Abstract
Cells from complementation groups A through G of the heritable sun-sensitive disorder xeroderma pigmentosum (XP) show defects in nucleotide excision repair of damaged DNA. Proteins representing groups A, B, C, D, F, and G are subunits of the core recognition and incision machinery of repair. XP group E (XP-E) is the mildest form of the disorder, and cells generally show about 50% of the normal repair level. We investigated two protein factors previously implicated in the XP-E defect, UV-damaged DNA binding protein (UV-DDB) and replication protein A (RPA). Three newly identified XP-E cell lines (XP23PV, XP25PV, and a line formerly classified as an XP variant) were defective in UV-DDB binding activity but had levels of RPA in the normal range. The XP-E cell extracts did not display a significant nucleotide excision repair defect in vitro, with either UV-irradiated DNA or a uniquely placed cisplatin lesion used as a substrate. Purified UV-DDB protein did not stimulate repair of naked DNA by DDB− XP-E cell extracts, but microinjection of the protein into DDB− XP-E cells could partially correct the repair defect. RPA stimulated repair in normal, XP-E, or complemented extracts from other XP groups, and so the effect of RPA was not specific for XP-E cell extracts. These data strengthen the connection between XP-E and UV-DDB. Coupled with previous results, the findings suggest that UV-DDB has a role in the repair of DNA in chromatin.
The heritable human disorder xeroderma pigmentosum (XP) is chiefly characterized by an increased incidence of benign and malignant skin lesions after exposure to sunlight. Affected individuals fall into one of eight different genetic complementation groups. Cells from the seven complementation groups A through G have reduced nucleotide excision repair (NER) of damaged DNA, while cells from the variant, or V, group are defective in a less-defined process of cellular recovery after DNA damage (11). Genes and proteins representing XP groups A (XP-A) B, C, D, F, and G have all been isolated and found to represent some of the subunits of the core NER recognition and incision machinery. XP-E is the mildest form of the disorder, and cells of this group generally have 40 to 60% of the normal repair level, as shown by autoradiographic measurement of unscheduled DNA synthesis (UDS) after UV irradiation. Cell fusion studies have assigned at least 16 individuals to this form of the disorder (6, 19, 23, 40).
There are several indications that a DNA damage binding protein denoted UV-DDB (or DDB) is involved in the primary XP-E defect. The protein has been detected in extracts of vertebrate cells as an activity that preferentially binds damaged oligonucleotides in electrophoretic mobility shift or filter binding assays. The protein has a particular affinity for (6-4) photoproducts in UV-irradiated DNA (10, 15, 16, 34, 41, 43), but UV-DDB also binds to DNA damaged by other agents, including cisplatin and nitrogen mustard (32). The activity has been purified as a single 127-kDa protein (2) and as a complex with two subunits of 127 and 48 kDa (21). Damage-binding activity is missing from some cells in the XP-E group, designated DDB−, but is present in other XP-E cell lines, designated DDB+ (3, 15, 19, 23). The genes encoding the p127 protein (7, 17, 39) and the p48 protein (7) have been isolated, but DNA sequence features have not yet yielded firm clues about their functions. Microinjection of purified UV-DDB into XP-E cells lacking UV-DDB activity substantially corrects the NER defect, as measured by UDS after UV irradiation, but UV-DDB+ cells are not corrected (22). Sequence alterations in the gene for p48 have been reported for several XP-E cell lines (29), and it is possible that these are causative mutations for XP-E.
There are also suggestions that the single-stranded DNA binding activity of replication protein A (RPA) is involved in the XP-E defect. RPA is a heterotrimer of three subunits with sizes of 70, 34, and 14 kDa that plays key roles in DNA replication, recombination, and DNA repair (44). It is one of the core components of the eukaryotic nucleotide excision-incision system (1, 12, 28). With regard to XP, it was recently reported that XP-E cell extracts are severely defective in NER in vitro and that RPA can specifically correct the repair defect of these extracts, but not those of extracts of other complementation groups (20). Moreover, it has been found that RPA copurifies to some extent with UV-DDB protein and that the two proteins interact, showing a tighter association with chromatin after UV irradiation of cells (31).
The availability of lymphoblastoid cell lines derived from three newly identified XP-E individuals has given us the opportunity to further investigate the possible relationships of UV-DDB and RPA to the molecular defect in XP-E and the influence of these proteins on NER.
MATERIALS AND METHODS
Cells.
Primary fibroblast cultures were established from skin biopsies from patients XP23PV and XP25PV. Both individuals had mild symptoms of XP which will be reported in detail elsewhere. Additional primary fibroblast strains from the Human Genetic Mutant Cell Repository (Coriell Institute, Camden, N.J.) were as follows: C3PV and GM00037 (normal), XP20PV (XP-A), XPCS2BA (XP-B), XP5PV (XP-C), TTD10VI (XP-D), XP2RO = GM02415 (XP-E), GM01389 (XP-E), XP2VO = GM04313 (XP-F), and XP2BI = GM03021 (XP-G). XP82TO cells (XP-E) were a generous gift from S. Kondo. The cells were grown by standard procedures in Ham’s F10 medium (GIBCO) supplemented with 10% fetal calf serum (Hyclone Europe) and subcultured by trypsinization.
Lymphoblastoid cell lines were established by Epstein-Barr virus immortalization of peripheral blood lymphocytes from the patients XP23PV and XP25PV. The cells were cultured in RPMI 1640 medium supplemented with 15% fetal calf serum. The lymphoblastoid cell line 705ori was similarly derived from lymphocytes of a normal, repair-proficient female by using cells supplied by Colin Arlett and Jane Cole (MRC Cell Mutation Unit, South Mimms, United Kingdom). GM02345 XP-A cells and GM01646 DDB− XP-E cells were from the Human Genetic Mutant Cell Repository (Coriell Institute). The lymphoblastoid XP-G cell line XPG83 and the simian virus 40-immortalized XP-G cell line XPG415A were previously described (30).
Genetic analysis.
Cell fusion was performed as previously reported (37, 38). Briefly, fibroblast strains used as partners in the fusion were grown for 3 days in medium containing latex beads of either 0.8 or 1.7 μm in diameter that were internalized as cytoplasmic markers. The cells were fused by using polyethylene glycol 4000 (Merck); after 48 h of incubation at 37°C, cells were UV irradiated (20 J/m2), incubated for 3 h in medium containing [3H]thymidine (3H-TdR; New England Nuclear; specific activity, 25 Ci/mmol), and processed for autoradiography. UDS was measured by counting the number of grains over nuclei in at least 25 homodikaryons and in 25 heterodikaryons (identified as binuclear cells containing beads of different sizes). Two cell strains were classified in the same group if no restoration of UDS level was observed in the heterodikaryons.
Proteins.
UV-DDB protein was purified from a 50-liter culture of HeLa cells as described previously (39), except that the pooled eluate from a first UV-DNA cellulose column was loaded onto a second UV-DNA cellulose column equilibrated with 10 mM Tris-HCl (pH 7.5), 700 mM NaCl, 1 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol (DTT), and 1 mM CHAPS {3-[(3-cholamidopropyl)-dimethyl-ammonia]-1-propanesulfonate}. Following stepwise elution with 0.8, 1.0, 1.5, and 2.0 M NaCl, the fractions containing the most UV-DDB activity were pooled; concentrated with a Macrosep 10 apparatus (Filtron Technology Corp.); adjusted to 10 mM Tris-HCl [pH 7.5], 350 mM NaCl, 1 mM EDTA, 1 mM MgCl2, 1 mM CHAPS, 1 mM DTT, and 10% glycerol; and stored at −80°C. Silver staining and immunostaining revealed p127 and p48 proteins, with p127 more prominent, at about 10 μg/ml.
Recombinant human RPA was purified after expression in Escherichia coli, as described previously (14), from a vector kindly provided by M. Wold. Human XPG protein produced from a recombinant baculovirus was purified as described previously (8), and recombinant human XPA protein was purified from E. coli (18).
DNA binding assays and immunoblotting.
The DNA binding assay was carried out as described previously (15, 33) with a UV-irradiated double-stranded 60-mer oligonucleotide, 60/54. Protein-DNA complexes were separated on a nondenaturing 6% polyacrylamide gel. Immunoblotting was performed with chemiluminescent detection (Tropix, Bedford, Mass.) with a primary polyclonal antibody against p127 (31) or monoclonal antibodies against RPA1 and RPA2 (Oncogene Science, Inc., Cambridge, Mass.).
In vitro DNA repair assays. (i) Repair synthesis with UV-damaged DNA.
The plasmids used were derivatives of pUC vectors: the 3.0-kb plasmid pBluescript KS+ (Stratagene) and the 3.7-kb plasmid pHM14. pBluescript KS+ was UV irradiated (450 J/m2). Both plasmids were treated with E. coli Nth protein, and closed-circular duplex DNA was purified from cesium chloride and sucrose gradients (46). Whole-cell extracts for repair were prepared as described previously (46). Reaction mixtures containing the indicated amounts of human cell extract protein in buffer D (45 mM HEPES-KOH [pH 7.8]; 70 mM KCl; 7.4 mM MgCl2; 0.9 mM DTT; 0.4 mM EDTA; 20 μM [each] dGTP, dCTP, and TTP; 8 μM dATP; 74 kBq of [α-32P]dATP [110 TBq/mmol]; 2 mM ATP; 22 mM phosphocreatine di-Tris salt; 2.5 μg of creatine phosphokinase; 3.4% glycerol; 18 μg of bovine serum albumin) were incubated at 30°C for 30 min, and then 250 ng of irradiated pBluescript KS+ and 250 ng of nonirradiated pHM14 were added to give a final reaction volume of 50 μl, and incubation continued for 3 h. Plasmid DNA was purified from the reaction mixtures, linearized (when appropriate), and loaded onto a 1% agarose gel containing ∼0.3 μg of ethidium bromide per ml. Data were analyzed by autoradiography with intensifying screens, densitometry, and liquid scintillation counting of the excised bands.
(ii) Dual-incision assay with cisplatin-damaged DNA.
Covalently closed-circular DNA containing a single 1,3-intrastrand d(GpTpG)-cisplatin cross-link (Pt-GTG) was produced, and the duplex form was purified as described previously (26). This DNA substrate was used to analyze the dual-incision process of NER, which leads to the displacement of platinated oligomer (24 to 32 nucleotides). The procedure involves incubation of cell extracts with Pt-GTG DNA (250 ng) in a 50-μl reaction mixture containing buffer D. After a 5-min preincubation at 30°C in the absence of DNA, DNA was added, and incubation was carried out for 30 min at 30°C. Where indicated, the DNA was purified and digested with XhoI and HindIII for 4 h at 37°C. The reactions were stopped by addition of formamide stop buffer containing bromophenol blue and xylene-cyanol. The DNA was denatured at 95°C for 5 min prior to being loaded on a 12% acrylamide gel. The bromophenol blue dye was allowed to migrate ∼30 cm from the wells before transfer of the DNA for 90 min onto a Hi-bond membrane soaked in 10× Tris-borate gel running buffer. The membrane was fixed in 0.4 M NaoH for 20 min, followed by a 2-min wash in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The fixed membrane was incubated at 42°C for 16 h in hybridization bottles containing 40 ml of buffer E (130 mM potassium phosphate [pH 7.0], 250 mM NaCl, 7% sodium dodecyl sulfate, 10% polyethylene glycol 8000) and 100 pmol of 32P-labelled 24-mer primer, which is complementary to the excised platinated oligomer (26). The membranes were washed for 10 min in 2× SSC buffer containing 0.1% SDS before exposure of the membrane to X-ray film.
RESULTS
UV-DDB activity defects in three newly identified XP-E cell lines.
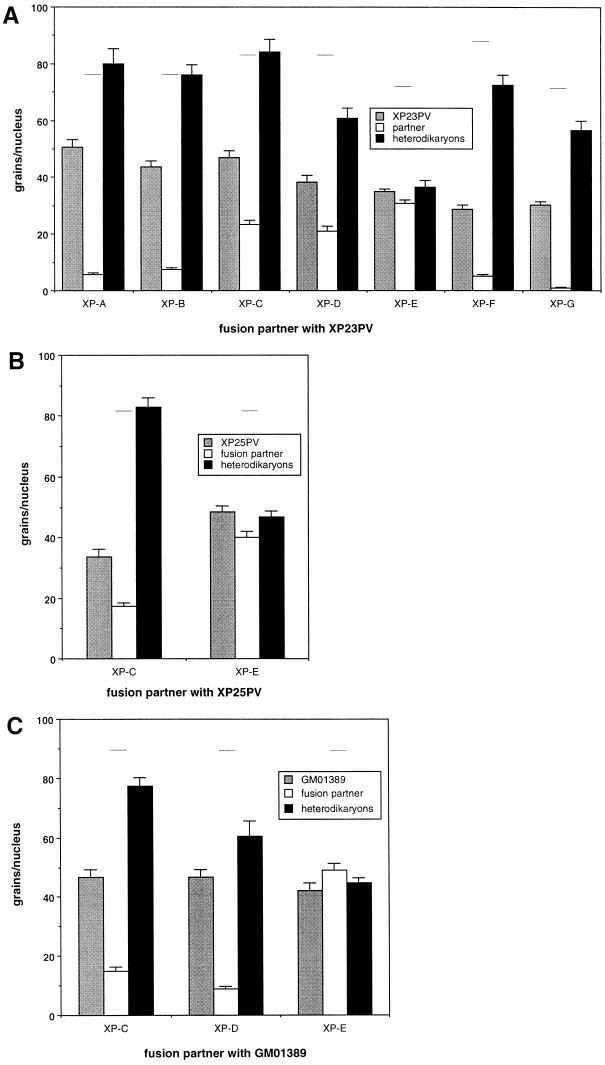
Fairly large numbers of XP-E cells are required in order to prepare whole-cell extracts for in vitro DNA repair studies, and lymphoblastoid cell lines are most convenient for this purpose. Cells from two previously undescribed Italian XP patients were found to be suitable. The individuals XP23PV and XP25PV are unrelated as far as can be determined and have XP cases of independent origin. A genetic complementation analysis of the repair defect in the cells from XP23PV and XP25PV revealed that both fell into XP-E, as shown in Fig. 1. The level of UV-induced DNA repair synthesis was analyzed in heterodikaryons obtained by fusion between each patient’s cells and XP cells belonging to different complementation groups. Fusion of XP23PV fibroblasts with cells from six XP groups (i.e., A to D and F to G) restored the capacity to perform repair synthesis in both nuclei, as expected if the repair defects are genetically different (Fig. 1A). In contrast, complementation was not observed in heterodikaryons between XP23PV and XP-E cells (XP2RO strain), with the XP23PV patient assigned to XP-E. Analogously, heterodikaryons of XP25PV cells resulting from fusion with the XP-E cells show no increase in UDS (Fig. 1B), indicating that patient XP25PV also falls into the XP-E group.
FIG. 1.
Complementation analysis in heterodikaryons. (A) Results obtained by fusion of XP23PV fibroblasts with cells representative of the seven excision repair-deficient XP groups (XP20PV, XP-A; XPCS2BA, XP-B; XP5PV, XP-C; TTD10VI, XP-D; XP2RO, XP-E; XP2VO, XP-F; and XP2BI, XP-G). The partners in each fusion were labelled with different-sized latex beads, and the level of UDS was analyzed 48 h following fusion. The columns indicate the mean number of autoradiographic grains per nucleus in homodikaryons of XP23PV (grey), the XP reference strain (white), and heterodikaryons (black). The bars indicate the standard error of the mean. The horizontal lines indicate the grain number per nucleus in normal C3PV cells analyzed in parallel. (B) Complementation analysis of heterodikaryons obtained by fusion of XP25PV fibroblasts with XP cells representative of XP-C (XP5PV) and XP-E (XP23PV). (C) Complementation analysis of heterodikaryons obtained by fusion of GM01389 fibroblasts (derived from the same patient who provided the GM01646 lymphoblastoid cells) with XP cells representative of XP-C (XP5PV), XP-D (XP17PV), and XP-E (XP23PV).
Cells from a third individual, represented by the primary fibroblast strain GM01389 and the lymphoblastoid cell line GM01646, were analyzed as described below and were found to lack UV-DDB activity. This was surprising, because the cells were initially reported to belong to the XP variant group (XP-V) when submitted to the Human Genetic Mutant Cell Repository, although published data supporting this assignment are not available. Complementation analysis showed that GM01389 cells have UDS that is about 60% of the normal level and that this repair deficiency can be corrected by fusion with XP-C or XP-D cells, but not with XP-E cells (Fig. 1C). It is clear that the cells from this individual are also in the XP-E group.
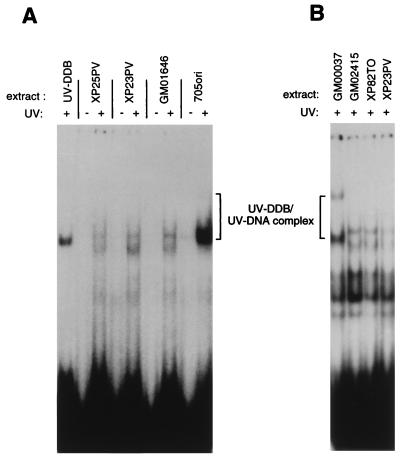
To characterize whether the newly identified XP-E cell lines contain UV-DDB protein able to bind damaged DNA, whole-cell extracts from XP23PV, XP25PV, and GM01646 lymphoblastoid cells were tested for binding to UV-damaged and undamaged DNA probes in an electrophoretic mobility shift assay (15, 33). All three extracts completely lacked specific binding activity (Fig. 2A). Two nonspecific bands, which were visible with less intensity when undamaged DNA was used and which migrated near the specific complex of UV-DDB and UV-irradiated DNA, were observed. Repair-proficient 705ori extract and a purified UV-DDB fraction run on the same gel were positive controls for damage-binding activity. Addition of more whole-cell extract protein from the XP-E cell lines (up to 10 μg) did not reveal any specific UV-DDB binding activity (not shown). To further confirm that the newly identified cell lines are DDB−, we compared extracts from XP23PV fibroblasts with extracts from the previously characterized DDB− XP-E cells from GM02415 and XP82TO. For this purpose, a procedure for making fibroblast extracts was used that requires fewer cells than are needed for a whole-cell extract (31). These extracts showed a UV-DDB-deficient profile (Fig. 2B) similar to that seen with XP23PV, XP25PV, and GM01646 lymphoblastoid extracts.
FIG. 2.
UV-DDB binding activity in three newly identified XP-E cell lines. An electrophoretic mobility shift assay for UV-DDB binding activity is shown, with UV-damaged and undamaged double-stranded 60-mer DNA oligonucleotides and whole-cell-extract protein (5 μg). The whole-cell extracts were prepared from the normal lymphoblastoid cell line 705ori or the XP-E lymphoblastoid cell lines GM01646, XP23PV, and XP25PV (A) or repair-proficient GM00037 fibroblasts and the XP-E fibroblast cell lines XP82TO and GM02415 (B). The UV-DDB–UV-DNA complex was separated on a nondenaturing 6% polyacrylamide gel. UV-DDB protein (0.1 ng) was used as a control.
NER by XP-E cell extracts and the effect of additional UV-DDB and RPA.
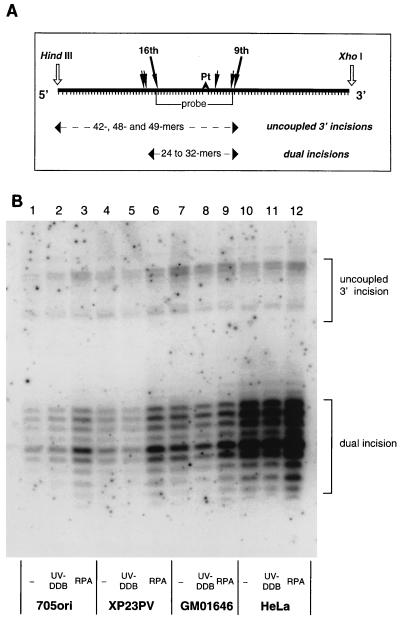
Extracts from 705ori and the three UV-DDB− cell lines were tested for their competence in NER by monitoring their ability to carry out dual incision of DNA containing a uniquely placed cisplatin adduct. All of the cell extracts could carry out repair of this adduct (Fig. 3, lanes 1, 4, 7, and 10, and data not shown), producing the characteristic pattern of 24- to 32-nucleotide excision fragments that is observed for this lesion when other human cell extracts are used (26). The amount of excision product increased with increasing amounts of DNA substrate in the reaction mixture (not shown), suggesting that under these conditions, the DNA lesion concentration is limiting. The results indicate that extracts from the newly characterized XP-E cell lines are not repair defective when naked DNA is the substrate.
FIG. 3.
(A) Schematic of the hybridization assay used to detect dual incisions and uncoupled 3′ incisions. The duplex DNA substrate containing the single cisplatin lesion (Pt) was cleaved with HindIII and XhoI before being probed with a labeled complementary strand overlapping the area around the lesion, indicated by “probe.” Dual incisions and uncoupled 3′ incisions are revealed by this method (9, 26). (B) NER by XP-E cell extracts and effect of additional RPA and UV-DDB. Reaction mixtures (50 μl) included 150 μg of normal (705ori), XP-E, or HeLa cell extract protein as indicated; 250 ng of closed-circular DNA containing a single 1,3-intrastrand d(GpTpG)-cisplatin adduct (1 nM adduct); and either 5 ng of UV-DDB protein or 50 ng of RPA. Incision products were detected by hybridization by the scheme in panel A and quantified with a phosphorimager.
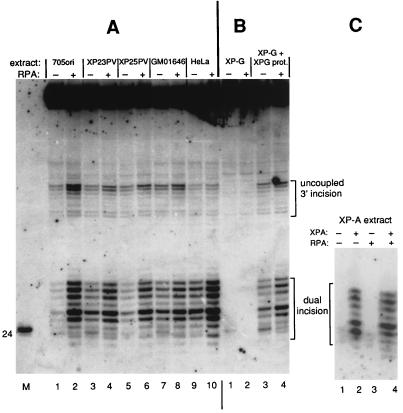
Various amounts of UV-DDB and RPA were added to reaction mixtures containing a limiting amount of DNA substrate (250 ng) in order to assess any effect of these proteins on repair by the extracts. An example is shown in Fig. 3. Other experiments (data not shown) led to the same conclusion as the one shown in the figure. Five nanograms (40 fmol) of UV-DDB protein was the maximum that could be added to reaction mixtures, but this addition had no discernible effect on the repair capacity of either normal (705ori and HeLa) or DDB− (GM01646 and XP23PV) extracts. Addition of 50 ng of RPA, on the other hand, enhanced repair synthesis in each case. The dual-incision products were quantified from a phosphorimage of the gel, and RPA stimulated repair by a factor of 2.2-fold for 705ori, 2.1-fold for XP23PV, 1.4-fold for GM01646, and 1.6-fold for HeLa. Addition of this amount of UV-DDB had no significant effect on the repair capacity of whole-cell extracts in these and additional experiments (the corresponding ratios were 1.1, 0.8, 0.9, and 1.0), and so we further explored the stimulation of repair by RPA.
Figure 4 shows results from an experiment in which the repair capacities of two non-XP cell extracts (705ori and HeLa) were compared with those of the three UV-DDB− extracts, XP23PV, XP25PV, and GM01646, with and without 100 ng of additional RPA. The extents of dual incision for all extracts were within the same range, and all were stimulated by RPA. The quantified extent of stimulation in three experiments varied from 1.5- to 6-fold and was greatest for extracts with the lowest intrinsic activity (705ori, in this case). These data strongly suggested that RPA has a generally stimulatory influence on the dual-incision reaction of NER. To confirm this general effect of RPA, we also examined the effect of this protein on repair reactions that were reconstituted by mixing completely repair-defective XP extracts with the cognate complementing repair protein. Figure 4B shows the outcome for XP-G cell extract complemented with XPG protein, and Fig. 4C shows the results for XP-A cell extract complemented with XPA protein. Neither the XP-G nor XP-A cell extract showed detectable dual (or uncoupled 3′)-incision activity, and as expected, addition of RPA to these extracts in the absence of respective complementing XPA or XPG protein had no effect. However, when repair in the extracts was restored by addition of complementing protein, it was enhanced by the additional RPA (Fig. 4B, lanes 3 and 4, and C, lanes 2 and 4).
FIG. 4.
RPA stimulates dual incision of DNA containing a cisplatin adduct. (A) Normal and XP-E cell extracts. Reaction mixtures included 250 ng of closed-circular DNA (∼1 nM lesion), cell extract protein (150 μg for 705ori, XP23PV, XP25PV, and GM01646; 100 μg for HeLa), and either 100 ng of additional RPA (+) or no (−) additional RPA. (B) XP-G cell extracts. Reaction mixtures included 250 ng of closed-circular DNA, 150 μg of extract protein from XPG83 XP-G cells (patient XP125LO), and 200 ng of XPG protein to correct the XP-G defect as indicated, with either 100 ng of additional RPA (+) or no (−) additional RPA. (C) XP-A cell extracts. Reaction mixtures included 250 ng of closed-circular DNA, 150 μg of extract protein from GM02345 XP-A cells (patient XP2OS), and 45 ng of XPA protein to correct the XP-A defect as indicated, with either 50 ng of additional RPA (+) or no (−) additional RPA.
NER synthesis in UV-irradiated DNA by normal and XP-E cell extracts.
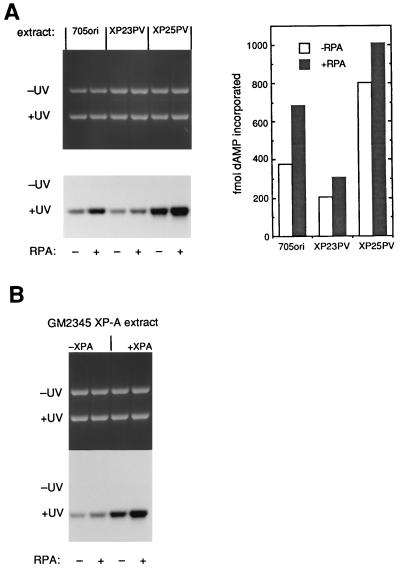
UV-DDB protein has a particular binding affinity for UV-induced (6-4) photoproducts in DNA, and so it was important to investigate the competence of the DDB− extracts for repair of these adducts and to check for any effect of RPA. An assay for damage-dependent repair synthesis carried out by whole-cell extracts with UV-irradiated plasmid DNA was performed, in which most of the repair synthesis arises from the repair of (6-4) photoproducts (45). The results of the repair synthesis experiment are shown in Fig. 5A. Normal extracts and each of the XP-E extracts (XP23PV, XP25PV, and GM01646) showed repair activity, and repair was stimulated in each case by addition of RPA (Fig. 5A and data not shown). The signal obtained in this assay is influenced not only by incision activity but also by the repair DNA synthesis activity of extracts, and so the magnitude of the signal can vary a few fold from one extract to another, precluding the use of error bars. Repair synthesis by cell extracts can also be used to detect complementation of repair-defective XP cell extracts (Fig. 5B). XPA protein-defective GM02345 extract by itself gives a low background signal which is not due to NER (18). Addition of XPA protein conferred NER capacity to the extract, and this XPA protein-complemented reaction was further stimulated by supplementation with RPA (Fig. 5B). This is consistent with the result obtained in Fig. 4 for repair of the cisplatin adduct. The background UV-dependent synthesis by XP-A extracts may represent residual base excision repair or synthesis of a small number of nicks in the UV-damaged template. This synthesis by XP-A extracts is proliferating cell nuclear antigen dependent (data not shown) and is therefore carried out by DNA polymerase delta or epsilon. RPA stimulates synthesis by DNA polymerase delta under some conditions (25), and this may be the reason that it also stimulates the background signal. Overall, the results in Fig. 3 to 5 show that additional RPA has the effect of increasing repair efficiency in vitro.
FIG. 5.
RPA stimulates repair synthesis in UV-irradiated DNA by normal and XP-E cell extracts. (A) Repair synthesis was measured by incubation of cell extracts with a mixture of undamaged (−UV) and UV-damaged (+UV) closed-circular plasmid DNA in a reaction mixture that includes [α-32P]dATP. Cell extract protein (150 μg) from the normal (705ori) or XP-E lines shown was incubated with (+) or without (−) 200 ng of additional RPA as indicated. Data were quantified and normalized for DNA recovery to give the results in the graph at the side. (B) Repair synthesis was measured in reaction mixtures as in panel A with 150 μg of protein from GM02345 XP-A cells (patient XP2OS). Where indicated, reaction mixtures included 45 ng of XPA protein to correct the repair defect of the GM2345 extracts and were supplemented with or without 200 ng of RPA.
Levels of RPA and UV-DDB proteins in XP-E cells.
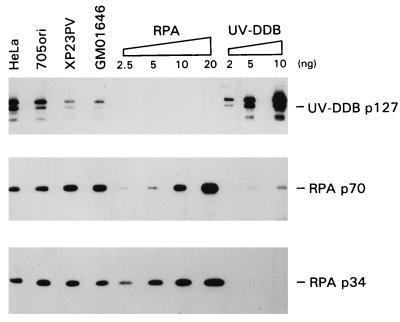
The findings presented above prompted us to examine the levels of RPA in cell extracts and to quantify the total amount of RPA in the reaction mixtures used in NER repair assays. Two of the subunits, RPA p34 and p70, were measured by immunodetection and were similar in normal and XP-E cell extracts (Fig. 6). The amount of RPA in whole-cell extracts was quantified by comparison to various amounts of recombinant human RPA, and the results are shown in Table 1. There is on the order of 100 ng of RPA heterotrimer per 150 μg of extract, the amount used in the experiments of Fig. 3 to 5, and an amount similar to that of the exogenous RPA added in those experiments. Immunoblotting of the purified UV-DDB fraction confirmed our earlier observation (31) that some RPA copurifies with UV-DDB. The RPA p70 subunit was predominantly detected in this fraction. Although partially degraded, about 10 ng of p70 per μl of purified UV-DDB was estimated.
FIG. 6.
Expression of RPA and UV-DDB p127 in normal and XP-E cells. Whole-cell extracts (20 μg of protein) from HeLa, 705ori, XP23PV, and GM01646 cells, RPA (2.5, 5, 10, and 20 ng), and purified UV-DDB (2, 5, and 10 ng) were separated by SDS–10% polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane, and probed with specific anti-p127, anti-p34, and anti-p70 antibodies. Data were quantified, and the amounts of RPA and UV-DDB in each cell line were calculated by calibration with the purified proteins (Table 1).
TABLE 1.
RPA and UV-DDB protein content of whole-cell extracts
| Cell extract | Protein content (ng/150 μg of extract protein)
|
||
|---|---|---|---|
| RPA p70 | RPA p34 | UV-DDB p127 | |
| HeLa | 57 | 30 | 37 |
| 705ori | 60 | 40 | 30 |
| GM01646 | 78 | 30 | 11 |
| XP23PV | 75 | 36 | 16 |
It was clearly of interest to determine the UV-DDB content of the DDB− XP-E cell extracts. In contrast to the nearly constant levels of RPA, the level of p127 protein in XP23PV and GM01646 was about one-half that in normal 705ori or HeLa cells. This result was reproducible for different extract preparations and was also found for XP25PV extracts (not shown). Other XP-E fibroblast extracts have also been observed to have reduced p127 levels compared to those of repair-proficient TC7 or HeLa cells (33a). We presently do not know if this difference is caused by a lower expression of UV-DDB or by an increased degradation of the protein. Immunoblotting revealed some products ascribed to degradation of p127 in all extracts (Fig. 6).
Microinjection of UV-DDB and RPA.
In view of the in vitro results, it was important to confirm that UV-DDB protein could actually enhance repair in vivo after microinjection into DDB− XP-E cells, as was reported for UV-DDB purified independently by a different method (22). Fibroblasts from a DDB− XP-E cell line were microinjected and assessed for UDS activity, with the results shown in Table 2. The XP-E fibroblasts had 57% of the UDS level of repair-proficient cells, a value in the anticipated range. Microinjection of UV-DDB protein indeed enhanced UDS in the XP-E cells, to about 67% of the normal level. The effect is less marked than in the previous report, perhaps because it was only possible to microinject about 0.1 additional cell equivalent of UV-DDB in these experiments. About 50 fl (∼0.5 cell equivalent) of purified RPA was also microinjected into cells, and no stimulatory effect was seen. In living cells, therefore, the amount of RPA may not be rate limiting for repair.
TABLE 2.
Stimulation of repair in XP-E cells by microinjection of UV-DDB
| Cells injected | Sample | No. of grains/nucleus | SE | No. of nuclei scored |
|---|---|---|---|---|
| XP2RO | Not injected | 41.2 | 1.1 | 54 |
| UV-DDB | 51.9 | 2.1 | 55 | |
| XP2RO | Not injected | 44.8 | 1.3 | 59 |
| RPA | 44.9 | 1.2 | 61 | |
| UV-DDB | 50.2 | 1.4 | 43 | |
| C5RO | Not injected | 75.8 | 3.1 | 38 |
DISCUSSION
Heterogeneity of XP-E.
A number of characteristics make group E unique among the NER-defective complementation groups of XP. Patients in the XP-E group have mild dermatological manifestations and are neurologically unaffected. Consistent with this, the repair defect in all known XP-E cells is also mild, showing 40 to 60% of normal repair and a modest sensitivity to UV light in comparison to the sensitivities of other XP groups. This moderate phenotype has probably been the major reason that a gene for XP-E has not been isolated by functional complementation of UV sensitivity.
A significant advance in the study of XP-E was made when it was recognized that extracts from the originally identified XP-E cell lines were lacking a strong DNA-damage binding activity, UV-DDB (3). However, group E is also remarkable for its heterogeneity, in that only a fraction of group E cell lines show a defect in UV-DDB binding activity. Of the 16 previously reported XP-E patients, 3 lack UV-DDB activity and 13 retain it. Several suggestions have been offered for this heterogeneity within XP-E, including UV-DDB mutations in some patients that do not affect in vitro DNA binding activity but alter some other aspect of its interaction with the repair complex or the existence of a separate gene that still shows a lack of complementation in cell fusion (23). All three of the additional XP-E cell lines reported here lack the activity, bringing the number of DDB− cell lines to 6 out of a total of 19 XP-E lines examined. This significantly strengthens the correlation between XP-E and UV-DDB activity. Moreover, with the availability of these three new lines, we and others can more definitively pursue the molecular cause of XP-E.
Differential effect of UV-DDB on XP-E in vivo and in vitro.
Further study of the UV-DDB protein has strongly suggested that it is functionally involved in the XP-E phenotype. Most directly, microinjection of purified UV-DDB containing approximately equimolar amounts of p127 and p48 subunits into DDB− XP-E cells can partially or fully correct their defect in repair in vivo, as measured by UDS (22). Correction was also seen with the UV-DDB preparation studied here, which contained more p127 subunit than p48 subunit of UV-DDB. The repair defect in the DDB+ class of XP-E cells is not corrected by microinjection of UV-DDB (22).
The UV-DDB protein in its p127 or p127-p48 form is not, however, a core component of the mammalian NER machinery, because it is not required to repair lesions in plasmid DNA with purified proteins in a soluble system (1, 28). Small amounts can stimulate repair a few fold in a purified system under appropriate conditions (1), although larger amounts of UV-DDB protein suppress repair. Taken together, the in vivo and in vitro results suggest that UV-DDB may have a specific role in repair of chromosomal DNA in the nuclear environment, which is not easily revealed in vitro. Indeed, studies of the binding of UV-DDB to chromatin show that it significantly varies in salt extractability after irradiation of cells, indicating translocation of its position in the nucleus to a tightly bound chromatin fraction (31). We hypothesize that the role of UV-DDB is to mobilize nucleosomes, freeing up damaged DNA for repair by the large catalytic “repairosome” complex (analogous to proteins which have or recruit histone acetylase activity, thereby facilitating the transcription of chromatinized DNA). Experiments to confirm this notion are in progress.
A number of reports indicate that the damaged-DNA binding activity of UV-DDB resides in the large p127 subunit. Surprisingly, analysis of the DNA sequence of the cloned p127 gene has not revealed any causative mutations for XP-E. However, in the three DDB− cases of XP-E examined to date, sequence changes different from normal have been detected in the p48 gene (29), and there is a real possibility that these are causative mutations for XP-E. Some evidence suggests that p48 can significantly modulate the damaged-DNA binding activity of the p127 subunit (29). A sequence analysis of p48 in the three XP-E patients analyzed here will be informative as to whether p48 is a causative gene for XP-E.
RPA stimulates NER in vitro, but is not specific for XPE extracts.
A different origin of the nucleotide excision repair defect in XP-E has also been proposed, in connection with the single-stranded DNA binding protein RPA. RPA is a core component of the human NER system and is necessary for incision of UV-irradiated DNA (35) and DNA containing other lesions (26, 27). RPA binds to XPA protein, and together these proteins have a substantial affinity for damaged DNA over nondamaged DNA (13) and form part of the damage recognition apparatus of NER.
Extracts from the three XP-E cell lines identified here were found to perform appreciable NER in vitro on both UV-irradiated DNA and DNA containing a single cisplatin lesion. These results demonstrate for the first time that there is no significant difference between repair of a naked DNA substrate with extracts from XP-E cells or normal cells, lending strong support to our hypothesis about the role of UV-DDB (i.e., UV-DDB, absent in the XP-E extracts, would only be required for a nucleosome-laden DNA substrate and not for a naked DNA substrate). UV-DDB binds strongly to (6-4) photoproducts, the lesions responsible for most of the repair synthesis seen here, as well as to cisplatin-treated DNA (32), and so any large effect of a UV-DDB defect on repair in vitro by whole-cell extracts would have been observed. An inherent variability between different extract preparations makes it difficult to gauge whether the XP-E extracts show slightly less repair than normal cell extracts. In contrast, extracts from several DDB+ and DDB− XP-E lines were reported to be severely defective in NER, but specifically correctable by RPA (20). These puzzling observations are not consistent with the results found here. We have repeatedly attempted to make repair extracts from the one XP-E lymphoblastoid cell line available from the Human Genetic Mutant Cell Repository, GM02450 (derived from patient XP3RO). However, this line grows too poorly to prepare enough cells for a reliable whole-cell extract. It is possible that the poor growth characteristics of this XP-E line and perhaps other XP-E lines often result in inactive or very-low-activity extracts.
We did find that additional RPA can stimulate NER in vitro on either UV-irradiated DNA or DNA containing a single cisplatin lesion. Stimulation of up to sixfold was found for XP-E extracts, extracts from normal cells, or extracts from cells of other XP groups complemented with the cognate XP protein. This enhancement of repair by added RPA is in agreement with previous studies in which exogenous RPA purified from HeLa cells increased repair several fold in UV-irradiated DNA or DNA containing an acetylaminofluorene lesion, as measured in a repair synthesis assay (4, 5). Thus, RPA is normally required for NER, and addition of RPA to normal cell extracts further stimulates this reaction in vitro. While the DNA substrates used here have a lower density of lesions than the substrate employed by Kazantsev et al. (20), this difference should not affect the results, since our substrates are entirely free of single-stranded DNA (26). Thus, RPA cannot be rate limiting in our experiments because of sequestration on single-stranded DNA. Increased repair by RPA is particularly evident under suboptimal reaction conditions with smaller amounts of DNA substrate, suggesting that RPA is rate limiting under these conditions. The most likely explanation is that RPA affects the damage-recognition step and that additional RPA increases the rate of damage recognition and hence the incision step. The stimulatory effect of RPA is confined to the incision stage of NER (4). Although RPA can increase the activity of isolated eukaryotic DNA polymerases under some conditions (42), it does not increase the overall extent of the gap-filling stage during NER (36).
As a core component of the NER reaction, the requirement for RPA is very specific in that other single-stranded DNA binding proteins cannot substitute. For example, neither the E. coli single-stranded DNA binding protein SSB nor adenovirus DNA binding protein could replace human RPA in reversing the effect of an anti-RPA antibody (4). Moreover, recent studies have revealed that the activity of human RPA in repair depends on specific domains in the protein that are not critical for strong binding to single-stranded DNA. A form of recombinant RPA with a deletion of the first 277 amino acids of the p70 subunit was produced by Lin et al. (24). The remaining residues 278 to 616 of the p70 subunit still form a tight heterotrimer with the p34 and p14 subunits, and the complex can be purified in soluble form. The mutant RPA complex binds single-stranded DNA with full avidity at low ionic strength (50 mM NaCl) and with 40% avidity at high ionic strength (0.5 M NaCl) (24). This RPA also binds nearly as well to simian virus 40 T antigen, as does the wild type, and stimulates human DNA polymerase delta as well as does intact RPA. However, the mutant RPA is completely unable to support NER, as measured by repair synthesis in UV-damaged DNA or incision of DNA containing the same cisplatin lesion as that used here (25). This mutant RPA also has no stimulatory or dominant negative effects when added to repair reaction mixtures containing nonmutated recombinant RPA (34a).
Finally, the total amount of RPA in XP-E cells appears normal (Fig. 6). These results, coupled with the lack of mutations in any of the RPA subunits (20), make it very unlikely that alterations in RPA are responsible for XP-E. Nevertheless, there is good evidence that RPA interacts with UV-DDB protein (31), and this interaction might be important during nucleotide excision repair of nuclear DNA in cells. We suggest that further studies of XP-E need to be focused on the UV-DDB protein complex and the mechanism of its action, because this avenue of study seems most likely to reveal the underlying cause of the repair defect.
ACKNOWLEDGMENTS
We thank Colin Arlett and Mahmud Shivji for invaluable assistance with reagents and Suzanne Rademakers for preparing fused cells for microinjection.
This work was supported by the National Institutes of Health, the Imperial Cancer Research Fund, and grants from the Associazione Italiana per la Ricerca sul Cancro.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Podust V N, Protić M, Hübscher U, Egly J-M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Abramic M, Levine A S, Protić M. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J Biol Chem. 1991;266:22493–22500. [PubMed] [Google Scholar]
- 3.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 4.Coverley D, Kenny M K, Lane D P, Wood R D. A role for the human single-stranded DNA binding protein HSSB/RPA in an early stage of nucleotide excision repair. Nucleic Acids Res. 1992;20:3873–3880. doi: 10.1093/nar/20.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coverley D, Kenny M K, Munn M, Rupp W D, Lane D P, Wood R D. Requirement for the replication protein SSB in human DNA excision repair. Nature. 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 6.de Weerd-Kastelein E, Keijzer W, Bootsma D. A third complementation group in xeroderma pigmentosum. Mutat Res. 1974;22:87–91. doi: 10.1016/0027-5107(74)90013-x. [DOI] [PubMed] [Google Scholar]
- 7.Dualan R, Brody T, Keeney S, Nichols A F, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA-binding protein. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 8.Evans E, Fellows J, Coffer A, Wood R D. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans E, Moggs J G, Hwang J R, Egly J-M, Wood R D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldberg R S, Grossman L. A DNA binding protein from human placenta specific for ultraviolet damaged DNA. Biochemistry. 1976;15:2402–2408. doi: 10.1021/bi00656a024. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 12.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision-repair with purified rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 13.He Z, Henricksen L A, Wold M S, Ingles C J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 14.Henricksen L, Umbricht C, Wold M. Recombinant replication protein A: expression, complex-formation, and functional-characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 15.Hirschfeld S, Levine A S, Ozato K, Protić M. A constitutive damage-specific DNA-binding protein is synthesized at higher levels in UV-irradiated primate cells. Mol Cell Biol. 1990;10:2041–2048. doi: 10.1128/mcb.10.5.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang B J, Chu G. Purification and characterization of a human protein that binds to damaged DNA. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- 17.Hwang B J, Liao J C, Chu G. Isolation of a cDNA encoding a UV-damaged DNA binding factor defective in xeroderma pigmentosum group E cells. Mutat Res DNA Repair. 1996;362:105–117. doi: 10.1016/0921-8777(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 18.Jones C J, Wood R D. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 19.Kataoka H, Fujiwara Y. UV damage-specific DNA-binding protein in xeroderma-pigmentosum complementation group E. Biochem Biophys Res Commun. 1991;175:1139–1143. doi: 10.1016/0006-291x(91)91684-5. [DOI] [PubMed] [Google Scholar]
- 20.Kazantsev A, Mu D, Nichols A F, Zhao X D, Linn S, Sancar A. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc Natl Acad Sci USA. 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeney S, Chang G J, Linn S. Characterization of a human DNA-damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 22.Keeney S, Eker A P M, Brody T, Vermeulen W, Bootsma D, Hoeijmakers J H J, Linn S. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage binding protein. Proc Natl Acad Sci USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney S, Wein H, Linn S. Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat Res. 1992;273:49–56. doi: 10.1016/0921-8777(92)90049-9. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y-L, Chen C, Keshav K F, Winchester E, Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y-L, Shivji M, Chen C, Kolodner R, Wood R, Dutta A. The evolutionarily conserved zinc finger motif in the largest sub-unit of human RPA is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem. 1998;273:1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 26.Moggs J G, Yarema K J, Essigmann J M, Wood R D. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- 27.Mu D, Hsu D S, Sancar A. Reaction-mechanism of human DNA-repair excision nuclease. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 28.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA-repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 29.Nichols A F, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E DDB− phenotype. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 30.O’Donovan A, Wood R D. Identical defects in DNA repair in xeroderma pigmentosum group G and rodent ERCC group 5. Nature. 1993;363:185–188. doi: 10.1038/363185a0. [DOI] [PubMed] [Google Scholar]
- 31.Otrin V, McLenigan M, Takao M, Levine A S, Protić M. Translocation of a UV-damaged DNA-binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J Cell Sci. 1997;110:1159–1168. doi: 10.1242/jcs.110.10.1159. [DOI] [PubMed] [Google Scholar]
- 32.Payne A, Chu G. Xeroderma-pigmentosum group E binding-factor recognizes a broad spectrum of DNA damage. Mutat Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 33.Protić M, Levine A S. Detection of DNA damage-recognition proteins using the band-shift assay and Southwestern hybridization. Electrophoresis. 1993;14:682–692. doi: 10.1002/elps.11501401109. [DOI] [PubMed] [Google Scholar]
- 33a.Rapić Otrin, V., and M. Protić. Unpublished data.
- 34.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J S, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts t[c,s]t, t[t,s]t, t[6-4]t, and t[dewar]t. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 34a.Shivji, M., and R. D. Wood. Unpublished results.
- 35.Shivji M K K, Kenny M K, Wood R D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 36.Shivji M K K, Podust V N, Hübscher U, Wood R D. Nucleotide excision repair DNA synthesis by DNA polymerase ɛ in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 37.Stefanini M, Giliani S, Nardo T, Marinoni S, Nazzaro V, Rizzo R, Trevisan G. DNA-repair investigations in 9 Italian patients affected by trichothiodystrophy. Mutat Res. 1992;273:119–125. doi: 10.1016/0921-8777(92)90073-c. [DOI] [PubMed] [Google Scholar]
- 38.Stefanini M, Vermeulen W, Weeda G, Giliani S, Nardo T, Mezzina M, Sarasin A, Harper J I, Arlett C F, Hoeijmakers J H J, Lehmann A R. A new nucleotide-excision-repair gene associated with the disorder trichothiodystrophy. Am J Hum Genet. 1993;53:817–821. [PMC free article] [PubMed] [Google Scholar]
- 39.Takao M, Abramic M, Moos M, Otrin V R, Wootton J C, McLenigan M, Levine A S, Protić M. A 127 kDa component of a UV-damaged DNA-binding complex, which is defective in some xeroderma pigmentosum group E patients, is homologous to a slime-mold protein. Nucleic Acids Res. 1993;21:4111–4118. doi: 10.1093/nar/21.17.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thielmann H W, Popanda O, Edler L, Jung E G. Clinical symptoms and DNA-repair characteristics of xeroderma pigmentosum patients from Germany. Cancer Res. 1991;51:3456–3470. [PubMed] [Google Scholar]
- 41.Treiber D K, Chen Z H, Essigmann J M. An ultraviolet light-damaged DNA recognition protein absent in xeroderma pigmentosum group E cells binds selectively to pyrimidine (6-4) pyrimidone photoproducts. Nucleic Acids Res. 1992;20:5805–5810. doi: 10.1093/nar/20.21.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, α and δ. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Assendelft G B, Rigney E M, Hickson I D. Purification of a HeLa cell nuclear protein that binds selectively to DNA irradiated with ultraviolet light. Nucleic Acids Res. 1993;21:3399–3404. doi: 10.1093/nar/21.15.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wold M S. Replication protein A: a heterotrimeric single-stranded DNA binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 45.Wood R D. Repair of pyrimidine dimer ultraviolet light photoproducts by human cell extracts. Biochemistry. 1989;28:8287–8292. doi: 10.1021/bi00447a005. [DOI] [PubMed] [Google Scholar]
- 46.Wood R D, Biggerstaff M, Shivji M K K. Detection and measurement of nucleotide excision repair synthesis by mammalian cell extracts in vitro. Methods. 1995;7:163–175. [Google Scholar]