Abstract
A biologically important human gene, cyclooxygenase-2 (COX-2), has been proposed to be regulated at many levels. While COX-1 is constitutively expressed in cells, COX-2 is inducible and is upregulated in response to many signals. Since increased transcriptional activity accounts for only part of the upregulation of COX-2, we chose to explore other RNA processing mechanisms in the regulation of this gene. We performed a comprehensive bioinformatics survey, the first of its kind known for human COX-2, which revealed that the human COX-2 gene has alternative polyadenylation (proximal and distal sites) and suggested that use of the alternative polyadenylation signals has tissue specificity. We experimentally established this in HepG2 and HT29 cells. We used an in vivo polyadenylation assay to examine the relative strength of the COX-2 proximal and distal polyadenylation signals, and have shown that the proximal polyadenylation signal is much weaker than the distal one. The efficiency of utilization of many suboptimal mammalian polyadenylation signals is affected by sequence elements located upstream of the AAUAAA, known as upstream efficiency elements (USEs). Here, we used in vivo polyadenylation assays in multiple cell lines to demonstrate that the COX-2 proximal polyadenylation signal contains USEs, mutation of the USEs substantially decreased usage of the proximal signal, and that USE spacing relative to the polyadenylation signal was significant. In addition, mutation of the COX-2 proximal polyadenylation signal to a more optimal sequence enhanced polyadenylation efficiency 3.5-fold. Our data suggest for the first time that alternative polyadenylation of COX-2 is an important post-transcriptional regulatory event.
INTRODUCTION
Cyclooxygenases (COX) are the key and rate-limiting enzymes in the production of prostaglandins [reviewed in (1) and references therein]. The first steps in prostanoid synthesis are the release of arachidonic acid from membrane phosopholipids by phospholipases and conversion to prostaglandin H2 by COX. Prostaglandins play a role in many biological processes, including but not limited to inflammation, bone formation, wound healing and pain perception. Inflammatory cells as well as other types of cells, including fibroblasts and epithelial cells produce prostaglandins (2,3).
Two separate COX genes have been identified, COX-1 and COX-2 (4–10). A spliced variant of COX-1 (COX-3) has also been identified (11). The proteins that these genes encode are ∼60% identical at the amino acid level. In contrast, the 3′-untranslated regions (3′-UTRs) of COX-1 and COX-2 are highly divergent. The most striking difference between these genes is in their regulation of expression; COX-1 is constitutively expressed while COX-2 is strongly induced in response to activation by hormones, pro-inflammatory cytokines, growth factors, oncogenes, carcinogens and tumor promoters (1–3,12–15).
The physiological or pathological outcomes of COX-2 activity depend upon its level of expression. COX-2 overexpression is associated with a number of conditions, including cancer, rheumatoid arthritis, seizures and inflammatory disorders [reviewed in (1,16–22)]. In addition, COX-2 upregulation contributes to pain; indeed, inhibition of COX-2 enzymatic activities is responsible for the anti-inflammatory properties of aspirin, indomethacin, ibuprofen and related NSAIDs, such as Vioxx (Rofecoxib) and Celebrex (Celecoxib). Molecular events leading to overexpression of COX-2 have not been definitively characterized. Some studies have clearly demonstrated increased levels of COX-2 mRNA in colorectal adenomas, colon cancer cell lines, adenocarcinomas, gastric cancer, breast cancer, certain ovarian and prostate cancers, and non-small lung cancer (22–33). Enhanced COX-2 mRNA transcription may play a role but enhanced COX-2 protein expression most probably requires post-transcriptional gene regulation events, such as are mediated through mRNA processing and mRNA turnover. Indeed, the kinetics of transcriptional activation alone cannot account for the sustained induction of COX-2 mRNA by interleukin 1 in vascular endothelial cells (14).
The 3′ end of nearly every fully processed eukaryotic mRNA has a poly(A) tail, which has been suggested to influence mRNA stability, translation and transport (reviewed in (34–37)]. Polyadenylation is a two-step process [reviewed in (38–41) and references therein], first involving specific endonucleolytic cleavage (42) at a site determined by binding of polyadenylation factors. The second step involves polymerization of the adenosine (A) tail to an average length of ∼200 residues. Most mammalian polyadenylation signals contain the consensus sequence AAUAAA or a close variant between 10 and 35 nt upstream of the actual cleavage and polyadenylation site. AAUAAA is associated with a frequency of ∼53% of all human polyadenylation signals and ∼59% of all mouse polyadenylation signals (43). This hexamer sequence serves as a binding site for the basal polyadenylation factor cleavage and polyadenylation specificity factor (CPSF) (38–41). Sequences ∼14–35 nt downstream of the polyadenylation signal are also known to be involved in directing polyadenylation by serving as a binding site for the basal polyadenylation factor cleavage stimulation factor (CstF) [(38–41,44–52) and references therein]. Elements upstream of the AAUAAA sequences, known as upstream efficiency elements (USEs), have also been characterized that can enhance polyadenylation efficiency, and have been identified in viral and cellular systems [(53–66) and references therein]. Spacing between the AAUAAA and the USE significantly influences USE efficiency in enhancing polyadenylation (65). Although USEs are polyadenylation efficiency elements, they may also provide additional functions in proper processing.
As has been appreciated in recent years, 3′ end formation is interconnected to mRNA processing events, as well as to mRNA transcription and transcription termination (40,67–69). This interconnection and execution of a functional mRNA likely results from recognition and utilization of cis- and trans-acting signals.
The 3′-UTR of an mRNA can have a major influence on developmental and tissue-specific regulation of gene expression. In fact, the 3′-UTR has recently been called ‘a molecular hotspot for pathology’ (70). Regulation of gene expression through the 3′-UTR can include alternative polyadenylation, translational control and differential mRNA stability. Preliminary evidence suggests that regulated polyadenylation and mRNA stability may play a pivotal role in COX-2 expression.
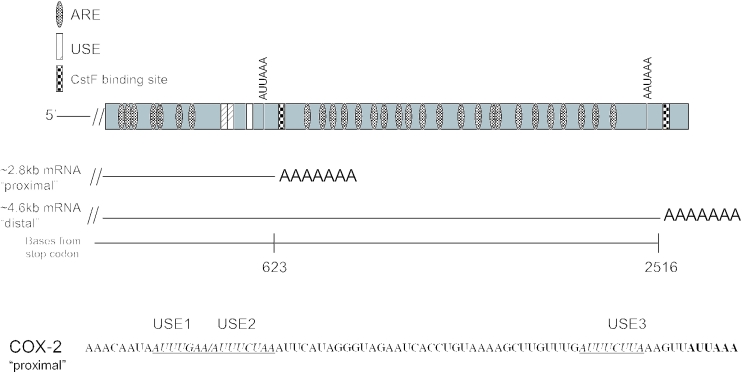
The COX-2 gene is made up of 10 exons; the 3′-UTR is contained within exon 10 (71). The COX-2 3′-UTR is larger than average, encompassing ∼2.5 kb, and has many interesting features. It has several polyadenylation signals, only two of which are commonly used, resulting in mRNAs of ∼2.8 kb using the proximal (with regard to the stop codon) polyadenylation signal or ∼4.6 kb using the distal polyadenylation signal (Figure 1). The proximal polyadenylation signal has a non-consensus CPSF binding site (AUUAAA) yet it is used. It is likely that regulation occurs here, resulting in two mRNAs with different RNA metabolism. Curiously, the mRNA that uses the proximal polyadenylation signal contains putative USEs which are similar in sequence and location to those we and others described previously (64–66). The 3′-UTR also has 22 repeats of an AU-rich motif resembling those known to be involved in regulation of mRNA stability (72,73). Recent studies have found that HuR, or a close variant, binds to these AU-rich elements (AREs) (74–76). Other studies have shown additional RNA binding proteins also may bind to the COX-2 3′-UTR (77–79). Additionally, it has been suggested, but not proven, that tissue specificity might play a role in COX-2 polyadenylation site choice (80,81). The significance of the different tissue distributions of these isoforms has not yet been addressed, and most probably will provide significant revelations with regard to COX-2 expression control.
Figure 1.
Schematic representation of the human COX-2 3′-UTR. Diagram of the 3′-UTR of the human COX-2 gene highlighting the major polyadenylation signals; resulting mRNAs are also depicted. Putative auxiliary USEs are represented by hatched boxes; checkered boxes represent CstF binding sites; AREs are represented by dotted ovals. Bottom: sequence of a portion of the COX-2 proximal polyadenylation signal. Putative USEs are italicized, underlined and noted above the sequence.
Alternative polyadenylation of the COX-2 3′-UTR results in two mRNAs differing only in their 3′ ends. Therefore, the choice of a particular polyadenylation site likely involves specific cis- and trans-acting factors, resulting in RNAs with different metabolism, and ultimately will influence the cytoplasmic fate of the mRNA. This study examines the regulation of alternative 3′ end formation in the human COX-2 pre-mRNA. We demonstrate that alternative polyadenylation does occur in the COX-2 mRNA and that the two major, utilized polyadenylation signals (proximal and distal) are different in strength and composition. We also show that alternative polyadenylation of the COX-2 mRNA occurs in a tissue-specific fashion in cells that endogenously overexpress COX-2. The proximal polyadenylation signal contains auxiliary USEs that are important for its efficient usage. These experiments suggest that alternative polyadenylation of COX-2 mRNA is an important level of gene expression regulation, because polyadenylation signal choice will include or exclude additional regulatory elements that may influence the metabolic fate of the mRNA.
MATERIALS AND METHODS
Plasmids
Primers for PCR to amplify the proximal and distal polyadenylation signals of the human COX-2 3′-UTR were prepared by the Molecular Resource Facility of UMDNJ-NJMS, and had the sequences as listed in Supplementary Table 2. The COX-2 proximal and distal primers contained BamHI (forward) and PstI (reverse) restriction sites to allow insertion into appropriately digested vectors. Human genomic DNA was used as a template to amplify by PCR the appropriate COX-2 proximal and distal polyadenylation signals and flanking sequences for insertion into pGem4 and pCβS vectors. The pCβS vector (a gift from David Fritz, UMDNJ) (64) has a multiple cloning site downstream of the CMV promoter and upstream of the bovine growth hormone (BGH) polyadenylation signal. This vector also includes intron 1 of the rabbit β-globin gene accompanied by the splice donor and acceptor sites. Thus, pCβS-proximal and pCβS-distal were created.
The USE mutants, the hexamer mutant and the non-specific mutant were made by PCR-based site-directed mutagenesis using the Stratagene QuikChange kit as per the manufacturer's protocols using the pCβS-proximal vector as the template. The USE and non-specific mutant were designed to create BglII restriction sites for ease in screening. The COX-2 proximal 5′ deletion (Δ) mutant was made by PCR using primers described in Supplementary Table 2. This construct is lacking 115 bases upstream of the COX-2 proximal polyadenylation signal.
The USE mutant 1 has the sequence AAGATCAAA instead of the wild-type sequence AATTTGAA; the USE mutant 2 has the sequence AAGATCTAA instead of the wild-type sequence AATTTCTAA; the USE mutant 3 has the sequence GAGATCTTA instead of the wild-type GATTTCTTA. The USE mutants 1,3 and 2,3 had both USEs mutated as described above.
The pCβS-proximal–distal-BGH construct was made in the following manner. The COX-2 distal polyadenylation signal was removed from pCβS-distal by BamHI and PstI, then the ends were blunted and inserted into pCβS-proximal which had been digested with EcoRV. Therefore, pCβS-proximal–distal-BGH has three polyadenylation signals in tandem. The pCβS-proximal–distal construct was made by digesting pCβS-proximal–distal-BGH with XhoI and KpnI to remove the BGH polyadenylation signal. Therefore, pCβS-proximal–distal has both COX-2 polyadenylation signals in tandem.
All constructs were transformed into Escherichia coli XL1-Blue cells. Positive clones were sequenced and assayed for expression of the appropriately sized insert. Constructs were verified by sequencing (Molecular Resource Facility, UMDNJ-NJMS). SVL RNA was used as described previously (66).
Rapid site-directed mutagenesis using two PCR-generated DNA fragments
The USE double (USE mut 1,2) and triple (USE mut 1,2,3) mutations were generated by standard PCR using KOD polymerase and using the upstream (5′) wild-type COX-2 proximal primer containing a BamHI restriction site and a downstream primer (3′; USE 1,2 R) containing the respective site-directed mutagenic base pair changes. Then the upstream mutagenic primer (5′; USE 1,2 F) was used in standard PCR using KOD polymerase and the wild-type COX-2 proximal downstream primer (3′) containing a PstI restriction site. The USE 1,2 mut was prepared using COX-2 proximal wild-type DNA template in the PCR, while the USE 1,2,3 mut was prepared using USE 3 mut DNA template. The products of the above two sets of PCR steps were re-amplified using the 5′ and 3′ wild-type COX-2 proximal primers and the resulting 268 bp fragment was cloned into the BamHI/PstI sites of the pCβS vector.
Mammalian cell culture
HeLa, MDA-MD231 and HepG2 cells were maintained in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin–streptomycin (Life Technologies). HT29 cells were maintained in RPMI-1640 Medium (Life Technologies) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.
Transfection
HeLa or MDA-MB-231 cells (7 × 105 cells/well) were seeded in 60 mm plates ∼12 h before transfection. When cells reached 80% confluency, they were transfected using LT-1 reagent (Mirus). Plasmid DNA (2.8 μg) was diluted in 180 μl of serum-free medium to which 6 μl of LT-1 was added and the mixture was incubated at room temperature for 20 min. Following the addition of 1 ml complete medium to the transfection mixture, the medium on the cells was removed and replaced with the entire transfection cocktail. After 24 h, cells were washed once with 1× phosphate-buffered saline (PBS). Cells were scraped and collected into 1 ml PBS and transferred into microcentrifuge tubes. Cells were then centrifuged at 7000 r.p.m. for 5 min (4500 g in an Eppendorf microcentrifuge). The PBS was aspirated and total RNA was extracted immediately from the cell pellet as described below.
Total RNA isolation and RNase protection assay
To determine polyadenylation signal use in vivo, total RNA from transfected cells was assayed by RNase protection. Total RNA was extracted from the cell pellet using either the TRIZOL method (Invitrogen) or the RNeasy Mini Kit (Qiagen) according to the manufacturer's spin protocol for isolation of total RNA from animal cells. For RNase protection assay reactions, 5 μg of total RNA was used per reaction. Probe RNA for the in vivo polyadenylation assay was prepared as described below. The reporter and/or endogenous RNA levels were determined by RNase protection using the RPAIII kit (Ambion Inc., Austin, TX). The RNA was then analyzed on 5% polyacrylamide–8 M urea gels as described previously (64). The ratio of the reporter assay and endogenous band detection was quantified using a Typhoon PhosphorImager and ImageQuant software.
In vitro transcription of RNA substrates
RNA transcripts for in vitro polyadenylation, in vivo polyadenylation assays and RNase protection assays were synthesized by use of SP6 or T7 RNA polymerase according to the supplier (Promega) in the presence of 50 μCi of [α-32P]UTP (Amersham Pharmacia Biosciences or Perkin Elmer Biosciences). RNAs were purified from 5% polyacrylamide–8 M urea–TBE gels by overnight crush elution in high salt buffer (0.4 M NaCl, 50 mM Tris, pH 8.0 and 0.1% SDS). Prior to use in reactions, eluted RNAs were ethanol precipitated and resuspended in water.
Linearization of all pCβS and pGEM4 DNA constructs at the BamHI site and transcription using T7 RNA polymerase as mentioned above generates antisense RNAs. Transcription of CβS COX-2 ‘proximal’ yielded a 610 base RNA, of COX-2 ‘distal’ an 854 base RNA for use in the in vivo polyadenylation assay. SVL was linearized with DraI as described previously (66).
In vitro polyadenylation assays
HeLa nuclear extracts were prepared as described [(64) and references therein] using HeLa cells purchased from the National Cell Culture Center (Minneapolis, MN) or grown in our laboratory. In vitro polyadenylation assays using HeLa nuclear extract, the SV40 late polyadenylation signal pre-mRNA and COX-2 USE RNA or non-specific oligoribonucleotides were performed as described previously (64). Briefly, the in vitro polyadenylation reactions contain a final concentration of 58% (v/v) HeLa nuclear extract, 16 mM phosphocreatine (Sigma), 0.8 mM ATP (Amersham Biosciences), 2.6% polyvinyl alcohol, and 1 × 105 c.p.m. of 32P-labeled SVL substrate RNA (∼50 fmol) in a total volume of 12.5 μl. These reactions are incubated at 30°C for 1 h. The COX-2 USE 3 RNA oligoribonucleotide was synthesized by Dharmacon Research, Inc. (Lafayette, CO) and had the sequence UUGUUUGAUUUCUUAAAGU. The non-specific RNA oligoribonucleotide was described previously (64).
Immunoblot analysis
Cell lysates were prepared by incubating 1.5 × 106 cells on ice for 30 min in a solution containing 1% NP-40, 150 mM NaCl and 50 mM Tris–HCl (pH 8.0), plus phenylmethylsulfonyl fluoride (50 μg/ml), leupeptin, aprotinin and pepstatin A (each at 1 μg/ml). Protein from cleared lysate (75 μg) was separated by 10% SDS–PAGE, then blotted onto nitrocellulose membrane and blocked in 5% non-fat dry milk. Primary antibody incubations were performed at a dilution of 1:100 in blocking solution for 3 h at room temperature. Mouse anti-human COX-2 monoclonal antibody and recombinant human COX-2 protein were purchased from Cayman Chemical (Ann Arbor, MI). Goat-antimouse horseradish peroxidase-conjugated secondary antibody (ICN) was used at a dilution of 1:5000 for 1 h at room temperature. Visualization of bound antibodies was accomplished through chemiluminescence using an ECL kit (Amersham Pharmacia Biosciences) and autoradiography.
Bioinformatics
Expressed sequence tag (EST) sequences corresponding to the human COX-2 gene and their tissue information were obtained from dbEST (March 2004 version; National Center for Biotechnology Information, NCBI), according to the UniGene database (March 2004 version; NCBI). Sequences were aligned to the human genome and polyadenylation sites were determined by using a method described previously [(43) and references therein]. ESTs with poly(A) tail sequence were used to infer polyadenylation sites.
Statistical analyses
Results are expressed as ±SD of the mean, and analyses were performed by two-sample one-tailed Student's t-test. P-values <0.05 were considered significant.
RESULTS
EST surveys suggest existence of multiple polyadenylation signals for the human COX-2 gene with tissue specificity
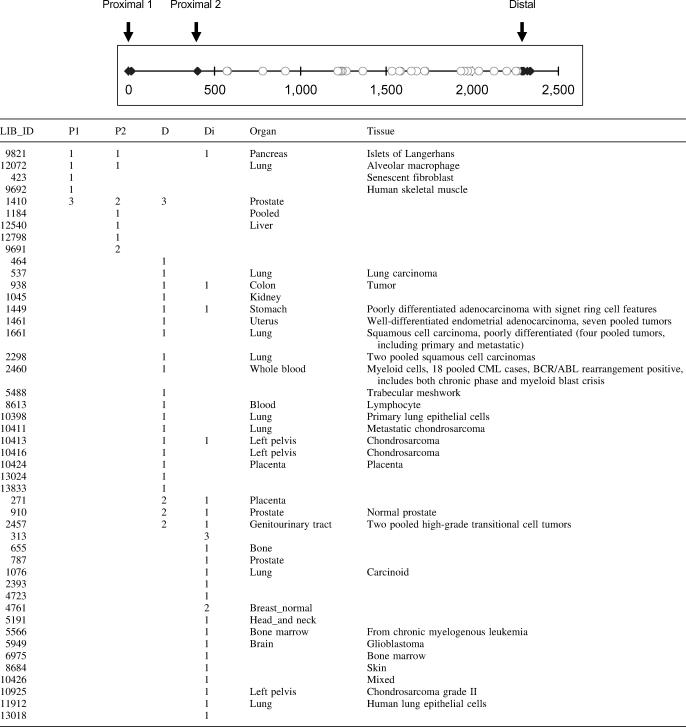
Because alternative polyadenylation is potentially a key regulator of human COX-2 expression, we systematically investigated the mechanism controlling alternative 3′ end formation. Using a bioinformatics approach, we have surveyed the current EST database for human COX-2 mRNAs that would represent utilization of either the proximal or the distal COX-2 polyadenylation signal. Using ESTs with poly(A) tail sequence (see Materials and Methods; B. Tian and C. S. Lutz, unpublished data), we identified three COX-2 polyadenylation signals, named proximal 1, proximal 2 and distal, respectively (Table 1, top). The closed diamonds represent ESTs containing poly(A) tail sequence. The open circles denote ESTs that do not have poly(A) tail sequence. Since these open circles are located after proximal 2, they indirectly support the use of the distal polyadenylation signal, assuming there is no other poly(A) signal between proximal 2 and distal. The distance between proximal 1 and proximal 2 is ∼400 nt, and between proximal 2 and distal is a little less than 2000 nt. Since proximal 1 lacks a definable CstF binding site and previous northern blot data only support transcript sizes resulting from proximal 2 and distal signal usage (80,81), we focused on proximal 2, which we will refer to simply as ‘proximal’ throughout the rest of our study. The distance between proximal and distal from this EST survey is in good agreement with those reported in previous studies [(80,81) and Figure 1]. Also in Table 1 are cDNA library IDs for the ESTs found (first column in Table 1) and tissue and organ information if known on the biological source of the cDNA libraries (last two columns in Table 1). Use of the COX-2 proximal and distal polyadenylation signals are noted by the numbers of ESTs that appeared from each library.
Table 1.
Expressed sequence tag survey result of COX-2 polyadenylation signals
Top: genomic sequence of COX-2 3′-UTR containing three polyadenylation signals is depicted schematically. The position of proximal 1 is arbitrarily set as 0. Closed diamonds denote the locations of poly(A) signals supported by ESTs with poly(A) tail sequence. Open circles are the locations of the 3′ end of ESTs without poly(A) tail sequence. Bottom: LIB_ID corresponds to cDNA library ID in the dbEST database. P1, proximal 1; P2, proximal 2; D, distal; and D1, distal supported by ESTs without poly(A) tail sequence (explained in the text).
To our knowledge, this is the first data for COX-2 examined in this manner. Moreover, our data are comprehensive; all available ESTs corresponding to human COX-2 were used. We would like to highlight that (i) both COX-2 polyadenylation signals are used in vivo; (ii) there are different tissue patterns/distributions and (iii) this is a much larger-scale analysis than has previously been reported. Importantly, these tissue-specific distribution patterns and the polyadenylation signals utilized closely match smaller scale, reported tissue specificity (80,81), with use of the proximal form notably predominating liver, while the distal form notably predominates in colon. Our data underscore the biological relevance of our study and highlight the need for understanding the mechanisms of regulation that are involved.
Endogenous COX-2 mRNAs are polyadenylated at different sites in cell lines representing different tissues
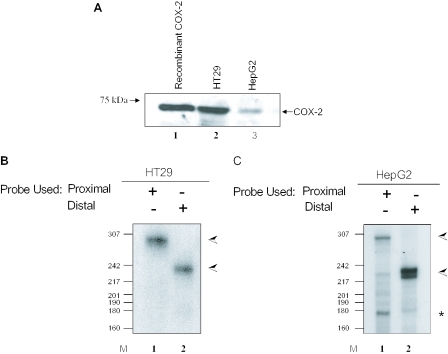
To further evaluate the biological usage of both polyadenylation signals with regard to the tissue specificity noted by our bioinformatics analysis, we chose two established human cancer cell lines, HT29 (colon) and HepG2 (liver). Like many colon cancer-derived cell lines (75), HT29 cells constitutively express COX-2 since endogenous COX-2 protein can be readily detected by western blotting in HT29 extracts (Figure 2A, lane 2). The constitutive expression of COX-2 in hepatocellular carcinoma cells such as HepG2 has been debated in the literature. In our hands, COX-2 protein was detected in HepG2 extract indicating that COX-2 is expressing in this hepatocellular carcinoma cell line (Figure 2A, lane 3). The levels of endogenous COX-2 protein appear to be lower in HepG2 cells than in HT29 cells. As a loading control, we probed the same western blot with an antibody specific for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and found GAPDH to be equivalent in each lane (data not shown).
Figure 2.
Tissue-specific alternative polyadenylation of COX-2 in HT29 and HepG2 cells. (A) Western blot of 75 μg total protein cell extract from HT29 and HepG2 cells using an anti-COX-2 monoclonal antibody. Lane 1, 1 μg recombinant COX-2 protein. (B and C) RNase protection assays using endogenous RNA from HT29 (B) and HepG2 (C) cells along with COX-2 proximal and/or distal probes. The data shown in (B), lane 2, illustrate that HT29 cells primarily use the COX-2 distal polyadenylation signal. (C) HepG2 cells utilizing the COX-2 proximal signal in lane 1 and the distal signal in lane 2. The band corresponding to utilization of the COX-2 proximal polyadenylation signal is marked with an asterisk; the band corresponding to utilization of the COX-2 distal polyadenylation signal is marked with an arrow.
It has been reported that the distal COX-2 polyadenylation signal is used in primary colon samples and that the proximal polyadenylation signal is used in liver samples as assayed by northern blots, but this conclusion was based purely on transcript size (81). We therefore decided to perform our assay based on a more precise method of analyzing polyadenylation signal usage. This assay allows us to specifically identify use of the proximal and distal polyadenylation signals (Figure 2B; see also Figure 1). Total RNA was extracted from HT29 and HepG2 cells and this RNA was used in standard RNase protection assays using specific COX-2 proximal and distal probes. pGEM4-distal and pGEM4-proximal antisense probes protected the specific bases corresponding to the proximal and distal polyadenylation signals. The data in Figure 2B show that HT29 cells primarily use the COX-2 distal polyadenylation signal, giving a protected fragment of 235 bp. HepG2 cells utilize both the COX-2 proximal and distal signals, giving protected fragments of 168 and 235 bp, respectively (Figure 2C). The proximal probe is also able to bind to and protect a 268 bp fragment in the distal mRNA, which corresponds to the entire, uncleaved and unused proximal polyadenylation signal (lane 1 in Figure 2B and C). The larger size reflects the fact that sequences downstream of the proximal cleavage site are contained in the distal mRNA. Our data reveal that there is regulated, differential, tissue-specific polyadenylation of COX-2.
In vivo polyadenylation of COX-2 using reporter assays
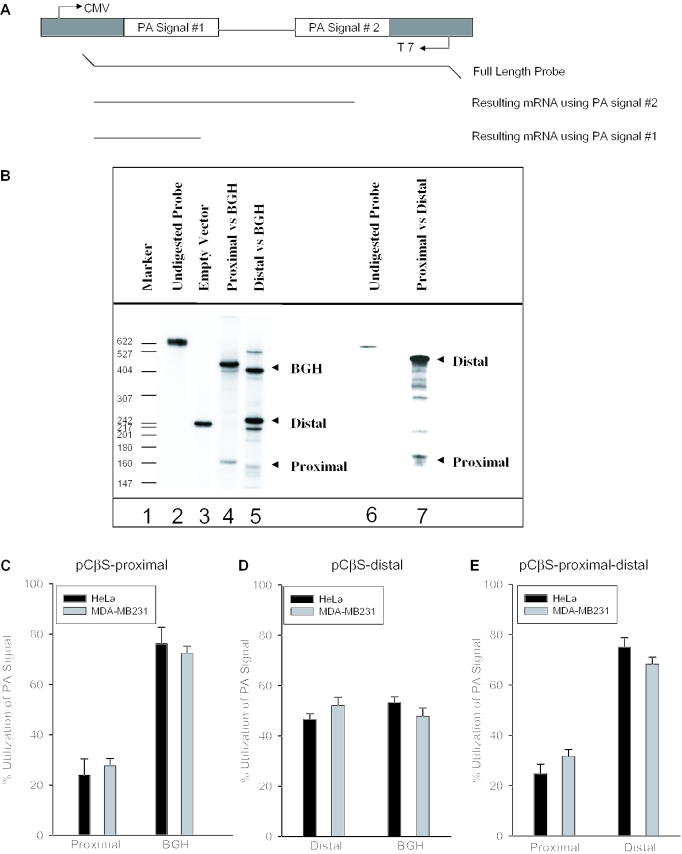
After confirming that both COX-2 polyadenylation signals are used in vivo, we next applied our in vivo polyadenylation assays (64) to address directly the mechanism controlling use of the COX-2 proximal and distal polyadenylation signals. We first addressed how the suboptimal AUUAAA proximal COX-2 polyadenylation signal behaved in our system. The apparent weakness of this polyadenylation signal was postulated based upon its divergence from the consensus AAUAAA CPSF binding site. We cloned a 268 bp fragment by PCR including the human COX-2 proximal polyadenylation signal and flanking sequences into our two polyadenylation signal reporter vector pCβS, diagrammed in Figure 3A. In this series of experiments, the proximal signal was inserted into the signal 1 site while the BGH signal occupied the signal 2 site. This pCβS-COX-2-proximal construct was then transformed into HeLa or MDA-MB-231 (breast cancer) cells. Total RNA was isolated after 24 h, RNase protection assays were performed using the antisense probe to the pCβS-COX-2-proximal construct (Figure 3A), and the results were quantified using a PhosphorImager and ImageQuant software. RNase protection assays will produce a protected fragment of 168 bases if the COX-2 proximal polyadenylation signal is used; utilization of the BGH signal will produce a protected fragment of 410 bases. We found that polyadenylation at the COX-2 proximal signal compared with that at the BGH signal occurred in a ratio of 1:3, indicating that the proximal COX-2 signal is indeed suboptimal (Figure 3B, lane 4 and Figure 3C).
Figure 3.
In vivo polyadenylation assays demonstrate the relative strength of the COX-2 proximal and distal polyadenylation signals. (A) Schematic representation of the pCβS reporter construct used in the in vivo polyadenylation assay. The relative position of each polyadenylation signal is diagramed. The BGH polyadenylation signal is always located at poly(A) site 2. The proximal and distal constructs have those respective sequences cloned into poly(A) site 1. Only the pCβS-proximal-distal-BGH and pCβS-proximal-distal constructs vary from this scheme. (B) Representative RNase protection data for in vivo polyadenylation assays using these constructs. Lane 1, size markers from pBR322 cut with MspI and 5′ end labeled with [γ-32P]ATP as indicated; lane 2, undigested probe; lane 3, pCβS empty vector; lanes 4 and 5, COX-2 constructs in pCβS as indicated above the lane; lane 6, undigested probe; and lane 7, pCβS-proximal-distal. (C) pCβS-proximal was transfected into HeLa (closed bars) or MDA-MB-231 cells (gray bars), RNA was isolated after 24 h, and RNase protection assays were performed. Quantification of three independent experiments is shown here. Constructs are shown on the x-axis; percent use of either the proximal or BGH is shown on the y-axis. The percent utilization was calculated based on the ratio of each signal being utilized within each experiment. Error bars represent SD. All P-values are <1.9 × 10−13. (D) pCβS-distal was transfected into HeLa (closed bars) or MDA-MB-231 cells (gray bars), RNA was isolated after 24 h, and RNase protection assays were performed. Quantification of three independent experiments is shown here. Constructs are shown on the x-axis; percent use either the distal or BGH polyadenylation signal is shown on the y-axis. The percent utilization was calculated based on the ratio of each signal being utilized within each experiment. Error bars represent SD. All P-values are <0.0008. (E) pCβS-proximal-distal was transfected into HeLa (closed bars) or MDA-MB-231 cells (gray bars), RNA was isolated after 24 h, and RNase protection assays were performed. Quantification of three independent experiments is shown here. Percent use of either the proximal or distal polyadenylation signal is shown on the y-axis. The percent utilization was calculated based on the ratio of each signal being utilized within each experiment. Error bars represent SD. All P-values were <0.005.
We next compared polyadenylation using the COX-2 distal polyadenylation signal to BGH by the same type of assay. We cloned a 454 bp fragment by PCR containing the COX-2 distal polyadenylation signal into the pCβS vector, with the COX-2 distal signal occupying the polyadenylation signal 1 site in the reporter vector. Use of the COX-2 distal polyadenylation signal in the RNase protection assay will protect a fragment of 235 bases. Because the distal polyadenylation signal has a consensus AAUAAA, we predicted that this signal would be stronger than the proximal polyadenylation signal of COX-2. Indeed, the COX-2 distal polyadenylation signal was stronger than the COX-2 proximal signal, and was used ∼45% of the time compared with polyadenylation at the BGH signal of ∼55% (Figure 3B, lane 5 and Figure 3D). In this set of experiments, the band corresponding to BGH migrated slightly faster than expected. This result was reproducible in this assay, and has been observed previously using the same assay and same vector (64). These assays were performed in HeLa or MDA-MB-231 cells where there is no detectable endogenous COX-2 protein or mRNA (data not shown). These data demonstrate that although both COX-2 polyadenylation signals are used, the COX-2 proximal signal is indeed weak and the COX-2 distal signal is stronger as compared with the internal control.
Since the COX-2 polyadenylation signals are normally in tandem in the COX-2 3′-UTR, and therefore may be in competition with each other, we measured their relative use in a similar assay as described above. Here, the COX-2 proximal and distal polyadenylation signals were both placed in their respective order in the pCβS reporter plasmid while keeping the BGH polyadenylation signal intact (pCβS-proximal-distal-BGH). After transfection into HeLa cells, RNase protection assays were performed as described above. In this context, the relative use of COX-2 proximal signal was 14.3%, the relative use of the COX-2 distal signal was 53.5%, and the relative use of the BGH polyadenylation signal was 32.2%, giving a ratio of proximal/distal/BGH of 1:3.8:2.3 (data not shown). When the BGH polyadenylation signal was removed to produce a tandem construct (pCβS-proximal-distal), transfected into HeLa or MDA-MB-231 cells, RNA isolated and RNase protection assays performed, the ratio of proximal/distal polyadenylation signal usage was 1:3, not appreciably different from when the BGH was present (Figure 3B, lane 7 and Figure 3E). Note that the creation of the pCβS-proximal-distal construct caused the resulting protected fragments to differ in size slightly from the results obtained using either the pCβS-proximal or the pCβS-distal alone. These data demonstrate that the COX-2 proximal polyadenylation signal does not compete significantly with the distal polyadenylation signal in tandem.
Effect of mutation of core and auxiliary polyadenylation elements in the suboptimal COX-2 proximal polyadenylation signal
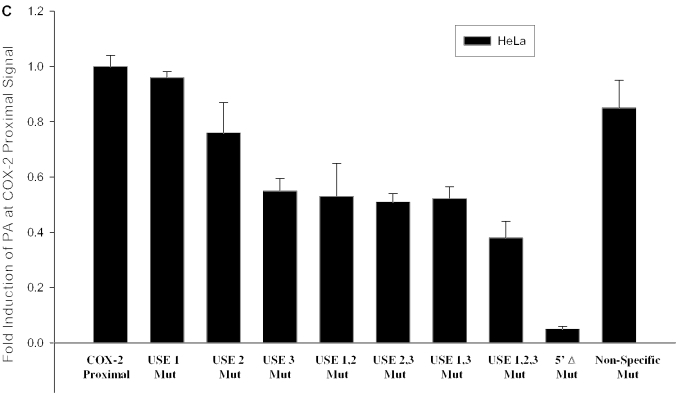
Since the COX-2 proximal core polyadenylation signal is AUUAAA, and therefore differs from the consensus AAUAAA, we showed that it was suboptimal in vivo (Figure 3B, lanes 4 and 7). Previous in vitro studies in the Wickens and Shenk laboratories had mutated the consensus AAUAAA to every base at each position (82,83). These studies had shown in vitro that while AUUAAA worked better than any other derivative of AAUAAA, it still did not perform as well as the consensus AAUAAA. To further prove this in vivo, we mutated the AUUAAA to the consensus sequence AAUAAA using site-directed mutagenesis (see Figure 4A for diagram). We then measured polyadenylation signal use (relative to BGH polyadenylation signal use) by RNase protection assays as described above. Here, we have calculated fold-induction at the proximal polyadenylation signal; so utilization at the wild-type proximal signal was set at a value of 1. We found that this single change, AUUAAA to AAUAAA mutation, enhanced polyadenylation by 3.5-fold over the level of polyadenylation at the COX-2 proximal polyadenylation signal (Figure 4B), thus acting as a positive mutant. Similar results were found using MDA-MB-231 cells (data not shown). These results confirmed directly for the first time in vivo that the difference between an ‘optimal’ AAUAAA and a ‘suboptimal’ AUUAAA core polyadenylation signal is operationally real.
Figure 4.
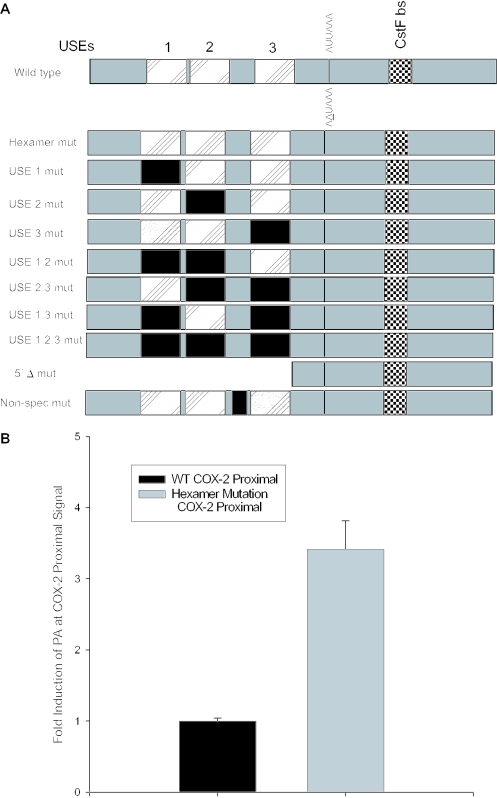
A positive mutant and mutation of the USEs influence use of the COX-2 proximal polyadenylation signal. (A) Diagram showing COX-2 proximal polyadenylation signal and mutations prepared. Closed rectangles represent mutations; hatched rectangles represent USEs; and checkered boxes represent the CstF binding site. (B) Mutants were prepared of the COX-2 proximal polyadenylation signal by site-directed mutagenesis and were cloned into pCβS, transfected into HeLa cells, RNA was isolated after 24 h, and RNase protection assays were performed. Quantification of four independent experiments is shown here. Constructs are shown on the x-axis; fold-induction of polyadenylation at the COX-2 proximal signal is shown on the y-axis. Error bars represent SD. All P-values were <0.0029. (C) USE single mutants, double mutants and the triple mutant, as well as the 5′Δ mutant were prepared as described, transfected into HeLa cells, RNA was isolated after 24 h, and RNase protection assays were performed. Quantification of three independent experiments is shown here. Constructs are shown on the x-axis; fold-induction of polyadenylation at the COX-2 proximal signal is shown on the y-axis. Error bars represent SD. All P-values were <0.0017.
Polyadenylation efficiency can be enhanced by the presence of auxiliary elements located upstream of the CPSF binding site, known as USEs (64–66). Sequences upstream of the COX-2 proximal polyadenylation signal that we identified bear striking similarity to the consensus UAU2–5GUNA USE sequence previously studied in our laboratory. The COX-2 USEs are located 58, 49 and 6 bases upstream of the proximal polyadenylation signal (Figure 4A). We predicted that these putative USEs would affect polyadenylation efficiency of the suboptimal COX-2 proximal polyadenylation signal. In order to experimentally address this prediction, we mutated the USEs in the context of our in vivo polyadenylation construct. Mutants were prepared by PCR-based site-directed mutagenesis in each of the three putative USEs, as well as a non-specific mutant in a non-USE region upstream of the COX-2 proximal polyadenylation signal (Figure 4A). These constructs were then transfected into HeLa (or MDA-MB-231 cells; data not shown), total RNA was isolated and RNase protection assays were performed as described above. The results are shown in Figure 4C. Mutation of each USE was increasingly more deleterious as the USE mutation approached the suboptimal core polyadenylation hexamer element AUUAAA, with USE 3 mutation alone decreasing in vivo polyadenylation to approximately half of the wild-type level (Figure 4C). Mutation of the USEs in duplicate (USE 1,2 mut; USE 2,3 mut; USE 1,3 mut) resulted in approximately the same level of reduced polyadenylation efficiency as the USE 3 mut alone (Figure 4C). However, the triple USE mutation (USE 1,2,3 mut) resulted in a significant decrease in polyadenylation signal utilization (Figure 4C). Deletion of the entire USE-containing region in the 5′Δ mutant almost abolished polyadenylation at the COX-2 proximal polyadenylation signal (Figure 4C). Taken together, these data suggest that the USEs may have additive or synergistic effects. These data also suggest that the USEs are important polyadenylation efficiency elements in the COX-2 proximal polyadenylation signal, and perhaps ensure that the proximal signal is utilized.
USE competition for polyadenylation suggests that a trans-acting factor could be influencing polyadenylation at signals containing USEs
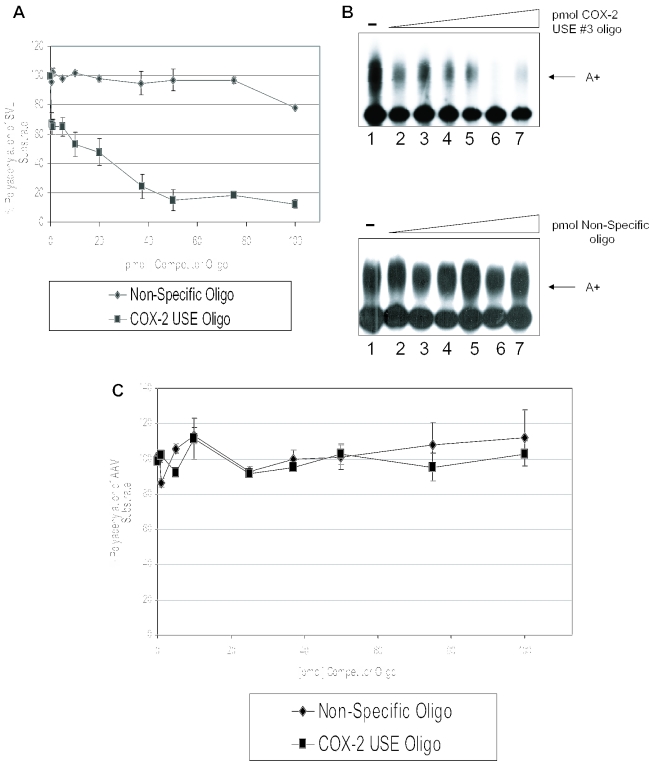
To further define a functional role of the USEs as auxiliary efficiency elements, we performed in vitro polyadenylation assays using specific competitors. Previously, we have shown that in vitro polyadenylation reactions containing an SV40 late polyadenylation signal substrate RNA (SVL) could be inhibited specifically by oligoribonucleotides representing the SV40 USE motifs (66), and that polyadenylation in vitro of human COL1A2 substrate RNA can be inhibited by a COL1A2 USE oligoribonucleotide (64). We now prepared an oligoribonucleotide that represents COX-2 USE 3, and added this oligoribonucleotide to in vitro polyadenylation reactions containing HeLa nuclear extract and a 32P-labeled SVL prepared by in vitro transcription using SP6 polymerase. This substrate RNA was chosen because it works well for in vitro polyadenylation, and because the USEs in SV40 are similar to those found in the proximal COX-2 polyadenylation signal. The data are shown in Figure 5A and B. Figure 5A demonstrates graphically that the COX-2 USE oligoribonucleotide specifically inhibited SVL polyadenylation in vitro, whereas a non-specific oligoribonucleotide had no significant effect on SVL polyadenylation. An additional non-specific oligonucleotide also had no effect (data not shown) (64). Figure 5B shows representative experiments. Lane 1 in Figure 5B, both upper and lower panels, represents a reaction performed in the absence of competitor oligoribonucleotides and demonstrates that the SVL RNA was efficiently polyadenylated in the in vitro system. Interestingly, 50 pmol of the COX-2 USE 3 oligoribonucleotide specifically inhibited polyadenylation to <10% of the control reaction (Figure 5B, upper panel). This amount is similar to other USE oligoribonucleotide inhibition reactions (64,66). The non-specific oligo did not inhibit in vitro polyadenylation (Figure 5B, lower panel). In vitro polyadenylation of a non-USE containing polyadenylation signal, AAV, was also not inhibited by the addition of the COX-2 USE 3 at similar concentrations as expected (Figure 5C). Polyadenylation of AAV was also not affected by addition of the non-specific oligonucleotides as expected (Figure 5C). Taken together, these data demonstrate that the COX-2 USE 3 oligoribonucleotide specifically binds and sequesters a putative common factor(s) important for polyadenylation, and suggest that the similarity to the SV40 USE motifs is functionally significant.
Figure 5.
Competition studies suggest USE binding factors influence the processing efficiency of COX-2 proximal polyadenylation signal. (A) Graphical representation of in vitro competitions using SVL and either COX-2 USE oligoribonucleotide (squares) or non-specific oligoribonucleotide (diamonds). Numbers on the x-axis represent pmol of competitor added, numbers on the y-axis represent percent polyadenylation. This quantification represents the average of three independent experiments. Error bars represent SD. (B) Representative in vitro polyadenylation reactions using SV40 late polyadenylation signal RNA as the substrate RNA and increasing amounts of either COX-2 USE oligoribonucleotide as competitor (upper panel) and non-specific oligoribonucleotide as competitor (lower panel). Amounts of competitor used in both sets of reactions are as follows: lane 1s, no competitor; lane 2s, 1 pmol competitor; lane 3s, 10 pmol; lane 4s, 20 pmol; lane 5s, 37 pmol; lane 6s, 50 pmol; and lane 7s, 75 pmol. (C) Graphical representation of in vitro competitions using AAV substrate RNA and either COX-2 USE oligoribonucleotide (squares) or non-specific oligoribonucleotide (diamonds).
DISCUSSION
Polyadenylation is a useful and underexplored mechanism of regulating gene expression, and this regulation is partially implied by the complex nature of this process. Alternative polyadenylation can generate mature transcripts that only differ in their 3′ ends. Efficiency of processing is another level at which regulation can occur, since most pre-mRNAs in the cell are not efficiently processed, and therefore, even small changes in the overall processing efficiency of a particular pre-mRNA may have a great effect on ultimate protein levels. Experimental evidence has demonstrated that poly(A) signal strength directly influences the amount of mature, exported mRNA (84,85).
Our objective in this study was to accurately define and characterize alternative polyadenylation as it applied to the human COX-2 3′-UTR. Interestingly, the size and scheme of the 3′-UTR is conserved among human, chimpanzee, mouse and rat COX-2 genes, along with the two polyadenylation signals, suggesting that the importance of alternative polyadenylation plays a key regulatory function (B. Tian, H. Zhang, and C. S. Lutz, unpublished data). Indeed, the sequences for USEs 1 and 2 are exact matches in the 3′-UTR of mouse COX-2 homolog (B. Tian, H. Zhang, and C. S. Lutz, unpublished data). Choice of a particular polyadenylation signal may result in mRNAs with different stability or different translation efficiency, which has direct consequences for how much protein product is ultimately produced. Therefore, polyadenylation signal choice can be a key step in mRNA maturation. Our work examined the relative strength and composition of the COX-2 proximal and distal polyadenylation signals. We have shown that the COX-2 proximal and COX-2 distal polyadenylation signals are utilized in an in vivo polyadenylation assay. We further demonstrated that the use of these COX-2 polyadenylation signals is regulated in a tissue-specific manner. We propose that while basal polyadenylation of these signals does occur experimentally in cell types that do not normally express COX-2 mRNA (HeLa and MDA-MB-231), the cell-type specific utilization of COX-2 proximal and distal polyadenylation signals may require additional as yet unidentified factors.
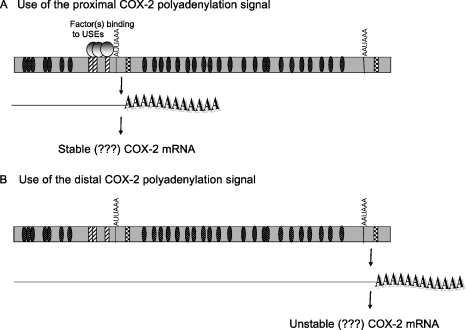
Additionally, we have demonstrated that a suboptimal CPSF binding site (AUUAAA) is truly weaker than the canonical CPSF binding site, AAUAAA. Taken together, the data suggest a working hypothesis, shown in Figure 6, for polyadenylation signal choice, as well as for possible correlations between polyadenylation signal choice and resulting mRNA stability as postulated (81).
Figure 6.
Working model of alternative polyadenylation in COX-2.
Our model (Figure 6) suggests that use of the COX-2 proximal polyadenylation signal results in a shorter mRNA, while use of the COX-2 distal polyadenylation signal results in a longer mRNA. Use of the proximal polyadenylation signal would exclude a portion of the 3′-UTR, which may contain important regulatory elements, for example, elements that regulate mRNA stability. The best characterized 3′-UTR sequences that cause mRNA instability are the AREs found in many short-lived mRNAs such as cytokines, growth factors and proto-oncogenes (72,73). The 3′-UTR of COX-2 is complex and has many AREs (Figure 1). Currently, data in the literature conflict with regard to which form of COX-2 is more stable. It has been shown that removal of some sequences upstream of the COX-2 proximal polyadenylation signal containing some of the AREs confers increased stability to a heterologous reporter (86), while other data have shown that this same region can transfer instability to a heterologous reporter (87,88). Additionally, the longer COX-2 mRNA has been reported to be the more unstable form as revealed by northern blotting of the COX-2 mRNA while the shorter form is generally thought to be the more stable form (81). While our study has not addressed these questions of stability directly, our study underscores the importance of understanding how alternative polyadenylation of COX-2 is regulated.
We have also demonstrated here that the USEs present in the suboptimal COX-2 proximal polyadenylation signal have a significant effect on in vivo polyadenylation of that signal, since mutation or deletion of the USEs has a substantial effect (Figure 4C). We suggest that the presence of the USEs in the COX-2 proximal polyadenylation signal ensure its usage, possibly by supporting and enhancing assembly of the basal polyadenylation machinery on the polyadenylation signal, or by serving as a trans-acting factor binding site which may recruit or enhance interactions with the basal polyadenylation machinery. These explanations are not mutually exclusive. Additionally, tissue-specific factors may be involved since we have shown tissue-specific utilization occurs at the COX-2 proximal polyadenylation signal. These details remain to be explored.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank the members of the Lutz, O'Connor, Rogers, Bellofatto, Modak and Wilusz laboratories for helpful experimental suggestions and comments on this manuscript. Special thanks to David Fritz for helpful suggestions and Matias Elijovich for technical assistance. This work was funded by American Cancer Society Grant RPG-00-265-01-GMC and NSF award MCB-0426195 to C.S.L. Funding to pay the Open Access publication charges for this article was provided by NSF award MCB-0426195.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hla T., Bishop-Bailey D., Liu C.H., Schaefers H.J., Trifan O.C. Cyclooxygenase-1 and -2 isoenzymes. Int. J. Biochem. Cell Biol. 1999;31:551–557. doi: 10.1016/s1357-2725(98)00152-6. [DOI] [PubMed] [Google Scholar]
- 2.Kelley D.J., Mestre J.R., Subbaramaiah K., Sacks P.G., Schantz T., Tanabe T., Inoue H., Ramonetti J.T., Dannenberg A.J. Benzo[a]pyrene up-regulates cyclooxygenase-2 gene expression in oral epithelial cells. Carcinogeneis. 1997;18:795–799. doi: 10.1093/carcin/18.4.795. [DOI] [PubMed] [Google Scholar]
- 3.DuBois R.N., Awad J., Morrow J., Roberts L.J., Bishop P.R. Regulation of eicosanoid production and mitogenesis in rat intestinal epithelial cells by transforming growth factor alpha and phorbol ester. J. Clin. Invest. 1994;93:493–498. doi: 10.1172/JCI116998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hla T., Farrell M., Kumar A., Bailey J.M. Isolation of the cDNA for human prostaglandin H synthase. Prostaglandins. 1986;32:829–845. doi: 10.1016/0090-6980(86)90093-6. [DOI] [PubMed] [Google Scholar]
- 5.DeWitt D.L., Smith W.L. Primary structure of prostaglandin G/H synthase from sheep vesicular gland determined from complementary DNA sequence. Proc. Natl Acad. Sci. USA. 1988;85:1412–1416. doi: 10.1073/pnas.85.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merlie J.P., Fagan D., Mudd J., Needleman P. Isolation and characterization of the complementary DNA for sheep seminal vesicle prostaglandin endoperoxidase synthase (cyclooxygenase) J. Biol. Chem. 1988;263:3550–3553. [PubMed] [Google Scholar]
- 7.Hla T., Neilson K. Human cyclooxygenase-2 cDNA. Proc. Natl Acad. Sci. USA. 1992;89:7384–7388. doi: 10.1073/pnas.89.16.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Banion M.K., Winn V.D., Yong D.A. cDNA cloning and functional activity of a glucocorticoid-regulated inflammatory cyclooxygenase. Proc. Natl Acad. Sci. USA. 1992;89:4888–4892. doi: 10.1073/pnas.89.11.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie W., Cipman J.G., Robertson D.L., Erikson R.L., Simmons D.L. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc. Natl Acad. Sci. USA. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garavito R., Mulichak A. The structure of mammalian cyclooxygenases. Annu. Rev. Biomol. Struct. 2002;32:183–206. doi: 10.1146/annurev.biophys.32.110601.141906. [DOI] [PubMed] [Google Scholar]
- 11.Chandrasekharan N.V., Dai H., Roos K.L.T., Evanson N.K., Tomsik J., Elton T.S., Simmons D.L. COX-3, a cyclooxygenase-1 variant inhibitied by acetaminophen and other analgesic/antipyretic drugs: cloning, structure and expression. Proc. Natl Acad. Sci. USA. 2002;99:13926–13931. doi: 10.1073/pnas.162468699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazan N.G., Fletcher B.S., Herschmann H.R., Mukherjee P.K. Platelet-activating factor and retinoic acid synergistically activate the inducible prostaglandin synthase gene. Proc. Natl Acad. Sci. USA. 1994;91:5252–5256. doi: 10.1073/pnas.91.12.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hla T., Maciag T. Cyclooxygenase gene expression is down-regulated by heparin-binding (acidic fibroblast) growth factor-1 in human endothelial cells. J. Biol. Chem. 1991;266:24059–24063. [PubMed] [Google Scholar]
- 14.Ristimaki A., Garfinkel S., Wessendorf J., Maciag T., Hla T. Induction of cyclooxygenase-2 by interleukin-1 alpha. Evidence for post-transcriptional regulation. J. Biol. Chem. 1994;269:11769–11775. [PubMed] [Google Scholar]
- 15.O'Banion M.K., Miller J.C., Chang J.W., Kaplan M.D., Coleman P.D. Interleukin-1 beta induces prostaglandin G/H synthase-2 (cyclooxygenase-2) in primary murine astrocyte cultures. J. Neurochem. 1996;66:2532–40. doi: 10.1046/j.1471-4159.1996.66062532.x. [DOI] [PubMed] [Google Scholar]
- 16.Bazan N.G. COX-2 as a multifunctional neuronal modulator. Nature Med. 2001;7:414–415. doi: 10.1038/86477. [DOI] [PubMed] [Google Scholar]
- 17.Bishop-Bailey D., Calatayud S., Warner T.D., Hla T., Mitchell J.A. Prostaglandins and the regulation of tumor growth. J. Environ. Pathol. Toxicol. Oncol. 2002;21:93–101. [PubMed] [Google Scholar]
- 18.Hla T., Ristimaki A., Appleby S., Barriocanal J.G. Cyclooxygenase gene expression in inflammation and angiogenesis. Ann. N.Y. Acad. Sci. 1993;696:197–204. doi: 10.1111/j.1749-6632.1993.tb17152.x. [DOI] [PubMed] [Google Scholar]
- 19.Williams C.S., Mann M., DuBois R.N. The role of cyclooxygenases in inflammation, cancer and development. Oncogene. 1999;18:7908–7916. doi: 10.1038/sj.onc.1203286. [DOI] [PubMed] [Google Scholar]
- 20.Prescott S.M. Is cyclooxygenase-2 the alpha and omega in cancer? J. Clin. Invest. 2000;105:1511–1513. doi: 10.1172/JCI10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott S.M., Fitzpatrick F.A. Cyclooxygenase-2 and carcinogenesis. Biochim. Biophys. Acta. 2000;1470:69–78. doi: 10.1016/s0304-419x(00)00006-8. [DOI] [PubMed] [Google Scholar]
- 22.Dixon D.A. Dysregulated post-transcriptional control of COX-2 gene expression in cancer. Curr. Pharm. Des. 2004;10:635–646. doi: 10.2174/1381612043453171. [DOI] [PubMed] [Google Scholar]
- 23.Hwang D., Scollard D., Byrne J., Levine E. Expression of cyclooxygenase-1 and cyclooxygenase-2 in human breast cancer. J. Natl Cancer Inst. 1998;90:455–460. doi: 10.1093/jnci/90.6.455. [DOI] [PubMed] [Google Scholar]
- 24.Eberhart C.E., Coffey R.J., Radhkia A., Giardiello F.M., Ferrenback S., Dubois R.N. Upregulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183–1188. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 25.Hida T., Leyton J., Makheja A.N., Ben A.P., Hla T., Martinez A., Mulshine J., Malkani S., Chung P., Moody T.W. Non-small cell lung cancer cyclooxygenase activity and proliferation are inhibited by non-steroidal anti-inflammatory drugs. Anticancer Res. 1998;18:775–782. [PubMed] [Google Scholar]
- 26.Kutchera W., Jones D.A., Matsunami N., Groden J., McIntyre T.M., Zimmerman G.A., White R.L., Prescott S.M. Prostaglandin synthase 2 is abnormally expressed in human colon cancer: evidence for a transcriptional effect. Proc. Natl Acad. Sci. USA. 1996;93:4816–4820. doi: 10.1073/pnas.93.10.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimberg J., Samuelsson A., Hugander A.M., Soderkvist P. Gene expression of cyclooxygenase-2, group,II and cytosolic phospholipase A2 in human colorectal cancer. Anticancer Res. 1998;18:3283–3287. [PubMed] [Google Scholar]
- 28.San H., Kawahito Y., Wilder R.L., Hashiramoto A., Mukai S., Asai K., Kimura S., Kato H., Kondo M., Hla T. Expression of cyclooxygenase-1 and -2 in human colorectal cancer. Cancer Res. 1995;55:3785–3789. [PubMed] [Google Scholar]
- 29.Williams C.S., Tsujii M., Reese J., Dey S.K., DuBois R.N. Host cyclooxygenase-2 modulates carcinoma growth. J. Clin. Invest. 2000;105:1589–1594. doi: 10.1172/JCI9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W., Reinmuth N., Stoeltzing O., Parikh A.A., Tellez C., Williams S., Jung Y.D., Takeda A., Akagi M., Bar-Eli M., et al. Cyclooxygenase-2 is up-regulated by interleukin-1 beta in human colorectal cancer cells via multiple signaling pathways. Cancer Res. 2003;63:3632–3636. [PubMed] [Google Scholar]
- 31.Xiong B., Sun T.J., Yuan H.Y., Hu M.B., Hu W.D., Cheng F.L. Cyclooxygenase-2 expression and angiogenesis in colorectal cancer. World J. Gastroenterol. 2003;9:1237–1240. doi: 10.3748/wjg.v9.i6.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denkert C., Winzer K.J., Muller B.M., Weichert W., Pest S., Kobel M., Kristiansen G., Reles A., Siegert A., Guski H., Hauptmann S. Elevated expression of cyclooxygenase-2 is a negative prognostic factor for disease free survival and overall survival in patients with breast carcinoma. Cancer. 2003;97:2978–2987. doi: 10.1002/cncr.11437. [DOI] [PubMed] [Google Scholar]
- 33.Landen C.N., Mathur S.P., Richardson M.S., Creasman W.T. Expression of cyclooxygenase-2 in cervical, endometrial, and ovarian malignancies. Am. J. Obstet. Gynecol. 2003;188:1174–1176. doi: 10.1067/mob.2003.284. [DOI] [PubMed] [Google Scholar]
- 34.Lewis J., Gunderson S., Mattaj I.W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J. Cell Sci. 1995;19(Suppl.):13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson A., Peltz S.W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 36.Sachs A.B., Sarnow P., Hentze M.W. Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 37.Wickens M., Anderson P., Jackson R.J. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 38.Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 39.Edmonds M. A history of polyA sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 40.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Wahle E., Kuhn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog. Nucleic Acids Res. Mol. Biol. 1997;57:41–71. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- 42.Ryan K., Calvo O., Manley J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian B., Hu J., Zhang H., Lutz C.S. A large scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen F., MacDonald C.C., Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen F., Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gil A., Proudfoot N.J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3′ end formation. Nature. 1984;312:473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- 47.Gil A., Proudfoot N.J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 48.McDevitt M.A., Imperiale M.J., Ali H., Nevins J.R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984;37:993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- 49.Sadofsky M., Connelly S., Manley J.L., Alwine J.C. Identification of a sequence element on the 3′ side of AAUAAA which is necessary for simian virus 40 late mRNA 3′ end processing. Mol. Cell. Biol. 1985;5:2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway L., Wickens M. A sequence downstream of AAUAAA is required for formation of simian virus 40 late mRNA in 3′ termini in frog oocytes. Proc. Natl Acad. Sci. USA. 1985;82:3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilusz J., Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA crosslinking and functionally substitutes for the downstream element of the polyadenylation signal. Mol. Cell. Biol. 1990;10:6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryner L.C., Takagaki Y., Manley J.L. Sequences downstream of AAUAAA signals affect pre-mRNA cleavage and polyadenylation in vitro both directly and indirectly. Mol. Cell. Biol. 1989;9:1759–1771. doi: 10.1128/mcb.9.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown P.H., Tiley L.S., Cullen B.R. Efficient polyadenylation within the human immunodeficiency type 1 long terminal repeat inhibits polyadenylation of its own pre-mRNA. J. Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carswell S., Alwine J.C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol. Cell. Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeZazzo J.D., Kilpatrick J.E., Imperiale M.J. Involvement of long terminal repeat U3 sequences overlapping the transcriptional control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol. Cell. Biol. 1991;11:1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sachchithananthan M., Stasinopoulos S., Wilusz J., Medcalf R. The relationship between the prothrombin upstream sequence element and the G20210A polymorphism: the influence of a competitive environment for mRNA 3′ end formation. Nucleic Acids Res. 2005;33:1010–1020. doi: 10.1093/nar/gki245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valsamakis A., Schek N., Alwine J.C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol. Cell. Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilmartin G.M., Fleming E.S., Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilmartin G.M., Fleming E.S., Oetjen J., Gravely B.R. CPSF recognition of and HIV-1 mRNA 3′ processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 60.Sanfacon H., Brodmann P., Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991;5:141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- 61.Russnak R. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-dependent, and additive. Nucleic Acids Res. 1991;19:6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreira A., Wollerton M., Monks J., Proudfoot N.J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreira A., Takagaki Y., Brackenridge S., Wollerton M., Manley J.L., Proudfoot N.J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Natalizio B.J., Muniz L.C., Arhin G.K., Wilusz J., Lutz C.S. Upstream elements present in the 3′ UTR of collagen gene influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 65.Schek N., Cooke C., Alwine J.C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol. Cell. Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lutz C.S., Alwine J.C. Direct interaction of the U1snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 67.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 68.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conne B., Stutz A., Vassali J.D. The 3′ untranslated region of messenger RNA: a molecular hotspot for pathology? Nature Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 71.Appleby S.B., Ristimaki A., Neilson K., Narko K., Hla T. Structure of the human cyclo-oxygenase-2 gene. Biochem. J. 1994;302:723–727. doi: 10.1042/bj3020723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaw G., Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 73.Chen C.Y., Shyu A.B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 74.Dean J.L., Sully G., Wait R., Rawlinson L., Clark A.R., Saklatvala J. Identification of a novel AU-rich element-binding protein which is related to AUF-1. Biochem. J. 2002;366:709–719. doi: 10.1042/BJ20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dixon D.A., Tolley N.D., King P.H., Nabors L.B., McIntyre T.M., Zimmerman G.A., Prescott S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nabors L.B., Gillespie G.Y., Harkins L., King P.H. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 77.Sengupta S., Jan B.C., Wu M.T., Paik J.H., Furneaux H., Hla T. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J. Biol. Chem. 2003;278:25227–25233. doi: 10.1074/jbc.M301813200. [DOI] [PubMed] [Google Scholar]
- 78.Sawaoka H., Dixon D.A., Oates J.A., Boutard O. Tristetraprolin binds to the 3′-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 79.Dixon D.A., Balch G.C., Kedersha N., Anderson P., Zimmerman G.A., Beauchamp R.D., Prescott S.M. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng L., Sun W., Xia Y., Tang W.W., Chanmugam P., Soyoola E., Wilson C.B., Hwang D. Cloning two isoforms of rat cyclooxygenase: differential regulation of their expression. Arch. Biochem. Biophys. 1993;307:361–368. doi: 10.1006/abbi.1993.1601. [DOI] [PubMed] [Google Scholar]
- 81.Lukiw W.J., Bazan N.G. Cyclooxygenase 2 RNA message abundance, stability and hypervariability in sporadic Alzheimer neocortex. J. Neurosci. Res. 1997;50:937–945. doi: 10.1002/(SICI)1097-4547(19971215)50:6<937::AID-JNR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 82.Sheets M.D., Ogg S.C., Wickens M.P. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilusz J., Pettine S.M., Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1998;17:3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Denome R.M., Cole C.N. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol. Cell. Biol. 1988;8:4829–4839. doi: 10.1128/mcb.8.11.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edwalds-Gilbert G., Prescott J., Falck-Pederson E. 3′ RNA processing efficiency plays a primary role in generating termination-competent RNA polymerase II elongation complexes. Mol. Cell. Biol. 1993;13:3472–3480. doi: 10.1128/mcb.13.6.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cok S.J., Morrsion A.R. The 3′ untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J. Biol. Chem. 2001;276:23179–23185. doi: 10.1074/jbc.M008461200. [DOI] [PubMed] [Google Scholar]
- 87.Dixon D.A., Kaplan C.D., McIntyre T.M., Zimmerman G.A., Prescott S.M. Post-transcriptional control of cyclooxygenase gene expression. J. Biol. Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 88.Gou Q., Liu C.H., Ben-Av P., Hla T. Dissociation of basal turnover and cytokine-induced transcript stabilization of the human cyclooxygenase-2 mRNA by mutagenesis of the 3′-untranslated region. Biochem. Biophys. Res. Commun. 1998;242:508–512. doi: 10.1006/bbrc.1997.7994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.