Abstract
The Gcn4p activation domain contains seven clusters of hydrophobic residues that make additive contributions to transcriptional activation in vivo. We observed efficient binding of a glutathione S-transferase (GST)–Gcn4p fusion protein to components of three different coactivator complexes in Saccharomyces cerevisiae cell extracts, including subunits of transcription factor IID (TFIID) (yeast TAFII20 [yTAFII20], yTAFII60, and yTAFII90), the holoenzyme mediator (Srb2p, Srb4p, and Srb7p), and the Adap-Gcn5p complex (Ada2p and Ada3p). The binding to these coactivator subunits was completely dependent on the hydrophobic clusters in the Gcn4p activation domain. Alanine substitutions in single clusters led to moderate reductions in binding, double-cluster substitutions generally led to greater reductions in binding than the corresponding single-cluster mutations, and mutations in four or more clusters reduced binding to all of the coactivator proteins to background levels. The additive effects of these mutations on binding of coactivator proteins correlated with their cumulative effects on transcriptional activation by Gcn4p in vivo, particularly with Ada3p, suggesting that recruitment of these coactivator complexes to the promoter is a cardinal function of the Gcn4p activation domain. As judged by immunoprecipitation analysis, components of the mediator were not associated with constituents of TFIID and Adap-Gcn5p in the extracts, implying that GST-Gcn4p interacted with the mediator independently of these other coactivators. Unexpectedly, a proportion of Ada2p coimmunoprecipitated with yTAFII90, and the yTAFII20, -60, and -90 proteins were coimmunoprecipitated with Ada3p, revealing a stable interaction between components of TFIID and the Adap-Gcn5p complex. Because GST-Gcn4p did not bind specifically to highly purified TFIID, Gcn4p may interact with TFIID via the Adap-Gcn5p complex or some other adapter proteins. The ability of Gcn4p to interact with several distinct coactivator complexes that are physically and genetically linked to TATA box-binding protein can provide an explanation for the observation that yTAFII proteins are dispensable for activation by Gcn4p in vivo.
Transcription initiation by RNA polymerase II (Pol II) requires assembly of a large complex consisting of Pol II and general transcription factors (GTFs) at the promoter. It has been proposed that assembly of this complex begins when TFIID, consisting of TATA box-binding protein (TBP) and its associated factors (TAFII proteins), binds to the core promoter, followed by sequential binding of other GTFs and Pol II itself (9). In another scenario, Pol II, certain GTFs, and coactivator proteins bind to the promoter as a preformed holoenzyme complex (46). Transcriptional activators bind to the promoter, generally upstream of the TATA element, and stimulate the assembly or function of the transcription initiation complex. Binding of TFIID to the core promoter appears to be rate limiting for initiation (12, 43, 88), and certain activators stimulate this step in initiation complex formation (3, 11, 21, 39, 40, 50, 91). Several activators bind TBP in vitro in a manner that depends on amino acids in the activation domain that are critical for transcriptional activation in vivo (7, 11, 26, 35, 38, 51, 61–63), suggesting that direct interactions between the activator and TBP are involved in recruiting TFIID to the core promoter. Certain activation domains also bind TFIIB in vitro in a sequence-specific manner (4, 7, 14, 41, 56, 91) and may stimulate recruitment of this GTF to the initiation complex (15, 41, 55, 56).
Other studies suggest that activator function is mediated by one or more of the TAFII coactivator proteins associated with TBP in TFIID. Different activators may require specific TAFII proteins for activation (13, 74–76), and indeed, certain activation domains bind preferentially to specific TAFII proteins in vitro (24, 37, 57, 83). The interactions between activators and TAFII proteins may serve primarily to recruit TFIID to the promoter (75). The human TAFII250 subunit (and its Saccharomyces cerevisiae homolog yTAFII130) has histone acetyltransferase (HAT) activity that may also promote initiation complex formation by destabilizing a repressive nucleosome structure at the promoter (64). A yeast Pol II-TAFII complex was shown to be required for transcriptional activation of a Gcn4p-regulated promoter in vitro (44); however, recent studies indicate that yTAFII proteins are not essential for transcriptional activation in vivo by Gcn4p and by several other yeast activator proteins (65, 85).
Activators can interact with coactivator proteins besides TAFII proteins to stimulate transcription initiation. The VP16 activation domain was shown to interact with one or more constituents of the mediator complex associated with a holoenzyme form of yeast RNA Pol II (27). The mediator is a multisubunit complex composed of numerous SRB proteins and Gal11p that functionally and physically interacts with the carboxy-terminal domain of the largest subunit of Pol II (42, 47). The mediator can support activated transcription in vitro by VP16 and Gcn4p (42, 47), and at least some of its components (Srb2p, Srb10p, and Srb11p) are required in vivo for transcriptional activation by Gal4p (45, 48, 53). In fact, it appears that virtually all Pol II transcription is dependent on Srb4p and Srb6p, which are essential for viability (82).
VP16 also binds specifically to the yeast coactivator protein Ada2p, and this interaction appears to be important for transcriptional activation by VP16 in yeast (5, 79). Ada2p, Ada3p, Gcn5p, Ada1p, and Spt20p (Ada5p) are required for high-level transcription by several yeast activators, including Gcn4p (6, 14, 22, 23, 34, 58, 67), and these proteins appear to be associated with one another (10, 22, 33, 34, 58, 59, 73) in a high-molecular-weight complex of ca. 1.8 mDa (25). This large complex also contains Spt3p and Spt7p, which together with Spt20p (Ada5p) functionally interact with TBP (19). Ada2p, Ada3p, and Gcn5p have also been found in lower-molecular-weight complexes devoid of the SPT proteins (25, 73), and it is not known whether a given activator can interact with both kinds of Adap-Gcn5p complexes. Gcn5p has HAT activity that is important for its coactivator function in vivo (86); thus, recruitment of an Adap-Gcn5p complex by an activator may serve to remove a repressive chromatin structure at the promoter. In addition, there is both genetic (34, 58) and biochemical evidence (5, 73) that the Adap-Gcn5p complex can bridge an interaction between activation domains and TBP in yeast, possibly providing a means of recruiting TBP to the promoter independently of the TAFII proteins in TFIID.
Several activators contain multiple activation domains and are able to promote high-level transcription with only a subset of these domains. This complexity was first demonstrated for yeast Gcn4p (30) and also applies to VP16 (70, 84). It has been proposed that the multiple activation domains in these proteins have redundant functions and that efficient activation requires only a critical number of domains to be present in the activator. This interpretation is supported by the fact that wild-type levels of activation can be achieved with artificial activators containing reiterated copies of a single domain (7, 77, 81). There is also evidence that a single activator can function in multiple ways to stimulate transcription. As mentioned above, VP16 has been implicated in direct interactions with TBP, TFIIB, dTAFII40, the mediator complex of yeast holoenzyme, and Ada2p. One way to explain these findings is to propose that many different GTFs and coactivators contain similar surfaces that can interact with the same residues in a given activation domain. Alternatively, the structure of an activation domain may be induced upon interaction with its target proteins, allowing it to assume different conformations when it interacts with different factors or different segments of the same factor (17, 52, 54). In either case, a single activation domain may act sequentially at different steps in the initiation pathway, exchanging one interaction for another as the initiation complex is assembled and activated (89). The notions that activators can interact with multiple GTFs or coactivator proteins and that these interactions provide redundant means of stimulating assembly of the initiation complex are consistent with the fact that tethering either TBP (12, 43, 88), TAFII proteins (2, 44), or a component of the holoenzyme mediator (20a, 80) to a promoter by fusing it to a DNA-binding domain can bypass the requirement for an activator. Presumably, bringing any one of these factors to the promoter is sufficient to nucleate assembly of the transcription initiation complex.
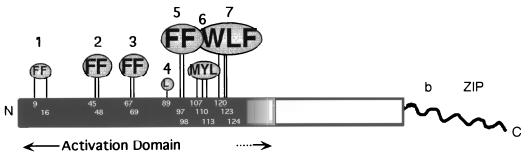
Gcn4p is a transcriptional activator of multiple genes encoding amino acid biosynthetic enzymes in S. cerevisiae. Expression of GCN4 is regulated at the translational level such that high levels of the protein are produced only in nutrient-deprived cells where its activation function is required (reviewed in reference 29). Gcn4p binds to DNA as a homodimer (32) and its DNA binding and dimerization (bZIP) domains are located at the extreme C terminus of the protein (20, 31). In previous studies, we showed that Gcn4p contains seven clusters of aromatic or bulky hydrophobic residues spanning ca. 125 residues in the N-terminal half of the protein which make additive contributions to transcriptional activation by Gcn4p in vivo (18, 36) (Fig. 1). None of these hydrophobic clusters was essential for high-level activation, and with one exception, a substantial reduction in GCN4 function required simultaneous inactivation of three of the seven clusters.
FIG. 1.
Locations and relative importance of hydrophobic clusters in the Gcn4p activation domain. A diagram of Gcn4p protein is depicted with the DNA-binding (b) and dimerization (ZIP) domains at the extreme C terminus shown in α-helical conformation, as has been predicted from X-ray crystallography (20), and with the rest of the protein shown as a rectangular box. The critical hydrophobic residues identified in our previous studies (18, 36) are shown above the sequence at the appropriate positions in the protein by the single-letter code. The critical residues are grouped in seven hydrophobic clusters, symbolized by shaded ovals. The size and height of the lettering for each cluster is proportional to the reduction in GCN4 function seen in response to Ala substitutions at that site. The functions of these hydrophobic clusters are redundant; thus, several must be inactivated simultaneously to destroy GCN4 function. Mutation of F97 and F98 (cluster 5) and W120, L123, and F124 (cluster 7) produced the only situation where GCN4 function in vivo was greatly impaired by substitutions in only two of the seven clusters. At the other extreme, mutation of clusters 1 to 4 produced the only situation where a substantial amount of GCN4 function occurred with only three clusters left intact (18, 36).
It was reported that a C-terminal segment of the Gcn4p activation domain interacts with TBP; however, this interaction is weak compared to that observed between Gal4p and TBP and was not shown to be dependent on specific residues or segments of the activation domain (61). Ada2p interacted with the Gcn4p activation domain in cell extracts; however, it was not determined whether this interaction was direct or if it required the hydrophobic residues needed for activation by Gcn4p in vivo (5). There is also in vitro evidence that activation by Gcn4p is mediated by the RNA Pol II holoenzyme (42) and by TFIID (44). Together, these findings suggest that Gcn4p may interact with multiple GTFs and coactivator proteins in order to stimulate transcription. It was important to address whether these and other potential interactions between Gcn4p and various proteins in the transcription initiation machinery are dependent on the critical hydrophobic residues identified in the Gcn4p activation domain. In addition, we wished to determine whether each hydrophobic cluster was optimized for interaction with a particular protein or, instead, could promote binding to any one of several factors with similar efficiencies. To answer this question, we examined physical interactions between Gcn4p and coactivator proteins in yeast whole-cell extracts and determined which of these interactions were impaired by Ala substitutions in the critical residues of the activation domain. The results of our analysis provide evidence that Gcn4p interacts specifically with TFIID, the Adap-Gcn5p coactivator complex, and the holoenzyme mediator complex.
MATERIALS AND METHODS
Plasmids.
Plasmids pCD35, pCD350, pCD322, pCD355, and pCD60 were described previously (18), as were plasmids p2195, p1705, p2053, p2241, p2067, p1890, p2052, and p2072 (36). Plasmid p1930 was constructed by first modifying pGEX-5x-3 (Pharmacia) to replace sequences between the BamHI and NotI sites with the oligonucleotide 5′ GATCACTCTCGAGAATCGAGAACTTAAG 3′, thereby destroying the BamHI and NotI sites in the polylinker and introducing XhoI and AflII sites, creating plasmid p1946. The 981-nucleotide (nt) XhoI-to-AflII fragment from pCD35 (18) was inserted between the corresponding sites in p1946. The resulting plasmid (p1933) was digested with XhoI, filled in with T4 DNA polymerase, and religated, creating a plasmid, p1930, that encodes a fusion between glutathione S-transferase (GST) and Gcn4p separated by a series of amino acids, IEGRGITLDRENKLNTNK, of which the first four correspond to the factor Xa cleavage site. p1927 was constructed by using PCR to replace the XhoI-to-BamHI fragment from p1933 with a nearly identical sequence (the difference being that 5′ AGA TCT 3′ [encoding Arg-Ser] was inserted between codons 17 and 18 of the Gcn4p coding sequences). Subsequently, this plasmid (p1928) was digested with XhoI, filled in with T4 DNA polymerase, and religated to create p1927, carrying a gene encoding a GST-Gcn4p fusion protein with the same linker described above for p1930. The following plasmids were constructed by replacing the 788-nt BamHI-AflII fragment in p1930 with the corresponding sequences from previously constructed plasmids (here shown in parentheses immediately following the newly constructed plasmid): p1949 (pCD350), p2144 (p2195), p2528 (pCD60), p2530 (p2053), p2531 (p1705), p2532 (p2067), p2533 (p2241), p2534 (pCD355), and p2535 (pCD322). p2292 was constructed by replacing the XhoI-KpnI fragment of p1928 with the XhoI-KpnI fragment of p2240. The resulting plasmid, p2291, was digested with XhoI, filled in with T4 DNA polymerase, and religated to generate p2292, carrying a gene encoding a GST-Gcn4p fusion protein with the same linker as that described above for p1930. Plasmid p2240 was constructed by replacing the SalI-BamHI fragment of p2195 with the corresponding fragment from p2631, which in turn was constructed by replacing the BstEII-BamHI fragment of pCD60 with the corresponding fragment synthesized by PCR with mutagenic primers and p1885 (36) as the template to introduce the amino acid substitutions F9A, F16A, F45A, and F48A. p2529 was constructed by replacing the 788-nt BamHI-AflII fragment in p1927 with sequences from pCD60. p2593 was constructed by replacing the 108-nt BglII-BamHI fragment in p1927 with sequences from p1890. p2591 was constructed by replacing the 288-nt BamHI-KpnI fragment in p1927 with sequences from p2052. p2595 was constructed by replacing the 288-nt BamHI-KpnI fragment in p2593 with sequences from p2052. p2536 was constructed by recombinant PCR replacing the 188-nt XhoI-BamHI fragment in p1933 with the same sequence but with Gcn4p codon changes to introduce the substitutions F9A, F16A, F45A, and F48A. (The GST and Gcn4p coding sequences are out of frame in construct p2536.) p2589 was then constructed by replacing the 788-nt BamHI-AflII fragment in p2536 with sequences from p2052. The resulting plasmid, p2537, was digested with XhoI, filled in with Klenow fragment, and religated, placing the GST and Gcn4p coding sequences in frame with a linker consisting of the amino acid sequence GITLDRENKLNTNK. p2598 was constructed by replacing the 288-nt BamHI-KpnI fragment in p2536 with sequences from p2072. The resulting plasmid, p2596, was digested with XhoI, filled in with Klenow fragment, and religated, placing the GST and Gcn4p coding sequences in frame with a linker consisting of the amino acid sequence GITLDRENKLNTNK. The GST-VP16 constructs were constructed previously (55).
Strains.
Yeast strains YBY181, YBY40-8, DPY213, and DPY107 expressing hemagglutinin (HA)-tagged forms of yTAFII60, yTAFII90, yTAFII130, and Mot1p, respectively, were described previously (69). These strains have the genotype MATα leu2-3,112 ura3-52 his3-Δ200 suc2-Δ9 ade2-1 lys2-801 Δtaf::TRP1 and contain the corresponding HA-yTAFII allele on HIS3 plasmid pRS313. SY6-2 (MATa ura3-52 ade2-101 trp1-Δ1 lys2-801 his3-Δ200 leu2::PET56 ada3::TRP1::HA-ADA3::URA3) expressing HA-Ada3p (60), strain Z687 (MATa his3-198,200 leu2-3,112 ura3-52 srb10-198-1::hisG), and its isogenic SRB10 parent Z719 (53) also were described previously. Strain RMY10 (MATα his3-609 trp1-63 leu2-3,112 ura3-52 srb2::HIS3) was constructed by transforming strain H2451 with a 3.3-kb EcoRI fragment of pTK33 containing the srb2::HIS3 deletion-insertion allele (45). His+ transformants were selected on synthetic complete medium lacking histidine (78) and screened for slow growth at 30°C and inositol auxotrophy, phenotypes characteristic of srb2 mutants. The phenotypes of one such transformant were suppressed by a plasmid containing SRB2 (pCT24) (45), and the srb2 deletion-disruption was confirmed by PCR with primers flanking SRB2 (53). Strains KNY104 (MATα leu2-3,112 ura3-52 ino1 trp1-Δ63 ada2Δ::hisG::URA3::hisG) and KNY105 (MATα leu2-3,112 ura3-52 ino1 trp1-Δ63 ada3Δ::TRP1), kindly provided by K. Natarajan, were constructed from strain H1511 (MATα leu2-3,112 ura3-52 ino1 trp1-Δ63) as follows. H1511 was transformed to Ura+ with the XhoI-BamHI fragment from the ADA2 disruption plasmid ADA2KO (6). Strains carrying ada2Δ::hisG::URA3::hisG were identified by screening the Ura+ transformants for a 3-aminotriazole (3-AT)-sensitivity phenotype. The presence of the ada2 disruption in transformant KNY104 was verified by its inability to complement the 3-AT sensitivity of an ada2Δ tester and to be complemented by a plasmid containing wild-type ADA2. To construct KNY105, a plasmid bearing the ada3::TRP1 allele (8) was digested with HindIII and integrated at ADA3 in H1511 by selecting for Ura+ transformants. Derivatives devoid of plasmid sequences in which ada3::TRP1 replaced the ADA3 sequences were selected on 5-fluoroorotic acid medium and identified by their 3-AT-sensitivity phenotype. The ada3Δ in KNY105 was confirmed by complementation for its 3-AT-sensitivity phenotype by introduction of a plasmid bearing ADA3.
Antibodies.
Polyclonal antiserum against Gcn4p (18) and monoclonal antibodies against Pab1p (1) were described previously, as were the polyclonal antibodies against Srb2p, Srb4p (82), Srb7p (27), and Srb10p (53). Mouse 12CA5 monoclonal HA antibodies purchased from Boehringer Mannheim (catalog no. 1583-816) were used to probe immunoblots of GST binding reaction mixtures, whereas 12CA5 antibodies purified from mouse ascites fluid (16) were used as immunoprecipitating antibodies and the resulting immune complexes were probed for HA with polyclonal rabbit antiserum HA.11 from Babco (catalog no. PRB-101C). Rabbit polyclonal antibodies against yTAFII90 were raised against segments containing residues 1 to 311 and 459 to 798 expressed in Escherichia coli. yTAFII130 rabbit antibodies were generated against the full-length protein expressed in Sf9 insect cells from a baculovirus vector. The yTAFII20 and TBP rabbit polyclonal antibodies were raised against the full-length proteins expressed in E. coli. Polyclonal antibodies against Ada2p were kindly provided by Shelley Berger.
Preparation of cell extracts and purified TFIID.
The preparation of yeast cell extracts was carried out essentially as described previously (87). Two different preparations of purified TFIID were used in pull-down reaction mixtures with GST-Gcn4p fusion proteins. For the experiment shown in Fig. 8A, we used a highly purified fraction prepared by affinity chromatography and with anti-TBP immunoglobulin G (as described in the legend to Fig. 1A of reference 68). The experiments shown in Fig. 8B to F involved a more highly concentrated fraction that was purified from strain DPY213 expressing HA-tagged yTAFII130 with a 12CA5 antibody-affinity column and elution of the bound TFIID with synthetic HA peptide (as described in the legend to Fig. 3C of reference 68). Bacterial cell extracts containing GST fusion proteins were prepared from transformants of strain DH10B. The transformants were grown and fusion proteins were induced under conditions recommended by Pharmacia, and cells were resuspended in 2.5 ml of lysis buffer (50 mM HEPES [pH 7.3], 200 mM potassium acetate, 12.5 mM magnesium acetate, 1 mM EGTA, 10% glycerol, 0.001 mM dithiothreitol, 0.01% Nonidet P-40) containing 1× Boehringer Mannheim complete protease inhibitor cocktail at ca. 80 μg/ml, 0.4 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, and 2.0 nM pepstatin. Cells were sonicated, and lysates were clarified by centrifugation at 14,000 rpm for 15 min in an Eppendorf 5415C microcentrifuge. We estimated that the GST fusion proteins were expressed at 1 to 2% of total bacterial protein.
FIG. 8.
yTAFII proteins in purified TFIID did not interact specifically with GST-Gcn4p. Components of purified TFIID were tested for the ability to interact specifically with GST-Gcn4p fusion proteins by mixing a fixed amount of purified TFIID with two different amounts of bacterial extract containing GST-Gcn4p fusion proteins bearing the wild-type activation domain or mutant activation domains with alanine substitutions in the hydrophobic clusters (numbered as shown in Fig. 1). The GST fusion proteins were precipitated and subjected to immunoblot analysis with polyclonal antibodies against the proteins indicated to the right of each panel, exactly as described for Fig. 3. (A and B) TFIID immunoaffinity purified with anti-TBP antibodies was incubated at a fixed concentration (60 ng per reaction mixture) with 100 or 200 μg of bacterial extracts containing GST proteins and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to ca. 300 μg. Lane 1 contained GST alone. Lanes 2 to 7 contained GST-Gcn4p fusion proteins with either wild-type (+) or mutant activation domains, with the clusters in which there were mutations being indicated by numbers in brackets across the top of the figure. Lanes 8 and 9 contained binding assay mixtures with the bacterial extract containing wild-type GST-Gcn4p and 1,200 μg of whole-cell extract (WCE) from yeast strain DPY213. Lane 10 contained 2/3 of the amount of TFIID used in the binding reaction mixtures in lanes 1 to 7 (40 ng), and lane 11 contained 1/30 of the yeast WCE used in the binding reaction mixtures shown in lanes 8 and 9. (C to G) TFIID bearing HA-yTAFII130 immunoaffinity purified with anti-HA antibodies was incubated at 300 ng per reaction mixture with 40 or 80 μg of bacterial extracts containing GST proteins and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of protein in each reaction mixture to ca. 80 μg. Lane 1 contained GST alone, and lanes 2 to 9 contained GST-Gcn4p fusion proteins with wild-type (+) or mutant activation domains (with cluster numbers indicated in brackets across the top of the blots). Lane 10 contained one-sixth of the TFIID used in the binding reaction mixtures in lanes 1 to 9. Lanes 12 and 13 contained a binding assay mixture with the bacterial extract containing wild-type GST-Gcn4p and 600 μg of WCE from strain DPY213. Lane 14 contained 1/15 of the yeast WCE used in the binding reaction mixtures in lanes 12 and 13. Lane 11 contained no sample.
FIG. 3.
Additive effects of mutations in hydrophobic clusters 5 to 7 of the activation domain in GST-Gcn4p fusion proteins on binding of yTAFII20, yTAFII60, and yTAFII90 in cell extracts. A fixed amount of yeast extract (containing 1,500 μg of protein) prepared from strain DPY213 expressing HA-yTAFII130 was incubated with three different amounts of bacterial extracts for each GST-Gcn4p fusion containing ca. 5, 10, and 20 μg of total bacterial protein and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to 23 μg. GST-Gcn4p fusion proteins contained the wild-type Gcn4p activation domain (+) (lanes 1 to 3, 14 to 16, and 27 to 29) or mutant activation domains with alanine substitutions in the hydrophobic clusters shown in brackets across the top of the figure. The designations are those adopted for Fig. 1. The GST fusion proteins were precipitated, resolved by SDS-PAGE, and subjected to immunoblot analysis with monoclonal anti-HA antibodies (to detect HA-yTAFII130) and then with polyclonal antibodies against the proteins indicated to the left of each panel. An enhanced-chemiluminescence system was used to detect the immune complexes. Lanes 13, 26, and 39 contain 1/20 of the input (In) amount of yeast extract employed in each of the binding reaction mixtures.
Binding reactions with GST fusion proteins.
Aliquots of bacterial extracts predetermined to contain equivalent amounts of each GST fusion protein were added to an appropriate amount of control extract prepared from a transformant of strain DH10B bearing the empty vector pUC19 to produce a mixture containing a fixed amount of total bacterial protein for a given panel of binding reactions (see below). To this mixture was added 4 μl of ethidium bromide (10 mg/ml), 20 μl of 5× binding buffer containing protease inhibitors (described above), an aliquot of yeast cell extract generally containing 1,500 μg of total protein, and water to a final volume of 100 μl. Tubes were incubated on ice for 1 h and then mixed with 15 μl of glutathione-Sepharose 4B resin (Pharmacia) resuspended in 1× binding buffer. The resulting mixture was gently agitated at 4°C for 1 h. The resin was collected by centrifugation at 4,000 rpm for 4 min at 4°C in an Eppendorf 5415C microcentrifuge and washed three times by adding 500 μl of 1× binding buffer with gentle mixing. The final pellets were resuspended in 3× Laemmli sample buffer (49) and stored at −70°C. Samples were boiled for 3 min and cooled on ice prior to fractionation by sodium dodecyl sulfate (SDS)–8 to 16% polyacrylamide gel electrophoresis (PAGE). The resolved proteins were analyzed by immunoblotting as described previously (18). Quantitation of the band intensities was carried out by scanning the films with a Lacie Silverscanner III and the National Institutes of Health Image version 1.61 software.
Immunoprecipitations.
Aliquots of yeast extract containing 1,250 μg of protein from strains containing the HA-tagged proteins of interest, prepared as described above, were mixed with 2 μl of ethidium bromide (10 mg/ml) and 5 μl of anti-HA antibody purified from mouse ascites fluid (16) in a final volume of 50 μl of the binding buffer described above (including the protease inhibitors) and incubated for 2 h on ice. Twenty-five microliters of protein A-Sepharose was added, and the resulting mixture was gently agitated for 1 h at 4°C. The mixture was centrifuged at maximum speed in an Eppendorf microcentrifuge for 5 s, the supernatant was reserved, and the pellet was washed three times with 100 μl of binding buffer. The proteins were resolved by SDS-PAGE and analyzed by immunoblotting as described previously (18).
RESULTS
The hydrophobic clusters make additive contributions to binding of yTAFII proteins, SRB proteins, and Ada3p to GST-Gcn4p in cell extracts.
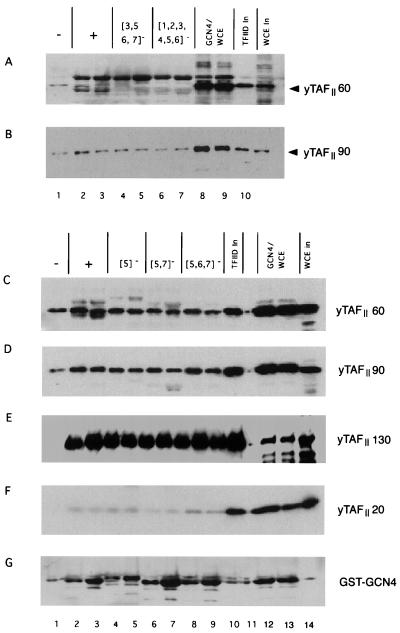
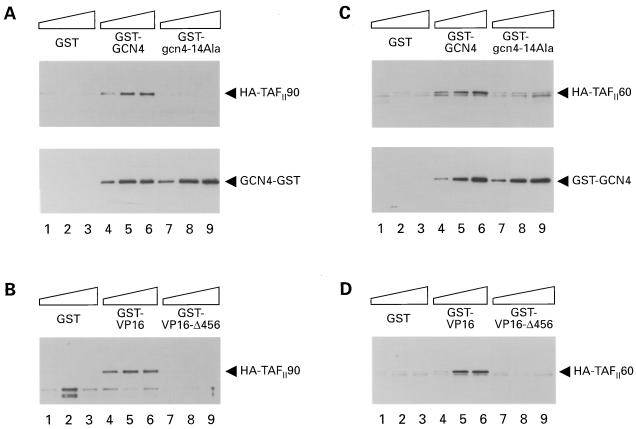
Our previous mutational analyses of the Gcn4p activation domain led to the identification of seven clusters of hydrophobic residues which make additive and redundant contributions to transcriptional activation of the HIS3 and HIS4 genes in vivo (Fig. 1). Table 1 summarizes the phenotypes of a representative set of gcn4 alleles containing different combinations of alanine substitutions in the seven hydrophobic clusters of the activation domain. To determine whether the hydrophobic clusters mediate physical interactions between Gcn4p and transcriptional coactivators, we expressed in E. coli fusion proteins between GST and either wild-type Gcn4p or the Ala-substituted mutant proteins listed in Table 1 and tested them for interactions with various coactivator proteins in a yeast cell extract. After the appropriate bacterial and yeast cell extracts were mixed together, the GST-Gcn4p proteins were precipitated with glutathione-Sepharose beads and the precipitates were probed by immunoblot analysis with antibodies against the coactivator proteins of interest. We first examined interactions between Gcn4p and two components of the TFIID complex, yTAFII60 and yTAFII90, by using the wild-type GST-Gcn4p fusion and a mutant protein containing 14 Ala substitutions in six of the seven hydrophobic clusters (14-Ala) that render GCN4 nonfunctional in vivo (Table 1). For comparison, we examined a GST fusion containing residues 413 to 490 from the VP16 activation domain and a truncation of this protein C terminal to residue 456, which eliminates the ability of VP16 to activate transcription in yeast (5). The yeast cell extracts were prepared from strains containing HA epitope-tagged versions of yTAFII60 or yTAFII90. Both HA-yTAFII60 and HA-yTAFII90 were specifically precipitated with the wild-type GST-Gcn4p protein at levels greater than that observed with the 14-Ala mutant or with GST alone (Fig. 2A and C, upper blots). Immunoblot analysis with Gcn4p antibodies confirmed that similar levels of wild-type and 14-Ala GST-Gcn4p proteins were recovered in the precipitated fractions (Fig. 2A and C, lower blots). Similar results were obtained for the wild-type GST-VP16 protein which showed greater binding to HA-yTAFII60 and HA-yTAFII90 than did GST-VP16Δ456 or GST alone (Fig. 2B and D). These results suggested that GST-Gcn4p interacted with HA-yTAFII60 and HA-yTAFII90 in a manner that depended on hydrophobic residues in the Gcn4p activation domain.
TABLE 1.
Summary of the effects of Ala substitutions in critical hydrophobic residues in the Gcn4p activation domain on transcriptional activation of HIS3 in vivoa
| GCN4 allele | Ala substitutions in Gcn4p residues | In vivo activation of HIS3 | GST-Gcn4p construct |
|---|---|---|---|
| Wild type | None (wild type) | 4+ | p1930, p1927,b p2528,c p2529d |
| {2}− | F45, F48 | 4+ | p2593b |
| {3}− | F67, F69 | 4+ | p2591d |
| {5}− | F97, F98 | 4+ | p2532c |
| {6}− | M107, Y110, L113 | 4+ | p2531e |
| {7}− | W120, L123, F124 | 4+ | p2530c |
| {2, 3}− | F45, F48, F67, F69 | 3+ | p2595d |
| {5, 6}− | F97, F98, M107, Y110, L113 | 3+ | p2535e |
| {5, 7}− | F97, F98, W120, L123, F124 | −/+ | p2534c |
| {6, 7}− | M107, Y110, L113, W120, L123, F124 | 3+ | p2533 |
| {1, 2, 3}− | F9, F16, F45, F48, F67, F69 | 2+ | p2589d |
| {5, 6, 7}− | F97, F98, M107, Y110, L113, W120, L123, F-124 | − | p1949c |
| {1, 2, 3, 5}− | F9, F16, F45, F48, F67, F69, F97, F98 | − | p2598d |
| {3, 5, 6, 7}− | F67, F69, F97, F98, M107, Y110, L113, W120, L123, F124 | − | p2144c |
| {1, 2, 3, 5, 6, 7}− 14-Ala | F9, F16, F45, F48, F67, F69, F97, F98, M107, Y110, L113, W120, L123, F124 | − | p2292d |
The GCN4 alleles are designated according to the seven hydrophobic clusters in the activation domain, numbered from the N to the C terminus, that contain alanine substitutions in the critical residues. In vivo activation of HIS3 expression was assayed by measuring resistance to 3-AT of transformants of a gcn4Δ strain bearing the indicated GCN4 alleles on single-copy-number plasmids. 3-AT is a competitive inhibitor of the HIS3 product, and growth on minimal medium containing this inhibitor at a concentration of 30 mM is absolutely dependent on transcriptional activation of HIS3 by Gcn4p. The data summarized in column 3 were published previously (18, 36).
Contains the triplets AGA and TCT inserted between codons 17 and 18 of the Gcn4p coding sequences.
Contains the triplets AGA and TCT inserted between codons 100 and 101 of the Gcn4p coding sequences.
Contains the triplets AGA and TCT inserted between codons 17 and 18 and between codons 100 and 101 of the Gcn4p coding sequences.
Contains the triplets AGA and TCT inserted between codons 100 and 101 and between codons 117 and 118 of the Gcn4p coding sequences.
FIG. 2.
Binding of yTAFII90 and yTAFII60 in cell extracts to a GST-Gcn4p fusion protein is dependent on the critical hydrophobic residues in the Gcn4p activation domain. (A and C) Aliquots of bacterial extracts containing approximately 25, 50, or 75 μg of total protein from strains expressing different GST fusion proteins were mixed with the appropriate amount of a control bacterial extract lacking a GST fusion protein (to bring the amount of bacterial protein in each reaction mixture to ca. 75 μg) and an aliquot of yeast extract containing 500 to 1,000 μg of protein from strains YBY40-8 and YBY181 expressing HA-yTAFII90 (A) or HA-yTAFII60 (C) as described in Materials and Methods. GST-GCN4 contains the wild-type Gcn4p activation domain, whereas GST–gcn4–14 Ala contains the 14 Ala substitutions in hydrophobic clusters 1 to 3 and 5 to 6 shown in Table 1. The GST fusion proteins were precipitated from the reaction mixtures with glutathione-Sepharose 4B resin, and the precipitated proteins were resolved by SDS-PAGE and subjected to immunoblot analysis with monoclonal antibodies against the HA epitope to detect the HA-tagged proteins (upper blots) with an enhanced-chemiluminescence system to detect immune complexes. Subsequently, the blots were stripped and reprobed with polyclonal antibodies against Gcn4p to detect the GST-Gcn4p proteins (lower blots). (B and D) The same yeast cell extracts were incubated with bacterial extracts containing GST-VP16 (bearing the wild-type VP16 activation domain), GST-VP16 Δ456 (bearing the truncated activation domain), or GST alone and processed exactly as described for panels A and C, except that the immunoblots were probed only with anti-HA antibodies.
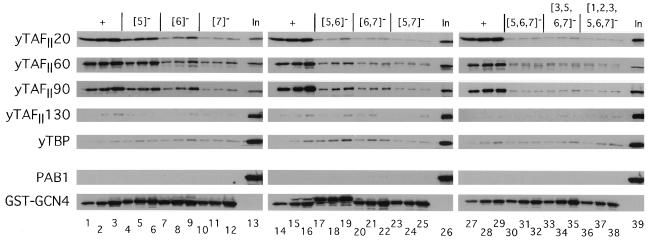
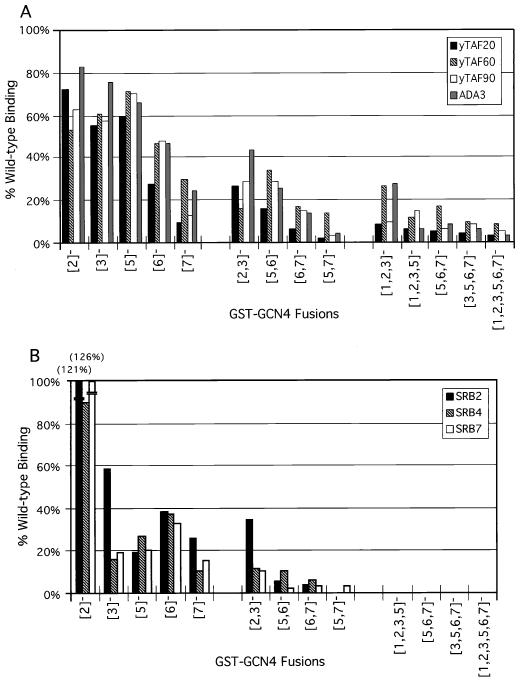
We proceeded to carry out additional binding experiments with a panel of GST-Gcn4p fusions containing mutations in the fifth, sixth, and seventh hydrophobic clusters (numbered from the N terminus as shown in Fig. 1 and Table 1) and yeast cell extracts from a strain expressing HA-tagged yTAFII130 (Fig. 3). Quantification of these data (Fig. 4A) showed that the cluster 5 mutations had the smallest effect, that cluster 6 mutations had somewhat greater effects, and that cluster 7 mutations had the largest effects on binding of yTAFII20, yTAFII60, and yTAFII90 to GST-Gcn4p. The combination of mutations in clusters 5 and 6 ({5, 6}−) reduced binding to a greater extent than did either the {5}− or {6}− single-cluster mutations, whereas the {6, 7}− combination led to essentially the same reduction in yTAFII20, -60, and -90 binding caused by the {7}− single-cluster mutation. The {5, 7}− mutation appeared to be the most severe double-cluster mutation, reducing binding to only 2 to 13% of the wild-type levels (Fig. 3 and 4A). The more extensively mutated proteins analyzed in Fig. 3 showed low-level binding to the three yTAFII proteins, similar to what was seen for the {5, 7}− mutant protein (Fig. 3 and 4A) and GST alone (Fig. 2 and data not shown). It is noteworthy that the different mutations in Gcn4p reduced binding of yTAFII20, yTAFII60, and yTAFII90 by similar amounts, consistent with the idea that these proteins interact with Gcn4p as components of the same complex (Fig. 4A).
FIG. 4.
Quantitation of the effects of mutations in the hydrophobic clusters in GST-Gcn4p fusions on binding to yTAFII20, -60, and -90; Ada3p; and Srb2p, -4p, and -7p in yeast cell extracts. The amounts of the different coactivator proteins that were precipitated with each GST-Gcn4p fusion protein were determined by densitometric scanning of the immunoblots shown in Fig. 3, 5, 6, and 7. The band intensities measured for the three binding reaction mixtures with different quantities of the GST fusion protein were summed and averaged for each mutant GST-Gcn4p construct, and these averages were divided by the corresponding value determined for the wild-type GST-Gcn4p analyzed in parallel. The resulting ratios were plotted on the y axis for each mutant GST-Gcn4p protein listed on the x axis. The designations are those adopted for Table 1.
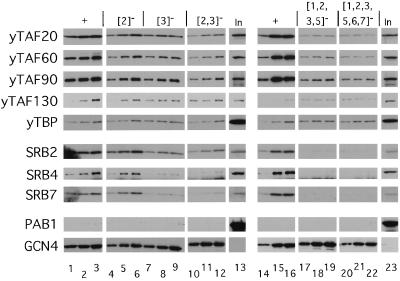
We obtained much different results when the same precipitated proteins were probed with anti-HA antibodies to visualize HA-tagged yTAFII130 or with antibodies against TBP. In the experiments described above, binding of yTAFII20, -60, and -90 to wild-type GST-GCN4 reached maximum values of 10 to 35% of the input amounts of these proteins in the extract. In contrast, only 1 to 2% of the HA-yTAFII130 and TBP in the extract was precipitated with wild-type GST-Gcn4p and there was little or no reduction in binding by either protein in response to single or multiple mutations in the hydrophobic clusters (Fig. 3 and 4A). In these respects, the results for HA-yTAFII130 and TBP more closely resembled those obtained for the negative control protein Pab1p than for the other yTAFII proteins (Fig. 3). These results are surprising in suggesting that, at least in cell extracts, yTAFII20, -60, and -90 can exist in stable TFIID subcomplexes lacking yTAFII130 and TBP that are competent for sequence-specific interactions with the Gcn4p activation domain.
To investigate whether Gcn4p interacts with holoenzyme mediator components, the protein precipitates analyzed for binding to yTAFII proteins and TBP described in the legend to Fig. 3 were also probed with antibodies against Srb2p, Srb4p, and Srb7p. The results (Fig. 5) were similar to those described above for yTAFII20, -60, and -90 in showing relatively high-level binding to the wild-type GST-Gcn4p fusion, reaching 10 to 15% of the input amounts of SRB proteins at the highest levels of GST-Gcn4p added to the reaction mixtures. In addition, binding of Srb2p, Srb4p, and Srb7p was completely dependent on the hydrophobic clusters in the Gcn4p activation domain and a complete loss in binding required the introduction of mutations in multiple clusters (Fig. 5). From quantitation of the binding data, it appeared that the {5}− single-cluster mutation and the {5, 6}− and {6, 7}− double-cluster mutations had greater effects on the interaction with SRB proteins than on the interaction with yTAFII proteins (compare Fig. 4A and B). As observed for the three yTAFII proteins, the {5, 7}− mutations reduced binding of Srb2p, -4p, and -7p to the low background levels observed for the more extensively mutated proteins. The fact that the mutations in Gcn4p led to similar reductions in the binding of all three SRB proteins is consistent with the idea that Srb2p, -4p, and -7p interacted with GST-Gcn4p as subunits of the mediator complex. The observation that certain combinations of mutations in clusters 5 to 7 had greater effects on the binding of Srb2p, -4p, and -7p than that of yTAFII20, -60, and -90 is consistent with the idea that the mediator and TFIID complexes bind independently to GST-Gcn4p.
FIG. 5.
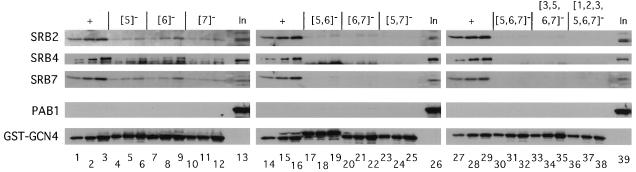
Additive effects of mutations in hydrophobic clusters 5 to 7 of the activation domain in GST-Gcn4p fusion proteins on binding of Srb2p, Srb4p, and Srb7p in yeast cell extracts. A fixed amount of yeast extract (containing 1,500 μg of protein) prepared from strain DPY213 expressing HA-yTAFII130 was incubated with three different amounts of bacterial extracts for each GST-Gcn4p fusion containing ca. 5, 10, and 20 μg of total bacterial protein and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to 23 μg. GST-Gcn4p fusion proteins contained the wild-type Gcn4p activation domain (+) (lanes 1 to 3, 14 to 16, and 27 to 29) or mutant activation domains with substitutions in the hydrophobic clusters shown in brackets across the top. The GST fusion proteins were precipitated and subjected to immunoblot analysis with polyclonal antibodies against the proteins indicated to the left of each panel, exactly as described for Fig. 3. Lanes 13, 26, and 39 contain 1/20 of the input (In) amount of yeast extract employed in each binding reaction mixture.
Figure 6 shows the results obtained for a set of GST-Gcn4p fusions containing mutations in hydrophobic clusters 1, 2, and 3. The {2}− and {3}− single-cluster mutations reduced binding of yTAFII20, -60, and -90 by modest amounts, whereas combining these mutations had an additive effect and reduced binding of all three yTAFII proteins to 16 to 29% (Fig. 4A, bar {2, 3}−, and Fig. 6). Combining the {1}− and {5}− mutations with the {2, 3}− double mutation led to an additional reduction in binding compared to that produced by {2, 3}− alone (Fig. 4A and 6). The results for Srb2p, -4p, and -7p were similar to those obtained for yTAFII20, -60, and -90, except that the {3}− and {2, 3}− mutations led to greater reductions in binding of Srb4p and Srb7p than those produced in the three yTAFII proteins (Fig. 4). This last result may indicate a particularly strong requirement for cluster 3 in the retention of Srb4p and Srb7p in mediator–GST-Gcn4p complexes.
FIG. 6.
Additive effects of mutations in hydrophobic clusters 1 to 3 of the activation domain in GST-Gcn4p fusion proteins on binding of yTAFII20, -60, and -90 and Srb2p, -4p, and -7p in yeast cell extracts. A fixed amount of yeast extract (containing 1,500 μg of protein) prepared from strain DPY213 expressing HA-yTAFII130 was incubated with three different amounts of bacterial extracts for each GST-Gcn4p fusion containing ca. 2, 4, and 8 μg of total bacterial protein and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to 9 μg. Shown are the GST-Gcn4p fusion proteins containing the wild-type Gcn4p activation domain (+) (lanes 1 to 3 and 14 to 16) or mutant activation domains with substitutions in the hydrophobic clusters shown in brackets across the top. The GST fusion proteins were precipitated and subjected to immunoblot analysis with polyclonal antibodies against the proteins indicated to the left of each panel, exactly as described for Fig. 3. Lanes 13 and 23 contain 1/20 of the input (In) amount of yeast extract employed in each binding reaction mixture.
It was shown previously that a GST fusion protein containing the Gcn4p activation domain can interact with Ada2p in a yeast nuclear extract (5). To investigate whether this interaction is dependent on the critical hydrophobic residues in Gcn4p, we employed the same set of GST-Gcn4p fusion proteins described above for binding reactions with a yeast cell extract prepared from a strain expressing an HA-tagged form of Ada3p. As described above, the binding of HA-Ada3p progressively declined as we combined mutations in clusters 5, 6, and 7 (Fig. 4A and 7A) and in clusters 1, 2, and 3 (Fig. 4A and 7B). The binding of HA-Ada3p seemed to resemble more closely that seen for yTAFII20, -60, and -90 than that seen for the SRB proteins in being relatively less affected by the {5}− and {5, 6}− mutations than were Srb2p, -4p, and -7p (Fig. 4). We also observed specific binding of Ada2p in the extracts prepared from the HA-ADA3 strain to the GST-Gcn4p fusion proteins (Fig. 7).
FIG. 7.
Additive effects of mutations in hydrophobic clusters 1 to 3 and 5 to 7 of the activation domain in GST-Gcn4p fusion proteins on binding of Ada3p in yeast cell extracts. (A) A fixed amount of yeast extract (containing 1,500 μg of protein) prepared from strain SY6-2 expressing HA-Ada3p was incubated with three different amounts of bacterial extracts for each GST-Gcn4p fusion containing ca. 5, 10, and 20 μg of total bacterial protein and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to 23 μg. (B) Aliquots of yeast extract containing 1,500 μg of protein from strain SY6-2 were incubated with three different amounts of bacterial extracts for each GST-Gcn4p fusion containing ca. 2, 4, and 8 μg of total bacterial protein and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to 9 μg. Shown are the GST-Gcn4p fusion proteins bearing the wild-type Gcn4p activation domain (+) (lanes 1 to 3, 14 to 16, and 27 to 29 in panel A and lanes 1 to 3 and 14 to 16 in panel B) or the mutant activation domains, with substitutions in the hydrophobic clusters shown in brackets across the top of each panel. The GST fusion proteins were precipitated and subjected to immunoblot analysis with monoclonal anti-HA antibodies to detect HA-Ada3p or polyclonal antibodies against the other proteins indicated to the left of each panel, exactly as described for Fig. 3. Lanes 13, 26, and 39 (A and B) contain 1/20 of the input (In) amount of yeast extract employed in each binding reaction mixture.
GST-Gcn4p does not bind specifically to yTAFII proteins in purified TFIID.
To determine whether the Gcn4p activation domain interacts directly with TFIID, we asked whether yTAFII proteins in highly purified preparations of TFIID could bind specifically to the GST-Gcn4p fusion proteins. TFIID was affinity purified with TBP antibodies or, for the strain expressing HA-tagged yTAFII130, with anti-HA antibodies and elution with HA peptide. Examination of the latter by SDS-PAGE and silver staining revealed that yTAFII150, yTAFII90, yTAFII60, yTAFII40, yTAFII30, and yTAFII25 all copurified with HA-yTAFII130 (see Fig. 3C in reference 68). Immunoblot analysis confirmed that this preparation was highly enriched for yTAFII20, -60, -90, and -130 and TBP but devoid of detectable amounts of Ada2p, Srb2p and Srb7p (data not shown). Using the TFIID fraction purified with anti-TBP antibodies, we observed little or no binding of yTAFII60 or yTAFII90 to wild-type GST-Gcn4p above the background levels for GST alone (Fig. 8A and B), even at much higher concentrations of GST-Gcn4p than were needed to detect interactions with these yTAFII proteins in cell extracts (e.g., Fig. 3). Using the TFIID fraction purified from the HA-yTAFII130 strain in binding reaction mixtures at approximately fivefold higher concentrations than were used in the experiments shown in Fig. 8A and B, we observed a higher level of binding by yTAFII60, yTAFII90, and yTAFII130 to GST-Gcn4p. However, the amounts of nonspecific binding to GST alone also increased for yTAFII60 and yTAFII90, and the binding was not diminished by double- or triple-cluster mutations in the Gcn4p activation domain for any of the four yTAFII proteins examined (Fig. 8C to G). Thus, we failed to obtain evidence that Gcn4p interacts specifically with any of these yTAFII proteins in purified TFIID. Accordingly, the specific interactions detected between GST-Gcn4p and yTAFII proteins present in cell extracts may require additional proteins absent in purified TFIID.
Ada2p and Ada3p are not required for binding of GST-Gcn4p to yTAFII or SRB proteins in cell extracts.
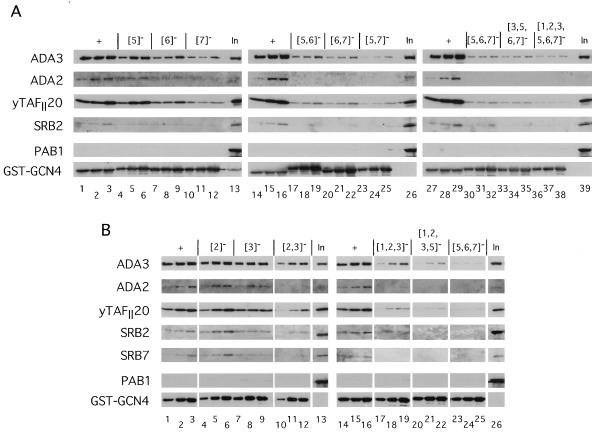
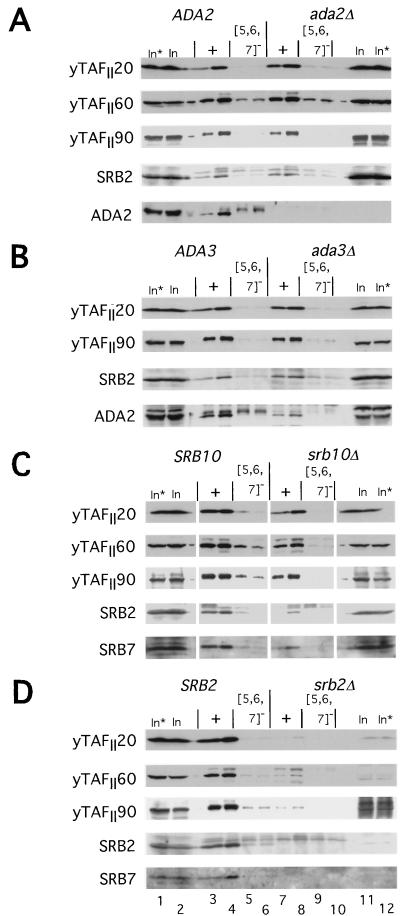
We considered the possibility that Gcn4p interacts directly with only a single coactivator protein or complex in the extracts, which functions as an adapter to mediate the interactions between Gcn4p and all other transcription factors. One or more subunits of the Adap-Gcn5p complex would be good candidates for such adapter proteins, because they are required for high-level activation by Gcn4p in vivo (6, 18, 23, 34, 58, 67). To investigate whether Ada2p or Ada3p is required for the interactions between GST-Gcn4p and the TFIID or mediator complexes, we prepared yeast extracts from isogenic wild-type, ada2Δ, and ada3Δ strains for in vitro binding experiments with the wild-type and {5, 6, 7}− GST-Gcn4p fusions. As shown in Fig. 9A and B, we saw no significant effect of deleting ADA2 or ADA3 on the binding of yTAFII20, -60, and -90 or Srb2p to the wild-type GST-Gcn4p fusion protein (compare + lanes for results with ADA2 and ada2Δ extracts). Less binding of Ada2p to wild-type GST-Gcn4p was observed in the ada3Δ than in the ADA3 extract (Fig. 9B, lanes 7 and 8 versus lanes 3 and 4); however, this finding appears to reflect a reduction in the abundance of Ada2p in the ada3Δ extract. Since Ada2p and Ada3p reside in the same Adap-Gcn5p complexes, Ada2p may be less stable in the absence of the Ada3p subunit.
FIG. 9.
Binding of yTAFII20, -60, and -90 and Srb2p in yeast extracts to wild-type GST-Gcn4p is not dependent on Ada2p, Ada3p, or Srb10p. Aliquots of yeast extract containing 1,500 μg of protein from pairs of isogenic mutant and wild-type strains were incubated with two different amounts of bacterial extracts containing 3 and 6 μg of protein and the appropriate amounts of a control bacterial extract lacking a GST fusion protein to bring the total amount of bacterial protein in each reaction mixture to 215 μg. The GST-Gcn4p fusion proteins contained either the wild-type activation domain (+; lanes 3 to 4 and 7 to 8) or the {5, 6, 7}− mutant activation domain (lanes 5 to 6 and 9 to 10). The GST fusion proteins were precipitated and subjected to immunoblot analysis with polyclonal antibodies against the proteins indicated to the left of each panel, exactly as described for Fig. 3. Lanes 1 and 12 contain 1/20 of the input (In) amount of yeast extract employed in each binding reaction mixture without incubation; lanes 2 and 11 contain the same amounts of yeast extract after incubation under reaction conditions in the absence of a GST fusion protein. (A) Yeast extracts derived from strains H1511 (ADA2) and KNY104 (ada2Δ); (B) yeast extracts derived from strains H1511 (ADA3) and KNY105 (ada3Δ); (C) yeast extracts derived from strains Z719 (SRB10) and Z687 (srb10Δ); (D) yeast extracts derived from strains H2451 (SRB2) and RMY10 (srb2Δ).
Similar results were obtained from a comparison of the binding of yTAFII20, -60, and -90 to GST-Gcn4p fusion proteins in extracts prepared from isogenic SRB10 and srb10Δ strains (Fig. 9C). The amounts of Srb2p and Srb7p that bound to GST-Gcn4p were reduced by the srb10Δ mutation (compare + lanes for results with SRB10 and srb10Δ extracts), and at least with Srb7p, this result could not be explained by a reduction in the level of the protein in the starting extract (Fig. 9C, compare In and In* lanes for results with SRB10 and srb10Δ extracts). This finding supports the idea that Srb2p and Srb7p bind to GST-Gcn4p as constituents of the mediator complex and that Srb10p is required for stability of this complex or its efficient interaction with GST-Gcn4p. We observed reductions in the binding of yTAFII20, -60, and -90 in an extract prepared from an srb2Δ mutant versus that made from the isogenic SRB2 strain; however, this result could be explained by reduced amounts of these yTAFII proteins in the starting srb2Δ extract (Fig. 9D, In and In* lanes). Perhaps the srb2Δ mutation leads to reduced transcription of the TAF genes. In fact, we detected sequence-specific binding to GST-Gcn4p by the residual amounts of these proteins present in the srb2Δ extract. We conclude that the absence of Srb2p or Srb10p in the extracts has little or no effect on the efficiency of binding to GST-Gcn4p by yTAFII20, -60, and -90. The results in Fig. 9 are in accordance with the idea that yTAFII20, -60, and -90 (TFIID) and Srb2p, -4p, and -7p (mediator) bind independently to Gcn4p, and they provide no evidence that Ada2p or Ada3p is required for the interactions between these complexes and the Gcn4p activation domain.
Identification of a stable complex containing Ada2p, Ada3p, and yTAFII20, -60, and -90.
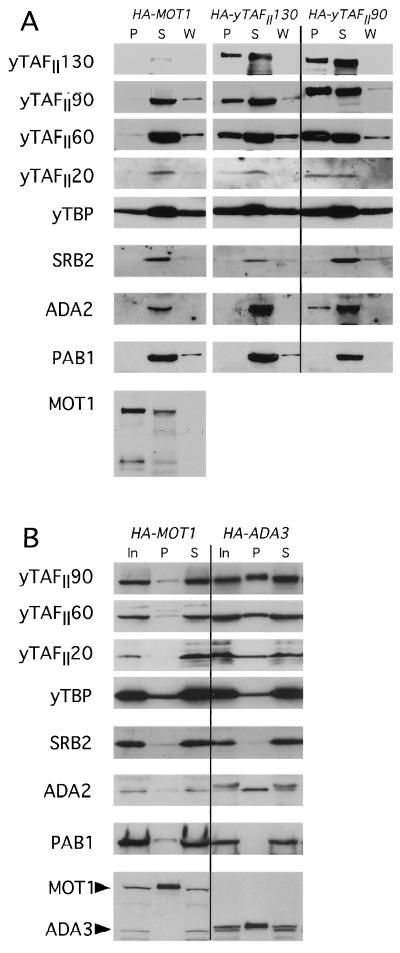
The binding of yTAFII20, -60, and -90 to GST-Gcn4p occurred at high levels and was fully dependent on the hydrophobic clusters in the Gcn4p activation domain, whereas the binding of yTAFII130 and TBP occurred at much lower levels and was independent of these residues (Fig. 3 to 6). Accordingly, we used coimmunoprecipitation analysis to determine whether yTAFII130 and TBP were associated with yTAFII20, -60, and -90 in the HA-yTAFII130 extracts used for the binding experiments described above. We also immunoprecipitated extracts from isogenic strains expressing HA-tagged forms of yTAFII60, yTAFII90, or Mot1p, a TBP-associated protein that does not appear to be a subunit of TFIID (69). yTAFII20, yTAFII60, and yTAFII90 were all coimmunoprecipitated with HA-yTAFII130 by using anti-HA antibodies (Fig. 10A, blots HA-yTAFII130), whereas, as expected, little or none of these three proteins was coimmunoprecipitated with HA-Mot1p (Fig. 10, blots HA-MOT1). Although we immunoprecipitated only a fraction of the total HA-yTAFII130, comparable proportions of HA-yTAFII130, yTAFII20, yTAFII60, and yTAFII90 were coimmunoprecipitated with HA antibodies from the HA-yTAFII130 extract. These findings are consistent with the idea that the majority of all four yTAFII proteins reside in the same complexes in our extracts. Immunoprecipitating with anti-HA antibodies from the HA-yTAFII90 extract gave similar results except that the yield of yTAFII130 in the immunoprecipitates was somewhat less than that seen for yTAFII20, -60, and -90 (Fig. 10A). This discrepancy might indicate that yTAFII20, yTAFII60, and yTAFII90 can exist in stable complexes which lack yTAFII130. Whether such yTAFII130-depleted subcomplexes are responsible for the specific binding to GST-Gcn4p described above for yTAFII20, -60, and -90, or whether intact TFIID binds to GST-Gcn4p and yTAFII130 and TBP dissociate from the complex, cannot be resolved without additional experimentation.
FIG. 10.
Coimmunoprecipitation analysis of components of TFIID, mediator, and Adap-Gcn5p complexes in yeast cell extracts. Aliquots of cell extracts containing 1,250 μg of total protein were immunoprecipitated with mouse monoclonal anti-HA antibodies from strains DPY107 (HA-MOT1), DPY213 (HA-yTAFII130), YBY40-8 (HA-yTAFII90), and SY6-2 (HA-ADA3). (A) The proteins in 100% of the immune complexes (lanes P), 40% of the supernatants (lanes S), and 20% of the washes from each immunoprecipitation (lanes W) were resolved by SDS-PAGE and subjected to immunoblot analysis with antibodies against the proteins indicated to the left of each blot, except for the Mot1p blot, which was probed with rabbit polyclonal anti-HA antibodies. (B) Proteins in 10% of the input extracts (lanes In), 100% of the immune complexes (lanes P), and 20% of the supernatants (lanes S) were analyzed by immunoblotting as described for panel A, again by probing the last blot at the bottom with polyclonal anti-HA antibodies to detect HA-Mot1p or HA-Ada3p.
We also probed the immune complexes isolated from the HA-yTAFII90 and HA-yTAFII130 extracts with antibodies against Srb2p and Ada2p. Srb2p was not stably associated with the yTAFII proteins (Fig. 10A), indicating that GST-Gcn4p interacted independently with the TFIID and mediator complexes in the experiments described above. In contrast, we found that a fraction of Ada2p in the cell extracts was coimmunoprecipitated with HA-yTAFII90 but not with HA-yTAFII130 or with HA-Mot1p (Fig. 10A). To confirm this unexpected interaction between Ada2p and yTAFII proteins, we immunoprecipitated Ada3p with anti-HA antibodies from the HA-ADA3 extract. As expected, the majority of Ada2p, but little or no Srb2p, was coimmunoprecipitated with HA-Ada3p, whereas significant fractions of yTAFII20, -60, and -90 and a small fraction of TBP were specifically coimmunoprecipitated with HA-Ada3p (Fig. 10B). Based on the recoveries of HA-Ada3p, Ada2p, and the yTAFII proteins, we estimate that about 10% of yTAFII20, -60, and -90 were physically associated with Ada2p and HA-Ada3p in the HA-ADA3 extract. The results shown in Fig. 10A suggest that a comparable proportion of Ada2p was associated with the yTAFII20, -60, and -90 proteins.
DISCUSSION
Evidence that the hydrophobic clusters in Gcn4p are capable of recruiting several coactivator complexes to the promoter.
We observed efficient binding of yTAFII20, -60, and -90; Srb2p, -4p, and -7p; and Ada2p and -3p in whole-cell extracts to a GST-Gcn4p fusion protein containing the wild-type activation domain. The double-cluster substitutions {5, 6}−, {5, 7}−, {6, 7}−, and {2, 3}− led to greater reductions in the binding of yTAFII20, -60, and -90; Ada2p and -3p; and Srb2p, -4p, and -7p than did the corresponding single mutations. This additive effect of combining mutations in different clusters on coactivator binding in vitro parallels the observation that inactivation of two clusters is required for a measurable reduction in transcriptional activation by Gcn4p in vivo. It is also noteworthy that among the double-cluster mutations, {5, 7}− led to the greatest reductions in coactivator binding in vitro and to activation of HIS3 transcription in vivo. Moreover, at least with Ada3p, the {1, 2, 3}− mutation was less deleterious to binding than was the {5, 6, 7}− mutation and adding the cluster 5 mutation to those in clusters 1 to 3 reduced the binding to GST-Gcn4p below that seen for the {1, 2, 3}− mutation alone. Simultaneously inactivating four different clusters reduced the binding of all the coactivators to background levels. In these respects, the additive effects of the mutations on coactivator binding in vitro (Fig. 4A) generally paralleled their cumulative effects on activation by Gcn4p in vivo (Table 1). This correlation is consistent with the idea that Gcn4p is capable of recruiting TFIID, the Adap-Gcn5p coactivator complex, and the holoenzyme mediator to the promoter as a means of stimulating transcription initiation in vivo (Fig. 11).
FIG. 11.
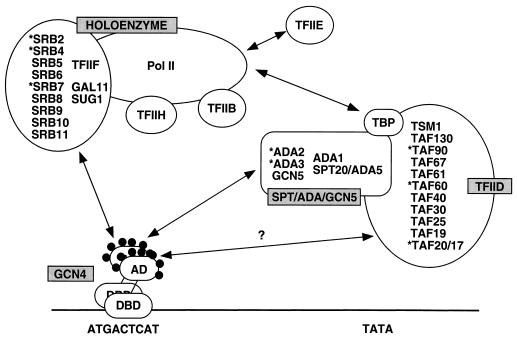
Model summarizing the in vitro interactions between the Gcn4p activation domain and three different coactivator complexes. A dimer of Gcn4p is depicted bound to a Gcn4p binding site located upstream from the TATA element in a Gcn4p-regulated promoter. The activation domain (AD) of Gcn4p bearing seven hydrophobic clusters (filled circles) is shown interacting independently with the mediator complex of RNA Pol II holoenzyme (containing SRB-encoded proteins, TFIIF, Gal11p, and Sug1p) (46) and the Sptp-Adap-Gcn5p complex (see the text for references). The interaction with TFIID (66, 68) may be indirect and may occur only in the context of an Sptp-Adap-Gcn5p–TFIID composite complex. Asterisks indicate subunits of the complexes in cell extracts that bound to recombinant GST-Gcn4p proteins in a manner that depended on the hydrophobic clusters in the Gcn4p activation domain. The Sptp-Adap-Gcn5p and TFIID complexes are shown physically interacting with one another based on our observations that fractions of yTAFII20, -60, and -90 in cell extracts specifically coimmunoprecipitated with HA-Ada3p and that a fraction of Ada2p coimmunoprecipitated with HA-TAFII90. TBP is shown as a component of TFIID but also as physically interacting with the Adap-Gcn5p complex and holoenzyme (see the text for details and additional references). DBD, DNA-binding domain.
Although the single-cluster mutations had no effect on HIS3 transcription in vivo (Table 1), they did lead to significant reductions in binding to GST-Gcn4p by Ada, yTAFII, and SRB proteins in vitro. In addition, the {5, 6}− and {6, 7}− mutations appeared to reduce protein binding in vitro more than they affected HIS3 transcription in vivo. To account for these discrepancies it could be proposed that the concentrations of the interacting proteins are much higher in vivo than in our in vitro experiments and that the in vivo concentrations are high enough to overcome reductions in affinity caused by single-cluster mutations in the Gcn4p activation domain. In addition, Gcn4p target-promoters typically contain multiple Gcn4p binding sites (29), so that the probability that a given coactivator is bound to a promoter containing several molecules of Gcn4p should be greater than the probability that any given Gcn4p molecule is in contact with the coactivator. Finally, protein-protein interactions among different coactivators and GTFs may allow for cooperative binding of coactivators to different molecules of Gcn4p bound at the same promoter.
The in vitro binding profiles of yTAFII20, -60, and -90 and Ada3p were quite similar for the panel of Gcn4p mutants we examined. In addition, a fraction of these TAF proteins coimmunoprecipitated with HA-Ada3p and a fraction of Ada2p coimmunoprecipitated with HA-TAFII90. These findings, combined with our inability to demonstrate a specific interaction between GST-Gcn4p and a highly purified TFIID fraction devoid of Ada2p and Ada3p, could be explained if Gcn4p interacts with TFIID in the context of a composite complex containing components of Adap-Gcn5p and TFIID. This interpretation is ostensibly at odds with our finding that the binding of yTAFII90 to GST-Gcn4p was not diminished by the absence of Ada2p or Ada3p in the extracts. The fact that Ada2p contains binding domains for Ada3p and Gcn5p (10) suggests that the Ada2p-Ada3p-Gcn5p complex is disrupted in the ada2Δ extract, and, indeed, deletion of ADA2 or ADA3 does eliminate the HAT activity of Gcn5p associated with the high-molecular-weight complexes containing these proteins (25). Moreover, we found that the level of Ada2p was diminished in an ada3Δ strain (Fig. 9B), suggesting that removal of Ada3p from the Adap-Gcn5p complex may destabilize Ada2p. However, it is possible that the TFIID-Ada composite complex we detected by coimmunoprecipitation contains Ada1p and Spt20p (Ada5p) in addition to Ada2p-Ada3p-Gcn5p and that this complex remains intact in the absence of the Ada2p or Ada3p subunits. If so, partial Sptp-Adap-Gcn5p complexes lacking only Ada2p or Ada3p may mediate the binding of yTAFII proteins to GST-Gcn4p observed in the ada2Δ or ada3Δ extracts, as well as the binding of Ada2p in the ada3Δ extract (Fig. 9B). In fact, there is genetic evidence that coactivator complexes containing Ada1p and Spt20p (Ada5p) persist in the absence of Ada2p, Ada3p, or Gcn5p (25, 34, 58, 72). Additional experiments will be required to determine whether the binding of yTAFII proteins to GST-Gcn4p is dependent on Spt20p (Ada5p), Ada1p, or some other uncharacterized adapter proteins.
Biochemical links between the Adap-Gcn5p proteins and TBP have come from observations that Ada2p can be isolated from nuclear extracts bound to recombinant GST-TBP, that Ada2p binds directly to the VP16 activation domain, and that deletion of Ada2p abolishes interaction between VP16 and TBP in yeast extracts (5, 79). Mutations in SPT20-ADA5 and ADA1 have numerous phenotypes in common with certain mutations affecting TBP (spt15 alleles), and it was shown that TBP can specifically interact with recombinant GST-Spt20p (71). Moreover, native TBP has been coimmunoprecipitated from cell extracts with Ada3p (73), a finding confirmed in this study (Fig. 10B). These observations suggest that the Sptp-Adap-Gcn5p complex is physically associated with TBP and able to bridge interactions between an activator and TBP. If so, physical interactions between Gcn4p and subunits of Sptp-Adap-Gcn5p complexes may serve to bring TBP to the promoter in addition to recruiting the HAT activity of Gcn5p.
Our GST-Gcn4p binding data imply that a TFIID subcomplex containing yTAFII20, -60, and -90 (and perhaps other yTAFIIs) can exist in the absence of yTAFII130 and TBP. This subcomplex may arise artifactually from proteolysis of yTAFII130 in the extracts or by dissociation of yTAFII130 and TBP upon binding to GST-Gcn4p. It is noteworthy that the Drosophila homologs of yTAFII60 and yTAFII20 (dTAFII62 and dTAFII42) have N-terminal segments that are folded into a histone-like motif and, like H3 and H4, can stably interact with one another in vitro in the absence of other TAFII proteins (90). Additional experiments are required to determine whether the yTAFII subcomplex we detected in vitro has any functional significance in vivo.
Evidence that Gcn4p interacts with holoenzyme mediator independently of the Adap-Gcn5p and TFIID complexes.
Our results suggest that recruitment of the holoenzyme to the promoter through interactions with mediator components is another important aspect of transcriptional activation by Gcn4p (Fig. 11). This conclusion is consistent with the fact that the purified holoenzyme mediator stimulated transcription from a Gcn4p-dependent promoter in vitro (42). In addition, we found recently that SRB2 and SRB10 are important for activation by Gcn4p in vivo (60a). With the exception of yTAFII30, which is shared by TFIID and holoenzyme in yeast (28), the polypeptide components of these two complexes appear to be completely distinct (40, 68, 86a). The fact that we could not coimmunoprecipitate Srb2p with various HA-yTAFII proteins or with HA-Ada3p suggests that the holoenzyme mediator is not tightly associated with the TFIID or Adap-Gcn5p complexes in our extracts. Consequently, the mediator complex probably bound to GST-Gcn4p independently of these other complexes. This conclusion is in accordance with the fact that the profile of binding of Srb2p, -4p, and -7p to the panel of GST-Gcn4p mutant proteins differed in several respects from that obtained for yTAFII20, -60, and -90 and Ada3p. In addition, the absence of Srb2p or Srb10p in the extracts did not significantly reduce the binding of yTAFII proteins to GST-Gcn4p. We conclude, therefore, that the Gcn4p activation domain is capable of interacting independently with the holoenzyme mediator and the TFIID and Adap-Gcn5p complexes.
The fact that mutations in clusters 1 to 3 and 5 to 7 in the Gcn4p activation domain reduced binding of Srb2p, -4p, and -7p; yTAFII20, -60, and -90; and Ada2p and -3p to GST-Gcn4p eliminates the possibility that each hydrophobic cluster is dedicated to interactions with a specific coactivator. Rather, it appears that each cluster contributes to the binding of all three coactivator complexes analyzed here. On the other hand, it seemed that certain clusters were more critical than others for interactions with a given coactivator. Thus, clusters 3, 5, and 7 appeared to be equally important for binding to Srb2p, -4p, and -7p whereas binding of Ada3p and yTAFII20, -60, and -90 was more dependent on cluster 7 than on clusters 3 and 5. The fact that all of the clusters contributed to binding of each coactivator, coupled with the unique requirements of each coactivator complex for efficient interaction with GST-Gcn4p, suggests a model in which the hydrophobic clusters in Gcn4p independently interact with different sites on each coactivator, with certain cluster contacts making a larger contribution to overall binding than others. The multiple sites of interaction with the hydrophobic clusters in Gcn4p may reside on different subunits of the coactivator or at different sites within a single subunit of the complex.
Although it was reported that TFIID is required for activation of a Gcn4p-dependent promoter in vitro (44), there is evidence that yTAFII proteins are dispensable for activation by Gcn4p in vivo (65). Given the functional interactions detected between TBP and Spt20p (Ada5p), Spt7p, and Spt3p, it is possible that Gcn4p can recruit TBP by interacting with Sptp-Adap-Gcn5p complexes in the absence of yTAFII proteins. There is also evidence for physical interactions between holoenzyme mediator components and TBP (45, 82). Thus, binding to mediator constituents may provide another pathway for Gcn4p to recruit TBP to the promoter in the absence of yTAFII proteins (Fig. 11). The fact that Gcn4p interacts with holoenzyme mediator, TFIID, and the Adap-Gcn5p complex may provide a simple explanation for why yTAFII proteins are dispensable for transcriptional activation by Gcn4p in yeast.
ACKNOWLEDGMENTS
We thank Christoph Hengartner and Richard Young for generous gifts of antibodies against SRB proteins, the srb10 mutant strain, and the construct for deleting SRB2. We also thank Chris Brandl for the HA-ADA3 strain and Shelley Berger for the Ada2p antibodies.
This work was supported in part by NIH grant GM52461 and training grant CA09385 (to P.A.W.).
C.M.D. and B.M.J. contributed equally to this work.
REFERENCES
- 1.Anderson J T, Paddy M R, Swanson M S. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 3.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M J, O’Malley B W. Interaction of human thyroid hormone receptor α with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 6.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcription adaptor required for function of certain acidic domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 7.Blair W B, Bogerd H P, Madore S J, Cullen B R. Mutational analysis of the transcriptional activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandl C J, Furlanetto A M, Martens J A, Hamilton K S. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 1993;12:5255–5265. doi: 10.1002/j.1460-2075.1993.tb06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994;77:1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 10.Candau R, Berger S L. Structural and functional analysis of yeast putative adaptors. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 11.Caron C, Rousset R, Beraud C, Moncollin V, Egly J M, Jalinot P. Functional and biochemical interaction of the HTLV-1 Tax1 transactivator with TBP. EMBO J. 1993;12:4269–4278. doi: 10.1002/j.1460-2075.1993.tb06111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 14.Chiang Y-C, Komarnitsky P, Chase D, Denis C L. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 15.Choy B, Green M R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993;366:531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 16.Cigan A M, Bushman J L, Boal T R, Hinnebusch A G. A protein complex of translational regulators of GCN4 is the guanine nucleotide exchange factor for eIF-2 in yeast. Proc Natl Acad Sci USA. 1993;90:5350–5354. doi: 10.1073/pnas.90.11.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahlman-Wright K, Baumann H, McEwan I J, Almlof T, Wright A P H, Gustafsson J A, Hard T. Structural characterization of a minimal function transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci USA. 1995;92:1699–1703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drysdale C M, Duenas E, Jackson B M, Reusser U, Braus G H, Hinnebusch A G. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 20.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 20a.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 21.Geisberg J V, Lee W S, Berk A J, Ricciardi R P. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc Natl Acad Sci USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgakopoulos T, Gounalaki N, Thireos G. Genetic evidence for the interaction of the yeast transcriptional co-activator proteins GCN5 and ADA2. Mol Gen Genet. 1995;246:723–728. doi: 10.1007/BF00290718. [DOI] [PubMed] [Google Scholar]
- 23.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 26.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 28.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 29.Hinnebusch A G. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Jones E W, Broach J R, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- 30.Hope I A, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 31.Hope I A, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 32.Hope I A, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horiuchi J, Silverman N, Piña B, Marcus G A, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reducing binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 36.Jackson B M, Drysdale C M, Natarajan K, Hinnebusch A G. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 38.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator Tat. Nature. 1994;367:295–300. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 39.Kerr L D, Ransone L J, Wamsley P, Schmitt M J, Boyer T G, Zhou Q, Berk A J, Verma I M. Association between proto-oncoprotein Rel and TATA-binding protein mediates transcriptional activation by NF-κB. Nature. 1993;365:412–419. doi: 10.1038/365412a0. [DOI] [PubMed] [Google Scholar]
- 40.Kim T K, Hashimoto S, Kelleher R J, Flanagan P M, Kornberg R D, Horikoshi M, Roeder R G. Effects of activation-defective TBP mutations on transcription initiation in yeast. Nature. 1994;369:252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- 41.Kim T K, Roeder R G. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci USA. 1994;91:4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 43.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 44.Klebanow E R, Poon D, Zhou S, Weil P A. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1996;271:13706–13715. doi: 10.1074/jbc.271.23.13706. [DOI] [PubMed] [Google Scholar]
- 45.Koleske A J, Buratowski S, Nonet M, Young R A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- 46.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 47.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 48.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Genetics. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 50.Lee M, Struhl K. Mutations on the DNA-binding surface of TATA-binding protein can specifically impair the response to acidic activators in vivo. Mol Cell Biol. 1995;15:5461–5469. doi: 10.1128/mcb.15.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 52.Leuther K K, Salmeron J M, Johnston S A. Genetic evidence that an activation domain of GAL4 does not require acidity and may form a β sheet. Cell. 1993;72:575–585. doi: 10.1016/0092-8674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- 53.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viijoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA in the polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 54.Lienhard Schmitz M, Dos Santos Silva M A, Altmann H, Czisch M, Holak T A, Baeuerle P A. Structural and functional analysis of the NF-κB p65 terminus. An acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 55.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 56.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 57.Lu H, Levine A J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martens J A, Genereaux J, Saleh A, Brandl C J. Transcriptional activation by yeast PDR1p is inhibited by its association with NGG1p/ADA3p. J Biol Chem. 1996;271:15884–15890. doi: 10.1074/jbc.271.27.15884. [DOI] [PubMed] [Google Scholar]
- 60a.McVeigh, R., and A. G. Hinnebusch. Unpublished observations.
- 61.Melcher K, Johnston S A. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Metz R, Bannister A J, Sutherland J A, Hagemeier C, O’Rourke E C, Cook A, Bravo R, Kouzarides T. c-Fos-induced activation of a TATA-box-containing promoter involves direct contact with TATA-box-binding protein. Mol Cell Biol. 1994;14:6021–6029. doi: 10.1128/mcb.14.9.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Metz R, Kouzarides T, Bravo R. A C-terminal domain in FosB, absent in FosB/SF and Fra-1, which is able to interact with the TATA binding protein, is required for altered cell growth. EMBO J. 1994;13:3832–3842. doi: 10.1002/j.1460-2075.1994.tb06694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 65.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 66.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piña B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poon D, Campbell A M, Bai Y, Weil P A. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 70.Regier J L, Shen F, Triezenberg S J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saleh A, Lang V, Cook R, Brandl J. Identification of native complexes containing the yeast coactivator/repressor proteins NCGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 74.Sauer F, Hansen S K, Tjian R. DNA template and activator-coactivator requirements for transcriptional synergism by Drosophila bicoid. Science. 1995;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- 75.Sauer F, Hansen S K, Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 76.Sauer F, Wassarman D A, Rubin G M, Tjian R. TAFIIs mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 77.Seipel K, Georgiev O, Schaffner W. A minimal transcription activation domain consisting of a specific array of aspartic acid and leucine residues. J Biol Chem. 1994;375:463–470. doi: 10.1515/bchm3.1994.375.7.463. [DOI] [PubMed] [Google Scholar]
- 78.Sherman F, Fink G R, Lawrence C W. Methods of yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 79.Silverman N, Agapite J, Guarente L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka M, Herr W. Reconstitution of transcriptional activation domains by reiteration of short peptide segments reveals the modular organization of a glutamine-rich activation domain. Mol Cell Biol. 1994;14:6056–6067. doi: 10.1128/mcb.14.9.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 83.Thut C J, Chen J L, Klemm R, Tjian R. P53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 84.Walker S, Greaves R, O’Hare P. Transcriptional activation by the acidic domain of Vmw65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol Cell Biol. 1993;13:5233–5244. doi: 10.1128/mcb.13.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 86.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86a.Weil, P. A. Unpublished observations.
- 87.Woontner M, Wade P A, Bonner J, Jaehning J A. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 91.Xu X, Prorock C, Ishikawa H, Maldonado E, Ito Y, Gelinas C. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol Cell Biol. 1993;13:6733–6741. doi: 10.1128/mcb.13.11.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]