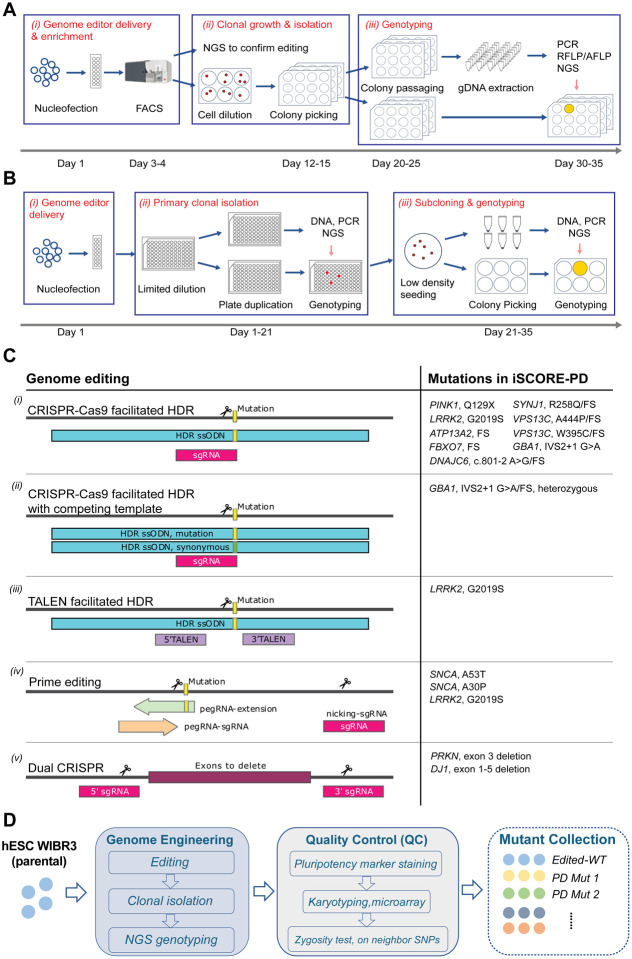
Figure 3. Gene editing workflow to generate iSCORE-PD Collection.
(A) Schematic illustrating genome editing pipeline A. This approach involves (i) FACS-based enrichment of nucleofected cells containing the gene editing reagents including a fluorescent reporter, (ii) the isolation of clonally expanded cell lines and (iii) the NGS-based genotyping to identify correctly edited cell lines.
(B) Schematic illustrating genome editing pipeline B. This approach utilizes a high-throughput cell isolation system. This approach includes (i) nucleofection of the gene editing reagents, (ii) the plating of cells in a limited dilution (~10 cells/well) to isolate wells containing correctly targeted cells by NGS and (iii) subcloning, expansion and NGS-based genotyping to isolate correctly targeted clonal cell line.
(C) Table summarizing the gene editing strategies used to generate the iSCORE-PD collection. These include: (i) CRISPR/Cas9 facilitated homology directed repair (HDR) using ssODNs containing the desired genetic modification as repair template for CRISPR/Cas9 induced double strand break. (ii) The use of competing HDR templates (ssODNs) containing synonymous mutations in the gRNA-target site to favor the generation of heterozygous over homozygous mutations. (iii) TALEN-facilitated HDR using ssODNs containing the desired genetic modification as repair template for CRISPR/Cas9 induced double strand break. (iv) Prime editing approach to insert the PD-associated point mutations into hESCs. (v) Dual CRISPR approach using 3’ and 5’ sgRNAs flanking the desired deletion to recreated large genomic structural alterations identified in PD patients.
(D) Overview depicting genome engineering and quality control steps in the generation of the iSCORE-PD collection.