Abstract
We have identified a novel serine/threonine kinase, designated ZIP kinase, which mediates apoptosis. ZIP kinase contains a leucine zipper structure at its C terminus, in addition to a kinase domain at its N terminus. ZIP kinase physically binds to ATF4, a member of the activating transcription factor/cyclic AMP-responsive element-binding protein (ATF/CREB) family, through interaction between their leucine zippers. The leucine zipper domain is necessary for the homodimerization of ZIP kinase as well as for the activation of kinase. Immunostaining study showed that ZIP kinase localizes in the nuclei. Overexpression of intact ZIP kinase but not catalytically inactive kinase mutants led to the morphological changes of apoptosis in NIH 3T3 cells, suggesting that the cell death-inducing activity of ZIP kinase depends on its intrinsic kinase activity. Interestingly, the catalytic domain of ZIP kinase is closely related to that of death-associated protein kinase (DAP kinase), which is a mediator of apoptosis induced by gamma interferon. Therefore, both ZIP and DAP kinases represent a novel kinase family, which mediates apoptosis through their catalytic activities.
Apoptosis, a genetically controlled cell death to eliminate unwanted cells from an organism, plays a central role in embryogenesis and tissue homeostasis, but its deregulation is associated with various human diseases including autoimmune diseases, neurodegenerative disorders, and cancers (7, 27, 30, 32). Apoptosis is also triggered when cells are exposed to external stimuli including cytokines, X rays, heat shock, UV irradiation, and certain drugs. Apoptotic cell death is characterized by a series of morphological changes such as cytoplasmic shrinkage, chromatin condensation, membrane blebbing, endonucleolytic degradation of genomic DNA, and the formation of apoptotic bodies (21). Although the morphological changes are well defined, a precise mechanism which regulates the process of apoptosis is still unclear. In some cases, it appears that apoptosis is initiated by a cascade of protease activation followed by the cleavage of a specific substrate(s) (25). This proteolytic pathway is mediated by the conserved group of cysteine proteases, now designated caspases (2, 26). This family has at least 10 members, and they are related to the mammalian interleukin-1β-converting enzyme and to the nematode Caenorhabditis elegans CED-3, which is necessary for apoptotic cell death during development (40). Ectopic expression of caspases promotes apoptosis, while inhibition of caspases by specific tetrapeptides blocks apoptosis. These findings demonstrate that caspases are essential for the induction of apoptosis.
In addition to activation of caspases, signaling events such as increases in cytosolic Ca2+ concentrations (22), activation of protein kinases (3), and production of ceramide (14, 17, 31, 36) are involved in apoptosis. Studies on responses to cytotoxic stresses have implicated the catalytic activities of Jun N-terminal kinases (JNKs) in signaling for apoptosis (10, 19). JNKs are rapidly activated in response to stresses such as UV irradiation, X ray, nerve growth factor withdrawal, tumor necrosis factor alpha (TNF-α), and Fas (13, 36, 38, 39). Furthermore, inhibitors of the JNK pathway block apoptosis induced by such stresses (36). In addition, other kinases such as ASK1 and RIP have been shown to induce apoptosis (20, 29). However, at present only a few kinases which mediate apoptosis have been identified, and there is no clear picture of the apoptotic pathway mediated by such protein kinases.
In this report, we describe the cloning and characterization of ZIP kinase, a novel serine/threonine kinase which is a mediator of apoptosis. ZIP kinase contains an N-terminal kinase domain and a C-terminal leucine zipper domain that is necessary for homodimerization as well as for heterodimerization with ATF4, a member of the activating transcription factor/cyclic AMP-responsive element-binding protein (ATF/CREB) family of transcription factors (15, 33, 35). Overexpression of ZIP kinase induces the morphological changes of apoptosis in mammalian cells, suggesting that it may play an important role in the induction of apoptosis.
MATERIALS AND METHODS
Two-hybrid screening.
Yeast two-hybrid screening was performed by the Matchmaker two-hybrid system (Clontech). To construct a bait plasmid, the leucine zipper domain of murine ATF4 (amino acids [aa] 298 to 349) amplified from mouse spleen cDNA by reverse transcription-PCR was cloned in frame into the DNA binding domain of GAL4 in the pAS2-1 plasmid. Saccharomyces cerevisiae Y190 cells were transformed with the ATF4 bait plasmid by a modified lithium acetate method, and the transformants were selected on synthetic dextrose medium lacking tryptophan (SD −Trp). The transformants grown on SD −Trp were further transformed with a mouse brain or a human placenta cDNA library fused to the GAL4 transactivation domain in pACT2 (Clontech). A total of 2 × 106 transformants were screened on plates lacking tryptophan, leucine, and histidine but containing 25 mM 3-aminotriazole (nacalai tesque) and were then assayed for β-galactosidase activity. Positive clones were picked, and the pACT2 library plasmids were recovered from individual clones and expanded in Escherichia coli. The cDNA inserts obtained in the plasmid were characterized by nucleotide sequence analysis with an automated DNA sequence analyzer (Applied Biosystems model 377).
Plasmid construction.
Expression plasmids for hemagglutinin (HA)-tagged ZIP kinase, FLAG-tagged ZIP kinase (mouse; aa 1 to 448), Myc-tagged ZIP kinase (mouse; aa 309 to 448), and FLAG-tagged ATF4 (human; full length) were constructed as follows. An expression plasmid, pEF-BOS, was digested with XbaI to remove the stuffer sequence, blunted with T4 polymerase, and ligated with SalI linker (24). Fragments which were epitope tagged at their NH2 terminus were generated by PCR, digested with SalI, and inserted into pEF-BOS. The sequence of each primer will be provided upon request.
To construct the deletion mutants containing ZIP kinase fused to the GAL4 DNA binding domain or transactivation domain, fragments were amplified by PCR and inserted in frame into the BamHI site of pAS2-1 or pACT2.
The mutant expression vectors, pEF-BOS-HA-ZIP kinase K42A and HA-ZIP kinase LA, were constructed by site-directed mutagenesis with a Transformer site-directed mutagenesis kit (Clontech). The mutagenic primer sequences are as follows: HA-ZIP kinase K42A, 5′-GGCATGGAGTATGCAGCTGCGTTCATCAAGAAGCGGCGC-3′; HA-ZIP kinase LA, 5′-GACGCGCTAGCCGCTCAGGCGGCCGCTGAGGTGCAATTCGCGCGCGACCTGGTGCGTGCGGC GGAGCAGGAACGGCTGCAG-3′. Other mutant expression vectors, pEF-BOS-FLAG-ZIP kinase K42A, pEF-BOS-FLAG-ZIP kinase K42A ΔLZ (aa 1 to 397), and pEF-BOS-FLAG-ZIP kinase K42A KD (aa 1 to 275), were constructed by PCR from pEF-BOS-HA-ZIP kinase K42A as the template.
The sequences of DNA fragments obtained by PCR were confirmed by DNA sequencing.
Transfection, immunoprecipitation, and Western blot analysis.
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% bovine calf serum. The COS-7 cells were transiently transfected with 5 μg of either pEF-BOS-mock, pEF-BOS-FLAG-ATF4, pEF-BOS-Myc-ZIP kinase, or a combination by lipofection as specified by the manufacturer (TaKaRa). For immunoprecipitation, 106 cells were harvested 36 h after transfection and lysed in 0.5% Nonidet P-40 lysis buffer (0.5% Nonidet P-40, 150 mM NaCl, 10 mM Tris HCl [pH 7.5], 1 mM EDTA). Cell lysates were precleared with protein A-Sepharose beads (Pharmacia) for several hours and then incubated with protein A-Sepharose together with either 10 μg of anti-FLAG M2 monoclonal antibody (MAb) (Eastman Kodak Co.) per ml or 4.0 μg of anti-Myc 9E10 MAb (Genosys Biotechnologies) per ml for 12 h by rotation. The beads were washed four times with lysis buffer. Immune complexes were eluted with Laemmli sample buffer, separated on a 4 to 20% polyacrylamide gradient gel, and transferred onto a nitrocellulose filter membrane. The filter was incubated with anti-FLAG MAb or anti-Myc MAb for 1 h before being washed three times in TBS-T (25 mM Tris HCl [pH 7.4], 137 mM NaCl, 2.7 mM KCl, 0.1% Tween 20), and then incubated with sheep anti-mouse peroxidase-conjugated secondary antibody (Amersham). After further washing with TBS-T, peroxidase activity was detected with the enhanced chemiluminescence system (Dupont).
Northern blot analysis.
Murine ZIP kinase cDNA was radiolabeled by the random-priming method and assayed by mouse multiple-tissue Northern blotting (Clontech) as specified by the manufacturer.
In vitro kinase assay.
COS-7 cells transiently transfected with HA epitope-tagged murine ZIP kinase or ZIP kinase mutants were lysed with 0.5% Nonidet P-40 lysis buffer. Following preclearing, the lysates were immunoprecipitated with 5 μg of anti-HA MAb 12CA5 (Boehringer Mannheim) per ml plus protein G-Sepharose beads (Pharmacia). The immunoprecipitates were washed four times with lysis buffer and once with kinase reaction buffer (10 mM MgCl2, 3 mM MnCl2, 10 mM Tris HCl [pH 7.2]). The in vitro kinase reaction was performed for 10 min at 30°C with kinase reaction buffer containing 10 μCi of [γ-32P]ATP (Amersham). Laemmli sample buffer was added to terminate the kinase reaction, and after being boiled, the samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Phosphorylated proteins were visualized by autoradiography.
Immunostaining of cells.
Transfected COS-7 cells were seeded on glass plates at a density of 20,000 cells/ml. After 48 h, the cells were washed twice with phosphate-buffered saline (PBS) before being incubated in a mixture of 3% paraformaldehyde and 0.3% Triton X-100 in PBS for 5 min for simultaneous fixing and permeabilization and then in 3% paraformaldehyde for 20 min. The cells were washed three times in PBS and blocked with 3% bovine serum albumin in PBS for 60 min. The cells were incubated with anti-FLAG M2 MAb (1:300) for 60 min. They were then washed three times in PBS and incubated with goat anti-mouse fluorescein-conjugated secondary antibodies (Tago) and 0.5 μg of 4′,6-diamidino-2-phenylindole (DAPI; Wako) per ml for an additional 30 min. The glass plates were washed three times in PBS and drained. Microscopy was carried out under fluorescent light.
X-Gal staining of cells.
NIH 3T3 cells were seeded at a density of 106 cells/100-mm dish in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine calf serum. After 12 h, the cells were cotransfected with 9.0 μg of a ZIP kinase expression vector (pEF-BOS-HA-ZIP kinase, pEF-BOS-HA-ZIP kinase K42A, or pEF-BOS-HA-ZIP kinase LA) and 1.0 μg of a β-galactosidase expression vector (pEF-BOS-lacZ) by lipofection. To identify β-galactosidase activity, the cells were washed once with PBS and fixed with 2.0% formaldehyde plus 0.2% glutaraldehyde in PBS for 5 min at 4°C. After being further washed with PBS, the cells were overlaid with a histochemical reaction mixture containing 1 mg of 5-chloro-4-bromo-3-indolyl-β-d-galactopyranoside (X-Gal) per ml, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 2 mM MgCl2 in PBS. Photographs of stained cells were taken with an Olympus IX70 microscope.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL and GenBank nucleotide sequence databases with accession no. AB007143 (mouse ZIP kinase) and AB007144 (human ZIP kinase).
RESULTS
Screening of proteins interacting with ATF4.
To identify proteins that interact with ATF4, we performed a yeast two-hybrid screening procedure with the leucine zipper portion of mouse ATF4 fused to the DNA binding domain of GAL4 as bait (11). We screened mouse brain and human placenta cDNA libraries that expressed proteins fused to the GAL4 transcriptional activation domain. From 2 × 106 yeast transformants, we identified about 100 clones that could activate reporter genes. As expected, most of these clones encoded bZip transcription factors. These include C/EBPβ (NF-IL6), which is known to associate with ATF4 in mammalian cells (1, 35), C/EBPα, C/EBPδ (NF-IL6β), CHOP, JunB, JunD, and ATF3 (15, 16, 28). These results indicate that this bait plasmid can specifically detect interactions between ATF4 and its heterodimerizing partners within yeast cells. The yeast two-hybrid screening also yielded seven cDNA clones from a mouse brain cDNA library and two from a human placenta cDNA library with novel nucleotide sequences. These clones contained independent cDNAs derived from the same mRNA. Full-length cDNA clones were isolated from mouse liver and human placenta λgt11 libraries. Their nucleotide sequences and conceptual translations are shown in Fig. 1. The lengths of the cDNA clones obtained matched those of the mRNAs detected by Northern blotting, although no preceding in-frame stop codons were noted. Taken together, these results suggest that these sequences harbor full-length cDNAs. The mouse ZIP kinase mRNA contains an open reading frame of 1,344 bp and is predicted to encode a protein of 448 aa with a calculated molecular mass of 51.4 kDa (Fig. 1A). On the other hand, the human ZIP kinase cDNA is 1,362 bp and encodes a protein of 454 aa with a calculated molecular mass of 52.5 kDa (Fig. 1B). In the 3′ untranslated region of both ZIP kinases, a typical polyadenylation signal, AATAAA, is found upstream of the poly(A) tail. A comparison study reveals 85% identity between mouse and human ZIP kinases at the amino acid level (Fig. 1C).
FIG. 1.
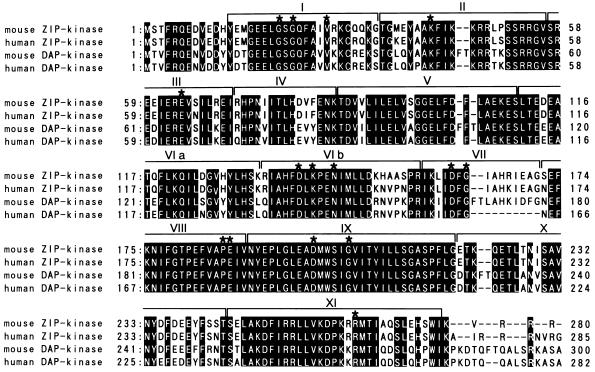
(A and B) Nucleotide and deduced amino acid sequences of murine ZIP kinase (A) and human ZIP kinase (B). Initiation (ATG) and stop (TGA) codons are boxed. Polyadenylation signals (AATAAA) are underlined. (C) Comparison of the amino acid sequences of murine and human ZIP kinases. Identical amino acid residues are indicated by boxes. The putative kinase domains and the regions of the leucine zipper domains are indicated.
ZIP kinase is a serine/threonine kinase and carries the leucine zipper domain in the C-terminal region.
Comparative analysis of the deduced amino acid sequences of the murine and human ZIP kinases revealed that ZIP kinase contained several known domains. The N-terminal region showed homology to the catalytic domain of serine/threonine kinases. The domain spans 263 aa from positions 13 to 275 in both mouse and human ZIP kinases (Fig. 1C). Furthermore, this domain is classically composed of 11 subdomains and contains all of the residues that are normally conserved in serine/threonine kinases (18) (Fig. 2).
FIG. 2.
Alignment of the catalytic domain of ZIP kinase with that of DAP kinase. A catalytic domain of the mouse ZIP kinase is aligned with the corresponding domain of the human ZIP kinase, mouse DAP kinase, and human DAP kinase. The kinase subdomains numbered I to XI are indicated. The conserved amino acid residues within the kinase domain are indicated by asterisks. The solid backgrounds indicate identical amino acid residues.
Secondary-structure predictions showed that the C terminus of ZIP kinase forms an α-helical structure from positions 408 to 445 in the mouse ZIP kinase and 413 to 450 in human ZIP kinase. These portions showed homology to myosin heavy chain, which contains coiled-coil structures (6). In addition, these domains contained heptad repeats of three hydrophobic amino acid residues with a potential leucine zipper. These residues are conserved within the mouse and human ZIP kinases (Fig. 1C). Thus, the leucine zipper structure in the ZIP kinase may be important for interactions with ATF4 and other leucine zipper proteins or for homodimerization. Therefore, we termed this novel serine/threonine kinase zipper-interacting protein kinase (ZIP kinase).
A catalytic domain of ZIP kinase is closely related to that of DAP kinase, which is a mediator of cell death induced by gamma interferon.
When aligned with sequences from the protein sequence database by using the BLAST and FASTA programs, the ZIP kinase catalytic domain showed the highest similarity to that of the recently cloned death-associated protein kinase (DAP kinase) (9). DAP kinase was initially identified as the protein encoded by the gene whose reduced expression by antisense cDNA transfection protected HeLa cells from gamma interferon-induced cell death. A catalytic domain of mouse ZIP kinase has 79.0% of its amino acid sequence identical to that of mouse DAP kinase, and human ZIP kinase has 79.5% of its sequence identical to that of human DAP kinase (Fig. 2). In subdomain IX, all the residues were identical between ZIP kinase and DAP kinase. The deduced amino acid structure of DAP kinase predicts several functional motifs. The N terminus is composed of a serine/threonine kinase domain followed by a region with homology to the calmodulin regulatory domains. However, the region immediately C-terminal to the catalytic domain of ZIP kinase showed no homology to any calmodulin regulatory proteins. In addition, it is interesting that both genes have distinct domains in their C-terminal regions. ZIP kinase has a leucine zipper domain, in contrast to DAP kinase, which contains an ankyrin repeat motif and a death domain. Both proteins are expected to dimerize with other proteins through these domains.
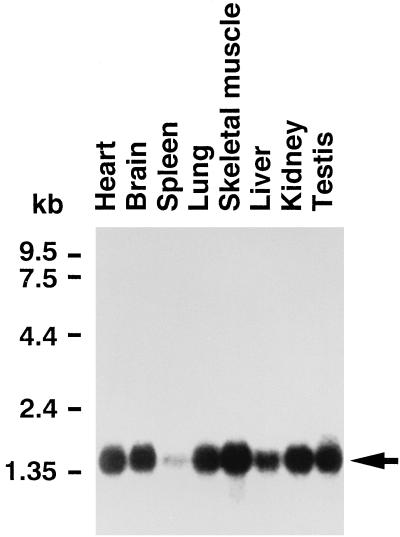
The ZIP kinase expression profile was examined by Northern blot analysis. As shown in Fig. 3, the single prominent 1.4-kbp band of mouse ZIP kinase mRNA was detected ubiquitously in various tissues, although the level of ZIP kinase mRNA was lower in spleen.
FIG. 3.
Expression profile of ZIP kinase mRNA by Northern blot analysis. Poly(A) RNAs (2 μg) from various murine tissues were loaded in each lane. The filter was hybridized with the radiolabeled mouse ZIP kinase probe and exposed to X-ray film. The major transcript of 1.4 kbp is indicated by an arrow.
ZIP kinase interacts with ATF4 in vitro and in vivo.
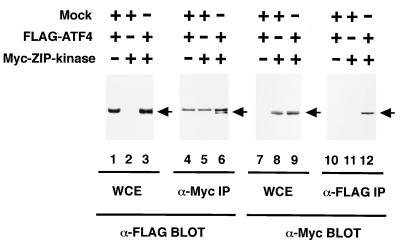
To confirm that ZIP kinase associates with ATF4 in mammalian cells, COS-7 cells were transiently transfected with expression plasmids for a Myc epitope-tagged ZIP kinase (Myc-ZIP kinase) and FLAG-tagged ATF4 (FLAG-ATF4). COS-7 cells were lysed, immunoprecipitated with anti-Myc or anti-FLAG MAb, and blotted with anti-FLAG or anti-Myc MAb. Western blot analysis with anti-FLAG MAb revealed that FLAG-ATF4 was coimmunoprecipitated with anti-Myc MAb when the cells were cotransfected with FLAG-ATF4 and Myc-ZIP kinase expression vectors (Fig. 4). To verify the interaction further, we carried out reciprocal experiments. The results of an immunoblotting study with anti-Myc MAb indicate that Myc-ZIP kinase was also coimmunoprecipitated with anti-FLAG MAb.
FIG. 4.
ZIP kinase interacts with ATF4 in mammalian cells. COS-7 cells were transiently transfected with 5 μg of the plasmids indicated. The cells were harvested 36 h after transfection and lysed in the lysis buffer. Whole-cell extracts (WCE) were separated by SDS-PAGE and then immunoblotted with anti-FLAG M2 MAb (lanes 1 through 3) or anti-Myc 9E10 MAb (lanes 7 through 9). The same lysates were immunoprecipitated with either anti-Myc MAb or anti-FLAG MAb, and the immunoprecipitates were blotted with either anti-FLAG MAb (lane 4 through 6) or anti-Myc MAb (lanes 10 through 12). The bands corresponding to Myc-ZIP kinase and FLAG-ATF4 proteins are indicated by arrows.
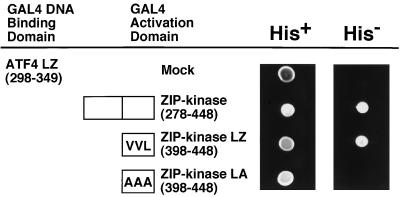
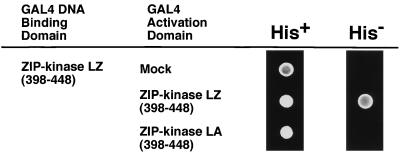
To determine which portions of the ZIP kinase are necessary for interaction with ATF4, we determined the ability of ATF4 to interact with several ZIP kinase mutants in the two-hybrid system. As shown in Fig. 5, ATF4 interacts with the leucine zipper domain of ZIP kinase (ZIP kinase LZ), but not when there are mutations within this domain (ZIP kinase LA, in which valine 422, valine 429, and leucine 436 are changed to alanines). These results demonstrate that ZIP kinase interacts with ATF4 in vivo via its leucine zipper domain.
FIG. 5.
ZIP kinase and ATF4 interact via their leucine zippers. The C-terminal region (mouse ZIP kinase 278–448), the leucine zipper domain (ZIP kinase LZ), and the mutant with substitutions in the leucine zipper domain (ZIP kinase LA) were constructed in pACT2 to express the fusion protein with the GAL4 transactivation domain. Each construct was cotransformed into yeast Y190 cells with a plasmid expressing the leucine zipper domain of ATF4 fused to the GAL4 DNA binding domain. Growth of yeast on a synthetic dextrose agar plate lacking leucine, tryptophan, and histidine (His−) is indicative of a protein-protein interaction, while growth in the absence of leucine and tryptophan (His+) indicates a control.
The leucine zipper domain is required for self-association of ZIP kinase.
The C-terminal region of ZIP kinase contains a leucine zipper motif which mediates protein-protein interactions. The yeast two-hybrid assay was used to investigate whether ZIP kinase can self-associate through the C-terminal regions. The reporter genes were activated when the ZIP kinase leucine zipper bait was expressed together with the ZIP kinase leucine zipper domain. On the other hand, the ZIP kinase LA mutant construct could not activate the reporter genes when cotransformed with the ZIP kinase leucine zipper bait plasmid (Fig. 6). Taken together, these results demonstrate that ZIP kinase can self-associate and that this ability is dependent on the intact structure of the leucine zipper motif.
FIG. 6.
Leucine zipper-mediated self-association of ZIP kinase. A plasmid expressing the leucine zipper domain of murine ZIP kinase fused to the GAL4 DNA binding domain was cotransformed with a plasmid expressing either the ZIP kinase LZ or ZIP kinase LA mutant fused to the GAL4 transactivation domain. Growth in the absence of leucine, tryptophan, and histidine (His−) is indicative of the interaction, while growth in the absence of leucine and tryptophan (His+) indicates a control.
An in vitro kinase assay confirmed that ZIP kinase possesses a kinase activity.
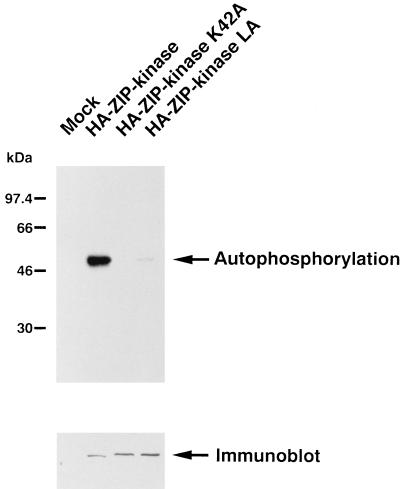
To begin to assess the biochemical functions and regulation of ZIP kinase, we developed an in vitro kinase assay. To express ZIP kinase, an expression vector for an HA-tagged ZIP kinase cloned into pEF-BOS was prepared. Two versions of ZIP kinase mutants were also prepared in this assay. The ZIP kinase K42A mutant carried a mutation in which a conserved lysine in the kinase subdomain II was changed to alanine. A point mutation of this portion in a number of kinases was shown to block the phosphotransfer reaction, giving rise to a catalytically inactive protein kinase (18). Another mutant version, a ZIP kinase LA mutant, had valine 422, valine 429, and leucine 436 changed to alanines in the leucine zipper domain, which would result in the loss of homodimerization. All the constructs were transiently transfected into COS-7 cells by lipofection. Lysates of the transfected cells were immunoprecipitated with anti-HA MAb and subjected to an in vitro kinase reaction in the presence of Mg2+ and Mn2+. A single prominent band at the expected size of 52 kDa was detected by SDS-PAGE only in the cell lysate transfected with the wild-type ZIP kinase (Fig. 7, upper panel). In contrast, phosphorylation of the ZIP kinase K42A mutant was not detected, and the in vitro kinase activity of the ZIP kinase LA mutant was dramatically decreased compared with that of wild-type ZIP kinase. These results indicate that ZIP kinase displays an intrinsic kinase activity, undergoes autophosphorylation, and acts as an active kinase when it homodimerizes through its leucine zipper structure. The amounts of protein loaded were determined by Western blot analysis with anti-HA MAb (Fig. 7, lower panel).
FIG. 7.
Kinase activity of ZIP kinase. HA-tagged ZIP kinase and its mutants were transiently transfected into COS-7 cells. Cell lysates were immunoprecipitated with anti-HA MAb (12CA5) and assayed for in vitro kinase activity. The upper panel shows the autophosphorylation of ZIP kinase. Relative molecular mass standards are indicated on the left. The amounts of HA-ZIP kinase and mutant proteins were shown to be the same by Western blotting with anti-HA MAb (lower panel).
ZIP kinase is localized to the nuclei.
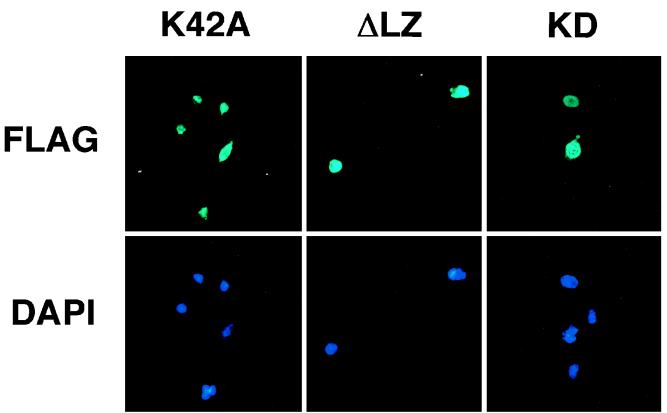
To define the subcellular localization of ZIP kinase, we transiently transfected the FLAG-tagged ZIP kinase K42A mutant expression vector into COS-7 cells. The ZIP kinase K42A construct was chosen to avoid morphological changes of the cells. Transfected COS-7 cells were double stained with anti-FLAG M2 MAb (for ZIP kinase) and DAPI (for nuclei). Subsequent microscopy analysis demonstrated that the FLAG-ZIP kinase K42A protein was localized exclusively to the nucleus (Fig. 8, K42A). Furthermore, we also tested the cellular localization of two deletion mutants and found that the deletion mutant lacking the leucine zipper domain (ZIP kinase K42A aa 1 to 397), which is not able to form the homodimer, was localized to the nuclei, as in the case of full-length ZIP kinase (Fig. 8, ΔLZ). Furthermore, the nuclear localization was also detected in cells which expressed only the kinase domain (ZIP kinase K42A aa 1 to 275), suggesting that the kinase domain is sufficient to direct the nuclear localization in cells (Fig. 8, KD). These results demonstrate that ZIP kinase is a nuclear serine/threonine kinase and that catalytic activity or leucine zipper-mediated homodimerization is not required for nuclear localization.
FIG. 8.
Cellular localization of ZIP kinase. COS-7 cells transiently transfected with the indicated FLAG-tagged ZIP kinase mutant plasmids were indirectly immunostained with anti-FLAG MAb and fluorescein-conjugated secondary antibody and simultaneously stained with DAPI to locate the nucleus. ΔLZ, mouse ZIP-kinase K42A aa 1 to 397; KD, ZIP kinase K42A aa 1 to 275.
Ectopic expression of ZIP kinase induces cell death in the NIH 3T3 line.
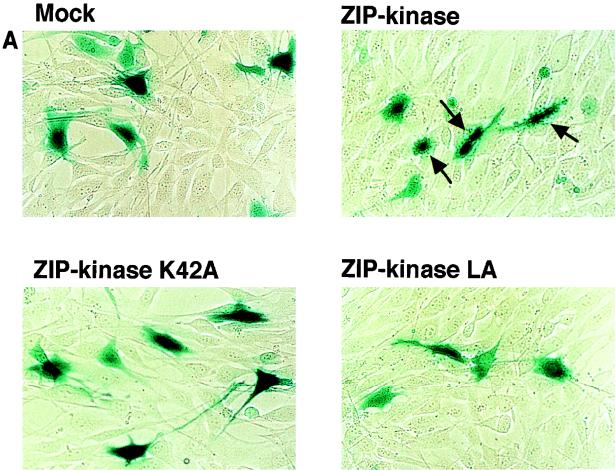
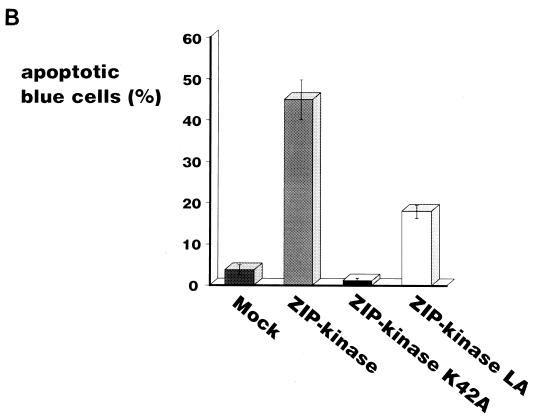
DAP kinase was first identified as a positive mediator of cell death. This was based on the findings that the reduced expression of DAP kinase by antisense mRNA protected HeLa cells from apoptotic cell death induced by gamma interferon (9) and that an elevated level of DAP kinase expression induced the death of HeLa cells without external stimuli (8). For this reason, it was very interesting to examine whether ectopic expression of ZIP kinase directly causes cell death. To express ZIP kinase in mammalian cells, we prepared expression vectors for the HA epitope-tagged wild-type ZIP kinase (HA-ZIP kinase), the catalytically inactive form of ZIP kinase (HA-ZIP kinase K42A), and the mutant with alterations in the leucine zipper domain so that it was not able to self-associate (HA-ZIP kinase LA). NIH 3T3 fibroblasts were transiently cotransfected with each ZIP kinase construct and a plasmid containing the β-galactosidase gene (pEF-BOS-lacZ). Transfected cells were identified by histochemical staining with an X-Gal solution. As shown in Fig. 9A, it was found that the blue-stained cells transfected with wild-type ZIP kinase exhibited the morphological changes of apoptotic cells, as characterized by their shrunken appearance, condensed chromatin, and membrane blebbing (44.9% of blue cells [Fig. 9B]). By contrast, a background level of 3.7% apoptotic cells was detected in the mock plasmid-transfected control. This indicates that ZIP kinase positively regulates apoptotic cell death. The transfectants with the ZIP kinase K42A mutant displayed mainly normal-appearing cells, and the number of apoptotic cells was almost the same (1.0%) as that of the mock transfectants. This result suggests that the catalytic activity of ZIP kinase is required for its ability to mediate apoptotic cell death. Furthermore, the number of apoptotic cells (17.8% of blue cells) was slightly decreased when NIH 3T3 cells were transfected with the ZIP kinase LA mutant plasmid, indicating that the leucine zipper-mediated homodimerization of ZIP kinase was required for the induction of apoptosis. These results strongly suggest that the catalytic activity of ZIP kinase correlates with the induction of cell death.
FIG. 9.
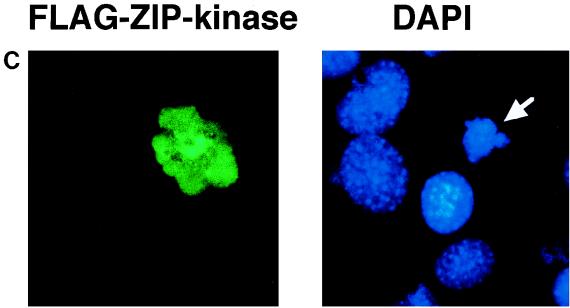
Ectopic expression of ZIP kinase causes the morphological changes of apoptotic cell death in NIH 3T3 cells. (A) An expression plasmid of ZIP kinase or ZIP kinase mutants was transiently cotransfected together with a plasmid expressing the β-galactosidase gene into NIH 3T3 cells. After 36 h of transfection, the cells were stained with a X-Gal solution to visualize the cells expressing the β-galactosidase gene. Cells with the apoptotic phenotype are indicated by the arrows. (B) Quantitative representation of ZIP kinase-induced cell death in NIH 3T3 cells. The data presented shows the percentage of blue-stained cells with the apoptotic phenotype relative to the total number of blue cells counted. The results are from three independent experiments. (C) NIH 3T3 cells transiently transfected with wild-type FLAG-ZIP kinase were doubly stained with anti-FLAG MAb and DAPI 48 h after transfection. The nuclei of transfected cells were significantly condensed (arrow).
To investigate the nuclear morphology of cells expressing wild-type ZIP kinase, we doubly stained the cells with anti-FLAG MAb (for ZIP kinase) and DAPI (for nuclei). As seen in Fig. 9C, the nuclei of cells transiently transfected with active ZIP kinase were significantly condensed compared with those of nontransfected cells, indicating the apoptotic changes in the nuclei.
DISCUSSION
The known function of the leucine zipper domain is to mediate homodimerization or heterodimerization with other proteins containing this domain. We performed a yeast two-hybrid screening assay with the leucine zipper domain of ATF4, a member of the ATF/CREB family, to identify novel ATF-interacting proteins. Here we report the cloning and characterization of a novel leucine zipper protein, ZIP kinase, that is a regulator of cell death. ZIP kinase is composed of a C-terminal leucine zipper domain and an N-terminal catalytic domain of serine/threonine protein kinase. Mutation studies indicate that an intact leucine zipper structure is required for homodimerization or heterodimerization with ATF4. An in vitro kinase assay shows that ZIP kinase has an intrinsic kinase activity. This catalytic activity is abolished by a point mutation in which a critical lysine residue in subdomain II is changed to alanine. Moreover, the activity is also abolished by mutations within the leucine zipper domain, indicating that leucine zipper-mediated homodimerization is necessary for activation of the kinase. We have found that ectopic expression of ZIP kinase induces the morphological changes of apoptosis in NIH 3T3 cells. This indicates that the function of this gene is linked to cell death. The kinase-negative ZIP kinase mutant with substitutions in the catalytic domain does not promote cell death, indicating that the catalytic activity is critical for the induction of cell death. Interestingly, the catalytic domain of ZIP kinase is closely related to that of the recently identified DAP kinase (9). DAP kinase was initially identified as being encoded by a gene whose reduced expression, mediated by antisense cDNA transfection, protects HeLa cells from gamma interferon-induced cell death. Overexpression of DAP kinase induces apoptosis in HeLa cells, while the kinase-negative mutant protects HeLa cells from gamma interferon-induced cell death (8). Thus, both ZIP kinase and DAP kinase represent a novel class of cell death-related kinases. Although both kinases are closely related in their catalytic domains, other regions are distinct and do not share any amino acid homology. DAP kinase is a Ca2+/calmodulin-dependent serine/threonine protein kinase and possesses two known major domains characterized by eight ankyrin repeats and a death domain but lacks the leucine zipper domain. The domains may mediate the interaction with putative effector molecules or downstream substrates. Although ZIP kinase homodimerizes, it is not known whether DAP kinase can self-associate through its own ankyrin repeats or its death domain. In addition, these kinases differ in their patterns of intracellular localization. ZIP kinase localizes in the nuclei, suggesting that nuclear proteins are the potential downstream substrates. In contrast, DAP kinase associates tightly with cytoskeletal elements. These results suggest that they play distinct roles in the regulation of apoptosis in vivo and associate with distinct downstream substrates or regulators. Regarding the nuclear localization of ZIP kinase, it is noteworthy that there is a stretch of basic residues in subdomain II of ZIP kinase which was conserved between ZIP kinase and DAP kinase. In fact, a deletion mutant lacking the C-terminal region of ZIP kinase was localized to the nucleus. Furthermore, deletion studies of DAP kinase demonstrated that the N-terminal kinase domain, but not the full-length kinase, was localized exclusively to the nuclei (8). It is likely that the highly conserved sequences in subdomain II are sufficient for the nuclear localization, although further mutagenesis experiments are required for confirmation.
Although there are many kinases which mediate cell growth, only a few protein kinases related to apoptosis, besides DAP and ZIP kinase, have been identified. Recently, JNKs were shown to be involved in apoptosis. Apoptotic stimuli such as TNF-α, Fas, ceramide, and UV irradiation activate JNKs (36). Expression of the molecular inhibitor of the JNK pathway could block apoptosis (36, 38). A major substrate of JNK, c-Jun, promotes apoptosis of NIH 3T3 cells (5). ASK1 is also a protein kinase involved in apoptosis (20). ASK1 was identified as a member of the mitogen-activated protein kinase kinase kinase (MAPKKK) family that activated two distinct signals involved in SEK1-JNK and MKK3/MAPKK6-p38 pathways. Inducible expression of ASK1 by stable transfection with metallothionein promoter-based expression plasmids induced apoptotic cell death. Moreover, ASK1 was activated by proinflammatory cytokines such as TNF-α, and a catalytically inactive form of ASK1 prevented TNF-α-induced cell death.
A major challenge for the future is to clarify the signaling pathway mediated by ZIP kinase, i.e., to identify the specific substrates for ZIP kinase which are phosphorylated during cell death as well as the signaling molecules upstream of ZIP kinase. Furthermore, considering that ZIP kinase homodimerizes to be activated, it is necessary to understand how formation of the homodimer might be controlled. One hypothesis is that the ZIP kinase normally interacts with a regulator(s) that interferes with the homodimerization. The inhibitory action of the regulator(s) may be blocked by events such as ligand binding, phosphorylation of a certain residue(s), or cleavage by a protease. ATF4 may be one of the candidates that participates in the negative regulation of ZIP kinase, since ATF4 binds to the leucine zipper domain of ZIP kinase and may interfere with the homodimerization of the kinase. In fact, both proteins colocalize to the nucleus. Cotransfection of the expression vectors for these proteins, along with the neomycin resistance gene, showed that ATF4 expression increased the number of neomycin-resistant colonies, suggesting that ATF4 blocks the apoptotic activity mediated by ZIP kinase (unpublished data). However, we do not have any direct evidence indicating that ATF4 is a physiological inhibitor of ZIP kinase in vivo.
It has been implied that tumorigenesis is controlled by the mutations of death-controlling genes such as bcl-2 and p53 (23, 34, 37). Furthermore, deregulated c-Myc can induce apoptosis in certain cancer cell lines (4, 12). Further expression studies in different cancer cell lines and chromosomal mapping of ZIP kinase should be performed to investigate its possible role in tumorigenesis. With a view to the role of ZIP kinase in the regulation of apoptosis, it seems important to design drugs that can activate ZIP kinase, since such drugs could be used for cancer therapy by inducing apoptosis of cancer cells.
ACKNOWLEDGMENTS
We thank Kin-ichi Nakashima for various suggestions and K. Ohgishi and T. Aoki for excellent secretarial assistance.
This work was supported by CREST (Core Research for Evolutional Science and Technology) of Japan Science and Technology Corporation (JST) and grants from the Ministry of Education of Japan.
REFERENCES
- 1.Akira S, Issiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P. Kinase cascades regulating entry into apoptosis. Microbiol Mol Biol Rev. 1997;61:33–46. doi: 10.1128/mmbr.61.1.33-46.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askew D, Ashmun R, Simmons B, Cleveland J. Constitutive c-myc expression in an IL-3-dependent myeloid cell line supresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 5.Bossy-Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen C, Parry D A D. α-helical coiled-coils: a wide-spread motif in proteins. Trends Biochem Sci. 1986;11:245–248. [Google Scholar]
- 7.Cohen J J, Duke R C, Fadok V A, Sellins K S. Apoptosis and programmed cell death in immunity. Annu Rev Immunol. 1992;10:267–293. doi: 10.1146/annurev.iy.10.040192.001411. [DOI] [PubMed] [Google Scholar]
- 8.Cohen O, Feinstein E, Kimchi A. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J. 1997;16:998–1008. doi: 10.1093/emboj/16.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deiss L P, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the γ interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 10.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 11.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type I catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 12.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 13.Goillot E, Raingeaud J, Ranger A, Tepper R I, Davis R J, Harlow E, Sanchez I. Mitogen-activated protein kinase-mediated Fas apoptotic signaling pathway. Proc Natl Acad Sci USA. 1997;94:3302–3307. doi: 10.1073/pnas.94.7.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulbins E R, Bissonnette R, Mahboubi A, Martin S, Nishioka W, Brunner T, Baier G, Baier-Bitterlich G, Byrd C, Lang F, Kolesnick R, Altman A, Green D. Fas-induced apoptosis is mediated via a ceramide-initiated Ras signaling pathway. Immunity. 1995;2:341–351. doi: 10.1016/1074-7613(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 15.Hai T, Liu F, Coukos W J, Green M R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 16.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haimovitz-Friedman A, Kan C C, Ehleiter D, Persaud R S, McLoughlin M, Fuks Z, Kolensnick R N. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. J Exp Med. 1994;180:525–535. doi: 10.1084/jem.180.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 19.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 20.Ichijo H, Nishida E, Irie K, Dijke P T, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 21.Kerr J F R, Wyllie A H, Currie A R. Apoptosis: a basic biological phenomenon with ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Christakos S, Small M B. Apoptosis and signal transduction: clues to molecular mechanism. Curr Opin Cell Biol. 1993;5:286–291. doi: 10.1016/0955-0674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 23.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. p53 is required for radiation induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 27.Raff M C, Barres B A, Burne J F, Coles H S, Ishizaki Y, Jacobson M D. Programmed cell death and the control of cell survival: lessons from the nervous system. Science. 1993;262:695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 28.Ron D, Habener J F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 29.Stanger B Z, Leder P, Lee T-H, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 30.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 31.Tepper C G, Jayadev S, Liu B, Bielawska A, Wolff R, Yonehara S, Hannun Y A, Seldin M F. Role for ceramide as an endogenous mediator of Fas-induced cytotoxicity. Proc Natl Acad Sci USA. 1995;92:8443–8447. doi: 10.1073/pnas.92.18.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson C B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 33.Tsujimoto A, Nyunoya H, Morita T, Sato T, Shimotohno K. Isolation of cDNAs for DNA-binding proteins which specifically bind to a tax-responsive enhancer element in the long terminal repeat of human T-cell leukemia virus type I. J Virol. 1991;65:1420–1426. doi: 10.1128/jvi.65.3.1420-1426.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsujimoto Y, Cossman J, Jaffe E, Croce C M. Involvement of bcl-2 in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 35.Vallejo M, Ron D, Miller C P, Habener J F. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc Natl Acad Sci USA. 1993;90:4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Haimovitz-Freidman A, Fuks Z, Kolesnick R N. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 37.White E. Life, death and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan J, Shaham L, Ledoux S, Ellis H M, Horvitz H R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]