, were labeled with [32P]dATP (6,000 Ci/mmol; New England Nuclear) using the Klenow fragment of DNA polymerase I. Equal quantities of nuclear extracts normalized for protein content were incubated with the labeled BS2 probe or with a second labeled oligonucleotide (M3 probe) containing a nonameric ETS-1 or ETS-2 binding site (CCCGGAAGT) not bound by DMP1 (17). For competition experiments, a 100-fold excess of unlabeled BS2 or M3 oligonucleotide was added to the reaction mixtures before addition of the labeled probes. To verify that complexes formed using extracts from untransfected cells contained endogenous DMP1, reaction mixtures were preincubated with nonimmune rabbit serum (NRS) or with an antiserum (AF) directed to the DMP1 carboxyl terminus before their electrophoretic resolution on nondenaturing 4% polyacrylamide gels (17).
Transactivation assay.
Transactivation assays were performed as described previously (17). Briefly, NIH 3T3 cells were transfected with 8 μg of a BS2-containing reporter plasmid encoding DMP1-responsive luciferase, with or without 3 μg of pFLEX-DMP1. Where indicated, cells were cotransfected with 12 μg of mammalian expression vectors encoding the indicated cyclins, CDKs, or CDK inhibitors. A plasmid (4 μg) encoding secreted alkaline phosphatase driven by the β-actin promoter was routinely cotransfected to normalize transfection efficiencies (5, 34a). Sixteen hours after transfection, cells were washed twice with PBS, cultured in complete medium for 24 h, and then serum starved for an additional 18 h before lysates and supernatants were prepared and assayed for luciferase and alkaline phosphatase activities as described previously (5, 34a). Where indicated, cells were maintained in serum for 42 h prior to assay.
BrdU incorporation assay and immunofluorescence.
NIH 3T3 cells were seeded on coverslips 16 h prior to transfection. Cells were transfected with pFLEX-DMP1 or its mutants, and 14 h later, they were washed with PBS and incubation continued for 8 h in complete medium containing 10% FBS. Where indicated, the cells were washed twice with PBS and starved for 24 h in medium containing 0.1% fetal calf serum, thereby rendering the cells quiescent in G0 phase. Growth-arrested cells were restimulated to enter the cell cycle synchronously with DMEM plus 10% FBS, and bromodeoxyuridine (BrdU) was added to the medium. Cells were fixed 22 h later (corresponding to one complete cell cycle) in ice-cold methanol-acetone (1:1) for 10 min at −20°C. For staining of Flag-tagged DMP-1 proteins, the coverslips were incubated with the M2 monoclonal antibodies (6 μg/ml; Kodak) in Tris-buffered saline (TBS) containing 1 mM CaCl2 for 1 h at room temperature. Coverslips were washed in TBS, followed by incubation for 30 min with biotinylated antibodies to mouse immunoglobulin G (1:500 dilution; Vector Laboratories, Burlingame, Calif.) in TBS containing 5% FBS as a blocking agent. The coverslips were washed and incubated under the same conditions with Texas red-conjugated streptavidin (1:500; Amersham). Staining with antibodies to BrdU (1:12; Vector Laboratories) was performed as described previously, and sites of BrdU incorporation into DNA were detected with fluorescein-conjugated antibodies (green) (35). Finally, after being washed in TBS, coverslips were incubated for 1 min in Hoechst 33258 dye (Sigma) in TBS (blue). Coverslips were mounted on glass slides with Vectashield medium (Vector Laboratories), and stained cells were visualized with an Olympus BX50 microscope fitted with appropriate fluorescence filters.
RESULTS
Mapping functional domains of DMP1.
DMP1 contains two acidic regions (N-terminal residues 4 to 169 and C-terminal residues 579 to 756) flanking three tandem myb-like repeats residing between residues 224 and 392. In an attempt to map functional domains within the protein, we prepared the series of deletion mutants schematized in Fig. 1A. Given that a conserved sequence in myb repeat 2 of DMP1 (K319QCRXXWXN) corresponds to the region where the c-myb protein contacts its DNA binding site (34), we also mutated lysine 319 to glutamic acid (K319E) (mutant M11). All of the mutant cDNAs were tagged at their N terminus, so that they could be precipitated with the M2 monoclonal antibody to the Flag epitope, and they were then cloned into both baculovirus and mammalian expression vectors.
Insect Sf9 cells infected with a baculovirus vector encoding wild-type DMP1 produce a family of ∼125-kDa proteins, representing different phosphorylated forms of DMP1. When separated on denaturing polyacrylamide gels containing SDS, each of these DMP1 species is reduced to a single major band of higher electrophoretic mobility following treatment with calf intestinal phosphatase, although the apparent molecular mass of the dephosphorylated protein is anomalous and still remains significantly greater than that predicted from the DMP1 cDNA sequence (∼85 kDa) (17). Given these considerations, all deletion mutants were well expressed and migrated electrophoretically at their expected relative positions (Fig. 1B). Those mutant proteins retaining distal C-terminal residues 661 to 761 (M1, M5 to M9, M11, and M15) exhibited markedly more heterogeneous mobilities on gels than those lacking this segment (M2 to M4, M10, and M12 to M14), implying that multiple sites of phosphorylation were clustered at the C terminus.
Nuclear lysates from NIH 3T3 cells transfected with vectors expressing wild-type or mutant forms of DMP1 were used for EMSA with a 32P-labeled oligonucleotide probe (previously designated BS2) that contains a nonameric DMP1 DNA binding sequence (CCCGTATGT). In the absence of any unlabeled competing oligonucleotide, full-length DMP1 and deletion mutants M1, M2, M3, M5, and M10 generated readily detectable probe-containing complexes (Fig. 2 [lanes marked with minus signs]). These were competed with the unlabeled BS2 oligonucleotide (lanes marked with plus signs) but not by three other probes containing point mutations throughout the consensus DMP1 binding sequence (data not shown). Taken together, these data indicate that the domain required for DNA binding includes the three myb-like repeats and at least some of the flanking sequences contained in mutant M10 (residues 87 to 458). The 5′ boundary of this region resides between amino acids 87 and 170 (compare results with mutant M5 and those with M6), and the 3′ boundary between residues 380 and 458 (M3 versus M4). Importantly for some of the studies that follow, the K319E mutation within this region (mutant M11) was itself sufficient to abrogate DNA binding. Data identical to those obtained with NIH 3T3 nuclear extracts were also obtained with recombinant proteins expressed in Sf9 lysates (data not shown).
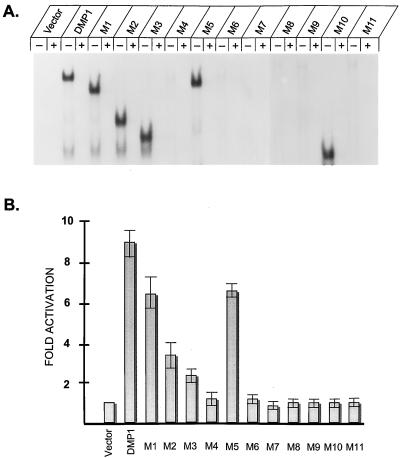
FIG. 2.
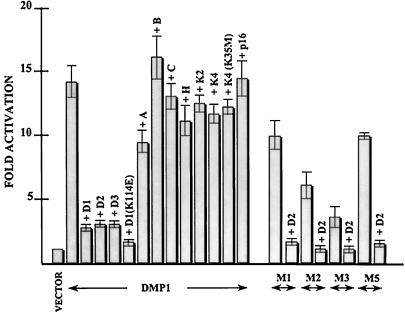
Identification of DNA binding and transactivation domains of DMP1. (A) EMSAs performed with DMP1 proteins produced in NIH 3T3 cells and with a radiolabeled DMP1-specific (BS2) probe containing the consensus binding sequence CCCGTATGT. NIH 3T3 cells were transfected with expression vectors encoding the indicated wild-type or mutant DMP1 proteins, and nuclear lysates were used for EMSA. Binding assays were performed in the absence (−) or presence (+) of excess competing oligonucleotide. (B) Results of transactivation assays performed in serum-starved NIH 3T3 cells using expression vectors encoding a DMP1-responsive luciferase reporter gene together with vectors specifying wild-type or mutant DMP1 proteins. Luciferase assays were performed 60 h after transfection, and cells were starved for serum for 18 h before assay. Error bars indicate standard deviations from the mean from multiple experiments.
With this information in hand, we used a luciferase reporter plasmid containing tandem DMP1 binding sites 5′ to the simian virus 40 minimal promoter to assay the DMP1 mutants for their ability to activate transcription. Proliferating mouse NIH 3T3 cells transfected with the reporter plasmid showed an approximately fivefold increase in luciferase activity when cotransfected with an expression vector encoding wild-type DMP1. However, an 8- to 15-fold increase in transactivation was observed in a subsequent series of experiments in which the transfected cells were serum starved for 18 h prior to performing the luciferase assay (Fig. 2B). Under these conditions, the absolute basal activity of the reporter gene was unchanged. Consistent with the idea that DMP1 may act as a more potent transactivator in nonproliferating cells, cotransfection of NIH 3T3 cells with the CDK inhibitor p27Kip1, at conditions under which it induces growth arrest of cells maintained in serum, also led reproducibly to increased DMP1-mediated promoter activity (10- to 15-fold basal levels) equivalent to that observed in serum-starved, quiescent cells (data not shown).
With the exception of M10, all DMP1 mutants that could bind to DNA (Fig. 2A) activated transcription (Fig. 2B). The processive C-terminal mutants, M1 to M3, were less-potent transactivators than the full-length protein, as was the M5 mutant, which lacked the extreme N terminus. Therefore, both the acidic N- and C-terminal domains of DMP1 contribute to transactivation, although the role of the C-terminal sequences (residues 458 to 761) appears to be more significant. In agreement, the minimal DNA binding domain (M10) lacking the latter sequences was completely unable to activate transcription (Fig. 2B).
DMP1 was initially isolated in a two-hybrid interactive screen using cyclin D2 as bait, and the recombinant protein can physically associate with D-type cyclins in vitro and when coexpressed with them in Sf9 cells (17). To map the minimal domain of DMP1 that interacts with D-type cyclins, we coexpressed cyclin D2 together with Flag-tagged DMP1 mutants in Sf9 cells. After metabolically labeling the cells with [35S]methionine and adjusting the protein inputs based on methionine content, DMP1 was precipitated with antibodies to the Flag tag and the precipitates were assayed for the presence of cyclin D2. As shown in a representative experiment (Fig. 3A), radiolabeled cyclin D2 failed to efficiently coprecipitate with the M9 mutant, which lacked N-terminal residues 1 to 224, whereas this region alone (mutant M12, residues 1 to 237) was sufficient for cyclin binding. In agreement, mutants lacking portions of the polypeptide between residues 237 and the C terminus (M3, M4, and M8) were able to bind cyclin D2. Residues 1 to 87 were not required for binding (mutants M5 and M13), and the remaining N-terminal sequences between residues 87 and 237 (mutant M14) were sufficient. Although some of the variations in D2 signal strength shown in Fig. 3A reflect experimental variations, the partial reduction in binding observed with mutants M5 and M14 was reproducibly seen, leaving open the possibility that some residues N terminal to amino acid 87 might contribute to the efficiency of cyclin D binding. It is important to note that the cyclin D-interacting segment falls within the portion of DMP1 that is required for DNA binding (mutant M10) (Fig. 2A and 3A); however, at least two, and possibly all three, of the myb repeats (i.e., sequences C terminal to residue 237) are dispensable for the association with D cyclin (mutants M8, M12, and M14 in Fig. 3A). Moreover, mutant M11 bound D cyclins as efficiently as wild-type DMP1. Virtually identical data were obtained by immunoblotting rather than radiolabeling to detect cyclin D2 in complexes with Flag-tagged DMP1 mutants or in experiments in which D1 was used instead of D2 (data not shown).
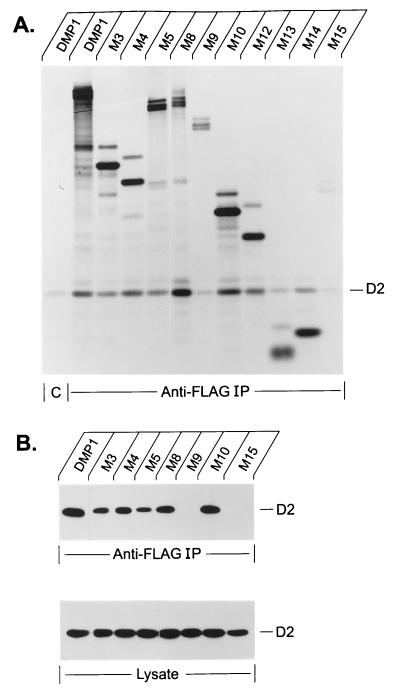
FIG. 3.
Mapping the cyclin D binding domains in DMP1. (A) Interactions between DMP1 and cyclin D2 in insect cells. Sf9 cells were coinfected with baculoviruses encoding either wild-type or mutant DMP1 together with murine cyclin D2. Cells metabolically labeled with [35S]methionine were disrupted, and cleared lysates were precipitated with anti-Flag M2 beads or with a control antibody (designated C), as indicated at the bottom of the panel. The position of coprecipitating cyclin D2 is indicated in the right margin. DMP1 mutants were normalized for methionine content to ensure roughly equivalent protein inputs. The one exception was mutant M13, which contains only a single methionine residue and was overloaded; despite the high input concentration of M13, it did not interact strongly with cyclin D2, reinforcing the central conclusions (see text). (B) Binding assays performed in transfected NIH 3T3 cells. NIH 3T3 cells cotransfected with pFLEX-DMP1 and pBJ5-cyclin D2 expression vectors were lysed in EBC buffer, and cleared lysates immunoprecipitated with M2 beads as indicated below the panel were immunoblotted with monoclonal antibodies to cyclin D2 (top). Lysates from the same transfections (one-sixth of the amounts used for immunoprecipitations) were directly blotted with anti-D2 to confirm equal expression of the cyclin in all transfections (bottom). The position of cyclin D2 is indicated to the right.
In NIH 3T3 cells cotransfected with expression vectors encoding DMP1 mutants and cyclin D2, the cyclin was coprecipitated with mutant proteins containing N-terminal residues 87 to 233 (M3 to M5, M8, and M10) but not with M9 or M15, which lack this region (Fig. 3B, upper panel). In this case, cyclin D2 binding was detected by immunoblotting of proteins precipitated with antibodies to the Flag epitope. As in Sf9 cells, mutant M5 interacted less well with cyclin D2 than those mutants that retained residues 1 to 87. The lower panel of Fig. 3B shows the amount of cyclin D2 detected in aliquots of the same lysates (loading one-sixth of the amounts taken for immunoprecipitation), confirming that approximately equal amounts of cyclin D2 were expressed in each of the transfections shown. In these experiments, fourfold more cyclin D2 vector than DMP1 vector was transfected into the cells (see Materials and Methods), so that cyclin D2 was produced in approximately fourfold molar excess over DMP1 (not shown). By comparing the amounts of bound (top) to aliquots (one-sixth) of total (bottom) cyclin D2, we can conclude that even under these conditions, a significant portion of the expressed cyclin D2 (>10%) moved into complexes with wild-type DMP1. Again, residues 87 to 237 were observed to contain the minimal D cyclin binding site(s). Similar data were obtained with cyclin D1 in lieu of D2 (data not shown).
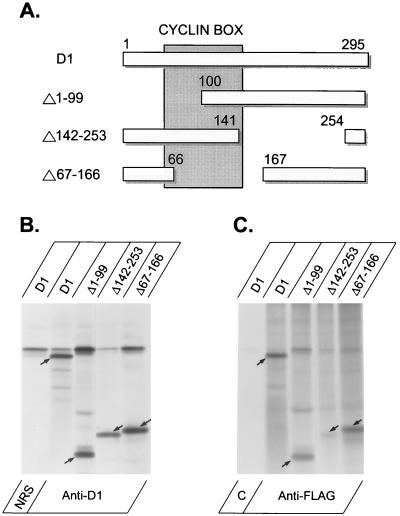
A separate series of experiments was undertaken to globally map regions within cyclin D1 which are required for the interaction with DMP1. The cyclin D1 deletion mutants schematized in Fig. 4A were coexpressed with Flag-tagged DMP1 in metabolically labeled Sf9 cells, and following precipitation of DMP1 with antibodies to the Flag epitope, coprecipitating cyclin was visualized on gels. The total quantities of labeled cyclins expressed in Sf9 lysates precipitated with antiserum to cyclin D1 (Fig. 4B) are compared to the amounts precipitated from equal aliquots of the lysates using antibody to Flag-tagged DMP1 (Fig. 4C). Although wild-type cyclin D1 and mutants D1(Δ1–99) and D1(Δ67–166) readily associated with DMP1 (Fig. 4C), D1(Δ142–253) did not. Based on homology with other cyclins whose structures have been determined, D1 amino acids 56 to 152 comprise the first cyclin fold (the so-called cyclin box), which contains all the residues that contact CDKs in the binary holoenzymes (Fig. 4A) (19, 23). Therefore, our data suggest that the regions in D1 which interact with CDK4 and DMP1 are distinct.
FIG. 4.
Localization of the DMP1-interacting domain in cyclin D1. (A) Deletions within cyclin D1. The so-called cyclin box containing the residues which are predicted to directly contact CDK4 is indicated by the shaded box, and deleted amino acid segments are numerically indicated. Cells infected with baculovirus expression vectors encoding wild-type DMP1 and the indicated cyclin D1 mutants were metabolically labeled with [35S]methionine, lysed, divided into equal aliquots, and immunoprecipitated with antiserum to cyclin D1 (B) or monoclonal antibodies to the DMP1 Flag tag (C). Control immunoprecipitations (left lanes in both panels) were performed with NRS or an irrelevant isotype-matched control monoclonal antibody (designated C), as indicated below the panels. The positions of wild-type D1 and deletion mutants are indicated by the arrows in each lane. Only D1(Δ142–253) showed impaired binding.
In summary, the DNA binding domain of DMP1 is contained within residues 87 to 458, and its function is completely disrupted by the K319E point mutation. The C-terminal acidic domain (residues 458 to 761) and, to a much lesser extent, the N-terminal acidic domain (residues 1 to 86) contribute to transactivation. The N terminus (residues 87 to 233) is also crucial for the association of DMP1 with D-type cyclins, overlaps or abuts the DNA binding domain, and, with the possible exception of residues 224 to 237, does not include the myb repeats. In turn, D-type cyclins likely use different contact residues to interact with DMP1 and CDK4.
D-type cyclins inhibit DMP1-mediated transactivation independently of CDK4.
Coexpression of D-type cyclins with DMP1 in transformed 293T cells inhibits its ability to activate transcription (17). These cells lack a functional Rb protein, underscoring the concept that the observed inhibitory effect of D-type cyclins does not depend upon CDK-mediated Rb phosphorylation. Figure 5 shows that each of the three D-type cyclins also interfered with DMP1-dependent transactivation in Rb-positive NIH 3T3 cells, whereas cyclins A, B, C, and H were unable to do so. As expected from observations described above that D-type cyclins interact with the DMP1 N-terminal domain, the residual transactivating potential of DMP1 mutants M1, M2, and M3 lacking C-terminal residues remained sensitive to cyclin D2-induced inhibition. Although mutant M5 was less potent than full-length DMP1 as a transactivator (Fig. 1B) and appeared to bind cyclin D2 somewhat less efficiently than mutants retaining residues 1 to 87 (Fig. 3), its transactivating activity was still inhibited by cyclin D2 (Fig. 5). Similar results were obtained when cells were transfected and maintained in serum, although the magnitude of DMP1-induced reporter gene expression in the absence of ectopically expressed D-type cyclins was again only four- to fivefold above basal levels (see above).
FIG. 5.
Effects of cyclins, CDKs, and p16INK4a on DMP1-mediated transactivation. NIH 3T3 cells were cotransfected with the DMP1-responsive reporter plasmid and with pFLEX-DMP1, either with or without a fourfold excess of expression plasmids encoding the indicated cyclins (D1, D2, D3, A, B, C, or H), CDK4 (K4), CDK2 (K2), or p16INK4a. The D1 (K114E) mutant does not assemble with CDK4, whereas the CDK4 (K35M) mutant is catalytically inactive. Similar studies were performed with DMP1 mutants M1 to M3 and M5, each of which binds to DNA and is only partially handicapped in transactivation (Fig. 2B). Luciferase assays were performed 60 h after transfection, and cells were starved for serum for 18 h before assay. Error bars indicate standard deviations from the mean taken from multiple experiments.
Surprisingly, CDK4 and CDK2 subunits, whether wild type or catalytically inactive (e.g., CDK4 [K35M]) had no effect on DMP1-mediated transcription, either alone (Fig. 5) or when cotransfected together with D cyclins by using equivalent levels of input cyclin D and CDK4 expression plasmids (data not shown). Consistent with these findings, cotransfection of the specific CDK4 inhibitors, p16INK4a (Fig. 5) or p19INK4d (not shown), did not alter the extent of DMP1-mediated transactivation. Conversely, a D1 cyclin box mutant (K114E), which fails to bind to CDK4, was even more potent than wild-type D1 in reversing the ability of DMP1 to transactivate reporter gene expression. We previously observed that D-type cyclins entered into mutually exclusive complexes with either CDK4 or DMP1, whereas ternary complexes containing the three proteins could not be detected (17). It is therefore conceivable that competition for cyclin D binding by endogenous CDK4 might limit the inhibitory effect of the cyclin on DMP1-mediated transactivation. If so, cyclin D1 mutants that are unable to bind CDK4 might be somewhat more potent DMP1 inhibitors than the wild-type cyclin, consistent with data in Fig. 5. However, the explanation for the increased potency of the K114E mutant may lie elsewhere, since cotransfection of CDK4 with D cyclins, at roughly equal ratios, did not counter the inhibitory effects of the cyclins on DMP1-mediated reporter gene expression. Given the total quantities of plasmid DNA that could be transfected, there were technical limitations in raising the ratio of CDK4 to D1 plasmids in such experiments, which were not pursued further. Together, the critical conclusions from such experiments are that inhibition of DMP1-mediated transcription by D-type cyclins does not depend upon the ability of cyclin D to regulate CDK4 activity or, in turn, to modify DMP1 via CDK4-induced phosphorylation.
Cyclin D1-DMP1 complexes do not bind DNA.
The above-mentioned data were consistent with the hypothesis that cyclin D1 might act as a corepressor, interacting with DNA-bound DMP1 and thereby canceling its function as a transcriptional activator. However, we have only negative evidence for interactions between D-type cyclins and DMP1 on DNA (17). First, extracts from Sf9 cells coexpressing DMP1 and D-type cyclins generated EMSA complexes whose mobilities on nondenaturing gels could not be distinguished from those formed with Sf9 lysates containing DMP1 alone. Similar data were obtained using DNA-binding, cyclin-interacting DMP1 deletion mutants M1 to M3 when they were expressed alone or together with D-type cyclins. Secondly, both polyvalent and monoclonal antibodies to different D-type cyclins were unable to supershift DMP1-oligonucleotide complexes formed in the presence of the cyclins under conditions in which the mobility of these complexes was readily altered by antibodies to DMP1. Thirdly, EMSA complexes formed with endogenous DMP1 present in mammalian cells had the same mobility on nondenaturing gels as that formed with the recombinant protein in Sf9 cells, and again, these complexes could not be supershifted with antibodies to D-type cyclins. Finally, EMSA complexes with sizes similar to those from proliferating cells (D cyclins abundant) (17) (see below) were detected with lysates from quiescent mammalian cells (D cyclin low or absent).
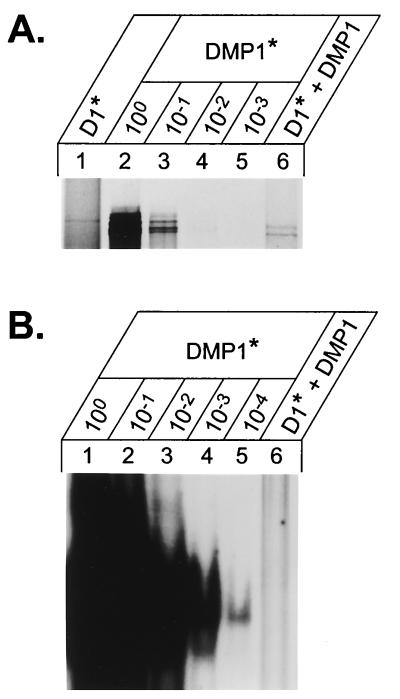
To approach this issue in another way, Sf9 cells were infected with baculoviruses encoding Flag-tagged DMP1 or with vectors encoding Flag-tagged cyclin D1 with or without untagged DMP1. Cells were labeled 24 h postinfection with [35S]methionine, and metabolically labeled lysates were precipitated with agarose-bound antibodies to the Flag epitope. Flag-tagged proteins were then eluted from washed beads by using excess Flag peptide and resolved on denaturing gels (Fig. 6A). No labeled bands with the mobility of DMP1 were recovered from cells expressing Flag-D1 alone (lane 1), whereas DMP1 was readily isolated from cells infected with Flag-DMP1 alone (lane 2). Densitometric analysis revealed that the quantity of DMP1 recovered in complexes with Flag-tagged cyclin D1 (Fig. 6A, lane 6) was equivalent to ∼2% of the recovered Flag-tagged DMP1 (also see below). The recovered Flag-tagged DMP1 protein was diluted serially 10-fold (Fig. 6A, lanes 3 to 5), mixed with a radiolabeled DMP1 binding site probe, and subjected to EMSA. Free Flag-tagged DMP1 generated readily detectable complexes even after 10,000-fold dilution (Fig. 6B, lanes 2 to 5). In contrast, DMP1-D1 complexes containing a quantity of DMP1 equivalent to a 50- to 100-fold dilution of the unbound protein (Fig. 6A, compare lanes 4 and 6) generated no detectable complexes (Fig. 6B, lane 6). Therefore, cyclin D1 appears to sequester the transcription factor in a form that can no longer bind to DNA. Given that the cyclin D binding domain (residues 87 to 237) overlaps the DNA binding domain (residues 87 to 458), the simplest idea is that D1 binding occludes the DNA binding site.
FIG. 6.
Cyclin D1-DMP1 complexes do not bind DNA. (A) Purification of Flag-tagged DMP1 and untagged DMP1 complexed to Flag-tagged cyclin D1. Sf9 cells were infected with baculoviruses encoding Flag-D1 (lane 1), Flag-DMP1 (lane 2), or Flag-D1 and untagged DMP1 (lane 6). Flag-tagged proteins in cell lysates were bound to M2 beads and eluted with a competing peptide. Purified Flag-DMP1 was subjected to serial 10-fold dilution (lanes 3 to 5), and equal aliquots of the purified proteins were electrophoretically separated on denaturing gels. Densitometric analysis indicated that the quantity of untagged DMP1 copurified with Flag-D1 was ∼2% of that of Flag-DMP1. As expected, no protein migrating at the position of DMP1 was detected in cells infected with Flag-D1 alone (lane 1). (B) Equal aliquots of the purified protein samples were subjected to EMSA using a 32P-labeled DMP1-specific oligonucleotide probe. Whereas DNA-probe complexes were detected in 10,000-fold diluted DMP1 samples, no binding was detected when cyclin D1-bound DMP1 was used.
DMP1 induces cell cycle arrest.
The levels of DMP1 protein expressed in mammalian fibroblasts are low, and the protein was only revealed after sequential immunoprecipitation and blotting using 125I-protein A for detection with prolonged autoradiographic exposures (9 days) (17). Correspondingly low levels of DMP1 mRNA and protein were detected in quiescent and proliferating macrophages and fibroblasts without significant oscillations throughout the cell cycle. The inability to readily detect the endogenous DMP1 protein has so far precluded all attempts to demonstrate a direct association between DMP1 and D-type cyclins in a physiologic in vivo setting. Moreover, because complexes between DMP1 and D-type cyclins do not bind to DNA (see above), we could not use sensitive EMSAs to score for the presence of putative complexes. Nonetheless, the fact that DMP1-mediated transactivation was significantly higher in growth-arrested versus proliferating cells and that its transcriptional activity was antagonized by D-type cyclins (see above) both suggested that DMP1 functions preferentially in quiescent cells and that its activity may be canceled as cells enter the cycle. We therefore compared the levels of endogenous DMP1 DNA binding activity in EMSAs performed with extracts prepared from untransfected quiescent and proliferating NIH 3T3 cells.
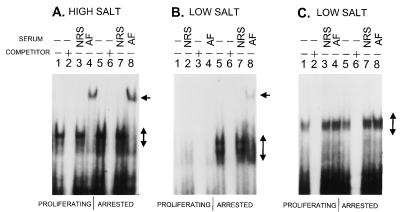
Using standard high-salt buffers to extract nuclear proteins and with equivalent inputs of total protein for EMSAs, we saw little difference in the total endogenous DMP1 binding activity recovered from quiescent or proliferating cells (Fig. 7A). With the DMP1-specific (BS2) probe, we detected equivalent levels of complexes (Fig. 7A, lanes 1 and 5), which were disrupted by an excess of competing BS2 oligonucleotide (lanes 2 and 6) and were supershifted with antibodies to DMP1 (lanes 4 and 8) but not by nonimmune serum (lanes 3 and 7). However, using lower salt conditions for extraction, significantly more binding activity was detected in extracts from quiescent cells (Fig. 7B, lanes 5 to 8) than from proliferating cells (lanes 1 to 4). Again, the complexes recovered from quiescent cells were specifically supershifted with antibodies to DMP1 (lanes 8). As a further control for protein recovery and specificity using the low-salt extraction protocol, we incubated the same extracts with an ETS-specific probe (M3) that contains a similar binding sequence (CCCGGAAGT versus CCCGTATGT). In this case, roughly equivalent amounts of probe-bound complexes were detected in low-salt extracts from quiescent and proliferating cells (Fig. 7C), and as expected, these were competed by the unlabeled cognate probe (lanes 2 and 6) but were not supershifted with antibodies to DMP1 (lanes 4 and 8). Dilution of the extracts confirmed that the probes were present in excess (data not shown). Using low-salt conditions, then, DMP1 extracted from proliferating cells was inhibited in its ability to bind to DNA (Fig. 7B), even though approximately equivalent amounts of latent DNA binding activity were present (Fig. 7A). It appears that DMP1 binding is masked by a salt-dependent association with another factor that either retains DMP1 in the nucleus during extraction or prevents its association with DNA. Whatever the mechanism, DMP1 DNA binding and transactivating activity are more robust in growth-arrested cells.
FIG. 7.
Differential, salt-dependent recovery of endogenous DMP1 binding activity from quiescent and proliferating cells. (A) Nuclear extracts from proliferating (lanes 1 to 4) or quiescent (lanes 5 to 8) NIH 3T3 cells were prepared with standard high-salt extraction buffer and incubated with a radiolabeled BS2 probe containing the nonameric DMP1 consensus binding sequence. Extracts were preincubated with cognate oligonucleotide competitor, NRS, and antiserum to DMP1 (AF) as indicated at the top of the panel. The positions of the DMP1-BS2 complex and a supershifted form are indicated by brackets and arrows, respectively, to the right of the panel. (B) The experiment shown in panel A was replicated with nuclear extracts generated with low-salt buffer. The quantities of input protein and probe were identical to those used for panel A. (C) The same quantities of nuclear extracts used for the experiment shown in panel B were incubated with a radiolabled M3 probe containing a nonameric ETS DNA binding sequence that does not react with DMP1. Positions of complexes containing ETS proteins are indicated by brackets to the right of the panel. All radiographic exposure times are 8 h.
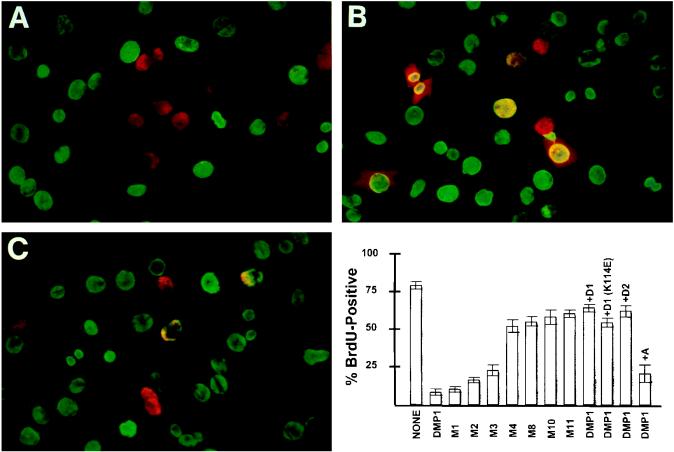
To determine whether DMP1 might negatively regulate cell proliferation per se, we ectopically expressed the protein in quiescent cells and examined its effects on their ability to subsequently enter S phase. Cells transfected with the DMP1 expression vector or with various Flag-tagged DMP1 mutants were serum starved to induce growth arrest and then restimulated to synchronously enter the cycle. BrdU was added to the medium together with serum, and cells were fixed 22 h later and stained for BrdU incorporation (green nuclear fluorescence) and for DMP1 expression with antibodies to the Flag epitope (red nuclear fluorescence). Under these conditions, a significant percentage of cells expressing DMP1 remained in G0/G1 and failed to incorporate BrdU (see the representative two-color fluorescence data in Fig. 8A [quantitative data in bar graph at lower right]). By contrast, cells expressing the M11 (K319E) mutant entered S phase (red plus green fluorescence yielded yellow stained nuclei in Fig. 8B), indicating that DNA binding was required for DMP1-induced G1 phase arrest. Other mutants defective in DNA binding (e.g., M4 and M8) were also unable to prevent S phase entry (bar graph), even though they retain the ability to interact with D-type cyclins (Fig. 3). The N- and C-terminal transactivating domains of DMP1 were required for cell cycle arrest, and the relative potency of the relevant mutants in inhibiting S phase entry correlated with their transactivating activities. (See results with mutants M1 to M3 in the bar graph.) In agreement with the ability of D-type cyclins to antagonize DMP1-induced transcription, cotransfection of cyclin D2 with DMP1 overrode its growth-inhibitory effects (Fig. 8C and bar graph). Cyclin D1 was as potent as cyclin D2, whereas cyclin A was not able to cancel DMP1-mediated growth arrest. The D1 (K114E) mutant, which cannot bind to CDK4 but still inhibited DMP1-mediated transactivation (Fig. 5), also retained its ability to counteract DMP1-induced cell cycle arrest (bar graph).
FIG. 8.
DMP1 arrests cell cycle progression in G1. NIH 3T3 cells transfected with expression vectors encoding either wild-type DMP1 (A), the DMP1 (K319E) mutant M11 (B), or wild-type DMP1 plus cyclin D2 (C) were made quiescent by serum starvation for 24 h. Cells restimulated with FBS to enter the cell cycle were labeled with BrdU for 22 h and then fixed and stained for the DMP1 Flag epitope (red) and BrdU (green). Whereas DMP1-transfected cells did not replicate their cellular DNA (A), cells infected with a DMP1 mutant that cannot bind to DNA (B) or those cotransfected with cyclin D2 (C) entered S phase; colocalization of DMP1 in BrdU-labeled cells is indicated by yellow fluorescence. The relative potency of DMP1 mutants in inhibiting S phase entry is quantitated in the bar graph at the lower right; 200 DMP1-transfected cells were counted per slide. Error bars indicate standard deviations from the mean, averaged from multiple independent experiments. Where indicated, cells were cotransfected with plasmids encoding wild-type D-type cyclins (+D1, +D2), a D1 (K114E) point mutant that does not bind to CDK4, or cyclin A (+A).
To determine whether DMP1 could arrest cells that were already in cycle, we introduced the expression vector into proliferating cells and, 14 h after transfection, recultured the cells in complete medium for 32 h. BrdU was then added to the culture medium for an additional 22 h. Under these conditions, 80% of cells transfected with a control vector remained in cycle and incorporated BrdU, whereas wild-type DMP1 prevented 70% of cells from entering S phase. Again, the M11 mutant was without effect, while cyclin D2 was able to completely override DMP1-induced growth arrest (data not shown). Thus, under both experimental conditions, DMP1 was able to induce growth arrest, and this activity was antagonized by D-type cyclins.
DISCUSSION
The activity of a novel transcription factor, DMP1, is antagonized by its interaction with D-type cyclins in a CDK4-independent manner. Deletion mutagenesis showed that the DNA binding domain of DMP1 was confined to its central region, which includes the three tandem myb-like repeats. Substitution of glutamic acid for lysine 319, a residue in the second myb repeat which was predicted to contact DNA (34), was sufficient in itself to abrogate both DNA binding and transactivation of a DMP1-responsive reporter gene. Although this central domain is sufficient to bind canonical DMP1 recognition sites in DNA, other acidic sequences at both the N and C-termini of the protein are required for transcriptional activation. Residues at the protein C terminus seem to be more important in this regard, but they must be widely distributed over this domain, because processive C-terminal deletions (mutants M1 to M3) led to a stepwise loss in transcriptional activation potential. In addition, the distal C terminus minimally includes that subset of phosphorylation sites that contribute to the heterogeneous migration of the protein on denaturing polyacrylamide gels. None of these conform to ideal sites for CDK phosphorylation ([S/T]PX[K/R]), arguing that other classes of protein kinases likely contribute to these modifications. In contrast, the N-terminal domain of DMP1 is crucial for its interaction with D-type cyclins, with residues 87 to 237 being sufficient to confer binding. Because removal of residues 1 to 86 reduced the ability of D-type cyclins to associate with DMP1, amino acids within this segment may contribute to optimized interactions. Together, these data suggest that regions required for transactivation and cyclin D binding are distinct, whereas the cyclin D binding domain abuts or overlaps the region of DMP1 that binds to DNA. In agreement, the transactivation potentials of all DMP1 mutants that retained the DNA binding domain were sensitive to cyclin D-mediated inhibition. Although D-type cyclins and DMP1 can directly bind to one another in vitro (17), we cannot exclude the possibility that in yeast, insect, or mammalian cells, other proteins contribute to the formation or stability of these complexes. However, CDKs appear not to be involved (see below).
Cotransfection of D-type cyclins with DMP1 abrogated its ability to activate transcription of a luciferase reporter gene driven by a minimal simian virus 40 promoter containing tandem 5′ DMP1 binding sites. Cyclins D1, D2, and D3 exerted similar effects in this assay, whereas cyclins A, B, C, and H lacked such activity. The D-type cyclins also antagonized the residual activity of those DMP1 deletion mutants (M1 to M3 and M5) that were partially handicapped in transactivation but which retained functional DNA binding and cyclin D-interactive domains. Cotransfection of CDK4 or CDK2 with DMP1 was without effect on reporter gene activity, and D-type cyclins inhibited DMP1-dependent transactivation whether or not exogenous CDK-encoding vectors were included. In transfected cells that had subsequently been rendered quiescent by serum starvation, the introduction of vectors encoding CDK4 inhibitors, including p16INK4a or p19INK4d did not affect the outcome. Importantly, a cyclin D1 mutant (K114E) that fails to bind or activate CDK4 was highly potent in antagonizing DMP1 activity. Therefore, the negative effects of D-type cyclins on DMP1-mediated transactivation do not directly depend upon their catalytic partners, and phosphorylation by CDK4 does not account for the ability of cyclin D to abrogate DMP1-mediated transcriptional activation.
Previous experiments indicated that D-type cyclins entered into mutually exclusive complexes with DMP1 and CDK4 whether the latter was catalytically active or not. Although sequences in the first cyclin fold (the cyclin box) are predicted to contain the critical cyclin residues for contacting their respective CDKs (19, 23), our studies suggest that sequences C terminal to this region are the crucial ones for DMP1 binding. Observations that a cyclin D1 mutant (K114E) that does not bind to CDK4 was still able to antagonize DMP1-dependent transcriptional activation also support the conclusion that CDK4 and DMP1 binding sites are distinct. However, from the available data, we cannot discern why ternary complexes between cyclin D, DMP1, and CDK4 do not form. A possibility raised by these results is that CDK4 and DMP1 might compete for cyclin D binding in living cells. In those transactivation experiments in which four expression plasmids (DMP1, DMP1-responsive reporter, cyclin D, and CDK4) were cointroduced into NIH 3T3 cells, cyclin D and CDK4 plasmids were transfected with equal quantities of input DNAs, neither of which was in great excess over that of the DMP1 expression plasmid. We would be unlikely to see significant competition under such circumstances, but in principle, higher ratios of CDK4 to DMP1 might quench the inhibitory effects of cyclin D. Given the inherently nonphysiologic nature of such experiments, and for technical reasons outlined in Results, we chose not to pursue this issue further.
Binding of D-type cyclins to DMP1 prevents its interaction with DNA. This is likely due to the fact that the cyclin D binding site (within residues 87 to 237) abuts or overlaps the DNA binding domain (within residues 87 to 458). Even under optimal conditions, lysates of insect cells coexpressing DMP1 and D-type cyclins contain a proportion of free DMP1 molecules, and it is only these that interact with labeled oligonucleotide probes to form the gel-shifted complexes visualized in EMSAs. D-type cyclins were not detected in these complexes, and the mobility of the complexes was not altered by exposing them to many different polyvalent or monoclonal antibodies to the D cyclins; in clear contrast, a number of different antibodies to DMP1 readily supershifted these species (17). To further analyze this issue, we purified DMP1 in complexes with Flag-tagged cyclin D1 and tested them for their ability to interact with a labeled oligonucleotide containing the DMP1 binding sequence. Comparatively low quantities of free DMP1 interacted with the probe and produced readily detectable EMSA complexes, but much greater quantities of cyclin D-bound DMP1 appeared inert for DNA binding. Together, these data argue against the idea that D-type cyclins act as DMP1 corepressors on DNA and instead suggest that D-type cyclins squelch DMP1 activity by titrating it into complexes that can no longer interact with DNA. This implies that inhibition of DMP1 activity should not be mediated by D-type cyclins in vivo under circumstances in which DMP1 is present in excess. In most cell lines so far surveyed, DMP1 has proven to be nonabundant and invariant throughout the cell cycle. In cycling cells, the effects of DMP1 might well be overridden by D-type cyclins, which accumulate to much higher levels (30). Conversely, in quiescent G0 cells in which D-type cyclin expression is low or entirely absent, free DMP1 would be available to regulate gene expression.
In this regard, it is intriguing that DMP1 was significantly more active in fibroblasts that had been made quiescent by either serum starvation or by transfection with CDK inhibitors, such as p27Kip1. This was a consistent finding observed in many transfected cell lines other than NIH 3T3. When endogenous DMP1 was extracted with high-salt buffer, the total DNA binding activities recovered from proliferating and quiescent cells were similar. Low-salt extraction of endogenous DMP1 yielded significantly more specific DNA binding activity from quiescent than from proliferating fibroblasts, while similar amounts of ETS DNA binding activity were recovered from both extracts. Therefore, in agreement with previous measurements, roughly equivalent amounts of DMP1 are found in quiescent and proliferating fibroblasts (17), but under conditions of low-salt extraction, a substantial fraction of DMP1 either is not released from the nuclei of proliferating cells or is recovered in an inactive form. Noncovalent interactions between DMP1 and other molecules that mask its recovery or activity might well be salt dependent, while posttranslational modifications of DMP1 would be less likely to account for the observed behavior. Although it is tempting to speculate that endogenous cyclin D might itself mask the ability of DMP1 to bind DNA and regulate gene expression, we have no evidence that this can occur in a physiologic setting. The major limitation is that the low levels of DMP1 protein that are normally expressed in fibroblasts have so far precluded attempts to demonstrate direct interactions between it and other proteins.
Given the increased activity of DMP1 in quiescent cells, we were motivated to test whether DMP1 might itself potentiate exit from the cell cycle. Surprisingly, proliferating cells transfected with DMP1 underwent growth arrest when maintained in serum-containing medium, whereas DMP1 mutants that could not bind DNA or activate transcription were unable to halt cell proliferation. Moreover, when DMP1 was transfected into cells that were subsequently made quiescent by depriving the cultures of serum, cells expressing DMP1 did not reenter S phase after serum restimulation, but untransfected cells from the same cultures replicated their DNA. When DMP1 mutants were used, there was a strict concordance between their ability to promote gene expression and induce cell cycle arrest under either of these experimental conditions. For example, a DMP1 point mutant (K319E) that does not bind DNA was completely unable to stop cell proliferation. Mutants M1 to M3 that were partially defective in transactivation were also less able to induce cell cycle arrest. Conversely, cotransfection of D-type cyclins overrode DMP1-induced arrest. A cyclin D1 (K114E) mutant that does not bind CDK4 but still blocked DMP1-mediated gene expression retained the ability to override DMP1-induced cell cycle arrest. This again provides a strong argument that the ability of D cyclins to antagonize DMP1 does not rely on CDK4-mediated phosphorylation. Perhaps, DMP1 normally serves to maintain cells in a quiescent state, and, in a manner conceptually analogous to that of MyoD1 (41, 51), its activity is overridden by D-type cyclins as cells are brought into cycle.
What gene products are responsible for this activity of DMP1? The factor binds to canonical ETS sites that are commonly embedded in many promoters. In addition, DMP1 can bind to a similar sequence (i.e., CCCGTATGT) which lacks the GGA core that is normally required by ETS proteins for DNA binding (55). Therefore, at least in principle, many genes are likely to prove to be DMP1 responsive. Nonetheless, the simplest hypothesis is that DMP1 induces the expression of a gene product, such as a CDK inhibitor, that enforces cell cycle arrest.
ACKNOWLEDGMENTS
We thank Hiroshi Hirai and Martine Roussel for helpful advice and discussion, Hiroshi Hirai for communicating unpublished data, Frederique Zindy for supplying the cyclin D1 deletion mutants, J. Alan Diehl for preparing the D1 (K114E) mutant, Richard Bram for supplying pFLEX1 and pBJ5 plasmids, and Jill Lahti for supplying cyclins B and C. We also thank Carol Bockhold, Joe Watson, and Shawn Hawkins for excellent technical assistance and other members of our laboratory for their criticisms and support.
This work was supported in part by Cancer Center CORE grant CA21765, by Leukemia Program Project grant CA-20180, and by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
REFERENCES
- 1.Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 2.Bram R J, Crabtree G R. Calcium signaling in T cells stimulated by a cyclophilin B-binding protein. Nature. 1994;371:355–358. doi: 10.1038/371355a0. [DOI] [PubMed] [Google Scholar]
- 3.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 4.Connell-Crowley L, Harper J W, Goodrich D W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis J N, Roussel M F. Cloning and expression of murine Elf-1 cDNA. Gene. 1996;171:265–269. doi: 10.1016/0378-1119(96)00013-3. [DOI] [PubMed] [Google Scholar]
- 6.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 7.Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 8.Farnham P J. Transcriptional control of cell growth: the E2F family. New York, N.Y: Springer Verlag; 1996. [Google Scholar]
- 9.Flemington E K, Speck S H, Kaelin W G., Jr E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan K-L, Jenkins C W, Li Y, Nichols M A, Wu X, O’Keefe C L, Matera A G, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 11.Hamel P A, Gill R M, Phillips R A, Gallie B L. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannon G J, Beach D. p15INK4b is a potential effector of cell cycle arrest mediated by TGF-β. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 13.Hatakeyama M, Brill J A, Fink G R, Weinberg R A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 14.Herrera R E, Sah V P, Williams B O, Mäkelä T P, Weinberg R A, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 16.Hirai H, Roussel M F, Kato J-Y, Ashmun R A, Sherr C J. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirai H, Sherr C J. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16:6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 19.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 20.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase, CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 21.Kato J, Sherr C J. Inhibition of granulocyte differentiation by G1 cyclins D2 and D3 but not D1. Proc Natl Acad Sci USA. 1993;90:11513–11517. doi: 10.1073/pnas.90.24.11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato J, Matsuoka M, Polyak K, Massagué J, Sherr C J. Cyclic AMP-induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase-4 activation. Cell. 1994;79:487–496. doi: 10.1016/0092-8674(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 23.Kim K K, Chamberlin H M, Morgan D O, Kim S H. Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase. Nat Struct Biol. 1996;3:849–855. doi: 10.1038/nsb1096-849. [DOI] [PubMed] [Google Scholar]
- 24.Koh J, Enders G H, Dynlacht B D, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 25.Lam E W-F, Watson R J. An E2F binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells, independent of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma protein-dependent cell cycle inhibition by the tumor suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 28.Matsushime H, Ewen M E, Strom D K, Kato J, Hanks S K, Roussel M F, Sherr C J. Identification and properties of an atypical catalytic subunit (p34PSKJ3/CDK4) for mammalian D-type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 29.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 31.Medema R H, Herrera R E, Lam F, Weinberg R A. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyerson M, Harlow E. Identification of a G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuman E, Ladha M H, Lin N, Upton T M, Miller S J, Direnzo J, Pestell R G, Hinds P W, Dowdy S F, Brown M, Ewen M E. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 34a.Owen R D, Bortner D M, Ostrowski M C. ras oncogene activation of a VL30 transcriptional element is linked to transformation. Mol Cell Biol. 1990;10:1–9. doi: 10.1128/mcb.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagano M, Theodoras A M, Tam S W, Draetta G F. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994;8:1627–1639. doi: 10.1101/gad.8.14.1627. [DOI] [PubMed] [Google Scholar]
- 36.Pardee A B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 37.Polyak K, Kato J, Solomon M J, Sherr C J, Massague J, Roberts J M, Koff A. p27Kip1, a cyclin-cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 38.Polyak K, Lee M, Erdjument-Bromage H, Koff A, Roberts J M, Tempst P, Massagué J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 39.Quelle D E, Ashmun R A, Shurtleff S A, Kato J, Bar-Sagi D, Roussel M F, Sherr C J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 40.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Cancer-associated mutations at the INK4a locus cancel cell cycle arest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc Natl Acad Sci USA. 1997;94:3436–3440. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao S S, Chu C, Kohtz D S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic-loop-helix regulators. Mol Cell Biol. 1994;14:5259–5267. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Resnitzky D, Reed S I. Differential effects of cyclins D1 and E on pRb phosphorylation. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynisdóttir I, Massagué J. The subcellular locations of p15INK4b and p27Kip1 coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 45.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Kip/Cip and Ink4 cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 46.Serrano M, Gomez-Lahoz E, DePinho R A, Beach D, Bar-Sagi D. Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 47.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 48.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 49.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 50.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 51.Skapek S X, Rhee J, Spicer D B, Lassar A B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 52.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin/cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 53.Vallance S J, Lee H, Roussel M F, Shurtleff S A, Kato J, Strom D K, Sherr C J. Monoclonal antibodies to mammalian D-type G1 cyclins. Hybridoma. 1993;13:37–44. doi: 10.1089/hyb.1994.13.37. [DOI] [PubMed] [Google Scholar]
- 54.Wang T C, Cardiff R D, Zukerberg L, Lees E, Arnold A, Schmidt E V. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 55.Wasylyk B, Hahn S L, Giovane A. The ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 57.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 58.Zwijsen R M L, Wientijens E, Klompmaker R, van der Sman J, Bernards R, Michalides R J A M. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]