Abstract
Crohn’s disease (CD) is the chronic inflammation of the ileum and colon triggered by bacteria, but insights into molecular perturbations at the bacteria-epithelium interface are limited. We report that membrane mucin MUC17 protects small intestinal enterocytes against commensal and pathogenic bacteria. In non-inflamed CD ileum, reduced MUC17 levels correlated with a compromised glycocalyx, allowing bacterial contact with enterocytes. Muc17 deletion in mice rendered the small intestine prone to atypical infection while maintaining resistance to colitis. The loss of Muc17 resulted in spontaneous deterioration of epithelial homeostasis and extra-intestinal translocation of bacteria. Finally, Muc17-deficient mice harbored specific small intestinal bacterial taxa observed in CD. Our findings highlight MUC17 as an essential line of defense in the small intestine with relevance for early epithelial defects in CD.
One-sentence summary:
Membrane mucin MUC17 protects enterocytes against bacterial attachment and constitutes an early defect in Crohn’s disease.
Introduction
Mucins are critical for the gastrointestinal defense system (1, 2). In the mouse distal colon, the gel-forming Muc2 forms an attached impenetrable mucus layer that separates bacteria from the epithelium (3). Defects in core mucus components constitute early steps in the pathogenesis of ulcerative colitis, a form of inflammatory bowel disease (IBD) in humans (4). In contrast, the mucus of the small intestine is non-attached and permeable to luminal material including bacteria (5). This regiospecific difference demands a distinct defense mechanism to manage host-microbiota interactions. In this context, the role of membrane mucins in small intestinal enterocytes is unexplored, due to limited insight into their gene sequences (6) and difficulties preserving them in fixed tissues. MUC17, alongside MUC13, is a conserved heavily O-glycosylated membrane mucin positioned at the apical brush border of differentiated enterocytes (7, 8) (Fig. 1A). In the mouse small intestine, the homeostatic cytokine IL-22 induces Muc17 expression and the establishment of a surface glycocalyx that in ex vivo experiments prevents direct attachment of bacteria to the brush border (9). Crohn’s disease (CD) is an IBD subtype characterized by patchy chronic inflammation involving dysregulated immune responses to the gut bacteria (10). Unlike UC that is limited to the colon, CD affects both the small and large intestines, with a third of patients displaying inflammation exclusively in the terminal ileum (11). Mutations in the intracellular peptidoglycan sensor NOD2 and autophagy protein ATG16L1 cause defects in the detection and degradation of intracellular bacteria and have been linked to susceptibility to CD (12–14). However, functional CD-associated defects upstream of intracellular innate immunity against bacteria have not been identified. Studies of ileal CD have reported transcriptional alterations associated with the enterocyte brush border (15). MUC17 is both transcriptionally and structurally associated with the brush border (9), but the role of MUC17 in epithelial defense and whether MUC17 expression and glycocalyx integrity are compromised in CD are not known.
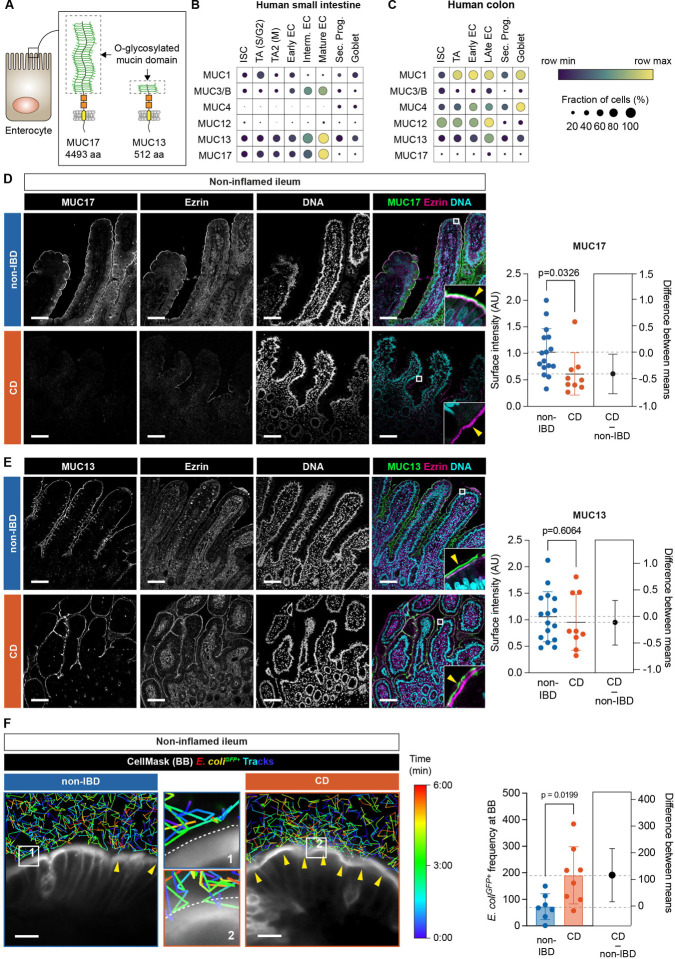
Fig. 1. Non-inflamed ileum of CD patients expresses reduced levels of MUC17 and carries a glycocalyx permeable to luminal bacteria.
(A) Schematic illustration of human MUC17 and MUC13 position at the enterocyte brush border. Dashed boxes show the O-glycosylated mucin domain in each mucin. aa, amino acids. (B) Dot plot showing mRNA expression of membrane mucin genes at single-cell resolution in epithelial cell types of the human small intestine. EC, enterocyte; ISC, intestinal stem cell; TA, transit amplifying; Sec. Prog., secretory progenitor; Goblet, goblet cell. S/G2, S- and G2-phase; M, M-phase of cell cycle. (C) Dot plot showing mRNA expression of membrane mucin genes in human colonic epithelial cell types. The color scheme shows row maximum and minimum representing the relative expression of each gene among all cell types in the intestinal segment. (D) Immunohistochemistry of MUC17 (green), Ezrin (magenta), and DNA (cyan) in histological sections of non-inflamed ileal biopsies from non-IBD control and CD patients, alongside semiquantitative analysis of MUC17 levels in the brush border. Each channel is also shown in grayscale. The yellow arrow points to the apical brush border. Scale bar 100 μm. n=16 for non-IBD, n=9 for CD. (E) Immunohistochemistry of MUC13 (green), Ezrin (magenta), and DNA (cyan) in histological sections of noninflamed ileal biopsies from non-IBD control and CD patients, alongside semiquantitative analysis of MUC13 in the brush border. Each channel is also shown in grayscale. The yellow arrow points to the apical brush border. Scale bar 100 μm. n=16 for non-IBD, n=9 for CD. (F) Time-lapse glycocalyx permeability assay in non-inflamed ileal biopsies from non-IBD and CD patients, stained with CellMask, and incubated with E. coliGFP+ tracked over the course of 6 min. Yellow arrows show points of contact between E. coliGFP+ and CellMask. Dashed white lite marks the brush border. Magnifications are labeled with numbers. Scale bar 10 μm. The pseudo color scale indicates the time point during the tracking of individual bacterial cells. The bar graph depicts the frequency of E. coliGFP+ attached to the brush border in each cohort. n=7 for non-IBD, n= 8 for CD. Data are means ± SD. Significance was determined by unpaired t-test (D-F). The estimation plot shows the difference between CD and non-IBD means with a 95% confidence interval.
Here, we analyzed non-inflamed ileal biopsies from CD patients and non-IBD controls together with a new genetic mouse model to define the function of MUC17 in the intestine. CD patients displayed decreased MUC17 levels and a glycocalyx that was more permeable to bacteria. In mice, intestine-specific deletion of the Muc17 gene was dispensable for colonic inflammation whilst rendering the small intestine abnormally sensitive to infection by the attaching effacing bacterium Citrobacter rodentium (C. rodentium). Muc17ΔIEC mice also suffered spontaneous deterioration of the epithelial cell barrier and displayed translocation of viable bacteria to extra-intestinal tissues. Lastly, we showed that Muc17 regulates the abundance of specific bacterial taxa in the small intestine, including bacteria enriched in CD patients. Thus, the Muc17ΔIEC model reproduces fundamental manifestations of CD, suggesting that MUC17 is essential for protecting the small intestine against commensal and pathogenic bacteria.
Decreased MUC17 levels and glycocalyx integrity in ileal Crohn’s disease
In humans, MUC17 is exclusively expressed in differentiated enterocytes of the small intestine (Fig. 1B–C). We analyzed the expression of MUC17 in histological sections from the non-inflamed terminal ileum of CD patients and a control group with non-IBD-related pathologies (table S1). MUC17 levels were significantly reduced in the enterocyte brush border of the CD compared to non-IBD ileum, which showed a typical MUC17 staining in the brush border marked by Ezrin (Fig. 1D). Membrane mucin MUC13 was not altered between the two cohorts (Fig. E). Next, we analyzed the ileal biopsies with an ex vivo timelapse glycocalyx permeability assay (9), which measures the ability of the glycocalyx to separate GFP-expressing Escherichia coli (E. coliGFP+) from the brush border (see materials and methods). In line with decreased apical MUC17 in CD, the frequency of E. coliGFP+ that approached the brush border was significantly higher in CD compared to non-IBD, indicating reduced glycocalyx barrier integrity in CD patients (Fig. 1F, fig. S1, Movie S1). Thus, the non-inflamed CD ileum exhibits an impaired glycocalyx barrier due to reduced MUC17 levels in the apical brush border.
The Muc17ΔIEC mouse lacks a small intestinal glycocalyx
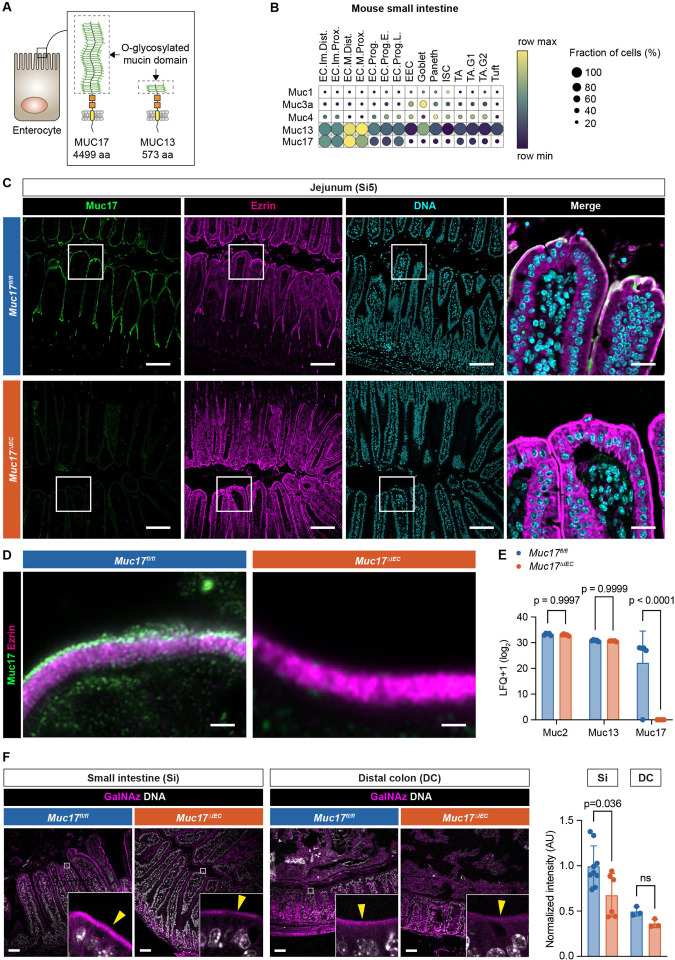
The loss of glycocalyx integrity in CD ileum suggests that the loss of apical MUC17 is an early defect resulting in increased bacterial contact with the brush border. To test this hypothesis, we generated a conditional Muc17ΔIEC mouse by crossing Muc17fl/fl and Vil1-Cre mice expressing the Cre recombinase in intestinal epithelial cells (fig. S2A–C). Unlike human MUC17, murine Muc17 is expressed in both the small intestine and colon (Fig. 2A–B, fig. S3, fig. S4). The successful deletion of Muc17 was thus confirmed in the duodenum, jejunum (Si5), ileum (Si8), proximal colon (PC), and distal colon (DC) (Fig. 2C, fig. S3, fig. S4). High-resolution microscopy of small intestinal enterocytes revealed that MUC17 was lacking from the tips of microvilli in Muc17ΔIEC mice. (Fig. 2D). Muc17 deletion was also demonstrated in microvillus-derived luminal vesicles isolated from the small intestinal (16, 17) (fig. S2D). To evaluate the impact of Muc17 deletion on other mucins, we performed proteomic analysis of isolated small intestinal epithelial cells from Muc17fl/fl and Muc17ΔIEC cohoused littermates. Deletion of Muc17 did not result in a compensatory change in the abundance of Muc13 or the gel-forming Muc2 (Fig. 2E, table S2). Moreover, Muc17ΔIEC mice did not exhibit alterations in tissue morphology of the small intestine or distal colon (fig. S2E). Notably, the deletion of Muc17 did not cause abnormalities in brush border morphology or cellular polarity, evaluated by the brush border markers Ezrin and Cdhr5, and basolateral EpCAM (fig. S2F–G). Mucin O-glycans convert the glycocalyx into a size-selective diffusion barrier and shield against bacterial degradation (1, 9). To determine if the loss of Muc17 impacted the density of glycocalyx O-glycans, we employed in vivo bioorthogonal labeling using GalNAz, the azide derivative of N-Acetylgalactosamine (GalNAc), the first glycan added to serines or threonines during mucin-type O-glycosylation (18). GalNAz intensity was significantly reduced in the small intestinal brush border of Muc17ΔIEC mice compared to controls but remained unchanged in the colonic brush border (Fig. 2F). The O-glycan deficit of the small intestinal glycocalyx was further supported by decreased staining with Aleuria aurantia lectin (AAL) and wheat germ agglutinin (WGA), which bind fucose and N-acetylglucosamine (fig. S2H). Thus, under baseline conditions Muc17 deletion results in an explicit loss of the glycocalyx without impacting the overall tissue and cell morphology of the small intestine.
Fig. 2. Epithelial-specific deletion of Muc17 results in the loss of Muc17 expression in the mouse small intestine.
(A) Schematic illustration of murine Muc17 and Muc13. Dashed boxes show the O-glycosylated mucin domain in each mucin. aa, amino acids. (B) Expression of membrane mucin transcripts in epithelial cell types of the mouse small intestine. The color scheme shows row maximum and minimum representing the relative expression of each gene among all cell types. EC, enterocyte; EEC, enteroendocrine cell; ISC, intestinal stem cell; TA, transit amplifying; Im, immature; M, mature; Prox, proximal; Dist, distal; Prog, progenitor; E, early; L, late; G1, G1-phase; G2, G2-phase of cell cycle. (C) Immunohistochemistry of Muc17 (green), Ezrin (magenta), and DNA (cyan) in histological sections of the jejunum (Si5) from Muc17fl/fl and Muc17ΔIEC mice. Scale bar 50 μm. Scale bar in insets 10 μm. (D) Airyscan high-resolution microscopy of Muc17 (green) and Ezrin (magenta) in the small intestinal brush border of Muc17fl/fl and Muc17ΔIEC mice. Scale bar 1 μm. (E) Bar graph representing the abundance of detected mucins in the proteome of small intestinal epithelial cells from Muc17fl/fl and Muc17ΔIEC mice. n= 5 for each group. (F) Representative confocal micrographs of small intestine (Si) and distal colon (DC) from Muc17fl/fl and Muc17ΔIEC mice, stained for bioorthogonally labeled O-glycans (GalNAz, magenta) and DNA (white). Scale bar 50 μm. Bar graphs represent the quantification of GalNAz intensity in the apical brush border of the small intestine (Si) and DC in each group. Si5: n=6–9 per group. DC: n=3 per group. Significance was determined by two-way ANOVA followed by Šidák correction (D) and Mann-Whitney test (E).
Deletion of Muc17 causes abnormal small intestinal susceptibility to Citrobacter Rodentium infection
Since we did not observe any baseline phenotype in Muc17ΔIEC mice, cohoused Muc17fl/fl and Muc17ΔIEC littermates were infected with C. rodentium, a natural murine pathogen that normally causes a self-limiting infection in the distal colon (19). Infection kinetics in each group were monitored over a period of 21 days post-infection (dpi) (Fig. 3A). Regardless of genotype, fecal C. rodentium counts peaked at 7–9 dpi and declined below the limit of detection (LOD) by 17 dpi (Fig. 3B). Both groups exhibited a slight but comparable weight loss during the initial infection phase (1–3 dpi) (Fig. 3C). Next, we quantified C. rodentium colonization in the luminal and mucosal compartments of the jejunum (Si5), ileum (Si8) and distal colon (DC) at 3 days post-infection. As expected, Muc17fl/fl mice exhibited a robust colonization in DC (Fig. 3D–E). In Muc17ΔIEC Si5, however, we observed three orders of magnitude higher C. rodentium counts compared to the control group (Fig. 3D–E). The abnormal C. rodentium infection of Si5 was not limited to the luminal compartment but was also detected in the mucosa, indicating that the small intestinal epithelium was more susceptible to C. rodentium infection in the absence of Muc17 and a glycocalyx. Greater pathogen counts were also observed in the mucosa of Muc17ΔIEC DC compared to the infected controls (Fig. 3E). There was also a trend towards increased translocation of C. rodentium to the peripheral tissues in Muc17ΔIEC mice at 7 dpi (Fig. 3F). Notably, across all tissues and time points post-infection, a larger fraction of Muc17ΔIEC mice carried a detectible pathogen burden compared to Muc17fl/fl mice (fig. S5). At 7 days post-infection, 91±6 % of all Muc17ΔIEC mice exhibited a small intestinal infection compared to only 54±13 % of the Muc17fl/fl mice. A similar analysis of colonic pathogen burden did not reveal any significant difference between the groups. Accordingly, the relative risk of C. rodentium infection in Muc17ΔIEC mice peaked in Si5 and decreased towards the DC, suggesting that Muc17 primarily serves as a small intestinal defense mechanism during bacterial infection (Fig. 3G). Muc17 deletion also resulted in a higher relative risk of pathogen translocation to the peripheral tissues (Fig. 3H). Histological analysis 3 days post-infection revealed numerous C. rodentiumGFP+ cells in contact with the epithelium at villus apexes, intervillar regions, and the crypt compartment of Muc17ΔIEC Si5, whereas C. rodentiumGFP+ was not detected in Muc17fl/fl (Fig. 3I). Assessment of the distal colon showed C. rodentiumGFP+ at the surface epithelium and in the crypts of Muc17ΔIEC mice (Fig. 3J–K). Collectively, the absence of Muc17 caused an abnormal small intestinal susceptibility to C. rodentium, indicating that Muc17 is essential for protecting the small intestinal epithelium against pathogenic bacteria.
Fig. 3. Deletion of Muc17 results in abnormal small intestinal susceptibility to C. rodentium infection.
(A) Schematic illustration of the C. rodentium infection protocol and sampling time points. Si5, jejunum; Si8, ileum; DC, distal colon. (B) Colony forming units (CFU) of C. rodentium in fecal samples collected from Muc17fl/fl and Muc17ΔIEC mice at various days post-infection (dpi). (C) Weight loss of infected Muc17fl/fl and Muc17ΔIEC mice, depicted as percent of the initial weight at the start of the challenge (time point 0 dpi). (D) CFU of C. rodentium in the luminal compartments of the jejunum (Si5), ileum (Si8), and distal colon (DC) at 3 dpi. (E) CFU of C. rodentium the mucosal compartments of the jejunum (Si5), ileum (Si8), and distal colon (DC) at 3 dpi. (F) CFU of C. rodentium in the mesenteric lymph nodes (MLN), Spleen, and Liver at 7 dpi. The circle charts represent the proportion of mice with each genotype carrying C. rodentium CFU above the limit of detection (LOD) in each intestinal segment or tissue site. (G) Relative risk of carrying C. rodentium CFU above LOD in Si5, Si8, and DC when Muc17ΔIEC and Muc17fl/fl mice are challenged with C. rodentium. (H) Relative risk of carrying C. rodentium CFU above LOD in MLN, Spleen, and liver when Muc17ΔIEC and Muc17fl/fl mice are infected with C. rodentium. n=6–10 per group. (I) Immunohistochemistry of C. rodentiumGFP+ (magenta) in relation to the epithelium (EpCAM, green) in Si5 of Muc17fl/fl and Muc17ΔIEC mice 3 days post-infection. Yellow arrows point to bacterial cells. Scale bar 50 μm. (J) Immunohistochemistry of C. rodentiumGFP+ (magenta) in relation to the epithelium (EpCAM, green) in DC of Muc17fl/fl and Muc17ΔIEC mice 3 days post-infection. Yellow arrows point to bacterial cells. Scale bar 50 μm. (K) Visualization of C. rodentiumGFP+ (magenta) in relation to the surface epithelium (DNA, cyan) in explants of Muc17fl/fl and Muc17ΔIEC DC 3 days post-infection. Upper panels show a top view (xy plane), and lower panels show an extended orthogonal view (xz plane) of the boxed region (orange). Penetration of C. rodentiumGFP+ (magenta) into the colonic crypts (yellow dashed lines) is highlighted (yellow arrows). Scale bar 100 μm. Significance was determined by Mann-Whitney test (D-F).
Muc17ΔIEC mice display spontaneous loss of epithelial homeostasis
Given that Muc17 protected the small intestine against pathogenic infection, we reasoned that Muc17 deletion might also allow commensal bacteria to make direct contact with enterocytes and thereby disrupt epithelial homeostasis. Thus, we turned our attention to unchallenged 6–8-week-old cohoused Muc17fl/fl and Muc17ΔIEC littermates maintained under standard pathogen-free (SPF) conditions. Histological analysis of Si5 did not reveal any genotype-dependent differences in crypt number, villus length, or goblet cell frequency (Fig. 4A, fig. S2E). Next, Si5 explants were flushed, and fixed to preserve the mucosal compartment, and the distribution of bacteria in relation to the brush border was assessed using confocal microscopy and automatic image segmentation. Compared to Muc17fl/fl, significantly higher numbers of bacteria were directly bound to the brush border of Muc17ΔIEC mice (Fig. 4B). Findings were supported by fluorescence in situ hybridization (FISH) with a universal eubacterial probe, showing bacteria in direct contact with the Muc17ΔIEC brush border (Fig. 4C). Uncontrolled bacterial interactions with the epithelium can trigger increased epithelial proliferation and cell death (20, 21). Correspondingly, we observed a significant expansion of proliferative mKi67+ epithelial cells within the crypt compartment of Muc17ΔIEC Si5 (Fig. 4D). Free nuclear DNA 3′-OH termini generated by single-strand breaks are hallmarks of apoptosis. Compared to controls, Muc17ΔIEC Si5 displayed a higher number of TUNELpositive apoptotic cells at villus apexes (Fig. 4E). Next, we used mass spectrometry to evaluate the impact of Muc17 deletion on epithelial homeostasis. Label-free protein quantification in epithelial cells from 5-, 8- and 36-weeks-old Muc17fl/fl and Muc17ΔIEC mice revealed altered expression in 0.25–1.00% of the proteome, mainly affecting brush border-associated proteins (Fig. 4F, table S2). Specifically, epithelial cells in 8-week-old Muc17ΔIEC mice overexpressed Rho guanine nucleotide exchange factor 26 (Arhgef26), which participates in enterocyte membrane ruffling during bacterial invasion (22) (Fig. 4F, fig. S6). In line with the persistent bacterial challenges of the epithelium, we also detected higher abundance of the antimicrobial proteins Lyz1 and Reg3b in 36-week-old Muc17ΔIEC mice (Fig. 4F, fig. S6).
Fig. 4. Epithelial barrier dysfunction in the small intestine of Muc17ΔIEC mice housed under baseline SFP conditions.
(A) Quantification of the number of villi per mm2 of histological section, villus length, and goblet cells (GCs) per villus in Muc17fl/fl and Muc17ΔIEC jejunum (Si5). n=5 for Muc17fl/fl, n=3 for Muc17ΔIEC. (B) Representative visualization of commensal bacteria (red) at the Si5 epithelium (white). Bacteria in direct contact with the epithelium are shown in yellow. Scale bar 100 μm. The quantification shows frequency of bacteria attached to the brush border in each cohort. N=6 for Muc17fl/fl, n=6 for Muc17ΔIEC. (C) Confocal micrographs of Muc17fl/fl and Muc17ΔIEC Si5 stained for bacteria (EUB338, green), epithelium and mucus (WGA, magenta), and DNA (cyan). Yellow arrows point to bacteria. Scale bar 25 μm. Scale bar in insets 10 μm. (D) Immunohistochemistry of mKi67 (magenta) and DNA (blue) in Muc17fl/fl and Muc17ΔIEC Si5. Each channel is also shown in grayscale. Yellow arrows point to mKi67+ cells with maximum migration along the villus-crypt axis. The dashed yellow line depicts the crypt bottom. Scale bar 50 μm. Quantification of the migration of mKi67+ cells along the crypt-villus axis and absolute numbers of mKi67+ per villus in Muc17fl/fl and Muc17ΔIEC Si5. n=3–6 villi per mouse, 3 mice per group. (E) Immunohistochemistry of TUNEL+ nuclei (magenta) and DNA (cyan) in Muc17fl/fl and Muc17ΔIEC Si5. Each channel is also shown in grayscale. Scale bar 50 μm. Quantification of TUNEL+ cells per villus in Muc17fl/fl and Muc17ΔIEC mice. n=4 for Muc17fl/fl, n=4 for Muc17ΔIEC. (F) Volcano plots showing the fold change of protein expression in epithelial cells of the jejunum (Si5) from 5-, 8-, and 36-week-old Muc17fl/fl compared to Muc17ΔIEC mice. Differentially expressed proteins (fold change ≥ 2, p-value < 0.05) are highlighted with filled circles. Specific proteins are labeled with their protein names. n=5 for each genotype in each age group. Significance was determined by Mann-Whitney test (A) and unpaired t-test (B, D, E).
Muc17 is dispensable for protection against chemically induced colitis
To define the protective role of Muc17 in the murine distal colon, cohoused Muc17fl/fl and Muc17ΔIEC littermates were subjected to 7-day ad libitum administration of dextran sodium sulfate (DSS), a chemically induced model of human ulcerative colitis (fig. S7A). Muc17ΔIEC mice displayed a reduction in weight compared to wild-type mice, but only at the end of the intervention (day 6). (fig. S7B). While there was a significant shortening of the colon in Muc17ΔIEC mice after 7 days (fig. S7C), only a moderately increased susceptibility to DSS was measured by stool consistency, fecal blood score, overall disease activity index, and survival (fig. S7D–G). Our observations were supported by histological analysis of Si5 and DC, which corroborated that Muc17fl/fl and Muc17ΔIEC mice responded similarly to DSSinduced colitis (fig. S7H–I). The late onset of moderate inflammation in the absence of Muc17 contrasted acute induction of severe colitis in C3GnT−/−, Vamp8−/−, and Tgm3−/−mice with defects in Muc2 glycosylation, secretion, and crosslinking (23–25). Thus, we concluded that Muc17 does not play a critical protective role in the distal colon.
Further assessment of Muc17ΔIEC mice under baseline conditions did not show any deviations in the number of crypts, crypt length, or goblet cell frequency in the distal colon (fig. S8A, fig. S2E). Next, we quantified the barrier properties of the inner mucus layer (IML) separating bacteria from the epithelium (26). FISH showed a defined separation of bacteria from the epithelium in 6–8-week-old Muc17fl/fl and Muc17ΔIEC mice (fig. S8B). Ex vivo mucus penetrability analysis in viable explants from 6–8- or 36–40 week-old mice, evaluating the penetration of microbeads through the IML, showed that both Muc17ΔIEC and Muc17fl/fl mice maintain an impenetrable IML typically observed in wild-type mice (3) (fig. S8C–D). Muc17ΔIEC mice did however display elevated occurrence of shed cells (fig. S8D) and increased epithelial cell proliferation measured by mKi67+ (fig. S8E). Together, our findings confirm that Muc17 is not required for the protection of the distal colon against commensal bacteria.
Muc17 deletion alters the small intestinal microbiota and elicits extra-intestinal translocation of bacteria
Muc17 deletion resulted in susceptibility to C. rodentium and increased contact between commensal bacteria and small intestinal enterocytes, but the precise effect of Muc17 on the regiospecific composition of the microbiota is not understood. We thus analyzed bacterial genomic DNA from the luminal compartment of the small intestine and colon of cohoused Muc17fl/fl and Muc17ΔIEC littermates by 16S rRNA gene sequencing. While there was no significant difference in species richness and evenness (alpha diversity) between the two groups (Fig. 5A and 5D), the beta diversity based on Bray-Curtis distances revealed distinct patterns in the microbiota of Muc17fl/fl and Muc17ΔIEC mice (Fig. 5B and 5E). The small intestinal community of Muc17ΔIEC mice was characterized by a higher abundance of the classes of Actinobacteria, Bacteroidia, Coriobacteria, and gammaproteobacteria (Fig. 5C), while the colonic microbiota shifted towards Clostridia, Coriobacteria, and Desulfovibrionia (Fig. 5F). Linear discriminant analysis effect size (LEfSe) analysis showed a depletion of Firmicutes, Bacilli and the genus Dubosiella belonging to the Erysipelotrichales from the Muc17ΔIEC small intestine (Fig. 5G, fig. S9A). The small intestinal alterations in composition were largely reproduced in the Muc17fl/fl colon, with the enrichment of Erysipelotrichales and Dubosiella alongside Lactobacillus (Fig. 5G, fig. S9B). In addition, Coriobacteriales, Lachnoclostridium, and Marvinbryantia were enriched in the Muc17ΔIEC colon (Fig. 5G, fig. S9B). Combined, the affected genera comprised an average of 15%−20% of the detected operational taxonomic units. Since the microbiota composition varies along the intestine (27), we assessed the regional distribution of the differentially enriched bacterial taxa. Bacilli, Coriobacteriia, and Erysipelotrichales, had a primarily small intestinal niche, with Erysipelotrichales dominating the jejunum (Si5) (Fig. 5G), thus suggesting a transmission of compositional changes from the small intestine to colon. Collectively, Muc17 deletion induced a substantial shift in the abundance of specific bacteria that colonize the small intestine (fig. S9C), suggesting that Muc17 regulates the regiospecific selection of commensal bacteria.
Fig. 5. Deletion of Muc17 results in alterations in luminal microbiota composition and translocation of commensal bacteria to peripheral tissues.
(A) α-diversity (Shannon index) of luminal bacterial communities in the small intestine (Si) of Muc17fl/fl (n=11) and Muc17ΔIEC (n=8) mice. (B) Principal coordinate analysis of β-diversity (Bray-Curtis dissimilarity) of luminal bacterial communities of Muc17fl/fl and Muc17ΔIEC Si. (C) Relative frequency of major classes found in Muc17fl/fl and Muc17ΔIEC Si. (D) α-diversity (Shannon index) of luminal bacterial communities in the colon of Muc17fl/fl (n=15) and Muc17ΔIEC (n=13) mice. (E) Principal coordinate analysis of β-diversity (Bray-Curtis dissimilarity) of luminal bacterial communities in Muc17fl/fl and Muc17ΔIEC colon. (F) Relative frequency of major classes found in Muc17fl/fl and Muc17ΔIEC colon. (G) Linear discriminant analysis (LDA) effect size identification of small intestinal and colonic taxa that differentiate between Muc17fl/fl (blue) and Muc17ΔIEC (red) mice. The heat map shows the row z-score representing the relative abundance of the selected taxa in different gastrointestinal segments. Stom, stomach; Duo, duodenum; Si5, jejunum; Si8, ileum; Ce, cecum; C, colon; F, feces; ND, not detected. (H) Total weight of cohoused Muc17fl/fl (n=19) and Muc17ΔIEC (n=13) littermate mice. Box plots show the median and minimum to maximum values for each group. (I) Representative image of the opened abdomen of 36–40-week-old Muc17fl/fl and Muc17ΔIEC mice. Scale bar 1 cm. (J) Quantification of bacterial 16S copy number in mesenteric lymph nodes (MLN), spleen, and Liver tissue by qPCR. n= 5 for each group. (K) Quantification of bacterial growth in tissue homogenates of MLN and spleen on BHIS agar plates under anaerobic conditions. (L) Representative image representing bacterial growth in tissue homogenates of MLN on BHIS agar under anaerobic conditions. (M) Identification of isolated bacterial colonies identified by 16S rDNA sequencing. Significance was determined by unpaired t-test (A) and Mann-Whitney test (C).
In ileal Crohn’s disease, displacement of mucosa-associated bacteria from the site of inflammation to mesenteric adipose tissue induces the expansion of encapsulating “creeping fat” (28). Bacterially induced enlargement of visceral adipose tissue also occurs in mice that fail to control microbial translocation (29). Under baseline SPF conditions, Muc17ΔIEC mice displayed a higher total body mass (Fig. 5H) and larger abdominal fat pads compared to agematched Muc17fl/fl cohoused littermates (Fig. 5I). Subsequently, we asked if Muc17ΔIEC mice displayed translocation of commensal bacteria to extra-intestinal tissues. Absolute 16S rRNA gene quantification uncovered significantly higher counts in the MLNs and spleen of Muc17ΔIEC mice compared to wild-types (Fig. 5J). Bacterial quantification was complemented with anaerobic culturing of viable bacteria from the affected tissues. Heavy bacterial growth was observed in 92% of Muc17ΔIEC MLNs, with the remaining 8% exhibiting light growth (Fig. 5K–L). In Muc17fl/fl mice, 33% of MLNs were clear of bacteria and 67% exhibited only light growth. We also observed a higher number of Muc17ΔIEC spleens with heavy bacterial growth compared to control tissues (Fig. 5K). Sequencing of the 16S gene identified the translocated bacteria as Escherichia coli, Staphylococcus spp., and Ligilactobacillus murinus (Fig. 5M), the latter described as a translocating strain (30). Thus, we concluded that viable commensal bacteria penetrate the intestinal epithelial cell barrier in the absence of Muc17, highlighting its key role in small intestinal barrier function.
Discussion
It is increasingly understood that the defensive properties of gel-forming mucins are important in protection against the development of human IBD (4), but the role of membrane mucins still remains elusive. In this study, we used a conditional mouse model to identify Muc17 as essential for the regiospecific protection of the small intestine against commensal and pathogenic bacteria. Significantly, a reduction in MUC17 levels in the enterocyte brush border leading to impaired glycocalyx protection against bacteria, was observed already in the non-inflamed ileum of CD patients. Our findings suggests that membrane mucin dysfunction precedes inflammation in CD.
The mechanisms underlying CD include genetic factors, such as NOD2 mutations that impair the sensing and removal of intracellular bacteria (10). These defects in turn trigger immune responses that evoke chronic inflammation. While there are no known disease-associated mutations in the MUC17 gene, studies have reported an altered glycocalyx ultrastructure in ileal CD (15). However, the implications of a compromised glycocalyx and the loss of its major component MUC17 in CD have not been addressed. In this study, we identified decreased MUC17 levels and a weakened glycocalyx in ileal CD to allow for increased bacteria-epithelium interactions. The results find mechanistic support in our earlier studies of murine small intestinal explants demonstrating that Muc17 forms the enterocyte glycocalyx (9). We also showed that the formation of the glycocalyx by Muc17 is induced by IL-22, a critical regulator of antibacterial defenses, including antimicrobial peptides (Reg3b and Reg3g) and mucin fucosylation via FUT2 (31). Notably, many IBD risk genes, such as IL23R, JAK1/2, TYK2, and STAT3, participate in IL-22 signaling (31) and the frequency of IL-22-producing type 3 innate lymphoid cells is lower in CD patients (32).
Human MUC17 is confined to small intestinal enterocytes, while murine MUC17 is expressed in both the small and large intestines. We disentangled the segment-specific function using two interventions with distinct mechanisms of action. DSS disrupts the mucus layer separating gut bacteria from the epithelium (33). Accordingly, DSS-induced colitis is rapid and severe in mouse models with mucus defects (23–25). Since glycocalyx-deficient Muc17ΔIEC mice still produced a functional colonic mucus barrier, they were equally sensitive to colitis as wild-type controls. In an alternative intervention, we used C. rodentium, which shares virulence factors with enteropathogenic Escherichia coli (EPEC), a major human pathogen that targets the small intestine. While C. rodentium is typically used to study pathogen-host interactions in the mouse distal colon (34), it surprisingly colonized the jejunum, including its mucosa, in Muc17-deficient mice. This finding highlights the crucial role of MUC17 in defending the small intestine against bacterial infections, even in the presence of a functioning mucus barrier. Interestingly, the ileum of Muc17-deficient mice was more resistant to colonization by C. rodentium than the jejunum. This difference can be explained by the lower expression of antimicrobial peptides in the mouse jejunum compared to the ileum, making it easier for the bacteria to colonize the jejunum (35). Given the link between CD and reduced antimicrobial peptide levels (36, 37), the murine jejunum emerges as a relevant model for ileal CD in humans.
Defects in intestinal defenses frequently result in the translocation of gut bacteria to peripheral tissues (26, 29). Bacterial translocation has also recently been linked the expansion of “creeping fat” in CD (28). Under baseline conditions, Muc17ΔIEC mice exhibited a spontaneous deterioration of epithelial homeostasis in the small intestine, involving elevated levels of host proteins participating in bacterial invasion and antimicrobial defenses. That Muc17ΔIEC mice displayed greater body mass, enlargement of abdominal fat, and viable commensal bacteria in peripheral tissues further supports the importance of the glycocalyx-forming MUC17 in limiting pathology related to CD.
Gel-forming mucins not only serve as a physical barrier against bacteria but also provide a habitat for bacteria that utilize mucins as a carbon source (35, 38). However, such a role for membrane mucins has not been described. Our work shows that Muc17 regulates gut microbiota composition and signifies that Muc17-dependent changes in the bacterial community propagate from the small intestine, where Muc17 exerts its function, distally towards the colon. Importantly, increased abundance of Coriobacteriia and Lachnoclostridium, enriched in Muc17ΔIEC mice, has also been reported in CD patients (39–41). Thus, our study suggests that MUC17 fulfills two purposes; forming a glycocalyx that blocks bacteria and regulating the selection of bacteria by the host. We postulate that the latter function depends on the apical localization of MUC17 on microvilli, which release luminal vesicles that bind and limit bacterial growth in the lumen (16). The absence of Muc17 from microvillus-derived luminal vesicles could alter vesicle interactions with bacteria, thereby impacting the microbial community.
In conclusion, we uncovered a role for the MUC17-based glycocalyx as an important defense system of the small intestine. Our findings shed light on the regiospecific role of membrane mucins as innate defense components that create a critical interface between the host epithelium and the gut microbiota. The disruption of this system in the non-inflamed ileum of CD patients suggests that defective MUC17 biosynthesis or trafficking is an early epithelial defect that precedes inflammation. Given the limited number of mouse models for small intestinal inflammation, the Muc17ΔIEC model provides opportunities for understanding the molecular mechanisms underlying epithelial cell dysfunction in Crohn’s disease.
Supplementary Material
Acknowledgements:
The authors acknowledge support from the National Genomics Infrastructure in Genomics Production Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council (2017.2008), and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We also thank the Bioinformatics and Data Centre at the Sahlgrenska Academy and Clinical Genomics Gothenburg at SciLifeLab for bioinformatics support. The authors also thank all the helpful colonoscopists at the Endoscopy Unit of the Sahlgrenska University Hospital.
Funding:
Swedish Society for Medical Research (Svenska Sällskapet för Medicinsk Forskning) S17-0005 (TP)
National Institutes of Health 5U01AI095542-08-WU-19-95, 5U01AI095542-09-WU-20-77 (TP)
Wenner-Gren Stiftelserna FT2017-0002 (TP)
Jeansson Foundations JS2017-0003 (TP)
Åke Wiberg Foundation M17-0062, M21-0022 (TP)
Biocodex Microbiota Foundation (TP)
Stiftelsen Clas Groschinskys Minnesfond M2254 (TP)
Sahlgrenska Academy International Starting Grant E2015/521 (TP)
Grants under the ALF agreement 236501 between the Swedish Government and the county councils, ALFGBG-991311, ALFGBG-440741 (TP)
Wenner-Gren Foundations UPD2018-0065, WUP2017-0005 (ELH)
Wilhelm och Martina Lundgrens Vetenskapsfond 2020-3647, 2021-3880, 2022-4051 (ELH)
Sahlgrenska Academy Core Facilities Elisabeth “Bollan” Lindén Stipend (ELH)
B.A.V. is the Children with Intestinal and Liver Disorders Foundation Chair in Pediatric Gastroenterology
Funding Statement
Swedish Society for Medical Research (Svenska Sällskapet för Medicinsk Forskning) S17-0005 (TP)
National Institutes of Health 5U01AI095542-08-WU-19-95, 5U01AI095542-09-WU-20-77 (TP)
Wenner-Gren Stiftelserna FT2017-0002 (TP)
Jeansson Foundations JS2017-0003 (TP)
Åke Wiberg Foundation M17-0062, M21-0022 (TP)
Biocodex Microbiota Foundation (TP)
Stiftelsen Clas Groschinskys Minnesfond M2254 (TP)
Sahlgrenska Academy International Starting Grant E2015/521 (TP)
Grants under the ALF agreement 236501 between the Swedish Government and the county councils, ALFGBG-991311, ALFGBG-440741 (TP)
Wenner-Gren Foundations UPD2018-0065, WUP2017-0005 (ELH)
Wilhelm och Martina Lundgrens Vetenskapsfond 2020-3647, 2021-3880, 2022-4051 (ELH)
Sahlgrenska Academy Core Facilities Elisabeth “Bollan” Lindén Stipend (ELH)
B.A.V. is the Children with Intestinal and Liver Disorders Foundation Chair in Pediatric Gastroenterology
Footnotes
Data and materials availability:
All data are available in the main text or the supplementary material. Primary data for all the described experiments are reported in table S4.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXDXXXX.
References
- 1.Pelaseyed T., Bergstrom J. H., Gustafsson J. K., Ermund A., Birchenough G. M., Schutte A., van der Post S., Svensson F., Rodriguez-Pineiro A. M., Nystrom E. E., Wising C., Johansson M. E., Hansson G. C., The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev 260, 8–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gustafsson J. K., Johansson M. E. V., The role of goblet cells and mucus in intestinal homeostasis. Nat Rev Gastroenterol Hepatol 19, 785–803 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Johansson M. E. V., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C., The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105, 15064–15069–15064–15069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Post S., Jabbar K. S., Birchenough G., Arike L., Akhtar N., Sjovall H., Johansson M. E. V., Hansson G. C., Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 68, 2142–2151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermund A., Schutte A., Johansson M. E., Gustafsson J. K., Hansson G. C., Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol 305, G341–347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang T., Pelaseyed T., Discovery of a MUC3B gene reconstructs the membrane mucin gene cluster on human chromosome 7. PLoS One 17, e0275671 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelaseyed T., Gustafsson J. K., Gustafsson I. J., Ermund A., Hansson G. C., Carbachol-induced MUC17 endocytosis is concomitant with NHE3 internalization and CFTR membrane recruitment in enterocytes. American journal of physiology. Cell physiology 305, C457–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malmberg E. K., Pelaseyed T., Petersson A. C., Seidler U. E., De Jonge H., Riordan J. R., Hansson G. C., The C-terminus of the transmembrane mucin MUC17 binds to the scaffold protein PDZK1 that stably localizes it to the enterocyte apical membrane in the small intestine. Biochem J 410, 283–289 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Layunta E., Javerfelt S., Dolan B., Arike L., Pelaseyed T., IL-22 promotes the formation of a MUC17 glycocalyx barrier in the postnatal small intestine during weaning. Cell Rep 34, 108757 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Khor B., Gardet A., Xavier R. J., Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crohn B. B., Ginzburg L., Oppenheimer G. D., Regional ileitis; a pathologic and clinical entity. Am J Med 13, 583–590 (1952). [DOI] [PubMed] [Google Scholar]
- 12.Martin H. M., Campbell B. J., Hart C. A., Mpofu C., Nayar M., Singh R., Englyst H., Williams H. F., Rhodes J. M., Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127, 80–93 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A. L., Barnich N., Bringer M. A., Swidsinski A., Beaugerie L., Colombel J. F., High prevalence of adherentinvasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F. M., Briggs J., Gunther S., Prescott N. J., Onnie C. M., Hasler R., Sipos B., Folsch U. R., Lengauer T., Platzer M., Mathew C. G., Krawczak M., Schreiber S., A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 39, 207–211 (2007). [DOI] [PubMed] [Google Scholar]
- 15.VanDussen K. L., Stojmirovic A., Li K., Liu T. C., Kimes P. K., Muegge B. D., Simpson K. F., Ciorba M. A., Perrigoue J. G., Friedman J. R., Towne J. E., Head R. D., Stappenbeck T. S., Abnormal Small Intestinal Epithelial Microvilli in Patients With Crohn’s Disease. Gastroenterology 155, 815–828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shifrin D. A. Jr., McConnell R. E., Nambiar R., Higginbotham J. N., Coffey R. J., Tyska M. J., Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr Biol 22, 627–631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell R. E., Higginbotham J. N., Shifrin D. A. Jr, Tabb D. L., Coffey R. J., Tyska M. J., The enterocyte microvillus is a vesicle-generating organelle. J Cell Biol 185, 1285–1298 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider H., Pelaseyed T., Svensson F., Johansson M. E. V., Study of mucin turnover in the small intestine by in vivo labeling. Sci Rep 8, 5760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhinder G., Sham H. P., Chan J. M., Morampudi V., Jacobson K., Vallance B. A., The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. J Vis Exp, e50222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abrams G. D., Bauer H., Sprinz H., Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest 12, 355–364 (1963). [PubMed] [Google Scholar]
- 21.Lesher S., Walburg H. E. Jr., Sacher G. A. Jr., Generation Cycle in the Duodenal Crypt Cells of Germ-Free and Conventional Mice. Nature 202, 884–886 (1964). [DOI] [PubMed] [Google Scholar]
- 22.Bourgeois J. S., Wang L., Rabino A. F., Everitt J., Alvarez M. I., Awadia S., Wittchen E. S., Garcia-Mata R., Ko D. C., ARHGEF26 enhances Salmonella invasion and inflammation in cells and mice. PLoS Pathog 17, e1009713 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An G., Wei B., Xia B., McDaniel J. M., Ju T., Cummings R. D., Braun J., Xia L., Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med 204, 1417–1429 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornick S., Kumar M., Moreau F., Gaisano H., Chadee K., VAMP8-mediated MUC2 mucin exocytosis from colonic goblet cells maintains innate intestinal homeostasis. Nat Commun 10, 4306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharpen J. D. A., Dolan B., Nystrom E. E. L., Birchenough G. M. H., Arike L., Martinez-Abad B., Johansson M. E. V., Hansson G. C., Recktenwald C. V., Transglutaminase 3 crosslinks the secreted gel-forming mucus component Mucin-2 and stabilizes the colonic mucus layer. Nat Commun 13, 45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson M. E. V., Gustafsson J. K., Holmén-Larsson J., Jabbar K. S., Xia L., Xu H., Ghishan F. K., Carvalho F. A., Gewirtz A. T., Sjövall H., Hansson G. C., Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lkhagva E., Chung H. J., Hong J., Tang W. H. W., Lee S. I., Hong S. T., Lee S., The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice. BMC Microbiol 21, 44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ha C. W. Y., Martin A., Sepich-Poore G. D., Shi B., Wang Y., Gouin K., Humphrey G., Sanders K., Ratnayake Y., Chan K. S. L., Hendrick G., Caldera J. R., Arias C., Moskowitz J. E., Ho Sui S. J., Yang S., Underhill D., Brady M. J., Knott S., Kaihara K., Steinbaugh M. J., Li H., McGovern D. P. B., Knight R., Fleshner P., Devkota S., Translocation of Viable Gut Microbiota to Mesenteric Adipose Drives Formation of Creeping Fat in Humans. Cell 183, 666–683 e617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergstrom J. H., Birchenough G. M., Katona G., Schroeder B. O., Schutte A., Ermund A., Johansson M. E., Hansson G. C., Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc Natl Acad Sci U S A 113, 13833–13838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L., Deitch E., Specian R., Steffen E., Berg R., Translocation of Lactobacillus-Murinus from the Gastrointestinal-Tract. Curr Microbiol 20, 177–184 (1990). [Google Scholar]
- 31.Keir M., Yi Y., Lu T., Ghilardi N., The role of IL-22 in intestinal health and disease. J Exp Med 217, e20192195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokkinou E., Soini T., Pandey R. V., van Acker A., Theorell J., Czarnewski P., Kvedaraite E., Vandamme N., Lourda M., Sorini C., Weigel W., Carrasco A., Tibbitt C. A., Schlums H., Lindforss U., Nordenvall C., Ljunggren M., Idestrom M., Svensson M., Henter J. I., Villablanca E. J., Bryceson Y. T., Rolandsdotter H., Mjosberg J., The single-cell transcriptional landscape of innate and adaptive lymphocytes in pediatric-onset colitis. Cell Rep Med 4, 101038 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson M. E. V., Gustafsson J. K., Sjöberg K. E., Petersson J., Holm L., Sjövall H., Hansson G. C., Bacteria Penetrate the Inner Mucus Layer before Inflammation in the Dextran Sulfate Colitis Model. PLoS ONE 5, e12238–e12238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nataro J. P., Kaper J. B., Diarrheagenic Escherichia coli. Clin Microbiol Rev 11, 142–201 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birchenough G. M. H., Schroeder B. O., Sharba S., Arike L., Recktenwald C. V., Puertolas-Balint F., Subramani M. V., Hansson K. T., Yilmaz B., Linden S. K., Backhed F., Hansson G. C., Muc2-dependent microbial colonization of the jejunal mucus layer is diet sensitive and confers local resistance to enteric pathogen infection. Cell Rep 42, 112084 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamas B., Richard M. L., Leducq V., Pham H. P., Michel M. L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T. W., Natividad J. M., Brot L., Taleb S., Couturier-Maillard A., Nion-Larmurier I., Merabtene F., Seksik P., Bourrier A., Cosnes J., Ryffel B., Beaugerie L., Launay J. M., Langella P., Xavier R. J., Sokol H., CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22, 598–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wehkamp J., Wang G., Kubler I., Nuding S., Gregorieff A., Schnabel A., Kays R. J., Fellermann K., Burk O., Schwab M., Clevers H., Bevins C. L., Stange E. F., The Paneth cell alpha-defensin deficiency of ileal Crohn’s disease is linked to Wnt/Tcf-4. J Immunol 179, 3109–3118 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Raba G., Luis A. S., Mucin utilization by gut microbiota: recent advances on characterization of key enzymes. Essays Biochem 67, 345–353 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alam M. T., Amos G. C. A., Murphy A. R. J., Murch S., Wellington E. M. H., Arasaradnam R. P., Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog 12, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raygoza Garay J. A., Turpin W., Lee S. H., Smith M. I., Goethel A., Griffiths A. M., Moayyedi P., Espin-Garcia O., Abreu M., Aumais G. L., Bernstein C. N., Biron I. A., Cino M., Deslandres C., Dotan I., El-Matary W., Feagan B., Guttman D. S., Huynh H., Dieleman L. A., Hyams J. S., Jacobson K., Mack D., Marshall J. K., Otley A., Panaccione R., Ropeleski M., Silverberg M. S., Steinhart A. H., Turner D., Yerushalmi B., Paterson A. D., Xu W., Consortium C. G. P. R., Croitoru K., Gut Microbiome Composition Is Associated With Future Onset of Crohn’s Disease in Healthy First-Degree Relatives. Gastroenterology 165, 670–681 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Chiodini R. J., Dowd S. E., Chamberlin W. M., Galandiuk S., Davis B., Glassing A., Microbial Population Differentials between Mucosal and Submucosal Intestinal Tissues in Advanced Crohn’s Disease of the Ileum. PLoS One 10, e0134382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary material. Primary data for all the described experiments are reported in table S4.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXDXXXX.