Abstract
Osteogenic differentiation is essential for bone development and metabolism, but the underlying gene regulatory networks have not been well investigated. We differentiated mesenchymal stem cells, derived from 20 human induced pluripotent stem cell lines, into preosteoblasts and osteoblasts, and performed systematic RNA-seq analyses of 60 samples for differential gene expression. We noted a highly significant correlation in expression patterns and genomic proximity among transcription factor (TF) and long noncoding RNA (lncRNA) genes. We identified TF-TF regulatory networks, regulatory roles of lncRNAs on their neighboring coding genes for TFs and splicing factors, and differential splicing of TF, lncRNA, and splicing factor genes. TF-TF regulatory and gene co-expression network analyses suggested an inhibitory role of TF KLF16 in osteogenic differentiation. We demonstrate that in vitro overexpression of human KLF16 inhibits osteogenic differentiation and mineralization, and in vivo Klf16+/− mice exhibit increased bone mineral density, trabecular number, and cortical bone area. Thus, our model system highlights the regulatory complexity of osteogenic differentiation and identifies novel osteogenic genes.
Keywords: bone, induced pluripotent stem cell, mesenchymal stem cell, osteoblast, RNA sequencing, differential gene expression, transcription factor, long noncoding RNA, splicing, single-cell RNA-seq, network analysis, systems biology
Introduction
Mesenchymal stem cells (MSCs) are multipotent cells that can differentiate into a variety of lineages, including osteoblasts (OBs), chondrocytes, and adipocytes (Pittenger et al., 1999). Osteoblast differentiation from MSCs plays a pivotal role in bone development, homeostasis, and fracture repair. Given their critical roles, exploring the molecular mechanisms underlying MSC differentiation into OBs is of significant clinical importance.
Osteogenic differentiation is highly regulated by multiple mechanisms, and previous research has highlighted the roles of various genes, including those coding for transcription factors (TFs), long noncoding RNAs (lncRNAs), alternative spliceoforms, and signaling pathways such as TGF-β/BMP, Notch, Hedgehog, Wnt, Hippo, and estrogen receptor signaling (Thomas and Jaganathan, 2022). Out of over 1,600 identified TFs (Lambert et al., 2018), only a small subset has been linked to osteogenic differentiation (Chan et al., 2021; Thomas and Jaganathan, 2022; Rauch et al., 2019), underscoring the need to explore other TFs’ roles (Kang et al., 2019). Precise and robust transcriptional regulation is required during osteoblast differentiation, achieved by networks of transcriptional regulators. For example, RUNX2 is a master transcription factor in early osteoblast differentiation (Komori et al., 1997; Schroeder et al., 2005), but its expression does not increase in later stages. Concurrently, Notch signaling initially decreases but later represses RUNX2, boosting terminal differentiation and mineralization (Canalis et al., 2013; Ji et al., 2017; Shao et al., 2018). TFs ATF4, TEAD4, and KLF4 work alongside RUNX2 to regulate osteogenesis (Suo et al., 2020; Yang et al., 2004; Yu et al., 2021). However, the broader TF interplay in osteoblast differentiation remains largely uncharted, even with insights from ENCODE (Encyclopedia of DNA Elements) data that reveal the co-association of human TFs in a combinatorial and context-specific manner (Gerstein et al., 2012).
LncRNAs are vital in regulating cellular growth and differentiation (Guttman et al., 2011; Ransohoff et al., 2018), and nearly 30,000 human IncRNA genes have been annotated (Hon et al., 2017). Though our understanding of their role in osteogenesis is emerging, only about 60 lncRNAs are known to influence osteogenic differentiation, affecting TFs, pathway genes, or acting as molecular sponges for miRNAs (Ghafouri-Fard et al., 2021; Lanzillotti et al., 2021). For instance, lncRNA H19 promotes osteoblast differentiation via the TGF-β1/SMAD3/HDAC pathway by encoding miR-675 (Huang et al., 2015), while lncRNA MALAT1 enhances osteoblast differentiation through SMAD4 up-regulation by sequestering miR-204 (Xiao et al., 2017). Numerous studies have independently investigated both human coding and lncRNA gene expression profiles during osteogenic differentiation (Abd Rahman, 2021; Håkelien et al., 2014; Hong et al., 2020; Huang et al., 2017; Khodabandehloo et al., 2020; Liu et al., 2020; Park et al., 2015; Qiu et al., 2017; Shaik et al., 2019; Skårn et al., 2014; Song et al., 2015; Sun et al., 2019; Twine et al., 2014; Wang et al., 2015; Xia et al., 2020; Zhang et al., 2017; Zheng et al., 2018). However, many of these studies are limited by such factors as reliance on microarrays, small sample sizes, and brief osteogenic differentiation periods. In-depth exploration of lncRNA profiles and their association with adjacent protein-coding genes would therefore be advantageous.
Around 4,000 genes were identified to have alternative splicing (AS) during osteogenic differentiation using microarray technology (Shang et al., 2016), while almost all human genes are expected to exhibit AS, producing multiple transcript isoforms (Wang et al., 2008). AS serves as a widespread regulatory mechanism in gene expression and is implicated in various processes, including MSC osteogenic differentiation (Aprile et al., 2018; Makita et al., 2008). Abnormal splicing, as seen in the mutant COL1A1 gene, can lead to conditions such as osteogenesis imperfecta (Gug et al., 2020). Despite these findings, the role of splicing factors (SF) and differential splicing (DS) in osteogenic differentiation remains globally unexplored.
Unraveling the osteogenic differentiation of MSCs demands a systems biology approach, using a comprehensive transcriptome atlas and combined analysis of TFs, lncRNAs, and spliceoforms. Due to the invasiveness required to obtain human primary MSC and the suboptimal conditions for preserving available samples, large-scale human transcriptomics projects, such as GTEx, exclude data from bone and cartilage. Consequently, studies focusing on these tissues are often limited to disease contexts. Induced pluripotent stem cell (iPSC) derived systems, given their robust pluripotency and differentiation, offer an avenue for studying skeletal development. iPSC-derived sclerotome models have been used to recapitulate endochondral ossification (Nakajima, et al., 2018; Lamandé et al., 2023; Tani, et al., 2023). However, previous studies for osteoblast differentiation from iPSC-derived MSCs included only 1–2 MSC cell lines (Rauch et al., 2019; Matsuda, et al., 2020), limiting the power for comprehensive transcriptomic analyses. In this study, we used 20 human iPSC lines, each from a healthy individual, to investigate osteogenic differentiation of MSCs by RNA sequencing at three key stages: MSC, preosteoblast (preOB), and OB. Our findings highlight dynamic gene expression changes and the intricate interactions of lncRNAs and TFs. We spotlight the significance of alternative splicing during osteogenic differentiation. Finally, we identify the regulatory role of TF KLF16 as a potential inhibitor of osteogenic differentiation. This work provides insights into osteogenic differentiation and sheds light on novel therapeutic targets for bone diseases.
Results
Transcriptome profiles of human iPSC-derived MSCs and OBs are similar to that of primary MSCs and OBs.
We established a workflow to generate iPSCs from peripheral blood mononuclear cells (PBMCs) or skin fibroblasts from 20 healthy individuals (9 males and 11 females) and differentiated them into MSC, preOB, and OB stages (Fig. 1a, Supplementary Fig. 1a, 1b, and 1h; Supplementary Table 1). To abrogate the memory of their somatic cell origin and attenuate in vitro transcriptional and differentiation heterogeneity, the established iPSC lines were cultured for at least 16 passages and underwent quality control (Polo et al., 2010). The lines then underwent MSC differentiation for three weeks and purification by sorting for CD105+/CD45- markers (Giuliani et al., 2011; Kang et al., 2015). No significant differences were found in the percentage of this cell population, irrespective of erythroblast or fibroblast origins (Supplementary Fig. 1c and 1d). After two weeks of expansion, our iPSC-derived MSCs were confirmed to have mesenchymal characteristics by prominently expressing positive MSC surface markers (CD29, CD73, CD90, and CD105) and by barely detecting negative markers (CD31, CD34, and CD45) at both the protein level by flow cytometry (Supplementary Fig. 1e and 1f) and mRNA level by RNA-seq (Supplementary Fig. 1g). After osteogenic differentiation for 21 days, the OBs demonstrated alkaline phosphatase (ALPL) enzyme activity and mineralization with alizarin red and von Kossa staining, reflective of functional OBs (Fig. 1b).
Fig. 1. Generation of healthy human iPSCs and osteogenic differentiation transcriptomic data.
a Flowchart of iPSC establishment, iPSC-derived MSC generation, MSC to OB differentiation (preOBs, preosteoblasts; OBs, osteoblasts), and RNA-seq data generation and analyses. b In vitro osteogenic differentiation of MSCs (Day 0), preOBs (Day 7), and OBs (Day 21) stained with alkaline phosphatase (ALP), alizarin red, and von Kossa. c Expression level of known osteogenic genes at the three osteogenic stages. Data are shown as mean ± SEM. * MSC vs preOB, # preOB vs OB, or + MSC vs OB, adjusted p value < 0.0001. d Principal component analysis (PCA) of RNA-seq data from our iPSC-derived MSCs and differentiated OBs as well as previously published human primary MSCs and OBs, iPSCs, and other tissues in GTEx.
A total of 60 RNA-seq libraries were generated from the 20 individuals at the three stages (Fig. 1a; Supplementary Table 1). We initially employed bulk RNA-seq rather than single-cell RNA-seq (scRNAseq) because the latter recovers only sparse transcriptional data from individual cells, thus limiting comprehensive expression analysis and the development of robust prediction models for regulatory and gene co-expression networks (Wang and Li, 2021). These data revealed increased expression of OB lineage markers (ALPL, COL1A1, RUNX2, SPARC, OMD, and OGN) with characteristic temporal expression patterns during osteogenic differentiation (Fig. 1c). We further compared our MSC and OB RNA-seq datasets with previously published human primary MSC and OB datasets, human iPSC datasets, and GTEx datasets for different tissues derived from the three germ layers (ectoderm, endoderm, and mesoderm) and germ cells (Al-Rekabi et al., 2016; Ardlie et al., 2015; Ma et al., 2019; Roforth et al., 2015; Rojas-Peña et al., 2014). By principal component analysis (PCA), our MSC and OB datasets clustered with those of published primary MSCs and OBs, respectively, and were discrete from iPSCs and other tissue datasets, further validating our MSC and OB cell lines (Fig. 1d).
Differential transcription profiles of human iPSC-derived MSCs, preOBs, and OBs.
During in vitro osteogenic differentiation, we detected the expression of 17,795 unique genes across all stages, of which 79% were protein-coding and 21% were noncoding (Supplementary Fig. 2a). Seventy percent or 9,724 of the protein-coding genes were differentially expressed (DE) between the osteogenic stages (MSC to preOB or preOB to OB) (fold change ⩾ 1.2 and adjusted p value ⩽ 0.05). Sixty percent (2,297) of the noncoding genes also were DE (Supplementary Fig. 2a), of which 2,249 (98% of DE noncoding) were lncRNA genes. By intersecting with the identified 1,600 TFs (Lambert et al., 2018), we found that nine percent (840 genes) of the DE coding genes (Supplementary Fig. 2b) and six percent (257 genes) of the non-DE coding genes coded for TFs (Supplementary Fig. 2a). And a chi-square test revealed a significant correlation between TFs and differentially expressed genes (DEGs) (X2 = 27.4, p < 0.0001), indicating TF genes were more likely than non-TF coding genes to be differentially expressed during osteogenic differentiation. There were 5,715 up-regulated genes and 4,409 down-regulated genes from the MSC to the preOB stage and 3,342 up-regulated genes and 2,835 down-regulated genes from the preOB to the OB stage, revealing that the majority of the DEGs were up-regulated during osteogenic differentiation (Supplementary Fig. 2c and 2d; Supplementary Table 2). Among the top statistically significant up- or down-regulated genes, known osteogenesis-associated genes, including a SMAD ubiquitination regulatory factor SMURF2 (Kushioka et al., 2020), natriuretic peptide precursor B NPPB (Aza-Carmona et al., 2014), hypoxia inducible factor 3 subunit alpha HIF3A (Zhu et al., 2014), a TF ZBTB16 (Onizuka et al., 2016), a Wnt signaling pathway family member WNT7B (Yu et al., 2020a), osteoinductive factor OGN, a SFRP family (modulators of Wnt signaling) member SFRP2 (Yang et al., 2020a), and a glycoprotein member of the glycosyl hydrolase 18 family CHI3L (Chen et al., 2017), were identified. There were also many genes with no defined function in bone, including those that play critical roles in RNA binding activity (EBNA1BP2), maintenance of protein homeostasis (PSMD2), arachidonic acid metabolism (PRXL2B), hemostasis and antimicrobial host defense (FGB), embryonic and induced pluripotent stem cells (PCSK2), as well as cell proliferation, differentiation and migration (FGFBP1). A larger number of DEGs with larger changes in gene expression was identified from the MSC to the preOB stage than from the preOB to the OB stage, even though there was half the differentiation interval (one week) between the earlier two stages than between the latter two stages (two weeks) (Fig. 2a, Supplementary Fig. 2c, 2d, and 2e).
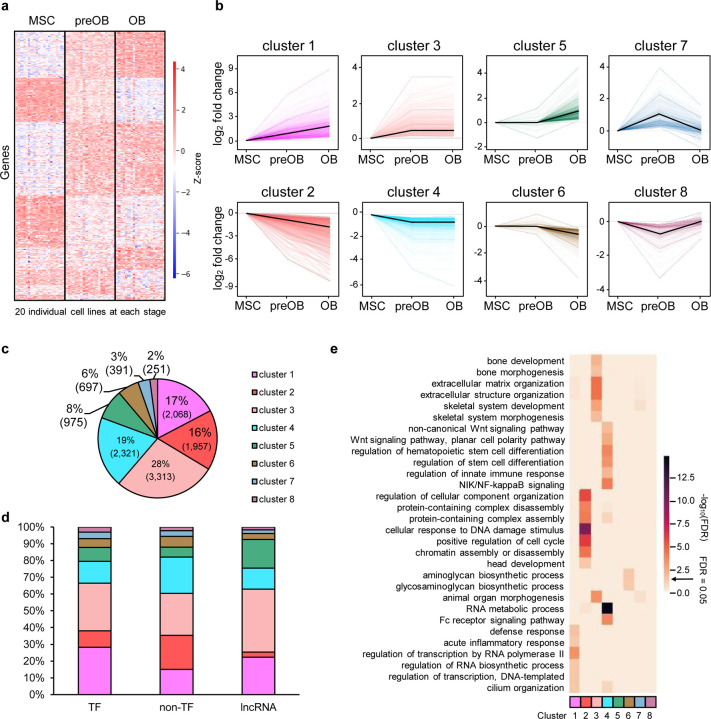
Fig. 2. Gene expression profile during osteogenic differentiation.
a Heatmap showing hierarchical clustering of 60 RNA-seq datasets from 20 iPSC-derived MSC, preOB, and OB lines (columns) and significant differentially expressed genes (DEGs) (rows), fold change ≥ 1.2 and adjusted p ≤ 0.05. Up-regulated and down-regulated gene expression is colored in red and blue, respectively. b Clustering of DEGs according to their expression profiles during differentiation, visualized with the TiCoNE Cytoscape app (Wiwie et al., 2019). In each plot, colored lines represent individual genes, and the black line represents cluster means. c Pie chart showing the number and percent of DEGs for each cluster. d Histogram chart showing the percentage distribution of transcription factor (TF), non-TF, and long noncoding (lnc) RNA DEGs in the eight clusters. Distribution correlation coefficient ρ = 0.9 between TF and lncRNA, 0.7 between TF and non-TF protein-coding genes, and 0.6 between lncRNA and non-TF coding genes. e Top significantly enriched gene ontology (GO) biological process (BP) terms of DEGs for each cluster.
Correlation of gene expression patterns of TFs and lncRNAs during human osteogenic differentiation.
Distinguishable gene expression profiles for each of the three osteogenic stages were displayed by a heatmap of Z-score normalized gene-expression values (Fig. 2a). We further characterized the DEGs by clustering them into eight possible patterns based on their expression up/down changes across the three osteogenic stages, MSC to preOB and preOB to OB (Fig. 2b; Supplementary Table 3). Cluster 1 and cluster 2 genes increased or decreased expression, respectively, throughout differentiation. The expression of genes for all other pairs of clusters also displayed opposite stage-specific changes. Genes in cluster 3 and cluster 4 increased or decreased expression, respectively, from the MSC to the preOB stage and remained relatively stable from the preOB to the OB stage. Genes in clusters 5 and 6 displayed no significant change In expression from the MSC to the preOB stage, but their expression increased or decreased, respectively, from the preOB to the OB stage. Genes in cluster 7 increased and in cluster 8 decreased in their expression from the MSC to the preOB stage, and in cluster 7 decreased and in cluster 8 increased from the preOB to the OB stage (Fig. 2b; Supplementary Table 3). When comparing the pairs of clusters, other than clusters 7 and 8, the number of genes with increased expression was consistently greater than that with decreased expression among stages (e.g., cluster 3 had 3,313 genes up-regulated while cluster 4 had 2,321 genes down-regulated from the MSC to the preOB stage) (Fig. 2b). 81% of all DEGs were in either pair of clusters 1 and 2 or 3 and 4, with almost half of all DEGs in clusters 3 and 4 (Fig. 2b and 2c). Also, each cluster contains various proportions of different types of genes, including TF, non-TF protein-coding, and lncRNA genes. Cluster 1 contained the highest percentage of TF genes (11%), cluster 2 contained the highest percentage of non-TF protein-coding genes (92%), and cluster 5 contained the highest percentage of lncRNA genes (40%) (Supplementary Fig. 2f). We determined the proportions of TF, non-TF protein-coding, and lncRNA genes in the eight clusters (Fig. 2d). We then calculated correlation coefficients to measure the similarity between the distribution of the proportion of a pair of the gene types. Interestingly, we found that TF and lncRNA genes were the most highly correlated with a correlation coefficient of 0.9 (p value = 0.003), compared to those of TF and non-TF protein-coding genes (ρ = 0.7, p value = 0.04), or lncRNA and non-TF coding genes (ρ = 0.6, p value = 0.11), suggesting that there is tighter regulatory control between TF and lncRNA genes, compared to the other pairs.
Gene ontology (GO) biological process (BP) overrepresentation tests revealed that each cluster was enriched for various functions that are distinct from other clusters (FDR ≤ 0.05, Fig. 2e; Supplementary Table 3). For example, cluster 3 was enriched for bone development, bone morphogenesis, ECM organization, skeletal system development, and skeletal system morphogenesis. Cluster 4 was enriched for Wnt signaling pathway planar cell polarity pathway, non-canonical Wnt signaling pathway, regulation of hematopoietic stem cell differentiation, regulation of innate immune response, regulation of stem cell differentiation, NIK/NF-kappaB signaling, RNA metabolic process, and Fc receptor signaling pathway. Cluster 5 had no significantly enriched BP terms, which may be explained by its high content of lncRNA genes (Supplementary Fig. 2f), of which biological functions are largely unknown and not annotated in the GO resource database (Carbon et al., 2019). Clusters 7 and 8 contained relatively small numbers of DEGs and showed no significantly overrepresented terms (Fig. 2c and 2e).
Transcription factor regulatory network during human osteogenic differentiation.
By PCA, we found that the expression profiles for all the DEGs segregated all of the samples by their differentiation stage, with MSCs being most distinct from preOBs and OBs, which is consistent with the larger number of DEGs present between the MSC and preOB stages than the later stages (Fig. 3a and Supplementary Fig. 2c and 2d). TFs were similar to other protein-coding genes in gene expression levels at each stage and their fold changes between stages (Supplementary Fig. 3a and 3b). Of note, when we used only the 840 differentially expressed TFs, their expression profiles also segregated the three osteogenic stages (Fig. 3b).
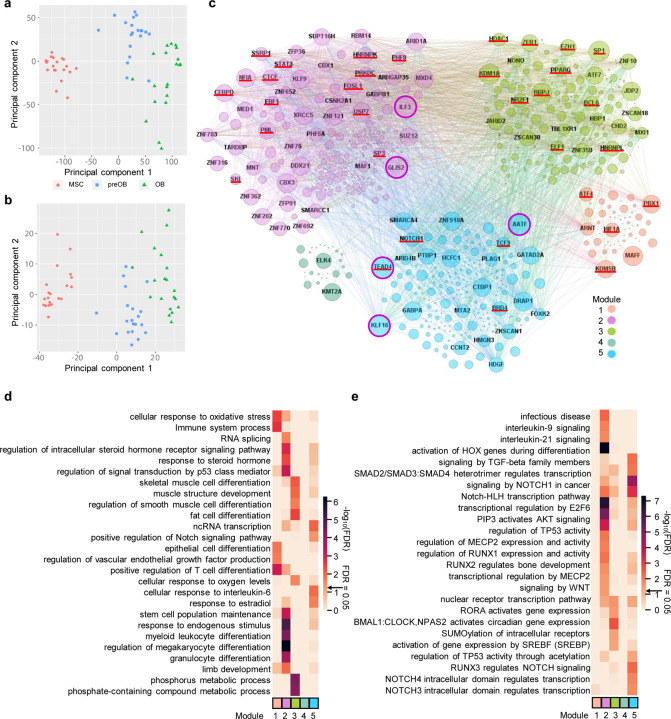
Fig. 3. Complex TF-TF regulatory network in osteogenic differentiation.
a Principal component analysis (PCA) of all 20 healthy cell lines at three stages of osteogenic differentiation using all differentially expressed genes (DEGs). b PCA using only differentially expressed TF genes. c Transcriptional regulator, mainly TF networks, during osteogenic differentiation. Each node represents a transcriptional regulator, with known bone formation associated regulators underlined in red. Two nodes are connected by a line where ReMap data suggest regulation and our RNA-seq data suggest the association between them. Nodes labeled with the gene name represent the top 100 strongest transcriptional regulators based on betweenness centrality. The size of the nodes reflects the regulation strength of the regulator, with the top 5 strongest circled in pink. d and e Top significantly enriched GO BP terms in d and Reactome pathways in e of transcriptional regulators in each network module.
TFs not only regulate the transcription of other protein-coding genes and noncoding RNA genes but also of TF genes themselves. We focused on the TF biological cooperativity with respect to TF-TF regulation because it has not been investigated globally in osteogenic differentiation. To predict the TF-TF regulatory network during osteogenic lineage differentiation, we constructed a network using the database ReMap, which integrated and analyzed 5,798 human ChIP-seq and ChIP-Exo datasets from public sources to establish a transcriptional regulatory repertoire to predict target genes (Chèneby et al., 2020), rather than more specific datasets as comprehensive MSC, preOB, or OB DNA-binding data are not available, in combination with our RNA-seq data. ReMap covers 1,135 transcriptional regulators, including mainly TFs and a few coactivators, corepressors, and chromatin-remodeling factors with a catalog of 165 million binding peaks. We filtered transcriptional regulators that did not show associations in ReMap, given that the transcription regulators are less likely to have real regulatory relationships in osteogenic differentiation if they do not display their associations in ReMap. To further assure the reliability and accuracy of the regulatory relationship in our osteogenic differentiation datasets, we then assessed regulations among them by computing correlation coefficients of gene expression, which resulted in 451 TFs preserved in the network. After the application of a partitioning algorithm, the generated network was organized into five interconnected modules (Bastian et al., 2009) (Fig. 3c; Supplementary Table 4). The network showed that TFs might regulate other TFs both internal and external to their respective modules, revealing a highly complex network of transcriptional regulators. We also identified the top 100 transcriptional regulators determined by their betweenness centrality, which coincided with their power to regulate others in the network (Fig. 3c; Supplementary Table 4) (Bastian et al., 2009). Among them, TF genes known to regulate osteogenic differentiation were present in different modules, such as ATF4 in module 1 (Yang et al., 2004), FOSL1 in module 2 (Krum et al., 2010), ZEB1 in module 3 (Fu et al., 2020), and TEAD4 in module 5 (Suo et al., 2020) (Supplementary Table 4). Of interest, TF KLF16 in module 5, which previously was not demonstrated to be involved in bone formation and bone development, was revealed to have an important role in the osteogenic network by its ranking as the fifth top regulator directly interacting with second top regulator TEAD4, and its high degree of association with 315 transcriptional regulators (Supplementary Table 4).
GO and Reactome pathway (RP) analyses revealed regulatory functions and pathways specific to each module (FDR ≤ 0.05; Fig. 3d and 3e; Supplementary Table 4). As examples for the GO BP terms, module 2 was enriched for RNA splicing. Module 3 was enriched for skeletal muscle cell differentiation and muscle structure development. These two terms were associated with six (EGR1, NR4A1, EGR2, SIX4, FOS, ATF3) and 15 (ZBTB18, MEF2A, EGR1, TCF7L2, EGR2, EPAS1, SRF, ZBTB42, ETV1, FOS, RBPJ, NR4A1, SIX4, ID3, ATF3) transcriptional regulator genes, respectively. Also, this module was enriched for the term fat cell differentiation and six associated TF genes (ATF2, NR4A1, TCF7L2, EGR2, PPARG, GLIS1). Module 5, containing KLF16, was enriched for terms ncRNA transcription and positive regulation of the Notch signaling pathway, which is essential in bone development and metabolism (Fig. 3d) (Liu et al., 2016).
In addition, each of the modules was enriched for GO cellular component (CC) terms of various complexes. Module 2 was enriched for the term ESC/E(Z) complex, a multimeric protein complex that can interact with several noncoding RNAs, a vital gene silencing complex regulating transcription, and an effector of response to ovarian steroids (Dubey et al., 2017) (Supplementary Fig. 4a). The modules were enriched for different GO molecular function (MF) terms of receptor binding of various hormones, including androgen, estrogen, glucocorticoid, and thyroid hormone receptor binding (Supplementary Fig. 4b), providing more evidence of the crosstalk between bone and gonads (Oury, 2012). With RP analysis, Module 2 was associated with activation of HOX genes during differentiation, consistent with its established important role in osteogenic differentiation of MSCs and skeletogenesis (Seifert, 2015). Module 5 was associated with signaling by TGF-beta family members, and the SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription (Fig. 3e). These results revealed the complex and robust nature of TF regulatory networks during osteogenic differentiation.
Correlation in genomic proximity of lncRNA and TF genes during human osteogenic differentiation.
Given a large number of lncRNA DEGs during osteogenic differentiation (Supplementary Fig. 2a and 2b) and the similarity of their distribution to that of regulatory TF genes in the defined clusters of expression patterns per the coefficient calculation above (Fig. 2d), we further investigated the characteristics of lncRNAs during osteogenic differentiation. Our data showed that among all lncRNA genes, there were near equal amounts with one exon (24%) or two exons (22%), and the remaining 54% had more than two exons, of which each category contributed no more than 15% (Supplementary Fig. 3c). The proportion of lncRNA genes with the various numbers of exon differed from TFs and non-TFs (Supplementary Fig. 3c), as well as from previous studies that demonstrated that lncRNA genes have a tendency to have only two exons (42%) while each category of the remaining lncRNA genes with other than two exons contributed less than 27% on average over a wide range of human organs (GENCODE consortium, Derrien et al., 2012). The expression level of DE lncRNA genes was lower than TF and non-TF coding genes at each osteogenic stage (Supplementary Fig. 3a). However, the expression fold changes of the lncRNA genes among the osteogenic stages were larger than that of TF and non-TF coding genes (Supplementary Fig. 3b). We also performed PCA using the expression profiles of DE lncRNA genes from all 60 samples. PCA segregated them into three groups corresponding to their osteogenic stage, indicating the differentiation stages also can be defined by lncRNA expression profiles (Fig. 4a).
Fig. 4. lncRNA expression profiles discriminate osteogenic differentiation stages.
a PCA of differentially expressed lncRNA genes from all 20 healthy cell lines at three stages of osteogenic differentiation. b and c Bean plots showing the expression fold change (FC) of the protein-coding DEGs near up-regulated, down-regulated, and non-differentially expressed lncRNA genes from MSC to preOB stages (p < 0.0001, by Kruskal-Wallis rank sum test) in b and from preOB to OB stages (p < 0.0001) in c. The individual observations are shown as small horizontal lines in a one-dimensional scatterplot, and the average is indicated as the long horizontal line. The estimated density of the distributions is visible. d Density of the correlations of expression of DE lncRNAs and their pairwise DE neighboring protein-coding genes (pink) and that of DE lncRNAs and their randomly shuffled pairwise DE protein-coding genes outside the neighboring area (blue). e Venn diagram showing the numbers of protein-coding DEGs near up-regulated and down-regulated lncRNA genes specifically from the MSC to preOB stages as in b (blue) or from the preOB to OB stages as in c (orange) and their overlap. Top significantly enriched GO BP terms of the neighboring differentially expressed protein-coding genes are shown in the lower panel. f Expression of lncRNAs AC005261.1 and FTX during osteogenic differentiation. Data are shown as mean + SEM. * MSC vs. preOB, # preOB vs. OB, or + MSC vs. OB, adjusted p value < 0.001.
Unlike many coding genes that have well-known biological functions, most lncRNA genes have little functional annotation. Given that a major function of lncRNAs is to regulate their neighboring coding genes and that a classic way to assign cis biological functions to a set of lncRNA genes is to analyze their nearby genes’ functional annotations, we matched the lncRNAs to their nearby coding genes (Statello et al., 2021) (see Methods). We identified 441 neighboring protein-coding genes that were also differentially expressed (Supplementary Table 5). The expression fold changes for coding DEGs near either up-regulated or down-regulated lncRNA genes were larger than the expression fold changes for coding DEGs near non-differentially expressed lncRNA genes both from MSC to preOB and from preOB to OB stage (p < 0.0001 for both comparisons, by Kruskal-Wallis rank sum test), respectively (Fig. 4b and 4c). lncRNAs have been reported to display either positive (Kim et al., 2010; Ørom et al., 2010) or negative (Brockdorff et al., 1992; Nagano et al., 2008) regulation of their neighboring protein-coding genes. We found that both positive (25.3% vs. 5.1%, 0.8 > ρ ≥ 0.5, Chi-Square test, p < 0.0001) and negative (5.8% vs. 2.8%, −0.5 ≤ ρ > −0.8, Chi-Square test, p < 0.05) expression correlations, as well as very strong positive expression correlations (3.0% vs. 0.5%, ρ ≥ 0.8, Chi-Square test p < 0.01) of DE lncRNA genes with their pairwise neighboring coding DEGs, were significantly stronger than with randomly shuffled coding DEGs (Fig. 4d). This indicates a dual-directional regulation of lncRNA genes to their nearby coding genes in osteogenic differentiation. There was no significant difference in very strong negative expression correlations (ρ ≤ −0.8). Among the coding DEGs associated with DE lncRNA genes, we found 55 TF genes, including several members of the TEAD and ZNF families known to be involved in osteogenesis, 16 SF genes (e.g., SYF2 and SRSF1), and 59 DS genes (Supplementary Table 5). A chi-square test revealed a significant correlation of DE lncRNA genes with DE TF genes (X2 = 8.6, p < 0.01), which indicated that DE TF genes were more likely than DE non-TF coding genes to be neighbors of lncRNA genes, providing more evidence regarding a tight regulatory control between lncRNA and TF genes. We did not see a significant correlation of DE lncRNA genes with DE SF genes or with DE DS genes.
We further performed GO BP enrichment analysis on these coding DEGs near DE lncRNA genes to investigate functional annotations. It was not surprising to see these coding DEGs were enriched for regulation of transcription and gene expression because of the significant correlation of lncRNA and TF genes in both gene expression pattern and neighborship. Eight percent (36 genes) were identified to be involved in skeletal system development or regulation of ossification, 14% involved in the regulation of cell differentiation, and 25% involved in the regulation of cell communication (FDR ≤ 0.05, Fig. 4e, Supplementary Table 5). 12% of the lncRNA neighboring protein-coding genes differentially expressed from the preOB to OB stage were associated with regulation of phosphorus and phosphate metabolic processes, which are essential for bone mineral deposition and bone growth (Penido and Alon, 2012).
Among all genes, noncoding as well as coding, two lncRNA genes, AC005261.1 and FTX, were the first and fifth most significantly up-regulated genes from the preOB to OB stage (Supplementary Fig. 2c and 2d). The expression of AC005261.1 increased only at the later stage of OB differentiation (cluster 5), and FTX increased throughout differentiation (cluster 1) (Fig. 4f). AC005261.1 is antisense to a KRAB-containing zinc finger transcription gene ZNF304, which plays a fundamental role in T- cell differentiation (Sabater et al., 2002). FTX, an X-inactive-specific transcript regulator, previously was demonstrated to have a regulatory function in cell proliferation of multiple tumor types (Huang et al., 2020), and its down-regulation promotes the differentiation of osteoclasts in osteoporosis (Yu et al., 2020b). Taken together, these results demonstrate that dynamic expression of lncRNA genes is an integral component of the transcriptional profiles of osteogenic differentiation and may contribute to the regulation of osteogenic differentiation by regulating proximal coding genes.
Differential splicing of TF and lncRNA genes during human osteogenic differentiation.
To systematically investigate the role of RNA alternative splicing during osteogenic differentiation, we first identified 287 DE SFs by intersecting our DEGs with an SF list curated from published literature (Fig. 5a and 5b; Supplementary Table 6) (Piva, et al., 2012; Giulietti, et al., 2013; Sveen, et al., 2011; Seiler, et al., 2018; Hegele, et al., 2012; Koedoot, et al., 2019). Thirty-six percent of them showed stage-specific expression changes, with 86 DE SFs from MSC to preOB and 18 DE SFs from preOB to OB stage (Fig. 5b). The expression fold changes of the SF genes between the earlier two stages were greater than that between the latter two stages (Fig. 5b; Supplementary Table 6). To the best of our knowledge, only six SFs (U2AF1, SF3b4, HnRNPL, SRSF1, SRSF2, and DDX17) have been revealed to be associated with osteogenesis (He et al., 2022; Jia et al., 2019; Liu et al., 2021; Watanabe et al., 2007; Yi et al., 2019), and five of them except U2AF1 were found to be differentially expressed (top 62 up-regulated and top 12 down-regulated) during osteogenic differentiation in our datasets (Supplementary Table 6). U2AF1 was not a DEG, but U2AF1L4, predicted to be part of H2AF complex, was ranked as the top seventh down-regulated gene. The remaining DE SFs, including the top five up-regulated (CELF2, JUP, ALYREF, MELK, and SNRPB) and down-regulated (SEC31B, PCBP3, CLK4, LGALS3, and CLK1), have not been investigated in osteogenesis (Supplementary Table 6). We then examined differential splicing among stages using Leafcutter, which quantifies intron usage across samples by leveraging spliced reads that span an intron and calculates the percent spliced-in (PSI), which is the percentage of transcripts for a given gene in which the intron sequence is present (Li et al., 2018). Around 30% of the maximum changes of PSI for DS intron clusters of variably excised introns of a gene across osteogenic stages were greater than 10% (Fig. 5c). We identified 1,932 DS intron clusters for 1,750 genes from the MSC to preOB stage, and 254 clusters for 260 genes from the preOB to OB stage (Fig. 5d; Supplementary Table 6); the difference of the number of DS genes between these stages may be a reflection of the difference in the number and expression level changes of SFs (Fig. 5a and 5b). 31% and 25% of the DS genes contained at least one previously unannotated exon, from the MSC to the preOB stage and from the preOB to OB stage, respectively, indicating specific transcription profiling and the complexity and diversity of the human transcriptome of osteogenic differentiation (Supplementary Table 6).
Fig. 5. Differential splicing between osteogenic differentiation stages.
a Heatmap of the expression of 287 splicing factors (SFs) differentially expressed across the MSC, preOB, and OB stages. b Venn diagram showing the numbers of the differentially expressed SFs from the MSC to preOB stages (blue) and the preOB to OB stages (orange) and their overlap. Their expression fold changes for both up and down directions between stages are shown in the lower panel. Center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum values; crosses represent sample means. c Proportion of the maximum percent spliced-in (PSI) changes for intron clusters with significant (FDR ⩽ 0.05) differential splicing (DS) between stages identified by LeafCutter. d Venn diagram showing the numbers of the differentially spliced genes from the MSC to preOB stages (blue) and the preOB to OB stages (yellow) and their overlap. Their top enriched GO terms and Reactome pathways (BP, biological process; MF, molecular function; CC, cellular component; RP, Reactome pathway) are shown in the lower panel. e Overview of a significant DS intron cluster in TF NFIX associated with increased expression of the intron at chr19:13025552–13073047 at the preOB stage compared to the MSC stage (CTF_NFI_2, CCAAT box-binding transcription factor_nuclear factor I_2 domain). (Left) Increased or decreased intron usage from the MSC to the preOB stages are indicated in red and turquoise, respectively. The FDR-corrected p value for the significant DS is indicated in blue above the intron cluster when it is significant. (Right) Details of the significant DS intron cluster indicate the PSI for each intron at the MSC stage (red) and the preOB stage (fuchsia) and the changes in PSI (ΔPSI) (blue) between stages. The affected protein domain is highlighted in grey above the intron cluster. f Similar to e, but for the lncRNA gene CYTOR. The significant p value for the DS is indicated above the intron cluster.
We then performed GO and RP enrichment analyses of the DS genes to explore their potential biological functions during osteogenic differentiation (FDR ≤ 0.05, Fig. 5d and Supplementary Table 6). Several hundred of the DS genes were significantly enriched for various metabolic processes, and 52 DS genes were associated with ECM organization. Various sets of the DS genes were associated with chromosome organization, histone modification, transcription factor binding and activity, transcription coactivator activity and coregulator activity, and post-translational protein modification, suggesting that DS events regulate gene expression at multiple levels, including modulating TFs and TF-TF association during osteogenic differentiation. 48 DS genes were significantly enriched for RNA splicing regulation, indicating modulation of the functions of SFs themselves by DS events (Fig. 5d and Supplementary Table 6).
A considerable number of differential splicing events are predicted to produce different protein isoforms, and more than 40% (MSC to preOB, 798 domains affected in a total of 1,929 DS genes; preOB to OB,110 domains affected in a total of 254 genes) of the events potentially modify domains of protein-coding genes (Supplementary Table 6). There were many DS genes encoding proteins with known roles in ECM organization, mineralization, and bone formation, such as VCAN (Nakamura et al., 2005), FN1 (Yang et al., 2020b), and MACF1 (Hu et al., 2021) (Supplementary Fig. 5a, 5b, and 5c). A notable example of a DS TF was NFIX (Fig. 5e). NFIX, encoding nuclear factor I transcription factor family member X, plays a pivotal role in bone mineralization (Driller et al., 2007). NFIX at the preOB stage compared to the MSC stage, showed increased expression of isoforms containing the intron at chr19:13025552–13073047, where the N-terminal DNA-binding domain is located (Fig. 5e). This domain contains four Cys residues highly conserved in all species, which are required for its DNA-binding activity (Novak et al., 1992). We found that some lncRNA genes also had differential splicing among stages. For example, cytoskeleton regulator RNA gene (CYTOR), which promotes cell proliferation, invasion, and migration in various types of cancer (Zhu et al., 2020), showed increased inclusion of the exon at chr2:87455645–87521207 (Fig. 5f). Altogether, these findings suggest that alternative splicing plays a prominent role in osteogenic differentiation.
Identification of key regulators in the co-expression network of human osteogenic differentiation.
We performed multiscale embedded gene co-expression network analysis (MEGENA) to identify gene co-expression structures (i.e., modules) as well as key network regulators (Holmes et al., 2020; Song and Zhang, 2015). MEGENA was conducted on the RNA-seq data from the three osteogenic differentiation stages and identified 168 co-expressed gene modules (FDR ⩽ 0.05, Fig. 6a; Supplementary Table 7). We then identified potential key network regulators (KNR) that were predicted to modulate a large number of DEGs in the network by key driver analysis (Zhang et al., 2013; Zhang and Zhu, 2013). Many modules enriched for DEGs and their associated KNR recapitulated known osteogenic factors. For example, module M188, comprised of 99 genes, was significantly enriched for the up-regulated DEGs (MSC to preOB, fold enrichment = 1.64, adjusted p value = 6.65E-3; preOB to OB, fold enrichment = 2.04, adjusted p value = 1.31E-3) (Supplementary Fig. 6a and Supplementary Table 7). This module contained ten genes previously implicated in bone development or remodeling (Supplementary Table 7). Three of them, including SF DDX17 and lncRNA MALAT1, were identified as KNRs by key driver analysis (Supplementary Fig. 6b) (He et al., 2022; Xiao et al., 2017). Another SF (RNPC3) and two lncRNAs (MIR99AHG and ZKSCAN2-DT) were identified as KNRs among 11 DE SFs and 10 DE lncRNAs in the module (Supplementary Fig. 6b). As TFs are known to be fundamental to the osteogenic process, we particularly were interested in finding TFs with key regulatory roles new to osteogenesis. We focused on modules that were significantly enriched for DEGs and contained DE TF genes identified as key drivers (Supplementary Table 7). We identified 80 such TF key drivers, of which 60 have unknown biological functions in osteogenesis (Supplementary Table 7). The top 3 up-regulated (ZBTB16, HIF3A, and NR2F1) (Manikandan et al., 2018; Onizuka et al., 2016; Zhu et al., 2014) and the top 2 down-regulated (TEAD4, HMGA1) (Suo et al., 2020; Wu et al., 2021) KNR TFs have established roles in osteogenesis. The third most down-regulated KNR TF is KLF16 of module M204, which previously had little known involvement in osteogenesis (Supplementary Table 7).
Fig. 6. Gene co-expression network analysis reveals KLF16 plays an important role in osteogenic differentiation.
a Sunburst plots represent the hierarchy structure of the MEGENA co-expression network constructed on gene expression during the osteogenic differentiation. The structure is shown as concentric rings where the center ring represents the parent modules, and outer rings represent smaller child modules. Subnetwork modules are colored according to the enrichment of differential gene expression between stages (FDR < 0.05; top, MSC to preOB; bottom, preOB to OB; blue, down-regulated; red, up-regulated; cyan, both up- and down-regulated). The subnetwork branch for module M204 is outlined and labeled. b Co-expression network module M204. Diamonds indicate key driver genes, and circles indicate non-key driver genes. Blue indicates DEGs from MSC to preOB stages, red indicates DEGs from preOB to OB stages, and cyan indicates shared DEGs for both comparisons. Differentially expressed lncRNA and SF genes are underlined in fuchsia and green, respectively. Genes known to be related to bone are in red.
To further evaluate the expression patterns of the top candidate genes that we had discovered through our MEGENA as well as our DEG and TF-TF network analyses at the cell type level, we employed our previously published single-cell RNA-seq data. This data from iPSC-induced MSC osteogenic differentiations in six healthy individuals, were originally used to compare the human and chimpanzee skeleton (Housman et al., 2022). In that study, gene expression had been assessed at two stages, MSCs (day 0) and osteogenic cells (day 21) – analogous to our bulk MSC (day 0) and OB (day 21) stages, respectively. Although the differentiation culture conditions differed from this study, we found similar differential expression patterns in a pseudobulk analysis of the scRNA-seq time points for the top-5 gene sets of up- and down-regulated KNR-TFs (Supplementary Table 7), TF regulators based on TF-TF regulatory networks (Supplementary Table 5), up- and 5 down-regulated SFs (Supplementary Table 6), as well as 6 well known osteogenic markers (Fig. 1c). KLF16 was significantly differentially expressed, with its expression decreasing from the MSC to the OB stage in both datasets (Fig. 7a, Supplementary Fig. 7, Supplementary Table 9), validating our observations. We further analyzed pseudobulk expression data derived from five different osteogenic cell types and found that most genes of interest showed similar levels of expression among these cell types with the most differences occurring in mature osteocytes, including a notable reduction of RUNX2 expression as expected (Supplementary Table 10) (Thomas and Jaganathan, 2022).
Fig. 7. Inhibitory role of Klf16 in osteogenic differentiation.
a The expression of Klf16 at three human osteogenic differentiation stages. Data are shown as mean + SEM. * MSC vs. preOB, # preOB vs. OB, or + MSC vs. OB, adjusted p value < 0.001. b Analysis of Klf16 expression by RT-qPCR in MC3T3-E1 cells transduced vectors containing either stuffer sequence or Klf16 cDNA. Data are presented as the mean ± SEM (n=3, unpaired t-test p < 0.01). c Osteogenic differentiation of MC3T3-E1 cells without or with overexpression of Klf16, stained for ALP at day seven and alizarin red and von Kossa at day 14 and 21. d Length, fat mass, and lean mass of wild type (WT) and Klf16+/− mice. e DEXA analysis of whole body (head excluded) bone mineral content (BMC), bone area (B-area), and bone mineral density (BMD) of WT and Klf16+/− mice. f and g Representative microCT images of distal femur trabecular bone in f (left, top view; right, side view) and cortical bone in g from WT and Klf16+/− mice. Scale bar: 1 mm. h and i Graphs show trabecular bone volume/tissue volume (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular separation (Tb.Sp) in h; cortical bone area (Ct.Ar), cortical periosteal perimeter (Ct.Pe.Pm), and cortical endosteal perimeter (Ct.En.Pm) in i. For d, e, h, and i, data are presented using BoxPlotR (Spitzer, et al., 2014) as the mean ± SEM (WT n = 6, Klf16+/− n = 6, 3 males and 3 females for each group, aged 17 weeks, paired t-test, * p < 0.05, ** p <0.01).
KLF16, a key regulator of module M204 involved in human osteogenic differentiation.
We further focused on KNR TF, KLF16, as it is a key regulator in module M204. Multiple lines of evidence point to a potentially critical role of M204 in osteogenic differentiation. M204, comprised of 85 genes, was significantly enriched for down-regulated DEGs (MSC to preOB, fold enrichment = 1.80, adjusted p value = 1.58E-2; preOB to OB, fold enrichment = 2.29, adjusted p value = 1.08E-3) (Fig. 6a). Additionally, more than 9% (8 genes) of the DEGs in M204 are related to bone development and metabolism (Supplementary Table 7). Three (PHLDA2, RHOC, PFN1) out of the eight genes were identified as KNRs (Fig. 6b). The expression of PHLDA2 is associated with skeletal growth and childhood bone mass (Lewis et al., 2012). Knockdown of RHOC dramatically inhibits osteogenesis (Zheng et al., 2019). PFN1, a member of the profilin family of small actin-binding proteins, is known to regulate bone formation. The loss of function of PFN1, causes the early onset of Paget’s disease of bone, a disease with impaired osteoclast and osteoblast differentiation (Scotto di Carlo et al., 2020). We also examined overlap of the genes in M204 with the genes from genome-wide association studies with significant signals for bone area (B-area), bone mineral density (BMD), and hip geometry (Hsu et al., 2019; Morris et al., 2019; Styrkarsdottir et al., 2019), and found five genes, CYFIP1, MMD, PELO, PRSS23, and SNX13. Thus, based on our accumulative analyses, we hypothesized that KLF16 plays an important role in osteogenic differentiation.
Regulatory role of Klf16 in murine osteogenic differentiation in vitro.
We explored the role of Klf16 in osteogenic differentiation and bone mineral density and architecture. We overexpressed Klf16 in the preosteoblast murine cell line MC3T3-E1 using lentivirus vector-mediated gene transfer (Fig. 7b). We then performed three independent osteogenic differentiation experiments, each with three technical replicates for each differentiation stage (day 7, 14, and 21). Consistent with its continuously decreased expression across osteogenic stages, overexpression of Klf16 in the preosteoblast murine cell line MC3T3-E1 dramatically suppressed osteogenic differentiation at the early stage (day 7) with reduced matrix maturation detected by ALP staining, and at later stages (day 14 and 21) with reduced mineralization detected by alizarin red and von Kossa staining, compared to the control cells (Fig. 7c). The inhibitory function of Klf16 in osteogenic differentiation in vitro was observed consistently for each of the experimental and technical replicates.
Klf16+/− mice show increased bone mineral density and abnormal trabecular and cortical bone.
We explored the role of Klf16 in bone formation in vivo by studying a Klf16 knock-out mouse, generated by deleting a 444 bp segment in exon 1 using CRISPR technology by the Knockout Mouse Phenotyping Program (KOMP). This mutation deleted the Kozak sequence and ATG start, leaving the final 16 bp of exon one and the splice donor site. We examined heterozygous rather than homozygous mutants because of difficulties in breeding for the latter. No significant difference in the overall length between adult Klf16+/− and wild type (WT) control mice of the same C57BL/6 background and matched by gender and age were initially found (Fig. 7d). However, adult Klf16+/− mice had significantly increased lean mass (total tissue mass – fat mass, including bone mineral content) (p < 0.05), and no significant change in fat mass relative to WT mice (Fig. 7d). Furthermore, there was a dramatic increase in whole body bone mineral content (BMC), bone area, and bone mineral density in Klf16+/− mice compared to WT control mice revealed by DEXA scanning (p <0.01) (Fig. 7e). Likewise, μCT scanning of trabecular bone from adult Klf16+/− mice showed significantly increased bone to tissue volume (BV/TV) (p < 0.05) and trabecular thickness (Tb.th) (p < 0.01). There were no significant differences in trabecular number (Tb.N) and trabecular separation (Tb.Sp), indicating the maintenance of distances between trabeculae in the mutant (Fig. 7f and 7h). In addition, adult Klf16+/− mice showed significantly increased cortical bone area (Ct.Ar) (p < 0.05), cortical periosteal perimeter (Ct.Pe.Pm) (p < 0.01), and cortical endosteal perimeter (Ct.En.Pm) (p <0.01) compared to WT control mice (Fig. 7g and 7i). These findings further validated our predicted TF and co-expression networks and established a regulatory role of TF Klf16 in bone. Studying the bone features in MSC-specific Klf16 conditional knockout mice could offer additional confirmation of its importance in the osteoblast lineage, as Klf16 might have functions in both osteoblasts and osteoclasts, yet such a animal model is not available to date.
Discussion
Understanding the cellular and molecular mechanisms that drive both physiological and pathological bone formation requires knowledge of global transcriptional changes during osteogenic differentiation. To shed light on this, we utilized iPSC-derived MSCs as our tool. By profiling RNA expression in 20 healthy human lines at three osteogenic stages, MSC, preOB and OB, we observed the transcriptome profiles of our iPSC-derived MSCs and OBs were consistent with those of their primary counterparts, which suggests the potential applicability of our model system for osteogenic differentiation studies. Our differential gene expression analyses provided insights into transcriptional variations for each osteogenic stage, marked by changes in expression levels of numerous coding and non-coding genes. Patterns of dynamic gene expression identified clusters of genes implicated in different biological functions during differentiation. Many genes known to be implicated in bone function, such as bone development, bone morphogenesis, and extracellular matrix organization, were found to be significantly up-regulated only from the MSC to the preOB stage (cluster 3).
Our systems biology approach revealed complex networks of TFs, lncRNAs, and differential splicing events underlying osteogenic differentiation. A particular observation was the high degree of correlation between the dynamic expression patterns of TFs and lncRNAs. TF networks in mammalian cells often arise from TFs modulating other TFs (Wilkinson et al., 2017). Our research unveiled a multifaceted osteogenic differentiation TF network, segmented into five interactive modules. These modules, consistent with previous findings on functional modularity (Dittrich et al., 2008), showcased specific biological functions. For instance, Module 3 underscores TFs associated with fat cell differentiation, suggesting a balance between osteogenesis and adipogenesis within bone structures. This resonates with recent studies emphasizing the role of stem cell TFs in osteogenesis, which also act as checks on adipogenesis (Rauch et al., 2019). Additionally, this module identifies TFs integral to muscle differentiation, hinting at overlapping regulatory mechanisms for both bone and muscle growth. This concept is further supported by recent discoveries linking mesenchymal progenitors in muscle to bone formation (Julien et al., 2021). Further analyses discerned connections between TFs, RNA splicing, and ncRNA transcription across modules. This connectivity endorses known interactions between signaling pathways like TGF-β/BMP and Wnt (Hernández-Vega et al., 2021; Thomas and Jaganathan, 2022). Overall, our findings reveal intricate TF networks, pathway interactions, and offer deeper insights into osteogenic differentiation.
Our results offer a pioneering atlas of the expression of lncRNAs across three osteogenic stages: MSC, preOB, and OB. We discerned 2,249 lncRNAs with notable expression changes, surpassing even those of TFs. Concurrently, we identified 441 differentially expressed adjacent protein-coding genes, illustrating the bi-directional influence of lncRNAs during differentiation. Numerous neighboring coding DEGs were associated with skeletal development and mineral deposition processes. Fifty-five TFs, including several TEAD and ZNF TF family members were within these genes, and the correlation in genomic proximity of DE lncRNA and DE TF genes was statistically significant. The TEAD family, vital in bone remodeling, operates via the Hippo pathway (Tang and Weiss, 2017; Zhao et al., 2018), while the ZNF family finely tunes osteogenic differentiation with both inhibition and promotion by different members (Quach et al., 2011; Stains and Civitelli, 2003; Twine et al., 2016). Moreover, 16 SFs emerged in these findings, reinforcing the significant role lncRNAs play in osteogenic differentiation through various regulatory pathways.
In our study, we systematically identified differentially expressed SFs and DS genes during osteogenic differentiation. We discovered extensive differentially spliced genes, including ECM, TF, lncRNA, and SF, highlighting alternative splicing’s pivotal role in osteogenic differentiation. Some DS genes, like NFIX, are linked to human diseases. Mutations in NFIX that affect splicing lead to Marshall-Smith syndrome, marked by rapid bone maturation and other skeletal abnormalities (Malan et al., 2010; Martinez et al., 2015). These observations underscore the importance of correct splicing in physiological osteogenesis and the potential implications in pathological conditions, including bone cancer (Liu and Rabadan, 2021).
Our study explored the osteogenic gene co-expression network, highlighting potential gene modules and driver genes, like TFs, lncRNAs and SFs, that play an important role in differentiation. In this study we experimentally validated the regulatory role of KLF16 of module M204 in osteogenic differentiation as a proof-of-concept that our dataset provides a resource for the discovery of novel mechanisms in osteogenic differentiation. Future studies will be required to explore the roles of specific lncRNA and alternative splicing genes identified in this study. Module M204 was enriched for DEGs, many of which are related to bone metabolism. Three of these hinted at being KNR genes. When we focused on the transcription factor KLF16, not previously associated with bone formation, we observed some implications for its involvement in osteogenic differentiation. Overexpression of Klf16 reduced matrix maturation during MC3T3-E1 differentiation. Conversely, Klf16-haploinsufficient mice showed altered bone characteristics, including increases in whole body BMD, femoral bone volume to total tissue volume, trabecular bone thickness, and cortical bone area, which suggests KLF16’s regulatory function in restraining bone formation.
Encoded as a C2H2-type zinc finger transcription factor, KLF16 has affinity for GC and GT boxes, displacing TFs SP1 and SP3 in neural settings (Wang et al., 2016). These TFs, when attached to certain promoters, play roles in osteogenic differentiation (Le Mée et al., 2005). Furthermore, KLF16 targets specific metabolic genes, including estrogen-related receptor beta (ESRRB) and PPARA (Daftary et al., 2012; Sun et al., 2020). Also, our TF regulatory network found that KLF16 is associated with Notch signaling and TGFβ/BMP-SMAD signaling pathways, and regulation of ncRNA transcription. This paints a broad canvas of potential regulatory mechanisms for KLF16 in bone dynamics. Furthermore, other members from the KLF family, especially KLF4 (module 1), KLF5 (module 5), and KLF15 (module 3) (Supplementary Table 4), significantly influence skeletal development (Li et al., 2021; Shinoda et al., 2008; Song et al., 2017; Yu et al., 2021; Zakeri et al., 2022). KLF5, co-existing with KLF16 in module 5, curtails osteogenesis by inhibiting β-catenin through DNMT3B-induced hypermethylation (Li et al., 2021). KLF4’s modulation of osteogenic differentiation hinges on the BMP4-dependent pathway, resulting in reduced osteoblast numbers in deficient mice (Yu et al., 2021). Meanwhile, KLF15 enhances chondrogenic differentiation via the TF SOX9 promoter (Song et al., 2017). The roles of KLF6 and KLF9 (modules 3 and 2) in bone metabolism remain less defined (Zakeri et al., 2022). Collectively, these findings underline the KLF family’s complex involvement in osteogenesis.
Interestingly, KLF16 was upregulated in the MSCs of five elderly patients (79–94 years old) suffering from osteoporosis compared to five age-matched controls in a previous study (Benisch et al., 2012). A following study identified KLF16 as one of the key TFs whose targets were enriched from the DEGs in the MSCs of the osteoporosis patients versus controls (Liu et al., 2019). The inhibitory role of KLF16 in osteoblast differentiation identified in our study supports the hypothesis that the overexpression of KLF16 might contribute to osteoporosis, and KLF16 could be a potential therapeutic target for osteoporosis.
Taken together, the differentiation from iPSC-derived MSC to OB presents a promising model to explore the gene expression and networks in osteogenic differentiation. Our study offers insights into the intricate, layered, and diverse landscape of osteogenic differentiation regulation, encompassing TFs, lncRNAs, differential splicing, and their reciprocal regulations. This information serves as a foundational resource for delving deeper into physiological osteogenesis processes and decoding the complexities of pathological bone formation. Furthermore, this experimental model might hold potential for therapeutic research, possibly aiding treatments for conditions like osteoporosis and osteomalacia.
Methods
Human subjects
The twenty human subjects included in this study were in good general health. The study protocols and informed consent were approved by the Institutional Review Board (IRB) of Stanford University and the IRB of the Icahn School of Medicine at Mount Sinai. Each subject gave written informed consent for study participation. Height and weight for each subject were measured while wearing light clothing and with no shoes. See Supplementary Table 1 for complete demographic metadata.
Animal care and use
The Klf16+/− mouse strain was generated on the C57BL/6NJ genetic background by the Knockout Mouse Phenotyping Program (KOMP) at The Jackson Laboratory using CRISPR technology as described in the above text. Mouse procedures were in compliance with animal welfare guidelines mandated by the Institutional Animal Care and Use Committee (IACUC) and Institutional Biosafety Committee (IBC) at The Jackson Laboratory. Genotyping was performed by real-time PCR of DNA extracted from tail biopsies. A detailed protocol can be found on The Jackson Laboratory website.
Cell lines
MC3T3-E1 Subclone 4 line (ATCC, CRL-2593, sex undetermined) was purchased directly from the ATCC. Mouse Embryonic Feeders were purchased from GlobalStem (Cat# GSC-6001G). Human iPSC lines were generated in this study. The cells were maintained at 37°C with 5% CO2.
Peripheral blood mononuclear cell (PBMC) and dermal fibroblast isolation
The twenty human subjects included in this study were in good general health. The study protocols and informed consent were approved by the Institutional Review Board (IRB) of Stanford University and the IRB of the Icahn School of Medicine at Mount Sinai. Each subject gave written informed consent for study participation. Height and weight for each subject were measured while wearing light clothing and with no shoes. See Supplementary Table 1 for complete demographic metadata. PBMCs were isolated as we previously described (Carcamo-Orive et al., 2017). Briefly, blood was drawn in tubes containing sodium citrate anticoagulant solution (BD Vacutainer CPT mononuclear cell preparation tube, 362760). PBMCs were separated by gradient centrifugation. The cells were then frozen in RPMI medium (MilliporeSigma, R7388) supplemented with either 12.5% human serum albumin and 10% DMSO (MilliporeSigma, D2438) or 10% fetal bovine serum (FBS) (MilliporeSigma, F4135) and 10% DMSO, and stored in liquid nitrogen for later use. The detailed protocol for dermal fibroblast isolation can be found in our previous publication (Schaniel et al., 2021). Briefly, a skin sample was taken from each of the clinically healthy subjects using a 3 mm sterile disposable biopsy punch (Integra Miltex, 98PUN3–11). Each sample was cut into smaller pieces and placed into gelatin-coated tissue culture dishes with DMEM (Thermo Fisher Scientific, 10567022) supplemented with antibiotics, 20% FBS, non-essential amino acids (Thermo Fisher Scientific, 11140050), 2mM L-glutamine (Thermo Fisher Scientific, 25030081), 2 mM sodium pyruvate (Thermo Fisher Scientific, 11360070), and 100 μM 2-mercaptoethanol (MP Biomedicals, C194705) to establish fibroblast lines. Fibroblasts were harvested using TrypLE Express (Thermo Fisher Scientific, 12605010) and passaged at a 1 to 4 split ratio. Fibroblasts were cryopreserved in 40% DMEM, 50% FBS, and 10% DMSO.
iPSC generation
Erythroblast protocol
The detailed erythroblast reprogramming process can be found in our previous publication (Carcamo-Orive et al., 2017). Briefly, PBMCs were thawed, and the erythroblast population was expanded for 9–12 days until approximately 90% of cells expressed CD36 and CD71. These expanded erythroblasts were reprogrammed by transduction with Sendai viruses (SeV) expressing factors Oct3/4, Sox2, Klf4, and c-Myc using the CytoTune™-iPS 2.0 Sendai Reprogramming kit (Thermo Fisher Scientific, A16518) according to manufacturer’s protocol.
Fibroblast protocol
The detailed fibroblast reprogramming process can be found in our previous publication (Schaniel et al., 2021). Briefly, mycoplasma-free fibroblasts at passage number 3–5 were reprogrammed using the mRNA reprogramming Kit (Stemgent, 00–0071) in combination with the microRNA booster kit (Stemgent, 00–0073) according to the manufacturer’s protocol.
iPSCs expansion and characterization
To establish iPSCs, the cells were cultured on Matrigel (BD Bioscience, 354230–10, or Corning, 254248) in feeder-free conditions and maintained in mTeSR1 (Stem Cell Technologies, 05850). The iPSCs were passaged every 5–7 days after clones were picked. To ensure the high quality of the iPSC lines used in this study, those lines generated with Sendai virus first underwent virus clearance by continuous passaging. To measure the loss of SeV in the iPSCs generated from erythroblasts with SeV, quantitative RT-PCR was performed according to the SeV reprogramming kit manufacture protocol. For quality control, G-banded karyotyping, ALP staining, iPSC pluripotency marker immunocytochemistry, and embryoid body formation analysis (Yaffe et al., 2016) were performed. Differentiating colonies were routinely eliminated from the cultures. All the lines included in the current study had normal karyotypes, exhibited human embryonic stem cell morphology, and expression of ALP and pluripotency markers NANOG, SOX2, OCT4, SSEA4, and TRA-1–60. Further, embryoid body (EB) formation and differentiation assays demonstrated their competence for differentiation into all three germ layers. We excluded samples where any of these characterizations were abnormal (Daley et al., 2009; Yaffe et al., 2016).
Mycoplasma quality control
Mycoplasma testing of the cells was performed intermittently during culture. Cultures were grown in the absence of antibiotics for at least three days before testing. The culture medium or the DNA harvested from the cultures were tested for mycoplasma contamination using a MycoAlert Mycoplasma Detection Kit (Lonza, LT07–418) or e-Myco PLUS PCR Detection Kit (BOCA scientific, 25237).
Alkaline Phosphatase Staining
A high level of ALP expression can characterize the undifferentiated state of iPSCs. ALP is also an osteogenic marker. ALP staining was performed on iPSCs and to monitor the osteogenic differentiation process. Cells cultured on plates were briefly washed with phosphate-buffered saline (PBS), fixed in 4% PFA for 3 minutes at room temperature, and then stained with Alkaline phosphatase kit II (Stemgent, 00–0055) according to the manufacturer’s instructions.
iPSC embryoid body analysis
iPSC embryoid body formation and characterization were performed as previously described with modifications (Lin and Chen, 2008). Briefly, iPSCs were cultured in mTeSR1 medium on Matrigel-coated plates, and the medium was changed daily. On the day of EB formation, when the cells grew to 60–80% confluence, cells were washed once with PBS and then incubated in Accutase for 8–10 minutes to dissociate colonies to single cells and resuspended with mTeSR1 containing 2uM Thiazovivin. To form self-aggregated EBs, single iPSCs were transferred into an ultra-low attachment plate (Corning, CLS3471) via 1:1 or 1:2 passage. EBs were aggregated from iPSCs for three days, then were transferred to 0.1% gelatin-coated 12 well plates and maintained in embryoid body medium (DMEM/F12 supplemented with 10% FBS, 2mM L-glutamine, 0.1 mM non-essential amino acids, and 0.1mM 2–mercaptoethanol) for another ten days for the spontaneous generation of three germ layers with medium changed every other day. Fluorescent immunostaining was performed to detect the expression of markers of the three germ layers.
Fluorescence immunocytochemistry
The cells cultured on plates were briefly washed with PBS, fixed in 4% paraformaldehyde (PFA) in PBS for 15 minutes, washed with PBS three times, and then permeabilized with 0.2% Triton in PBS for 15 mins. The cells were then washed with PBS three times prior to blocking with 1% bovine serum albumin (BSA) (MilliporeSigma, A8412) in PBS for 1 hour at room temperature and incubating with primary antibody 1:100 to 1:900 dilutions in 0.1% BSA in PBS for overnight at 4°C. Washing three times was conducted with PBS followed by incubation with a corresponding secondary antibody in 1:500 dilution in 0.1% BSA for 1 hour at room temperature. After washing with PBS, 1:50,000 diluted Hoechst (Thermo Fisher Scientific, H1398) was added to stain nuclei for 5 minutes, followed by washing with PBS three times and imaging. The following primary antibodies were applied to detect the expression of iPSC markers: anti-TRA-1–60 (Invitrogen, 41–1000, 1/100), anti-SSEA4 (Invitrogen, 41–4000, 1/200), anti-NANOG (Abcam, ab109250, 1/100), anti-OCT-4 (Cell signaling technology, 2840S, 1/400), and anti-SOX2 (Abcam, ab97959, 1/900). To detect the expression of the markers of three germ layers, anti-AFP (Agilent, A0008, 1/100) for endoderm, anti-α-SMA (MilliporeSigma, A5228, 1/200) for mesoderm, and anti-TUBB3 (MilliporeSigma, T2200, 1/200) for ectoderm were used. All the Alexa-conjugated secondary antibodies used were from Invitrogen: anti-mouse Alexa Fluor 488 (A-11029), anti-rabbit Alexa 488 (A-21206), and anti-rabbit Alexa 594 (A-11037).
G-banding karyotyping
Karyotype analysis was performed on all iPSC lines by the Human Genetics Core facility at SEMA4. iPSCs were cultured on Matrigel-coated T25 flasks before karyotyping and reached approximately 50% confluence on the day of culture harvest for karyotyping. Twenty cells in metaphase were randomly chosen, and karyotypes were analyzed using the CytoVision software program (Version 3.92 Build 7, Applied Imaging).
Differentiation of iPSCs to MSCs
In vitro differentiation of iPSCs to MSCs was carried out with commercial cell culture media according to the manufacture protocols with modifications. Briefly, iPSC colonies were harvested with Accutase (Innovative Cell Technologies, AT-104) and seeded as single cells on a Matrigel-coated plate in mTeSR1 and supplemented with 2uM Thiazovivin. The next day, TeSR1 medium was replaced with STEMdiff mesoderm induction medium (MIM) (Stem Cell Technologies, 05221) when cells were at approximately 20 – 50% confluence. Cells were then fed daily and cultured in STEMdiff MIM for four days. On day 5, the culture medium was switched to MesenCult-ACF medium (Stem Cell Technologies, 05440/8) for the rest of the MSC induction duration. Cells were passaged as necessary using MesenCult-ACF Dissociation Kit (Stem Cell Technologies, 05426). Cells were subcultured onto Matrigel at the first passage to avoid loss of the differentiated cells before tolerating the switch to MesenCult-ACF attachment substrate (Stem Cell Technologies, 05448). For passage two or above, cells were subcultured on MesenCult-ACF attachment substrate. After three weeks of differentiation, the cells were sorted for the CD105 (BD Biosciences, 561443, 1/20) positive and CD45 (BD Biosciences, 555483, 1/5) negative MSC population (Giuliani et al., 2011; Kang et al., 2015) using BD FACSAria II in the Mount Sinai Flow Cytometry Core Facility, and then expanded in MSC medium consisting of low-glucose DMEM (Gibco, 10567022) containing 10% FBS (Sotiropoulou et al., 2006). After expansion, a few MSC lines were further examined by BD CantoII for expression of other MSC positive surface markers CD29 (Thermo Fisher Scientific, 17–0299, 1/20), CD73 (BD Biosciences, 561254, 1/20), CD90 (BD Biosciences, 555595,1/ 20), absence of other negative surface markers CD31 (BD Biosciences, 561653, 1/20), CD34 (BD Biosciences, 560940, 1/5), and retention of marker CD105 positivity and CD45 negativity as well.
Fluorescence activated cell sorting (FACS) and analysis
Single cells were washed with FACS buffer (PBS supplemented with 1% FBS and 25mM HEPES (Thermo Fisher Scientific, 1688449)) twice, resuspended to a concentration of 1×107 cells/ml in ice-cold FACS buffer, and incubated with the above conjugated antibodies for 30 minutes at 4° in the dark. Stained cells were then washed with FACS buffer three times before FACS or flow cytometry analysis. Flow cytometry data were analyzed with BD FACSDiva software.
Differentiation of iPSC-derived MSCs to osteoblasts
iPSC-derived MSCs were plated in a 6-well plate at a density of 3×103 /cm2 in MSC medium. After three days, the culture medium was switched to osteogenic differentiation medium (α MEM (Thermo Fisher Scientific, A1049001) supplemented with 10% FBS, 1% non-essential amino acids, 0.1 uM dexamethasone (MilliporeSigma, D4902), 10 mM β-glycerophosphate (MilliporeSigma, G9422), and 200 uM ascorbic acid (MilliporeSigma, A4544)) (Barberi et al., 2005; Holmes et al., 2020; Lee et al., 2015) and maintained in this medium for 21 days. The culture medium was changed every 2–3 days. Cells were harvested for total RNA isolation and RNA sequencing at day 0, day 7, and day 21 of differentiation, as indicated in the main text. ALP staining, and alizarin red staining and von Kossa staining, were employed to examine bone ALP expression and mineralization, respectively.
Alizarin red staining and von Kossa staining
Mineralization of osteoblast extracellular matrix was assessed by both alizarin red staining and von Kossa staining. Cells were washed briefly with PBS, fixed with 4% PFA for 15 minutes, and washed with deionized distilled water three times. For alizarin red staining, fixed cells were incubated in alizarin red stain solution (MilliporeSigma, TMS-008-C) with gentle shaking for 30 minutes, followed by washing with water four times for 5 minutes each with gentle shaking to remove non-specific alizarin red staining. For von Kossa staining, fixed cells were incubated in 5% silver nitrate solution (American Master Tech Scientific, NC9239431) while exposed to UV light for 20 minutes. Unreacted silver was removed by incubating in 5% sodium thiosulfate (American Master Tech Scientific, NC9239431) for 5 minutes, followed by washing with water twice. Mineralized nodules were identified as red spots by alizarin red staining and dark brown to black spots by von Kossa staining.
RNA-seq library preparation and sequencing
For total RNA isolation, cells were washed with PBS and harvested for RNA extraction using miRNeasy mini kit (QIAGEN, 217004) according to the manufacturer’s instruction. Illumina library preparation and sequencing were conducted by the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine (Baltimore, MD). RNA concentration and quality were determined using NanoDrop Spectrophotometer (Thermo Scientific, DE). RNA integrity (RNA Integrity Number, RIN) was verified using Agilent BioAnalyzer 2100 and the RNA Nano Kit prior to library creation. Illumina’s TruSeq Stranded Sample Prep kit was used to generate libraries. Specifically, ribosomal depleted RNA was converted to cDNA and size selected to 150 to 200 bp in length, then end-repaired and ligated with appropriate adaptors. Ligated fragments were subsequently size selected and underwent PCR amplification techniques to prepare the libraries with a median size of 150bp. Libraries were uniquely barcoded and pooled for sequencing. The BioAnalyzer was used for quality control of the libraries to ensure adequate concentration and appropriate fragment size. Sequencing was performed on an Illumina HiSeq 2500 instrument using standard protocols for paired-end 100bp sequencing. All the samples were processed in one batch.
RNA-seq pre-processing and gene differential expression analyses
Illumina HiSeq reads were processed through Illumina’s Real-Time Analysis (RTA) software generating base calls and corresponding base call quality scores. CIDRSeqSuite 7.1.0 was used to convert compressed bcl files into compressed fastq files. After adaptor removal with cutadapt (Martin, 2011) and base quality trimming to remove 3′ read sequences if more than 20 bases with Q ⩾ 20 were present, paired-end reads were mapped to the human GENCODE V29 reference genome using STAR (Dobin et al., 2013) and gene count summaries were generated using featureCounts (Liao et al., 2014). Raw fragment (i.e., paired-end read) counts were then combined into a numeric matrix, with genes in rows and experiments in columns, and used as input for differential gene expression analysis with the Bioconductor Limma package (Ritchie et al., 2015) after multiple filtering steps to remove low-expressed genes. First, gene counts were converted to FPKM (fragments per kb per million reads) using the RSEM package (Li and Dewey, 2011) with default settings in strand specific mode, and only genes with expression levels above 0.1 FPKM in at least 20% of samples were retained for further analysis. Additional filtering removed genes less than 200 nucleotides in length. Finally, normalization factors were computed on the filtered data matrix using the weighted trimmed mean of Mvalues (TMM) method, followed by voom (Law et al., 2014) mean-variance transformation in preparation for Limma linear modeling. The limma generalized linear model contained fixed effects for sex (male/female), and cell source (fibroblast/erythroblast) and a random effect term was included for each unique subject. Data were fitted to a design matrix containing all sample groups, and pairwise comparisons were performed between sample groups (i.e., MSC stage, preOB stage, and OB stage). eBayes adjusted p values were corrected for multiple testing using the Benjamin-Hochberg (BH) method and used to select genes with significant expression differences (adjusted p value ≤0.05).
Gene expression patterns
Significantly up- or down-regulated genes identified by the above DEG analyses were clustered into eight possible patterns based on their up/down expression changes between MSC to preOB and preOB to OB stages, including up-up, down-down, up-stable, down-stable, stable-up, stable-down, up-down, and down-up. Gene expression of the DEGs was normalized by Z-score transformation. Heatmap of Z-score normalized gene expression of the DEGs in the eight clusters was plotted using Seaborn in Python (Waskom et al., 2021). Dynamic gene expression change patterns were plotted with TiCoNE using the log2 fold change of gene expression from MSC to preOB and preOB to OB stages as input for each of the clusters.
Integration of RNA-Seq data of human primary MSCs and OBs, iPSCs, and tissues in GTEx
GTEx gene expression data (version 7) and metadata were downloaded from the GTEx website (Ardlie et al., 2015). Raw counts were extracted for the brain, heart, kidney, liver, lung, muscle, nerve, ovary, testis, and thyroid tissues. After filtering the lowly expressed genes (1CPM in less than 20% samples), normalization factors were computed using the weighted trimmed mean of M-values (TMM) method, followed by the voom mean-variance transformation (Law et al., 2014). The Human iPSC dataset was from our previous publication, deposited in the GEO database with accession number GSE79636 (Carcamo-Orive et al., 2017). Two datasets for each of primary human MSC and OB cell type were collected from the GEO accessions GSE94736 (Roforth et al., 2015) and GSE118808 (Kaczynski et al., 2003; Ma et al., 2019), and GSE55282 (Rojas-Peña et al., 2014) and GSE75524 (Al-Rekabi et al., 2016), respectively. All the datasets were filtered and normalized in the same way as the GTEx data. Genes from all the datasets were intersected to obtain shared genes. Gene expression levels of the shared genes were extracted from all the datasets. Principal component analysis (PCA) was performed on the extracted and combined gene expression matrix. Multi-dimensional scaling was used to visualize the top 2 principal components.
Gene ontology and reactome pathway enrichment analyses
Gene ontology (GO) biological process (BP), molecular function (MF), and cellular component (CC), and Reactome pathway enrichment analyses were performed using the PANTHER classification system (www.pantherdb.org) (Mi et al., 2019a). The statistical overrepresentation test in PANTHER was fulfilled by Fisher’s exact test together with FDR multiple test correction to identify enriched GO categories and Reactome Pathways among the input genes relative to the indicated reference list as stated in figure legends (Carbon et al., 2019; Mi et al., 2019b). We filtered enrichment tests using FDR ≤ 0.05.
Transcription factor network analysis
We downloaded the ChIP-seq and Chip-Exo datasets of DNA binding peaks of human transcription regulators (TRs), where the overwhelming majority are TFs, from ReMap 2020 (Chèneby et al., 2020). We extracted the genomic coordinates of the transcription start sites of human genes from Ensembl Human Genes GRCh37.p13, and then applied BedTools v2.3.0 to identify the target TR genes of a TR by checking the intersection of the latter TR’s binding peaks and the 2kb window before and after the transcription start sites of the target TR genes (Quinlan and Hall, 2010). We used the Python package scipy.stats.pearsonr to calculate the correlation coefficient between the expression of TR genes (Hao et al., 2015; Jiang et al., 2014; Virtanen et al., 2020; Zhu et al., 2015). The regulatory relationship between TRs was predicted using two criteria: (1) the binding site peaks of a TR are within the distance of 2kb upstream or downstream of the known transcription start sites of the target TRs; and (2) the absolute value of the correlation coefficient between the TR genes is greater than 0.6 and adjusted p value is less than 0.05 (Camacho et al., 2005). We then used Gephi 0.9.2 to generate and visualize the regulatory network, where each node was defined as a TR gene, and two nodes were connected by an edge when ReMap data demonstrated regulation between the two TRs, and our RNA-seq data also showed expression correlation between them. Furthermore, the network community detection hierarchical algorithm was applied to determine relationships between subsets of the whole network and define modules. Betweenness centrality for each of the nodes was calculated and then used to rank node sizes (Bastian et al., 2009).
LncRNA analysis
All the lncRNA and protein-coding genes detected in our dataset were used to identify their neighborships. The genomic coordinates of the lncRNA genes and the transcription start sites of the protein-coding genes were extracted from Ensembl Human Genes GRCh37.p13. We identified the neighboring coding genes of the lncRNA genes by intersecting the lncRNA genomic coordinates with the 10kb windows before and after the transcription start sites of the protein-coding genes using BedTools v2.3.0 (Cabili et al., 2011; Quinlan and Hall, 2010; Ransohoff et al., 2018). In each of the comparisons of the MSC to preOB and preOB to OB stages, the paired lncRNA and neighboring protein-coding genes were sorted into three groups: pairs with up-regulated lncRNAs, pairs with down-regulated lncRNAs, and pairs with non-DE lncRNAs. The expression changes of the paired neighboring coding genes of each group were then compared. In addition, we randomly paired DE lncRNAs with coding DEGs outside the 10kb neighboring area and analyzed the density of the correlations of expression of lncRNAs and their pairwise DEGs with Python seaborn.kdeplot (Waskom et al., 2021, Rosenblatt, 1956).
Analysis of local splicing differences
Local splicing analysis was performed in an annotation-free approach using LeafCutter as we previously described (Holmes et al., 2020). Briefly, we used LeafCutter to first find clusters of variably spliced introns across all samples and then to identify differential splicing between osteogenic stages, modeling intron clusters using the Dirichlet-Multinomial generalized linear model (GLM) (Li et al., 2018). We used LeafCutter to find intron clusters as follows: overlapping introns (i.e., exon junction-spanning reads) were clustered and filtered to keep intron clusters supported by at least 25 split reads across all samples, retaining introns of up to 100 kb and accounting for at least 1% of the total number of reads in the entire cluster. This yielded 17,270 clusters encompassing 32,542 introns in 9,505 genes from MSC to preOB stages and 33,033 introns in 9,517 genes from preOB to OB stages that were used for further analysis. This intron count file was then used in the differential splicing analysis. DS intron clusters were identified in pairwise analyses between sample groups (MSC stage compared to preOB stage and preOB stage compared to OB stage), using the same generalized linear model as used for differential gene expression analysis. After discarding introns that were not supported by at least one read in 5 or more samples, clusters were analyzed for DS if at least three samples in each comparison group had an overall coverage of 20 or more reads. P values were corrected for multiple testing using the BH method and used to select clusters with significant splicing differences (FDR ≤ 0.05).
Gene co-expression network analysis and key driver analysis
Gene co-expression network was constructed using the R package MEGENA v1.3.7 (Song and Zhang, 2015). The same filtered, normalized, and covariate-adjusted gene expression data matrix used in the differential gene expression analysis was used as input for MEGENA. Specifically, the Pearson correlation was used to calculate significant correlations between gene pairs among the 60 samples. With a cutoff of 0.05 FDR, significant correlations were identified by 100 permutations of the gene expression matrix. Next, planar filtered network construction and multi-scale clustering analysis were performed. Finally, significant modules were identified at a 5% FDR with 100 times of network permutation. Modules of smaller than 50 genes or larger than 5,000 genes were excluded from the downstream analyses. Enrichment analysis was performed between the modules and DEG signatures between MSC and preOB stages and between preOB and OB stages through Fisher’s exact test. BH procedure was applied to the p-values to correct for multiple-testing (Yoav Benjamin, 1995). Modules were visualized using the Cytoscape (Otasek et al., 2019). Sunburst plots of modules were visualized using the R package sunburstR v2.1.5. We used all the MEGENA nodes and edges as input in the key driver analysis to identify global network key drivers that were predicted to modulate a large number of downstream DEGs in the network (Zhang et al., 2013; Zhang and Zhu, 2013). For each gene in the MEGENA network, we tested whether the network neighborhood of the gene was enriched with the DEG signature. Specifically, we tested if the nodes within a path length of 6 steps of the candidate key driver gene were enriched for the DEGs using Fisher’s Exact test. The p-values were then corrected by the Bonferroni procedure to adjust for multiple comparisons.
Processing of comparative single-cell pseudobulk data
scRNA-seq data (GEO accession: GSE181744) from our previous publication (Housman et al., 2022), which includes mixtures of iPSC-derived, MSC osteogenic differentiations from different species, was used as a comparative dataset. To maximize the comparative utility of these data, we re-processed the raw scRNA-seq data to obtain pseudobulk expression values for all genes annotated in the human genome and for cell types of interest from the six humans and one human technical replicate included in this dataset. Briefly, reads were processed using standard 10X Genomics Cell Ranger 3.1.0 pipelines (Zheng et al. 2017) that extracted 10X cell barcodes and UMIs and aligned the remaining reads to the human genome (hg38). Human cells and specific cell types of interest (MSCs, osteogenic cells, preosteoblasts, osteoblasts, embedding osteoblasts, mineralizing osteoblasts, and maturing osteocytes) were isolated from the newly processed data using species assignments and cell classifications (Housman et al., 2022). Lastly, single-cell gene count data were consolidated to produce pseudobulk expression values for each unique grouping of individual, replicate, and cell classification. Specifically, these pseudobulk data were defined as the sum of raw single-cell UMI counts within each individual-replicate for a given cell classification. Computational scripts for these processing steps can be found on GitHub at https://github.com/ghousman/human-skeletal-scRNA. Pseudobulk data were filtered and normalized as described above.
Lentivirus stable cell line generation
Lentiviral transduction of MC3T3-E1 Subclone 4 cells (ATCC, CRL-2593) was performed as we previously described with modifications (Holmes et al., 2020). Cells were infected by incubating in lentivirus-containing cell culture medium at a multiplicity of infection (MOI) of 100 in the presence of 6 μg/ml polybrene for 24 hours. The selection was performed with 2 μg/ml puromycin for 12 days until EGFP expression was observed in 100% of cells, no further cell death was observed, and no live cells remained in non-transduction negative control dish. Selected cells were expanded and cryopreserved. EGFP expression was monitored routinely, and additional selection was performed when necessary.
MC3T3-E1 osteoblast differentiation assay
Osteoblast differentiation of MC3T3-E1 transfected with lentivirus vectors containing either Klf16 cDNA or a 300bp nonfunctional stuffer sequence was performed as follows: cells were plated in 6-well plates at a density of 3×103 /cm2 in MC3T3-E1 maintenance medium (αMEM supplemented with 10% FBS). After three days, the culture medium was switched to osteogenic differentiation medium (Barberi et al., 2005; Holmes et al., 2020; Lee et al., 2015), and cells were maintained in this medium, which was replenished every two days for 21 days. Triplicate experiments, each having triplicate wells, were carried out for both control and Klf16-overexpressing cells.
RT-qPCR
The RT-qPCR method was used to assess gene expression in stable lentiviral MC3T3-E1 cell lines. When cells were plated in 6 well plates for osteogenic differentiation as described above, one parallel well was used for RNA isolation after being cultured in MC3T3-E1 maintenance medium for three days before switching to osteogenic medium. Total RNA was extracted with the RNeasy Kit (Qiagen, 74106) according to the manufacturer’s protocol. cDNA was synthesized with AffinityScript One-Step RT-PCR Kit (Agilent, 600188). Each cDNA sample was amplified in triplicate using the SYBR Green and Platinum Taq polymerase (Thermo Fisher Scientific, S7567) on a 7900HT Real-Time PCR instrument (Thermo Fisher Scientific, 10966). The housekeeping gene Actb was used as the reference. mRNA relative expression was calculated by the ΔΔCt method. qPCR primers used are listed in the Supplementary Table 8.
Whole body bone mineral density
Bone mineral density (BMD) was scanned with a Dual Energy X-ray Absorptiometry (DEXA) Analyzer (Lunar, Piximus II, GE Medical System). Mouse weight and length were measured (nose to the beginning of tail) before scanning. Each unconscious mouse was placed in the DEXA analyzer. A scout-scan was performed. The analysis was conducted on a whole body scanning, excluding the head. The mouse was removed once the image was captured and placed on a heated mat set at 37°C in a cage and closely monitored until consciousness was regained.
Microcomputed tomography
Femurs were isolated by removing attached soft tissues, fixed in 4% PFA at 4°, and imaged with microcomputed tomography (microCT) scan (Skyscan 1172a, Skyscan, Belgium) at 50 kV and 200 μA using a 0.5 mm aluminum filter and an image voxel size of 4.5 μm isotropic. Images were captured every 0.7°, with 8× averaging, through 180° rotation of each bone and reconstructed using Skyscan nRecon software. Imaging analysis of the metaphyseal regions of each femur was performed by first determining a reference point of the most proximal slice where the growth plate had begun to disappear. Offsets of 101 slices and 501 slices from the point of reference in the growth plate were used for trabecular and cortical analyses, respectively, with Skyscan CTAn software. Volume visualization was performed in Avizo 2020.2 (Thermo Fisher Scientific). Data were reported in the standard format used by The American Society for Bone and Mineral Research (ASBMR) (Parfitt et al., 1987).
Statistical analyses
Statistical analyses were performed with Microsoft Excel using Student’s t-test to create graphs displaying mean values with error bars corresponding to the standard error of the mean (SEM) for Klf16 in vitro overexpression and animal experiments. p value ≤ 0.05 was considered significant. Sample sizes are indicated in figure legends. Image analyses were performed with Fiji (National Institutes of Health).
Supplementary Material
Supplementary Table 1, related to Fig. 1 Sample demographic metadata.
Supplementary Table 2, related to Fig. 2 and Supplementary Fig. 2 Differential gene expression between stages.
Supplementary Table 3, related to Fig. 2 Differential gene expression patterns and gene ontology enrichments.
Supplementary Table 4, related to Fig. 3b, 3c, 3d, 3e, and Supplementary Fig. 4 Transcription factor regulatory networks and gene ontology enrichments.
Supplementary Table 5, related to Fig. 4b, 4c, 4d, and 4e Differentially expressed (DE) neighboring protein-coding genes of DE lncRNA genes and gene ontology enrichments.
Supplementary Table 6, related to Fig. 5c, 5d, 5e, 5f, and Supplementary Fig. 5 Local splicing differences between differentiation stages and gene ontology enrichments.
Supplementary Table 7, related to Fig. 6 and Supplementary Fig. 6 Significant MEGENA modules.
Supplementary Table 8, related to Fig. 7 Primers used for Klf16 in vitro overexpression study.
Supplementary Table 9, related to Supplementary Fig. 7 Pseudobulk gene expression at osteogenic differentiation stages using single-cell RNA-seq data in Housman et al., 2022.
Supplementary Table 10. Gene expression in five cell types at 21 days of osteogenic differentiation using single-cell RNA-seq data in Housman et al., 2022.
Acknowledgements
We thank Paige Cundiff for support in iPSC derivation, Arvind Babu for chromosome analyses, Xuqiang Qiao for assistance in the Dean’s Flow Cytometry Core, and Jacqui White and Arie Mobley of The Jackson Laboratory Center for Biometric Analysis for the microCT imaging and DEXA scanning of the Klf16 mouse line. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD026880 and S10OD030463. This work was funded by NIH grant P01HD078233 (E.W.J.) and F32AR075397 (G.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing interests
Data availability
The RNA-seq data supporting the findings of this study are deposited in Gene Expression Omnibus (GEO) with accession number GSE200492. All the other relevant data presented in this paper will be made available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
- Abd Rahman F. (2021). Gene expression profiling on effect of aspirin on osteogenic differentiation of periodontal ligament stem cells. British Dental Journal Open 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Rekabi Z., Wheeler M.M., Leonard A., Fura A.M., Juhlin I., Frazar C., Smith J.D., Park S.S., Gustafson J.A., Clarke C.M., et al. (2016). Activation of the IGF1 pathway mediates changes in cellular contractility and motility in single-suture craniosynostosis. Journal of Cell Science 129, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprile M., Cataldi S., Ambrosio M.R., D’Esposito V., Lim K., Dietrich A., Blüher M., Savage D.B., Formisano P., Ciccodicola A., et al. (2018). PPARγΔ5, a naturally occurring dominant-negative splice isoform, impairs PPARγ function and adipocyte differentiation. Cell Reports 25, 1577–1592.e6. [DOI] [PubMed] [Google Scholar]
- Ardlie K.G., Deluca D.S., Segre A. V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., Lek M., et al. (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Carmona M., Barca-Tierno V., Hisado-Oliva A., Belinchón A., Blanco D.G., Rodriguez J.I., Benito-Sanz S., Campos-Barros A., and Heath K.E., (2014). NPPB and ACAN, two novel SHOX2 transcription targets implicated in skeletal development. pLoS One 9, e83104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T., Willis L.M., Socci N.D., and Studer L. (2005). Derivation of multipotent mesenchymal precursors from human embryonic stem cells. pLoS Medicine 2, 0554–0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M., Heymann S., and Jacomy M. (2009). Gephi: An open source software for exploring and manipulating networks. Third International AAAI Conference on Weblogs and Social Media 3, 361–362. [Google Scholar]
- Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S., (1992). The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526. [DOI] [PubMed] [Google Scholar]
- Cabili M., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., and Rinn J.L. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes and Development 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho D., Fuente A.D.L., and Mendes P. (2005). The origin of correlations in metabolomics data. Metabolomics, 1, 53–63. [Google Scholar]
- Carbon S., Douglass E., Dunn N., Good B., Harris N.L., Lewis S.E., Mungall C.J., Basu S., Chisholm R.L., Dodson R.J., et al. (2019). The gene ontology resource: 20 years and still gOing strong. Nucleic Acids Research 47, D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I., Hoffman G.E., Cundiff P., Beckmann N.D., D’Souza S.L., Knowles J.W., Patel A., Papatsenko D., Abbasi F., Reaven G.M., et al. (2017). Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell 20, 518–532.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.C.W., Tan Z., To M.K.T., and Chan D. (2021). Regulation and role of transcription factors in osteogenesis. International Journal of Molecular Sciences 22, 5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Q., Jiao J., He X.Q., Zhang J.P., Wang H., Xu Y.Q., and Jin T. (2017). CHI3L1 regulation of inflammation and the effects on osteogenesis in a Staphylococcusaureus-induced murine model of osteomyelitis. The FEBS Journal 284, 1738–1747. [DOI] [PubMed] [Google Scholar]
- Chèneby J., Ménétrier Z., Mestdagh M., Rosnet T., Douida A., Rhalloussi W., Bergon A., Lopez F., and Ballester B. (2020). ReMap 2020: A database of regulatory regions from an integrative analysis of human and 55 rogressives DNA-binding sequencing experiments. Nucleic Acids Research 48, D180–D188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftary G.S., Lomberk G.A., Buttar N.S., Allen T.W., Grzenda A., Zhang J., Zheng Y., Mathison A.J., Gada R.P., Calvo E., et al. (2012). Detailed structural-functional analysis of the Krüppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. Journal of Biological Chemistry 287, 7010–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q., Lensch M.W., Jaenisch R., Meissner A., Plath K., and Yamanaka S. (2009). Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell 4, 200–201. [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich M.T., Klau G.W., Rosenwald A., Dandekar T., and Müller T. (2008). Identifying functional modules in protein-protein interaction networks: An integrated exact approach. Bioinformatics 24, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driller K., Pagenstecher A., Uhl M., Omran H., Berlis A., Gründer A., and Sippel A.E. (2007). Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Molecular and Cellular Biology 27, 3855–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey N., Hoffman J.F., Schuebel K., Yuan Q., Martinez P.E., Nieman L.K., Rubinow D.R., Schmidt P.J., and Goldman D. (2017). The ESC/E(Z) complex, an effector of response to ovarian steroids, manifests an intrinsic difference in cells from women with premenstrual dysphoric disorder. Molecular Psychiatry 22, 1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R., Lv W.C., Xu Y., Gong M.Y., Chen X.J., Jiang N., Xu Y., Yao Q.Q., Di L., Lu T., et al. (2020). Endothelial ZEB1 promotes angiogenesis-dependent bone formation and reverses osteoporosis. Nature Communications 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R., et al. (2012). Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafouri-Fard S., Abak A., Avval S.T., Rahmani S., Shoorei H., Taheri M., Samadian M. (2021). Contribution of miRNAs and lncRNAs in osteogenesis and related disorders. Biomedicine & Pharmacotherapy 142, 111942. [DOI] [PubMed] [Google Scholar]
- Giuliani M., Oudrhiri N., Noman Z.M., Vernochet A., Chouaib S., Azzarone B., Durrbach A., and Bennaceur-Griscelli A. (2011). Human mesenchymal stem cells derived from induced pluripotent stem cells down-regulate NK-cell cytolytic machinery. Blood 118, 3254–3262. [DOI] [PubMed] [Google Scholar]
- Giulietti M., Piva F., D’Antonio M., “Onorio De Meo P., Paoletti D., Castrignanò T., “Erchia A.M., Picardi E., Zambelli F., Principato G., et al. (2013). SpliceAid-F: a database of human splicing factors and their RNA-binding sites. Nucleic acids research, 41(D1), D125–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gug C., Caba L., Mozos I., Stoian D., Atasie D., Gug M., and Gorduza E.V. (2020). Rare splicing mutation in COL1A1 gene identified by whole exomes sequencing in a patient with osteogenesis imperfecta type I followed by prenatal diagnosis: A case report and review of the literature. Gene 741, 144565. [DOI] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., et al. (2011). LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkelien A.M., Bryne J.C., Harstad K.G., Lorenz S., Paulsen J., Sun J., Mikkelsen T.S., Myklebost O., and Meza-Zepeda L.A. (2014). The regulatory landscape of osteogenic differentiation. STEM CELLS 32, 2780–2793. [DOI] [PubMed] [Google Scholar]
- Hao Y., Wu W., Shi F., Dalmolin R.J.S., Yan M., Tian F., Chen X., Chen G., and Cao W. (2015). Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer 15, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Liu M., Ding Q., Yang F., and Xu T. (2022). Upregulated miR-9–5p inhibits osteogenic differentiation of bone marrow mesenchymal stem cells under high glucose treatment. Journal of Bone and Mineral Metabolism 40, 208–219. [DOI] [PubMed] [Google Scholar]
- Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C.L., Pena V., Lührmann R., and Stelzl U. (2012). Dynamic protein-protein interaction wiring of the human spliceosome. Molecular Cell, 45, 567–580. [DOI] [PubMed] [Google Scholar]
- Hernández-Vega A. M., and Camacho-Arroyo I. (2021). Crosstalk between 17β-Estradiol and TGF-β Signaling Modulates Glioblastoma Progression. Brain Sciences, 11, 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G., Gonzalez-Reiche A.S., Lu N., Zhou X., Rivera J., Kriti D., Sebra R., Williams A.A., Donovan M.J., Potter S.S., et al. (2020). Integrated transcriptome and network analysis reveals spatiotemporal dynamics of calvarial suturogenesis. Cell Reports 32, 107871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Hu S., Kang Z., Liu Z., Yang W., Zhang Y., Yang D., Ruan W., Yu G., Sun L., et al. (2020). Identification of functional lncRNAs based on competing endogenous RNA network in osteoblast differentiation. Journal of Cellular Physiology 235, 2232–2244. [DOI] [PubMed] [Google Scholar]
- Housman G., Briscoe E., and Gilad Y. (2022). Evolutionary insights into primate skeletal gene regulation using a comparative cell culture model. pLoS Genetics 18, e1010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. H., Estrada K., Evangelou E., Ackert-Bicknell C., Akesson K., Beck T., Brown S. J., Capellini T., Carbone L., Cauley J., et al. (2019). Meta-Analysis of Genomewide Association Studies Reveals Genetic Variants for Hip Bone Geometry. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research, 34, 1284–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Yin C., Chen D., Wu Z., Liang S., Zhang Y., Huang Z., Liu S., Xu X., Chen Z., et al. (2021). MACF1 promotes osteoblast differentiation by sequestering repressors in cytoplasm. Cell Death and Differentiation 28, 2160–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Kang Y., Huang Z., Zhang Z., Meng F., Chen W., Fu M., Liao W., and Zhang Z. (2017). Identification and characterization of long non-coding RNAs in osteogenic differentiation of human adipose-derived stem cells. Cellular Physiology and Biochemistry 42, 1037–1050. [DOI] [PubMed] [Google Scholar]
- Huang S., Zhu X., Ke Y., Xiao D., Liang C., Chen J., and Chang Y. (2020). LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1. Cancer Biology and Therapy 21, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zheng Y., Jia L., and Li W. (2015). Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells 33, 3481–3492. [DOI] [PubMed] [Google Scholar]
- Jia X., Miron R.J., Yin C., Xu H., Luo T., Wang J., Jia R., Wu M., Zhang Y., and Li Y. (2019). HnRNPL inhibits the osteogenic differentiation of PDLCs stimulated by SrCl2 through repressing Setd2. Journal of Cellular and Molecular Medicine. 23, 2667–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Jia P., Zhao Z., and Shen B. (2014). Key regulators in prostate cancer identified by co-expression module analysis. BMC Genomics 15, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien A., Kanagalingam A., Martínez-Sarrà E., Megret J., Luka M., Ménager M., Relaix F., and Colnot C. (2021). Direct contribution of skeletal muscle mesenchymal progenitors to bone repair. Nature Communications 12, 2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J., Cook T., and Urrutia R. (2003). Protein family review Sp1- and Krüppel-like transcription factors. Genome Biology 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Zhou Y., Tan S., Zhou G., Aagaard L., Xie L., Bünger C., Bolund L., and Luo Y. (2015). Mesenchymal stem cells derived from human induced pluripotent stem cells retain adequate osteogenicity and chondrogenicity but less adipogenicity. Stem Cell Research and Therapy 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Sun Y., and Zhang Z. (2019). Identification of key transcription factors–- gene regulatory network related with osteogenic differentiation of human mesenchymal stem cells based on transcription factor prognosis system. Experimental and therapeutic medicine 17, 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodabandehloo F., Taleahmad S., Aflatoonian R., Rajaei F., Zandieh Z., Nassiri-Asl M., and Eslaminejad M.B. (2020). Microarray analysis identification of key pathways and interaction network of differential gene expressions during osteogenic differentiation. Human Genomics 14, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedoot E., Smid M., Foekens J. A., Martens J. W., Le Dévédec S. E., and van de Water B. (2019). Co-regulated gene expression of splicing factors as drivers of cancer progression. Scientific reports, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum S.A., Chang J., Miranda-Carboni G., and Wang C.Y. (2010). Novel functions for NFκB: Inhibition of bone formation. Nature Reviews Rheumatology 6, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushioka J., Kaito T., Okada R., Ishiguro H., Bal Z., Kodama J., Chijimatsu R., Pye M., Narimatsu M., Wrana J.L., et al. (2020). A novel negative regulatory mechanism of Smurf2 in BMP/Smad signaling in bone. Bone Research 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamandé S. R., Ng E. S., Cameron T. L., Kung L. H. W., Sampurno L., Rowley L., Lilianty J., Patria Y. N., Stenta T., Hanssen E., et al. (2023). Modeling human skeletal development using human pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America, 120(19), e2211510120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., and Weirauch M.T. (2018). The human transcription factors. Cell 172, 650–665. [DOI] [PubMed] [Google Scholar]
- Lanzillotti C., De Mattei M., Mazziotta C., Taraballi F., Rotondo J.C., Tognon M., and Martini F. (2021). Long non-coding RNAs and microRNAs interplay in osteogenic differentiation of mesenchymal stem cells. Frontiers in Cell and Developmental Biology 9, 646032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., and Smyth G.K. (2014). Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mée S., Fromigué O., and Marie P.J. (2005). Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Experimental Cell Research 302, 129–142. [DOI] [PubMed] [Google Scholar]
- Lee D.-F., Su J., Schaniel C., and Lemischka I.R. (2015). Modeling familial cancer with induced pluripotent stem cells. Cell 161, 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.M., Cleal J.K., Ntani G., Crozier S.R., Mahon P.A., Robinson S.M., Harvey N.C., Cooper C., Inskip H.M., Godfrey K.M., et al. (2012). Relationship between placental expression of the imprinted PHLDA2 gene, intrauterine skeletal growth and childhood bone mass. Bone 50, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang H., Chen X., Li X., Wang G., Jie Z., Zhao X., Sun X., Huang H., Fan S., et al. (2021). Oxidative stress-induced hypermethylation of KLF5 promoter mediated by DNMT3B impairs osteogenesis by diminishing the interaction with β-catenin. antioxidants & redox signaling, 35, 1–20. [DOI] [PubMed] [Google Scholar]
- Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., and Pritchard J.K. (2018). Annotation-free quantification of RNA splicing using LeafCutter. Nature Genetics 50, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin Y., and Chen G. (2008). Embryoid body formation from human pluripotent stem cells in chemically defined E8 media. StemBook 1–4. [PubMed] [Google Scholar]
- Liu Z. and Rabadan R. (2021). Computing the role of alternative splicing in cancer. Trends in Cancer 7, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Ping Y., Ma M., Zhang D., Liu C., Zaidi S., Gao S., Ji Y., Lou F., Yu F., et al. (2016). Anabolic actions of Notch on mature bone. Proceedings of the National Academy of Sciences of the United States of America 113, E2152–E2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li Y., Liu X., and Wang C.S. (2019). Investigation of transcriptome mechanism associated with osteoporosis explored by microarray analysis. Experimental and Therapeutic Medicine 17, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Dong X., Cao Z., Qiu S., Li Y., Zhong M., Xue Z., Xu Y., Xing H., Tang K., et al. (2021). Mutant U2AF1-induced differential alternative splicing causes an oxidative stress in bone marrow stromal cells. Experimental biology and medicine / Society for Experimental Biology and Medicine 246,1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xu S., Dao J., Gan Z., and Zeng X. (2020) Differential expression of lncRNA/miRNA/mRNA and their related functional networks during the osteogenic/odontogenic differentiation of dental pulp stem cells. Journal of Cellular Physiology 235, 3350–3361. [DOI] [PubMed] [Google Scholar]
- Ma J., Wu J., Han L., Jiang X., Yan L., Hao J., and Wang H. (2019). Comparative analysis of mesenchymal stem cells derive d from amniotic membrane, umbilical cord, and chorionic plate under serum-free condition. Stem Cell Research and Therapy 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita N., Suzuki M., Asami S., Takahata R., Kohzaki D., Kobayashi S., Hakamazuka T., and Hozumi N. (2008). Two of four alternatively spliced isoforms of RUNX2 control osteocalcin gene expression in human osteoblast cells. Gene 413, 8–17. [DOI] [PubMed] [Google Scholar]
- Malan V., Rajan D., Thomas S., Shaw A.C., Louis Dit Picard H., Layet V., Till M., Van Haeringen A., Mortier G., Nampoothiri S., et al. (2010). Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a sotos-like or a Marshall-Smith Syndrome. American Journal of Human Genetics 87, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikandan M., Abuelreich S., Elsafadi M., Alsalman H., Almalak H., Siyal A., Hashmi J.A., Aldahmash A., Kassem M., Alfayez M., et al. (2018). NR2F1 mediated down-regulation of osteoblast differentiation was rescued by bone morphogenetic protein-2 (BMP-2) in human MSC. Differentiation 104, 36–41. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high throughput sequencing reads. EMBnet Journal 17, 10–12. [Google Scholar]
- Martinez F., Marín-Reina P., Sanchis-Calvo A., Perez-Aytés A., Oltra S., Roselló M., Mayo S., Monfort S., Pantoja J., and Orellana C. (2015). Novel mutations of NFIX gene causing Marshall-Smith syndrome or Sotos-like syndrome: One gene, two phenotypes. Pediatric Research 78, 533–539. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Yamanaka Y., Uemura M., Osawa M., Saito M. K., Nagahashi A., Nishio M., Guo L., Ikegawa S., Sakurai S., et al. (2020). Recapitulating the human segmentation clock with pluripotent stem cells. Nature, 580(7801), 124–129. [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., and Thomas P.D. (2019a). PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., and Thomas P.D. (2019b). Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nature Protocols 14, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. A., Kemp J. P., Youlten S. E., Laurent L., Logan J. G., Chai R. C., Vulpescu N. A., Forgetta V., Kleinman A., Mohanty S. T., et al. (2019). An atlas of genetic influences on osteoporosis in humans and mice. Nature genetics, 51, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R., and Fraser P. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Shibata M., Nishio M., Nagata S., Alev C., Sakurai H., Toguchida J., and Ikeya M. (2018). Modeling human somite development and fibrodysplasia ossificans progressiva with induced pluripotent stem cells. Development (Cambridge, England), 145(16), dev165431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Takahashi I., Echigo S., and Sasano Y. (2005). Versican and ADAMTSs are involved in bone development. International Congress Series 1284, 336–337. [Google Scholar]
- Novak A., Goyal N., and Gronostajski R.M. (1992). Four conserved cysteine residues are required for the DNA binding activity of nuclear factor I. The Journal of Biological Chemistry 267,12986–12990. [PubMed] [Google Scholar]
- Onizuka S., Iwata T., Park S.J., Nakai K., Yamato M., Okano T., and Izumi Y. (2016). ZBTB16 as a downstream arget gene of osterix regulates osteoblastogenesis of human multipotent mesenchymal stromal cells. Journal of Cellular Biochemistry 117, 2423–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otasek D., Morris J.H., Bouças J., Pico A.R., and Demchak B. (2019). Cytoscape automation: Empowering workflow-based network analysis. Genome Biology 20, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oury F. (2012). A crosstalk between bone and gonads. Annals of the New York Academy of Sciences 1260, 1–7. [DOI] [PubMed] [Google Scholar]
- Parfitt A.M., Drezner M.K., Glorieux F.H., Kanis J.A., Malluche H., Meunier P.J., Ott S.M., and Recker R.R. (1987). Bone histomorphometry: Standardization of nomenclature, symbols, and units: Report of the ASBMR histomorphometry nomenclature committee. Journal of Bone and Mineral Research 2, 595–610. [DOI] [PubMed] [Google Scholar]
- Park S.J., Bae H.S., and Par J.C. (2015). Osteogenic differentiation and gene expression profile of human dental follicle cells induced by human dental pulp cells. Journal of Molecular Histology 46, 93–106 [DOI] [PubMed] [Google Scholar]
- Penido M.G.M.G., and Alon U.S. (2012). Phosphate homeostasis and its role in bone health. Pediatric Nephrology 27, 2039–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva F., Giulietti M., Burini A. B., and Principato G. (2012). SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Human mutation, 33, 81–85. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Liu S., Figueroa M.E., Kulalert W., Eminli S., Tan K.Y., Apostolou E., Stadtfeld M., Li Y., Shioda T., et al. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature Biotechnology 28, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Jia B., Sun X., Hu W., Chu H., Xu S., and Zhao J.J. (2017). The critical role of long noncoding RNA in osteogenic differentiation of human bone marrow mesenchymal stem cells. BioMed Research International 2017, 5045827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach J.M., Walker E.C., Allan E., Solano M., Yokoyama A., Kato S., Sims N.A., Gillespie M.T., and Martin T.J. (2011). Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. Journal of Biological Chemistry 286, 4186–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff J.D., Wei Y., and Khavari P.A. (2018). The functions and unique features of long intergenic non-coding RNA. Nature Reviews Molecular Cell Biology 19, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Haakonsson A.K., Madsen J.G.S., Larsen M., Forss I., Madsen M.R., Van Hauwaert E.L., Wiwie C., Jespersen N.Z., Tencerova M., et al. (2019). Osteogenesis depends on commissioning of a network of stem cell transcription factors that act as repressors of adipogenesis. Nature Genetics 51, 716–727. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roforth M.M., Farr J.N., Fujita K., McCready L.K., Atkinson E.J., Therneau T.M., Cunningham J.M., Drake M.T., Monroe D.G., and Khosla S. (2015). Global transcriptional profiling using RNA sequencing and DNA methylation patterns in highly enriched mesenchymal cells from young versus elderly women. Bone 76, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Peña M.L., Olivares-Navarrete R., Hyzy S., Arafat D., Schwartz Z., Boyan B.D., Williams J., and Gibson G. (2014). Characterization of distinct classes of differential gene expression in osteoblast cultures from non-syndromic craniosynostosis bone. Journal of Genomics 2, 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt M. (1956). Remarks on some nonparametric estimates of a density function. The Annals of Mathematical Statistics. 27, 832–837. [Google Scholar]
- Sabater L., Ashhab Y., Caro P., Kolkowski E.C., Pujol-Borrell R., and Domínguez O. (2002). Identification of a KRAB-containing zinc finger protein, ZNF304, by AU-motif-directed display method and initial characterization in lymphocyte activation. Biochemical and Biophysical Research Communications 293, 1066–1072. [DOI] [PubMed] [Google Scholar]
- Schaniel C., Dhanan P., Hu B., Xiong Y., Raghunandan T., Gonzalez D.M., Dariolli R., D’Souza S.L., Yadaw A.S., Hansen J., et al. (2021). A library of induced pluripotent stem cells from clinically well-characterized, diverse healthy human individuals. Stem Cell Reports 16, 3036–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto di Carlo F., Pazzaglia L., Esposito T., and Gianfrancesco F. (2020). The loss of profilin 1 causes early onset Paget’s disease of bone. Journal of Bone and Mineral Research 35, 1387–1398. [DOI] [PubMed] [Google Scholar]
- Seifert A. (2015). Role of Hox genes in stem cell differentiation. World Journal of Stem Cells 7, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M., Peng S., Agrawal A. A., Palacino J., Teng T., Zhu P., Smith P.G., Cancer Genome Atlas Research Network, Buonamici S., and Yu L. (2018). Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Reports, 23, 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik A., Martin E.C., Hayes D.J., Gimble J.M., and Devireddy R.V. (2019) Transcriptomic profiling of adipose derived stem cells undergoing osteogenesis by RNA-Seq. Scientific Reports 9, 11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wang H., Fan X., Shangguan L., and Liu H. (2016). A genome wide analysis of alternative splicing events during the osteogenic differentiation of human cartilage endplate-derived stem cells. Molecular medicine reports 14, 1389–96. [DOI] [PubMed] [Google Scholar]
- Shinoda Y., Ogata N., Higashikawa A., Manabe I., Shindo T., Yamada T., Kugimiya F., Ikeda T., Kawamura N., Kawasaki Y., et al. (2008). Kruppel-like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. Journal of Biological Chemistry, 283, 24682–24689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skårn M., Noordhuis P., Wang M.Y., Veuger M., Kresse S.H., Egeland E.V., Micci F., Namløs H.M., Håkelien A.M., Olafsrud S.M., et al. (2014). Generation and characterization of an immortalized human mesenchymal stromal cell line. Stem Cells and Development 23, 2377–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.M., and Zhang B. (2015). Multiscale embedded gene co-expression network analysis. PLoS Computational Biology 11, e1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.Q., Gu W.Q., Qian Y.B., Ma X., Mao Y.J., and Liu W.J. (2015). Identification of long non-coding RNA involved in osteogenic differentiation from mesenchymal stem cells using RNA-Seq data. Genetics and Molecular Research 14, 18268–18279. [DOI] [PubMed] [Google Scholar]
- Song Z., Lian X., Wang Y., Xiang Y., and Li G. (2017). KLF15 regulates in vitro chondrogenic differentiation of human mesenchymal stem cells by targeting SOX9. Biochemical and Biophysical Research Communications, 493, 1082–1088. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou P.A., Perez S.A., Salagianni M., Baxevanis C.N., and Papamichail M. (2006). Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells 24, 462–471. [DOI] [PubMed] [Google Scholar]
- Spitzer M., Wildenhain J., Rappsilber J., and Tyers M. (2014). BoxPlotR: a web tool for generation of box plots. Nature Methods 11, 121–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stains J.P., and Civitelli R. (2003). Genomic approaches to identifying transcriptional regulators of osteoblast differentiation. Genome Biology 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L., Guo C.J., Chen L.L., and Huarte M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nature Reviews Molecular Cell Biology 22, 96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkarsdottir U., Stefansson O. A., Gunnarsdottir K., Thorleifsson G., Lund S. H., Stefansdottir L., Juliusson K., Agustsdottir A. B., Zink F., Halldorsson G. H., et al. (2019). GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nature Communications, 10, 2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Shen C., Zhang L., Wu X., Yu Y., Yang X., Yang C., Zhong C., Gao Z., Miao W., et al. (2020). Hepatic Krüppel-like factor 16 (KLF16) targets PPARα to improve steatohepatitis and insulin resistance. Gut 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jia B., Qiu X.L., Chu H.X., Zhang Z.Q., Wang Z.P., and Zhao J.J. (2019). Potential functions of long non-coding RNAs in the osteogenic differentiation of human bone marrow mesenchymal stem cells. Molecular Medicine Reports 19, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J., Feng X., Li J., Wang J., Wang Z., Zhang L., and Zou W. (2020). VGLL4 promotes osteoblast differentiation by antagonizing TEADs-inhibited Runx2 transcription. Science Advances 6, eaba4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A., Ågesen T. H., Nesbakken A., Rognum T. O., Lothe R. A., and Skotheim R. I. (2011). Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Medicine, 3, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., and Weiss S.J. (2017). Snail/Slug-YAP/TAZ complexes cooperatively regulate mesenchymal stem cell function and bone formation. Cell Cycle 16, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani S., Okada H., Onodera S., Chijimatsu R., Seki M., Suzuki Y., Xin X., Rowe D. W., Saito T., Tanaka S., Chung U. I., Ohba S., & Hojo H. (2023). Stem cell-based modeling and single-cell multiomics reveal gene-regulatory mechanisms underlying human skeletal development. Cell Reports, 112276. [DOI] [PubMed] [Google Scholar]
- Thomas A., and Jaganathan B. G. (2022) Signaling network regulating osteogenesis in mesenchymal stem cells. Journal of Cell Communication and Signaling 16, 47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine N.A., Chen L., Pang C.N., Wilkins M.R., and Kassem M. (2014). Identification of differentiation-stage specific markers that define the ex vivo osteoblastic phenotype. Bone 67, 23–32. [DOI] [PubMed] [Google Scholar]
- Twine N.A., Harkness L., Kassem M., and Wilkins M.R. (2016). Transcription factor ZNF25 is associated with osteoblast differentiation of human skeletal stem cells. BMC Genomics 17, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature Methods 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Y. and Li X. (2021). From bulk, single-cell to spatial RNA sequencing. International Journal of Oral Science 13, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., and Burge C.B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Galvao J., Beach K.M., Luo W., Urrutia R.A., Goldberg J.L., and Otteson D.C. (2016). Novel roles and mechanism for krüppel-like factor 16 (KLF16) regulation of neurite outgrowth and ephrin receptor A5 (EphA5) expression in retinal ganglion cells. Journal of Biological Chemistry 291, 18084–18095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu S., Shi J., Liu H., Li J., Zhao S., and Yi Z. (2020). The role of lncRNAsin osteogenic differentiation of bone marrow mesenchymal stem cells. Current Stem Cell Research & Therapy 15, 243–249. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang Y., Li Z., Li Z., and Yu B. (2015). Differential expression of long noncoding ribonucleic acids during osteogenic differentiation of human bone marrow mesenchymal stem cells. International Orthopaedics 39, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao D., Zhu Y., Dong Y., and Liu Y. (2019). Long non-coding RNA GAS5 promotes osteogenic differentiation of bone marrow mesenchymal stem cells by regulating the miR-135a-5p/FOXO1 pathway. Molecular and Cellular Endocrinology, 496, 110534. [DOI] [PubMed] [Google Scholar]
- Wilkinson A.C., Nakauchi H., and Göttgens B. (2017). Mammalian transcription factor networks: Recent advances in interrogating biological complexity. Cell Systems 5, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom M.L. (2021). Seaborn: statistical data visualization. Journal of Open Source Software 6, 3021. [Google Scholar]
- Watanabe H., Shionyu M., Kimura T., Kimata K., and Watanabe H. (2007). Splicing factor 3b subunit 4 binds BMPR-IA and inhibits osteochondral cell differentiation. The Journal of Biological Chemistry 13, 20728–20738. [DOI] [PubMed] [Google Scholar]
- Wiwie C., Kuznetsova I., Mostafa A., Rauch A., Haakonsson A., Barrio-Hernandez I., Blagoev B., Mandrup S., Schmidt H.H.H.W., Pleschka S., et al. (2019). Time-resolved systems medicine reveals viral infection-modulating host targets. Systems Medicine 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Wang X., Shan L., Zhou J., Zhang X., Zhu E., Yuan H., and Wang B. (2021). High-mobility group AT-Hook 1 mediates the role of nuclear factor I/X in osteogenic differentiation through activating canonical Wnt signaling. Stem Cells 39, 1349–1361. [DOI] [PubMed] [Google Scholar]
- Xia K., Cen X., Yu L., Huang X., Sun W., Zhao Z., and Liu J. (2020). Long noncoding RNA expression profiles during the NEL-like 1 protein–induced osteogenic differentiation. Journal of Cellular Physiology 235, 6010–6022. [DOI] [PubMed] [Google Scholar]
- Xiao X., Zhou T., Guo S., Guo C., Zhang Q., Dong N., and Wang Y. (2017). LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. International Journal of Cardiology 243, 404–412. [DOI] [PubMed] [Google Scholar]
- Yaffe M.P., Noggle S.A., and Solomon S.L. (2016). Raising the standards of stem cell line quality. Nature Cell Biology 18, 236–237. [DOI] [PubMed] [Google Scholar]
- Yang C., Wang C., Zhou J., Liang Q., He F., Li F., Li Y., Chen J., Zhang F., Han C., et al. (2020b). Fibronectin1 activates WNT/β-catenin signaling to induce osteogenic differentiation via integrin β1 interaction. Laboratory Investigation 100, 1494–1502. [DOI] [PubMed] [Google Scholar]
- Yang H., Li G., Han N., Zhang X., Cao Y., Cao Y., Fan Z. (2020a). Secreted frizzled-related protein 2 promotes the osteo/odontogenic differentiation and paracrine potentials of stem cells from apical papilla under inflammation and hypoxia conditions. Cell Proliferation 53, e12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M., et al. (2004). ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: Implication for Coffin-Lowry syndrome. Cell 117, 387–398. [DOI] [PubMed] [Google Scholar]
- Yi Q, Liu H, Feng J, Wu Y, Sun W, Ou M, and Tang L. (2019). Splicing factor-modulated generation of mechano growth factor regulates physiological processes in osteoblasts under mechanical stimuli. Cell adhesion and migration 13, 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoav Benjamin Y.H. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 57, 289–300. [Google Scholar]
- Yu F., Wu F., Li F., Liao X., Wang Y., Li X., Wang C., Shi Y., and Ye L. (2020a). Wnt7b-induced Sox11 functions enhance self-renewal and osteogenic commitment of bone marrow mesenchymal stem cells. Stem Cells 38, 1020–1033. [DOI] [PubMed] [Google Scholar]
- Yu S., Guo J., Sun Z., Lin C., Tao H., Zhang Q., Cui Y., Zuo H., Lin Y., Chen S., et al. (2021). BMP2-dependent gene regulatory network analysis reveals Klf4 as a novel transcription factor of osteoblast differentiation. Cell Death & Disease, 12, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Yao P., Wang Z., and Xie W. (2020b). Down-regulation of FTX promotes the differentiation of osteoclasts in osteoporosis through the Notch1 signaling pathway by targeting miR-137. BMC Musculoskeletal Disorders 21, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri S., Aminian H., Sadeghi S., Esmaeilzadeh-Gharehdaghi E., and Razmara E. (2022). Krüppel-like factors in bone biology. Cellular signalling 93, 110308. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., et al. (2013). Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhu J. (2013) Identification of key causal regulators in gene networks. Proceedings of the word congress on engineering 2, 1309–1312. [Google Scholar]
- Zhang W., Dong R., Diao S., Du J., Fan Z., and Wang F. (2017). Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Research and Therapy 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Guan H., Song C., Wang Y., Liu C., Cai C., Zhu H., Liu H., Zhao L., and Xiao J. (2018). YAP1 is essential for osteoclastogenesis through a TEADs-dependent mechanism. Bone 110, 177–186. [DOI] [PubMed] [Google Scholar]
- Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. (2017). Massively Parallel Digital Transcriptional Profiling of Single Cells. Nature Communications 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Ramnaraign D., Anderson B.A., Tycksen E., Nunley R., and McAlinden A. (2019). MicroRNA-138 Inhibits Osteogenic Differentiation and Mineralization of Human Dedifferentiated Chondrocytes by Regulating RhoC and the Actin Cytoskeleton. JBMR Plus 3, e10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Li X., Huang Y., Jia L., and Li W. (2018). Time series clustering of mRNA and lncRNA expression during osteogenic differentiation of periodontal ligament stem cells. Peer J 6, e5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shan Y., Ge K., Lu J., Kong W., and Jia C. (2020). LncRNA CYTOR promotes pancreatic cancer cell proliferation and migration by sponging miR-205–5p. Pancreatology 20, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang Q., Yao Y., Fang J., Sun F., Ni Y., Shen Y., Wang H., and Shao S. (2015). Microarray analysis of long non-coding RNA expression profiles in human gastric cells and tissues with helicobacter pylori infection. BMC Medical Genomics 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang F., Zhao Y., Yang P., Chen J., Sun H., Liu L., Li W., Pan L., Guo Y., et al. (2014). A gain-of-function mutation in Tnni2 impeded bone development through increasing Hif3a expression in DA2B mice. Plos Genetics 10, e1004589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1, related to Fig. 1 Sample demographic metadata.
Supplementary Table 2, related to Fig. 2 and Supplementary Fig. 2 Differential gene expression between stages.
Supplementary Table 3, related to Fig. 2 Differential gene expression patterns and gene ontology enrichments.
Supplementary Table 4, related to Fig. 3b, 3c, 3d, 3e, and Supplementary Fig. 4 Transcription factor regulatory networks and gene ontology enrichments.
Supplementary Table 5, related to Fig. 4b, 4c, 4d, and 4e Differentially expressed (DE) neighboring protein-coding genes of DE lncRNA genes and gene ontology enrichments.
Supplementary Table 6, related to Fig. 5c, 5d, 5e, 5f, and Supplementary Fig. 5 Local splicing differences between differentiation stages and gene ontology enrichments.
Supplementary Table 7, related to Fig. 6 and Supplementary Fig. 6 Significant MEGENA modules.
Supplementary Table 8, related to Fig. 7 Primers used for Klf16 in vitro overexpression study.
Supplementary Table 9, related to Supplementary Fig. 7 Pseudobulk gene expression at osteogenic differentiation stages using single-cell RNA-seq data in Housman et al., 2022.
Supplementary Table 10. Gene expression in five cell types at 21 days of osteogenic differentiation using single-cell RNA-seq data in Housman et al., 2022.
Data Availability Statement
The RNA-seq data supporting the findings of this study are deposited in Gene Expression Omnibus (GEO) with accession number GSE200492. All the other relevant data presented in this paper will be made available from the corresponding author upon reasonable request. Source data are provided with this paper.