Short abstract
Recently, ideas from the field of ontology have been picked up by computer scientists as a basis for encoding knowledge and with the hope of achieving interoperability and intelligent system behavior. The use of anatomy ontologies to represent space in biological organisms, specifically mouse and human are reviewed here.
Abstract
Ontology has long been the preserve of philosophers and logicians. Recently, ideas from this field have been picked up by computer scientists as a basis for encoding knowledge and with the hope of achieving interoperability and intelligent system behavior. In bioinformatics, ontologies might allow hitherto impossible query and data-mining activities. We review the use of anatomy ontologies to represent space in biological organisms, specifically mouse and human.
Ontologies and biology
Biological science is a knowledge-intensive discipline. To become expert in any field in biology requires an extensive apprenticeship and a long experience in the field. Use of bioinformatic resources often requires similar expertise, and having both together is rare within a research group let alone in an individual. Ontologies are emerging as the key mechanism for encoding structured knowledge, and when used in the context of resources such as bioinformatics databases they open the possibility for more automated use of biological data.
Traditionally a subject of study in philosophy, ontologies are now a key topic for the development of the semantic web [1] - the next generation of the worldwide web - as well as for the semantic grid [2]. Here the term 'grid' refers to the extension of the more familiar worldwide web to include complex high-performance computing, databases and collaborative virtual organizations; and 'semantic' indicates that this next generation of the web will include structure that will convey meaning, rather than an amorphous mass of information. See Box 1 for a glossary of terms. The promise of semantic infrastructures lies in the automation they would allow. But for bioinformatics services to become automated, the knowledge that is to be used must be formalized and represented in a computationally accessible form. The aim of ontology research has therefore been to develop knowledge representations that can be shared and reused by machines as well as people; a modern definition is: "an ontology is a formal, explicit specification of a shared conceptualization" [3]. The constitution of an ontology is widely debated, however. For our purposes, we take the pragmatic view that an ontology is a structured and clearly defined encapsulation of knowledge about a field that can be used for annotation and reasoning within that domain of knowledge.
Box 1.

Although some of the conceptualization that is represented by an ontology will be independent of the domain of knowledge that is being considered - as exemplified by the Dublin Core Metadata Initiative, which provides "an open forum engaged in the development of interoperable online metadata standards that support a broad range of purposes and business models" [4] - domain-specific ontologies are needed to support particular areas, such as bioinformatics. In this context, the best known ontology is the gene ontology, GO, developed by the Gene Ontology Consortium [5], which describes molecular functions, biological processes and cell components. Various other bio-ontologies, including some for anatomy, can be found on the Open Biological Ontologies (OBO) website [6]. Under the umbrella of the group Standards and Ontologies for Functional Genomics (SOFG), a community effort is under way to integrate human and mouse anatomy ontologies [7]. Our experience is in the development of an anatomical ontology for the mouse, as part of a project to develop a database of mouse anatomy and gene expression [8], and it is to this example that we return throughout this article.
The representation of these ontologies varies greatly, ranging from fairly simple lists to complex structures expressed in specific ontology languages, such as OWL [9]. And tools have been created to support the development and management of ontologies; examples include OilEd, OntoEdit and Protege2000 (for a brief survey, see [10]). There are also bioinformatics-specific tools, such as DAG-Edit, COBrA and AmiGO (all described on the GO website [5]). An important goal for any ontology is standardization, at the syntactic as well as the semantic level. For computational systems to interact effectively, everyone concerned must agree on the representation and meaning of the concepts that form part of the computational interaction.
The basic components of an ontology are terms or symbols (usually words) that represent concepts plus the links or relationships between these terms. In a biological ontology each term represents a biological concept, such as 'heart' or 'branchial arch', in symbolic form; all specific examples of that concept - such as a real heart in a specific mouse - are instances of that concept. Terminologically we say that each example heart is an instance of the heart class as denoted by the ontological symbol 'heart'. Links then define relationships between terms that can allow inference or reasoning to generate a new relationship that is not directly represented in the ontology. In anatomical ontologies the two most common relationships are 'part-of' and 'type-of'. Both these relations are transitive: so, for example, if A is part-of B and B is part-of C then A is part-of C. In addition, both are directional and are said to be non-reflexive: in general, if A is part-of B then it is not true that B is part-of A. Directional or non-reflexive relationships are described as directed, so that if the set of terms is depicted graphically then the part-of links will generate a part-of hierarchy, also called a 'partonomic' hierarchy and the type-of link will generate a 'class' hierarchy. The term 'hierarchy' here refers to the fact that a concept may have several other concepts as its parts, and in turn these concepts may consist of a number of further concepts, and so on; similarly type-of links can be hierarchical. In most cases each anatomical term may be part of more than one parent structure and the resultant graph is termed a directed acyclic graph (DAG). Figure 1 shows a simple example of this from GO.
Figure 1.
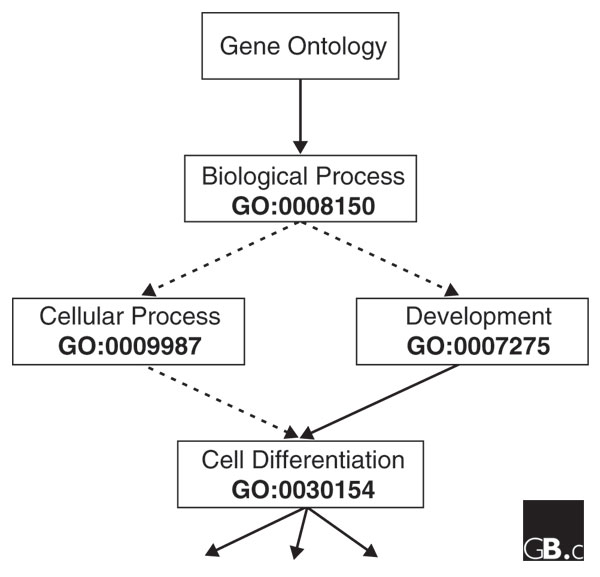
An example of a directed acyclic graph (DAG) taken from the gene ontology (GO). The solid arrows indicate the GO 'part-of' link and the dashed arrows the GO 'is-a' link. The GO unique identifiers (IDs) are printed below each term. The term 'Cell Differentiation' has two parents (Cellular Process and Development), which in turn link back to the same antecedent 'Biological Process' which is part-of the Gene Ontology. The unterminated arrows leading from Cell Differentiation indicate that it has a number of offspring terms.
Anatomy: parts and types
The formal study of anatomy is declining as an academic discipline. But with the development of atlas-type databases as reference frameworks for biomedical research, anatomy is witnessing a renaissance as attempts are made to capture the concepts of anatomy for use in database systems. Sets of anatomical terms have appeared in many 'ontologies' (see the SOFG website [1]). The purpose of these is to provide a controlled vocabulary for annotation and referencing and to capture anatomical relationships and knowledge. But, even within a single domain of knowledge, such as mouse embryonic development, there could be many possible ontologies, capturing the anatomy in different ways and with different interpretations for the same symbol. In Figure 2 these are represented by column (a) with an example from the Edinburgh Mouse Atlas [8]. Each ontology may have its own definitions in text or relationship terms and may also have a graphical representation.
Figure 2.
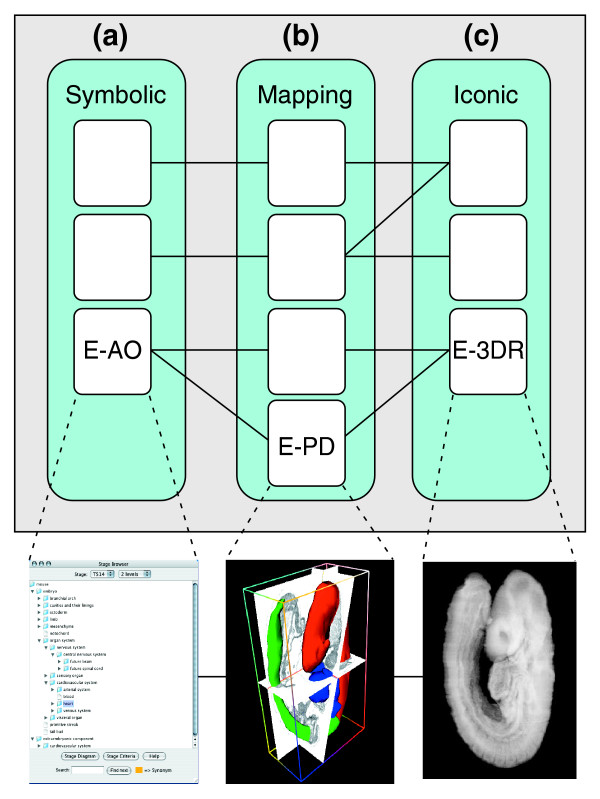
Extending the scope of an ontology. (a) Current anatomical ontologies are purely symbolic, providing a structured collection of terms each corresponding to a particular anatomical concept. An example is the EMAP Anatomy Ontology E-AO [8]. Symbolic ontologies define relationships such as 'part-of', 'is-a' or 'derives-from' (denoting a lineage). Ontologies with extended scope include graphical mapping (b) and iconic (c) representations; examples are the EMAP Painted domains (E-PD) and EMAP 3D Reconstructions (E-3DR) ontologies, respectively, from which the illustrations in (b,c) are taken. The lines between columns represent links, or mappings, between the concept symbols and other representations. A completely iconic representation of the embryo and, implicitly, of the corresponding anatomy is the reconstruction of the embryo as a three-dimensional grey-level voxel model (c) with a fully defined geometric space. This includes additional geometric and topological relationships such as 'volume', 'connected to', 'next-to', 'distance-from', and so on. The middle column (b) represents the step between concept and geometric space reconstruction and is an image representation we define in the same coordinate frame as the embryo reconstructions.
The graphical form, illustrated by column (b) in Figure 2, may also have a number of representations, but most importantly may include alternative views of the underlying concepts. This brings to the fore a critical development of the notion of what constitutes an ontology. By definition an ontology should be consistent, but here we try to capture alternative views of the underlying terms, so we need to build in inconsistency. Consistency is of course rescued by subdividing the concept into separate classes, such as 'hindbrain-expert-1' and 'hindbrain-expert-2' to denote views from two researchers, but the idea is to capture the current state of knowledge, which will evolve as understanding changes. At this point the ontology is almost a database. The ontology forms part of the theoretical framework for the field [11] and what was experimental data at one stage will be part of the current model or theory at a later stage.
The graphical representation is an extension of the definition of a concept to a graphical form. This definition may, however, be in terms of a particular individual. For example, in the case of the Mouse Atlas the graphical representation is part or all of a mouse embryo. The representation may be from a single animal or may be synthesized and averaged from a group of individuals. Either way, there is selection of a representative model within which the ontological concepts can be interpreted. The graphical representations of the parts is usually referred to as an atlas. Of course, there could be many such atlases, as indicated by column (c) in Figure 2. An atlas, therefore, consists of at least three parts, an ontology of terms (sometimes implicit, for example in the case of a list of countries, which need not be provided as an actual list but can still serve as one), a representative individual example on which to define the spatial extent and coordinates (which may include time), and a mapping, or interpretation, between the two.
A simple example of an anatomy ontology is the one we have developed as part of the Edinburgh Mouse Atlas Project (EMAP) [8,11-13]. This ontology is designed to capture the structural changes that occur during embryonic development and consists of a set of 26 hierarchies, one for each developmental stage, where a stage is characterized by the internal and external morphological features of an embryo recognizable during that period of development (as defined by Theiler [14]). The ontology can be displayed as a set of hierarchical trees, with each term subdivided into its constituent parts. There is no requirement that each anatomical term is divided into non-overlapping structures, or that each component has only one parent, so the ontology can be represented as a DAG. Each node represents the biological concept, such as heart, at that particular time. Many of the terms and structures are repeated at each stage and it is possible to collapse the set of terms onto a single large hierarchy that includes all of the terms from all stages. This large DAG is stage-independent (with a few exceptions) and is referred to as the 'abstract-mouse'; terms within the DAG now represent the biological concepts for all stages. Within the EMAP database the abstract mouse and stage terms can be independently referenced via unique identifiers. In addition, EMAP can include a 'derived-from' link as a putative lineage relationship between tissues. These link the stage-specific components so that it becomes possible to query the derivation (and destination) of any given tissue.
An anatomy ontology for the adult mouse that is compatible with the EMAP ontology has been developed for the Mouse Genome Informatics (MGI) databases at the Jackson Laboratory, USA [15]. A similar ontology was designed for human developmental anatomy [16], building on the work carried out by EMAP. Ontologies for adult human anatomy have been created as part of two projects, the General Architecture for Languages, Encyclopedias and Nomenclatures in Medicine (GALEN) [17] and the Digital Anatomist's Foundational Model (FMA) [18] projects. GALEN provides an ontology aimed at clinical applications, contains more than 10,000 anatomical concepts and uses the description logic language GRAIL (GALEN Representation and Integration Language) for representation. Relationship types between concepts are defined, including, for example, 'part-of', 'branch-of', 'contains' and 'connects'. Unlike the EMAP developmental anatomy, GALEN subdivides 'part-of into a number of different partonomic relationships. (A review of 10 years of experience developing GALEN has been published [19].) On the basis of work on the FMA, Rosse and Mejino [20] provide a comprehensive discussion of the ontological issues involved with developing an anatomical nomenclature. The FMA [18] uses a set of well defined principles and structures provided by Protégé-2000, a software tool for the creation of knowledge-based systems, developed by Stanford University [21]. As in the case of GALEN, the FMA not only supports the basic relationships of 'part-of and 'type-of, but also further subdivides these.
Although GALEN and FMA cover the same domain of knowledge, namely human adult anatomy, attempts to develop methods to align the two ontologies have enabled no more than 7% of FMA's and 17% of GALEN's concepts to be matched [22]. This should not be too surprising, however, considering that the creation of such ontologies not only requires the identification and naming of the concepts involved, but also often includes the identification of a set of attributes and a general definition describing the properties of these concepts. In addition, the relationships between concepts and rules for the propagation of properties need to be determined. Where all these activities are carried out independently by two groups, one should indeed expect to find significant differences - reflecting the purpose and expertise of each group - in the ontologies.
Whereas FMA and GALEN are text-based, Höhne et al. [23], within their Voxel-Man system of graphical human representation, have pioneered the use of sophisticated three-dimensional graphics and rendering to provide visual and interactive access to an atlas of anatomy including links to microscopic and functional data. (A voxel is the three-dimensional volume equivalent to a two-dimensional pixel.) Schubert and Höhne [24] discuss the specific challenges this has provided in terms of an anatomical partonomic hierarchy. As is the case for GALEN, they determine that certain properties can only be propagated along particular relationships and that this depends both on the nature of the data - they have microscopic, topographical, and functional information - and the type of part-of relationship. They use the six basic types of part-of relationships, developed by Gerstl and Pribbenow [25], extended to include a notion of topographical relationship, such as containment. Knowledge representation within the Voxel-Man system has similarities to the model presented in Figure 2. Its semantic network corresponds to a symbolic representation (Figure 2, column (a)) in our model view, and its image volume can be seen as an iconic representation (Figure 2, column (b)), whereas other attribute volumes are similar to the mappings discussed earlier. In our model, however, we recognize not only the possibility of multiple mappings but also the existence of multiple symbolic and iconic representations and the additional links across representations that follow from that.
An ontology that encompasses both the spatial mapping aspects discussed here (in two dimensions) and the notion of alternative interpretations of the 'same' term is provided by the Brainlnfo atlas [26]. Here, the authors have collated anatomical terms from a number of published brain atlases for mammalian brains, principally primate but with reference to rat and mouse; they provide a tool for navigating either via ontological terms or via location on standard views of the brain.
So far we have discussed anatomies that are expressed in the form of an ontology. Of course other sets of anatomical terms exist. The most methodical and complete is the Terminalogica Anatomica (formerly Anatomica Nomina) developed over many years by the Federative Committee on Anatomical Terminology (FCAT) [27]. This is an unstructured list, not in an open electronic form and is not widely used - so, for bioinformatics purposes it is not useful except as a set of reference terms. More structured and available is the Unified Medical Language System (UMLS) which provides a standardized set of terms, particularly with respect to medical and clinical terminology. As with other anatomies, however, it is not easy to use outside of the tools provided.
The ontologies discussed so far together undoubtedly provide an exhaustive set of terms that will, in principle, cover all bioinformatic requirements for a reference anatomy with a set of relationships to allow reasoning about anatomy and function. But, so far, the terms are not used anywhere except within the domains of application for which they were developed, unlike the Gene Ontology (GO) which has rapidly found widespread use. Why should this be the case? The answer seems to be partly accessibility and partly community. Useful ontologies must be easy to pick up and reuse and must include a sense that anybody with expertise can contribute. In addition, for many applications the complexity is a barrier. An example of an attempt to break down such barriers is the Standard Anatomy Entry List (SAEL) (see [7]) which is a small, unstructured list of anatomical terms, useful in particular for annotating genomic and proteomic data from gene-expression microarrays and serial analysis of gene expression (SAGE). Each of the terms in the SAEL will be mapped to the corresponding terms in the more detailed anatomy ontologies. Simplicity and accessibility are provided while retaining the links to more complex ontologies that can provide sophisticated reasoning capability.
Towards the next generation of anatomy ontologies
In this article we have discussed anatomy and how emerging ontologies are attempting to capture not only structural knowledge of anatomy but also some of the functional and spatial relationships between tissues. There are, however, some omissions in these attempts to formalize anatomical knowledge. The first is that they are only just beginning to become community enterprises that not only admit submissions from all parts of a scientific community but also allow alternative views of what purport to be the same biological concepts. How do we capture this knowledge? The task is large but no funds are available for bringing together the necessary expertise into a single project. A more plausible model is provided by the open-source software mechanism, which relies on contributions from committed experts in a distributed and altruistic fashion. In many cases the people collaborating will never meet. We need mechanisms to support such virtual organizations.
The second omission is that existing anatomy ontologies are basically about known concepts and are very limited for properties that are poorly expressed in words. A good example of such a property is geometry. The existing ontologies can to some extent encode something of the topological relationships - adjacency, overlap and enclosure - but are not useful for encoding distance, direction and spatial measures. For a proper understanding and modeling of development, as well as the simple capture of data such as phenotype, geometry is critical. To include geometry implies a representation of an 'individual' or standard specimen. This defines a real geometric space and the anatomical concepts can then be mapped into that space. In terms of a framework of understanding, the natural way to think of this is as an extension of the ontology to include geometry. Interestingly, informal feedback from a group of graduate students at the Human Genetics Unit in Edinburgh suggests that they found it perfectly natural to consider the geometric atlas with its associated anatomical domains linked to an anatomical nomenclature to be an ontology. Extending ontologies in a natural way to include more iconic forms of information is required.
A third omission, related to the other forms of information that are discussed above, is the issue of uncertainty. All scientific reasoning is ultimately based on an understanding of uncertainty. We need to manage and reason with uncertainty. It is clear that probability is the right language [28], but how do we merge this with the current logical approaches to ontologies? Finally, this discussion of anatomy has been founded on the underlying understanding of anatomy in the context of structure visualized by traditional dissection and histology. We now have a much more informative view of an organism's internal organization by looking at genetic activity. Now the 'structure' is also found in the high-dimensional gene-expression space, and the developmental trajectory is not only through the geometric space and time of the embryo but also through this 'gene space'. In spatiotemporal coordinates we know that the cellular trajectory is connected, since every cell has a parent. What do such paths or trajectories look like in gene-space? What can be considered 'close' in the 30,000-dimensional space of gene expression? These are questions to be answered as the structural view evolves to encompass the informational anatomy of gene expression and not just the morphological and functional anatomy derived from standard histology.
We are in need of a new generation of ontologies that go beyond the current preoccupation with predicate logic and expand into other representations of knowledge. This has echoes in many areas of understanding in science and touches on the basic meaning of scientific inference and scientific 'truth', an open philosophical debate that now has practical importance in the issue of encoding our current beliefs, even in such away as to allow limited reasoning capability within a highly constrained system. The attempt to make computers more useful in a practical sense is forcing to the foreground the basic meaning of biological knowledge and how can it be used computationally.
References
- Berners-Lee T, Hendler J, Lassila O. The semantic web. Sci Am Digital. 2001;284:34–43. [Google Scholar]
- de Roure D, Jennings N, Shadbolt N. The semantic grid: a future e-science infrastructure. In: Berman F, Fox G, Hey A, editor. Grid Computing - Making the Global Infrastructure a Reality. Hoboken NJ: John Wiley; 2003. pp. 437–470. [Google Scholar]
- Gruber T. A translation approach to portable ontology specifications. Knowledge Acquisition. 1993;5:199–220. doi: 10.1006/knac.1993.1008. [DOI] [Google Scholar]
- Dublin Core Metadata Initiative http://www.dublincore.org
- Gene Ontology http://www.geneontology.org
- Open Biological Ontologies http://obo.sourceforge.net
- SOFG - Standards and Ontologies for Functional Genomics http://www.sofg.org
- Edinburgh Mouse Atlas Project http://genex.hgu.mrc.ac.uk/
- World Wide Web Consortium (W3C) http://www.w3.org/
- Fensel D. Ontologies: A Silver Bullet for Knowledge Management and Electronic Commerce. Berlin: Springer; 2001. [Google Scholar]
- Davidson D, Baldock R. Bioinformatics beyond sequence: mapping gene function in the embryo. Nat Rev Genet. 2001;2:409–418. doi: 10.1038/35076500. [DOI] [PubMed] [Google Scholar]
- Baldock R, Bard J, Kaufman M, Davidson D. A real mouse for your computer. BioEssays. 1992;14:501–502. doi: 10.1002/bies.950140713. [DOI] [PubMed] [Google Scholar]
- Burger A, Davidson D, Baldock R. Formalization of mouse embryo anatomy. Bioinformatics. 2004;20:259–267. doi: 10.1093/bioinformatics/btg400. [DOI] [PubMed] [Google Scholar]
- Theiler K. The House Mouse. New York: Springer; 1989. [Google Scholar]
- Mouse Genome Informatics (MGI) http://www.informatics.jax.org/
- Hunter A, Kaufman MH, McKay A, Baldock R, Simmen MW, Bard JBL. An ontology of human developmental anatomy. J Anat. 2003;203:347–355. doi: 10.1046/j.1469-7580.2003.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OpenGalen http://www.opengalen.org
- Foundational Model of Anatomy http://sig.biostr.washington.edu/projects/fm
- Rogers J, Roberts A, Solomon D, van der Haring E, Wroe C, Zanstra P, Rector A. GALEN ten years on: tasks and supporting tools. MEDINFO. 2001;10:256–260. [PubMed] [Google Scholar]
- Rosse C, Mejino J. A reference ontology for biomedical informatics: the foundational model of anatomy. Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Protégé http://protege.stanford.edu
- Zhang S, Mork P, Bodenreider O. Lessons learned from aligning two representations of anatomy. In: Hahn U, editor. Proceedings of First International Workshop on Formal Biomedical Knowledge Representation. Aachen: Technical University of Aachen; 2004. pp. 102–108. [Google Scholar]
- Höhne KH, Pflesser B, Pommert A, Riemer M, Schiemann T, Schubert R, Tiede U. A new representation of knowledge concerning human anatomy and function. Nat Med. 1995;1:506–511. doi: 10.1038/nm0695-506. [DOI] [PubMed] [Google Scholar]
- Schubert R, Höhne KH. Partonomies for interactive explorable 3D-models of anatomy. In: Chute CG, editor. A Paradigm Shift in Health Care Information Systems: Clinical Infrastuctures for the 21st Century Proceedings 1998, AMIA Annual Fall Symposium. Orlando FL: American Medical Informatics Association; 1998. pp. 433–437. [PMC free article] [PubMed] [Google Scholar]
- Gerstl P, Pribbenow S. Midwinters, end games, and body parts: a classification of part-whole relations. Int J Hum-Comput Stud. 1995;43:865–889. doi: 10.1006/ijhc.1995.1079. [DOI] [Google Scholar]
- BrainInfo http://braininfo.rprc.washington.edu/
- Federative Committee on Anatomical Terminology . Terminologica Anatomica. Stuttgart: Thieme; 1998. [Google Scholar]
- Jaynes ET. Probability Theory: The Logic of Science. Cambridge: Cambridge University Press; 2003. [Google Scholar]


