Summary
Candida auris is an emerging multidrug-resistant yeast associated with invasive infection in healthcare settings. Recently, C auris cases in the United States have been detected in 11 states with the majority of cases in New York, New Jersey and Illinois. Rapid and accurate identification of C auris is critical for patient care and the implementation of public health measures to control the spread of infection. Our aim was to develop and validate a rapid DNA extraction method using the Roche MagNA Pure 96 instrument and a TaqMan real-time PCR assay for reliable, high-throughput identification of C auris. We evaluated 247 patient dermal swab samples previously analysed by culture/MALDI-TOF. The diagnostic sensitivity and specificity were 93.6% and 97.2%, respectively. The assay was highly reproducible with a detection limit of 1 C auris CFU/10 μL. A receiver operating characteristic curve analysis of the real-time PCR data showed an area of 0.982 under the curve, with a CT cut-off value of ≤37.0. The turnaround time from DNA extraction to real-time PCR results was approximately 200 samples/day. In conclusion, we successfully validated a rapid and high-throughput method for accurate and reproducible identification of C auris with a significantly reduced turnaround time compared to culture/MALDI-TOF based methods.
Keywords: Candida auris, high-throughput rapid DNA extraction, TaqMan real-time PCR
1 |. INTRODUCTION
Candida auris (C auris) is a multidrug-resistant fungus that causes invasive infection and is associated with nosocomial outbreaks in healthcare settings.1,2 First reported in Japan in 2009,3 C auris has been found in multiple countries, including the United States. Recently, C auris cases in the United States have been detected in 11 different states but are concentrated in the New York City, New Jersey and Chicago areas.4 As of September 2018, a total of 433 confirmed and 30 probable cases from 11 US States have been reported to the Centers for Disease Control and Prevention (CDC). The majority of confirmed cases were identified in New York (239), New Jersey (94) and Illinois (80), respectively.5 Whole genome analysis indicates the simultaneous emergence of all four clades of C auris on three continents.6 Candida auris is a growing public health threat and is associated with high mortality for patients in healthcare settings with serious illnesses.7,8 Most C auris isolates show high minimum inhibitory concentrations (MIC) to the antifungal drugs Fluconazole and Amphotericin B9,10 and reduced susceptibility to Voriconazole, Caspofungin and Flucytosine.11 It is reported that C auris can be misidentified as Candida haemulonii, Candida duobushaemulonii, Candida catenulata, Candida famata, Candida guilliermondii, Candida lusitaniae and Candida parapsilosis when using conventional microbiological methods for yeast identification, for example VITEK 2 YST, API 20C, BD Phoenix yeast identification system and MicroScan.11,12 Accurate identification is possible with matrix-assisted laser desorption ionisation-time of flight mass spectrometry (MALDI-TOF MS) but is very time consuming because growth of an isolate is needed for analysis. This method also requires a reference database for accurate identification.13,14 Rapid and accurate identification of C auris is critical for patient care and for timely implementation of public health measures to control the spread of infection.
Because the cell wall of yeast is so tough and rigid, special methods are required for DNA extraction. The most common methods are bead beating and enzymatic lysis. These additional processing steps are time consuming and labour intensive and so are not ideal for high-throughput DNA extraction. Our aim was to validate a reproducible and rapid DNA extraction method using the Roche MagNA Pure 96 instrument and pair it with a TaqMan real-time PCR assay for reliable, high-throughput identification of C auris. We evaluated this method with 247 patient dermal swab samples previously analysed by culture and MALDI-TOF by the Mycotic Diseases Branch, CDC (Atlanta, GA).
2 |. MATERIALS AND METHODS
2.1 |. Clinical samples
These studies were performed with the approval of the Institutional Research Human Subjects Review Board at the CDC (Atlanta, GA). Patient dermal swabs were collected using a single BD Eswab containing Amies buffer (Cat # 220245, BD Diagnostics, Franklin Lakes, NJ; USA) by healthcare facilities and submitted to the Mycotic Diseases Branch, CDC (Atlanta, GA) for C auris screening using culture and MALDI-TOF.15 We evaluated 247 de-identified dermal swab samples (Positive = 73; Negative = 174) provided by the Mycotic Diseases Branch, CDC (Atlanta, GA). The swab samples were previously analysed by culture and MALDI-TOF and stored at 4°C before DNA extraction and real-time PCR assay analysis.
2.2 |. DNA extraction
DNA extraction was performed with the MagNA Pure 96 automated extraction system (Roche Diagnostic, Indianapolis, IN; USA), using the DNA and Viral NA Small Volume Kit (Cat # 6543588001, Roche Diagnostic, Indianapolis, IN; USA). The samples were pretreated prior to the automated processing. For the pretreatment, a 100 μL volume from each patient swab or spiked sample was transferred to a tube containing 100 μL Bacterial Lysis Buffer (Cat # 04659180001; Roche Diagnostic, Indianapolis, IN; USA) and 20 μL Proteinase K (Cat # EO0492; ThermoFisher Scientific, Waltham, MA; USA). The tubes were vortexed for a few seconds and incubated for 10 minutes at 65°C while shaking at 250 RPM. Following incubation, 200 μL of each sample was transferred to a MagNA Pure 96 Processing Cartridge (Cat # 6374921001; Roche Diagnostic, Indianapolis, IN; USA) per the manufacturer’s instructions. DNA extraction was performed following the Pathogen Universal 200 3.1 protocol with an elution volume of 50 μL.
2.3 |. Real-time PCR assay for identification of C auris
Candida auris identification was performed by real-time PCR assay using the ABI 7500 Fast Dx qPCR system (Applied Biosystems Inc., Foster City, CA; USA.). We followed the previously published TaqMan procedure for identification of C auris.16 The C auris ITS2 gene-specific primers (CAURF: 5′-CAG ACG TGA ATC ATC GAA TCT-3′; CAURR: 5′-TTT CGT GCA AGC TGT AAT TT-3′), probe (CAURP: 5′-FAM-AAT CTT CGC-ZEN-GGT GGC GTT GCA TTC A -IFQ-3′) and Bicoid gene-specific primers (BICF: 5′-CAG CTT GCA GAC TCT TAG-3′; BICR: 5′-GAA TGA CTC GCT GTA GTG-3′), probe (BICP: 5′-Quas570-AAC GCT TTG ACT CCG TCA CCC A -IRQ-3′) used in this study were described previously.16 The primers and probes were obtained from the Biotechnology Core Facility Branch, CDC (Atlanta, GA), and Bicoid plasmid was obtained from Addgene (Cat # 34340; Addgene, Watertown, MA; USA). 1 μL (1 pg/μL) Bicoid DNA was added to each PCR reaction mixture as an inhibition control. Positive controls (C auris; B11220) and negative controls (Amies buffer) were included in each real-time PCR assay. All samples were analysed in triplicates. The threshold (CT) value of ≤37 was considered a positive and >37 was considered a negative result.
2.4 |. Evaluation of analytical sensitivity, specificity and reproducibility
Candida auris isolate B11220 was used to evaluate the analytical sensitivity of the real-time PCR assay. Candida auris was grown overnight on a Sabouraud’s Dextrose Agar plate. Approximately 10–15 colonies of C auris were collected from the plate and suspended in Amies buffer. An automated Cellometer K2 cell counter (Nexcelom Bioscience, Lawrence, MA; USA) was used for cell counting. Serial dilutions of the cell suspension (from 5 × 106 to 5 × 102 CFU/mL) were prepared for evaluation of instrument response. For analytical sensitivity, five dilutions (from 5 × 103 to 10 CFU/mL) were evaluated with twenty replicates of each dilutions. Reproducibility was evaluated by performing six independent DNA extractions and subsequent real-time PCR assays with 20 swab samples (positive = 10, negative = 10) on six different days by two individuals. The analytical specificity was evaluated by spiking C auris (final concentration 500 CFU/mL) in twelve swabs, previously analysed by MALDI-TOF, and known to contain Malassezia furfur, Candida orthopsilosis, Candida glabrata and Candida parapsilosis.
2.5 |. Evaluation of specificity with bacterial strains and human specimen control
We also evaluated analytical specificity of the real-time PCR assay with Escherichia coli (ATCC 25922), Proteus mirabilis (ATCC 12453), Staphylococcus epidermidis (ATCC 14990), Staphylococcus aureus (ATCC 29213) and Human Specimen Control (a non-infectious cultured human cell material). The cells from these strains and the HSC were spiked in Amies buffer.
3 |. RESULTS
3.1 |. Evaluation of analytical sensitivity, specificity and reproducibility
We have tested the real-time PCR assay on all four clades of C auris (Clade I—South Asia; Clade II—East Asia; Clade III—South Africa and Clade IV—South America), and this method is able to accurately identify all four clades as C auris does not differentiate between the four clades. We also assessed the instrument response for real-time PCR assay with 10-fold serially diluted C auris spiked samples. The instruments showed a mean 3.30-fold (standard deviation: 0.415) increase in CT value with every 10-fold dilution. The equation for the linear regression and R-value were y = −3.376x + 45.703 and 0.9981, respectively (Figure 1). The reproducibility study showed the co-efficient of variance (Cv) < 0.05 for all samples except one which had a slightly higher Cv of 0.059. The real-time PCR assay was highly reproducible and sensitive with a detection limit of 1 C auris CFU/10 μL (Figure 2). This method was highly specific, as none of the other Candida species and bacterial strains showed any cross-reactivity (See Materials and Methods for the list of microorganisms used for evaluation of specificity).
FIGURE 1.
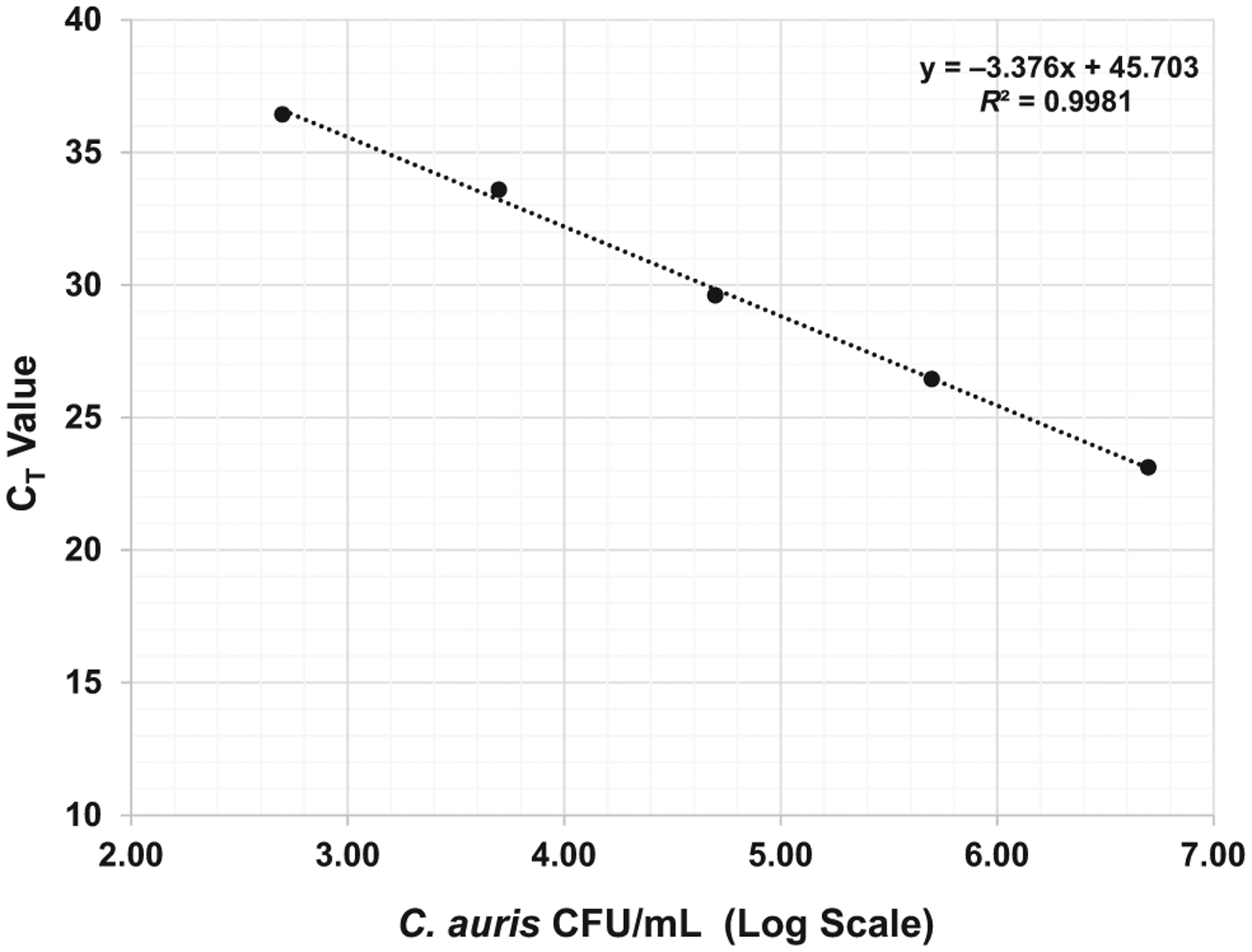
Instrument response: evaluation of instrument response for real-time PCR assay. Five dilutions (from 5 × 106 to 5 × 102 CFU/mL) were prepared for evaluation of the instrument response and analytical sensitivity of real-time PCR assay. The instruments showed mean 3.30-fold (standard deviation: 0.415) increase in CT value with every 10-fold dilution
FIGURE 2.
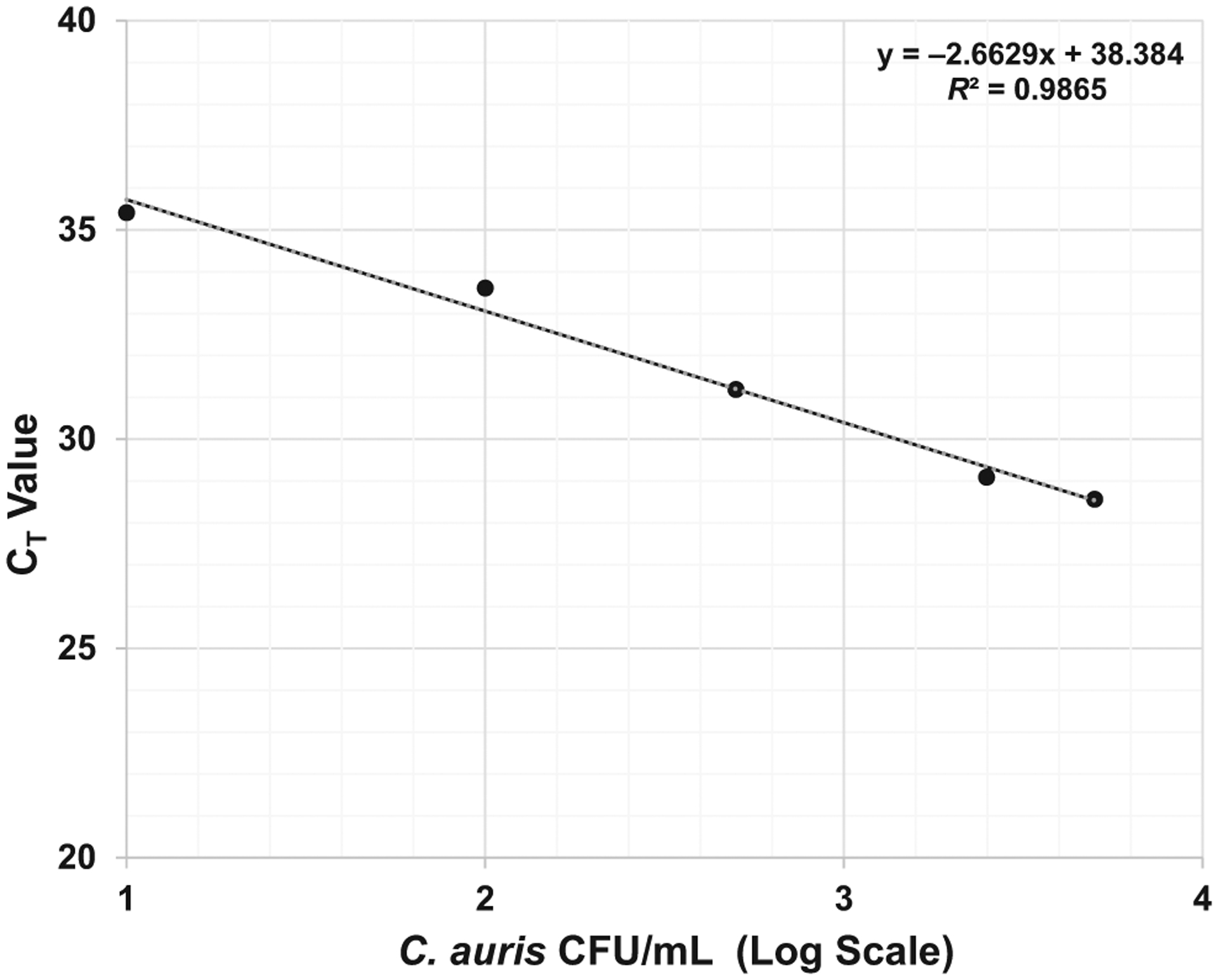
Assay sensitivity: real-time PCR assay sensitivity. Five dilutions (from 5 × 103 to 10 CFU/mL) were evaluated with twenty replicates of each dilutions. The assay was highly reproducible with a limit of detection (LOD) of 100 Candida auris CFU/mL or 1 C auris CFU/10 μL
3.2 |. Evaluation of dermal swabs with C auris real-time PCR assay
We evaluated 247 de-identified patient dermal swab samples. The swabs were previously cultured and analysed by MALDI-TOF for C auris (Positive = 73 and Negative = 174). The real-time PCR assay identified 68 true positive and 173 true negative results compared with the MALDI-TOF results. The analysis also identified 5 false positive and 5 false negative results (Table 1) compared with the MALDI-TOF results. The method achieved 96.1% detection accuracy compared with culture and MALDI-TOF results. The diagnostic sensitivity and specificity were 93.6% and 97.2%, respectively. The distribution of CT values of positive and negative samples was presented in Figure 3. A receiver operating characteristic (ROC) curve analysis of the real-time PCR assay data showed an area of 0.982 under the curve,17 with a CT cut-off value of ≤37.0 (Figure 4). The positive predictive value was 93.15 (95% Cl, 85.12%–97.00%), and the negative predictive value was 97.13 (95% Cl, 93.55%–98.75%).
TABLE 1.
Comparison of real-time results with culture and MALDI-TOF results. The dermal swab samples were previously analysed by culture and MALDI-TOF
| Real-time RT-PCR result | MALD-TOF/culture result | ||||
|---|---|---|---|---|---|
| Positive | Negative | Accuracy | Sensitivity | Specificity | |
| Positive | 68 | 5 | 96.1% | 93.6% | 97.2% |
| Negative | 5 | 169 | |||
FIGURE 3.
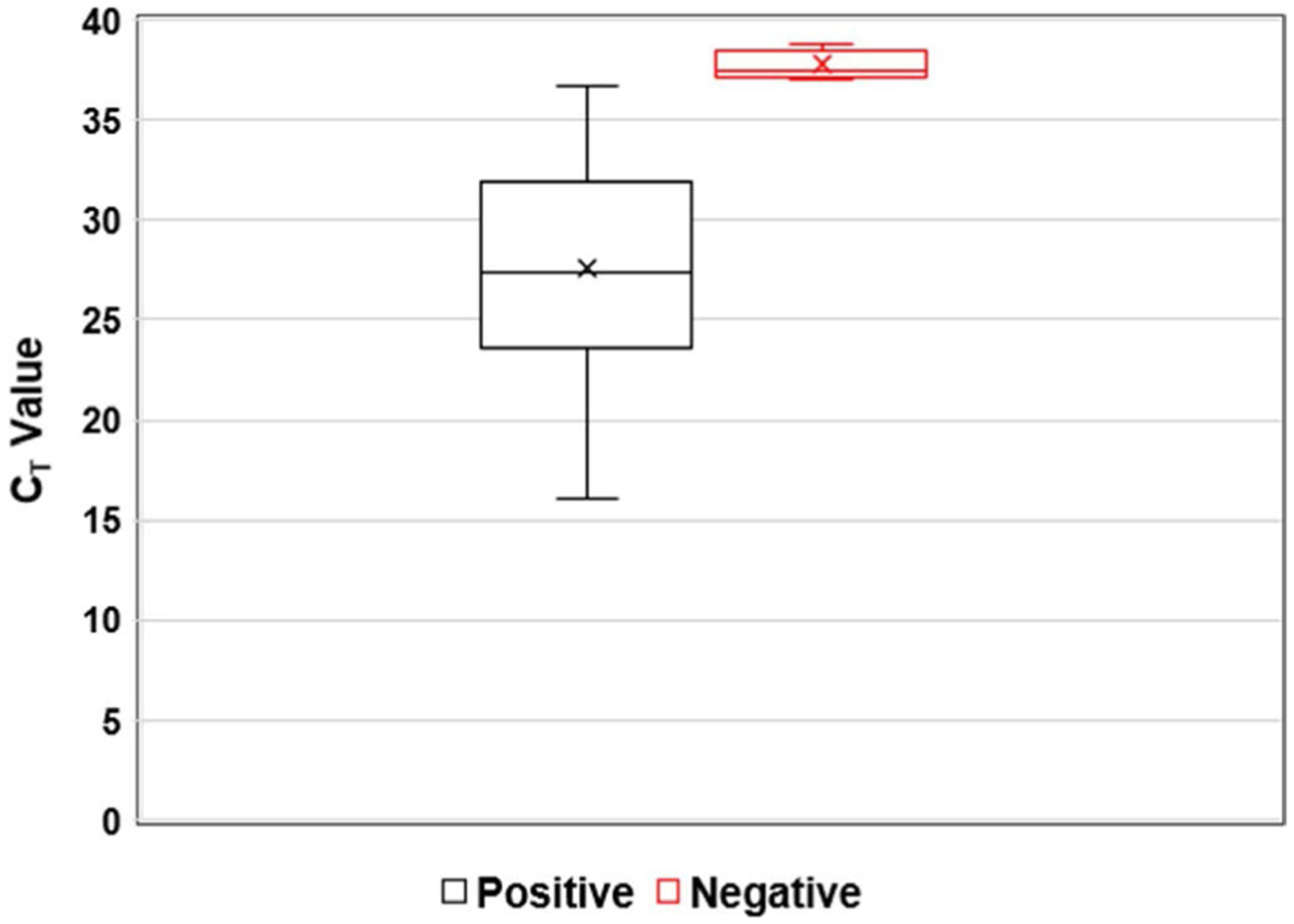
Distribution of CT value: CT value distribution for Candida auris positive and negative samples. A centerline across the boxes indicates the median. Lower and upper boxes indicating the 25th percentile to the 75th percentile. Whiskers caps represent the maximum and minimum CT values
FIGURE 4.
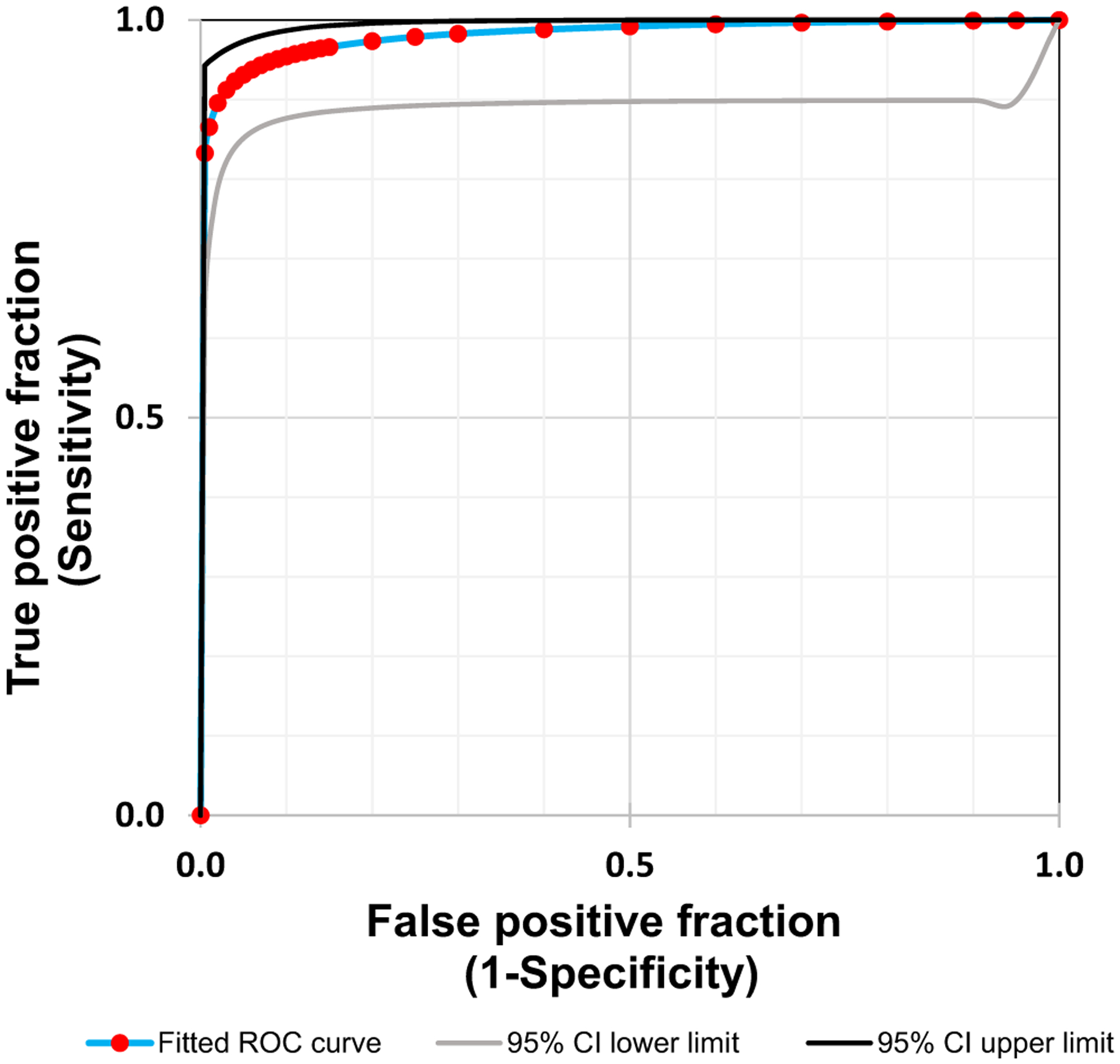
Receiver operating characteristic (ROC) curve: a ROC curve analysis of the real-time PCR data showed an area of 0.982 under the curve, with a CT value cut-off of ≤37.0
4 |. DISCUSSION
Over the last few decades, fungal infection has emerged as a serious concern for human health, especially for immunocompromised patients and those hospitalised with serious underlying diseases. Candida spp. are the most common fungal pathogens, are responsible for nosocomial candidiasis18,19 and are associated with prolonged hospitalisation and increased healthcare costs.20 The most recent nosocomial outbreak of Candida auris is ongoing. With cases continuing to be identified in multiple countries and several US states, Candida auris poses a serious global health threat. Major challenges to control the spread of C auris are (a) resistance to antifungal agents9,21–23 and (b) misidentification when using conventional microbiological methods for fungi identification The commonly used bead beating method for DNA extraction is laborious and time consuming and not very suitable for handling a large number of samples. Compared with more common and labour intensive bead beating and enzymatic lysis methods, the method we validated uses a high-throughput rapid DNA extraction method (approximately 200 samples/day) allowing for faster generation of highly reproducible results. The viability of samples was tested following the pretreatment with Bacterial Lysis Buffer (Cat # 04659180001; Roche Diagnostic, Indianapolis, IN; USA). All samples showed negative results following the incubation step (data not presented). The C auris identification was performed by real-time PCR assay as described previously.16 However, Leach et al16 used crude DNA extracts which increases the possibility of PCR inhibitors to be present. The results of 247 dermal swabs from patients showed a high-degree of concordance with the gold standard method, that is culture and MALDI-TOF for identification of C auris. The method achieved 96.1% detection accuracy with diagnostic sensitivity and specificity of 93.6% and 97.2%, respectively (Table 1). Leach et al16 reported a clinical sensitivity of 89% for surveillance swab samples. This could be due to the fact the Leach et al16 used crude DNA extracts, and it could also be due to the technical variability involved in the analyses. The real-time PCR assay was very sensitive, with a detection limit of 1 C auris CFU/10 μL or 100 CFU/mL (Figure 2). This is similar to the limit of detection reported by Leach et al,16 but is lower than the limit of detection for the SYBR Green method of real-time PCR reported at 4 C auris CFU/PCR reaction.24
Our assay detected both false positives and false negative when compared to the gold standard of culture and MALDI-TOF. Both of these could be due to the fact that only 100 μL of the 1 mL specimen is processed. For a low-density culture, this could mean that C auris cells were captured in either only culture or only RT-PCR and could account for some of the discrepant results. The false positives could also be due to the fact that the RT-PCR assay will detect dead C auris cells while culture will only detect live ones. As many of these patients are treated with topical antiseptics, the swab may contain dead C auris that the RT-PCR assay may be detecting rather than providing false positive results.
Using an automated DNA extraction method reduces DNA extraction time, as well as the potential for handling errors and variability when extracting from a large number of samples. Reproducible results were available from 92 samples in 4–5 hours with these high-throughput assay methods. FDA has approved a fully automated T2 Magnetic Resonance assay for C auris identification, which also provides results in 4–8 hours; however, this assay can only process up to 7 samples at a time.25 Recently, several different methods were published for identification of C auris.26–28 These methods are more laborious and time consuming, making them less suitable for screening a large number of samples, especially at the time of an outbreak.
Since we adopted the TaqMan real-time PCR assay from Leach et al,16 and the authors performed extensive cross-reactivity studies, we used a limited number of yeast species (Malassezia furfur, Candida orthopsilosis, Candida glabrata and Candida parapsilosis), bacterial strains E coli (ATCC 25922), P mirabilis (ATCC 12453), S epidermidis (ATCC 14990), S aureus (ATCC 29213) and Human Specimen Control (a non-infectious cultured human cell material) for cross-reactivity studies. None of these analytes were cross-reactive with the assay.
In conclusion, we successfully validated a rapid and high-throughput DNA extraction method and real-time PCR assay for accurate identification of C auris with a significantly reduced turnaround time. This method provides high-throughput analysis of samples that make it ideal for use in an outbreak situation where quick and reliable results are paramount for patient care and the implementation of public health measures to control the spread of infection.
Footnotes
CONFLIC T OF INTEREST
The authors declare that they have no conflict of interest.
DISCLAIMER
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The use of trademarks is for identification purposes only and does not imply endorsement by the U.S. Department of Health and Human Services.
REFERENCES
- 1.Ruiz-Gaitan A, Moret AM, Tasias-Pitarch M, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61(7):498–505. [DOI] [PubMed] [Google Scholar]
- 2.Belkin A, Gazit Z, Keller N, et al. Candida auris Infection Leading to Nosocomial Transmission, Israel, 2017. Emerg Infect Dis. 2018;24(4):801–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44. [DOI] [PubMed] [Google Scholar]
- 4.Vallabhaneni S, Kallen A, Tsay S, et al. Investigation of the first seven reported cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus-United States, May 2013-August 2016. Am J Transplant. 2017;17(1):296–299. [DOI] [PubMed] [Google Scholar]
- 5.CDC. Tracking Candida auris—August 2018. https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html. Accessed October 04, 2018.
- 6.Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S, Rai G, Tigga RA, et al. Candida auris in critically ill patients: emerging threat in intensive care unit of hospitals. J Mycol Med. 2018;28(3):514–518. [DOI] [PubMed] [Google Scholar]
- 8.Adams E, Quinn M, Tsay S, et al. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis. 2018;24(10):1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockhart SR, Jackson BR, Vallabhaneni S, Ostrosky-Zeichner L, Pappas PG, Chiller T. Thinking beyond the Common Candida Species: need for species-level identification of Candida due to the emergence of multidrug-resistant Candida auris. J Clin Microbiol. 2017;55(12):3324–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Ami R, Berman J, Novikov A, et al. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017;23(1):195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathuria S, Singh PK, Sharma C, et al. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J Clin Microbiol. 2015;53(6):1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC. Recommendations for Identification of Candida auris. https://www.cdc.gov/fungal/candida-auris/recommendations.html. Accessed October 04, 2018.
- 13.Girard V, Mailler S, Chetry M, et al. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59(8):535–538. [DOI] [PubMed] [Google Scholar]
- 14.Sterkel A, Bateman A, Valley A, Warshauer D. Viability of Candida auris and other Candida species after various matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry-based extraction protocols. J Clin Microbiol. 2018;56(9):e00886–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welsh RM, Bentz ML, Shams A, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017;55(10):2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leach L, Zhu Y, Chaturvedi S. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol. 2018;56(2):e01223–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eng J ROC analysis: web-based calculator for ROC curves. Johns Hopkins University, Baltimore. http://www.jrocfit.org. Accessed October 04, 2014. [Google Scholar]
- 18.Epelbaum O, Chasan R. Candidemia in the intensive care unit. Clin Chest Med. 2017;38(3):493–509. [DOI] [PubMed] [Google Scholar]
- 19.Wisplinghoff H, Ebbers J, Geurtz L, et al. Nosocomial bloodstream infections due to Candida spp. in the USA: species distribution, clinical features and antifungal susceptibilities. Int J Antimicrob Agents. 2014;43(1):78–81. [DOI] [PubMed] [Google Scholar]
- 20.Lockhart SR. Current epidemiology of Candida infection. Clin Microbiol Newsl. 2014;36(17):131–136. [Google Scholar]
- 21.Chowdhary A, Anil Kumar V, Sharma C, et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis. 2014;33(6):919–926. [DOI] [PubMed] [Google Scholar]
- 22.Lockhart SR, Berkow EL, Chow N, Welsh RM. Candida auris for the clinical microbiology laboratory: not your grandfather’s Candida species. Clin Microbiol Newsl. 2017;39(13):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidaud AL, Chowdhary A, Dannaoui E. Candida auris: an emerging drug resistant yeast - A mini-review. J Mycol Med. 2018;28(3):568–573. [DOI] [PubMed] [Google Scholar]
- 24.Sexton DJ, Kordalewska M, Bentz ML, Welsh RM, Perlin DS, Litvintseva AP. Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR Green qPCR assay. J Clin Microbiol. 2018;56(12):e01337–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sexton DJ, Bentz ML, Welsh RM, Litvintseva AP. Evaluation of a new T2 Magnetic Resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses. 2018;61(10):786–790. [DOI] [PubMed] [Google Scholar]
- 26.Arastehfar A, Fang W, Badali H, et al. Low-cost tetraplex PCR for the global spreading multi-drug resistant fungus, Candida auris and its phylogenetic relatives. Front Microbiol. 2018;9:1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol. 2017;55(8):2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto M, Alshahni MM, Tamura T, et al. Rapid detection of Candida auris based on loop-mediated isothermal amplification (LAMP). J Clin Microbiol. 2018;56(9):e00591–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


