Abstract
The multisubunit yeast transcription factor IIIC (TFIIIC) is a multifunctional protein required for promoter recognition, transcription factor IIIB recruitment, and chromatin antirepression. We report the isolation and characterization of TFC7, an essential gene encoding the 55-kDa polypeptide, τ55, present in affinity-purified TFIIIC. τ55 is a chimeric protein generated by an ancient chromosomal rearrangement. Its C-terminal half is essential for cell viability and sufficient to ensure TFIIIC function in DNA binding and transcription assays. The N-terminal half is nonessential and highly similar to a putative yeast protein encoded on another chromosome and to a cyanobacterial protein of unknown function. Partial deletions of the N-terminal domain impaired τ55 function at a high temperature or in media containing glycerol or ethanol, suggesting a link between PolIII transcription and metabolic pathways. Interestingly, τ55 was found, together with TFIIIC subunit τ95, in a protein complex which was distinct from TFIIIC and which may play a role in the regulation of PolIII transcription, possibly in relation to cell metabolism.
In eucaryotic cells, the transcription of a variety of small genes is conducted by RNA polymerase III (PolIII) and requires several auxiliary factors. For yeast tRNA gene (tDNA) activation, preinitiation complexes are assembled in a defined order within and upstream of the transcription unit (18, 27, 52). Transcription factor IIIC (TFIIIC) plays a primary role in this multistep complex assembly by binding to the intragenic promoter elements of tRNA genes (the A and B blocks). Yeast TFIIIC is a remarkably large multisubunit factor made of two protein subassemblies, named τA and τB, that have distinct DNA binding properties, that can be visualized by electron microscopy in a free or DNA-bound form (46), and that can be cleaved by limited proteolysis (37). The τB domain binds tightly to the B block (37) and has been shown to display all the properties of enhancer binding proteins (11). Binding of the τA domain to the A block is weaker and mostly B block dependent. Once bound, TFIIIC promotes the binding of transcription factor IIIB (TFIIIB) upstream of the transcription start site (6, 26, 28, 31). The process is similar for yeast 5S RNA gene activation, except that TFIIIC assembly is dependent upon the binding of transcription factor IIIA (TFIIIA) to the internal promoter sequence. TFIIIB by itself does not bind detectably to TATA-less PolIII genes but, once assembled via TFIIIC, interacts intimately with DNA and is sufficient, at least in the yeast system, for directing accurate initiation by PolIII during multiple rounds of transcription in vitro (28, 31). Hence, TFIIIB is the initiation factor required for the activation of all PolIII genes, whereas TFIIIC and TFIIIA act as assembly factors. However, it has been shown that TFIIIC is a multifunctional protein, involved not only in promoter recognition and TFIIIB recruitment but also in the displacement of nucleosomes to relieve the repression of transcription by chromatin (10).
The molecular structure of yeast TFIIIB and TFIIIC has been much investigated. Yeast TFIIIB comprises three components: TBP, the TATA box binding protein, which is also required for transcription by PolI and PolII, and two additional polypeptides, TFIIIB70/BRF1 and TFIIIB90/B", first identified by protein-DNA cross-linking (6). Together with TBP, TFIIIB70 is able to bind to TFIIIC-tDNA complexes (29). The resulting complex becomes competent to recruit PolIII after the assembly of TFIIIB90 (29, 30). Purified yeast TFIIIC comprises six polypeptides, of 138, 131, 95, 91, 60, and 55 kDa (5, 16, 43). Purification of TFIIIC to near homogeneity, protein-DNA cross-linking (5, 9, 16), and coimmunoprecipitation experiments (13, 44) suggested that the four largest polypeptides were subunits of TFIIIC, a suggestion which was confirmed by gene cloning (2, 34, 36, 44, 48). The 138- and 95-kDa components (τ138 and τ95), located in τB and τA, respectively, are thought to be DNA binding subunits, since they could be specifically cross-linked to a tDNA probe and were mapped at the level of the B block and the A block, respectively (5, 9, 13, 16). τ91 was recently shown to cooperate with τ138 for TFIIIC-tDNA binding (2) and was mapped at the most 3′ location of TFIIIC-5S RNA gene complexes (9). τ131 stands as the TFIIIB-assembling subunit based on its upstream gene location, shown by protein-DNA cross-linking (5, 6), and its direct interaction with both TFIIIB70 and TFIIIB90 (12, 32, 45). On the other hand, little is known about the smallest polypeptides, of 60 and 55 kDa, which reproducibly copurify with yeast TFIIIC activity. Both proteins were found among the six polypeptides isolated from TFIIIC-tDNA complexes (16). A 55-kDa polypeptide was located by photo-cross-linking experiments together with τ95, on opposite sides of the DNA helix, in the vicinity of the A block of tRNA genes (5, 6). The 60-kDa polypeptide was not cross-linked to DNA.
We report here the cloning and identification of TFC7, an essential gene encoding the 55-kDa subunit of TFIIIC. Analysis of deletion mutants showed that only the C-terminal half of τ55 is necessary for TFIIIC transcriptional activity. We found that τ55 interacts with τ95 and that these two subunits are present in at least two distinct protein complexes.
MATERIALS AND METHODS
Yeast strains and methods.
YNN281 × YNN282 (22) was used for gene disruption. Preparation of media, tetrad dissection, and other yeast methods were performed by standard techniques (3). Plasmids harboring modified alleles of the TFC7 gene were used to transform the YNM2 haploid strain (ade2-101 lys2-801 ura3-52 trp1-Δ1 his3-Δ200 tfc7-Δ::HIS3 pNM2). The modified copies of TFC7 were substituted for wild-type TFC7 by plasmid shuffling on plates containing 5-fluoro-orotic acid. Viable strains isolated at 30°C were also tested for growth at 37 and 16°C.
Purification and immunoprecipitation of TFIIIC.
TFIIIC was purified starting from 30 g (wet weight) of Saccharomyces cerevisiae cells following the procedure described by Huet et al. (25). Immunoprecipitation was performed as described by Ossipow et al. (39). Cells expressing wild-type or epitope-tagged versions of τ138, τ131, or τ95 (25, 34, 36) were harvested in the exponential phase, and crude extracts were prepared as described by Huet et al. (25), except that protease inhibitors (O-complete; Boehringer) and extraction buffer containing 20 mM HEPES (pH 7.5), 50 mM CH3COOK, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% glycerol were used. Proteins were precipitated with ammonium sulfate, resuspended in 5% of the original crude extract volume in dialysis buffer (25 mM HEPES [pH 7.5], 100 mM KCl, 0.1 mM EDTA, 0.25 mM DTT, 10% glycerol), and dialyzed twice for 2 h each time at 4°C against 250 volumes of the same buffer. Typically, 10 g (wet weight) of yeast cells yielded 2 ml of dialyzed extract containing 15 to 30 mg of protein/ml, as estimated by Bradford analysis (8). Per assay, 1.2 μg of mouse monoclonal antihemagglutinin (HA) antibodies (53) was incubated for 30 min at 10°C with 20 μl of magnetic beads (8 × 108 beads/ml in phosphate-buffered saline containing 0.1% bovine serum albumin [BSA]) coated with rat monoclonal antibodies directed against mouse immunoglobulin G2b (Dynal M450). After extensive washing in phosphate-buffered saline containing 0.1% BSA and then in dialysis buffer, the beads were incubated with gentle shaking at 10°C with 50 μl of dialyzed extract. After 3 h of incubation, the beads were washed three times with 200 μl of washing buffer (25 mM HEPES [pH 7.5], 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.1% Triton X-100). Proteins were eluted by incubation for 30 min at room temperature with 16 μl of washing buffer containing 2 mg of a synthetic peptide corresponding to the HA sequence per ml. Immunoprecipitated proteins were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Amino acid sequence determination.
TFIIIC was purified on a preparative scale following the immunopurification procedure described by Huet et al. (25). Affinity-purified fractions containing TFIIIC DNA binding activity were pooled (133 fractions, 66.5-ml final volume). Proteins were precipitated with cold trichloroacetic acid (10% final concentration) for 40 min in ice and centrifuged at 4°C for 4 h at 17,600 ×g. Pellets were washed with cold acetone, centrifuged (4°C, 20 min, 17,600 × g, and resuspended in 0.6 ml of SDS electrophoresis buffer (3). Proteins were separated by electrophoresis overnight through an 8% polyacrylamide–SDS gel and slightly stained with Coomassie blue. Polypeptides of 138, 131, 95, 91, 80, 75, 60, 55, and 50 kDa were revealed. Starting from 880 g (wet weight) of YCS7 cells expressing an HA-tagged version of τ95 (25), about 200 to 250 pmol of TFIIIC was obtained. A gel slice containing the 95-kDa subunit was used for anti-τ95 production (see below). A gel slice containing the 55-kDa polypeptide was excised, crushed, and incubated with a protease as described previously (48), except that proteinase K was used instead of trypsin. The resulting peptides were separated by reversed-phase high-pressure liquid chromatography and sequenced. Seven peptide sequences (YDNPRM, EIPVY, TYIPF, ELAFPN, ERLVGT, FASPF, and SDRKWV) were determined.
Cloning and disruption of TFC7.
Two degenerate oligonucleotides (Ol20 [5′CGGAATTCRTTNGGRAANGCNARYTC] and Ol8 [5′NNTAYGAYAAYCCNMGNATG]) designed from peptides ELAFPN and YDNPRM, respectively, were used to amplify a yeast genomic DNA fragment by “touchdown” PCR (14). A 509-bp DNA fragment was obtained, cloned into pBSKS (Stratagene), sequenced, and found to contain a continuous open reading frame (ORF) encoding the two initial peptides plus three others. The sequence of the entire TFC7 gene was found by searching the Munich Information Centre for Protein Sequences (MIPS) database (GenBank accession no. Z75018).
Disruption of the TFC7 gene was performed by a PCR method (7). Two 57-mer oligonucleotides harboring sequences complementary to the TFC7 gene and to the yeast HIS3 selectable marker were used to amplify by PCR an ≈1.1-kb DNA fragment that was directly introduced into the yeast YNN281 × YNN282 strain by transformation. In the resulting His+ transformants, one copy of the whole TFC7 ORF was replaced by the HIS3 gene, surrounded by stop codon modules, and inserted in the antisense direction with respect to TFC7. Diploid transformants were verified by PCR analysis. The heterozygous diploid strain was then transformed with the multicopy plasmid pNM2 harboring TFC7 and sporulated. One spore bearing the chromosomally deleted allele of TFC7 but containing the pNM2 plasmid was chosen to yield strain YNM2 used for plasmid shuffling.
Construction of plasmids.
The 2.6-kb EcoRI/KpnI DNA fragment from cosmid cospEOA273 containing the coding and flanking sequences of TFC7 was cloned into plasmid YEplac195 (19), creating pNM2. The sequence encoding a methionine residue followed by the YPYDVPDYA epitope (HA epitope) derived from the influenza virus HA protein (53) was added just before the initiation codon of TFC7 by PCR-mediated mutagenesis of plasmid pNM2. Two oligonucleotides, NM8 (5′-TCCTTTTCAATACATATGTATCCTTACGACGTTCCTGATTATGCCATGGTGGTGAACAC) and NM7 (5′-TCAGCGGGATCCTTACATAGGGCGGACATTGC), were used for mutagenesis. NM8 contains the epitope coding sequence (boldface letters) and nucleotides that are mostly complementary to TFC7 DNA and that create NdeI and NcoI restriction sites. NM7 is complementary to TFC7 and harbors a BamHI restriction site (boldfaced) just after the stop codon. The amplified DNA fragment was cloned into the pGEM-T vector (Promega), creating pNM8. The NdeI/BamHI or NcoI/BamHI fragments from pNM8 were cloned into pET28a (Novagen), pAS2, and pACT2 (21), creating pNM11, pAS-τ55, and pACT-τ55, respectively.
An NdeI restriction site was inserted in the TFC7 gene promoter by PCR-mediated mutagenesis of plasmid pNM2 with the oligonucleotides NM5 (5′-CAGCCATTGACCCCAAAATGAGAA) and NM9 (5′-CGTGTTCACCACCATATGTATTGAAAAGGA). The resulting PCR product was cloned into the pGEM-T vector, creating pNM10. The SphI/NdeI fragment from pNM10 and the NdeI/BamHI fragment from pNM8 were sequentially cloned into YCplac22 (19), creating pNM12. The τ55-ΔN1, τ55-ΔN2, τ55-ΔN3, and τ55-ΔC mutants were constructed by deletions of nucleotides 49 to 288, 286 to 834, 34 to 834, and 835 to 1335, respectively, of the epitope-tagged version of TFC7. These mutants were obtained by cleavage of pNM8 with BsaBI/XbaI, XbaI/EcoRV, StyI/EcoRV, and EcoRV/BamHI, respectively, treatment with Klenow DNA polymerase or mung bean nuclease, religation, and sequencing of the junction. The resulting NdeI/BamHI DNA fragments were cloned into pNM12, in place of the wild-type tagged version of TFC7, creating pNM24, pNM25, pNM26, and pNM15, respectively. The EcoRI/SalI DNA fragments from pNM12, pNM15, pNM24, pNM25, and pNM26 were cloned into pUN45 and used to transform YNM2 yeast cells.
Expression of TFC7 in Escherichia coli.
Recombinant TFC7 protein (rTFC7p) tagged at its N-terminal end with six histidines and with the HA epitope was expressed from plasmid pNM11 in E. coli BL21(pLysS). Crude extract preparation and protein purification on Ni2+-nitrilotriacetic acid (NTA)–agarose (Qiagen) under native conditions were performed as described by Chaussivert et al. (12).
Anti-τ55, -τ95, and -τ131 polyclonal antibodies.
rTFC7p and recombinant τ131ΔTPR2 (12) were expressed as hexahistidine fusions, purified on Ni2+-NTA–agarose, and loaded on a preparative SDS–8% polyacrylamide gel. Gel slices containing the recombinant τ55 or τ131 derivatives or the 95-kDa subunit from the preparative SDS-polyacrylamide gel used for the τ55 amino acid sequence determination (see above) were excised and injected into rabbits (three to six injections at 3-week intervals) for the production of antibodies. Preimmune antibodies or antibodies directed to TFIIIC subunits were purified on protein A-Sepharose as described previously (20), and the protein concentration was estimated by Bradford analysis (8).
Interaction of τ55 with 35S-labeled τ95.
Far-Western experiments were performed as described previously (24), except that rTFC7p was denatured-renatured on filters before incubation with the labeled probe. Plasmid pCS5, harboring the TFC1 gene encoding τ95 (48), was linearized with StuI. The gene was transcribed in vitro with T7 RNA polymerase in wheat germ extracts (Promega) in the presence of [35S]methionine. Recombinant τ55 was subjected to SDS-PAGE, blotted onto nitrocellulose, and denatured-renatured according to the method of Papavassiliou and Bohmann (42). The filters were incubated with 35S-labeled τ95, washed, and autoradiographed. Recombinant τ55 was located by use of anti-HA antibodies, and immune complexes were visualized with an ECL kit (Amersham).
Two-hybrid assays.
The expression of GAL4(1-147)-τ55 and GAL4(768-881)-τ55 fusion proteins from plasmids pAS-τ55 and pACT-τ55, respectively, was verified by Western blot analysis of yeast crude extracts with polyclonal antibodies directed against GAL4. GAL1-LacZ activation assays were performed as described previously (12) after transformation of a yeast strain with combinations of plasmids. Transcriptional activation of the lacZ reporter gene was assayed by growing the transformed cells on selective medium and overlaying them with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) agar. β-Galactosidase activity in yeast extracts was measured exactly as described previously (51), at 30°C for at least three independent transformants. The interaction between TFIIIB70 and τ131 (12) was used as a reference.
DNA binding and transcription assays.
The transcription factor-tDNA interaction was monitored by a gel retardation assay essentially as described previously (35). TFIIIC was partially purified from wild-type or mutant crude extracts by chromatography on an Ultrogel-heparin A4R (Sepracor) column as described previously (25). Heparin-purified TFIIIC fractions (1 μl, ≈0.5 μg of protein) were incubated with a 32P-labeled DNA probe (3 to 10 fmol; 3,000 to 10,000 cpm) in 15 μl of binding buffer containing 10 mM Tris-HCl (pH 8), 1 mM EDTA, 150 mM KCl, 10% glycerol, 50 μg of BSA, and 1 μg of competitor DNA. The probe was a 200-bp PCR-amplified fragment from plasmid pUC-Glu (17), carrying the yeast tRNA3Glu gene, or a 200-bp PCR-amplified fragment from plasmid pGE2 (4), harboring only the B block of the yeast tRNA3Leu gene. Complexes were analyzed by nondenaturing gel electrophoresis and revealed by autoradiography (25).
Transcription assays were performed with three different templates: pUC-Glu (17), harboring the tRNA3Glu gene; pRS316-SUP4, containing the yeast SUP4 tRNA gene (a gift from S. Shaaban); and pGE2 (4), harboring the yeast tRNA3Leu gene. Transcription reactions were carried out for 45 min at 25°C in 40-μl mixtures containing 20 mM HEPES-KOH (pH 7.9); 10% glycerol; 5 mM MgCl2; 90 mM KCl; 0.1 mM EDTA; 1 mM DTT; 1 U of RNasin (Amersham); 0.6 mM each ATP, GTP, and CTP; 0.03 mM [32P]UTP (2 to 10 Ci/mmol); 0.1 μg of plasmid DNA; heparin-purified TFIIIC fraction (1 μl, ≈0.5 μg); RNA PolIII (50 ng); recombinant TFIIIB70 (150 ng); recombinant TBP (40 ng); and B" fraction (400 ng). Protein fractions were prepared according to the method of Huet et al. (25). RNA transcripts were analyzed by polyacrylamide-urea gel electrophoresis and revealed by autoradiography (25).
Nucleotide sequence accession number.
The GenBank accession number for TFC7 is Z75018.
RESULTS
Isolation and disruption of the TFC7 gene.
Yeast TFIIIC is a multisubunit protein that comprises six polypeptides, of 138, 131, 95, 91, 60, and 55 kDa. In order to clone the gene encoding the 55-kDa subunit, TFIIIC was purified on a preparative scale from crude extracts by an immunoaffinity and DNA affinity purification procedure (25). TFIIIC components were separated by preparative SDS-PAGE and stained with Coomassie blue. The TFIIIC fraction contained the six polypeptides consistently found in affinity-purified fractions plus three additional polypeptides, of 80, 75, and 50 kDa. The 75-kDa polypeptide, occasionally found in purified TFIIIC fractions, was recently shown to be immunologically related to the 91-kDa protein (2). The 80- and 50-kDa polypeptides were found to be contaminants that can be easily separated from TFIIIC with a linear gradient of ammonium sulfate instead of a salt elution step for the DNA affinity column. The fraction containing the 50- and 80-kDa polypeptides had RNase activity that we did not explore further.
The 55-kDa polypeptide was excised from the gel, and several peptides were obtained after proteinase K digestion. The amino acid sequences of seven peptides were determined. Degenerate oligonucleotides designed from the amino acid sequences of two peptides were used as primers to amplify a genomic DNA fragment by “touchdown” PCR (14). A 509-bp DNA fragment was obtained and found to contain a continuous ORF that encoded three other peptides. At this stage, the sequence of the entire gene, hereafter named TFC7, was obtained from the MIPS database (GenBank accession no. Z75018). The TFC7 gene is located on chromosome XV.
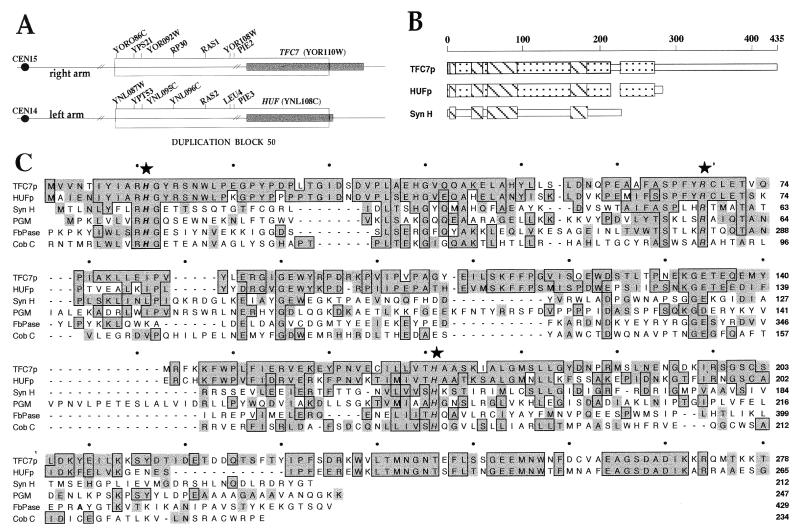
The TFC7 ORF encodes a 435-amino-acid protein with a predicted Mr of 49,000 and a theoretical pI of 5 and that contains the seven microsequenced peptides. Comparison of the TFC7 protein (TFC7p) sequence with the National Center for Biotechnology Information (NCBI) nonredundant database by use of the BLAST program server (1) revealed intriguing similarities between the N-terminal half of TFC7p and other proteins unrelated to transcription. First, the N-terminal domain of TFC7p shows 64% identity and 80% similarity (Fig. 1C) to an S. cerevisiae 30.7-kDa protein of unknown function and encoded by a gene located on chromosome XIV and provisionally named HUF (for homolog of unknown function). It appears that the TFC7 and HUF genes are part of one of two large blocks of gene duplication between chromosomes XIV and XV. As shown in Fig. 1A, the TFC7 and HUF genes are located just at the border of duplication block 50, as defined by Wolfe and Shields (54). The coding sequence for the N-terminal part of TFC7p belongs to duplication block 50, whereas the coding sequence for the C-terminal half is present only on chromosome XV.
FIG. 1.
Sequence analysis of TFC7. (A) Schematic representation of duplication block 50 as defined by Wolfe and Shields (54). The locations on chromosomes XIV and XV of the duplicated genes (TFC7 and HUF) are indicated by shading. (B) The regions of sequence similarities among TFC7p, HUFp, and Syn H are schematically represented. (C) Sequence similarities to TFC7p. The N-terminal part of TFC7p (amino acids 1 to 278) was aligned with the following protein sequences: HUFp, S. cerevisiae 30.7-kDa hypothetical protein; Syn H, Synechocystis hypothetical protein; PGM, S. cerevisiae PGM; FbPase, S. cerevisiae FbPase; and Cob C, S. typhimurium α-ribazole-5′-phosphate phosphatase (GenBank accession no. Z75018, Z71385, D64002, P00950, S42124, and U12808, respectively). Complete sequences are shown only for Syn H and PGM. The flanking portions of the other proteins, which showed no homology to TFC7p, are not included. The amino acid positions for each sequence are indicated on the right. Identical residues are boxed, and conserved substitutions are shaded. Conserved active-site residues of PGM, FbPase, and Cob C enzymes are indicated by stars.
The N-terminal part of TFC7p also shows regions of sequence similarity (Fig. 1B and C) to a Synechocystis 23.7-kDa protein (Syn H), again of unknown function, as well as to a family of phosphoglycerate mutase (PGM), fructose-2,6-biphosphatase (FbPase), and acid phosphatase enzymes from various species. These enzymes catalyze similar phosphotransfer reactions that involve a phosphohistidine intermediate. Their activity has been shown to be dependent on two highly conserved histidinyl residues and one arginyl residue (reference 40 and references therein). Figure 1C shows a comparison of TFC7p N-terminal half, HUF protein (HUFp), and Syn H sequences with sequences of PGM and FbPase enzymes from S. cerevisiae and the Cob C acid phosphatase from Salmonella typhimurium. The TFC7p N-terminal moiety showed approximately 20% identity and 40% similarity at the amino acid sequence level to both PGM and Cob C enzymes (and 15% identity and 30% similarity to the FbPase enzyme). The three highly conserved catalytic residues as well as the amino acids flanking the histidinyl residues were conserved in TFC7p. These sequence similarities to proteins clearly unrelated to transcription and to a cyanobacterial protein raised the question of the functional role of TFC7p, if any, in PolIII transcription. On the other hand, no similarity to existing sequences in databases could be found for the C-terminal half of TFC7p, which is not included in duplication block 50 (Fig. 1A).
To test whether TFC7 was essential for cell growth or viability, a DNA fragment harboring the yeast HIS3 gene surrounded by stop codons was inserted in place of the whole TFC7 ORF by a PCR method (7). The resulting diploid cells (YNM1 strain) had one of the chromosomal copies of the TFC7 gene deleted. Analysis of the sporulation products revealed two nonviable and two viable (always his−) spores per tetrad, showing that TFC7 was an essential gene. To confirm this result, strain YNM1 was transformed with multicopy plasmid pNM2, harboring TFC7, and sporulated. The resulting haploid strain with a chromosomal disruption but expressing the plasmidic TFC7 gene was viable. Thus, TFC7, like the genes encoding the four largest subunits of TFIIIC, was essential for cell viability. On the other hand, we found that HUF, the homolog of unknown function of the TFC7 gene, was expressed but was not essential for cell viability (results not shown).
TFC7 encodes the 55-kDa subunit of TFIIIC.
The TFC7 gene was engineered to add six histidine residues and an epitope derived from the influenza virus HA protein (HA epitope) at the N terminus of the protein. The tagged protein was expressed in E. coli cells and purified as a histidine fusion under nondenaturing conditions on Ni2+-NTA–agarose. Western blot analysis of the purified protein fraction was performed with monoclonal antibodies directed against the HA epitope. The HA-tagged recombinant protein (predicted Mr, 50,000) migrated on an SDS-polyacrylamide gel with an apparent size of ≈56 kDa, suggesting that TFC7p is not posttranslationally modified.
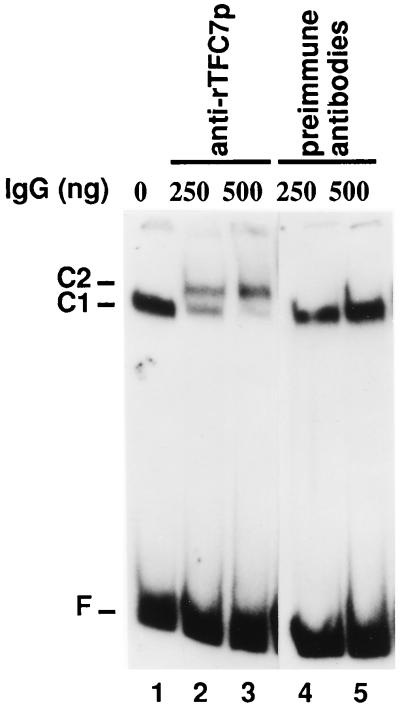
rTFC7p was injected into rabbits for antibody production. Immunoblotting performed with immune serum raised against rTFC7p specifically revealed the 55-kDa polypeptide present in the DNA affinity-purified TFIIIC fraction (data not shown). In order to investigate whether TFC7p was one of the TFIIIC subunits, purified anti-rTFC7p antibodies were used to alter the migration of TFIIIC-tDNA complexes in mobility shift assays (16, 48). Preformed TFIIIC-tDNA3Glu complexes were incubated with polyclonal anti-rTFC7p antibodies (or preimmune control immunoglobulins) and analyzed by electrophoresis on a polyacrylamide gel. As shown in Fig. 2, anti-rTFC7p antibodies were able to bind to TFIIIC-tDNA complexes (C1), thus converting them into larger species (C2) that migrated more slowly (lanes 2 and 3). On the other hand, the migration of TFIIIC-tDNA complexes was not affected by preimmune antibodies (Fig. 2, lanes 4 and 5). The supershift induced by anti-rTFC7p antibodies showed that TFC7p is part of TFIIIC-tDNA complexes and corresponds to τ55.
FIG. 2.
TFC7 encodes a subunit of TFIIIC. TFIIIC was purified by heparin chromatography and assayed by gel retardation with a labeled tDNA3Glu probe. Preformed TFIIIC-tDNA3Glu complexes (lane 1) were incubated with 250 or 500 ng of preimmune antibodies (lanes 4 and 5) or anti-rTFC7p antibodies (lanes 2 and 3). Complexes were separated by nondenaturing electrophoresis and revealed by autoradiography. C1, position of TFIIIC-tDNA complexes; C2, position of complexes bound by anti-rTFC7p antibodies; F, free labeled tDNA.
Analysis of τ55 mutants.
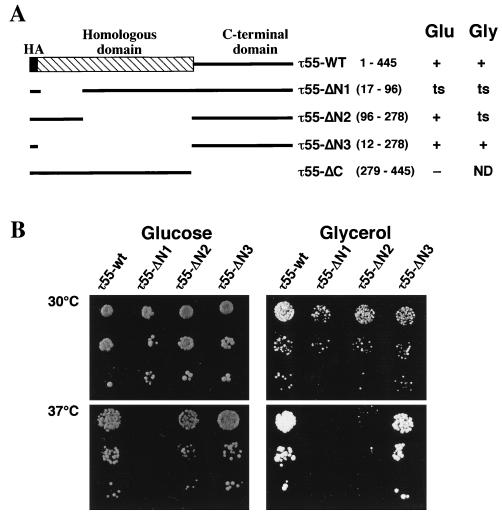
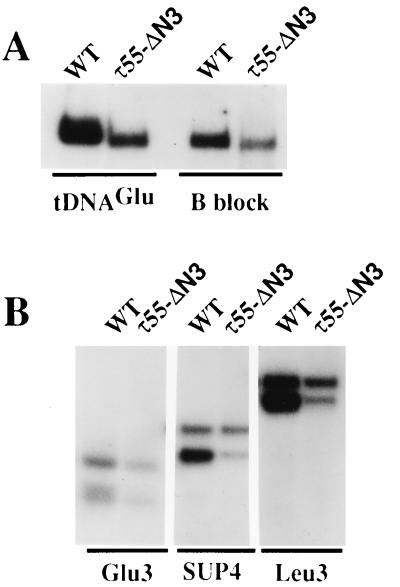
The similarity of the N-terminal half of TFC7p to a cyanobacterial protein or to enzymes related to the glycolytic pathway was intriguing. To define τ55 domains that were necessary for TFIIIC activity, N-terminal and C-terminal deletions (Fig. 3A) in an HA-tagged version of τ55 were generated as described in Materials and Methods. Centromeric plasmids harboring mutant copies of the TFC7 gene were tested for their ability to functionally replace a chromosomally disrupted copy of TFC7. The resulting strains were grown at different temperatures in medium containing either glucose or glycerol as a carbon source. As shown in Fig. 3A, deletion of the C-terminal domain (τ55-ΔC) was lethal, suggesting that this domain is essential for TFIIIC function. In contrast, deletion of the whole N-terminal domain (τ55-ΔN3), homologous to HUFp, resulted in a wild-type phenotype (Fig. 3B). This in vivo result was confirmed by in vitro studies. Wild-type and τ55-ΔN3 TFIIIC factors were purified by heparin chromatography and tested for tDNA binding activity by gel retardation assays or for their ability to promote the transcription of different templates in the presence of TFIIIB and RNA PolIII. Figure 4A shows that the deletion form of TFIIIC was able to form a complex with the tRNA3Glu gene. The migration of the TFIIIC-tDNA3Glu complexes formed could be altered by anti-HA antibodies, showing that epitope-tagged τ55-ΔN3 associated with the other subunits to form TFIIIC (data not shown). Deletion of the N-terminal half of τ55 noticeably increased the rate of migration of TFIIIC-tDNA3Glu complexes, and the same increase in migration rate was observed when a DNA fragment harboring only the B block of the tRNA3Leu gene was used as a probe (Fig. 4A). This difference in migration between the wild-type and mutant TFIIIC-tDNA complexes is not easily explained by the 5% difference in molecular mass between wild-type and mutant factors (30 kDa of ≈600 kDa) and could reflect different conformational states of the factor.
FIG. 3.
Deletion analysis of τ55. (A) Deletion mutants were constructed as described in Materials and Methods from an HA-tagged version of τ55 (τ55-WT). τ55-WT and τ55 deletions are schematically represented. The positions of deleted amino acids (inclusive) for each construct are indicated in parentheses. Centromeric plasmids harboring a deletion mutant copy of TFC7, expressed from its own promoter, were tested for their ability to functionally replace, at different temperatures and either in glucose (Glu)- or in glycerol (Gly)-containing medium, a chromosomally disrupted copy of TFC7. A summary of the viability and thermal sensitivity of the strains is shown; lethal (−), wild-type (+), and temperature-sensitive (ts) phenotypes are indicated; ND, not determined. (B) Viability and thermal sensitivity of yeast strains harboring wild-type τ55 or deletion variants of τ55. Growth at 30 and 37°C in glucose- or glycerol-containing medium of the wild-type or viable τ55 deletion strains is shown.
FIG. 4.
DNA binding and transcriptional activities of τ55-ΔN3 TFIIIC. TFIIIC was purified by heparin chromatography from the wild-type strain (WT) or the mutant strain expressing only the C-terminal part of τ55 (τ55-ΔN3). (A) Formation of TFIIIC-DNA complexes. Factor-DNA complexes were formed as described in Materials and Methods by incubating wild-type or τ55-ΔN3 TFIIIC with DNA probes harboring either the tRNA3Glu gene or the B block of the tRNA3Leu gene, as indicated. Complexes were analyzed by electrophoresis and autoradiography and quantified by scanning. (B) Transcription of various class III genes by τ55-ΔN3 TFIIIC. Wild-type or τ55-ΔN3 TFIIIC was incubated with three tRNA genes in reconstituted transcription mixtures as described in Materials and Methods. The different templates are indicated. The RNA products were isolated by electrophoresis, revealed by autoradiography, and quantified by scanning. For the Glu3 template, only the upper RNA band was taken into account, whereas for the SUP4 and Leu3 templates, both RNA bands, corresponding to primary and mature transcripts, were quantified. The different relative yields of primary and mature transcripts depended on the level of maturase activity present in the heparin-purified TFIIIC fraction.
Even though the TFIIIC fractions purified from wild-type and mutant strains were shown by Western blot analysis to contain similar amounts of TFIIIC, based on the 95-kDa subunit (data not shown), the yield of complexes obtained with the τ55-ΔN3 TFIIIC fraction was about twofold lower than that obtained with the wild-type factor. This result, obtained with two distinct preparations of the τ55-ΔN3 factor, suggested that the mutant form of TFIIIC was slightly defective in DNA binding or more unstable during purification than the wild-type factor.
Next, the τ55-ΔN3 factor was assayed for specific transcription of three different tRNA genes with or without an intron in a reconstituted transcription system in the presence of TFIIIB and PolIII. As shown in Fig. 4B, using the same amount of TFIIIC (based on the 95-kDa subunit), the deletion form of TFIIIC showed reduced levels of transcriptional activity compared with the wild type, about twofold, in keeping with the twofold-lower amount of TFIIIC-tDNA complexes formed with the mutant factor. The lower transcriptional activity of the mutant form of TFIIIC could therefore be accounted for by its deficiency in DNA binding. Furthermore, the deletion form of TFIIIC was able to assemble TFIIIB at a correct gene position, since the transcripts obtained with both factors migrated at the same level. We concluded, therefore, that the N-terminal half of τ55 may play a role in the stability and/or the conformation of TFIIIC but that the C-terminal domain is sufficient to support τ55 function in TFIIIC-tDNA binding and proper TFIIIB assembly.
In contrast to deletion of the whole N-terminal domain, homologous to HUFp, partial deletion of this domain of τ55 (τ55-ΔN1) resulted unexpectedly in a temperature-sensitive phenotype (Fig. 3B). This effect was specific to the τ55-ΔN1 deletion, since a different partial deletion, τ55-ΔN2, did not detectably affect cell growth at 37°C in glucose-containing medium. Since the N-terminal part of τ55 has regions of sequence similar to those of enzymes related to the glycolytic pathway (PGM or FbPase; Fig. 1), mutant strains were also grown in medium containing glycerol or ethanol. Cells growing in glycerol medium underwent glycolysis more slowly than did cells growing in glucose medium as a result of the poor uptake of glycerol by yeast cells, whereas cells growing in medium containing ethanol, a respiratory substrate, did not undergo glycolysis. One could imagine that if the N-terminal part of τ55 has an activity which interferes with metabolism, the growth of N-terminal deletion mutants in medium containing either glycerol or ethanol instead of glucose would be affected. As shown in Fig. 3B, the growth of τ55-ΔN2 mutant cells in glycerol-containing medium resulted in a temperature-sensitive phenotype that was not observed in glucose-containing medium. Furthermore, the doubling time of τ55-ΔN1 mutant cells grown at 30°C in glycerol-containing medium was increased (about twofold) compared to those of wild-type τ55, τ55-ΔN2, or τ55-ΔN3 mutant cells. Similar results were obtained when cells were grown in medium that contained ethanol (or ethanol plus glycerol) instead of glucose (data not shown). In all cases, the wild-type phenotype could be restored by transformation of the mutant cells with a centromeric plasmid harboring the wild-type TFC7 gene. We also verified that our mutant strains were rho+. We concluded from these results that the N-terminal domain, when partially deleted, can impair τ55 function. Furthermore, the reduced rate of growth of τ55-ΔN1 mutant cells at 30°C as well as the thermal sensitivity of τ55-ΔN2 mutant cells observed only when the cells were grown with glycerol or ethanol instead of glucose suggested a potential link between metabolic pathways and PolIII transcription.
τ55 interacts with τ95.
To gain some insight into the function of the 55-kDa subunit of TFIIIC, we used the two-hybrid system to study the interactions between τ55 and all the components of the PolIII system cloned so far. This method previously revealed interactions of τ131 with TFIIIB70 (12) as well as with TFIIIB90 (45). The TFC7 ORF was fused to the DNA binding domain [GAL4(1-147)] or to the transcriptional activation domain [GAL4(768-881)] of the yeast GAL4 protein. All combinations between these τ55 fusion proteins and the TFIIIB (TFIIIB70, TFIIIB90, and TBP), TFIIIC (τ138, τ131, τ95, and τ91), TFIIIA, and PolIII (C160, C128, C82, C53, AC40, C34, C31, C27, AC19, ABC10α, and ABC10β) complementary fusion proteins were assayed (2, 12, 51). Activation of the lacZ reporter gene was estimated by β-galactosidase assays of selected transformants. The interaction between TFIIIB70 and τ131 (12) was used as a reference. Significant levels of β-galactosidase activity were detected when τ55 fused to the transcriptional activation domain was assayed with the τ95 complementary fusion (Table 1). This observation suggested that τ55 and τ95 interacted in the cell. The reciprocal combination gave lower but significant levels of β-galactosidase activity (data not shown). Next, we investigated the interaction between τ95 and deletion versions of τ55. The truncated fragments of τ55 shown in Fig. 3A were fused to the DNA binding or transcriptional activation domain of GAL4 and assayed with the reciprocal τ95 fusions. Deletions of the N-terminal domain of τ55 did not alter significantly the τ55-τ95 interaction. On the other hand, deletion of the C-terminal domain of τ55 gave background β-galactosidase levels (data not shown). This result suggested that the C-terminal part of τ55 is essential for its interaction with τ95.
TABLE 1.
In vivo interaction of τ55 with τ95 in the two-hybrid system
| Fusion proteins | β-Galactosidase activitya | |
|---|---|---|
| GAL4(1-147) | GAL4(768-881)-τ55 | 6 |
| GAL4(1-147)-τ95 | GAL4(768-881) | 10 |
| GAL4(1-147)-τ95 | GAL4(768-881)-τ55 | 81 |
| GAL4(1-147)-TFIIIB70 | GAL4(768-881)-τ131 | 133 |
Nanomoles of o-nitrophenyl-β-d-galactoside hydrolyzed per minute per milligram of protein.
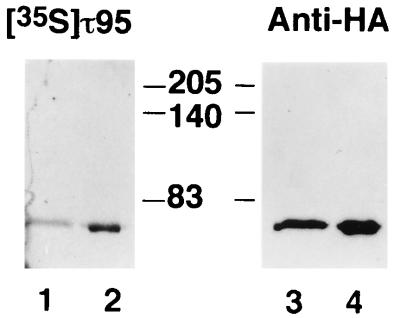
The interaction between τ55 and τ95 was confirmed in vitro with a far-Western blotting experiment (24). Partially purified rTFC7p was subjected to SDS-PAGE, transferred to membranes, denatured-renatured, and probed with 35S-labeled τ95 and then with antibodies directed to the HA epitope present at the N-terminal end of rTFC7p. As shown in Fig. 5, τ95 specifically bound at the level of τ55. No signal was obtained when the filters were incubated with another labeled protein (τ91) or in similar experiments in which τ95 was used as a labeled probe to interact with different blotted polypeptides (τ95, TBP, and TFIIIB70) (results not shown).
FIG. 5.
τ55 interacts with τ95. Recombinant τ55, expressed as an HA-tagged hexahistidine fusion, was purified from E. coli cells under native conditions. Eluted polypeptides (1 and 5 μg; respectively, lanes 1 and 2) were subjected to SDS-PAGE, transferred to a membrane, denatured-renatured, and probed with 35S-labeled τ95 as described in Materials and Methods. Labeled polypeptides were revealed by autoradiography (lanes 1 and 2). The same membrane was incubated with anti-HA antibodies (lanes 3 and 4), and immune complexes were visualized with an ECL kit. The molecular masses of marker polypeptides are indicated in kilodaltons.
Existence of a τ55-τ95 subcomplex distinct from TFIIIC.
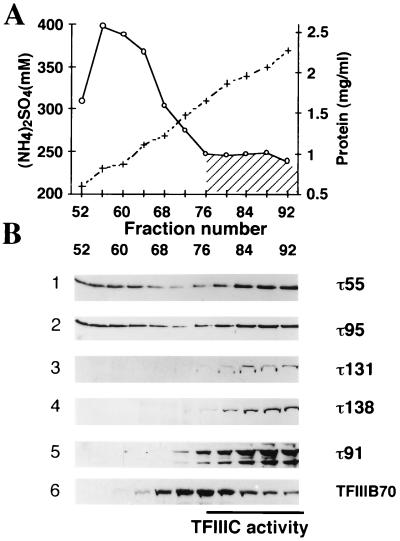
In other work on the purification of TFIIIC from yeast crude extracts, we observed that a β-galactosidase–τ95 fusion protein eluted in two peaks during heparin chromatography (12a). The 95-kDa subunit was present in fractions that contained the tDNA binding activity of TFIIIC but also in fractions eluting at lower salt concentrations and that contained no detectable tDNA binding activity. We decided to study more precisely the elution profile of the different TFIIIC subunits during heparin chromatography to correlate the presence of TFIIIC subunits to the factor tDNA binding activity. Extracts from wild-type or various epitope-tagged yeast strains were chromatographed on heparin columns, and proteins were eluted with an ammonium sulfate gradient. Fractions were assayed for salt and protein concentrations as well as for DNA binding activity on a 32P-labeled tDNA (Fig. 6A). Eluted proteins were then subjected to SDS-PAGE, transferred to nitrocellulose membranes, and probed with polyclonal antibodies directed against τ55 or τ95. Anti-HA antibodies were used to reveal epitope-tagged τ91, τ131, or τ138 subunits. As shown in Fig. 6B, τ55, τ91, τ95, τ131, and τ138 eluted in fractions (76 to 92) containing 300 to 350 mM ammonium sulfate, with the tDNA binding activity. In contrast to τ91, τ131, or τ138, both τ55 and τ95 also coeluted at lower salt concentrations (200 to 250 mM ammonium sulfate), in fractions 52 to 68, which did not contain detectable TFIIIC-tDNA binding activity. These results suggested that τ55 and τ95 exist in two forms: either associated with TFIIIC or as a subcomplex potentially containing other proteins but not TFIIIB. TFIIIB70 (Fig. 6B) and TFIIIB (data not shown) transcriptional activities coeluted in fractions 68 to 92, well after the τ55-τ95 complex (fractions 52 to 68).
FIG. 6.
Elution profile for TFIIIC subunits during chromatography on a heparin column. (A) Cell extracts from the wild-type strain or strains expressing HA-τ91, HA-τ131, or HA-τ138 were chromatographed on a heparin column as described previously (25). Eluted fractions were assayed for ammonium sulfate (+) or protein (○) concentrations. The DNA binding activity of TFIIIC assayed by gel retardation is indicated by the hatched zone. The data correspond to the results obtained with fractions from the wild-type strain. Similar results were obtained with fractions from the epitope-tagged strains. (B) Heparin fractions (20 μl) were subjected to SDS-PAGE, proteins were transferred to membranes, and filters were incubated with antibodies. Immune complexes were revealed with an ECL kit. Heparin fractions from the wild-type strain were probed with polyclonal antibodies directed against τ55 (row 1) and then with polyclonal antibodies directed against τ95 (row 2) or TFIIIB70 (row 6). Heparin fractions from HA-τ131 (row 3), HA-τ138 (row 4), or HA-τ91 (row 5) were probed with anti-HA antibodies.
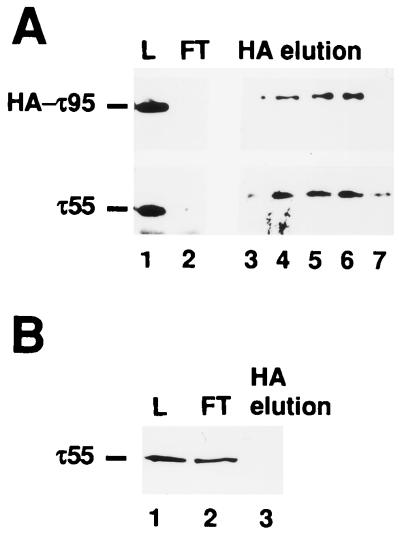
In order to verify the physical association of τ55 and τ95, the putative subcomplex was prepared from a strain expressing an HA-tagged version of τ95. After heparin chromatography, protein fractions that eluted at 200 to 250 mM ammonium sulfate and that contained both τ55 and HA-τ95 were pooled and further purified by immunoaffinity chromatography on an anti-HA column as described previously (25). Proteins eluted by competition with the HA peptide were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-HA antibodies or polyclonal antibodies directed to τ55 (Fig. 7A). As expected, HA-tagged τ95 bound to the anti-HA column (Fig. 7A, lanes 1 and 2) and was eluted by competition with the HA peptide (lanes 3 to 7). τ55 was also retained on the antibody column (Fig. 7A, lane 2) and was coeluted with τ95 (lanes 3 to 7). In order to verify that the coelution of τ55 with τ95 resulted from the stable association of these two subunits and was not due to the fortuitous binding of τ55 to the anti-HA column, a similar experiment was performed with a control heparin fraction prepared from a wild-type strain harboring untagged τ95. As shown in Fig. 7B, when τ95 was untagged, τ55 (as well as untagged τ95 [results not shown]) was fully recovered in the flowthrough fraction (Fig. 7B, lane 2) and was not detected in HA-eluted fractions (lane 3). These results confirmed the existence of a τ55-τ95 subcomplex that may include other polypeptides. When analyzed by silver staining, the eluted fractions contained both HA-τ95 and τ55 as well as several other polypeptides (data not shown).
FIG. 7.
τ55 is stably associated with τ95. Cell extracts from strains expressing HA-τ95 or wild-type τ95 were chromatographed on a heparin column as described previously (25). (A) Heparin fractions eluting at 200 to 250 mM ammonium sulfate and containing both HA-τ95 and τ55 were pooled (30 ml) and further chromatographed on a 4-ml anti-HA column as described previously (25). Proteins were eluted (1-ml fractions) by competition with the synthetic HA peptide (0.1 mg/ml). Protein samples (40 μl) were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-HA antibodies or polyclonal antibodies directed to τ55. Immune complexes were revealed with an ECL kit. Lanes: 1 (L), heparin fraction loaded onto the anti-HA column; 2 (FT), flowthrough fraction; 3 to 7, eluted fractions. (B) Heparin fractions (1.5 ml) containing untagged τ95 and τ55 were chromatographed on an anti-HA column (0.15 ml) as described above. Proteins were eluted batchwise (0.1-ml fraction) by competition with the synthetic HA peptide. Protein samples (40 μl) were analyzed by Western blotting with antibodies directed to τ55. Lanes are as in panel A.
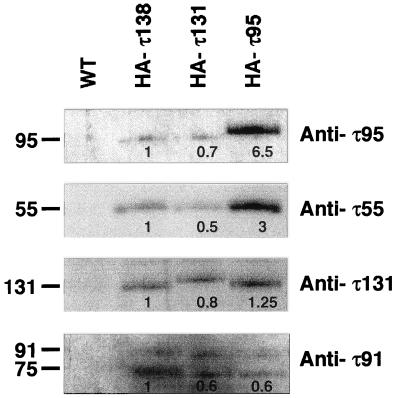
To determine whether this τ55-τ95 subcomplex was also present in a crude extract and to confirm that it was not due to partial TFIIIC dissociation during heparin chromatography, immunoprecipitations were performed with crude extracts prepared from a wild-type strain or from strains expressing an HA-tagged copy of τ138 (HA-τ138), τ131 (HA-τ131), or τ95 (HA-τ95). Anti-HA antibodies were used for immunopurification, and bound proteins were eluted by competition with a specific peptide antigen. Using the immunopurified proteins from HA-τ138, HA-τ131, or HA-τ95 crude extracts, we succeeded in reconstituting the transcription of a tRNA gene, and TFIIIC activity was recovered with similar efficiencies in all three cases (data not shown). Eluted TFIIIC fractions were analyzed by Western blotting and probed with polyclonal antibodies directed to τ95, τ55, τ91, or τ131. Immune complexes were revealed by chemiluminescence (Fig. 8). As expected, no TFIIIC subunit was detected when immunopurification was performed with a wild-type crude extract containing no HA-tagged polypeptide (Fig. 8, WT). In contrast, immunoprecipitation with crude extracts containing an HA-tagged subunit (Fig. 8, HA-τ138, HA-τ131, or HA-τ95) resulted in the copurification of 95-, 55-, 131-, and 91-kDa subunits, indicating the association of these polypeptides with each other.
FIG. 8.
Coimmunoprecipitation of TFIIIC subunits from crude extracts. Immunoprecipitations with anti-HA antibodies were performed as described in Materials and Methods with crude extracts from the wild-type (WT) strain or strains expressing HA-tagged versions of τ138, τ131, and τ95 (HA-τ138, HA-τ131, and HA-τ95, respectively). Proteins eluting with the synthetic peptides were subjected to SDS-PAGE, transferred to a membrane, and probed successively with polyclonal antibodies directed against τ95, τ55, τ131, and τ91. Immune complexes were revealed with an ECL kit and quantified by scanning. A value of 1 was arbitrarily given to the amounts of τ95, τ55, τ131, and τ91 subunits immunoprecipitated from the HA-τ138 crude extract and was used as a reference. On the left are indicated molecular masses (in kilodaltons) of the polypeptides probed by the antibodies noted on the right. The 75-kDa polypeptide revealed by anti-τ91 antibodies was immunologically related to τ91 (2).
The results of the immunopurification shown in Fig. 8 were quantified by scanning. The value of 1 was arbitrarily given to the amount of the τ95, τ55, τ131, or τ91 subunit immunopurified from the HA-τ138 crude extract and was used as a reference. As shown in Fig. 8, these four polypeptides were recovered with approximately the same efficiencies and with the same relative yields from the HA-τ131 crude extract. Note that HA-τ131 was not overrepresented, suggesting the absence of a significant pool of free τ131 subunit. In contrast, when the immunopurification procedure was applied to the HA-τ95 crude extract, the relative yields of the four TFIIIC subunits were markedly modified. The recoveries of τ131 and of τ91 remained at the same levels, but those of τ55 and of HA-τ95 were greatly increased, in the range of three- to sixfold, respectively. These results indicated that there was markedly more than one τ95 or τ55 subunit in yeast crude extracts relative to τ131 and τ91 and corroborated the existence of a τ55-τ95 subcomplex in yeast cells.
DISCUSSION
Yeast TFIIIC is a multifunctional protein required for promoter binding, TFIIIB recruitment, and chromatin antirepression. We have pursued the characterization of this multisubunit factor and report here the isolation of an essential gene, named TFC7, encoding its smallest subunit. It appears that this polypeptide is a chimeric protein that belongs to different protein complexes.
Based on biochemical data and gene cloning, it is now well established that the four largest polypeptides, of 138, 131, 95, and 91 kDa, contained in highly purified TFIIIC fractions are subunits of TFIIIC. The two smallest polypeptides (60 and 55 kDa) have also been consistently found in TFIIIC fractions from different laboratories (5, 16, 43). Using peptide sequences obtained from the gel-purified protein, we have identified the TFC7 gene encoding the 55-kDa polypeptide. Polyclonal antibodies directed to rTFC7p were able to supershift TFIIIC-tDNA complexes, thus confirming the presence of the 55-kDa polypeptide within the factor-tDNA complex. Like all the genes encoding components of the yeast PolIII transcription system and isolated so far, TFC7 is an essential gene.
Searches in databases revealed the chimeric structure of TFC7p. The N-terminal part of TFC7p showed intriguing similarities to other proteins unrelated to transcription and was highly similar to a yeast hypothetical protein of unknown function (named HUFp). On the other hand, no significant sequence similarities could be detected with the C-terminal part of TFC7p. In fact, the coding sequences for the N-terminal part of TFC7p and for HUFp are located just at the border of duplication block 50 present on both chromosome XIV and chromosome XV (54). One hypothesis is that the chimeric structure of τ55 resulted from a fusion between an ancestral transcription factor subunit (corresponding to the C-terminal part of τ55 that is not included in the duplication block) and another protein of still unknown function (encoded by ancestral HUF and corresponding to the N-terminal part of τ55). This hypothesis, which may explain the intriguing sequence similarities between a TFIIIC subunit and a cyanobacterial protein (Syn H), is supported by our results from τ55 deletion mutant analysis. Indeed, only the C-terminal part of τ55 was necessary for interaction with τ95 and was sufficient for transcription factor activity. Whereas the deletion of the C-terminal part of τ55 was lethal, the mutant form of TFIIIC entirely deprived of the N-terminal half of τ55 (τ55-ΔN3) supported normal cell growth, was able to bind to tDNA promoter sequences in vitro, and, once bound, recruited TFIIIB productively and as efficiently as the wild-type factor. However, although the whole N-terminal domain of τ55 was dispensable for TFIIIC activity, a partial deletion of this domain (τ55-ΔN1) impaired τ55 function at a high temperature. The residual presence of a truncated N-terminal fragment may have interfered with τ55 folding or led to a defect in TFIIIC assembly or stability.
The N-terminal part of τ55 showed sequence similarities to different enzymes that catalyze similar phosphotransfer reactions, more specifically, enzymes related to the glycolytic pathway (PGM or FbPase) and acid phosphatase. Since residues important for the catalytic activity of these enzymes were conserved in τ55, we assayed TFIIIC and rTFC7p for enzymatic activity. No PGM or phosphatase activities were detected (results not shown). Nevertheless, partial deletions of the N-terminal domain of τ55 resulted in a reduced growth rate at 30°C (τ55-ΔN1) or in a thermosensitive phenotype (τ55-ΔN1 and τ55-ΔN2) in glycerol- or ethanol-containing medium. These growth defects, which were revealed only in medium containing glycerol or ethanol instead of glucose, suggest a possible relationship between PolIII transcription and metabolic pathways which deserves to be investigated further.
The direct interaction between τ55 and τ95 observed in vitro and in vivo agrees well with the photo-cross-linking mapping of these two polypeptides on opposite sides of the DNA helix, in the vicinity of the A block of tRNA genes (5, 6). Our inability to alter the migration of τB-tDNA complexes with anti-τ55 antibodies (data not shown) further argues in favor of the localization of τ55 in τA, the TFIIIC domain that binds to the A block of tRNA genes. However, sequence analysis of τ55 did not reveal known DNA binding motifs, and our attempts to demonstrate, by gel retardation assays or Southwestern blotting, that τ55 or τ95 binds to DNA, even nonspecifically, failed (results not shown) (44, 48). One could imagine that the interaction of τA with the A block of tRNA genes requires the association of both τ95 and τ55. However, no tDNA binding activity or transcriptional activation (or inhibition) was found to be associated with a partially purified τ55-τ95 subcomplex (results not shown). From this point of view, the presence of two TFIIIC components in two distinct complexes is reminiscent of but not equivalent to the chromatographic separation of silkworm or human TFIIIC into distinct, complementary activities. TFIIIC activity from silkworm cells can be separated into two fractions, both of which are required to form a complex on a tRNA gene (41). For human TFIIIC, two fractions, named TFIIIC1 and TFIIIC2, are both necessary to reconstitute full TFIIIC DNA binding and transcriptional activities: TFIIIC2, a multisubunit protein (33, 50, 56), binds strongly by itself to the B block of tRNA genes, but A block binding and gene transcription require TFIIIC1 (50, 55). Furthermore, TFIIIC1 can be chromatographically separated from an additional activity, TFIIIC0, that is able to partially substitute for TFIIIC1 activity (38). The situation is different here, since the τ55-τ95 complex (with possible associated components) is not required for TFIIIC activity. The existence of a τ55-τ95 complex distinct from TFIIIC suggests a dual role for these two subunits. This complex is possibly endowed with regulatory functions or has other roles unrelated to transcription. The subcellular localization of this complex and the identification of associated proteins, if any, may shed some light on its function. It will also be of interest to uncover the phenotype(s) caused by the deletion of HUFp, which is highly similar to the N-terminal part of τ55. HUFp is distantly homologous to enzymes related to the glycolytic pathway. This similarity suggests a connection with cell metabolism that is also supported by the altered growth phenotype of τ55-ΔN1 and τ55-ΔN2 mutant cells in glycerol- or ethanol-containing medium. The level of PolIII transcription is known to vary according to the cell growth rate in human, mouse, and yeast cells (15, 23, 47, 49). TFIIIC components may play a role in such coordinated regulation.
ACKNOWLEDGMENTS
We thank Christophe Carles and Françoise Bouet for peptide sequence determination and Anny Ruet for help in raising rabbit polyclonal antibodies. We are grateful to Janine Huet and Emmanuel Favry for B", recombinant TBP, recombinant TFIIIB70, and PolIII preparations. We thank Jochen Rüth and Geneviève Dujardin for helpful discussions and Cathy Jackson for improving the manuscript.
This work was supported by a grant from the European Union BIOTECH program (to A.S.). N.M. was supported by a fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche, and R.A. was supported by a P.F.I. postdoctoral fellowship from the Spanish Ministerio de Educación y Cultura.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arrebola R, Manaud N, Rozenfeld S, Marsolier M C, Lefebvre O, Carles C, Thuriaux P, Conesa C, Sentenac A. τ91, an essential subunit of yeast transcription factor IIIC, cooperates with τ138 in DNA binding. Mol Cell Biol. 1998;18:1–9. doi: 10.1128/mcb.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Baker R E, Hall B D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984;3:2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Braun B R, Bartholomew B, Kassavetis G A, Geiduschek E P. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J Mol Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 10.Burnol A-F, Margottin F, Huet J, Almouzni G, Prioleau M-N, Méchali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993;362:475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- 11.Burnol A F, Margottin F, Schultz P, Marsolier M C, Oudet P, Sentenac A. Basal promoter and enhancer elements of yeast U6 snRNA gene. J Mol Biol. 1993;233:644–658. doi: 10.1006/jmbi.1993.1542. [DOI] [PubMed] [Google Scholar]
- 12.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 12a.Conesa, C. Unpublished results.
- 13.Conesa C, Swanson R N, Schultz P, Oudet P, Sentenac A. On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/τ. J Biol Chem. 1993;268:18047–18052. [PubMed] [Google Scholar]
- 14.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards D R, Parfett C L, Denhardt D T. Transcriptional regulation of two serum-induced RNAs in mouse fibroblasts: equivalence of one species to B2 repetitive elements. Mol Cell Biol. 1985;5:3280–3288. doi: 10.1128/mcb.5.11.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabrielsen O S, Marzouki N, Ruet A, Sentenac A, Fromageot P. Two polypeptide chains in yeast transcription factor τ interact with DNA. J Biol Chem. 1989;264:7505–7511. [PubMed] [Google Scholar]
- 17.Gabrielsen O S, Oyen T B. The requirement for the A block promoter in tRNA gene transcription in vitro depends on the ionic environment. Nucleic Acids Res. 1987;15:5699–5713. doi: 10.1093/nar/15.14.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S L, Yamamoto K R, editors. (Transcriptional regulation. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 19.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 22.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf D H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 23.Hoeffler W K, Roeder R G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985;41:955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- 24.Huet J, Conesa C, Manaud N, Chaussivert N, Sentenac A. Interactions between yeast TFIIIB components. Nucleic Acids Res. 1994;22:3433–3439. doi: 10.1093/nar/22.16.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 26.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 107–126. [Google Scholar]
- 28.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 29.Kassavetis G A, Joazeiro C A P, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 30.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 33.Kovelman R, Roeder R G. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267:24446–24456. [PubMed] [Google Scholar]
- 34.Lefebvre O, Carles C, Conesa C, Swanson R N, Bouet F, Riva M, Sentenac A. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor TFIIIC. Proc Natl Acad Sci USA. 1992;89:10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefebvre O, Rüth J, Sentenac A. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J Biol Chem. 1994;269:23374–23381. [PubMed] [Google Scholar]
- 36.Marck C, Lefebvre O, Carles C, Riva M, Chaussivert N, Ruet A, Sentenac A. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeat and basic helix-loop-helix motifs. Proc Natl Acad Sci USA. 1993;90:4027–4031. doi: 10.1073/pnas.90.9.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marzouki N, Camier S, Ruet A, Moenne A, Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor τ. Nature. 1986;323:176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- 38.Oettel S, Härtel F, Kober I, Iben S, Seifart K H. Human transcription factors IIIC2, IIIC1 and a novel component IIIC0 fulfill different aspects of DNA binding to various pol III genes. Nucleic Acids Res. 1997;25:2440–2447. doi: 10.1093/nar/25.12.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 40.O’Toole G A, Trzebiatowski J R, Escalante-Semerena J C. The cobC gene of Salmonella typhimurium codes for a novel phosphatase involved in the assembly of the nucleotide loop of cobalamin. J Biol Chem. 1994;269:26503–26511. [PubMed] [Google Scholar]
- 41.Ottonello S, Rivier D H, Doolittle G M, Young L S, Sprague K U. The properties of a new polymerase III transcription factor reveal that transcription complexes can assemble by more than one pathway. EMBO J. 1987;6:1921–1927. doi: 10.1002/j.1460-2075.1987.tb02452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papavassiliou A G, Bohmann D. Optimization of the signal-to-noise ratio in south-western assays by using lipid-free BSA as blocking reagent. Nucleic Acids Res. 1992;20:4365–4366. doi: 10.1093/nar/20.16.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsons M C, Weil P A. Purification and characterization of Saccharomyces cerevisiae transcription factor TFIIIC. Polypeptide composition defined with polyclonal antibodies. J Biol Chem. 1990;265:5095–5103. [PubMed] [Google Scholar]
- 44.Parsons M C, Weil P A. Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J Biol Chem. 1992;267:2894–2901. [PubMed] [Google Scholar]
- 45.Rüth J, Conesa C, Dieci G, Lefebvre O, Düsterhöft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz P, Marzouki N, Marck C, Ruet A, Oudet P, Sentenac A. The two DNA-binding domains of yeast transcription factor τ as observed by scanning transmission electron microscopy. EMBO J. 1989;8:3815–3824. doi: 10.1002/j.1460-2075.1989.tb08559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethy I, Moir R D, Librizzi M, Willis I M. In vitro evidence for growth regulation of tRNA gene transcription in yeast. A role for transcription factor (TF) IIIB70 and TFIIIC. J Biol Chem. 1995;270:28463–28470. doi: 10.1074/jbc.270.47.28463. [DOI] [PubMed] [Google Scholar]
- 48.Swanson R N, Conesa C, Lefebvre O, Carles C, Ruet A, Quemeneur E, Gagnon J, Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor τ (TFIIIC) Proc Natl Acad Sci USA. 1991;88:4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tower J, Sollner-Webb B. Polymerase III transcription factor B activity is reduced in extracts of growth-restricted cells. Mol Cell Biol. 1988;8:1001–1005. doi: 10.1128/mcb.8.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Roeder R G. Transcription factor IIIC1 (TFIIIC1) acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 52.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 53.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 54.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 55.Yoshinaga S K, Boulanger P A, Berk A J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci USA. 1987;84:3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshinaga S K, L’Etoile N D, Berk A J. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]