Abstract
Proteins of the basic helix-loop-helix (bHLH) family are required for a number of different developmental pathways, including neurogenesis, lymphopoiesis, myogenesis, and sex determination. Using a yeast two-hybrid screen, we have identified a new bHLH transcription factor, ABF-1, from a human B-cell cDNA library. Within the bHLH region, ABF-1 shows a remarkable conservation with other HLH proteins, including tal-1, NeuroD, and paraxis. Its expression pattern is restricted to a subset of lymphoid tissues, Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines, and activated human B cells. ABF-1 is capable of binding an E-box element either as a homodimer or as a heterodimer with E2A. Furthermore, a heterodimeric complex containing ABF-1 and E2A can be detected in EBV-immortalized lymphoblastoid cell lines. ABF-1 contains a transcriptional repression domain and is capable of inhibiting the transactivation capability of E47 in mammalian cells. ABF-1 represents the first example of a B-cell-restricted bHLH protein, and its expression pattern suggests that ABF-1 may play a role in regulating antigen-dependent B-cell differentiation.
The basic helix-loop-helix (bHLH) family of transcription factors is composed of a large number of proteins involved in a wide array of developmental processes, including cellular proliferation and differentiation (40, 41). These proteins share a common sequence motif consisting of a basic region and an adjacent helix-loop-helix (HLH) structure (42, 43). The basic region has been shown to be important for DNA binding, while the HLH domain mediates dimerization (42, 72). The DNA binding sites for bHLH proteins, known as E boxes, consist of the consensus sequence CANNTG. E-box elements were first identified in the immunoglobulin heavy-chain (IgH) intronic enhancer and have since been found in a large number of pancreatic-, lymphoid-, and muscle-specific promoter and enhancer elements (7, 11, 20, 40, 41, 74, 75). Mutational analysis of the E-box sites, in particular, the E2 box ([G/A]CAGNTG[T/G]), present in a variety of different regulatory elements has demonstrated their importance in regulating cell-type-specific gene transcription (40, 41).
The bHLH proteins have been categorized into different classes based on dimerization specificity and tissue distribution. Class I HLH members, also known as E proteins, include E12, E47, HEB, and E2-2 (40). Each is capable of homo- and/or heterodimerization and is widely expressed (40). Class II HLH proteins, including MyoD, myogenin, NeuroD, and members of the achaete-scute complex in Drosophila melanogaster, display a tissue-restricted expression pattern and bind DNA only as heterodimers with the class I HLH proteins (40, 41, 43). The class I HLH protein daughterless, for example, forms heterodimers with achaete-scute family members to activate common target genes that control sex determination and neurogenesis (9, 10, 12, 16).
A number of myogenic-specific bHLH proteins, including MyoD, Myf-5, and myogenin, interact with class I HLH proteins to activate genes that are required for proper vertebrate muscle development (25, 45, 56, 57, 74). NeuroD, a neuronal- and pancreatic-specific bHLH protein, has been shown to regulate terminal differentiation of neurons in Xenopus laevis (35). Surprisingly, NeuroD knockout mice display a dramatic pancreatic defect and develop diabetes, yet neuronal development appears unaffected (46). Studies with knockout mice have demonstrated that hematopoiesis is also regulated, in part, by bHLH proteins. Recently, it was shown that mice lacking the SCL/tal-1 bHLH gene exhibit a block in early hematopoiesis, affecting multipotent progenitors (49). The HEB gene has been shown to be required for proper thymocyte development, and the E2A gene, which encodes both E12 and E47, is absolutely essential for proper B- and T-cell development (1a, 3, 78, 79). In E2A-deficient mice, B-lineage development is blocked prior to the stage in which Ig rearrangements are normally initiated (3). Furthermore, B-cell development is inhibited in transgenic mice that overexpress the E2A inhibitor, Id1 (66). E2A-like molecules have also been suggested to play a role in Ig switching, since overexpression of Id1 in B-cell lines interfered with the ability of these cells to undergo isotype switching (22).
During thymocyte development, both E2A and HEB gene products which bind as heterodimers to the E2 box site are expressed (1a, 59). In B-lineage cells, both E2A and E2-2 gene products are expressed (2). In pre-B cells, both E47 and E2-2 bind the E2-box site. In mature B cells, E47 homodimers are the predominant E2-box-binding species (2, 44, 63). Interestingly, relatively high levels of E2A transcripts can be detected in germinal centers, suggesting that E2A may play a role later in B-lymphocyte development (52).
Several class II bHLH genes have been shown to be expressed in the hematopoietic compartment (4, 34, 71). The lyl-1 bHLH gene, for example, is expressed in a number of erythroid, myeloid, and B-cell lines (34, 71). To date, however, there has been no evidence supporting the existence of a B-cell-restricted bHLH protein. Here we report the isolation of a novel bHLH transcription factor, called ABF-1, from a B-cell cDNA library. ABF-1 displays a remarkable conservation within the tissue-specific class II bHLH proteins. We demonstrate that ABF-1 is a transcriptional repressor which is expressed in a number of B-cell lines, lymphoid tissues, and, strikingly, activated primary B cells. Like other class II bHLH proteins, we find that ABF-1 is capable of binding to an E-box hexanucleotide element as a heterodimer with the E2A proteins in nuclear extracts derived from immortalized lymphoblastoid cell lines (LCLs). The data presented here suggest that ABF-1 is a downstream target of signalling through the antigen receptor.
MATERIALS AND METHODS
Yeast two-hybrid screening.
Two-hybrid screening was performed with the reporter Saccharomyces cerevisiae strain y190 essentially as described previously (19). Details on construction of the human E-protein bait plasmids are available upon request.
Electrophoretic mobility shift assays (EMSA).
DNA binding assays with the μE5 probe were performed essentially as described previously (2). Complexes were resolved on 5% native polyacrylamide gels in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA). For gel shift assays using the μE4-OCT (octamer) and μE4 probes, the following reaction conditions were used: 12 mM HEPES (pH 7.9), 100 mM KCl, 10 μM ZnCl2, 12% glycerol, 0.05% Nonidet P-40, 50 μg of bovine serum albumin per ml, 25 μg of poly(dI-dC) per ml, 3 μl of in vitro-translated protein, 50,000 cpm of 32P-end-labeled probe, and 1 μl of antiserum, when appropriate. Proteins were translated in vitro by using a coupled transcription-translation kit (TNT) as instructed by the manufacturer (Promega). Nuclear extracts were prepared as previously described (17). The oligonucleotide probes used in this study were μE5 (42), μE4-OCT (5′-CCG AAT TCA CAC CAC CTG GGT AAT TTG CAT TTC-3′), and μE4 (5′-TCG AGA CAC CAC CTG GGT AAG-3′).
cDNA library screening, sequencing, and plasmid construction.
Several cDNA clones encoding the entire open reading frame of ABF-1 were isolated by screening the λACT B-cell library with the partial cDNA, clone 95, using standard techniques (58). The longest cDNA, designated 95-1A, was excised as an XhoI fragment and subcloned into pBSK+ (Stratagene) at the SalI site to generate pBSK-ABF-1. The ABF-1 cDNA was sequenced on both strands by using a Sequenase version 2.0 DNA sequencing kit as instructed by the manufacturer (Amersham). The entire coding region of ABF-1 was amplified by PCR with the primers ABF FLAG F1 (5′-ATA GGG ATC CGA GCT TCG GGG GCT GCA G-3′) and ABF FLAG R1 (5′-ATA GGA ATT CTT AAC GAA TAA TCC CAT CAA G-3′). The PCR product was ligated into pSP64FLAG (48) digested with BamHI and EcoRI to generate pFLAG-ABF-1. pGST-ABF-1 was constructed by subcloning an XhoI (blunted)/EcoRV-digested ABF-1 cDNA into pGEX2TK linearized with SmaI. The GAL4 DNA binding domain–ABF-1 fusions used in the repression assays were constructed as follows. The full coding region of ABF-1 was amplified by PCR with the primers ABF-GF1 (5′-ATA GGA ATT CAT GTT CAC GGG CTC GGT GAG T-3′) and ABF FLAG R1. The EcoRI-digested PCR fragment was ligated into the EcoRI site of pBXG1 (51) to create pGAL4-ABF-1(FL). The GAL4–ABF-1 deletion was created with the primers ABF-GF1 and ABF-GR1 (5′-ATA GGG ATT CTT ACC GCT GCG ACT GCT TGC ACT C-3′). The PCR product was subsequently cloned into pBXG1 at the EcoRI and BamHI sites to create pGAL4-ABF-1(ΔC). All constructs were sequenced to confirm the correct reading frame. pFLAG-ABF-1 was digested with EcoRI and HindIII to release the FLAG–ABF-1 cDNA. This fragment was made blunt with Klenow enzyme and ligated into pHβAneo (23) at the BamHI site (blunted). pHβA-E47 was kindly provided by Gretchen Bain.
Northern blot analysis.
Total RNA was isolated from cells by using Trizol reagent as instructed by the manufacturer (Gibco-BRL). RNA samples were separated on formaldehyde gels, transferred, and hybridized as described previously (58). All Northern blots were hybridized with a 32P-labeled 0.8-kb 3′ untranslated region fragment derived from BamHI digestion of pBSK-ABF-1.
Protein purification and generation of ABF-1-specific antiserum.
Host strain Escherichia coli BL21(DE3), harboring the pGST-ABF-1 expression vector, was grown to an optical density at 600 nm of 0.6 and induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 1.5 h at 37°C. Cells were subsequently harvested and lysed as described previously (38). Glutathione S-transferase (GST)–ABF-1 protein was solubilized from the pellet fraction in 6 M urea and subjected to dialysis in phosphate-buffered saline. GST–ABF-1, which precipitated out of solution during dialysis, was pelleted and resuspended in 1× sodium dodecyl sulfate (SDS) sample buffer. The sample (approximately 70% pure) was fractionated on an SDS–10% polyacrylamide gel and Coomassie blue stained to reveal the GST–ABF-1 fusion protein. The protein band was excised from the gel and prepared for injection into rabbits as described previously (24). Animals were immunized with 250 to 300 μg of protein, and serum was collected and purified by using standard techniques (24).
Western blotting.
Western blot analyses were performed as described previously (38). Briefly, 50 μg of nuclear extract was fractionated on SDS–12% polyacrylamide gel, transferred to an Immobilon-P membrane (Millipore), and incubated with affinity-purified polyclonal ABF-1 antiserum used at a 1:500 dilution. Bands were visualized by chemiluminescence with a horseradish peroxidase-conjugated goat anti-rabbit antibody.
Isolation and in vitro activation of human B cells.
Human B cells were purified from peripheral blood obtained from healthy donors. T cells were eliminated by complement-mediated lysis using an anti-CD3 antibody. Fluorescence-activated cell sorting analysis indicated that greater than 85% of the purified cells were CD19+ and less than 5% stained positive for OKT3. B cells were activated by culturing in a 1:10,000 dilution of Staphylococcus aureus Cowan 1 (SAC) (Calbiochem). Interleukin-2 (IL-2) was used at a final concentration of 50 U/ml. Cells were cultured in RPMI 1640 supplemented with 2.5% human serum, penicillin-streptomycin, and l-glutamine.
Cell culture and transfections.
HeLa S3 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, l-glutamine, and antibiotics. For transient transfections, 3 × 105 HeLa S3 cells were plated onto 6-cm-diameter dishes and transfected with Superfect reagent the next day as instructed by the manufacturer (Qiagen). Cells were harvested 48 h posttransfection and assayed for either chloramphenicol acetyltransferase (CAT) or luciferase activity as described previously (58).
RESULTS
Identification of a novel bHLH gene.
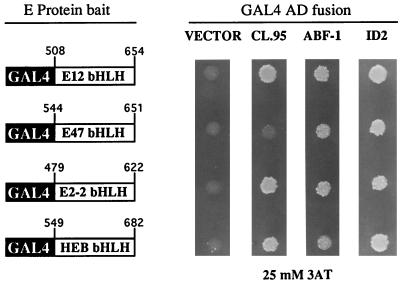
To identify potential B-cell-specific class II bHLH proteins, we performed a yeast two-hybrid screen. Using the bHLH region of E2-2 as bait, we screened a human B-cell GAL4 activation domain (AD)-cDNA fusion library for interacting proteins (Fig. 1) (19). From approximately 1.9 × 106 yeast transformants plated, we isolated a total of 249 clones that were capable of growth on 25 mM 3-aminotriazole (3AT), which indicates activation of the HIS3 reporter gene (19). Of these, 131 also activated a lacZ reporter gene and thus turned blue when assayed for β-galactosidase activity. Clones that could activate both reporter genes, which is indicative of a potential interaction between the E2-2 bait and a library-encoded GAL4 AD fusion protein, were analyzed further. Approximately 89% of the 131 clones failed to interact with the unrelated bait SNF2, lamin, p53, or CDK2, suggesting that these clones were highly specific for the E2-2 bait (data not shown). Subsequent characterization of the cDNAs isolated from the positive clones revealed that the vast majority were identical to human Id2 and Id3 (data not shown). One cDNA, clone 95, encoded a protein with significant homology to the tissue-specific class of bHLH transcription factors. Because of the expression pattern of clone 95 (discussed below), it will be hereafter referred to as ABF-1, for activated B-cell factor 1.
FIG. 1.
ABF-1 is capable of interacting with the E proteins in the two-hybrid system. Expression plasmids encoding the indicated GAL4 DNA binding domain fusions (E-protein bait) and GAL4 AD fusions were cotransformed into the reporter yeast strain y190, which contains an integrated GAL1-HIS3 reporter gene (19). Transformants were initially selected on synthetic medium lacking tryptophan and leucine and then spotted onto synthetic medium lacking histidine and containing 25 mM 3AT. Growth on the selective medium indicates an interaction between the E-protein bait and the GAL AD fusion protein (19). Yeast cells were allowed to grow for 4 days at 30°C and then photographed. Vector, pACT2; CL.95, clone 95; ABF-1, pACT2-ABF-1; ID2, pACT-ID2. The GAL4 AD–ABF-1(FL) fusion conferred a mild slow-growth phenotype in yeast strain y190.
Because we were unable to isolate ABF-1 by using other E-protein baits (38a), we further used the two-hybrid system to assess whether the gene product is capable of interacting with different E-protein family members. The original isolate, clone 95, was able to interact with the E2-2, HEB, and E12 bait proteins, as indicated by growth on selective medium containing 25 mM 3AT (Fig. 1). However, when the entire bHLH domain of ABF-1 was fused to the GAL4 AD, an interaction with all E proteins, including E47, could be detected (Fig. 1). These interactions were strong enough to support growth on 50 mM 3AT (data not shown). As a positive control, the E-protein baits were shown to interact with a GAL4 AD-Id2 fusion protein (Fig. 1).
ABF-1 encodes a novel HLH protein.
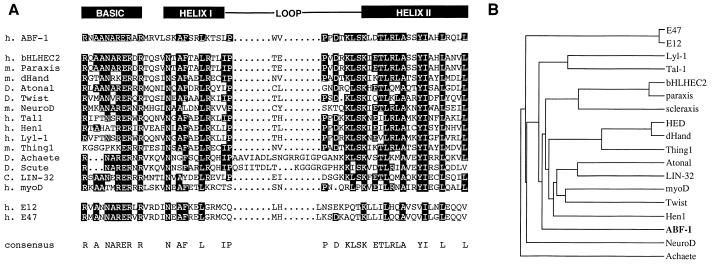
The data described above suggest that ABF-1 encodes a protein which has the ability to interact with class I HLH proteins. To examine whether ABF-1 encodes an HLH protein, a longer cDNA clone was isolated from a B-cell library and completely sequenced (Fig. 2). A computer database search using the ABF-1 nucleotide sequence and the BLAST algorithm revealed that ABF-1 indeed encodes a bHLH protein. Sequence alignment of ABF-1 with various class II family members revealed a remarkable conservation within the HLH region (Fig. 3A). ABF-1 is 60% identical to bHLHEC2 and paraxis and 53% identical to dHAND and Atonal within this domain. In addition, ABF-1 also shows significant identity to tal-1, NeuroD, and members of the achaete-scute family (Fig. 3A). Outside the bHLH region, however, ABF-1 shows no significant homology to any known proteins. To examine the evolutionary relationship of ABF-1 to other HLH proteins, a dendrogram was constructed (Fig. 3B). Interestingly, the ABF-1 protein is more closely related to class II HLH proteins, including the muscle- and pancreatic-specific HLH proteins, than to the class I HLH proteins E12 and E47 (Fig. 3B). These data indicate that a novel HLH protein closely related to the class II HLH proteins is expressed in hematopoietic lineage cells.
FIG. 2.
Nucleotide sequence of the human ABF-1 cDNA. The longest open reading frame encodes a protein 218 amino acids in length. Although not full length (see Fig. 4), the ∼1.9-kb cDNA contains two potential start codons, both with good Kozak sequences (58). The conceptual translation product predicts a 23.6-kDa protein with an estimated pI of 9.5. In addition to the bHLH motif (bold underline), there is a putative nuclear localization signal (dashed underline), a glycine-rich region (boldface), and a stretch of acidic residues (double underlined). The boxed leucine residue (position 130) represents the fusion point of clone 95 with the GAL4 AD. The asterisk denotes the stop codon.
FIG. 3.
Sequence alignment of ABF-1 with the class II family of bHLH proteins. (A) Multiple sequence alignment of the bHLH region of human (h.) ABF-1 with the most related class II bHLH proteins was created by using the PileUp and Pretty algorithms (Genetics Computer Group sequence analysis software package). Amino acids identical in at least half of the sequences are shown as blackened boxes. For reference, the bHLH regions of the class I proteins E12 and E47 are shown. m., mouse; D., Drosophila; C., Caenorhabditis elegans. (B) Dendrogram displaying a graphical output of the pairwise alignments of ABF-1 and related bHLH family members generated by PileUp.
ABF-1 is highly expressed in activated human B lymphocytes.
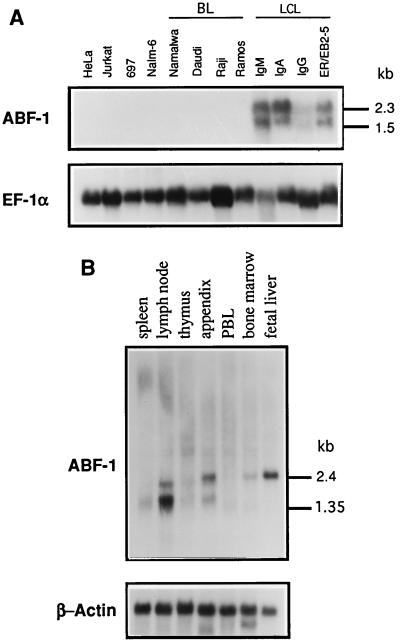
To determine the expression pattern of ABF-1, RNA derived from various cell lines and lymphoid tissues was isolated and analyzed by Northern blotting. A panel of human cell lines derived from both lymphoid and nonlymphoid origin was examined for the presence of ABF-1 transcripts (Fig. 4A). No expression of ABF-1 was detected in HeLa, Jurkat, and a number of Burkitt lymphoma B-cell lines, including Namalwa, Daudi, and Raji (Fig. 4A). Since ABF-1 was isolated from an Epstein-Barr virus (EBV)-transformed human B-cell cDNA library, we next analyzed RNA derived from a number of EBV-immortalized LCLs for the presence of ABF-1. ABF-1 mRNA was detected in all LCLs analyzed (Fig. 4A). Most LCLs expressed ABF-1 at relatively high levels. The mRNA consisted of a doublet running at 1.5 and 2.3 kb, which likely represents alternative splicing of ABF-1 transcripts (Fig. 4A). ABF-1 message is also highly abundant in ER/EB2-5, an EBV-transformed B-cell line expressing an estrogen receptor (ER)-EBNA2 fusion protein (Fig. 4A) (32).
FIG. 4.
Northern blot analysis of ABF-1 expression. (A) Expression pattern of ABF-1 in human cell lines. Ten micrograms of total RNA was isolated from each cell line and analyzed by Northern blotting. The blot was probed with the ABF-1 cDNA (top), stripped, and subsequently reprobed with the human elongation factor 1 alpha (EF-1α) cDNA (6) as a loading control (bottom). HeLa, carcinoma; Jurkat, T-cell leukemia; 697, pre-B ALL harboring a t(1;19) translocation; Nalm-6, pre-B; BL, Burkitt lymphoma; LCL, lymphoblastoid cell line of the indicated Ig isotype. The ER/EB2-5 cell line was grown in the presence of 1 μM β-estradiol (27). (B) A human tissue Northern blot (Clontech) containing 2 μg of poly(A)+ RNA per lane was sequentially hybridized with ABF-1 (top) and β-actin (bottom). PBL, peripheral blood lymphocyte.
To examine for the presence of ABF-1 in primary cells, a Northern blot containing poly(A)+ RNA isolated from human lymphoid tissues was probed with a radiolabeled ABF-1 cDNA (Fig. 4B). Interestingly, ABF-1 transcripts were detected in the lymph node, appendix, fetal liver, and to a lesser degree bone marrow (Fig. 4B). However, no expression was detected in peripheral blood lymphocytes (Fig. 4B). Taken together, these data indicate that ABF-1 is expressed both in a subset of human B-cell lines and in primary cells present in lymphoid tissues.
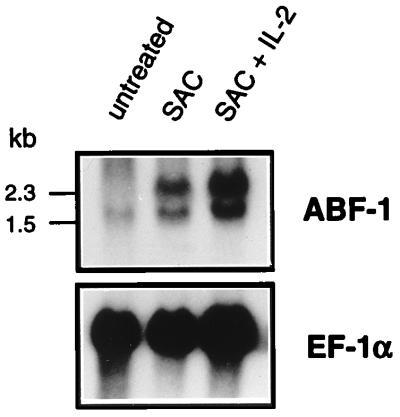
Since EBV-transformed B-cell lines resemble activated human B lymphocytes (8, 15, 53, 69, 73), we wished to address whether ABF-1 expression could be induced in mitogen-treated peripheral blood B lymphocytes. Human B cells were purified from peripheral blood and activated in vitro with SAC or SAC plus IL-2. B cells left untreated showed little, if any, expression of ABF-1 (Fig. 5), consistent with the lack of ABF-1 mRNA in the peripheral blood lymphocyte sample (Fig. 4B). However, treatment with SAC alone caused a dramatic upregulation of ABF-1 which could be further induced by addition of IL-2 (Fig. 5). These data indicate that ABF-1 expression is induced upon B-cell activation. Although ABF-1 transcripts were not detected in the thymus or the T-cell line Jurkat, we wished to determine whether ABF-1 is expressed in activated human T cells. Northern blot analysis revealed that mitogen-activated T cells do not express detectable levels of ABF-1 mRNA (38a). Thus, in lymphoid cells, activation of ABF-1 expression is restricted to B-lineage-derived cells.
FIG. 5.
ABF-1 mRNA is highly abundant in activated human B cells. Northern blot analysis of 10 μg of total RNA isolated from purified peripheral blood B lymphocytes activated in vitro. B cells were treated with SAC for 3 days or with SAC and IL-2 for 6 days or were left untreated. The Northern blot was probed with the ABF-1 cDNA (top), stripped, and then reprobed with elongation factor 1 alpha (EF-1α; bottom) as a control.
ABF-1 has the ability to bind either as a homodimer or as a heterodimer to DNA.
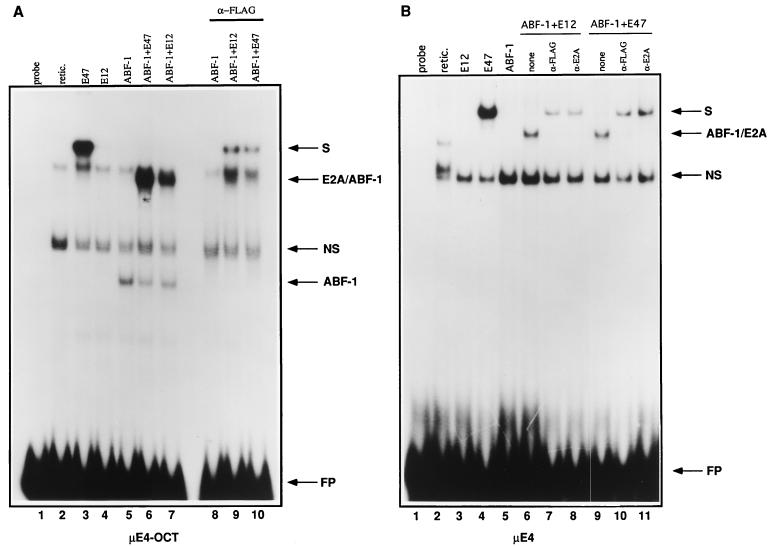
To assess whether ABF-1 has the ability to bind the E2-box site, we performed an EMSA. E12, E47, and FLAG-tagged ABF-1 were translated in vitro and analyzed by SDS-gel electrophoresis. Each of the gene products was translated efficiently (data not shown). Subsequently, the in vitro-translated proteins were incubated with a 32P-labeled μE4-OCT-containing oligonucleotide probe derived from the IgH intronic enhancer and analyzed by EMSA. The μE4 site is closely related to the μE5 and κE2 sites and binds with high affinity to E47 (5) and (38a). As predicted, E47 was capable of binding to the μE4 site as a homodimer (Fig. 6A, lane 3). As expected, E12 was not able to bind efficiently to the site, as it has been shown to contain an inhibitory domain which interferes with homodimerization (Fig. 6A, lane 4) (67). In contrast to E12, ABF-1 has the ability to bind the μE4-OCT site alone (Fig. 6A, lane 5). This complex was eliminated by the addition of an anti-FLAG antibody and thus likely represents a homodimer of ABF-1 (Fig. 6A, lane 8). When ABF-1 was cotranslated with either E12 or E47, a complex of intermediate mobility was formed (Fig. 7A, lanes 6 and 7). This complex, which could be supershifted with an anti-FLAG antibody, most likely represents a heterodimer composed of ABF-1 and E2A (Fig. 6A; compare lanes 6 and 7 with lanes 9 and 10). Alternatively, incubation with the anti-FLAG antibody may inhibit DNA binding of ABF-1–E2A heterodimers, thus allowing E47 homodimers to form on the μE4 probe (Fig. 6).
FIG. 6.
ABF-1 binds to an E box in vitro. (A) EMSA analysis of FLAG ABF-1 homo- and heterodimeric complexes formed on the μE4-OCT probe. Arrows indicate the position of ABF-1 homodimers and ABF-1–E2A heterodimers. retic., reticulocyte lysate; FP, free probe; NS, nonspecific complex; S, supershifted complex. (B) ABF-1 binds as a heterodimer to the μE4 probe. Addition of a nonspecific control antibody had no effect on complex formation (not shown).
FIG. 7.
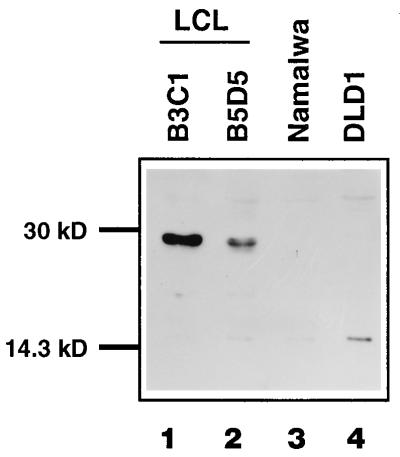
Western blot analysis of ABF-1 in nuclear extracts isolated from several human cell lines. Lane 1, EBV-immortalized LCL B3C1; lane 2, B5D5 (LCL); lane 3, Namalwa (Burkitt lymphoma); lane 4, DLD1 (colon carcinoma). ABF-1 was detected by using a polyclonal antibody as described in Materials and Methods.
To confirm that ABF-1 bound to the E-box site, we performed an EMSA with a minimal oligonucleotide containing only the μE4 site. In the presence of either E12 or E47, ABF-1 could bind the E-box site as a heterodimer (Fig. 6B, lanes 6 and 9). However, ABF-1 could not bind as a homodimer to the E-box site (Fig. 6B, lane 5). As expected, the heterodimeric complexes could be supershifted with both an anti-FLAG and an anti-E2A antibody, confirming that ABF-1 with either E12 or E47 has the ability to bind as heterodimers to DNA (Fig. 6B, lanes 7, 8, 10, and 11). E2-2 and HEB are also able to interact with ABF-1 in the two-hybrid system as demonstrated above (Fig. 1). However, gel shift analysis failed to detect any binding of HEB- or E2-2–ABF-1 heterodimers to the μE4 site (not shown).
Since the μE4 and μE4-OCT oligonucleotide probes contain the same E-box element, we postulated that the octamer site may be involved in facilitating the binding of ABF-1 homodimers. To test if the octamer site is required, we generated a μE4-OCT mutant probe in which the octamer site has been destroyed. ABF-1 homodimers were equally capable of forming on both the wild-type and mutant μE4-OCT oligonucleotide probes, demonstrating that the octamer site is not required (38a).
ABF-1 forms heterodimers with E2A proteins in vivo.
To determine whether ABF-1 protein is present in nuclear extracts derived from B-cell lines, we generated a polyclonal antibody specific for ABF-1. Nuclear extracts derived from a human B-cell line, Namalwa, and two EBV-transformed LCLs were analyzed by immunoblotting. ABF-1 protein was detected in nuclear extracts derived from EBV-immortalized B-cell lines but not from the Burkitt lymphoma line Namalwa or the colon carcinoma line DLD1 (Fig. 7). Both in vitro-translated and endogenous ABF-1 proteins have an apparent molecular mass of 29 kDa, as revealed by Western blot analysis using an ABF-1-specific polyclonal antibody (Fig. 7 and data not shown). Therefore, as suggested by the Northern blot analysis described above, ABF-1 protein is expressed in EBV-transformed LCLs.
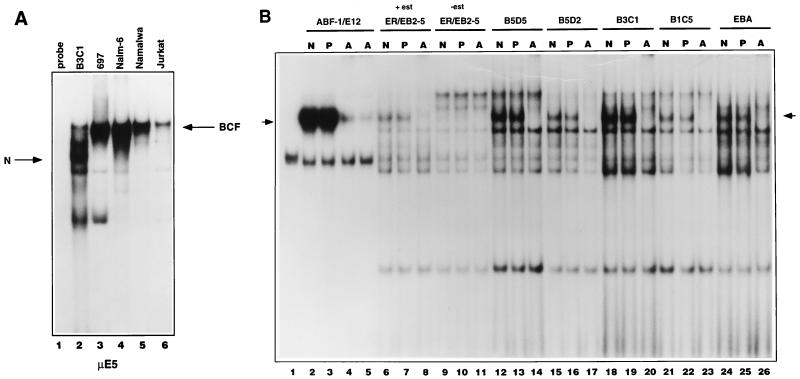
Previously, we have shown that two B-cell-specific μE5 binding complexes are present in nuclear extracts from a wide variety of pre-B- and mature B-cell lines, designated BCF1 and BCF2. The BCF1 and BCF2 complexes are composed mainly of homodimers of E2-2 and E47 in pre-B cells, whereas in mature B cells the binding complex consists predominantly of E47 homodimers (2, 44, 63). To determine whether ABF-1 DNA binding activity could be detected in vivo, we examined a number of nuclear extracts derived from various B-cell lines by EMSA using the μE5 site as a probe. B-cell-specific complexes BCF1 and BCF2 were detected in the pre-B line Nalm-6, the pre-B acute lymphoblastic leukemia (ALL) line 697, the mature B line Namalwa, and to a lesser degree the T-cell ALL line Jurkat as described previously (Fig. 8A) (2, 44). Interestingly, the LCL B3C1 extract contained a nucleoprotein complex of faster mobility not seen in the other extracts (Fig. 8A, lane 2). To determine if the novel complex was a common feature of EBV-immortalized lines, we analyzed a number of nuclear extracts that were derived from various EBV-transformed B-cell lines. Of the seven lines examined, all contained the unique complex, albeit at various levels (Fig. 8B and data not shown). This group also included the EBNA2 conditional cell line ER/EB2-5, an estrogen-dependent LCL (32). Complex formation was clearly dependent on the presence of β-estradiol in the culture medium and thus required the presence of EBNA2 (Fig. 8B; compare lanes 6 and 9).
FIG. 8.
ABF-1 is part of an E-box binding complex present in EBV-immortalized LCLs. (A) Gel shift analysis with a μE5 probe reveals a novel complex (N) whose mobility differs from that of BCF present in the EBV-immortalized line B3C1 (lane 2). (B) The ABF-1-containing nucleoprotein complex, indicated by arrows, was detected in all EBV-immortalized LCLs (seven cell lines in total were analyzed). Lane 1, unprogrammed reticulocyte lysate; lanes 2 to 5, in vitro-cotranslated ABF-1 and E12 proteins; lanes 6 to 11, nuclear extract derived from the conditional cell line ER/EB2-5 (32) grown in the presence of 1 μM β-estradiol (+est) or estrogen starved for 48 h (−est); lanes 12 to 26, nuclear extract isolated from independent LCLs. N, no antibody; P, preimmune serum; A, ABF-1-specific antiserum. The free probe was run off the gel.
Interestingly, this complex comigrated with an in vitro-translated heterodimer of ABF-1 and E12 (Fig. 8B; compare lane 2 with lanes 6, 12, 15, 18, 21, and 24). To assess whether this complex contained ABF-1, preimmune serum or ABF-1-specific antiserum was added to the reactions. The preimmune sera had no effect on the complex, but addition of anti-ABF-1 antibody completely eliminated complex formation (Fig. 8B). Furthermore, addition of an E2A-specific monoclonal antibody supershifted the complex, indicating that the complex is composed of an ABF-1–E2A heterodimer (38a). Taken together, these data indicate that ABF-1 forms heterodimers with the E2A gene products in EBV-transformed lymphoblastoid B-cell lines.
ABF-1 is a transcriptional repressor.
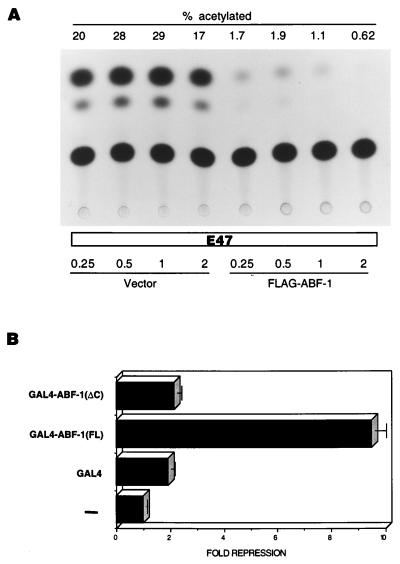
As shown above, ABF-1 has the ability to form heterodimers with E2A in vivo. This finding suggests that ABF-1 may be involved in modulating the transcriptional activity of E2A. We therefore checked whether coexpression of ABF-1 and E47 alters the transactivation properties of E47 in mammalian cells. An E47 expression vector was introduced into HeLa cells along with a CAT reporter gene driven by six copies of the μE5-μE2 E-box elements derived from the IgH gene enhancer (Fig. 9A) (27). Cotransfection of increasing amounts of a FLAG–ABF-1 expression plasmid resulted in a dramatic inhibition of E47-mediated transactivation (Fig. 9A). In fact, cotransfection of equal amounts of E47 and FLAG–ABF-1 expression vectors resulted in a 12-fold reduction of reporter gene activation (Fig. 9A). As expected, cotransfection of increasing amounts of empty expression vector had no effect on E47 transcriptional activity (Fig. 9A).
FIG. 9.
ABF-1 functions as a transcriptional repressor in mammalian cells. (A) ABF-1 interferes with the ability of E47 to activate transcription in mammalian cells. HeLa S3 cells were transiently transfected with 1 μg of (μE5-μE2)6CAT reporter plasmid, 0.25 μg of E47 expression vector (pHβA-E47), and increasing amounts (shown in micrograms) of empty expression vector (pHβAneo) or FLAG–ABF-1 expression vector (pHβA-FLAG-ABF-1) as indicated. The total amount of DNA used in each transfection experiment was adjusted to 5 μg by addition of pBSK; 30 μl of Superfect reagent was used for each transfection. Equal amounts of protein extracts were assayed for CAT activity. CAT activity is indicated by % acetylated, which represents the conversion of 14C-chloramphenicol to acetylated forms. (B) ABF-1 contains a transcriptional repression domain. HeLa S3 cells were transiently transfected as described above with 1 μg of 3XUASGALTK LUC reporter plasmid, 0.025 μg of pCMVβGAL, and 1 μg of the indicated GAL4 expression constructs. In the absence of effector plasmid, the 3XUASGALTK LUC reporter gene activity was 16,546 relative light units. Luciferase activity was normalized to β-galactosidase levels as a control for transfection efficiency.
To address the issue of how ABF-1 might inhibit E47-mediated transcriptional activation, we tested whether ABF-1 can function as a transcriptional repressor. The full-length ABF-1 protein [ABF1(FL)] was fused in-frame with the GAL4 DNA binding domain and introduced into HeLa cells with a luciferase reporter gene containing three GAL4 binding sites upstream of the thymidine kinase promoter (Fig. 9B) (26). This reporter, which is moderately active in HeLa cells, can be used to monitor transcriptional repression (26). The GAL4–ABF-1(FL) fusion was capable of repressing transcription ninefold (Fig. 9B). The deletion derivative GAL4–ABF-1(ΔC), which lacks the bHLH domain and C-terminal sequence, failed to repress the activity of the reporter gene (Fig. 9B). Therefore, ABF-1 contains a domain(s) present in the carboxy-terminal half of the protein which can function as a transcriptional repressor when fused to a heterologous DNA binding domain.
DISCUSSION
Transcription factors belonging to the bHLH family are known to regulate a variety of developmental programs, including neurogenesis, myogenesis, sex determination, and hematopoiesis (1a, 3, 40, 41, 45, 49, 57, 79). Many of these processes are controlled by a heterodimer composed of a tissue-restricted bHLH factor and an E protein. It appears, however, that B-lymphocyte development requires the activity of E2A homodimers (2, 3, 63, 79). Although formally possible, there has been a dearth of evidence supporting the notion of a B-cell-specific class II bHLH factor. In undertaking this study, we reasoned that B-lymphocyte development may parallel many other developmental processes in their requirement for a cell-type-specific bHLH protein.
We have named this gene ABF-1 because of its high expression levels in activated human B cells. ABF-1 expression was induced upon treatment of purified human B cells with the polyclonal activator SAC. SAC activates human B cells through cross-linking of the surface Ig (21, 54, 55, 60, 64). This finding suggests that ABF-1 may be a downstream target of the B-cell receptor signal transduction pathway and thus may be induced upon physiological encounter with antigen. The expression pattern in lymphoid tissues is consistent with a role in B-cell development. Significant levels of ABF-1 mRNA were found in the appendix and lymph node, sites where activated B cells are known to be localized. Since ABF-1 mRNA was also seen in fetal liver and bone marrow, however, we cannot rule out the possibility that ABF-1 also plays a role early in B-cell development. More extensive Northern blot analysis of poly(A)+ RNA derived from 50 different human tissues showed that low-level expression of ABF-1 was restricted to the lymph node and aorta (38a).
Intriguingly, the ABF-1 gene was highly expressed in a number of EBV-immortalized LCLs. These lines have many features in common with activated B lymphocytes, including increased cell size, proliferation, and expression of surface markers (8, 15, 53, 69, 73). Indeed, resting B cells stimulated by physiological agents or by EBV infection exhibit identical patterns of expression of several cell cycle control genes (28). Taken together, these data suggest that EBV infection of resting B cells may induce the normal activation program, thus bypassing the need for physiological signals (28). This may explain why ABF-1 mRNA is expressed in both LCLs and activated human B cells. Furthermore, it suggests that the ABF-1–E2A complex detected in nuclear extracts isolated from LCLs may also be present in physiologically activated B cells. In fact, a complex of similar mobility can be detected in extracts derived from human B cells activated with SAC and IL-2 (1). Surprisingly, the presence of EBV proteins is not sufficient to allow expression of ABF-1; all Burkitt lymphoma lines tested did not express ABF-1 mRNA.
A conditional LCL, ER/EB2-5, also contains high levels of ABF-1 transcripts. An ER-EBNA2 fusion protein renders this cell line dependent on estrogen for growth (32). Withdrawal of estrogen causes the cells to leave the cell cycle and arrest in G0 (32). Interestingly, expression of ABF-1 mRNA is also estrogen responsive. When ER/EB2-5 is estrogen starved, ABF-1 message becomes undetectable within 24 h (Fig. 4A and data not shown). This finding correlates well with the absence of ABF-1 transcripts in noncycling, resting human B cells. Upon addition of estrogen, ABF-1 mRNA becomes apparent within 10 h and is maintained at a steady state thereafter (38a). As expected from these observations, formation of an active ABF-1–E2A DNA binding complex in ER/EB2-5 also appears to be critically dependent on the presence of estrogen. Perhaps, then, further study of ABF-1 function in LCLs may lead to insights as to its role in physiologically activated B lymphocytes.
The E-box hexanucleotide sequence CANNTG has been identified in a number of cell-type-specific promoter and enhancer elements (40, 41). E boxes have long been implicated in the regulation of a number of B-lineage genes. For example, the IgH intronic enhancer, the 3′ IgH enhancer, and the kappa enhancer all contain E-box elements (47, 65). Mutational analysis has shown that the integrity of these elements is crucial for full transcriptional activity (65). We have shown here that ABF-1–E2A heterodimers are capable of binding at least two types of E-box sequences, the μE4 element (CACCTG) and μE5 (CAGGTG), thus raising the possibility that ABF-1 contributes to the regulation of Ig gene transcription.
It is not entirely clear why ABF-1 is capable of forming homodimers on the μE4-OCT probe but not on the μE4 probe. We conclude, however, that sequences flanking the core E-box element, CACCTG, present in the μE4-OCT oligonucleotide are important for ABF-1 homodimer formation. Clearly, binding site selection studies are necessary to address this issue further. Nevertheless, the ability of ABF-1 to bind DNA as a homodimer will have important implications for understanding its role in B-cell development.
After antigenic stimulation and in the presence of T-cell help, activated B lymphocytes can undergo the process of Ig isotype switching. Recently, it was shown by targeted deletion in mice that the 3′ IgH enhancer is required for the process of class switch recombination (CSR) (14). Interestingly, the 3′ IgH enhancer core contains an E-box element which has been shown to be important for enhancer activity (39). The switch regions present in the IgH gene consist of repetitive DNA elements in which recombination breakpoints lie (33, 76). It has been postulated that DNA binding factors may contribute to the generation of loop structures formed during the CSR process (33, 76, 77). The Sγ switch regions have been shown to contain E2-box-like elements to which a binding activity known as SNAP recognizes (37). It appears that E47 or a related factor is part of the SNAP hetero-oligomeric complex (37). Although it has yet to be demonstrated that these E-box elements are important for CSR, it is interesting to speculate that ABF-1 plays a role in this process. ABF-1 could participate in higher order nucleoprotein complexes postulated to regulate recombination within the switch regions.
Studies of IgE synthesis in human B cells have shown that two signals are required to mediate the process: an activation signal and the presence of a cytokine (13, 31, 70). For example, switching to IgE requires that the B cell be exposed to both mitogen and IL-4 (31, 70). Although IL-4 treatment alone induces germ line epsilon transcripts, Sμ/Sɛ deletional switch recombination does not occur and mature Cɛ mRNA is not produced (30, 61, 62). Interestingly, infection with EBV could provide the activation stimulus required for the switching event to take place (31, 61, 70). Perhaps ABF-1, which is known to be induced upon such a mitogenic signal, is involved in this development event.
We have demonstrated here that ABF-1 is capable of inhibiting E47-directed transcriptional activation from an E-box-containing reporter gene. Furthermore, we have shown that when tethered to DNA, ABF-1 strongly represses transcription. The ability of ABF-1 to repress transcription may be dependent on sequence context. The transcription factor Dorsal, for example, can function as either a transcriptional repressor or an activator in Drosophila (29, 68). Dorsal can activate transcription of the twist gene yet function as a repressor of the zen gene (29, 68). Dorsal can be converted into a transcriptional repressor by the protein DSP1, a member of the high-mobility-group protein family (36). Repression is dependent on a sequence element known as the negative regulatory element which is found adjacent to Dorsal binding sites in the zen promoter (18, 36). It is conceivable that the repressor activity of ABF-1 helps maintain the proliferative state of EBV-transformed LCLs and activated B cells. Recently, it has been shown that the cyclin-dependent kinase inhibitor, p21, contains E2A binding sites within its promoter element (50). E47 was shown to be capable of activating expression of a reporter gene driven by the p21 promoter element as well as activation of the endogenous p21 gene (50). This observation provides a possible model for growth inhibition conferred by overexpression of E2A, whereby increased expression of p21 interferes with cell cycle progression (50). The ability of ABF-1 to antagonize E47 transactivation suggests that ABF-1 may help promote proliferation of activated B cells and EBV-transformed LCLs by inhibiting E2A-directed p21 gene transcription. It will now be essential to generate mice lacking ABF-1 to determine its role in B-cell activation.
ACKNOWLEDGMENTS
We thank the following individuals for their contributions to this work: Ingrid Wolvers-Tettero for assistance with Northern blots; Maarten Stuiver and Gretchen Bain for valuable discussions; Barbara Kee for critical reading of the manuscript; Johan van Es for LCLs and advice; Bettina Kempkes for the conditional ER/EB2-5 cell line; Marieke Griffioen for the hEF-1α cDNA; Steve Elledge for two-hybrid reagents; and Michael G. Rosenfeld for the 3xUAS-TK LUC reporter plasmid.
This work was supported by the National Institutes of Health, the Council for Tobacco Research, and the Edward Mallinckrodt Jr. Foundation.
REFERENCES
- 1.Bain, G. Personal communication.
- 1a.Bain G, Engel I, Maandag E R, te Riele H, Voland J, Sharp L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in αβ T-cell development and to the rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain G, Gruenwald S, Murre C. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol Cell Biol. 1993;13:3522–3529. doi: 10.1128/mcb.13.6.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain G, Maandag E, Izon D, Amsen D, Kruisbeek A, Weintraub B, Krop I, Schlissel M, Feeney A, van Roon M, van der Valk M, te Riele H, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 4.Begley C, Aplan P, Denning S, Hayes B, Waldmann T, Kirsch I. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related motif. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell T, Weintraub H. Differences and similarities in DNA-binding preferences of myoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 6.Brands J, Maassen J, van Hemert F, Amons R, Moller W. The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur J Biochem. 1986;155:167–171. doi: 10.1111/j.1432-1033.1986.tb09472.x. [DOI] [PubMed] [Google Scholar]
- 7.Brennan T J, Olson E N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990;4:582–595. doi: 10.1101/gad.4.4.582. [DOI] [PubMed] [Google Scholar]
- 8.Calender A, Billaud M, Aubry J, Banchereau J, Vuillaume M, Lenoir G. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc Natl Acad Sci USA. 1987;84:8060–8064. doi: 10.1073/pnas.84.22.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudy M, Grell E H, Dambly-Chaudiere C, Ghysen A, Jan L Y, Jan Y N. The maternal sex determination gene daughterless has zygotic activity necessary for the formation of peripheral neurons in Drosophila. Genes Dev. 1988;2:843–852. doi: 10.1101/gad.2.7.843. [DOI] [PubMed] [Google Scholar]
- 10.Caudy M, Vaessin H, Brand M, Tuma R, Jan L Y, Jan Y N. daughterless, a gene essential for both neurogenesis and sex determination in Drosophila, has sequence similarities to myc and the achaete-scute complex. Cell. 1988;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 11.Church G M, Ephrussi A, Gilbert W, Tonegawa S. Cell type specific contacts to immunoglobulin enhancers in nuclei. Nature. 1985;313:798–801. doi: 10.1038/313798a0. [DOI] [PubMed] [Google Scholar]
- 12.Cline T W. The affairs of daughterless and the promiscuity of developmental regulators. Cell. 1989;59:231–234. doi: 10.1016/0092-8674(89)90280-8. [DOI] [PubMed] [Google Scholar]
- 13.Coffman R, Lebman D, Rothman P. Mechanism and regulation of immunoglobulin isotype switching. Adv Immunol. 1993;54:229–270. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 14.Cogne M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng H L, Alt F W. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Cordier M, Calender A, Billaud M, Zimber U, Rousselet G, Pavlish O, Banchereau J, Tursz T, Bornkamm G, Lenoir G. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cronmiller C, Schedl P, Cline T W. Molecular characterization of daughterless, a Drosophila sex determination gene with multiple roles in development. Genes Dev. 1988;2:1666–1676. doi: 10.1101/gad.2.12a.1666. [DOI] [PubMed] [Google Scholar]
- 17.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle H, Kraut R, Levine M. Spatial regulation of Zerknullt: a dorsal-ventral patterning gene in Drosophila. Genes Dev. 1989;3:1518–1533. doi: 10.1101/gad.3.10.1518. [DOI] [PubMed] [Google Scholar]
- 19.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A, Lee W-H, Elledge S. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 20.Ephrussi A, Church G M, Tonegawa S, Gilbert W. B-lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 21.Falkoff R, Zhu L, Fauci A. Separate signals for human B cell proliferation and differentiation in response to Staphylococcus aureus: evidence for a two-signal model of B cell activation. J Immunol. 1982;129:97–102. [PubMed] [Google Scholar]
- 22.Goldfarb A, Flores J, Lewandowska K. Involvement of the E2A basic helix-loop-helix protein in immunoglobulin heavy chain class switching. Mol Immunol. 1996;33:947–956. doi: 10.1016/s0161-5890(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 23.Gunning P, Leavitt J, Muscat G, Ng S, Kedes L. A human β-actin expression vector system directs high-level accumulation of anti-sense transcripts. Proc Natl Acad Sci USA. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 25.Hasty P, Bradley A, Morris J H, Edmondson D G, Venuti J M, Olson E N, Klein W H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 26.Heinzel T, Lavinsky R, Mullen T, Soderstrom M, Laherty C, Torchia J, Yang W, Brard G, Ngo S, Davie J, Seto E, Eisenman R, Rose D, Glass C, Rosenfeld M. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 27.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/κE2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 28.Hollyoake M, Stuhler A, Farrell P, Gordon J, Sinclair A. The normal cell cycle activation program is exploited during the infection of quiescent B lymphocytes by Epstein-Barr virus. Cancer Res. 1995;55:4784–4787. [PubMed] [Google Scholar]
- 29.Ip Y, Kraut R, Levine M, Rushlow C. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell. 1991;64:439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- 30.Jabara H, Ahern D, Vercelli D, Geha R. Hydrocortisone and IL-4 induce IgE isotype switching in human B cells. J Immunol. 1991;147:1557–1560. [PubMed] [Google Scholar]
- 31.Jabara H, Schneider L, Shapira S, Alfieri C, Moody C, Kieff E, Geha R, Vercelli D. Induction of germ-line and mature C epsilon transcripts in human B cells stimulated with rIL-4 and EBV. J Immunol. 1990;145:3468–3473. [PubMed] [Google Scholar]
- 32.Kempkes B, Spitkovsky D, Jansen-Durr P, Ellwart J, Kremmer E, Delecluse H-J, Rottenberger C, Bornkamm G, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenter A, Wuerffel R, Sen R, Jamieson C, Merkulov G. Switch recombination breakpoints occur at nonrandom positions in the S gamma tandem repeat. J Immunol. 1993;151:4718–4731. [PubMed] [Google Scholar]
- 34.Kuo S S, Mellentin J D, Copeland N G, Gilbert D J, Jenkins N A, Cleary M L. Structure, chromosome mapping, and expression of the mouse Lyl-1 gene. Oncogene. 1991;6:961–968. [PubMed] [Google Scholar]
- 35.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by neuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 36.Lehming N, Thanos D, Brickman J, Ma J, Maniatis T, Ptashne M. An HMG-like protein that can switch from a transcriptional activator to a repressor. Nature. 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 37.Ma L, Hu B, Kenter A. Ig S gamma-specific DNA binding protein SNAP is related to the helix-loop-helix transcription factor E47. Int Immunol. 1997;9:1021–1029. doi: 10.1093/intimm/9.7.1021. [DOI] [PubMed] [Google Scholar]
- 38.Massari M, Jennings P, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Massari, M. E., and C. Murre. Unpublished observations.
- 39.Meyer K, Skogberg M, Margenfeld C, Ireland J, Pettersson S. Repression of the immunoglobulin heavy chain 3′ enhancer by helix-loop-helix protein Id3 via a functionally important E47/E12 binding site: implications for developmental control of enhancer function. Eur J Immunol. 1995;25:1770–1777. doi: 10.1002/eji.1830250643. [DOI] [PubMed] [Google Scholar]
- 40.Murre C, Bain G, v. Dijk M A, Engel I, Furnari B A, Massari M E, Matthews J R, Quong M W, Rivera R R, Stuiver M H. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 41.Murre C, Baltimore D. The helix-loop-helix motif: structure and function. Vol. 2. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 42.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 43.Murre C, Mccaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 44.Murre C, Voronova A, Baltimore D. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defects. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 46.Naya F, Huang H-P, Qiu Y, Mutoh H, DeMayo F, Leiter A, Tsai M-J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelsen B, Sen R. Regulation of immunoglobulin gene transcription. Int Rev Cytol. 1992;133:121–149. doi: 10.1016/s0074-7696(08)61859-8. [DOI] [PubMed] [Google Scholar]
- 48.Peltenburg L T C, Murre C. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 1996;15:3385–3393. [PMC free article] [PubMed] [Google Scholar]
- 49.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt F, Orkin S. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 50.Prabhu S, Ignatova A, Park S, Sun X. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quong M W, Massari M E, Zwart R, Murre C. A new transcriptional activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts V J, Steenbergen R, Murre C. Localization of E2A mRNA expression in developing and adult rat tissues. Proc Natl Acad Sci USA. 1993;90:7583–7587. doi: 10.1073/pnas.90.16.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers R, Strominger J, Speck S. Epstein-Barr virus in B lymphocytes: viral gene expression and function in latency. Adv Cancer Res. 1992;58:1–26. doi: 10.1016/s0065-230x(08)60288-2. [DOI] [PubMed] [Google Scholar]
- 54.Romagnani S, Amadori A, Giudizi M, Biagiotti R, Maggi E, Ricci M. Different mitogenic activity of soluble and insoluble staphylococcal protein A (SPA) Immunology. 1978;35:471–478. [PMC free article] [PubMed] [Google Scholar]
- 55.Romagnani S, Giudizi M, Biagiotti R, Almerigogna F, Maggi E, Prete G D, Ricci M. Surface immunoglobulins are involved in the interaction of protein A with human B cells and in the triggering of B cell proliferation induced by protein A-containing Staphylococcus aureus. J Immunol. 1981;127:1307–1313. [PubMed] [Google Scholar]
- 56.Rudnicki M, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to an up-regulation of the myogenic HLH gene Myf5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 57.Rudnicki M, Schnegelsbery P, Stead R, Braun T, Arnold H, Jaenisch R. MyoD or Myf5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual, 2 ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sawada S, Littman D R. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuurman R, Gelfand E, Dosch H-M. Polyclonal activation of human lymphocytes in vitro. I. Characterization of the lymphocyte response to a T cell-independent B cell mitogen. J Immunol. 1980;125:820–826. [PubMed] [Google Scholar]
- 61.Shapira S, Jabara H, Thienes C, Ahern D, Vercelli D, Gould H, Geha R. Deletional switch recombination occurs in interleukin-4-induced isotype switching to IgE expression by human B cells. Proc Natl Acad Sci USA. 1991;88:7528–7532. doi: 10.1073/pnas.88.17.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shapira S, Vercelli D, Jabara H, Fu S, Geha R. Molecular analysis of the induction of immunoglobulin E synthesis in human B cells by interleukin 4 and engagement of CD40 antigen. J Exp Med. 1992;175:289–292. doi: 10.1084/jem.175.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen C P, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shokri F, Mageed R, Maziak B, Jefferis R. Expression of VHIII-associated cross-reactive idiotype on human B lymphocytes. Association with Staphylococcal protein A binding and Staphylococcus aureus Cowan I stimulation. J Immunol. 1991;146:936–940. [PubMed] [Google Scholar]
- 65.Staudt L, Lenardo M. Immunoglobulin gene transcription. Annu Rev Immunol. 1991;9:373–398. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- 66.Sun X-H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 67.Sun X-H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 68.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 69.Thorley-Lawson D, Mann K. Early events in Epstein-Barr virus infection provide a model for B cell activation. J Exp Med. 1985;162:45–59. doi: 10.1084/jem.162.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thyphronitis G, Tsokos G, June C, Levine A, Finkelman F. IgE secretion by Epstein-Barr virus-infected purified human B lymphocytes is stimulated by interleukin 4 and suppressed by interferon gamma. Proc Natl Acad Sci USA. 1989;86:5580–5584. doi: 10.1073/pnas.86.14.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Visvader J, Begley C, Adams J. Differential expression of the LYL, SCL and E2A helix-loop-helix genes within the hemopoietic system. Oncogene. 1991;6:187–194. [PubMed] [Google Scholar]
- 72.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang F, Gregory C, Rowe M, Rickinson A, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci USA. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 75.Whelan J, Cordle S R, Henderson E, Weil P A, Stein R. Identification of a pancreatic β-cell insulin gene transcription factor that binds to and appears to activate cell-type-specific gene expression: its possible relationship to other cellular factors that bind to a common insulin gene sequence. Mol Cell Biol. 1990;10:1564–1572. doi: 10.1128/mcb.10.4.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wuerffel R, Jamieson C, Morgan L, Merkulov G, Sen R, Kenter A. Switch recombination breakpoints are strictly correlated with DNA recognition motifs for immunoglobulin S gamma 3 DNA-binding proteins. J Exp Med. 1992;176:339–349. doi: 10.1084/jem.176.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wuerffel R, Kenter A. Protein recognition motifs of S gamma 3 DNA are statistically correlated with switch recombination breakpoints. Curr Top Microbiol Immunol. 1992;182:149–156. doi: 10.1007/978-3-642-77633-5_18. [DOI] [PubMed] [Google Scholar]
- 78.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]