Abstract
Early region 3 (E3) of group C human adenoviruses (Ad) encodes several inhibitors of tumor necrosis factor alpha (TNF-α) cytolysis, including an E3 14.7-kDa protein (E3-14.7K) and a heterodimer containing two polypeptides of 10.4 and 14.5 kDa. To understand the mechanism by which the viral proteins inhibit TNF-α functions, the E3-14.7K protein was used to screen a HeLa cell cDNA library to search for interacting proteins in the yeast two-hybrid system. A novel protein containing multiple leucine zipper domains without any significant homology with any known protein was identified and has been named FIP-2 (for 14.7K-interacting protein). FIP-2 interacted with E3-14.7K both in vitro and in vivo. It colocalized with Ad E3-14.7K in the cytoplasm, especially near the nuclear membrane, and caused redistribution of the viral protein. FIP-2 by itself does not cause cell death; however, it can reverse the protective effect of E3-14.7K on cell killing induced by overexpression of the intracellular domain of the 55-kDa TNF receptor or by RIP, a death protein involved in the TNF-α and Fas apoptosis pathways. Deletion analysis indicates that the reversal effect of FIP-2 depends on its interaction with E3-14.7K. Three major mRNA forms of FIP-2 have been detected in multiple human tissues, and expression of the transcripts was induced by TNF-α treatment in a time-dependent manner in two different cell lines. FIP-2 has consensus sequences for several potential posttranslational modifications. These data suggest that FIP-2 is one of the cellular targets for Ad E3-14.7K and that its mechanism of affecting cell death involves the TNF receptor, RIP, or a downstream molecule affected by either of these two molecules.
To effectively protect themselves against viral infections, animals have developed an array of complex immune responses. Among the defensive strategies, tumor necrosis factor alpha (TNF-α) plays a critical role. Concurrently, viruses have evolved to contain genes whose proteins regulate the activity of cytokines, and these cytokine-regulatory viral proteins are thought to facilitate acute infection or promote persistence in animal hosts (25). Group C human adenoviruses (Ad) of types 2 and 5 contain an early transcription region 3 (E3) that codes for three proteins that inhibit the cytolytic effects of TNF-α (44). For many cells, the susceptibility to TNF-α cytolysis requires sensitizing agents such as protein inhibitors (cycloheximide), cytochalasin E2, or the presence of another early Ad protein, coded in the E1A region. Under all of these conditions, the cytolytic action of TNF-α is inhibited by an Ad 14.7-kDa protein, designated Ad E3-14.7K, expressed either after viral infection or from transfected plasmids (13, 15, 16). Two other Ad E3 proteins of 10.4 and 14.5kDa, named E3-10.4K and E3-14.5K, respectively, can function as a heterodimer to inhibit TNF-α-mediated cytolysis (14). E3-10.4K can also accelerate the internalization of the epidermal growth factor receptor (5), which belongs to the TNF-α receptor family. In addition, another protein, Ad E1B-19K, encoded in early region 1 (E1), can also inhibit cell death induced by TNF-α (43). Ad E1B-19K is a structural and functional homolog of Bcl-2, a cellular protein that inhibits apoptosis (8). The presence of multiple anti-TNF-α proteins presumably suggests that the control of TNF-α cytolysis is important to the survival or life cycle of Ad.
In vivo effects of Ad E3-14.7K have been shown by using viral deletion mutants of the Ad E3-14K (plus the 10.4K and 14.5K) anti-TNF proteins. In cotton rats infected intranasally with Ad E3-14.7K deletion mutants, the pulmonary inflammatory response consisted of a peribronchial polymorphonuclear leukocyte infiltration, which replaced a mononuclear response that followed infection with wild-type Ad (12). However, in a murine pneumonia model, there was a markedly increased alveolar infiltration after Ad E3-14.7K deletion mutant infection in comparison to that with wild-type Ad type 5 (35). The effects of isolated Ad E3-14.7K were also demonstrated when its gene was expressed in vaccinia virus (VV) in various combinations with TNF-α (38). In this model of VV-induced pneumonia in mice in which TNF-α alone had been shown to be antiviral, it was shown that E3-14.7K antagonized the effects of TNF-α. This was measured by enhanced pathology such as pulmonary inflammation, as well as increased viral titers in lung tissue and mortality. Since the enhancing effects of Ad E3-14.7K on VV disease also could be observed in SCID mice, the experiments indicated that neither B nor T cells were necessary for these observed effects (39).
TNF-α is a proinflammatory cytokine which has a number of important biologic functions in addition to the control of viral infection (reviewed in reference 46). All the functions of TNF-α are transmitted through two specific receptors on the cell surface, which contain 55- and 70-kDa polypeptides, respectively (37). The 55-kDa TNF receptor (TR55) undergoes interactions to form a trimer that has extracellular, transmembrane, and short intracytoplasmic domains (33). Overexpression of the intracellular domain of the TR55 can cause cell death. Some of the molecules that interact with the cytoplasmic domain have been elucidated. These include a death-promoting molecule called TRADD, which was recently isolated by using the 55-kDa TR55 intracellular domain in the yeast two-hybrid system (18). Two other proteins, MORT-1/FADD and RIP (3, 7, 36), which have “death domain” homology with TRADD, were identified initially by their interactions with Fas/APOI, another member of the TNF-α receptor family (10). Several other molecules, such as TRAF1 and TRAF2 (28), TRAF3 (6, 19, 26), and TRAP1 and TRAP2 (34), have also been shown to interact with TNF-α receptors and are thought to be important for transmitting signals from the receptor to downstream targets. A protein called MACH/FLICE (2, 27), which has death domain homology with TRADD and the ICE protease family, has been identified as the downstream target of MORT-1/FADD and can mediate cell death. In addition, it was recently demonstrated that some of the proteins mentioned above are involved in apoptosis and/or NF-κB activation induced by TNF-α. Activation of NF-κB is associated with an inhibition of apoptosis (1, 17, 24, 40). Although the identification of these molecules has greatly enhanced our understanding of the early steps of TNF-α signal transduction, less is known about downstream steps that are presumably involved together with the proximal effectors of cell death (41).
Not much is known about the mechanism of Ad E3-14.7K inhibition of TNF-α cytolysis; however, this viral protein can prevent the TNF-α-stimulated release of arachidonic acid by the action of phospholipase A2 (47). The effect is indirect, as E3-14.7K does not inhibit the enzymatic activity of phospholipase A2. Ad E3-14.7K was also shown to inhibit the proteolysis of some indicator polypeptides and generally inhibited the appearance of the products of proteolysis (42). Ad E3-14.7K has no effect on the binding of TNF-α to either of the two TNF-α receptors (16), nor is there any available evidence that it inhibits the TNF-α transcriptional effects mediated through NF-κB (11). As discussed above, the anti-TNF function of E3-14.7K is independent of other viral proteins; therefore, it most likely inhibits TNF cytolysis by directly interacting with cellular proteins involved in TNF-α signaling pathways. The unique functional characteristics of the Ad E3-14.7K protein in interfering with the TNF-α cytolytic pathway may provide a useful assay for dissection of the cell death pathway.
The goal of the current studies was to define the host cell protein targets that bind to Ad E3-14.7K and eventually to determine their mechanism of action. The Ad E3-14.7K protein was utilized successfully in the yeast two-hybrid system to find four host-cell interacting proteins. This report describes one of these proteins, called FIP-2, which is a novel protein containing multiple leucine zipper domains. The results describing the structure and function of FIP-1, which is another of the cell proteins that binds to Ad E3-14.7K and is a low-molecular-weight GTP-binding protein with some homology to Ras, have recently been published (23).
(The data in this paper are from a thesis submitted by Yongan Li in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.)
MATERIALS AND METHODS
Cell lines.
The human embyonic kidney 293 cell line was maintained in RPMI medium supplemented with 10% fetal bovine serum, 50 U of penicillin per ml, and 50 μg of streptomycin per ml. The mouse fibroblast C3HA cell lines with or without constitutively expressed Ad E3-14.7K (obtained from Linda Gooding of Emory University) were maintained in Dulbecco’s modified Eagle’s medium with the same supplements as stated above.
Plasmid constructs.
The “bait” vector containing Ad E3-14.7K protein, the pGST-14.7K expression plasmid, and pcDNA-T7 were constructed as previously described (23). All mammalian expression constructs containing FIP-2 cDNA of various lengths, except full-length FIP-2 cDNA and pcDNA-FIP-2CΔ346, were released from pGAD-GH vectors with BamHI and XhoI and cloned into corresponding sites in pcDNA-T7. The FIP-2 full-length clone and FIP-2CΔ346 in a pGAD-GH vector were made as follows. Full-length cDNA of FIP-2 was obtained by a two-step PCR. The 5′ and 3′ ends of the FIP-2 cDNA were amplified by rapid amplification of cDNA ends (RACE) (described below) and regular PCR, respectively. The PCR products from two reactions were purified, mixed, and used as templates for a second PCR, which used 5′ and 3′ primers from previous RACE and normal PCR, respectively. The product from the second PCR was purified and cloned in frame into pGAD-GH and pcDNA-T7. The FIP-2CΔ346 clones in pGAD-GH and pcDNA-T7 were derived from the full-length clones by digestion of the parental plasmids with HpaI and XhoI and religation. The fidelity of the constructs was confirmed by sequencing.
For in vitro transcription and translation of FIP-2, the BamHI/XhoI-released FIP-2 DNA from the target vector was cloned into pCITE-4b (Novagen) at the corresponding sites. The plasmid for making the FIP-2 probe for the RNase protection assay (RPA) was constructed by first amplifying the FIP-2 region from base 727 to 898 (see Fig. 2) by PCR and then cloning the PCR product into pcDNA3.
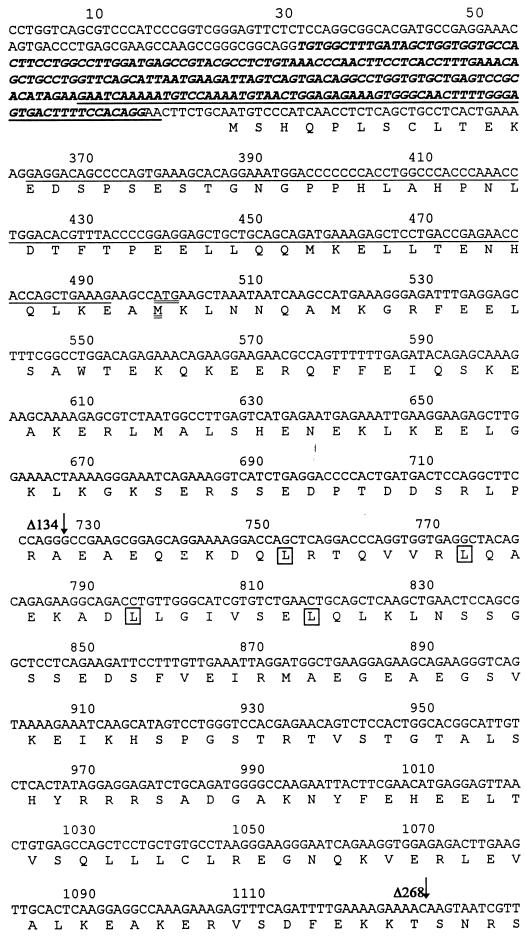
FIG. 2.
Polypeptide and cDNA sequences of FIP-2. The cDNA sequences were derived from clones from the yeast two-hybrid screening and RACE studies. Polypeptide sequences were deduced by using conventional genetic codons through a computerized program. Three different splicing forms are shown as follows: form I, unspliced; form II, the italicized sequence upstream of the first methionine is spliced out; and form III, the underlined sequences are spliced out. Forms I and II probably utilize the same start codon (bases 328 to 330), while form III utilizes the start codon at amino acid 58 (double-underlined Met at bases 499 to 501). The 5′ ends of three clones identified in the yeast two-hybrid screening are identified by arrows. The amino acids in the putative leucine zipper domains are boxed.
Construction of the FLAG-tagged E3-14.7K expression vector was done by incorporating the FLAG epitope into the first of the following PCR primers: 5′-GGAAAGCTTACCATGGACTACAAAGACGATGACGACAAGGATCCCCCGGGGAATTCGGTGGAGATGACTGAATCTCTA and 5′-CTCGCGGCCGCTTTATGTTAGTTGAATGGAAT. The HindIII/NotI-digested PCR product was cloned into pcDNA3 at the corresponding sites. The frame of the construct was confirmed by DNA sequencing, and its protein expression was confirmed by Western blotting.
Isolation of three alternatively spliced FIP-2 messages by RACE.
To obtain the 5′-end cDNA sequences which were missing in the two-hybrid cDNA clones of FIP-2, PCRs were performed with a gene-specific primer (5′-AGTGGAGACTGTTCTCGTGGACCC-3′) and an adapter primer (5′-CCATCCTAATACGACTCACTATAGGGC-3′) present in the RACE-ready heart cDNA template (CloneTech). The PCR conditions were as follows: 94°C for 1 min, 1 cycle; 94°C for 1 min and 70°C for 3 min, 5 cycles; 94°C for 1 min and 68°C for 3 min, 20 cycles; and 68°C for 7 min, 1 cycle.
PCR products were purified and ligated to T-vector (Promega). Ligation products were used to transform Library Efficiency DH5α cells (Gibco-BRL), and the transformed cells were plated on Luria-Bertani plates containing ampicillin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Blue colonies containing the cDNA inserts were identified by digestion with PvuII, transferred onto Hybond+ nylon membranes, and analyzed by Southern blotting with FIP-2 cDNA inserts as probes. The positive clones on Southern analysis were subjected to DNA sequencing.
Yeast two-hybrid screening and reagents for specificity testing.
The yeast two-hybrid screening, specificity test, and plasmids used in these assays have been previously described (23). From approximately 107 colonies screened, 21 clones were found to contain the FIP-2 insert. The HeLa cDNA library was a gift from Greg Hannon and David Beach of Cold Spring Harbor Laboratory. Bait plasmids containing lamin, TAD, basic helix-loop-helix (bHLH), and MaxI were kindly provided by Ron DePinho and described previously (32). Bcl-2, E1B-19K, and BIK-1 were generously made available to us by G. Chinnadurai of St. Louis University (4).
Co-immunofluorescent labeling of mouse cells containing constitutively expressed AdE3-14.7K and a transiently transfected T7–FIP-2 fusion protein.
Immunohistochemical colocalization studies in C3HA cells were done by previously reported procedures (23). Briefly, mouse C3HA cells containing the Ad E3-14.7K gene were grown on chamber slides (Novagen) and were transfected with 1 μg of pcDNA-T7-FIP-2Δ134 DNA per well by using the Lipofectamine technique. Patterns of colocalization of FIP-2 and Ad E3-14.7K were observed by double immunofluorescence (rhodamine and fluorescein) analyzed on a confocal microscope. The antibody to E3-14.7K was a generous gift from William Wold, St. Louis University.
Preparation and use of an Ad E3-14.7K–GST fusion protein.
The expression and absorption of the glutathione S-transferase (GST) fusion protein to glutathione-conjugated beads, in vitro labeling of FIP-2Δ134, and the in vitro protein-protein interaction assay were previously described (23). An aliquot of in vitro-labeled FIP-2 was incubated at 4°C for 2 h with either GST alone or GST–E3-14.7K preabsorbed on glutathione-conjugated beads. Following the incubation, the beads were washed three times with 150 mM NaCl-NETN buffer (9), twice with 500 mM NaCl-NETN buffer, and three times again with 150 mM NaCl-NETN buffer. After the washes, the beads were resuspended in Laemmli buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 mM dithiothreitol, 60 mM Tris (pH 6.8), 0.001% bromphenol blue), and equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Coimmunoprecipitation of FIP-2 and Ad E3-14.7K protein.
Human 293 cells that constitutively express simian virus 40 large T antigen were grown on 100-mm-diameter dishes for 24 h and were transfected with 2 μg each of pcDNA-T7-FIP-2Δ134 and pcDNA-FLAG-14.7 by using the Lipofectamine technique according to the protocol of the manufacturer (Gibco-BRL). Forty hours after transfection, cells were washed once with 1× phosphate-buffered saline (PBS) and disrupted with ice-cold Nonidet P-40 lysis buffer (150 mM NaCl, 50 mM Tris-Cl [pH 8], 1% Nonidet P-40, 150 μg of phenylmethylsulfonyl fluoride per ml, 1 μg [each] of aprotinin and leupeptin per ml). The lysate was cleared by centrifugation at maximum speed in a Microfuge, and 500 μl of each lysate was subjected to immunoprecipitation with either 2 μl of anti-FLAG M5 monoclonal antibody (Kodak) or anti-E3-gp19 monoclonal antibody. After rocking for 1 h at 4°C, 30 μl of 50% (vol/vol) protein A beads (Sigma) was added to the lysate and rocked for another hour. The beads were washed five times with the lysis buffer and resuspended in 30 μl of Laemmli buffer. After being boiled, the samples were subjected to SDS-PAGE. The gel was transblotted to nitrocellulose, and the blot was preblocked with 1× PBS–5% nonfat dry milk and subsequently interacted with T7 monoclonal antibody conjugated with horseradish peroxidase. After four 15-min washes with 1× PBS–0.1% Tween 20, immunoreactive proteins were detected with a chemiluminescence reagent (Boehringer).
RPA of FIP-2 mRNAs induced by TNF-α.
The 32P-labeled antisense RNA probe of FIP-2 and glyceraldehyde-3-phosphate dehydrogenase (internal control) from Ambion were generated by using the MaxiTranscript system (Ambion) according to the manufacturer’s protocol. Human 293 cells and adenocarcinoma MCF-7 cells were seeded onto 100-mm-diameter plates. They were treated with 200 ng of human TNF-α (BRL-Gibco) per ml for 0, 4, or 16 h. After treatment, the cells were used for total RNA purification with the TriReagent (MRC). The RPA assay was done by using the RPAII system (Ambion) with 15 μg of total RNAs and 105 cpm per probe according to the manufacturer’s protocol. The protected bands were analyzed by urea–8% PAGE and were visualized by autoradiography.
Studies of the effect of FIP-2 on E3-14.7K inhibition of TR55 killing.
Human 293 cells on six-well plates were transfected with the following plasmids in various combinations: 1.5 μg of pcDNA-FLAG-14.7K, 2.2 μg of pcDNA-T7-FIP-2 or its deletion mutants, 0.3 μg of pcDNA-TR55 (obtained from David Wallach of the Weizmann Institute), and an amount of pcDNA-T7 to bring the total amount of transfected DNA to 4 μg. All transfection mixtures contained 0.3 μg of pGLP (GreenLantern Protein; Gibco). The transfections were done by using Trans-LT2 (PanVera) according to the manufacturer’s protocol. Twenty-four hours after transfection, the cells were observed with a Nikon Zeiss Oxipat 1 fluorescence microscope and photographed through a fluorescein isothiocyanate filter.
RESULTS
Identification of FIP-2 as an Ad E3-14.7K-interacting protein by yeast two-hybrid screening and its expression pattern.
FIP-2 was isolated by using the yeast two-hybrid system in a search for the cellular proteins which interacted with the Ad E3-14.7K protein. The FIP-2 genes were recognized as a family of multiple overlapping clones which were identical at their 3′ ends but extended for various lengths toward the 5′ end of the gene. FIP-2 did not interact with a series of heterologous baits (hLamin-C, mMyc-TAD, mMyc-bHLH, or mMaxI) or with the Ad E1B-19K or Bcl-2 protein. These results indicate that the cell proteins interacting specifically with E3-14.7K did not overlap with the targets of the antiapoptotic Ad E1B or Bcl-2 protein (Table 1).
TABLE 1.
Specificity of FIP-2 protein interactions in the yeast two-hybrid systema
| LexA Hybrid (bait) | Gal4 hybrid (target) | β-Galactosidase activity (colony color) | His growthb |
|---|---|---|---|
| Ad E3-14.7K | FIP-2 | Blue | |
| Ad E3-14.7K plus: | |||
| hLamin-C | FIP-2 | White | − |
| mMyc-TAD | FIP-2 | White | − |
| mMyc-bHLH | FIP-2 | White | − |
| mMaxI | FIP-2 | White | − |
| Ad E1B-19K | BIK-1 | Blue | * |
| Bcl-2 | BIK-1 | Blue | * |
| Ad E1B-19K | FIP-2 | White | * |
| Bcl-2 | FIP-2 | White | * |
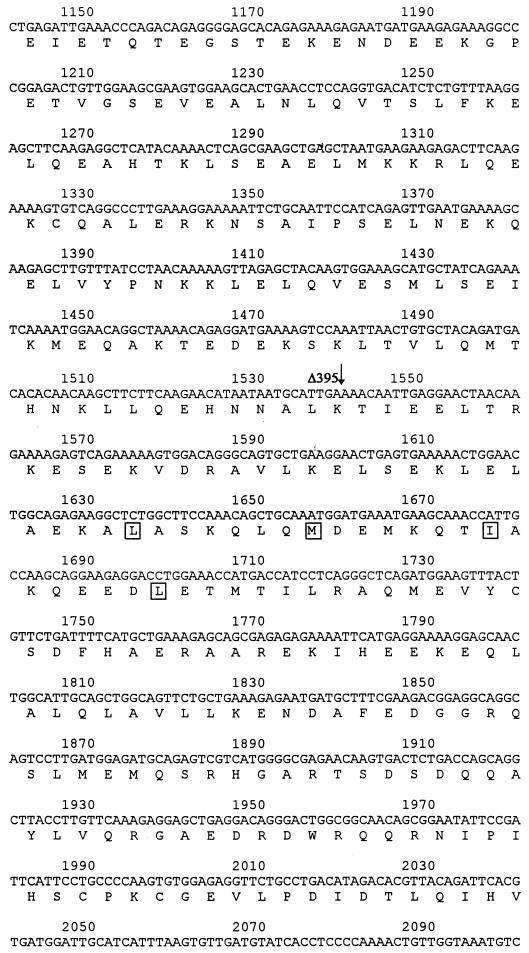
By Northern analysis with the isolated cDNA clones as probes, we have found that FIP-2 existed as at least three major message species, as shown by the three bands in the Northern blot (Fig. 1). Although the level of FIP-2 expression varied among the tissues analyzed, the three major forms seem to have the same relative abundance in the various tissues, except perhaps for brain. The differences between the sizes of FIP-2 mRNAs revealed by Northern analysis and the shorter lengths of cDNAs isolated in the yeast two-hybrid screening suggested that the cDNAs isolated by the latter procedure lacked various sequences at the 5′ ends. Subsequently, 5′-RACE reactions and sequencing of the RACE products confirmed the existence of the three FIP-2 message forms. Sequencing data suggested that the three message forms are likely the result of alternative splicing, and the major differences among the three forms were within the 5′ end of the gene. The sequence of FIP-2 is shown in Fig. 2. Sequencing analysis indicates that FIP-2 is a novel protein which has no significant homology with any protein in the databases and contains two leucine zipper domains. However, the presence of both leucine zipper domains is not required for the FIP-2–E3-14.7K interaction, because some clones isolated in the yeast two-hybrid screening lacked one of these domains.
FIG. 1.
Expression of FIP-2 mRNA in human tissues. A blot of mRNAs obtained from eight human organs as indicated was purchased from CloneTech and hybridized under stringent conditions as described by the supplier. The FIP-2 cDNA used as probe was labeled with [32P]dCTP (Amersham). (The top band of approximately 7.5 kb was also visible in some of the other organs as well as skeletal muscle on the original autoradiograph.) Numbers on the left are sizes in kilobases.
FIP-2 colocalizes with and causes redistribution of E3-14.7K.
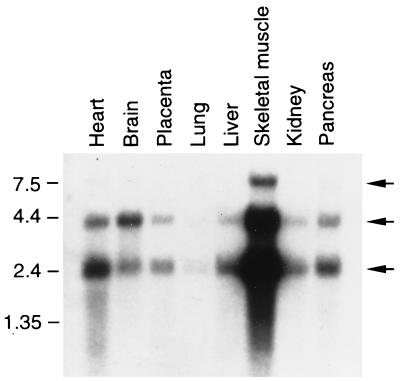
In addition to protein-protein interaction detected in the yeast two-hybrid system, FIP-2 interaction with the Ad E3-14.7K was demonstrated by three assays, including in vivo colocalization by immunocytochemistry. Figure 3A demonstrates the presence of Ad E3-14.7K in all cells of a C3HA murine cell line stably transfected with this Ad gene (16). The cell designated in Fig. 3A and C by the single asterisk stained only with reagents that detected Ad E3-14.7K but not with those that recognized FIP-2 (Fig. 3B, double asterisk). The single-asterisked cell shows the diffuse cytoplasmic pattern that was characteristic of the Ad E3-14.7K protein. One of the cells in Fig. 3A also contains FIP-2 as shown by staining for transient expression in the cell marked by the double asterisk (Fig. 3B). The colocalization of both Ad E3-14.7K and FIP-2 is most strikingly shown by the bright beaded structures present around the nuclear membrane. This colocalization of FIP-2 and Ad E3-14.7K was demonstrated better by the appearance of the perinuclear beaded structure in yellow by confocal microscopy (Fig. 3C), indicating the convergence of the Ad E3-14.7K stained red by rhodamine and the FIP-2 stained green by fluorescein. By comparing the expression patterns of E3-14.7K in the cells producing this protein alone or coexpressing both E3-14.7K and FIP-2 in Fig. 3A and B, it can be seen that overexpression of FIP-2 caused redistribution of the E3-14.7K. This suggests that FIP-2 directly interacts with E3-14.7K in vivo.
FIG. 3.
Intracellular colocalization of FIP-2 with Ad E3-14.7K. FIP-2 was cloned behind the cytomegalovirus promoter and coexpressed as a fusion protein with a T7 tag in the murine C3HA cell line constitutively expressing Ad E3-14.7K (16). (A) Ad E3-14.7K was visualized with a polyclonal rabbit antibody which recognized the viral protein either alone as a cytoplasmic protein (∗) or in cells also cotransfected with T7-FIP-2 (∗∗). (B) FIP-2 was visualized on identical cells with antibody to T7 (∗∗). The distribution of Ad E3-14.7K within individual cells in panel A was different in cells expressing E3-14.7K alone (∗) or overexpressing FIP-2 (∗∗) together with E3-14.7K, which appeared in bead-like perinuclear structures (arrows). (C) The colocalization of FIP-2 and Ad E3-14.7K is also highlighted by the yellow color, resulting from the convergence of the rhodamine image of panel A plus the fluorescence image of panel B. Bar, 10 μm.
In vitro and in vivo interaction between E3-14.7K and FIP-2.
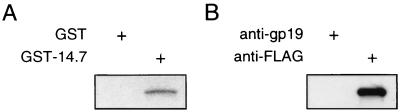
To study direct protein-protein interaction between E3-14.7K and FIP-2, FIP-2Δ134, the largest clone isolated from the yeast two-hybrid screening, was used for both in vitro and in vivo assays. The interaction was first shown by creating a GST–E3-14.7K fusion protein that was bound to glutathione-conjugated beads. The GST–E3-14.7K protein selectively bound to FIP-2, allowing the latter to absorb to the glutathione beads, whereas the protein derived from the GST vector alone failed to retain FIP-2 (Fig. 4A). The in vivo interaction between E3-14.7K and FIP-2Δ134 was shown by coimmunoprecipitation of the two proteins in the lysate from transfected cells. In Fig. 4B, it can be seen that FIP-2 can be coprecipitated by antibody against the epitope-tagged (FLAG) E3-14.7K protein (right lane) but not by the control anti-gp19 antibody (left lane) or the antibody to FLAG in the absence of cotransfection with the FLAG–E3-14.7K protein (data not shown). These in vitro and in vivo interactions further demonstrate the specificity of E3-14.7K binding to FIP-2 as detected in yeast.
FIG. 4.
In vitro and in vivo interaction between FIP-2-and E3-14.7K. (A) The in vitro interaction of radiolabeled FIP-2 with the Ad E3-14.7K–GST fusion protein or with GST alone was assayed as described in Materials and Methods. The amounts of FIP-2 absorbed and eluted from Ad E3-14.7K (GST-14.7) or from the GST protein alone as a negative control are shown after SDS-PAGE. (B) The in vivo interaction between E3-14.7K and FIP-2 is shown by coimmunoprecipitation from extracts of 293 cells transiently transfected with plasmids expressing T7-tagged FIP-2 (Δ134) and FLAG-tagged E3-14.7K proteins. Twenty hours after transfection, the cells were harvested and lysed. Cleared lysates were subjected to immunoprecipitation with either anti-FLAG (specific for E3-14.7K) or anti-gp19 (nonspecific) monoclonal antibody. The immunoprecipitates were analyzed by SDS-PAGE followed by Western blotting. FIP-2 was detected by anti-T7 monoclonal antibody.
FIP-2 reverses the protective effect of E3-14.7K on TNF receptor-induced cytolysis, and the reversal depends on the interaction of FIP-2 with E3-14.7K.
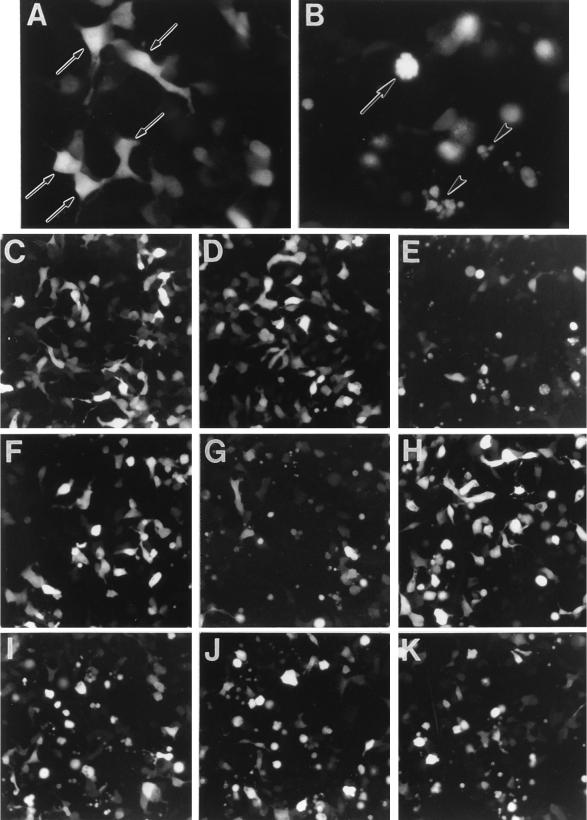
E3-14.7K was reported to be able to protect against TNF-α-induced cell killing (16). We recently found that E3-14.7K can also block the cell killing induced by overexpression of the intracellular domain of TR55. The green fluorescent protein (GFP) was used as an indicator for cells that were transiently cotransfected with other plasmids expressing TR55, E3-14.7K, and FIP-2 (wild type and deletion mutants). As demonstrated in Fig. 5B, TR55 is a potent inducer of apoptosis, as previously reported (18). All of the cells transfected with TR55 and pGFP were green and had become rounded. Some cells had cytoplasmic blebbing (Fig. 5B), and others were in an advanced stage of disintegration. In contrast, the cells transfected with empty plasmid or E3-14.7K (Fig. 5A) were flat and diffusely stained green by the GFP, with pseudopod-like projections. Their morphology was similar to that of the nontransfected, unstained cells in the monolayer.
FIG. 5.
FIP-2 reverses the protective effect of E3-14.7K on TNF receptor-induced cytolysis. Human embryonic kidney 293 cells in six-well plates were transfected with the following plasmids: (A) pcDNA-FLAG-14.7; (B) pcDNA-FLAG-TR55; (C) pcDNA-T7-FIP2; (D) pcDNA-FLAG-14.7; (E) pcDNA-TR55; (F) pcDNA-FLAG-14.7 plus pcDNA-TR55; (G) pcDNA-FLAG-14.7K plus pcDNA-TR55 plus pcDNA T7 FIP-2; (H) pcDNA-FIP-2 CΔ346 plus pcDNA-FLAG-14.7 plus pcDNA-TR55; (I) pcDNA FIP-2Δ134 plus pcDNA-FLAG-14.7 plus pcDNA-TR55; (J) pcDNA FIP-2Δ268 plus pcDNA-FLAG-14.7 plus pcDNA-TR55; (K) pcDNA-FIP-2Δ395 plus pcDNA FLAG-14.7 plus pcDNA TR55. All of these cells were cotransformed with a plasmid expressing the GFP gene (see Materials and Methods). Twenty-four hours after transfection, the cells were observed with a fluorescence microscope and photographed with a fluorescein isothiocyanate filter. Panels A and B were photographed through a 40× objective. The arrows in panel A point to transfected normal cells stained with GFP. In panel B, the arrow shows a rounded cell with cytoplasmic blebbing, and the arrowheads show completely disintegrated cells. Both morphologies are typical for apoptosis. Panels C to K were photographed through a 20× objective. Panels C, D, F, and H show morphologies that are normal or nearly normal. The other panels (E, G, I, J, and K) demonstrate various degrees of apoptosis. The percentages of cells in the panels that appeared normal after each transfection were as follows: C, 97%; D, 92%; E, 6%; F, 81%; G, 4%; H, 87%; I, 11%; J, 9%; and K, 14%.
Cells transiently transfected with FIP-2 also retained their normal appearance, as shown at lower magnification in Fig. 5C. The E3-14.7K or TR55 transfections at this magnification are also included in Fig. 5D and E, respectively, to demonstrate a larger microscopy field. E3-14.7K can substantially reverse the cytolytic effect of TR55 as shown by the increased number of normal cells (Fig. 5F); however, FIP-2 can strongly block the protective effect of E3-14.7K, as evidenced by the apoptotic rounded cells after FIP-2, E3-14.7K, and TR55 cotransfection (Fig. 5G).
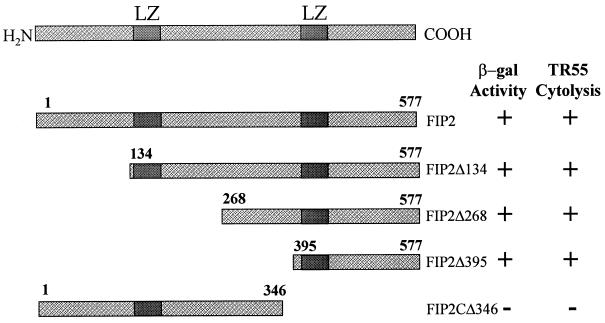
To understand whether the effect of FIP-2 on E3-14.7K inhibition of TR55 killing correlates with the interaction between FIP-2 and E3-14.7K, we used a series of FIP-2 deletion mutants to study the correlation of their interactions with E3-14.7K and their abilities to reverse E3-14.7K’s protective effect. As shown by the cell morphology assays (Fig. 5H to K) and the β-galactosidase activity (Fig. 6), the C terminus of FIP-2 is required for its interaction with E3-14.7K, while most of the N-terminal region is dispensable for the interaction. By performing TR55 cytolysis experiments (Fig. 5H to K) in the presence of E3-14.7K and each of the FIP-2 deletion mutants, we found that the C-terminal deletion mutant (FIP-2CΔ346), which did not interact with E3-14.7K, did not reverse the E3-14.7K inhibition of TR55 cytolysis (Fig. 5H). The amino-terminal mutants (FIP-2 Δ134, FIP-2 Δ268, and FIP-2 Δ395), which continued to interact with E3-14.7K, reversed the E3-14.7K protective effect and again resulted in TR55 cytolysis (Fig. 5I to K). The results clearly demonstrate that the FIP-2 effect on 14.7K function in the TR55 cytolysis assays was dependent on FIP-2–E3-14.7K interaction.
FIG. 6.
FIP-2 binding to E3-14.7K correlates with FIP-2 reversal of E3-14.7K inhibition of TR55 cytolysis. Deletion mutants of FIP-2 were cloned in appropriate vectors, and the ability of each mutant to interact with E3-14.7K in the yeast two-hybrid system (β-galactosidase [β-gal] activity) was compared with the reversal of the E3-14.7K inhibition on TR55 killing. +, full activity; −, no activity detected (see Materials and Methods); LZ, leucine zipper.
TNF-α treatment increases the mRNA level of FIP-2.
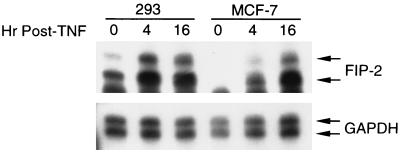
Since TNF-α can induce expression of a number of cellular genes (22, 45), we studied whether it can alter the expression level of FIP-2. When measured by RPA analysis, TNF-α could increase the mRNA level of FIP-2 in a time-dependent manner in two human cell lines, 293 (human embryonic kidney cells) and MCF-7 (human adenocarcinoma cells) (Fig. 7). In addition, the presence of an unexpected second protected band suggested that there was another spliced mRNA form which was not identified in the previous 5′-RACE analysis.
FIG. 7.
Induction of FIP-2 expression by TNF-α. Human embryonic kidney 293 cells and adenocarcinoma MCF-7 cells were treated with TNF-α as indicated in Materials and Methods. The purified total RNAs were used in RPAs with FIP-2 as a probe. The upper band was expected from sequence data, but the lower band probably represents an alternative splicing form of FIP-2. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control.
DISCUSSION
By using the yeast two-hybrid system, we have identified a cellular protein, FIP-2, which interacts with the Ad anti-TNF-α protein E3-14.7K. Several lines of in vitro and in vivo evidence demonstrate that the E3-14.7K–FIP-2 interaction detected in yeast is specific and biologically relevant. First, strong interaction in the yeast two-hybrid system was indicated by the isolation of 21 independent clones of FIP-2 of various lengths during the screening. All the clones that specifically interact with E3-14.7K do not bind to the heterologous baits tested. Second, the interaction can be demonstrated by both the in vitro GST protein binding assay and in vivo coimmunoprecipitation. Furthermore, FIP-2 not only colocalizes with E3-14.7K intracellularly but, most significantly, causes the redistribution of E3-14.7K to a perinuclear position within the cells.
Further functional studies suggest that FIP-2 is a component of the TNF-α signaling pathway. The expression of FIP-2 protein can block the protective effect of E3-14.7K on TR55 killing, and the inhibition is dependent on the interaction between FIP-2 and 14.7K. In addition, the expression of FIP-2 can be induced by TNF-α in a time-dependent manner. These data suggest that FIP-2 plays a role in TNF-α signaling. However, the yeast two-hybrid data, showing that FIP-2 did not interact with either E1B-19K or Bcl-2, indicate that Ad E1B-19K and E3-14.7K target different cellular proteins to block TNF-α cytolysis.
FIP-2 appears to be a novel protein with no significant homology with any known sequence in the databases. The sequence analysis did not yield any recognizable functional motifs except for two leucine zipper domains. The presence of the leucine zipper domains is interesting, since this motif is often found in transcriptional regulatory proteins (20, 21, 29). Similar domains are found in other recently identified TR55-interacting proteins, such as the zinc fingers in several of the TRAF family members (6, 26, 28). It is possible that these proteins may have functions similar to those of STAT or NF-κB, which can be activated by incoming signals and migrate into the nucleus to regulate the expression of certain genes (30). Although the exact functions of these two leucine zipper domains are unknown, it is clear that both were not required for the interaction between FIP2 and E3-14.7K. For example, the shortest clone of FIP-2 isolated from the two-hybrid screening lacked one of two leucine zipper domains. Deletion analysis indicates that the FIP-2 domain important for its interaction with E3-14.7K is located in the C-terminal 172 residues, but further deletions will be required to determine if the C-terminal leucine zipper is required for this protein-protein interaction.
We have not been able to show that overexpression of FIP-2 alone (in the absence of E3-14.7K and exogenous TR55) can activate the cell death signaling pathway as was shown for other known cell death proteins, such as RIP, TRADD, MORT/FADD, or MACH/FLICE. However, it is possible that normally in the absence of the Ad E3-14.7K, FIP-2 can utilize some inducible cellular cofactors to activate the cell death pathway. These may function either with TR55 or with RIP as a target, for we have also shown that RIP can substitute for TR55 in all of the cell-killing experiments shown in Fig. 5 (data not shown). RIP has a critical role at the junction of two pathways that can lead either to apoptosis or to the activation of NF-κB, which inhibits apoptosis (17, 24, 40). Ad E3-14.7K appears to be able to shift the equilibrium towards inhibition of apoptosis induced either by TR55 or by RIP. In contrast, FIP-2 can shift the equilibrium toward the induction of apoptosis, although this has been shown only in the presence of Ad E3-14.7K.
We have been unable to show that either FIP-2 or Ad E3-14.7K can bind directly to TR55 or to RIP when assayed in the yeast two-hybrid system or by protein-protein interaction in the coimmunoprecipitation assay (data not shown). However, we have isolated another novel E3-14.7K-interacting protein, FIP-3, which does bind to RIP and has homology with FIP-2 (unpublished data). Thus, there is a potential for complex formation that could bind Ad E3-14.7K to RIP through the mediation of FIP-3. Determination of whether FIP-2 could also be accommodated in this complex must await the definition of the binding domains for each set of proteins and attempts to assemble these complexes after transient transfection.
ACKNOWLEDGMENTS
We acknowledge the invaluable assistance of Michael Cammer of The Analytical Imaging Facility, a shared facility of the Albert Einstein College of Medicine Institutional Cancer Center. We thank David Wallach of the Weizmann Institute, Israel, for helpful discussions.
This research was supported by grants P30-CA13330 and RO1-CA72963 (to M.S.H.) and by National Institutes of Health training grant 5T32 CA 09060 (to Y.L.).
REFERENCES
- 1.Beg A A, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 2.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 3.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 4.Boyd J M, Gallo G J, Elangovan B, Houghton A B, Malstrom S, Avery B J, Ebb R G, Subramanian T, Chittenden T, Lutz R J, Chinnadurai G. Bik, a novel death-inducing protein, shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene. 1995;11:1921–1928. [PubMed] [Google Scholar]
- 5.Carlin C R, Tollefson A E, Brady H A, Hoffman B L, Wold W S M. Epidermal growth factor receptor is down-regulated by a 10,400 mw protein encoded by the E3 region of adenovirus. Cell. 1989;57:135–144. doi: 10.1016/0092-8674(89)90179-7. [DOI] [PubMed] [Google Scholar]
- 6.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 7.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 8.Chiou S K, Tseng C C, Rao L, White E. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J Virol. 1994;68:6553–6566. doi: 10.1128/jvi.68.10.6553-6566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chittenden T, Livingston D M, Kaelin W G., Jr The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell. 1991;65:1073–1082. doi: 10.1016/0092-8674(91)90559-h. [DOI] [PubMed] [Google Scholar]
- 10.Cleveland J L, Ihle J N. Contenders in FasL/TNF death signaling. Cell. 1995;81:479–482. doi: 10.1016/0092-8674(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrov, T., M. Hannick, and W. S. Wold. 1995. Personal communication.
- 12.Ginsberg H S, Lundholm-Beauchamp U, Horswood R L, Pernis B, Wold W S M, Chanock R M, Prince G A. Role of early region 3 (E3) in pathogenesis of adenovirus disease. Proc Natl Acad Sci USA. 1989;86:3823–3827. doi: 10.1073/pnas.86.10.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gooding L R, Elmore L W, Tollefson A E, Brady H A, Wold W S M. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell. 1988;53:341–346. doi: 10.1016/0092-8674(88)90154-7. [DOI] [PubMed] [Google Scholar]
- 14.Gooding L R, Ranheim T S, Tollefson A E, Aquino L, Duerksen-Hughes P J, Horton T M, Wold W S M. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J Virol. 1991;65:4114–4123. doi: 10.1128/jvi.65.8.4114-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gooding L R, Sofola I O, Tollefson A E, Duerksen-Hughes P J, Wold W S M. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J Immunol. 1990;145:3080–3086. [PubMed] [Google Scholar]
- 16.Horton T M, Ranheim T S, Aquino L, Kusher D I, Saha S K, Ware C F, Wold W S M, Gooding L R. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. J Virol. 1991;65:2629–2639. doi: 10.1128/jvi.65.5.2629-2639.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu H, Huang J, Shu H-B, Baichwal V, Goeddel D V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 18.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 19.Hu H M, O’Rourke K, Boguski M S, Dixit V M. A novel RING finger protein interacts with the cytoplasmic domain of CD40. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 20.Klug A. Gene regulatory proteins and their interaction with DNA. Ann NY Acad Sci. 1995;758:143–160. doi: 10.1111/j.1749-6632.1995.tb24814.x. [DOI] [PubMed] [Google Scholar]
- 21.Klug A, Schwabe J W. Protein motifs. 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 22.Kronke M, Schutze S, Scheurich P, Pfizenmaier K. TNF signal transduction and TNF-responsive genes. Immunol Ser. 1992;56:189–216. [PubMed] [Google Scholar]
- 23.Li Y, Kang J, Horwitz M S. Interaction of an adenovirus 14.7-kilodalton protein inhibitor of tumor necrosis factor alpha cytolysis with a new member of the GTPase superfamily of signal transducers. J Virol. 1997;71:1576–1582. doi: 10.1128/jvi.71.2.1576-1582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis, while NF-B activation prevents cell death. Cell. 1996;87:565–575. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 25.McFadden G. Getting to know you: viruses meet CD40 ligand. Nat Med. 1995;1:408–409. doi: 10.1038/nm0595-408. [DOI] [PubMed] [Google Scholar]
- 26.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M, Chinnaiyan A M, Kischkel F C, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, A novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 28.Rothe M, Wong S C, Henzel W J, Goeddel D V. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 29.Sassone-Corsi P. Goals for signal transduction pathways: linking up with transcriptional regulation. EMBO J. 1994;13:4717–4728. doi: 10.1002/j.1460-2075.1994.tb06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber-Agus N, Chin L, Chen K, Torres R, Thomson C T, Sacchettini J C, DePinho R A. Evolutionary relationships and functional conservation among vertebrate Max-associated proteins: the zebra fish homolog of Mxi1. Oncogene. 1994;9:3167–3177. [PubMed] [Google Scholar]
- 33.Smith C A, Farrah T, Goodwin R G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 34.Song H Y, Dunbar J D, Zhang Y X, Guo D, Donner D B. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J Biol Chem. 1995;270:3574–3581. [PubMed] [Google Scholar]
- 35.Sparer T, Tripp R A, Dilleha D L, Hermiston T W, Wold W S, Gooding L R. The role of human adenovirus early region 3 proteins (gp19K, 10.4K, 14.5K, and 14.7K) in a murine pneumonia model. J Virol. 1996;70:2431–2439. doi: 10.1128/jvi.70.4.2431-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanger B Z, Leder P, Lee T H, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 37.Tartaglia L A, Goeddel D V. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 38.Tufariello J, Cho S, Horwitz M S. The adenovirus E3 14.7-kilodalton protein which inhibits cytolysis by tumor necrosis factor increases the virulence of vaccinia virus in a murine pneumonia model. J Virol. 1994;68:453–462. doi: 10.1128/jvi.68.1.453-462.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tufariello J, Cho S, Horwitz M S. The adenovirus E3 14.7K protein, an antagonist of tumor necrosis factor cytolysis, increases the virulence of vaccinia virus in SCID mice. Proc Natl Acad Sci USA. 1994;91:10987–10991. doi: 10.1073/pnas.91.23.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 41.Vandenabeele P, Declercq W, Beyaert R, Fiers W. Two tumour necrosis factor receptors: structure and function. Trends Cell Biol. 1995;5:392–399. doi: 10.1016/s0962-8924(00)89088-1. [DOI] [PubMed] [Google Scholar]
- 42.Voelkel-Johnson C, Entingh A J, Wold W S M, Gooding L R, Laster S M. Activation of intracellular proteases is an early event in TNF-induced apoptosis. J Immunol. 1995;154:1707–1716. [PubMed] [Google Scholar]
- 43.White E, Sabbatini P, Debbas M, Wold W S M, Kusher D I, Gooding L R. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol Cell Biol. 1992;12:2570–2580. doi: 10.1128/mcb.12.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wold W S M, Hermiston T W, Tollefson A E. Adenovirus proteins that subvert host defenses. Trends Microbiol. 1994;2:437–443. doi: 10.1016/0966-842x(94)90801-x. [DOI] [PubMed] [Google Scholar]
- 45.Wong G H W, Goeddel D V. Induction of manganous superoxide dismutase by tumor necrosis factor: possible protective mechanism. Science. 1988;242:941–944. doi: 10.1126/science.3263703. [DOI] [PubMed] [Google Scholar]
- 46.Wong G H W, Kamb A, Goeddel D V. Antiviral properties of TNF. In: Beutler B, editor. Tumor necrosis factors: the molecules and their emerging role in medicine. New York, N.Y: Raven Press; 1992. pp. 371–381. [Google Scholar]
- 47.Zilli D, Voelkel-Johnson C, Skinner T, Laster S M. The adenovirus E3 region 14.7 kDa protein, heat and sodium arsinate inhibit the TNF-induced release of arachidonic acid. Biochem Biophys Res Commun. 1992;188:177–183. doi: 10.1016/0006-291x(92)92366-6. [DOI] [PubMed] [Google Scholar]