Abstract
INTRODUCTION:
The identification of risk factors for precursor lesions of colorectal cancer (CRC) holds great promise in the context of prevention. With this study, we aimed to identify patient characteristics associated with colorectal polyps (CPs) and polyp features of potential malignant progression. Furthermore, a potential association with gut microbiota in this context was investigated.
METHODS:
In this single-center study, a total of 162 patients with CPs and 91 control patients were included. Multiple variables including information on lifestyle, diet, serum parameters, and gut microbiota, analyzed by 16S-rRNA gene amplicon sequencing and functional imputations (Picrust2), were related to different aspects of CPs.
RESULTS:
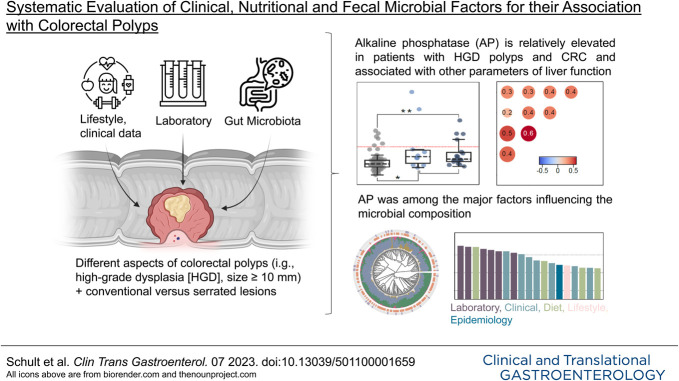
We observed that elevated serum alkaline phosphatase (AP) levels were significantly associated with the presence of high-grade dysplastic polyps. This association was further seen for patients with CRC. Thereby, AP correlated with other parameters of liver function. We did not observe significant changes in the gut microbiota between patients with CP and their respective controls. However, a trend toward a lower alpha-diversity was seen in patients with CRC. Interestingly, AP was identified as a possible clinical effect modifier of stool sample beta diversity.
DISCUSSION:
We show for the first time an increased AP in premalignant CP. Furthermore, AP showed a significant influence on the microbial composition of the intestine. Relatively elevated liver enzymes, especially AP, may contribute to the detection of precancerous dysplastic or neoplastic changes in colorectal lesions. The association between elevated AP, premalignant CP, and the microbiome merits further study.
KEYWORDS: colorectal polyp, colorectal cancer, microbiome, lifestyle, alkaline phosphatase
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide (1). CRC arises both on the ground of chronic inflammation and from nonmalignant colorectal polyps (CPs) (2). The 2 major subtypes of CPs are conventional adenomas (tubular/tubulovillous/villous adenomas) and serrated lesions and polyps (hyperplastic polyps, sessile serration lesions, and traditional serrated adenomas) (2). Removing polyps at an early stage can reduce the risk of CRC, making colonoscopy the gold-standard screening method. Several risk factors for malignant transformation have been defined for colorectal adenomas such as size ≥10 mm, the presence of at least 3 adenomas, villous histology, or high-grade dysplasia (HGD) (3). However, although early screening colonoscopy helped to lower the overall incidence of CRC, the incidence increases in patients younger than 50 years (4). Therefore, there is a persisting need to identify modifiable risks and augment existing screening methods.
In recent years, studies have attempted to identify the impact of lifestyle factors, such as diet, physical activity, and tobacco use on the risk of CRC development. Certain dietary habits, including a high consumption of red meat, are associated with an increased risk of CRC (5). Fewer studies have examined associations of dietary patterns with CP development. Among others, it was demonstrated that a high intake of simple carbohydrates was associated with increased risk for the occurrence of CP (6), whereas green vegetables were negatively associated with CP (7). Studies suggest that diet and lifestyle affect the development of CRC through an altered intestinal microbiome (5,8). Moreover, changes in the gut microbiota occur in patients with CP (9), and the microbiome seems to change along the adenoma-carcinoma sequence (10). However, the results from studies on gut microbial changes in CRC and CP are not consistent and, in some cases, contradictory (11–15).
Here, we examined multiple clinical factors for their association with different biological aspects of CP and further evaluated coincident changes to the gut bacterial composition in a large cohort of patients. The aim of this study was to provide data on diagnostic and potentially protective or risk factors associated with the occurrence of CP and their progression toward CRC to support preventive efforts.
METHODS
Patient recruitment and sample collection
Figure 1a and b provide an overview of the recruitment infrastructure and the cohorts. Participants underwent colonoscopy examination for screening purposes or polypectomy. The recruitment took place between July 2019 and January 2023 at the University Hospital Klinikum rechts der Isar, Technical University Munich, Germany. Patients aged between 18 and 80 years with suspected gastrointestinal disease were included. Patients who had contraindications to biopsy (e.g., thrombocytopenia <50,000/µL, partial thromboplastin time >50 seconds) and patients with poor general health (Eastern Cooperative Oncology Group >2) were excluded.
Figure 1.
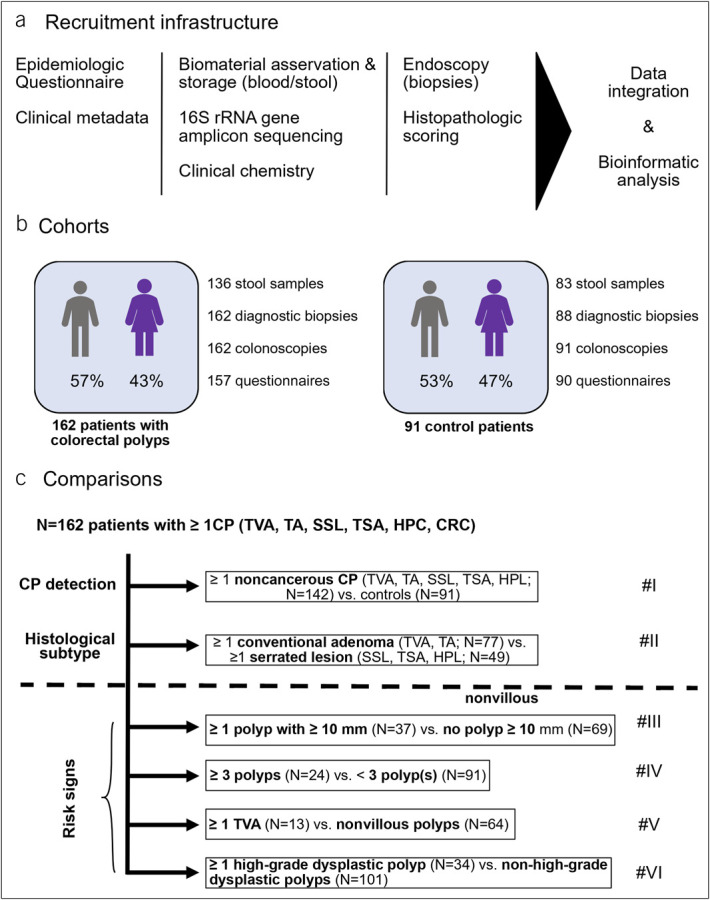
Recruitment infrastructure, cohort demographics, and analyses. (a) Overview of the study's infrastructure and recruitment process, (b) patient cohorts and biomaterial availability, and (c) overview of the respective comparisons in the study. In total, 162 patients had at least 1 CP. Six comparisons were performed (#I–#VI) covering detection of polyps, comparison of conventional adenomas vs serrated lesions and risk signs. CP, colorectal polyp; CRC, colorectal carcinoma; HPL, hyperplastic polyp; SSL, sessile serrated lesion; TA, tubular adenoma; TSA, traditional serrated adenoma; TVA, tubulovillous adenoma.
All patients provided stool samples. After sorting out the samples with insufficient quality, 219 stool samples were considered for analysis. No serial sampling or analysis of mucosal microbiota was performed. To prevent the bowel preparations for colonoscopy from affecting the microbiome, the stool sample was taken 4–6 weeks after bowel preparations (16). The sampling vessel contained a stabilizer, which was used to stabilize bacterial nucleic acids allowing storage at room temperature for at least 1 week (MaGix PBI medium; Microbiomix GmbH, Regensburg, Germany). Blood samples were collected on the day of colonoscopy and analyzed by the hospital's Department of Clinical Chemistry. Biopsies were stored in formalin and examined by at least 1 experienced gastrointestinal pathologist. For complete macroscopic inspection of the large intestine, ileum or caecum was inspected in every colonoscopy. A subset of the individuals from the control group have been published regarding their intestinal bacterial composition in another project of our research group (17).
Study population and CP subtypes
Patients in the CP cohort had at least 1 CP. Patients without known gastrointestinal disease and an inconspicuous colonoscopy, including unremarkable colorectal biopsies, served as healthy gut controls. Table 1 lists the patients clinical characteristics.
Table 1.
Characteristics of the study population
| Polyp cohort (N = 162) | Healthy gut controls (N = 91) | P value | |
| Sex, female:male | 70:92 | 43:48 | 0.535 |
| Age, yr, mean | 60.5 | 57.4 | 0.099 |
| BMI, mean | 26.6 | 25.4 | 0.080 |
| Stool samples, n | 136 | 83 | |
| Blood samples, n | 135 | 71 | |
| Comorbidities, n (%) | |||
| Diverticular disease | 60 (37) | 37 (40.7) | 0.592 |
| IBD | 16 (9.9) | 0 (0) | 0.0007 |
| Gastritis | 32 (19.8) | 26 (28.6) | 0.121 |
| Hepatitis | 8 (4.9) | 8 (8.8) | 0.283 |
| Liver cirrhosis | 7 (4.3) | 1 (1.1) | 0.265 |
| Liver metastases | 7 (4.3) | 1 (1.1) | 0.265 |
| Arterial hypertension | 75 (46.3) | 32 (35.2) | 0.111 |
| Chronic heart failure | 5 (3.09) | 1 (1.1) | 0.424 |
| Dysplipidemia | 67 (41.36) | 43 (47.3) | 0.428 |
| Atrial fibrillation | 13 (8) | 2 (2.2) | 0.093 |
| Type II diabetes | 32 (19.8) | 14 (15.4) | 0.497 |
| COPD | 8 (4.9) | 1 (1.1) | 0.163 |
| Hypothyroidism | 43 (26.5) | 17 (18.7) | 0.169 |
| Hyperthyroidism | 2 (1.2) | 2 (2.2) | 0.620 |
| Active cancer | 31 (19.1) | 3 (3.3) | 0.0002 |
| History of cancer | 43 (26.5) | 22 (24.2) | 0.765 |
| Chronic kidney disease | 9 (5.6) | 6 (6.6) | 0.785 |
| Medication, n (%) | |||
| Immunosuppressants | 23 (14.2) | 5 (5.5) | 0.037 |
| Laxatives | 7 (4.3) | 2 (2.2) | 0.496 |
| Pain killers | 32 (19.7) | 12 (13.2) | 0.227 |
| Heart medication | 67 (41.4) | 28 (30.7) | 0.106 |
| Antibiotics | 40 (24.7) | 20 (22) | 0.648 |
| Probiotics | 8 (4.9) | 8 (8.8) | 0.283 |
| Diet and lifestyle, n (%) | |||
| Whole grain bread | |||
| Frequent consumption | 98 (60.5) | 62 (68.1) | 0.277 |
| Occasional consumption | 34 (21) | 17 (18.7) | 0.745 |
| Rare consumption | 24 (14.8) | 9 (9.9) | 0.332 |
| NA | 6 (3.7) | 3 (3.3) | 1 |
| Vegetables, raw | |||
| Frequent consumption | 108 (66.7) | 60 (65.9) | 1 |
| Occasional consumption | 40 (24.7) | 20 (22) | 0.648 |
| Rare consumption | 8 (4.9) | 9 (9.9) | 0.189 |
| NA | 6 (3.7) | 2 (2.2) | 0.715 |
| Vegetables, cooked | |||
| Frequent consumption | 106 (65.4) | 61 (67) | 0.890 |
| Occasional consumption | 44 (27.2) | 23 (25.3) | 0.769 |
| Rare consumption | 5 (3.1) | 5 (5.5) | 0.503 |
| NA | 7 (4.3) | 2 (2.2) | 0.496 |
| Fruits | |||
| Frequent consumption | 114 (70.4) | 67 (73.6) | 0.664 |
| Occasional consumption | 32 (19.7) | 16 (17.6) | 0.740 |
| Rare consumption | 10 (6.2) | 6 (6.6) | 1 |
| NA | 6 (3.7) | 2 (2.2) | 0.715 |
| Red meat | |||
| Frequent consumption | 89 (54.9) | 45 (49.4) | 0.433 |
| Occasional consumption | 51 (31.5) | 29 (31.9) | 1 |
| Rare consumption | 16 (9.9) | 15 (16.5) | 0.161 |
| NA | 6 (3.7) | 2 (2.2) | 0.715 |
| Alcohol | |||
| Daily or several times a week | 53 (32.7) | 28 (30.7) | 0.780 |
| Once a week | 39 (24.1) | 21 (23.1) | 0.880 |
| Less than once a week or never | 63 (38.9) | 39 (42.9) | 0.594 |
| NA | 7 (4.3) | 3 (3.3) | 1 |
| Smoking | |||
| Yes | 94 (58) | 50 (54.9) | 0.692 |
| No | 61 (37.7) | 39 (42.9) | 0.425 |
| NA | 7 (4.3) | 2 (2.2) | 0.496 |
| Physical activity | |||
| Active | 105 (64.8) | 62 (68.1) | 0.679 |
| Not active | 51 (31.5) | 27 (29.7) | 0.779 |
| NA | 6 (3.7) | 2 (2.2) | 0.715 |
| Laboratory parameters (mean) | |||
| AP (40–129 U/L) | 84.8 | 68.8 | 0.003 |
| GGT (<39 U/L) | 51.6 | 32.1 | 0.091 |
| AST (10–35 U/L) | 31.9 | 34.5 | 0.257 |
| ALT (10–35 U/L) | 24.8 | 27.5 | 0.291 |
| LDH (<244 U/L) | 212.9 | 215.4 | 0.771 |
Frequent consumption = several times a week or daily, occasional consumption = several times a month or once a week, rare consumption = 1 time per month or less. Physically active = A minimum of 2 hours of physical exercise per week, or at least 30 minutes of cycling per day, or at least 1 hour of walking per day. For calculation of P value, Fisher exact test and paired t test were used.
ALT, alanine aminotransferase; AP, alkaline phosphatase; AST, aspartate aminotransferase; BMI, body mass index; COPD, chronic obstructive pulmonary disease; CRC, colorectal carcinoma; GGT, gamma-glutamyl transferase; HPL, hyperplastic polyp; IBD, inflammatory bowel disease; LDH, serum lactate dehydrogenase; NA, not available; SSL, sessile serrated lesion; TA, tubular adenoma; TSA, traditional serrated lesion; TVA, tubulovillous adenoma.
All lesions were histologically classified in accordance to the criteria given by the current World Health Organization classification of tumors of the digestive tract (18). The grade of dysplasia of conventional adenomas was subdivided into low-grade and high-grade subgroups based on the degree of architectural and cytomorphological abnormalities.
The full cohort comprised a total of 253 patients, 162 of whom were diagnosed with 1 or more CP and 91 healthy gut controls. A total of 350 CP were detected in 162 patients. In 77 (47.5%), 41 (25.3%), and 44 (27.2%) patients, respectively, 1, 2, and 3 or more polyps were detected on colonoscopy. Most of the patients (N = 122, 75.3%) carried exactly 1 histological polyp subtype even if more than 1 polyp was present. Patients with multiple histological subtypes (N = 40, 24.7%) were allowed to participate in the evaluation of each histological subtype of interest if all other inclusion criteria for the comparison were met. Further details are specified in Figure 1c and the results section for each of the outcome variables.
Patient data acquisition
Participants answered a questionnaire, developed by a team of experts in the field of the gut microbiome. The questionnaire included questions on (i) epidemiological and family background (e.g., origin, residence, marital status, family history, and employment), (ii) health (e.g., chronic diseases, stool irregularities, and medications), (iii) exercise (sports and work activity), (iv) smoking, (v) alcohol consumption, and (vi) diet (e.g., different types of meat and fish, various kinds of vegetables, fruits, dairy products, whole grain products, chewing gum, and coffee).
The questions addressed the patient's lifestyle of the past 6 months. However, some questions were also asked to clarify whether patients ever pursued a particular lifestyle (e.g., “have you ever smoked tobacco products?”).
For dietary questions, patients indicated frequency of consumption: 1 time per month or less (“rare”), several times a month or once a week (“occasional”), or several times a week or daily (“frequent”). Patients who met 1 or more of the following criteria were assigned to the physically active group: a minimum of 2 hours of physical exercise per week, or at least 30 minutes of cycling per day, or at least 1 hour of walking per day.
Medical information, such as comorbidities, continued to be collected during the educational interview and was extracted from our hospital's medical records. The respective answers to the patients' questionnaire can be found in detail in Supplementary Table 1 (see Supplementary Digital Content 1, http://links.lww.com/CTG/B47). A total of 60 patients reported having taken antibiotics and 16 patients reported having taken probiotics in the last month before stool collection. However, the exact time of intake could not be determined. Patients taking antibiotics and probiotics were not excluded to increase the statistical power of the analysis regarding multiple clinical variables. Furthermore, we aimed for a comprehensive analysis of possible risk or protective factors on the development of precancerous and cancerous polyps, without prior exclusion of single factors. However, we investigated the potential impact of antibiotics and probiotics on the gut microbiota (Figure 3d).
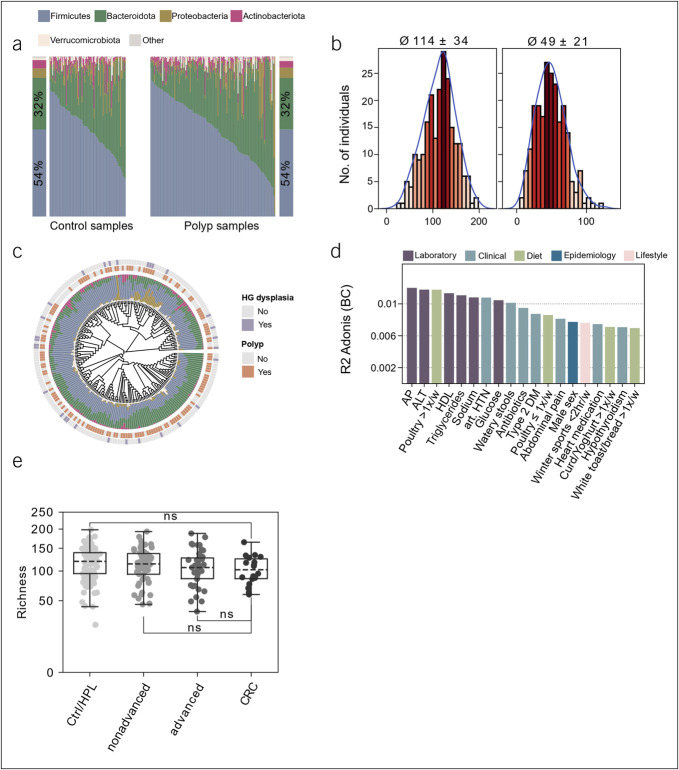
Figure 3.
Microbial properties of the study population. (a) Relative abundances of phyla across control individuals and patients with polyp. Samples are ordered according to increasing relative abundances of Firmicutes. (b) Alpha diversity of the fecal microbiota in the whole cohort. Richness (left; 114 ± 34) and Shannon effective number of species (right; 49 ± 21). (c) Phylogenetic distance tree calculated from generalized Unifrac distances for all microbial stool samples. Stacked barplots show taxonomic distribution on the phylum level. Inner label shows the presence of the indicated polyp features. (d) Explained variations in fecal microbiota composition by covariates. All variables shown had a significant influence (P ≤ 0.05), displayed as proportions of explained variations based on R2. (e) Association of the indicated patient groups (x-axis) with species richness (y-axis). Hypothesis testing was performed using a 2-tailed Mann-Whitney U test with post hoc adjustment of P values using the Benjamini-Hochberg method. In boxplots, the box ranges from Q1 (the first quartile) to Q3 (the third quartile) of the distribution and the range represents the IQR. The median is indicated by a dashed line across the box. The “whiskers” on box plots extend from Q1 and Q3 to 1.5 the IQR. **FDR ≤0.01; *FDR ≤0.05; ns, not significant. ALT, alanine aminotransferase; AP, alkaline phosphatase; CRC, colorectal carcinoma; DM, diabetes mellitus; FDR, false discovery rate; HDL, high-density lipoprotein; HTN, hypertension; HPL, hyperplastic polyp; IQR, interquartile range.
16S rRNA gene amplicon sequencing and analysis
Preparation and sequencing of microbial samples were performed as described previously (19). Briefly, sample preparation and paired-end sequencing were performed on an Illumina MiSeq targeting the V3V4 region of the 16S rRNA gene using primers 341F and 785R. Raw FASTQ files were processed using the NGSToolkit (https://github.com/TUM-Core-Facility-Microbiome/ngstoolkit) based on USEARCH to generate denoised zero-radius operational-taxonomic units (zOTUs). Assessment of alpha diversity and taxonomic binning were conducted using the Rhea software pipeline (20). Specifically, the following metrics were computed and compared between groups of interest: sample richness, normalized richness, Shannon, and Simpson index and effective. Similarity of microbial profiles, that is, beta diversity, was assessed using generalized UniFrac distances which were calculated using the GUniFrac R package v1.7 (21). Rarefaction curves of the sequencing data showed that all samples with ≥5,000 reads are sufficiently covering the diversity of the samples (median read count per sample: 20,000). Thus, we omitted samples below 5,000 reads. Read number was normalized to the lowest read number of a given sample in the sample set by rule of proportion/rule of 3 (i.e., 5,000 reads) following (20). For each of 40 samples, controls were included. These controls represented positive controls (ZymoBIOMICS Microbial Community Standard, Zymo Research, Germany) or mock controls (i.e., testing stabilizer tubes without a sample starting with DNA isolation) or negative controls (i.e., water controls for the polymerase chain reactions conducted). Positive controls showed very high repeatability between sequencing cartridges and batches. The variation of the sequencing cartridges from Illumina was found to be very minor in recent years. In addition, spurious taxa were filtered if below 5% relative abundance to account for nonmicrobial sequences, for example, due to crosstalk or contamination by environmental DNA. Thus, batch effects were considered negligible. To avoid adding noise in the analysis, the data were not rarefied prior analysis (22).
Statistical analysis
Statistical analysis of 16S rRNA profiles was conducted as described previously (19). Briefly, read counts were normalized, and differences in relative abundance of taxa and/or zOTUs were determined by a Kruskal-Wallis rank sum test for multiple groups and Mann-Whitney U tests for pairwise comparisons, respectively. P values were adjusted for testing multiple hypotheses using the Benjamini-Hochberg procedure, where an adjusted P value ≤0.05 was considered significant. Confounders and possible effect modifiers of microbial ecosystems were determined through a permutational multivariate analysis of variance as implemented in the adonis3 function from the GUniFrac R package v1.7 (21) using a matrix comprising generalized UniFrac distances. We used the Picrust2 (23) algorithm for prediction of functions from 16S sequences. Pathway abundances per sample were inferred using the Picrust2 (23) software (v2.5.2), and differential pathway abundance analysis was performed using the ALDEx2 R package (24).
For clinical, laboratory, dietary, and epidemiological covariates, univariate testing was performed using Mann-Whitney U and Fisher exact tests for continuous and categorical data, respectively. P values were adjusted for testing multiple hypotheses using the Benjamini-Hochberg procedure, where an adjusted P value ≤0.05 was considered significant. Significant findings underwent evaluation in a multivariate logistic regression model. The baseline model considered covariates with an established connection to colorectal carcinogenesis and known factors influencing the gut microbiota: the consumption of products rich in fiber (fruits, vegetables, and whole grain bread), red meat and alcohol, physical activity level, smoking habits, age, sex, and body mass index (8,25–28). Significant covariates identified in our study were then added to the model, and the direction and significance of their effect were compared with their univariate results. Table 1 presents the distribution of patients' diet and lifestyle factors studied in the multivariate model.
RESULTS
Frequency of individual polyp subtypes in our cohort and corresponding analyses
Owing to the occurrence of multiple polyps and histologies in some patients, we found at least 1 tubular adenoma (TA) in 90, at least 1 hyperplastic polyp (HPL) in 53, at least 1 sessile serrated lesion (SSL) or traditional serrated adenoma (TSA) in 29, at least 1 tubulovillous adenoma (TVA) in 16, and at least 1 colorectal carcinoma (CRC) in 20 patients. Drawing from this cohort of patients and a set of 91 control patients, we designed the following comparisons to evaluate possible clinical and microbial associations with CP detection and histology (Figure 1c): (I) patients with noncancerous CPs (N = 142) vs healthy gut controls (N = 91) and (II) patients with only conventional adenomas (TVA, TA, N = 77) vs those with only serrated polyps (SSL, TSA, HPL, N = 49).
Furthermore, we grouped colorectal adenomas according to risk signs of malignant transformation and necessity of close endoscopic follow-up (29,30) into adenomas or SSL/TSA ≥10 mm, ≥3 adenomas or SSL/TSA in 1 patient, tubulovillous histology, and the presence of HGD. The currently valid German guideline of management of colorectal carcinoma (29) recommends closer follow-up of patients carrying an adenoma exhibiting at least one of the specified risk signs. In this context, we considered the risk factors leading to the diagnosis of advanced adenoma (HGD, ≥10 mm, villous component) (3) separately. This allowed us to identify specific associations whose biological significance can be investigated in further studies. The guideline (29) further recommends endoscopic follow-up after removal of an SSL or a TSA analogous to conventional adenomas with features of risk. Therefore, we also applied these risk signs of conventional adenomas to SSL and TSA and studied these subtypes regarding these endpoints together with conventional adenomas. Owing to the lower potential of malignant transformation, HPL was not considered in the context of risk factors (3,31). Since CRC is not a precursor lesion, patients with CRC were included only in the analysis of HGD. This approach yielded another 3 comparisons including: (III) patients with at least 1 adenoma or SSL/TSA of size ≥10 mm (N = 37) vs those without (N = 69), (IV) patients with ≥3 adenomas or SSL/TSA (N = 24) vs those without (N = 91), and (V) patients with at least 1 tubulovillous polyp (N = 13) vs those without (N = 64). Of note, no adenomas with only villous morphology were detected. For comparisons III through VI, patients diagnosed with CRC and those carrying only 1 or more HPL were excluded. Finally, we evaluated (VI) patients in whom at least 1 CP with HGD or CRC (N = 34) was detected vs those without (N = 101). Of the entire 253 study participants, 111 (43.9%) contributed to only 1 comparison, whereas the remaining 142 (54.1%) were evaluated in 2 or more comparisons. Figure 1c shows the comparisons between the individual CP groups, covering detection of noncancerous polyps, comparison of conventional vs serrated lesions, and risk for malignant transformation in adenomas and SSL/TSA. A per individual summary of the outcome variables is provided in Supplementary Table 1 (see Supplementary Digital Content 1, http://links.lww.com/CTG/B47).
Alkaline phosphatase levels correlate with a higher occurrence of HGD in CPs
For each of the aforementioned comparisons, we evaluated associations between a comprehensive collection of clinical data, including comorbidities, medication, laboratory values, nutritional preferences, and epidemiological variables with polyp detection (comparison I), subtype (comparison II), and risk factor (comparison III–VI), respectively.
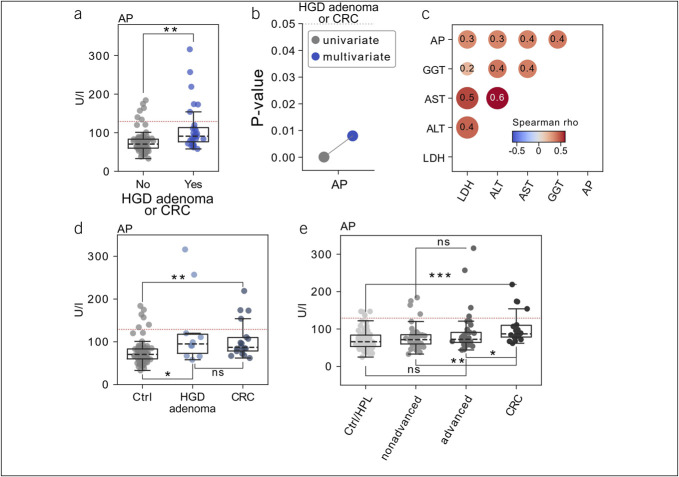
With the exception of the presence of HGD or CRC, no outcomes of our interest yielded significant associations with patients' clinical data after correcting for the testing of multiple hypotheses. Here, alkaline phosphatase (AP) levels were relatively elevated and significantly higher in patients carrying a polyp with high-grade dysplastic features or CRC (Mann-Whitney U test, false discovery rate [FDR] <0.01, Figure 2a). Next, we evaluated a multivariate logistic regression model accounting for clinical factors with well-described relationships to colon carcinogenesis (see Methods for further details), where AP levels retained their significant association (Figure 2b). Among clinical covariates, we observed significant positive correlation with other parameters of liver function testing such as gamma-glutamyltransferase as well as aspartate and alanine transaminase (Figure 2c), suggesting that AP elevations were related to liver damage. However, in the group of patients with HGD and CRC, liver disease (liver metastases together with other liver diseases, such as hepatitis or cirrhosis) was not observed more frequently when compared with controls (data not shown). To determine whether there was an association between high-grade dysplastic polyps and AP or whether the association was a result of patients with CRC, we next considered precancerous polyps and CRC separately. Interestingly, patients with high-grade dysplastic CP (N = 14) showed significantly increased levels of AP compared with respective controls (N = 101, FDR = 0.018). This association continued to be significant when only patients with CRC were considered (N = 20, FDR = 0.0017, Figure 2d). There was no significant difference in AP levels between patients carrying a high-grade dysplastic CP and patients diagnosed with CRC, indicating that the association of AP with HGD is not driven by patients with CRC. The specific examination of the individual factors of advanced adenoma (3) allowed us to better understand associations with the respective risk factor of malignant progression. In addition, we investigated whether AP is also relatively increased in patients with polyps covering any risk factor of malignant progression. For this purpose, we classified and compared the polyps in advanced adenoma (adenoma with HGD and/or villous component and/or size ≥10 mm, N = 42), nonadvanced adenoma (adenoma without these features of risk, N = 73), healthy gut controls and HPL (N = 118), and CRC (N = 20). With this approach, we found that AP levels were still shown to be elevated in CRC but not in advanced adenoma (Figure 2e). This underscores the finding of relatively elevated AP in high-grade dysplastic lesions.
Figure 2.
Clinical covariates related to risk factors of malignant progression in colorectal polyps. (a) Association of the presence of Association of the presence of adenoma with HGD feature or CRC (x-axis) with serum AP levels (y-axis). Hypothesis testing was performed using a 2-tailed Mann-Whitney U test. (b) Comparisons of P values derived from univariate assessment (grey) vs multivariate modeling (blue) for alkaline phosphatase levels. (c) Pairwise Spearman rank correlation between alkaline phosphatase levels and the indicated numeric covariates. (d) Association of the presence of HGD or CRC (x-axis) with serum alkaline phosphatase levels (y-axis). (e) Association of the indicated patient groups (x-axis) with serum alkaline phosphatase levels (y-axis). Hypothesis testing was performed using a 2-tailed Mann-Whitney U test with post hoc adjustment of P values using the Benjamini-Hochberg method. In boxplots, the box ranges from Q1 (the first quartile) to Q3 (the third quartile) of the distribution and the range represents the IQR. The median is indicated by a dashed line across the box. The “whiskers” on box plots extend from Q1 and Q3 to 1.5 the IQR. **FDR ≤0.01; *FDR ≤0.05; ns not significant. ALT, alanine aminotransferase; AP, alkaline phosphatase; AST, aspartate aminotransferase; CRC, colorectal carcinoma; FDR, false discovery rate; GGT, gamma-glutamyl transferase; HGD, high-grade dysplasia; HPL, hyperplastic polyps; IQR, interquartile range; LDH, serum lactate dehydrogenase.
Landscape of the gut microbiota in patients with CP
Matching, high-quality 16S rRNA stool profiles were available for a total of 219 individuals, that is, 83 healthy controls and 136 patients with CP detected on colonoscopy. Comparing individual microbiota compositions confirmed diverse ecosystems dominated by the 2 major phyla Firmicutes and Bacteroidetes (cumulative mean relative abundance, 86%) (Figure 3a). The cohort was characterized by an average individual richness of 114 ± 34 OTUs and 49 ± 21 Shannon effective number of species (Figure 3b). Unsupervised analysis based on generalized UniFrac distances did not show any clear relation to CP biology as exemplified for the presence of any polyp in general and the presence of a polyp with dysplastic histological features or carcinoma, respectively (Figure 3c). Multivariate permutational analysis of metadata with the fecal microbiota profiles identified 19 of 164 features with significant covariation including laboratory (e.g., AP levels), clinical (e.g., watery stool consistency), and dietary (e.g., poultry intake) information (Figure 3d), indicating an influence on the bacterial composition of the gut. Although the intake of antibiotics achieved significant results in this analysis, its frequency was balanced between all groups compared. Reported probiotic intake was also balanced across groups and had no significant effect on the gut microbiota (Table 1).
For each of our comparisons I through VI, we undertook an extensive evaluation of multiple measures of alpha diversity (e.g., species richness), and the abundance of more than 400 taxa, more than 500 zOTU, and more than 300 functional pathways, imputed by Picrust2 (23). However, we did not observe significant changes between any of our outcomes and the respective control group after correcting for the testing of multiple hypotheses. We then investigated whether the microbiota differed in patient with malignant lesions, namely CRC, and noncancerous CP and in patients with CRC and healthy controls, respectively. Here, an insignificant tendency for lower richness and alpha diversity was seen in patients with CRC compared with healthy gut controls. This trend was not seen in the comparison of CRC and noncancerous CP. Analysis of microbial differences between advanced adenoma, nonadvanced adenoma, controls, and CRC did not reveal significant differences in alpha diversity. Furthermore, there were no significant differences at the genus or phylum level. However, a previously described trend of lower alpha diversity in patients with CRC could be appreciated (Figure 3e).
DISCUSSION
Various modifiable risk factors of CRC are known, including smoking, alcohol consumption, and obesity (32). In addition, the gut microbiome seems to play a role in the development of CRC (10). Fewer studies have addressed lifestyle and dietary risk factors for CP (26,33–35) and revealed associations with the gut microbiome (36,37). In this study, we aimed to integrate various patients' characteristics to examine their association with different aspects of CP biology and concurrent changes in the gut microbiota.
In view of the analyzed clinical metadata, we observed a significant association of relatively elevated serum AP levels with the presence of HGD and CRC. In this regard, AP was significantly higher in both precancerous high-grade dysplastic polyps and patients with CRC compared with the respective control group. This implies that the relatively increased AP in precancerous lesions is not explained by metastases in patients with CRC. This association retained its significance when subjected to a multivariate logistic regression model. The analysis of AP levels comprises many distinct enzymes of the AP family in the body (38), among which the intestinal AP seems to modulate gut microbiota (39). Patients suffering from CRC showed a proportional increase in AP levels with disease progression (40), and AP levels showed a significant association with the stool microbial ecosystem diversity in our analysis. Hence, measuring AP levels may serve to support screening colonoscopy for the detection of premalignant and malignant colorectal lesions. Of note, in most cases, AP was not elevated above the upper limit of 129 U/L considered relevant in Germany, but rather, it was shown to be elevated relative to the control group (Figure 2a). We further observed a significant correlation of elevated AP levels with other parameters of liver function, suggesting that AP elevations were related to liver damage. The presence of any liver disease has been found to associate with CPs at risk for malignant progression (41), which led the authors to suggest more intensified screening colonoscopy for patients with liver disease and may be confirmed by our data. However, studies on the significance of liver enzymes in patients with CPs are scarce, and an association between elevated AP and polyps at risk for malignant transformation has not been described to our knowledge. Furthermore, the findings of elevated liver enzymes in patients with CRC are not consistent in the literature. For example, a large prospective study showed an inverse correlation of liver values at baseline with the incidence of CRC (42). In summary, further studies are needed on the connection of liver function tests, the pathogenesis of CPs, and the gut microbiome.
We did not detect significant associations for other clinical factors between patients with CP after correcting for multiple hypothesis testing. This included detailed questionnaire data, medical information, and serological analyses. We recognize that the relatively small number of patients in our subgroups (Figure 1c), relative to the large number of clinical and lifestyle parameters investigated, may negatively affect statistical power, and further studies are needed with targeted questions regarding diet, lifestyle, and development of CP.
We further investigated the microbial composition and bacterial metabolic function of the stool samples in different subgroups. Besides a trend toward decreased alpha diversity in patients with CRC compared with healthy gut controls, the microbiota showed no difference in their distribution or metabolic function in CP compared with the respective controls. However, previous studies have pointed to significant microbial changes accompanying CP formation. Exemplarily, a recent study revealed distinct stool microbial signatures between SSA and TA, and environmental factors link with the identified species (37). Another study demonstrated distinct taxa between patients with CP and controls, but no differences in microbial species richness or diversity. Furthermore, there were no significant differences regarding histological classifications (e.g., HGD) (11). Moreover, the changes in the microbiome in CP are in some cases even opposite. For example, Bacteroides was shown to be increased in fecal samples of patients with CRC (12) and CPs (13), whereas in other studies, Bacteroides was more common in healthy patients than in patients with CP (14,15). Similar divergent results have been described for Faecalibacterium spp. Although Chen et al (13) found a comparable abundance of Faecalibacterium spp. in mucosa samples from patients with CP and controls, others reported a higher abundance in patients with CP compared with controls (15). Furthermore, Feng et al (10) observed a greater richness in genes or genera in patients with more advanced adenoma or CRC, whereas Peters et al (9) observed the opposite, namely a lower species richness in stool samples from patients with CP, especially advanced CP, compared with controls. Possible explanations for the inconsistent findings are confounding factors on the microbiome and differences in sequencing methods. Because we did not see significant differences in microbiota between the groups studied, a confounding analysis was superfluous. However, we investigated the influence of different host factors on the microbial composition of the stool and observed that besides symptoms (e.g., watery stools and abdominal pain), secondary diseases (e.g., diabetes mellitus II) and diet (e.g., yogurt consumption) seem to have an influence (Figure 3d). Owing to the inconsistent findings in the microbiome in patients with CP, we believe that the microbiome cannot yet contribute to the diagnosis with sufficient certainty. Further studies taking into account possible confounders are needed.
We see important limitations to our study. First, it cannot be ruled out with certainty that individual taxa may be absent in the longer term because of bowel preparation (43). Nevertheless, we based our sampling time point 4–6 weeks after bowel preparation on previous studies, according to which a normalization of the gut microbial structure occurred 14 days to 6 weeks after bowel cleansing (16,44). Second, the timing of antibiotic and probiotic use cannot be determined. However, the distribution was similar in the studied groups, and since we did not see significant differences, the results do not seem to be influenced by antibiotics or probiotics. Third, our study includes a limited number of patients with CRC, which limits the analysis of microbial signatures in this important subgroup. Furthermore, we examine a variety of clinical and epidemiologic metadata in relatively small subgroups (Figure 1c). This affects the statistical power. Nevertheless, after correcting for multiple hypotheses testing, we report to the best of our knowledge for the first time a significant association of relatively elevated AP with CPs containing high-grade dysplastic features, which could represent a valuable addition to improve screening efforts. Interestingly, AP was among the factors that most strongly influenced microbial communities. The association between elevated AP, premalignant CP, and the microbiome should be investigated in further studies.
CONFLICTS OF INTEREST
Guarantor of the article: Moritz Middelhoff, MD.
Specific author contributions: D.S.: conceptualization: lead; project administration: lead; investigation: lead; methodology: equal; formal analysis: equal; visualization: supporting; validation: equal; manuscript—writing: lead; manuscript—review & editing: lead. H.C.M.: conceptualization: lead; project administration: lead; supervision: lead; investigation: equal; methodology: lead; formal analysis: lead; visualization: lead; validation: equal; manuscript—writing: lead; manuscript—review & editing: lead. M.F.: conceptualization: equal; project administration: equal; investigation: lead; methodology: equal; formal analysis: equal; visualization: supporting; validation: equal; manuscript—writing: lead, manuscript—review & editing: equal. M.R., U.M., J.U., M.H., S.R., T.L., C.H., C.G., N.T., T.W., K.S., M.J., and M.A.: investigation: supporting. S.R. and K.-P.J: resources: supporting; manuscript—review & editing: supporting. K.N.: resources: equal; validation: supporting; manuscript—review & editing: equal. M.Q. and D.H.: conceptualization: supporting, resources: equal; validation: supporting; funding acquisition: equal; manuscript—review & editing: equal. R.M.S.: resources: equal; funding acquisition: equal. M.M.: conceptualization: lead; project administration: lead; supervision: lead; funding acquisition: equal; manuscript—review & editing: lead.
Financial support: The study was funded by German Research Foundation (DFG) grant 395357507—collaborative research centre 1371 (Microbiome Signatures).
Potential competing interests: None to report.
Institutional review board approval statement: The study was conducted in accordance with the declaration of Helsinki and approved by the ethics committee of the Technical University Hospital of Munich (322/18 S; 2023-226-S-KH). All patient data are anonymized, and no individual information is disclosed.
Patients consent statement: All patients provided written informed consent.
Data availability and data transparency statement: FASTQ files of the 16S rRNA gene amplicon sequencing is available under SRA accession number PRJNA928694.
Study Highlights.
WHAT IS KNOWN
✓ Colorectal polyps (CPs) have different risks for malignant transformation.
✓ The interplay of gut microbiome, diet, lifestyle, and CP biology is not well understood.
✓ Early detection of premalignant polyps is an important goal of colorectal cancer prevention.
WHAT IS NEW HERE
✓ This study addresses the relationship between diet, lifestyle, and fecal microbiota with CP.
✓ Elevated liver enzymes, especially alkaline phosphatase (AP), were associated with the presence of high-grade dysplastic polyps and carcinoma.
✓ AP showed a significant effect on the microbial composition of the gut.
✓ AP may thus contribute to the screening efforts of precancerous polyps.
✓ The link between elevated AP, premalignant polyps, and the microbiome should be investigated in further studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lisa Fricke and Julia Horstmann from the ColoBAC register study for their excellent support in patient recruitment. Furthermore, we are grateful to Angela Sachsenhauser, Caroline Ziegler, and Lukas Mix from the Core Facility Microbiome of the ZIEL Institute for Food & Health for their excellent work in sample preparation and 16S rRNA gene amplicon sequencing.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/B47
David Schult, H. Carlo Maurer, and Marina Frolova contributed equally to this work.
Contributor Information
David Schult, Email: david.schult@mri.tum.de.
H. Carlo Maurer, Email: carlo.maurer@tum.de.
Marina Frolova, Email: frolova.marina06@gmail.com.
Marc Ringelhan, Email: marc.ringelhan@mri.tum.de.
Ulrich Mayr, Email: ulrich.mayr@mri.tum.de.
Jörg Ulrich, Email: joerg.ulrich@mri.tum.de.
Markus Heilmaier, Email: markus.heilmaier@mri.tum.de.
Sebastian Rasch, Email: sebastian.rasch@mri.tum.de.
Tobias Lahmer, Email: tobias.lahmer@mri.tum.de.
Sandra Reitmeier, Email: sa.reitmeier@gmail.com.
Chiara Hennig, Email: chiara.hennig@mri.tum.de.
Christina Gassner, Email: christina.gassner@mri.tum.de.
Niklas Thur, Email: niklasthur@gmail.com.
Theresa Will, Email: willtheresa09@gmail.com.
Klaus-Peter Janssen, Email: klaus-peter.janssen@tum.de.
Katja Steiger, Email: katja.steiger@tum.de.
Moritz Jesinghaus, Email: moritz.jesinghaus@uni-marburg.de.
Klaus Neuhaus, Email: neuhaus@tum.de.
Michael Quante, Email: michael.quante@uniklinik-freiburg.de.
Dirk Haller, Email: dirk.haller@tum.de.
Mohamed Abdelhafez, Email: mohamed.abdelhafez@mri.tum.de.
Roland M. Schmid, Email: rolandm.schmid@mri.tum.de.
REFERENCES
- 1.Morgan E, Arnold M, Gini A, et al. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023;72(2):338–44. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen LH, Goel A, Chung DC. Pathways of colorectal carcinogenesis. Gastroenterology 2020;158(2):291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieberman DA, Rex DK, Winawer SJ, et al. Guidelines for colonoscopy surveillance after screening and polypectomy: A consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012;143(3):844–57. [DOI] [PubMed] [Google Scholar]
- 4.Dwyer AJ, Murphy CC, Boland CR, et al. A summary of the fight colorectal cancer working meeting: Exploring risk factors and etiology of sporadic early-age onset colorectal cancer. Gastroenterology 2019;157(2):280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 2016;13(12):691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joh HK, Lee DH, Hur J, et al. Simple sugar and sugar-sweetened beverage intake during adolescence and risk of colorectal cancer precursors. Gastroenterology 2021;161(1):128–42.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tantamango YM, Knutsen SF, Beeson WL, et al. Foods and food groups associated with the incidence of colorectal polyps: The Adventist Health Study. Nutr Cancer 2011;63(4):565–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 2020;158(2):322–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters BA, Dominianni C, Shapiro JA, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 2016;4(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 11.Hale VL, Chen J, Johnson S, et al. Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomarkers Prev 2017;26(1):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 2013;66(2):462–70. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Niu M, Pan J, et al. Bacteroides, butyric acid and t10,c12-CLA changes in colorectal adenomatous polyp patients. Gut Pathog 2021;13(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brim H, Yooseph S, Zoetendal EG, et al. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS One 2013;8(12):e81352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010;1(3):138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata N, Tohya M, Fukuda S, et al. Effects of bowel preparation on the human gut microbiome and metabolome. Sci Rep 2019;9(1):4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schult D, Reitmeier S, Koyumdzhieva P, et al. Gut bacterial dysbiosis and instability is associated with the onset of complications and mortality in COVID-19. Gut Microbes 2022;14(1):2031840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagtegaal I, Arends M, Odze R, et al. Digestive System Tumours: Tumours of the Colon and Rectum. International Agency for Research on Cancer (IARC): Lyon, France, 2019. [Google Scholar]
- 19.Reitmeier S, Kiessling S, Neuhaus K, et al. Comparing circadian rhythmicity in the human gut microbiome. STAR Protoc 2020;1(3):100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagkouvardos I, Fischer S, Kumar N, et al. Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 2017;5:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012;28(16):2106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurdie PJ, Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput Biol 2014;10(4):e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas GM, Maffei VJ, Zaneveld JR, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 2020;38(6):685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandes AD, Reid JN, Macklaim JM, et al. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapelle N, Martel M, Toes-Zoutendijk E, et al. Recent advances in clinical practice: Colorectal cancer chemoprevention in the average-risk population. Gut 2020;69(12):2244–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman DA, Prindiville S, Weiss DG, et al. Risk factors for advanced colonic neoplasia and hyperplastic polyps in asymptomatic individuals. JAMA 2003;290(22):2959–67. [DOI] [PubMed] [Google Scholar]
- 27.Aune D, Chan DS, Lau R, et al. Dietary fibre, whole grains, and risk of colorectal cancer: Systematic review and dose-response meta-analysis of prospective studies. BMJ 2011;343:d6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White A, Ironmonger L, Steele RJC, et al. A review of sex-related differences in colorectal cancer incidence, screening uptake, routes to diagnosis, cancer stage and survival in the UK. BMC Cancer 2018;18(1):906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmiegel W, Buchberger B, Follmann M, et al. S3-Leitlinie: Kolorektales karzinom. Z Gastroenterol 2017;55(12):1344–498. [DOI] [PubMed] [Google Scholar]
- 30.Cross AJ, Robbins EC, Pack K, et al. Colorectal cancer risk following polypectomy in a multicentre, retrospective, cohort study: An evaluation of the 2020 UK post-polypectomy surveillance guidelines. Gut 2021;70(12):2307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien MJ, Zhao Q, Yang S. Colorectal serrated pathway cancers and precursors. Histopathology 2015;66(1):49–65. [DOI] [PubMed] [Google Scholar]
- 32.Keum N, Giovannucci E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16(12):713–32. [DOI] [PubMed] [Google Scholar]
- 33.He X, Wu K, Ogino S, et al. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology 2018;155(2):355–73.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davenport JR, Su T, Zhao Z, et al. Modifiable lifestyle factors associated with risk of sessile serrated polyps, conventional adenomas and hyperplastic polyps. Gut 2018;67(3):456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben Q, Sun Y, Chai R, et al. Dietary fiber intake reduces risk for colorectal adenoma: A meta-analysis. Gastroenterology 2014;146(3):689–99.e6. [DOI] [PubMed] [Google Scholar]
- 36.Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015;6:8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JWJ, Plichta DR, Asher S, et al. Association of distinct microbial signatures with premalignant colorectal adenomas. Cell Host Microbe 2023;31(5):827–38.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fawley J, Gourlay DM. Intestinal alkaline phosphatase: A summary of its role in clinical disease. J Surg Res 2016;202(1):225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malo MS, Alam SN, Mostafa G, et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut 2010;59(11):1476–84. [DOI] [PubMed] [Google Scholar]
- 40.Saif MW, Alexander D, Wicox CM. Serum alkaline phosphatase level as a prognostic tool in colorectal cancer: A study of 105 patients. J Appl Res 2005;5(1):88–95. [PMC free article] [PubMed] [Google Scholar]
- 41.Troschel AS, Miks A, Troschel FM, et al. Chronic liver disease promotes lesions of the colorectal adenoma-carcinoma sequence, independent of liver cirrhosis. United European Gastroenterol J 2019;7(5):662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He MM, Fang Z, Hang D, et al. Circulating liver function markers and colorectal cancer risk: A prospective cohort study in the UK Biobank. Int J Cancer 2021;148(8):1867–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tropini C, Moss EL, Merrill BD, et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell 2018;173(7):1742–54.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powles STR, Gallagher KI, Chong LWL, et al. Effects of bowel preparation on intestinal bacterial associated urine and faecal metabolites and the associated faecal microbiome. BMC Gastroenterol 2022;22(1):240. [DOI] [PMC free article] [PubMed] [Google Scholar]