Abstract
Keratin (K) is a major protein component of hair and is involved in hair growth and development. In this study, we analysed the expression, localization, and polymorphism of the K84 gene (KRT84) in Gansu Alpine Fine-wool sheep using immunofluorescence, RT-qPCR, and PARMS (penta-primer amplification refractory mutation system). Haplotypes of KRT84 were also constructed and their relationship with wool traits analysed. It was revealed that KRT84 was highly expressed in hair follicles, including the inner root sheath, outer root sheath, and hair medulla and at all six lamb ages investigated from 1 to 270 days of age. Three SNPs were detected in KRT84 exon 1, and they formed three haplotypes (named H1, H2, and H3) and six genotypes. Analyses revealed an association between haplotype combinations (diplotypes) and the mean fibre curvature, mean staple length, mean staple strength, mean fibre diameter, the coefficient of variation of fibre diameter, and comfort factor for these sheep. These results suggest that KRT84 is of importance in determining several key traits in Gansu Alpine Fine-wool sheep and that the gene could possibly be used as a genetic marker for wool trait selection in these sheep.
Keywords: Gansu Alpine Fine-wool sheep, keratin, SNPs, spatiotemporal expression
1. Introduction
Keratinous hair is unique to mammalian skin. It is a terminally differentiated tissue that forms in hair follicles and grows from the inner to the outer part of the epidermis [1]. The hair follicle contains both mesenchymal and epithelial components, separated by a basement membrane. In the follicle bulb, the dermal papilla (DP) consists of mesenchymal cells that function in the regulation of hair growth. The DP contains blood vessels and nerve endings that supply oxygen and nutrients to the root of the hair and is associated with the epithelial progenitor populations of the follicle. The bulb produces several distinct cell layers, including the outer root sheath, the inner root sheath, and the hair shaft [2]. Maria et al. [3] divided the process of hair follicle development into three phases: follicular substrate formation, organogenesis, and cell differentiation, whereas Saxena et al. [4] divided the stages of hair follicle development in more detail, based on the differentiation of different types of cells within the follicle.
Keratins (Ks) are fibrous proteins found in various tissues. They typically serve a protective function, being found in a variety of animal hairs, hoof tissues, feathers, and scales, and they are the major structural proteins in hair follicle cells. In sheep, keratins are produced in the wool follicle, and they are intrinsically involved in determining the structural characteristics of wool fibres. Accordingly, keratin genes are candidate genes for determining variation in wool fibre characteristics and biology [5]. In wool, keratin proteins form complex structures through the interaction of their amino acid side chains with other proteins. Of these interactions, the formation of strong covalent disulfide bonds is prominent [6]. While the internal structures of different mammalian hairs are similar, there are notable differences in morphology, with these in part attributable to variation in the keratin proteins and their molecular variability [7].
The keratins can be classified as epithelial keratins and hair keratins, and they have been divided into type I and type II families based on their sites of expression and functions. These keratin families are differentially expressed during hair fibre development [5,8]. Type I keratin genes (KRTs) are 4 to 5 kilobase pairs in length and have six introns, while the type II KRTs are larger, ranging from 7 to 9 kilobase pairs in length with eight introns. McLaren et al. [9] used linkage analyses to reveal that the type I and type II genes are located on chromosomes 11 and 3, respectively, in sheep, and both types of proteins are encoded by multiple genes [9,10].
Of the different keratins, K84 has been revealed to be expressed at high levels in plucked hairs collected from domestic horses (Equus ferus caballus) [11]. In cashmere goats (Capra hircus), it was found that KRT84 promoter activity can be affected by HOXC13 [12], while in sheep (Ovis aries), it was revealed that KRT84 transcripts were present only in the fibrous cuticle, and that this may have resulted in a 3:2 ratio of type I to type II in the epibulbar region of the follicle, compared to a ratio of 2:1 in humans [13]. To date, there are no reports on the role of KRT84 in wool follicle growth and development or whether the gene affects wool traits. To explore KRT84’s role in wool production in Gansu Alpine Fine-wool sheep from China, the spatiotemporal expression of KRT84 was investigated, sequence variation was revealed in the gene, and haplotypes of the gene were tested to ascertain if they were associated with variation in key wool traits.
2. Materials and Methods
2.1. Sheep and Sampling
Two hundred and twenty-five Gansu Alpine Fine-wool sheep ewes of around three years in age with the same feeding levels and living environment were selected for the penta-primer amplification refractory mutation system (PARMS) and genetic association analyses. Blood was collected from the jugular vein of each sheep using a 5 mL sodium heparin tube, numbered, and stored at −20 °C until needed. Wool samples from the mid-flank of each sheep were collected by clipping against the skin and sealed in self-sealing bags with identification details. The mean fibre diameter (MFD), fibre diameter standard deviation (FDSD), coefficient of variation of fibre diameter (CVFD), mean staple length (MSL), mean fibre curvature (MFC), comfort factor (CF), and mean staple strength (MSS) for each wool sample were assessed. All wool testing was performed by New Zealand Pastoral Measurements LTD (Ahuriri, Napier, New Zealand) using International Wool Textile Organisation (IWTO; https://iwto.org/)-endorsed methods (https://iwto.org/resources/wool-testing-resources/; accessed on 15 September 2023).
Three new-born Gansu Alpine Fine-wool ewe lambs were selected for the immunofluorescence analysis. Skin tissues were collected from a 5 cm2 area of the posterior border of their scapulae at 1 day (d), 30 d, 60 d, 90 d, 180 d, and 270 d of age, using a skin sampler (0.88 cm in diameter). The skin samples were collected after localized injection of lidocaine hydrochloride (Jiangxi Roaching Longboat Veterinary Medicine Co., Ltd., Nanchang, China) at the site of collection for anaesthesia, and penicillin (Suicheng Pharmaceutical Co., Ltd., Zhengzhou, China) and Yunnan Baiyao (Yunnan Baiyao Group Co., Ltd., Kunming, China) were applied to the wound after the sampling to prevent infections.
After being cleaned with diethylpyrocarbonate-treated water, a portion of the skin tissue was placed in a tube containing 4% paraformaldehyde and stored at 4 °C. Separate portions of the skin samples were also placed in tubes containing an RNA protective solution (Servicebio Biotechnology Co., Ltd., Wuhan, China), which was allowed to penetrate the tissue to stabilize and protect the cellular RNA. These samples were stored in liquid nitrogen.
2.2. DNA Extraction
DNA was extracted using blood genomic DNA extraction kits (Tiangen Biochemical Technology Co., Ltd., Beijing, China). After the DNA was extracted, it was stored in TE buffer and quantified using a NanoDropTM 2000 spectrophotometer (ThermoFisher, Shanghai, China). The quality of the DNA was checked with agarose gel electrophoresis, and it was stored at −20 °C for subsequent analysis.
2.3. RNA Extraction
RNA was extracted from the skin tissue following the instructions provided with the Trizol reagent (Shanghai Yuanyye Bio-Technology Co., Ltd., Shanghai, China). The concentration of the extracted RNA was determined using ultraviolet spectrophotometry and the RNA samples were stored at −80 °C until needed.
2.4. Detecting KRT84 Sequence Variaiton
Based on the sequence variation reported for the KRT84 reference sequence ENSOARG00020013379 in the Ensembl database (https://www.ensembl.org), exon 1 was chosen for sequence variation analysis. Primers were designed using Oligo 7 software (https://www.oligo.net/) to amplify the entire exon 1 coding sequence. The PCR primer sequences are shown in Table 1.
Table 1.
Primer sequences and annealing temperatures used for PCR and RT-qPCR.
| Gene | Primer Sequences (5′–3′) | Size (bp) | Annealing Temperature (°C) |
Use |
|---|---|---|---|---|
| KRT84 | F: CCATCATGTCTTGCCGCTCCT | 587 | 60 | PCR |
| R: ACACCCCAGTGCATTTAGCTC | ||||
| KRT84 | F: ACGATCTTCAGTGGCCCAAG | 104 | 60 | RT-qPCR |
| R: GGGGTTAAACCAACACCCCA | ||||
| ACTB | F: AGCCTTCCTTCCTGGGCATGGA | 113 | 60 | RT-qPCR |
| R: GGACAGCACCGTGTTGGCGTAGA |
The PCR amplifications were carried out in a 20 μL reaction, containing 100 ng genomic DNA, 0.5 U of Taq DNA polymerase (Takara, Dalian, China), 2.5 mM Mg2+, 150 μM of each dNTP (Takara), and 0.25 μM of each primer. The thermal cycle parameters for amplification consisted of an initial denaturation incubation at 95 °C for 2 min, followed by 35 cycles of incubation for 30 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C, and completed with a final extension incubation at 72 °C for 5 min. Amplicons were then sequenced in both directions.
2.5. Genotyping Using PARMs-PCR and Mass Spectrometry
All the 225 sheep DNA samples were genotyped for individual SNPs using a PARMS–polymerase chain reaction (PARMS-PCR) approach, following the procedure described by Sun et al. [14]. For each SNP, three primers were designed, a common forward primer and two reverse primers that contained either FAM or HEX fluorescent sequences at the 5’ end. The sequence and fluorescent signal type information are shown in Table 2, and they were synthesized by Gentides Technology Co., Ltd. (Wuhan, China).
Table 2.
PARMS-PCR primer sequences and fluorescent signal types.
| SNP | Primer | Primer Sequences (5′–3′) | Fluorescent Signal | Genotype |
|---|---|---|---|---|
| c.97A/G | 1Rt | GAAGGTGACCAAGTTCATGCTCCTGCAGGAGACAGAGCTGAT | FAM | A |
| 1Rc | GAAGGTCGGAGTCAACGGATTCCTGCAGGAGACAGAGCTGAC | HEX | G | |
| 1F | CCCCTCAGAACCTGAACCG | / | / | |
| c.162C/T | 2Rg | GAAGGTGACCAAGTTCATGCTACGATCCAAAGCTGATGACG | FAM | C |
| 2Ra | GAAGGTCGGAGTCAACGGATTCACGATCCAAAGCTGATGACA | HEX | T | |
| 2F | CCCCTCAGAACCTGAACCG | / | / | |
| c.286G/A | 3Rc | GAAGGTGACCAAGTTCATGCTGGCTGCAGAAGCCATAGCC | FAM | G |
| 3Rt | GAAGGTCGGAGTCAACGGATTGGGCTGCAGAAGCCATAGCT | HEX | A | |
| 3F | GGGGTTTAGAGCCAGAAGTGG | / | / |
The PARMS-PCR amplification thermal cycle was pre-denaturation at 94 °C for 15 min, denaturation at 94 °C for 20 s, and a 65 °C (−0.7 °C per cycle) annealing and extension step for 1 min, for 10 cycles. This was followed by denaturation at 94 °C for 20 s and a 57 °C annealing and extension step for 1 min, for 35 cycles. The fluorescence was detected with a TECAN Infinite F200 Enzyme Labeler (AiYan Biotechnology Co., Ltd., Shanghai, China) and then parsed and converted into fluorescence signals using the online software SNP Decoder (http://www.snpway.com/snpdecoder/; accessed on 5 October 2023), which outputted the genotype results. Unique haplotypes were prepared and then sequenced by Gentides Technology Co., Ltd. (Wuhan, China).
2.6. RT-qPCR
The sheep KRT84 mRNA sequence (XM_004006329.5) was used to design a pair of PCR primers to amplify a fragment of 3′-UTR from the cDNA. The β-actin gene (ACTB) (NM_001009784) was used as a reference to calibrate the level of gene expression. These primer sequences are shown in Table 1. Approximately 0.4 μg of total RNA was used for reverse transcription and complementary DNA (cDNA) was prepared using a PrimeScriptTM RT kit (Nanjing Vazyme Biotechnology Co., Ltd., Nanjing, China) according to the kit instructions. The qPCR reactions were performed using a Biosystems QuantStudioR 6 Flex (Thermo Fisher Scientific Inc, Waltham, MA, USA) platform with the annealing temperatures described in Table 1, the cDNA from above as the template, and SYBR Green Pro Taq HS qPCR kits (Hunan Accurate Biotechnology, Changsha, China). In 20 μL reactions, the thermal cycle parameters were as follows: initial denaturation at 95 °C for 10 min, followed by 45 cycles of 15 s at 95 °C and 60 °C annealing and extension for 1 min. The cDNAs were quantified in triplicate, with four technical replicates.
2.7. Immunofluorescence Analysis
Diao’s immunofluorescence method was used in this experiment [15]. The primary antibody used in the experiment was K84 (TD10116, rabbit antibody) and the secondary antibody was CY3 (GB21303, goat anti-rabbit IgG), provided by Servicebio Technology Co., Ltd. (Wuhan, China). The immunofluorescence results were scanned and imaged using a PANNORAMIC panoramic slice scanner (3DHISTECH, Budapest, Hungary). The outputted scan files were opened with the CaseViewer 2.2 software using 200× magnification. The integral optical density (IOD) and the corresponding positive pixel area (area) of three fields of view for each slice were measured separately using Image-Pro Plus 6.0 software (Media Cybernetics Inc., Rockville, MD, USA) to calculate the average optical density (AOD) using IOD/Area = AOD. The scoring was performed on three fields of view for the corneum, dermal papilla, sebaceous gland, outer root sheath, inner root sheath, and hair matrix for each slice. AOD was used to indicate the degree of positive intensity.
2.8. Statistical Analyses
The genotype and haplotype frequencies of the KRT84 SNPs were counted using Excel 2019 software, and an online calculator (http://www.msrcall.com/Gdicall.aspx; accessed on 12 October 2023) was used to calculate homozygosity (Ho), heterozygosity (He), polymorphism information content (PIC), and effective allele numbers (Ne). The Hardy–Weinberg equilibrium of the haplotypes was tested using the chi-square test in the online calculator. The SNP chain imbalance analysis and haplotype analysis were performed using Haploview 4.2 software, after eliminating the results from sheep that could not be typed.
General linear models (IBM SPSS 26.0) were used to analyse the association of the different haplotypes revealed with the measured wool traits MFD, CVFD, MSL, MFC, CF, and MSS in the Gansu Alpine Fine-wool sheep. The model was yijk = µ + Hi + eijk, where yijk indicates the trait phenotype value, µ denotes the overall mean, Hi indicates a haplotype combination, and eijk denotes the random error. The results were output as mean ± standard error (S.E.), with p < 0.05 as the criteria for accepting significant differences.
The RT-qPCR results were calculated in Excel using the 2−ΔΔCT [16] method and correlation analysis with wool trait data.
3. Results
3.1. Localization of K84 mRNA in the Skin of Gansu Alpine Fine-Wool Lambs
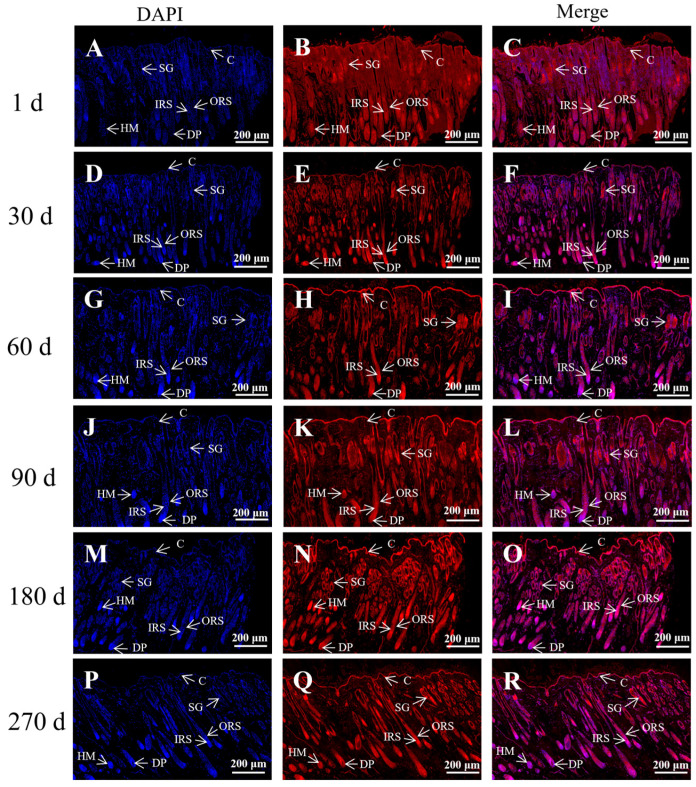
The immunofluorescence analysis of the distribution of K84 in the skin tissues of Gansu Alpine Fine-wool sheep at different lamb ages (Figure 1) revealed that K84 was highly expressed in hair follicles such as the inner root sheath (IRS), outer root sheath (ORS), and hair medulla (HM) and at all six lamb ages. The dermal papilla at 1, 60, and 90 days of age had strong expression of K84; and skin tissues at 30, 180, and 270 days of age had strong K84 expression.
Figure 1.
Immunofluorescence staining for K84 in the skin of Gansu Alpine Fine-wool sheep at different times. Immunofluorescence staining of K84 (5×). (A–C) 1d; (D–F) 30 d; (G–I) 60 d; (J–L) 90 d; (M–O) 180 d; (P–R) 270 d. The blue tissue in the figure is the DAPI-labelled nuclear fluorescence staining and the red tissue is the fluorescence staining of K84. C: corneum; DP: dermal papilla; SG: sebaceous gland; ORS: outer root sheath; IRS: inner root sheath; HM: hair matrix.
3.2. K84 Levels in the Skin of Gansu Alpine Fine-Wool Sheep at Different Lamb Ages
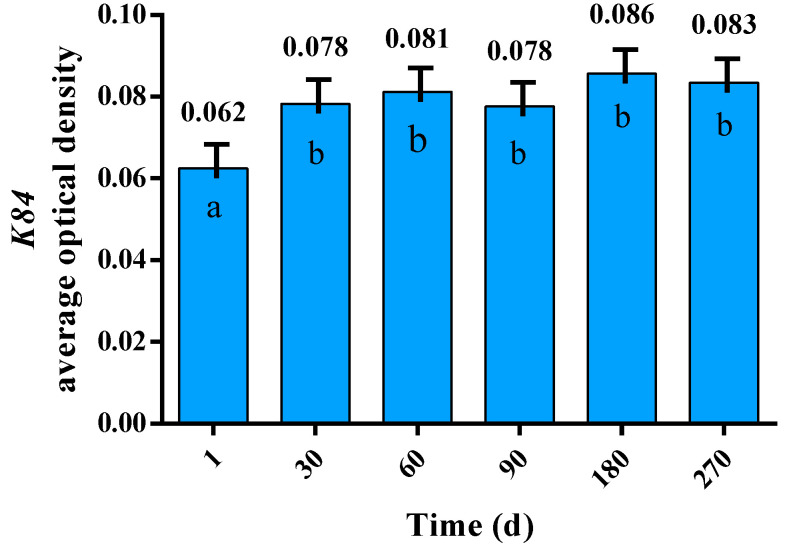
The average optical density of the tissue sections is revealed in Figure 2. The protein K84 was found at different levels in the skin of lambs of differing age, with an overall trend towards an increase. The amount of K84 was significantly higher at 30, 60, 90, 180, and 270 days, when compared to the level at birth (Day 1; p < 0.05).
Figure 2.
Levels of K84 in the skin of Gansu Alpine Fine-wool sheep at different times. The data are expressed as means ± S.E. Different letters indicate significant differences between lambs of differing age (p < 0.05).
3.3. SNP Detection and Genotyping
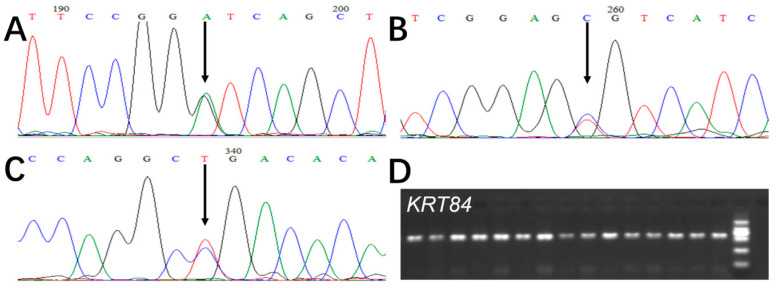
In Figure 3, the DNA sequencing of exon 1 amplicons revealed three SNPs in Gansu Alpine Fine-wool sheep, and these were named c.97A/G, c.162C/T, and c.286G/A, according to their position in the gene.
Figure 3.
PCR amplification and sequencing of KRT84 in Gansu Alpine Fine-wool sheep. (A) c.97A/G, (B) c.162C/T, (C) c.286G/A (noting that the overlapping peaks indicated by arrows in A, B, and C are SNPs and that the numbering on the electropherograms reflects the location in the sequencing reaction and not the position of the SNP in the gene). Sequences in A and B are from the forward strand whereas the sequence in C is from the reverse strand. (D) D illustrates the PCR amplification of KRT84.
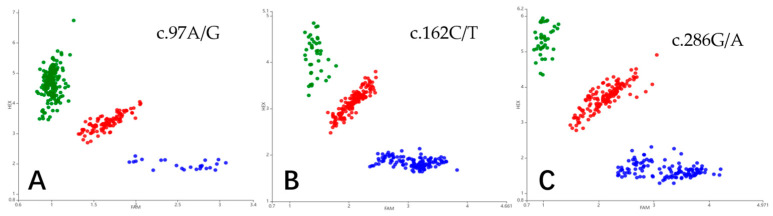
In Figure 4, the PARMS labelling assay produced scatterplots, where the horizontal coordinate is the FAM fluorescence value, and the vertical coordinate is the HEX fluorescence value. The heterozygous genotypes are in the middle of the scatterplot (indicated with red dots), while the homozygous sheep are located at the edges of the scatterplot and are indicated by blue and green dots.
Figure 4.
Genotyping of KRT84 in Gansu Alpine Fine-wool sheep using PARMS analysis. (A) The red, green, and blue dots indicate GA, GG, and AA, respectively; (B) The red, green, and blue indicate CT, TT, and CC, respectively; (C) The red, green, and blue dots indicate GA, GG, and AA, respectively.
3.4. Analysis of the Sequence Variation in KRT84
The polymorphic information content (PIC), homozygosity (Ho), heterozygosity (HE), and number of effective alleles (Ne) of KRT84 are shown in Table 3. The PICs of the three SNPs (c.97A/G, c.162C/T, and c.286G/A) were 0.3577, 0.6956, and 0.6965, respectively. All the SNPs were in Hardy–Weinberg equilibrium in this population (p > 0.05), with this suggesting that these Gansu Alpine Fine-wool sheep were not inbred or subject to some form of selection pressure for KRT84 variation.
Table 3.
The population genetic diversity of the three SNPs in exon 1 of ovine KRT84.
| SNP | Ho | He | PIC | Ne | H |
|---|---|---|---|---|---|
| c.97A/G | 0.5335 | 0.4665 | 0.3577 | 1.8743 | 0.6592 |
| c.162C/T | 0.2738 | 0.7262 | 0.6956 | 3.6528 | 0.7173 |
| c.286G/A | 0.2735 | 0.7265 | 0.6965 | 3.6566 | 0.7156 |
Ho: homozygosity; He: heterozygosity; PIC: polymorphic information content; Ne: number of effective alleles; H: Hardy–Weinberg equilibrium; PIC < 0.25 low polymorphism, 0.25 < PIC < 0.5 for moderate polymorphism, PIC > 0.5 for highly polymorphic.
The SNP haplotype frequencies and genotype frequencies are revealed in Table 4. For c.97A/G, G was the most common nucleotide, with a frequency of 0.757, and GG was the most common genotype, with a genotype frequency of 0.589. For c.162C/T, C was the most common nucleotide with a frequency of 0.664 and CC was the most common genotype, with a frequency of 0.444. At c.286G/A, G was the more common nucleotide, with a frequency of 0.664, and GA was the most common genotype, with a frequency of 0.449.
Table 4.
SNP genotype frequencies and nucleotide frequencies for exon 1 of KRT84.
| SNP | SNP Genotype Frequency (Number) |
Nucleotide Frequency | X2 | p | |||
|---|---|---|---|---|---|---|---|
| c.97A/G | GA | GG | AA | G | A | 0.8304 | 0.660 |
| 0.335(75) | 0.589(132) | 0.076(17) | 0.757 | 0.243 | |||
| c.162C/T | CT | CC | TT | C | T | 0.0181 | 0.991 |
| 0.440(99) | 0.444(100) | 0.116(26) | 0.664 | 0.336 | |||
| c.286G/A | GA | GG | AA | G | A | 0.0035 | 0.006 |
| 0.449(101) | 0.440(99) | 0.111(25) | 0.664 | 0.336 | |||
Note: The figures in parentheses are the number of individuals that were genotyped.
3.5. Haplotype Construction for the KRT84 Exon 1 SNPs and Association Analyses between These Haplotypes and Wool Traits
Linkage disequilibrium analysis was performed on the three SNPs in KRT84. Haplotypes were constructed for the three SNPs, with reporting of those haplotype combinations (diplotypes). This allowed the construction of a haplotype block (Figure 5), and the SNPs were in a strongly interlocking state (R2 = 1), with three haplotypes detected that were named H1-H3 (Table 5). The haplotype with the highest frequency in the Gansu Alpine Fine-wool sheep was H1. Combinatorial analysis of the three haplotypes yielded a total of six diplotypes with frequencies greater than 0.05 (Table 5), with the H1H2 diplotype having the highest frequency (28.25%).
Figure 5.
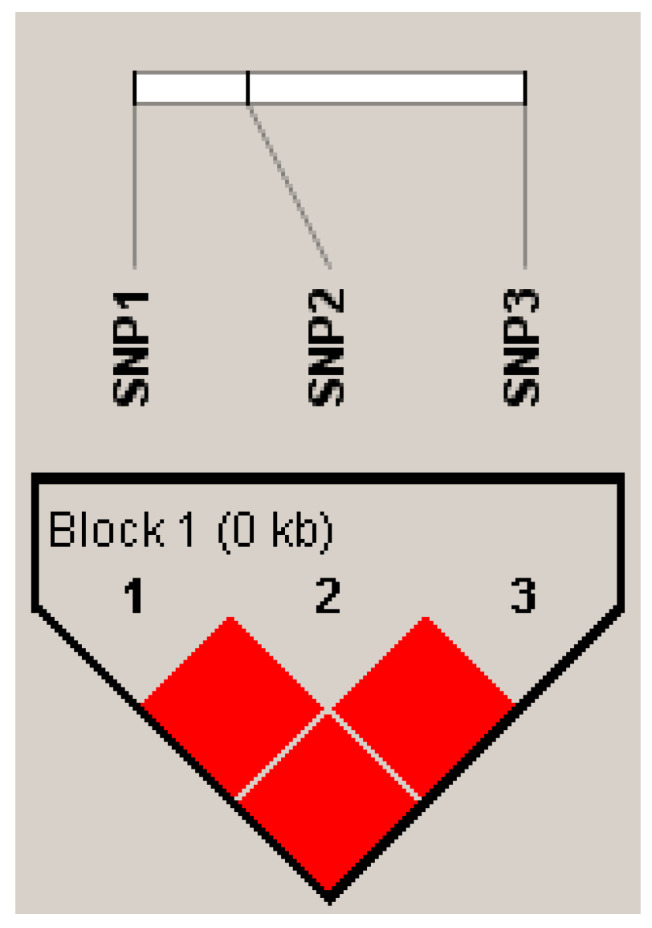
Linkage disequilibrium analysis of the KRT84 SNPs in Gansu Alpine Fine-wool sheep. The red square in the figure has an R2 value of 1, suggesting a strongly interlocking state.
Table 5.
Frequency of haplotypes and diplotypes for the SNPs in exon 1 of KRT84.
| Haplotype | c.97A/G | c.162C/T | c.286G/A | Frequency (%) | Diplotype | Frequency (%) |
|---|---|---|---|---|---|---|
| H1 | G | C | G | 42.30 | H1H1 | 19.28 |
| H2 | G | T | A | 33.50 | H1H2 | 28.25 |
| H3 | A | C | G | 24.20 | H1H3 | 17.49 |
| H2H2 | 11.21 | |||||
| H2H3 | 16.14 | |||||
| H3H3 | 7.62 |
Association analyses of these six diplotypes with wool traits (Table 6) suggested an effect of KRT84 variation on MFD, CVFD, MSL, MFC, CF, and MSS at a significance level of p < 0.05.
Table 6.
Relationship between KRT84 haplotypes and selected key wool traits.
| Diplotype | MFD (Microns) |
FDSD (Microns) |
CVFD | MSL (mm) |
MFC (°/mm) |
CF (%) |
MSS (cN/dT) |
|---|---|---|---|---|---|---|---|
| H1 H1 | 21.4 ± 2.56 b | 5.4 ± 1.12 | 24.9 ± 3.36 b | 77.4 ± 15.05 a | 103.9 ± 12.48 b | 91.4 ± 9.93 a | 14.0 ± 6.75 b |
| H1 H2 | 22.1 ± 2.95 ab | 5.5 ± 1.06 | 25.0 ± 2.99 ab | 74.0 ± 11.80 ab | 105.6 ± 9.94 b | 88.6 ± 11.18 ab | 13.6 ± 5.22 b |
| H1 H3 | 22.9 ± 2.81 a | 5.8 ± 1.13 | 25.4 ± 2.95 a | 70.9 ± 14.87 b | 104.7 ± 12.54 b | 85.9 ± 11.72 b | 13.7 ± 5.05 b |
| H2 H2 | 22.3 ± 3.20 ab | 5.5 ± 0.94 | 24.6 ± 2.60 b | 68.2 ± 13.90 b | 111.5 ± 11.15 a | 87.6 ± 11.62 ab | 15.3 ± 6.15 ab |
| H2 H3 | 22.4 ± 2.83 ab | 5.6 ± 0.92 | 25.0 ± 2.42 ab | 71.3 ± 13.56 ab | 104.9 ± 11.15 b | 87.5 ± 11.98 ab | 14.1 ± 6.60 b |
| H3 H3 | 23.2 ± 3.63 a | 6.0 ± 0.78 | 26.3 ± 3.43 a | 71.9 ± 16.94 b | 106.5 ± 13.71 ab | 82.4 ± 15.60 b | 17.5 ± 6.60 a |
Different letters in the columns indicate significant differences (p < 0.05). The results were expressed as means ± standard errors (S.E.). MFD: mean fibre diameter, FDSD: fibre diameter standard deviation, CVFD: coefficient of variation of fibre diameter, MSL: mean staple length, MFC: mean fibre curvature, CF: comfort factor, MSS: mean staple strength.
The MFD of the homozygous H1H1 sheep was lower than the H3H3 or H1H3 sheep (p < 0.05), while the CVFD of the homozygous H1H1 sheep was also lower than the H3H3 or H1H3 sheep (p < 0.05). The MSL of the homozygous H1H1 sheep was greater than the H3H3 or H1H3 sheep (p < 0.05), yet the MFC of the homozygous H2H2 sheep was higher than H1H1, H1H2, H1H3, and H2H3 (p < 0.05). The CF of H1H1 sheep was higher than H1H3 and H3H3 (p < 0.05), while the MSS of the homozygous H3H3 sheep was higher than the H1H1, H1H2, H1H3, and H2H3 (p < 0.05). Diplotype did not appear to affect FDSD (p > 0.05).
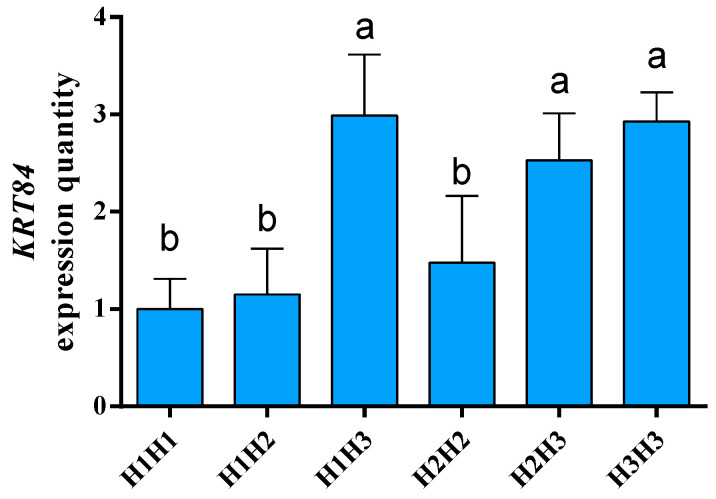
Quantitative fluorescence analysis (Figure 6) suggested that the KRT84 mRNA levels of H1H3, H2H3, and H3H3 were higher than the levels of H1H1, H1H2, and H2H2.
Figure 6.
The mRNA levels of KRT84 in the skin of Gansu Alpine Fine-wool sheep. The β-actin gene (ACTB) was used as a reference gene to calibrate the level of gene expression. The genotype expression levels in skin are presented as means ± S.E. and significant differences (p < 0.05) are denoted by different letters.
4. Discussion
4.1. Variation in KRT84
It is not unprecedented for ovine keratin genes to contain nucleotide sequence variation, with evidence for this being presented in analyses of the whole genome and studies of individual genes such as KRT31 [17], KRT33A [18], and KRT83 [19]. Analysis of KRT84 in Ensembl (release 110, 2023) reveals a sequence in the Rambouillet v1.0 genome construct CM008474.1 (a fine wool sheep breed), identified as ENSOARG00020013379 (GenBank GENE ID: 10056798), which is near another keratin gene KRT82 (EN-SOART00020020319). The KRT84 sequence has nine exons, with six containing SNPs. Some SNPs in exons 1, 2, 7, and 9 are non-synonymous, with splice region variants also described upstream of exons 6 and 9. Exon 1 encodes the head region of KRT84, while exons 2–8 encode the intermediate filament rod domain. The identified coding sequence is 1782 nucleotides in length with the ATG start site in exon 1 and a predicted length of 593 amino acids.
The SNPs c.97A/G and c.286G/T are both missense, and while c.97A/G (p.Ile33Val) is a conservative change, c.286G/T is far less so, with glycine putatively being substituted with cysteine. In keratins, cysteine is known to cross-link with both other keratins and the keratin-associated proteins; hence, this SNP potentially changes the ability of K84 to interact with other proteins in the wool fibre, and thus possibly the wool fibre structure and function. In this respect, if expressed, this amino change would be in the head region of K84, a region that is important in the formation of the keratin dimers and tetramers that arrange into protofilaments. In the report of Harland et al. [20], fibers from Romney sheep were subjected to stretching or to their breaking point under wet or dry conditions to detect disulfide bonds breakage. Many of the identified cysteine residues were located close to the terminal ends of the keratins (head or tail domains), where they likely affect keratin-associated protein interactions and keratin interactions.
4.2. Association Analysis between Haplotypes of KRT84 and Wool Traits for Gansu Alpine Fine-Wool Sheep Wool
The discovery of variation in wool keratin genes has led to the question of whether it affects the growth and development of wool fibres. In that respect, nucleotide sequence variation in KRT31 has been associated with variation in greasy fleece weight (GFW), clean fleece weight (CFW), and MSL in Merino-cross sheep [17], while variation in KRT33A has been associated with variation in fibre crimp frequency, MFC, wool bulk, fleece weight, MSL, and washing yield (CFW/GFW × 100%) in Perendale sheep [18]. Within half-sib families of Merino and Merino-cross sheep, variation in KRT33A was associated with FDSD, MSS, and wool colour in one family, and with MSL in another family [21]. In 2017 Chai et al. [19] revealed variation in KRT83 to be associated with variation in FDSD, CVFD, MFD, MFC, PF, and a decrease in washing yield, and in 2022, Chai et al. [22] reported that variation in the promotor region of KRT34 was associated with MFD, FDSD, and MSL.
The KRT84 SNPs in Gansu Alpine Fine-wool sheep described here were also associated with several traits, including wool MFD, CVFD, MSL, MFC, CF, and MSS. Many of these are important wool traits, not least MFD, which typically is the key determinant of the value proposition for wool. This indicates that the other traits like CVFD, MSL, MSS, FDSD, and MFC all can affect wool value too. In respect to MFC, and given there was nearly an 8 degree difference in curvature difference between H1H1 and H2H2 sheep, research with mice has revealed that two missense mutations in the first exon of mouse KRT71 lead to the production of wavy hair [23] and Kuramoto et al. [24] demonstrated that in rats, the Rex mutation in KRT71 (a 7 bp deletion at the splice acceptor site of intron 1) causes curly hair.
While the associations between variation in keratin genes and wool fibres traits are noteworthy, it should also be noted that several studies have linked variation in the keratin-associated proteins with variation in wool traits. These associations are in part summarized in Gong et al. [25]. It must also be remembered that wool growth is influenced by dietary factors, for example with Coetzee et al. [26] illustrating how infusion of the amino acids methionine, lysine, and rumen-protected methionine derivates affect wool growth rate. Future studies could therefore investigate the relationship, if it exists, between specific dietary factors or regimes and the expression of the keratin genes.
4.3. Localization of KRT84 Gene in the Skin of Gansu Alpine Fine-Wool Sheep at Different Lamb Ages
In this experiment, the presence of the wool keratin protein K84 was investigated in the skin and wool follicles at different stages of post-natal growth, using a K84 specific antibody and immunofluorescent detection methods. This revealed that the protein was present in the follicle IRS, the ORS, and the HM over the different ages of the lambs. This contrasts with some of the findings of Yu et al. [27]. They reported that KRT84 could be amplified from total sheep skin RNA from the Wiltshire breed (a shedding sheep breed) using primers designed from the bovine KRT84 ortholog, but they only obtained a weak signal using an RT-PCR approach. Analysis of where the gene appeared to be expressed suggested expression in the fibre cuticle, the medulla, and what is called the ‘brush end’. The function of the IRS may be to determine the shape of the fibre during hair growth, and its growth characteristics and defects may affect the shape of the hair, resulting in hairs with curved traits. Yu et al. [27] revealed that the KRT32, KRT35 and KRT85 genes were expressed in both the IRS and fibre cuticle, while KRT36 and KRT87 were expressed only in cortical layers. Only KRT34 and KRT36 had high levels of expression in the HM, but KRT82 was widely expressed from the middle of the hair bulb to the upper part of the cuticle.
In a study of Merino sheep wool, Hynd et al. [28] found that different growth rates of cortical cells in the upper part of the wool bulb resulted in a curved trait. It was found that some keratin genes were asymmetrically expressed, including KRT32, KRT35, and KRT85, with these genes expressed in the cuticle and fibre cortex and where they appear to be involved in the differentiation of the cortex. This asymmetry was not obvious for KRT35 and KRT85 in the work of Yu et al. [27], but KRT32 may have been asymmetrically distributed and KRT38 appeared to be asymmetrically distributed in the cortex. While the distribution and location of KRT expression appears inconsistent, which may reflect differences between different animals and breeds, Hynd et al. [28] have suggested that the associations between cortical properties and fibre crimp are not consistent and that they therefore may not reflect the underlying causation of fibre curvature. They instead suggested that variations in fibre crimp could be accounted for by quantitative differences in both the degree of mitotic asymmetry in follicle bulbs and the distance from the bulb to the point at which keratinization is completed.
Based on the effect of KRT84 variation on several wool traits in Gansu Alpine Fine-wool sheep in this study, it is hypothesized that the gene plays an important role in the development of the wool follicles and therefore the fibre, where it appears to affect wool traits. However, further studies on the relationship between KRT84 (and other KRTs and keratin-associated protein genes) and wool traits are needed to fully understand how these genes affect fibre properties, which ultimately may enable the development of hair disease therapies, keratin-based cosmetic ingredients, and for sheep selection to improve wool quality. In this respect, improving the quality of the wool produced by Gansu Alpine Fine-wool sheep would not only better meet the diversified needs of markets but also likely increase the income of farmers.
5. Conclusions
Expression of the KRT84 gene appears to vary during post-natal growth (days 0 to 270) and vary across different locations in the skin. This lays a foundation for more research into the level and location of expression of other keratin and keratin-associated protein genes. The discovery of associations between KRT84 diplotypes and wool traits suggests that KRT84 affects MFD, MSL, MFC, CF and MSS, with sheep with the H3H3 diplotype appearing to have increased MFD, CVFD and MSS and reduced MSL and CF compared to H1H1 sheep. The gene KRT84 might therefore be useful as a gene-marker for breeding Gansu Alpine Fine-wool sheep and other breeds too, if that same variation is found in them.
Author Contributions
X.Y. wrote the draft manuscript; B.S. undertook the data curation; Z.Z. performed various technical analyses; S.L. obtained the research funding, administered the project, and managed resources; J.H. and Z.H. performed some of the investigation; X.L. and M.L. undertook some of the analyses; F.Z. analysed the data with the various software packages; J.W. undertook various validations; H.Z. undertook the results visualization; J.G.H.H. undertook the manuscript writing, reviewing, and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study was reviewed and approved by the Faculty Animal Policy and Welfare Committee of Gansu Agricultural University (Ethic approval file No. GSAU-LC-2020-019), 15 April 2020, and the Faculty Animal Policy and Welfare Committee of Gansu Agricultural University (Ethic approval file No. GSAU-ETH-AST-2021-028), 2 March 2021.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This study was financially supported by the National Natural Science Foundation of China (32060140), the Distinguished Young Scholars Fund of Gansu Province (21JR7RA857), Discipline Team Project of Gansu Agricultural University (GAU-XKTD-2022-21), Fuxi Young Talents Fund of Gansu Agricultural University (Gaufx-03Y04), and Gansu Provincial Key R&D Program Projects (22YF7WA115).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Salas P.J., Forteza R., Mashukova A. Multiple roles for keratin intermediate filaments in the regulation of epithelial barrier function and apico-basal polarity. Tissue Barriers. 2016;4:e1178368. doi: 10.1080/21688370.2016.1178368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimomura Y., Cristiano A.M. Biology and genetics of hair. Annu. Rev. Genom. Hum. Gen. 2010;11:109–132. doi: 10.1146/annurev-genom-021610-131501. [DOI] [PubMed] [Google Scholar]
- 3.Forni M.F., Trombetta-Lima M., Sogayar M.C. Stem cells in embryonic skin development. Biol. Res. 2012;45:215–222. doi: 10.4067/S0716-97602012000300003. [DOI] [PubMed] [Google Scholar]
- 4.Saxena N., Mok K.-W., Rendl M. An updated classification of hair follicle morphogenesis. Exp. Dermatol. 2019;28:332–344. doi: 10.1111/exd.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langbein L., Rogers M.A., Winter H., Praetzel S., Beckhaus H.-R., Schweizer J. The catalog of human hair keratins. I. Expression of the nine type I members in the hair follicle. J. Biol. Chem. 1999;274:19874–19884. doi: 10.1074/jbc.274.28.19874. [DOI] [PubMed] [Google Scholar]
- 6.Eckhart L., Dalla Valle L., Jaeger K. Identification of reptilian genes encoding hair keratin-like proteins suggests a new scenario for the evolutionary origin of hair. Proc. Natl. Acad. Sci. USA. 2008;105:18419–18423. doi: 10.1073/pnas.0805154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frenkel M.J., Powell B.J., Ward K.A., Sleigh M.J., Rogers G.E. The keratin BIIIB gene family: Isolation of cDNA clones and structure of a gene and a related pseudogene. Genomics. 1989;4:182–191. doi: 10.1016/0888-7543(89)90298-X. [DOI] [PubMed] [Google Scholar]
- 8.Langbein L., Rogers M.A., Winter H., Praetzel S., Schweizer J. The catalog of human hair keratins. II. Expression of the six type II members in the hair follicle and the combined catalog of human type I and II keratins. J. Biol. Chem. 2001;276:35123–35132. doi: 10.1074/jbc.M103305200. [DOI] [PubMed] [Google Scholar]
- 9.McLaren R.J., Rogers G.R., Davies K.P., Maddox J.F., Montgomery G.W. Linkage mapping of wool keratin and keratin-associated protein genes in sheep. Mamm. Genome. 1997;8:938–940. doi: 10.1007/s003359900616. [DOI] [PubMed] [Google Scholar]
- 10.Gortari M.J.D., Freking B.A., Kappes S.M., Leymaster K.A., Stone R.T., Beattie C.W., Crawford A.M. Extensive genomic conservation of cattle microsatellite heterozygosity in sheep. Anim. Genet. 1997;28:274–290. doi: 10.1111/j.1365-2052.1997.00153.x. [DOI] [PubMed] [Google Scholar]
- 11.Naboulsi R., Cieślak J., Headon D., Jouni A., Negro J.J., Andersson G., Lindgren G. The Enrichment of Specific Hair Follicle-Associated Cell Populations in Plucked Hairs Offers an Opportunity to Study Gene Expression Underlying Hair Traits. Int. J. Mol. Sci. 2022;29:561. doi: 10.3390/ijms24010561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Luo Z., Zhang Y., Yuan D., Ge W., Wang X. The inconsistent regulation of HOXC13 on different keratins and the regulation mechanism on HOXC13 in cashmere goat (Capra hircus) BMC Genom. 2018;19:630. doi: 10.1186/s12864-018-5011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langbein L., Rogers M.A., Praetzel-Wunder S., Böckler D., Schirmacher P., Schweizer J. Novel type I hair keratins K39 and K40 are the last to be expressed in differentiation of the hair: Completion of the human hair keratin catalog. J. Investig. Dermatol. 2007;127:1532–1535. doi: 10.1038/sj.jid.5700734. [DOI] [PubMed] [Google Scholar]
- 14.Sun H., He Z., Xi Q., Zhao F., Hu J., Wang J., Liu X., Zhao Z., Li M., Luo Y., et al. Lef1 and Dlx3 May Facilitate the Maturation of Secondary Hair Follicles in the Skin of Gansu Alpine Merino. Genes. 2022;13:1326. doi: 10.3390/genes13081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao X., Yao L., Wang X., Li S., Qin J., Yang L., He L., Zhang W. Hair Follicle Development and Cashmere Traits in Albas Goat Kids. Animals. 2023;13:617. doi: 10.3390/ani13040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–4088. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Chai W., Zhou H., Gong H., Wang J., Hickford J.G.H. Nucleotide variation in the ovine KRT31 promoter region and its association with variation in wool traits in Merino-cross lambs. J. Agric. Sci. 2019;157:182–188. doi: 10.1017/S0021859619000406. [DOI] [Google Scholar]
- 18.Sumner R.M.W., Forrest R.H.J., Zhou H., Henderson H.V., Hickford J.G.H. Association of the KRT33A (formerly KRT1.2) gene with live-weight and wool characteristics in yearing Perendale sheep. Proc. New Zealand Soc. Anim. Prod. 2013;73:158–164. [Google Scholar]
- 19.Chai W., Zhou H., Forrest R.H.J., Gong H., Hodge S., Hickford J.G.H. Polymorphism of KRT83 and its association with selected wool traits in Merino-cross lambs. Small Rumin. Res. 2017;155:6–11. doi: 10.1016/j.smallrumres.2017.08.019. [DOI] [Google Scholar]
- 20.Harland D.P., Popescu C., Richena M., Deb-Choudhury S., Wichlatz C., Lee E., Plowman J.E. The susceptibility of disulfide bonds to modification in keratin fibers undergoing tensile stress. Biophys. J. 2022;121:2168–2179. doi: 10.1016/j.bpj.2022.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itenge T.O., Hickford J., Forrest R., McKenzie G., Frampton C. Association of variation in the ovine KAP1.1, KAP1.3 and K33 genes with wool traits. Int. J. Sheep Wool Sci. 2010;58:1–19. [Google Scholar]
- 22.Chai W., Zhou H., Gong H., Hickford J.G.H. Variation in the ovine KRT34 promoter region affects wool traits. Small Rumin. Res. 2022;206:106586. doi: 10.1016/j.smallrumres.2021.106586. [DOI] [Google Scholar]
- 23.Kikkawa Y., Oyama A., Ishii R., Miura I., Amano T., Ishii Y., Yoshikawa Y., Masuya H., Wakana S., Shiroishi T., et al. A small deletion hotspot in the type II keratin gene mK6irs1/Krt2-6g on mouse chromosome 15, a candidate for causing the wavy hair of the caracul (Ca) mutation. Genetics. 2003;165:721–733. doi: 10.1093/genetics/165.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuramoto T., Hirano R., Kuwamura M., Serikawa T. Identification of the rat Rex mutation as a 7-bp deletion at splicing acceptor site of the Krt71 gene. J. Vet. Med. Sci. 2010;72:909–912. doi: 10.1292/jvms.09-0554. [DOI] [PubMed] [Google Scholar]
- 25.Gong H., Zhou H., Forrest R.H.J., Li S., Wang J., Dyer J.M., Luo Y., Hickford J.G.H. Wool keratin-associated protein genes in sheep—A review. Genes. 2016;7:24. doi: 10.3390/genes7060024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coetzee J., de Wet P.J., Burger W.J. Effects of infused methionine, lysine and rumen-protected methionine derivatives on nitrogen retention and wool growth of Merino wethers. S. Afr. J. Anim. Sci. 1995;25:87–94. [Google Scholar]
- 27.Yu Z., Wildermoth J.E., Wallace O.A., Gordon S.W., Maqbool N.J., Maclean P.H., Nixon A.J., Pearson A.J. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Exp. Dermatol. 2011;20:582–588. doi: 10.1111/j.1600-0625.2011.01274.x. [DOI] [PubMed] [Google Scholar]
- 28.Hynd P.I., Edwards N.M., Hebart M., McDowall M., Clark S.A. Wool fibre crimp is determined by mitotic asymmetry and position of final keratinisation and not ortho- and para-cortical cell segmentation. Animal. 2009;3:838–843. doi: 10.1017/S1751731109003966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.