Abstract
Eukaryotic precursor (pre)-tRNAs are processed at both ends prior to maturation. Pre-tRNAs and other nascent transcripts synthesized by RNA polymerase III are bound at their 3′ ends at the sequence motif UUUOH [3′ oligo(U)] by the La antigen, a conserved phosphoprotein whose role in RNA processing has been associated previously with 3′-end maturation only. We show that in addition to its role in tRNA 3′-end maturation, human La protein can also modulate 5′ processing of pre-tRNAs. Both the La antigen’s N-terminal RNA-binding domain and its C-terminal basic region are required for attenuation of pre-tRNA 5′ processing. RNA binding and nuclease protection assays with a variety of pre-tRNA substrates and mutant La proteins indicate that 5′ protection is a highly selective activity of La. This activity is dependent on 3′ oligo(U) in the pre-tRNA for interaction with the N-terminal RNA binding domain of La and interaction of the C-terminal basic region of La with the 5′ triphosphate end of nascent pre-tRNA. Phosphorylation of La is known to occur on serine 366, adjacent to the C-terminal basic region. We show that this modification interferes with the La antigen’s ability to protect pre-tRNAiMet from 5′ processing either by HeLa extract or purified RNase P but that it does not affect interaction with the 3′ end of pre-tRNA. These findings provide the first evidence to indicate that tRNA 5′-end maturation may be regulated in eukaryotes. Implications of triphosphate recognition is discussed as is a role for La phosphoprotein in controlling transcriptional and posttranscriptional events in the biogenesis of polymerase III transcripts.
Studies of pre-tRNA processing in Escherichia coli have revealed that although the order of 5′ and 3′ processing events can vary among substrates, many eukaryotic precursor (pre)-tRNAs are processed in a preferred order (21). Classical eukaryotic tRNA genes are monocistronic and compared to most of their prokaryotic counterparts their primary transcripts are short and simple (18, 33). Although many features of tRNA maturation, including 5′ processing by RNase P, have been conserved from bacteria to humans, mechanisms of 3′-end maturation were not. Like other transcripts synthesized by RNA polymerase III (Pol III), pre-tRNAs are terminated at their 3′ end by the sequence motif UUUOH, hereafter referred to as 3′ oligo(U), which comprises a high-affinity binding site for the La protein (34). This oligo(U) tract, together with a short sequence stretch proximal to it, comprise the 3′ trailer of eukaryotic pre-tRNAs which must be removed prior to enzymatic addition of CCA (8).
Evolutionary conservation of La phosphoprotein and its interaction with 3′ oligo(U) indicate the importance of this protein in the biogenesis of Pol III transcripts (38, 42). Indeed, La can modulate pre-tRNA 3′-end metabolism in Saccharomyces cerevisiae (43). Yeast cells can process pre-tRNA 3′ ends by either exonuclease- or endonuclease-mediated pathways, and La can influence which pathway is used (43).
La’s involvement in RNA biogenesis is almost certainly not limited to tRNAs since La is also found associated with the precursors of 5S rRNA, U6 snRNA, 7SL RNA, Alu retroposon RNAs, and all other nascent transcripts synthesized by Pol III (see reference 35). Experiments performed in cell extracts have revealed activities for human La in transcript release and Pol III reinitiation, as well as B1-Alu RNA 3′ end metabolism (12, 23, 25). Thus, while specificity for 3′ oligo(U) reflects La’s activity as a transcription termination factor (13, 14), this interaction is also a means by which La remains associated with nascent Pol III transcripts after their synthesis (16, 30, 35). Investigating La’s role in tRNA expression should also provide insight into the biogenesis and maturation of this broad class of eukaryotic RNAs.
An earlier study suggested that a Pol III transcription-coupled factor could modulate 3′-end maturation of B1-Alu transcripts in a manner that is sensitive to the sequence context of the Pol III terminator (24). The La protein was then identified as the Pol III termination factor that protected B1-Alu RNA from 3′ processing (25). Human La is 408 amino acids long and contains an N-terminal domain that binds RNA and a phosphoserine-containing C-terminal domain that is not required for general RNA binding (5, 6, 12, 19, 29). It is therefore significant that alterations in the C-terminal domain of La were found to disrupt RNA recognition by La and render nascent B1-Alu transcripts susceptible to processing (12). This same basic region in the C-terminal domain is also required for the transcription factor activity of human La protein (12), which can be regulated by phosphorylation and dephosphorylation of serine 366, which resides within a conserved casein kinase II (CKII) site (10). Involvement of the C-terminal basic region of La in RNA recognition and transcription factor activity presumably reflects a mechanistic coupling between the Pol III termination and reinitiation cofactor activities of La (23).
The appearance of a B1-Alu intermediate RNA in the presence of certain C-terminal altered forms of La suggested that the N- and C-terminal domains of La may mediate distinct RNA recognition events (12). We have therefore extended our investigation to a previously studied pathway of human pre-tRNAiMet processing, using a cell-free system to define the determinants in the La protein (and the RNA substrate) involved in protection of pre-tRNAiMet from processing. Interaction of the N-terminal domain of La with the 3′ oligo(U) tract of pre-tRNAiMet is required for La’s C-terminal-domain-mediated protection of the 5′-end region of pre-tRNA from processing by RNase P. Site-specific phosphorylation of La on serine 366, adjacent to the C-terminal basic region, modulates this 5′ protection activity specifically.
MATERIALS AND METHODS
Pol III transcription-processing reactions.
In vitro Pol III transcription reaction mixtures (40 μl) contained 200 ng of plasmid DNA [270-bp human pre-tRNAiMet gene 2 fragment in pBluescript(SK+), designated ptRNAiMet270] and either 2 μl of unfractionated S100 extract or 4 μl of TFIIIB-Pol III, 3 μl of TFIIIC1, and 2 μl of TFIIIIC2 (see Fig. 1B). In subsequent experiments, 4 μl of each TF was used (see Fig. 5A and 8A). Reactions were carried out in mixtures containing 25 mM HEPES (pH 7.9); 5 mM MgCl2; 90 mM KCl; 1 mM dithiothreitol (DTT), 0.5 μl of RNase inhibitor (Promega); 5% glycerol; 0.5 mM each of ATP, CTP, and UTP; 25 mM of GTP, 2 μCi of [α-32P]GTP (New England Nuclear), and either 2 pmol (100 ng) of purified La protein or the amount indicated. All components except the nucleoside triphosphates (NTPs) were premixed and incubated on ice for 5 min. Reactions were started by addition of NTPs and incubation at 30°C. The time course experiment (Fig. 1A) utilized streptavidin agarose-immobilized biotinylated ptRNAiMet270 DNA. After a pulse of [α-32P]GTP incorporation, excess unlabeled GTP was added and the supernatant was separated from the template by centrifugation. Aliquots were removed at specified times thereafter and analyzed.
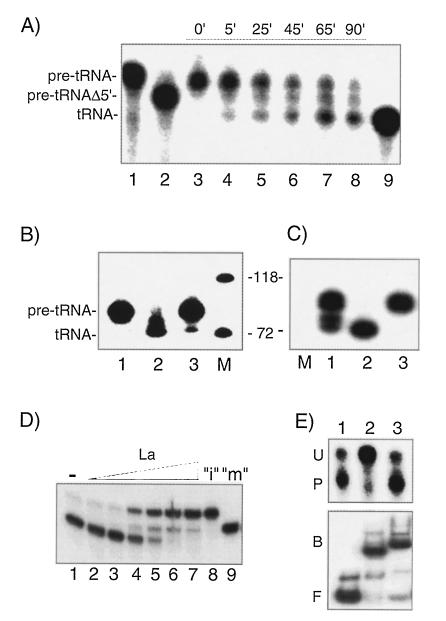
FIG. 1.
La inhibits processing of pre-tRNAiMet in a homologous in vitro system. (A) After assembly of transcription complexes from fractionated HeLa components and partial elongation of Pol III in the presence of CTP, ATP, and [α-32P]GTP, a pulse of synthesis was allowed to occur in the presence of all four NTPs. The supernatant was then separated from the immobilized template by centrifugation, and aliquots were removed from it at various times thereafter as indicated (lanes 3 to 8). Lanes 1, 2, and 9 contain T7 RNAP-synthesized transcripts representing pre-tRNAiMet of 89 nucleotides (lane 1), pre-tRNAiMet that lacks the 5′ leader sequence (pre-tRNAΔ5′, lane 2), and the mature tRNAiMet of 72 nucleotides (lane 9), as determined by Zasloff et al. (45). (B) A fraction containing TFIIIB and Pol III, along with TFIIIC1 and TFIIIC2 fractions (including trace amounts of La), reconstitute transcription of the human tRNAiMet gene but yield a product of 72 nucleotides (lane 2). Supplementation of the reconstituted reaction mixture with La protein to 50 nM (lane 1) or use of unfractionated S100 extract in a reaction mixture that contains 50 nM endogenous La (lane 3) produces a larger transcript. Products were analyzed by denaturing 6% polyacrylamide gel electrophoresis and autoradiography. (C) Pre-tRNAiMet processing was examined in reactions that were immunodepleted with anti-La immunoglobulin G immobilized on protein A-Sepharose and after repletion with buffer alone (lane 2) or La (lane 3). RNA synthesized in the nondepleted reaction is shown in lane 1. (D) 32P-labeled pre-tRNAiMet was synthesized by T7 RNAP, purified, and incubated with fractionated extract, 100 ng of E. coli 5S rRNA nonspecific competitor, and either buffer alone (lane 1) or increasing amounts of La (0.1, 0.3, 0.5, 1.0, 2.0, and 5.0 pmol; lanes 2 to 7, respectively) and examined for processing. Lane 8 contains input RNA (“i”) that was not subjected to extract and lane 9 contains a T7 RNAP-synthesized 32P-labeled tRNAiMetΔ5′ (“m”) corresponding to the 5′ processed intermediate as described earlier (45) which served as size markers. (E) Processing (upper panel) and binding (lower panel) of T7 RNAP-synthesized pre-tRNAiMet were examined in 10-μl reaction mixtures containing 100 ng of 5S rRNA and buffer alone (lane 1) or 3 pmol of La (lane 2) or 3 pmol of E. coli single-strand binding protein (lane 3). HeLa fractions were omitted from the EMSA reactions. For the upper panel, the positions of unprocessed pre-tRNAiMet (U) and processed (P) RNAs are shown; for the lower panel, the positions of bound (B) and free (F) pre-tRNAiMet probe are shown.
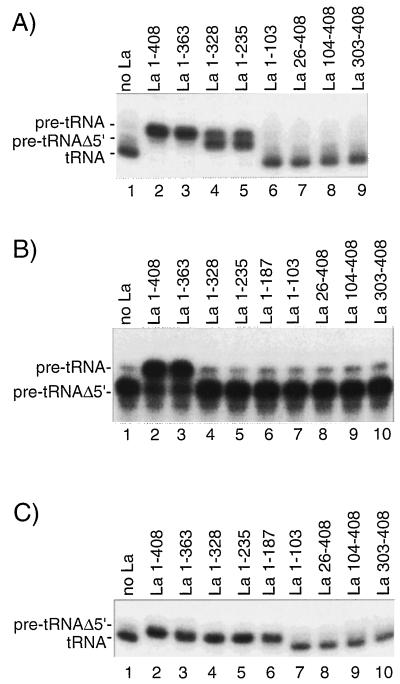
FIG. 5.
Dissection of the regions of La responsible for protection of pre-tRNAiMet from processing. (A) The Pol III transcription-processing assay was performed in the absence of added La (lane 1) or in the presence of the truncated La proteins as indicated above the lanes. The positions of the T7-synthesized markers pre-tRNAiMet, pre-tRNAiMetΔ5′, and tRNAiMet are indicated. (B) T7 RNAP-synthesized pre-tRNAiMet was subjected to RNase P in the presence of the La proteins indicated above the lanes. Portions (1.00 μl) of RNase P were used for each reaction, and the incubation time was 15 min. Positions of input and processed RNAs are indicated to the left. (C) T7-synthesized pre-tRNAiMetΔ5′ was assayed for processing in reactions containing HeLa fractions and the La proteins indicated above the lanes. The positions of input and processed RNAs are indicated to the left.
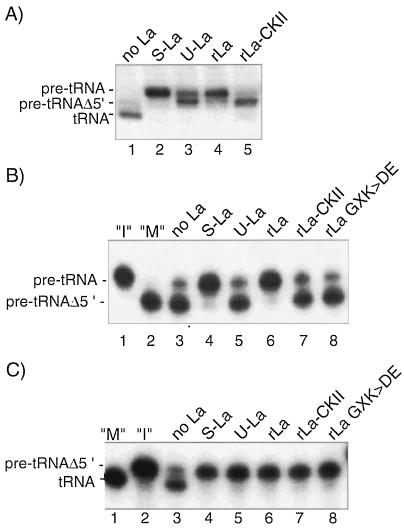
FIG. 8.
Phosphorylation of La on serine 366 interferes with protection of pre-tRNAiMet from 5′ processing. (A) The human tRNAiMet gene was expressed in the Pol III transcription-processing system with reconstituted fractions alone (lane 1) or with the added La proteins indicated (lanes 2 to 5). (B) RNase P (1.50 μl) was incubated with pre-tRNAiMet and the La proteins indicated. Lanes 1 and 2 show the pre-tRNAiMet input (“I”) substrate and a pre-tRNAiMetΔ5′ size marker (“M”) in the absence of RNase P. (C) The proteins indicated above the lanes (lanes 3 to 8) were examined for protection of pre-tRNAiMetΔ5′ from processing by HeLa fractions. Lanes 1 and 2 contain the RNA size markers indicated to the left.
Fractionated HeLa cell extract.
HeLa cell-derived cytoplasmic S100 was fractionated in buffer A (20 mM HEPES, pH 7.9; 20% glycerol; 0.2 mM EDTA; 0.5 mM DTT; 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) containing 100 mM KCl by phosphocellulose (P11) chromatography into PC-A, -B, and -C (10). Crude PC-B was dialyzed in buffer A containing 100 mM KCl and refractionated on P11 phosphocellulose; fractions eluting at 0.25 to 0.3 M KCl contained TFIIIB-Pol III and were used as they were. PC-C was dialyzed as described above and fractionated on Mono Q (44). Fractions eluting at 0.2 to 0.28 M KCl were pooled and rechromatographed on Mono Q; TFIIIC1 eluted at 0.26 M KCl, was dialyzed as described above, and was fractionated on heparin-Sepharose. The resulting TFIIIC1 activity was fractionated on Mono S and eluted at 0.45 to 0.5 M KCl. TFIIIC2 activity that eluted from the first Mono Q column at 0.35 to 0.55 M KCl was adjusted to 1 M NH4SO4 in buffer A and fractionated on phenyl-Sepharose, eluting at 0.05 to 0 M NH4SO4. This was adjusted to 0.1 M KCl and fractionated on B-box-DNA affinity resin (7). The eluted TFIIIC2 activity was dialyzed as described above, fractionated on Mono Q, step eluted with 0.35 M KCl, and collected as TFIIIC2. Activities of TFIIIC1, TFIIIC2, and TFIIIB-Pol III were monitored by in vitro transcription. For most reactions, 20 times the amount of La present in the TFIIIB-Pol III fraction was used for reconstitution. This amount of La was used because it represents the amount found in the unfractionated S100.
La proteins.
C-terminal histidine (His)-tagged La proteins, derived from E. coli, as well as HeLa-derived U-La and S-La, were as previously described (10). After phosphorylation by recombinant CKII, the His-tagged La was affinity purified by using nickel agarose as described previously (10).
T7 RNA polymerase (RNAP)-synthesized transcripts.
T7 RNAP-synthesized transcripts were produced from templates generated by PCR (22) designed to yield RNAs corresponding precisely to pre-tRNAiMet, the 5′ processed pre-tRNAiMetΔ5′, and the fully processed tRNAiMet as described previously (45). For some experiments, T7-synthesized 32P-labeled RNAs were incubated in 200 μl of calf intestinal phosphatase (CIP) buffer and 45 U of CIP (Boehringer Mannheim) for 30 min at 37°C. All 32P-labeled RNAs were gel purified prior to use.
RNA electrophoretic mobility shift assays (EMSAs).
EMSAs were performed as described earlier (10) in 10 μl with poly(rG) or 100 ng of 5S rRNA competitor as described below (Fig. 1E).
RNA processing.
In vitro processing of T7-synthesized RNA was carried out in 10-μl reaction mixtures containing 3,000 or 6,000 cpm of 32P-labeled RNA; 0.5 μl of RNasin (Promega); 100 ng of E. coli 5S rRNA carrier (Boehringer Mannheim); 25 mM HEPES (pH 7.9); 5 mM MgCl2; 120 mM KCl; 1 mM DTT, 6% glycerol; 0.5 mM concentrations each of ATP, CTP, and UTP; 25 mM GTP; 3 pmole of La protein or the amount indicated; and 1 μl each of TFIIIB-Pol III, TFIIIC1, and TFIIIC2 (these conditions were used because they are similar to the Pol III transcription-processing reactions). After addition of all three TF fractions, reaction mixtures were incubated at 30°C for 45 min and then stopped, and the RNA was purified.
Primer extension.
The primer extension was performed as described earlier (11) with 32P-5′-end-labeled oligoDNA 5′-TAGCAGAGGATGGTTTCGATCCATCGACCTCT-3′, which is complementary to the last 32 nucleotides of 3′-processed tRNAiMet (45). This primer was chosen because it is complementary to a region within the 5′-processed intermediate, the 3′-processed intermediate, and the fully processed tRNA (5′ and 3′ processed) and therefore detects all three species. Negative control reactions in which NTPs were omitted from the Pol III transcription reaction confirmed that our purified fractions were not contaminated by endogenous tRNAiMet species (not shown). Use of T7 RNAP-synthesized marker species corresponding to pre-tRNAiMet and 5′-processed pre-tRNAiMet provided positive controls. With these controls in place, it was clear that this assay distinguished the unprocessed and 5′-processed pre-tRNAiMet species in our processing reactions.
RNase P protection assay.
Nuclear RNase P, purified from S. cerevisiae, was separated from all contaminating nuclease activities by a multistep chromatography protocol that included Mono Q fast protein liquid chromatography as described (4). Reactions were carried out in 10 μl containing ∼6,000 cpm of T7-synthesized 32P-labeled RNA substrate, 100 ng of 5S rRNA carrier, 3 pmol of purified La protein or buffer alone, 0.2 μl of RNasin, RNase P (diluted 10-fold immediately prior to use), 20 mM HEPES (pH 7.9), 10 mM MgCl2, and 110 mM KCl.
RESULTS
La protects human pre-tRNAiMet from processing in a homologous cell-free system.
The nascent transcript synthesized from the human pre-tRNAiMet gene is processed by removal of the 8-nucleotide leader and the ∼10-nucleotide trailer (heterogeneity is found in the number of U residues at the 3′ end [45]). Using a fractionated Pol III transcription system that contains trace residual LA under pulse-chase conditions, we demonstrated a precursor-product relationship for the transcripts produced from this human tRNAiMet gene (Fig. 1A, lanes 3 to 8). T7 RNAP-synthesized 32P-labeled RNAs corresponding to pre-tRNAiMet (lane 1), pre-tRNAiMet lacking its 5′ leader (pre-tRNAiMetΔ5′; lane 2), and fully processed tRNAiMet (lane 9), as determined by Zasloff et al. (45), were used as size markers to show that the slow- and fast-migrating Pol III-synthesized transcripts corresponded in size to pre-tRNAiMet and tRNAiMet, respectively. Analysis by primer extension (see Materials and Methods) confirmed that the tRNAiMet-size product had had its 5′ leader removed as described previously (45; data not shown). A reproducible band that migrated between the precursor and fully processed product (Fig. 1A, lanes 6 to 8) suggested that processing occurred in two steps as expected for distinct 5′- and 3′-processing events described for this and other eukaryotic pre-tRNAs (8, 45).
When recombinant La protein was added to these reactions at 50 nM, the precursor transcript remained unprocessed after a 60-min incubation (Fig. 1B, compare lanes 1 and 2). Reactions with unfractionated HeLa S100 containing endogenous La at 50 nM (2 pmol) also yielded primarily unprocessed precursor (lane 3), a finding consistent with previous results (45).
Because the lack of processing in S100 might be due to an inhibitory protein other than La, we performed reactions that were immunodepleted of La and after repletion with purified La (Fig. 1C). Depletion of La led to the tRNAiMet-size transcript as the major product (lane 2). Most importantly, protection of pre-tRNA from processing could be recovered by adding highly purified La back into the immunodepleted reaction mixture (lane 3). In contrast to depletion with anti-La, mock depletion with normal human immunoglobulin G produced unprocessed pre-tRNAiMet (not shown). This established that La is inhibitory to pre-tRNAiMet processing at the concentration found in reactions with HeLa cell extract. Analysis by primer extension confirmed that the 5′ leader had been removed in the absence of La but left uncleaved in the presence of La (not shown; see Materials and Methods). These results, in conjunction with the comigration of these species with our T7 RNAP-synthesized-tRNAiMet-derived markers, establish that this system generates the precursor and tRNAiMet products previously described (45). Analyses of a tRNAArg gene, as well as of two other tRNAiMet genes (Xenopus and human genes), revealed that La also prevented processing of a two-step processing pathway, which is characteristic of the human tRNAiMet gene 2 shown in Fig. 1 (data not shown). Presumably, the 3′ processing seen in this system is due to a 3′-5′ exonuclease.
Although the transcription system used here contains detectable amounts of La (9), this amount is not sufficient for stable association or to provide lasting protection of the nascent transcript from processing (see Discussion). It is not known why reconstitution with La sometimes led to an increase in the amount of RNA recovered from the reaction mixtures in addition to an increase in protection from terminal processing (compare Fig. 1B, lanes 1 and 2). Terminal protection required a 10-fold-higher concentration of La than that required to increase the amount of RNA (not shown), suggesting that La may stimulate transcription at a concentration lower than that required for stable association with the nascent transcript. These higher concentrations of La are closer to what is found in reactions with HeLa extract and were used in the rest of the study.
Next, we used purified 32P-labeled pre-tRNAiMet synthesized with T7 RNAP to examine processing in the fractionated system directly, uncoupled from Pol III transcription (Fig. 1D and E). 32P-labeled pre-tRNAiMet added to the reconstituted reactions was protected from processing by La in a dose-dependent manner (Fig. 1D). T7-synthesized pre-tRNAiMet required a higher concentration of La than nascent Pol III-synthesized RNA for protection (not shown). Several additional points regarding Fig. 1D are noteworthy. First, inhibition of processing occurs between 30 and 50 nM La (lanes 3 and 4), values that are not too far from the La antigen’s Kd for oligo(U)-containing RNAs previously reported at 5 to 10 nM (6, 12), yet more closely comparable to the concentration of La in standard reactions with HeLa cell extract (9). Second, the persistence of pre-tRNAiMet in the presence of La (lanes 4 to 7) but not in its absence (lane 1), coupled with the appearance of an intermediate band, suggests that La protects against two processing events (5′ and 3′), in a way similar to the Pol III transcription reactions with the same fractions. Furthermore, both the intermediate and the unprocessed precursor were found together at relatively low concentrations of La (lane 4). Since protection from 3′ processing is a specific activity of La, this suggests that protection from the second processing event also occurs as a result of specific high-affinity interactions. Finally, we note that at 1 pmol (100 nM; lane 5), La protected most of the precursor from processing, which is similar to the coupled transcription-processing reactions.
We examined protection of and binding to pre-tRNAiMet by La in parallel (upper and lower panels of Fig. 1E, respectively), using the same concentrations of La, pre-tRNAiMet, and competitor RNA for both assays. Most of the precursor remained unprocessed (lane 2, upper panel) and stably bound to La (lane 2, lower panel). It is important to note that under these conditions a single major complex was formed (lower panel, lane 2, band B) which exhibited the same mobility as had the first complex formed in titration experiments (not shown). This demonstrated that significant amounts of low-affinity, nonspecific complexes were not formed. Fig. 1E also showed that the protective action of La was specific, since other nucleic-acid-binding proteins did not protect this substrate from processing. For example, although E. coli single-strand binding protein interacted with the substrate (Fig. 1E, lane 3, lower panel), it protected very little if any of the substrate from processing (Fig. 1E, lane 3, upper panel). Recombinant U1 RNA-associated protein, U1-A (obtained from D. Kenan, Duke University), also interacted with this substrate but could not protect it (data not shown). We conclude that the ability of La to protect the tRNAiMet precursor from processing is specific. Below we show that this ability is dependent on the well-known characteristic of La: recognition of the 3′ oligo(U) tract of the nascent RNA.
The 3′ oligo(U) tract of pre-tRNAiMet is required for protection from 5′ and 3′ processing by La.
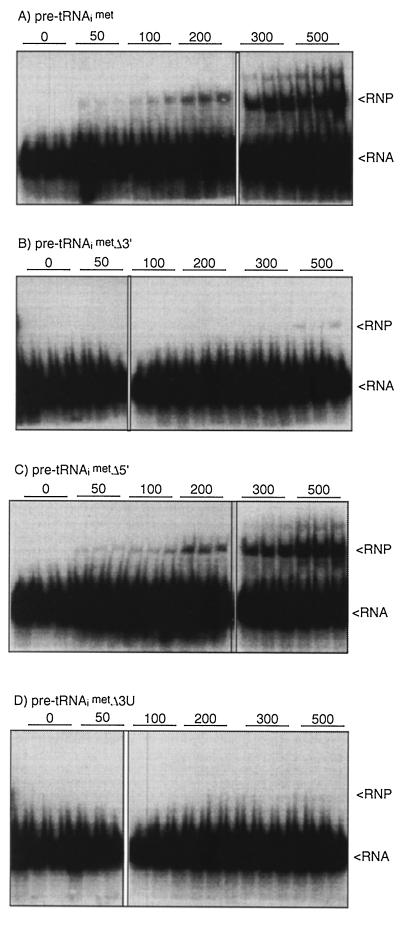
Presynthesized transcripts corresponding to precursor and singly processed pre-tRNAiMet intermediates were previously used to examine the individual 5′- and 3′-processing events (2, 3). We used T7 RNAP-synthesized pre-tRNAiMet transcripts carrying deletions or substitutions at either the 3′ end or the 5′ end to examine processing (Fig. 2). Conversion of the nonmutated, full-length pre-tRNAiMet to the tRNAiMet-size product is inhibited by La (Fig. 2, lanes 1 to 3). The substrate pre-tRNAiMetΔ3′ lacks the entire 3′ trailer, including the oligo(U) tract, and was used to monitor 5′ processing. This substrate (lane 4) was processed to the stable tRNAiMet-size product in the absence of La (lane 5) and, as expected, could not be protected by La (lane 6). The substrate shown in lane 7, pre-tRNAiMetΔ5′, contains an intact 3′ trailer but lacks the 5′ leader and has been used previously to monitor 3′ processing (3). This substrate was processed to the stable tRNAiMet-size product in the absence of La (lane 8) but was protected in the presence of La (lane 9). These data suggested that the 3′ trailer is required for La-mediated protection from two distinct processing events.
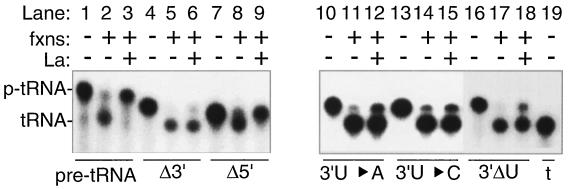
FIG. 2.
La-mediated protection of pre-tRNA is specific and dependent on the RNA 3′ oligo(U) terminus. Effects of La on processing of T7 RNAP-synthesized pre-tRNAiMet (lanes 1 to 3), pre-tRNAiMet-derived transcripts bearing deletion of the 3′ trailer (Δ3′, lanes 4 to 6), deletion of the 5′ leader (Δ5′, lanes 7 to 9), substitution of the terminal 4 Us (3′U→A, lanes 10 to 12; 3′U→C, lanes 13 to 15), and deletion of the terminal 4 Us (3′ΔU, lanes 16 to 18) as indicated below the lanes. Lanes 1, 4, 7, 10, 13, and 16 show the input RNAs for each reaction. Lane 19 shows the T7 RNAP-synthesized mature tRNAiMet transcript of 72 nucleotides. 5S rRNA (100 ng) was included as a nonspecific competitor in all reactions; other components including HeLa fractions (fxns) are indicated above the lanes.
In order to further map the region of the trailer that was required for protection and to examine it for sequence specificity, 3′ mutants of pre-tRNAiMet were used to monitor the nuclease protection activity of La. In reactions parallel to those described above, pre-tRNAiMet transcripts that were substituted at the four terminal uridylates with either A4 (Fig. 2, lanes 10 to 12) or C4 (lanes 13 to 15) produced substrates that were not well protected by La. Likewise, deletion of the terminal uridylates to create pre-tRNAiMetΔ3′U produced a substrate that is not well protected by La (lanes 16 to 18). These data demonstrated that protection was dependent on the presence of the terminal 3′ oligo(U) tract in pre-tRNAiMet. Thus, although La can interact with RNAs that do not contain 3′ oligo(U) in general RNA binding assays (1, 6, 20, 28), dependence on the 3′ oligo(U) tract in the functional assays used here suggest that general RNA binding, if it occurs in these reactions, is not sufficient for protection from processing. Rather, the protective activities characterized here are dependent on the most specific RNA binding modality known for La, i.e., 3′ oligo(U) recognition.
Specific, high-affinity binding of La to the 3′ oligo(U) tract of pre-tRNAiMet.
Although our nuclease protection assays indicated an interaction between La and the terminus of pre-tRNAiMet, it was important to demonstrate specificity for the 3′ oligo(U) tract of pre-tRNAiMet in a direct binding assay. Four tRNAiMet-derived transcripts carrying terminal mutations were examined in parallel and in triplicate by EMSA with various concentrations of La. At all of the concentrations tested, La formed stable complexes with pre-tRNAiMet (Fig. 3A) and pre-tRNAiMetΔ5′ (Fig. 3C), both of which carry oligo(U) 3′ termini. In contrast to these substrates pre-tRNAiMetΔ3′ (Fig. 3B) and pre-tRNAiMetΔ3′U (Fig. 3D), both of which lack oligo(U) 3′ termini, were bound less avidly, requiring a substantially higher concentration of La for complex formation.
FIG. 3.
High-affinity binding of La to pre-tRNAiMet is dependent on the 3′ oligo(U) terminus. EMSAs with wild-type and pre-tRNAiMet-derived transcripts bearing various terminal mutations and increasing amounts of La were performed in parallel with 0.1 ng of probe and 20 ng of poly(rG) competitor. Each concentration was tested in triplicate as shown in three lanes. Amounts of La are indicated above the lanes in femtomoles. All reactions were performed and analyzed in parallel on the same autoradiogram.
These EMSA conditions differ from the protection assay conditions in that only 20 ng of poly(rG) RNA was used as a relatively weak competitor in the presence of pure La for EMSA rather than the 100 ng of E. coli 5S rRNA that was included to provide broader competition in the HeLa extract processing reactions. It was demonstrated in Fig. 1E that concentrations of substrate, 5S rRNA competitor, and La protein used in processing assays revealed a single predominant complex, indicating that La did not form nonspecific higher-order complexes under processing assay conditions. We conclude that protective activity in our assays is not due to a general RNA-binding activity of La but rather instead requires specific recognition of 3′ oligo(U).
The C-terminal basic region of La is required to protect pre-tRNAiMet from 5′ processing.
A panel of His-tagged, affinity-purified, mutant La proteins (12) was used to determine which regions of La are required for pre-tRNAiMet protection. A diagram illustrating a working model of human La struture is shown in Fig. 4, along with a representation of the La constructs used in this study and a summary of the results obtained. La deletion constructs were examined in three assays: (i) Pol III transcription-processing, (ii) RNase P-mediated processing, and (iii) 3′ processing of pre-tRNAiMetΔ5′ (Fig. 5). In the transcription-processing assay, the tRNAiMet-size product was generated in the absence of La (Fig. 5A, lane 1), whereas La 1-408 (i.e., La amino acid residues 1 to 408) and La 1-363 protected pre-tRNAiMet from any detectable processing (lanes 2 and 3). La 1-328 and La 1-235 allowed processing of approximately 50% of pre-tRNAiMet to an intermediate band (lanes 4 and 5). La 1-187 also reproducibly allowed processing to the intermediate band only (not shown). These data suggest that residues 329 to 363 contribute to the protective action of La. Although a significant amount of pre-tRNAiMet remained unprocessed in the presence of La 1-235 and La 1-328, these proteins did not appear to be generally defective since a processing intermediate, but not the final processed product, accumulated in their presence. If these proteins were generally defective we would have expected a distribution of all three bands.
FIG. 4.
A schematic representation of human La. Deletion and substitution mutants used for the experiments described below are shown according to the three-RRM model of human La (12, 19). +++, C-terminal basic region. Phosphoserine 366 is indicated by the encircled P (10). A His tag is present on the C termini of the recombinant constructs but is absent from the native HeLa proteins S-La and U-La (10). Results obtained in the RNase P (5′) and the pre-tRNAiMetΔ5′ (3′) processing protection assays for these proteins are summarized to the right.
La 1-103 and the N-terminal truncated proteins La 26-408, La 104-408, and La 303-408 exhibited no protective activity since they allowed complete processing (Fig. 5, lanes 6 to 9). These proteins do not exhibit high-affinity binding to 3′ oligo(U)-containing RNA (12) and, as described below, do not even protect the oligo(U)-containing substrate pre-tRNAiMetΔ5′ from 3′ processing.
We used primer extension to compare the 5′ termini of the precursor, intermediate, and mature transcripts produced in the presence of appropriate La mutants. By this analysis, the intermediate and mature transcripts exhibited processed 5′ termini, whereas the precursor exhibited an unprocessed 5′ terminus (not shown; see Materials and Methods). These data supported the contention that La protects pre-tRNAiMet from two events, 5′ and 3′ processing, with the C-terminal domain of La required only for protection from 5′ processing. We examine below the protective activity of La in reactions in which only one processing event occurs.
Both a C-terminal basic region and the N-terminal RNA-binding domain of La are required for protection of pre-tRNAiMet from 5′ processing by RNase P.
The data suggested that the N-terminal domain of La associates with the 3′ oligo(U) tract of a pre-tRNA and that this promotes blockage of both processing sites on the substrate, with the C-terminal domain blocking 5′ processing specifically. There is no evidence to indicate and we think it unlikely that La interacted with either RNase P or another processing nuclease in our fractions to inhibit these enzymes directly (see Discussion). This model is consistent with all of our results, including those described below that were obtained with a preparation of highly purified, yeast nuclear RNase P.
The experimental results shown in Fig. 5B reflect the ability of human La protein to protect pre-tRNAiMet from cleavage by yeast nuclear RNase P. La 1-408 and La 1-363 protected pre-tRNAiMet in this assay (lanes 2 to 3). By contrast, the more truncated proteins exhibited no protection from RNase P (lanes 4 to 7) even though some, e.g., La 1-328 and La 1-235, protected pre-tRNAiMetΔ5′ from 3′ processing as described below. Primer extension analysis of the RNase P reaction products confirmed that the yeast enzyme indeed cleaved the 5′ leader of the human pre-tRNAiMet to produce the major product observed in the absence of La (not shown, but see Materials and Methods). The specificity of this assay was further demonstrated by the facts that pre-tRNAiMetΔ3′ could not be protected from RNase P by La and that pre-tRNAiMetΔ5′ was not processed by RNase P in the absence or presence of La (not shown). Proteins that contain an intact C-terminal region but lack the N-terminal residues required for high-affinity RNA binding did not prevent 5′ processing (lanes 8 to 10), allowing the conclusion that the La antigen’s C-terminal region does not inhibit RNase P in trans. These results are in agreement with those given above with the HeLa processing activities.
The mutant La proteins were also tested for their ability to protect pre-tRNAiMetΔ5′ from 3′ processing. This transcript represents a pre-tRNAiMet that has been cleaved at its 5′ end but that retains its oligo(U)-containing 3′ trailer (Fig. 5C). As expected, La 1-408 and La 1-363 protected this substrate from 3′ processing (lanes 2 and 3). However, in contrast to the results obtained with RNase P (Fig. 5B), this substrate was protected by La 1-328, La 1-235, and La 1-187 (Fig. 5C, lanes 4 to 6). Therefore, this substrate was protected by La proteins that extend from amino acids 1 to 187 or beyond (lanes 2 to 6), but not by La 1-103 (lane 7) or any of the proteins that lacked the first 25 or more N-terminal residues (lanes 8 to 10). These data support the idea that the highly conserved residues found near the N terminus of La confer high-affinity interaction with 3′ oligo(U) (19). Since La 1-328, La 1-235, and La 1-187 protected pre-tRNAiMetΔ5′ from 3′ processing, it is likely that the inability of these proteins to protect pre-tRNAiMet from 5′ processing by RNase P is a specific deficiency that is not due to their failure to interact with the 3′ oligo(U)-containing substrate.
Evidence of an interaction between the nascent 5′ triphosphate of pre-tRNAiMet and La.
A T7 RNAP-synthesized nascent transcript lacking the 5′ leader of pre-tRNAiMet exhibited high-affinity for La (Fig. 3C), suggesting that La does not interact with the leader in a sequence-specific manner. The basic region of human La was previously noted to contain the sequence 333GRRFKGKG340, representing a potential NTP binding site of the consensus sequence GXXXXGKX found in a number of ATP-binding proteins (reference 37 and references therein). This observation, in conjunction with this region’s overall basic nature, suggested that La might recognize the 5′ triphosphate as a distinct determinant of a newly synthesized RNA. This mode of contact allows specific recognition of the 5′ ends of a variety of pre-tRNAs despite their lack of sequence homology (see Discussion). This model is consistent with efficient binding of La to pre-tRNAiMetΔ5′ which, because of synthesis by T7 RNAP, contains a nascent pppG 5′ terminus.
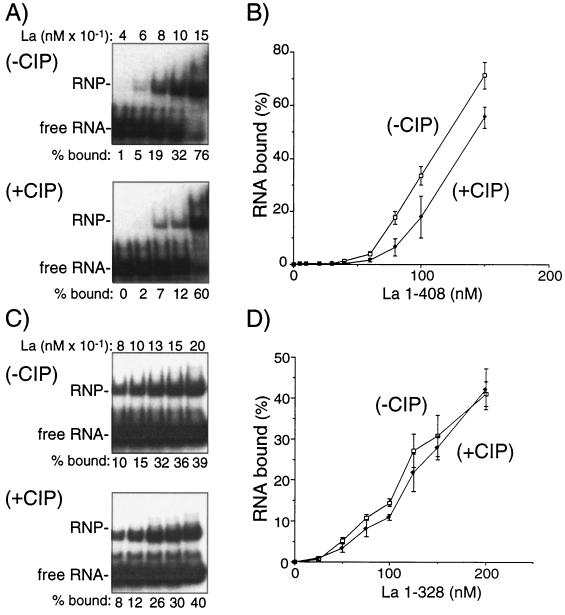
We compared La’s ability to interact with nascent pre-tRNAiMet with its ability to interact with pre-tRNAiMet whose 5′ end was dephosphorylated by CIP. For this we used the direct, albeit general, RNA EMSA (Fig. 6), as well as the specific RNase P protection assay (Fig. 7). In preliminary binding experiments, La bound more nascent full-length pre-tRNAiMet than dephosphorylated pre-tRNAiMet (not shown). We suspected that the contribution to La binding made by the 5′ triphosphate might be small relative to that made by 3′ oligo(U). We therefore used pre-tRNAiMetΔ3′U, which lacks terminal uridylates, to focus the effects of the triphosphate on binding (Fig. 6). At subsaturating concentrations, La bound approximately three times more nascent RNA than dephosphorylated RNA (Fig. 6A, compare −CIP and +CIP panels). Under our EMSA conditions, threefold more binding was considered to be significant as demonstrated by performing the EMSAs in triplicate, confirming that there was loss of affinity between La and the nascent pre-tRNA when the latter was dephosphorylated (Fig. 6B). These results provide direct evidence to suggest that a 5′ triphosphate increases the affinity of an RNA for La (see Discussion).
FIG. 6.
Direct binding evidence that La recognizes the 5′ triphosphate of nascent pre-tRNA. (A) EMSA with nascent pre-tRNA3′ΔU (−CIP, upper panel) or dephosphorylated pre-tRNA3′ΔU (+CIP, lower panel) and increasing amounts of full-length La as indicated. Reaction mixtures were 10 μl and contained a small amount (2 ng) of the nonspecific competitor poly(rG). The fraction of bound RNA (percent bound = [bound/bound + unbound] × 100) was quantitated and is shown below the lanes. Positions of free RNA and RNA-protein (RNP) are indicated to the left. (B) EMSAs as described in panel A were performed in triplicate. The bound versus unbound RNA was then quantitated by phosphorimager analysis, and the bound RNA (percent bound = [bound/bound + unbound] × 100) was plotted against the La concentration; error bars represent the range of three experiments. Equilibrium Kd values under these conditions are 120 nM for nascent pre-tRNAiMet3′ΔU and 142 nM for CIP-treated tRNAiMet3′ΔU (see text). (C) Same as for panel B except that La 1-328 was used. Note that concentrations of La 1-328 (indicated above the upper panel) span a different range than in panel A (see text). Experiments represented in panels A and C were performed in parallel and quantitated from the same exposure by phosphorimager analysis (not shown). The CIP-treated and mock-treated probes were nearly indistinguishable in their migration in these assays and migrated as indicated by “free RNA” in panels A and C when coelectrophoresed in the same gels (not shown). (D) Same as for panel B except that La 1-328 was used. Note that both axes are on scales different from that in panel B (see text). Experiments represented in panels B and D were performed and analyzed in parallel.
FIG. 7.
RNase protection evidence that La recognizes the 5′ triphosphate of nascent pre-tRNA. (A) Nascent pre-tRNAiMet (lanes 2 and 3) and pre-tRNAiMet dephosphorylated by CIP (lanes 4 and 5) were compared in the RNase P protection assay in the presence and absence of La as indicated. The CIP-treated pre-tRNA substrate is presented in lane 1 to show its integrity. RNase P (1.00 μl) was incubated with the substrates for 45 min. (B) The data from three additional experiments performed as described in panel A were quantitated by phosphorimager analysis and converted to percent processed ([processed/processed + unprocessed] × 100) and plotted as bars. Error bars reflect the range of three datum points.
As an additional control we performed the binding assay in parallel with La 1-328 which lacks the C-terminal basic region (Fig. 6C and D). Two observations are noteworthy. First, La 1-328 appeared to exhibit an overall lower affinity for the RNAs than did full-length La, a finding consistent with an interaction between the basic region and the RNA. This can be seen by comparing the binding at intermediate concentrations of La; La 1-328 bound less of the triphosphate-containing substrate (−CIP, 15% bound) than did full-length La (−CIP, 32% bound). Since the La 1-328 binding reactions did not reach saturation, the data cannot be used to determine precise affinities, but they are nonetheless consistent with the conclusion that La 1-328 exhibits lower affinity for pre-tRNAiMet than did La 1-408. Second, and more importantly, in contrast to the full-length protein, La 1-328 exhibited no significant preference for either nascent or dephosphorylated RNA. Thus, although the preference of La 1-408 for the nascent RNA was not found to be large by this assay, it is significant and specific as demonstrated by La 1-328, which exhibits no preference.
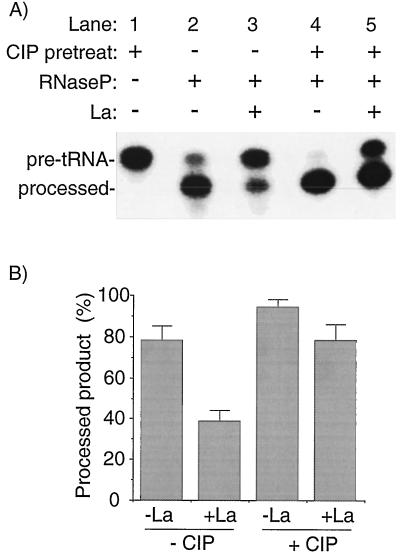
La-mediated nuclease protection is a more specific assay of La RNA recognition than is EMSA (12). We therefore examined nascent pre-tRNAiMet and CIP-treated pre-tRNAiMet for La-mediated protection from RNase P. CIP-treated RNA was a slightly better substrate than mock-treated RNA, as indicated by comparing the amount of precursor remaining in lane 2 versus lane 4, although we do not have an explanation for this. A more substantial difference was seen in the presence of La. CIP treatment decreased the ability of La to protect pre-tRNAiMet from cleavage by RNase P (Fig. 7A, compare lanes 3 and 5). Although La protected some CIP-treated substrate from RNase P digestion, this was a significantly smaller fraction than with the mock-treated substrate. Quantitative data from three additional experiments are presented in Fig. 7B. These provided reproducible evidence to indicate that La’s ability to protect the pre-tRNA leader is dependent to a significant degree on the 5′ triphosphate of the nascent transcript.
In summary, both RNA binding and nuclease protection indicate that the 5′ triphosphate is a significant, positive determinant of pre-tRNAiMet recognition by La. While it is unknown whether the interaction is simply ionic or highly specific, the data provide additional evidence to support the conclusion that La residues 329 to 408 contribute to RNA recognition (12) and exhibit some affinity for the 5′-triphosphate end of the RNA.
Phosphorylation of La serine 366 interferes with protection of pre-tRNAiMet from 5′ processing.
Native HeLa La can be separated into a serine 366-phosphorylated form (U-La) and an unphosphorylated form (S-La) (10). Since human recombinant La can be phosphorylated in vitro by CKII specifically at this site and subsequently purified (10), we could also examine recombinant His-tagged La 1-408 in both phosphorylated and unphosphorylated forms in the Pol III transcription-processing assay (Fig. 8A), the RNase P protection assay (Fig. 8B), and the 3′ nuclease protection assay (Fig. 8C), along with the native proteins U-La and S-La.
Pol III transcription-processing reactions produced the fully processed, tRNAiMet-size product in the absence of La (Fig. 8A, lane 1). S-La prevented processing (lane 2), while U-La allowed a significant amount of processing but only to the intermediate band (lane 3). Likewise, pre-tRNAiMet synthesized by Pol III in the presence of La 1-408 remained mostly unprocessed (lane 4), whereas CKII-phosphorylated La 1-408 protected very little of the substrate from conversion to the intermediate band (lane 5). The data suggested that phosphorylated La allowed 5′ processing while preventing 3′ processing.
The results presented in Fig. 8B confirmed that processing by RNase P proceeded in the presence of phosphorylated but not unphosphorylated La. The phosphoproteins U-La and CKII-phosphorylated La 1-408 conferred little if any protection of nascent pre-tRNAiMet from processing by RNase P (Fig. 8B, lanes 5 and 7) compared to the control (lane 3), while their unphosphorylated counterparts S-La and La 1-408 conferred efficient protection (lanes 4 and 6). These reactions contain highly purified components and provide direct evidence that phosphoserine 366 interferes with La’s ability to protect pre-tRNA from 5′ cleavage by RNase P. We also examined the La mutant La GXK>DE, in which residues 328 to 344 were substituted with acidic residues (12). Although this protein interacts with RNA in a general binding assay (12) and can protect pre-tRNAiMetΔ5′ from 3′ processing (see below), it reproducibly failed to protect pre-tRNAiMet from 5′ processing (Fig. 8, lane 8).
Since native phosphorylated U-La and in vitro-phosphorylated La 1-408 allowed processing to an intermediate but not the fully processed product (Fig. 8A), the data suggested that these proteins protected the 3′ end of the nascent transcript from processing. Consistent with this conclusion, pre-tRNAiMetΔ5′ was efficiently protected by U-La and CKII-La in the 3′-nuclease protection assay (Fig. 8C). Not only does this indicate a selective loss of pre-tRNAiMet 5′-end protection by U-La and CKII-La, it also suggests that 5′ processing occurs while the pre-tRNAiMet substrate is bound at its 3′ end by native La phosphoprotein. It appears that the phosphorylated form of La antigen stably interacts with the 3′ oligo(U) tract on nascent pre-tRNA while allowing 5′ processing to occur, whereas unphosphorylated La antigen blocks 5′ processing by RNase P.
DISCUSSION
The results reported here suggest that 5′ processing of tRNA precursors may be regulatable in eukaryotes. La antigen has previously been shown to modulate 3′ processing of tRNA precursors in yeast (43). Human tRNAiMet maturation occurs along an ordered pathway with 5′ processing preceding 3′ processing (3, 45), which is consistent with the pre-tRNA maturation pathways elucidated in yeast cells (38, 43). The data reported here extend our understanding of La’s role in tRNA metabolism by showing that in addition to modulating 3′ processing, La can also modulate 5′ processing of pre-tRNA. In addition, a new binding modality for La was identified, the ability to recognize the 5′ triphosphate of a nascent transcript. Because of its unique position on newly synthesized RNAs, the triphosphate moiety would provide La with the ability to interact with the initiation ends of various nascent transcripts while 3′ oligo(U) recognition promotes interaction with the termination ends. Triphosphate binding may also help explain other activities that have been reported for La as discussed below. Finally, regions within both La and the pre-tRNA required for 5′ and 3′ protection were identified.
We wish to emphasize that 3′ processing of pre-tRNAs can proceed by either of two pathways (43). In our system, 3′ processing is presumably mediated by an exonuclease, one similar to the 3′ exonuclease that matures pre-tRNA in S. cerevisiae cells that have been depleted of La (43).
Several considerations suggest that 5′ protection by La occurs in vivo. First, protection occurred at a concentration of La found in unfractionated S100. La immunodepletion and repletion experiments confirmed that La is a pre-tRNA processing inhibitory factor. Second, the effects of La in this system were shown to be specific by several criteria. Concordance of specificity in binding and protection assays demonstrated that La functionally interacts with pre-tRNAiMet via the 3′ oligo(U) tract, a well-known characteristic of La. Modulation of 5′ processing by La’s basic region was also shown to be specific, since proteins altered in this region remain active for high-affinity binding to oligo(U)-containing RNAs (10, 12), as well as for 3′ protection (Fig. 5A and C), while losing 5′ protection activity specifically (Fig. 5B). Likewise, loss of 5′ protection by phosphorylated La (Fig. 8A and B) is specific, since this modification does not interfere with La’s ability to bind oligo(U)-containing RNAs (10) and to protect the oligo(U)-containing substrate, pre-tRNAiMetΔ5′, from 3′ processing (Fig. 8C). Expression of human La in La-deleted S. pombe cells corrects aberrant tRNA 3′-end maturation but also leads to a disproportionately large amount of pre-tRNA (relative to mature tRNA) that retains its 5′ leader (38). Although other explanations are possible, this suggests that human La protects S. pombe pre-tRNAs from 5′ processing because it is not phosphorylated efficiently when overexpressed in S. pombe. Finally, previous results suggested that 5′ processed tRNAiMet intermediates are associated with La in vivo (30) and therefore support the notion that a La-pre-tRNA complex is a substrate for RNase P (Fig. 5 and 8). The structure of this complex appears to be altered by the phosphorylation status of La serine 366.
Phosphorylation and dephosphorylation control La activity.
Although a small amount of La is present in the TFIIIB-Pol III fraction used for transcription, this amount of La is clearly insufficient to protect pre-tRNA from processing. This endogenous La is phosphorylated by endogenous CKII, or a CKII-like activity, during our transcription-processing reactions (9). Therefore, pre-tRNA processing and La phosphorylation both appear to be occurring in our Pol III transcription-processing reactions. Addition of exogenous La overwhelms the phosphorylation capacity of these reactions and protects against 5′ processing. Consistent with this, exogenous CKII added to these reactions reverses the 5′ protection activity of exogenous La (9).
La protects the pre-tRNA as opposed to inhibiting RNase P directly.
The C-terminal basic region of La does not inhibit RNase P directly or in a general way, as demonstrated by the N-terminal truncated proteins, La 26-408, La 104-408 and La 303-408, which contain the C-terminal basic region but do not inhibit RNase P (Fig. 5B, lanes 8 to 10). Thus, the C-terminal basic region exhibits 5′ protective activity only when present in cis with the RRM-containing N-terminal domain of La.
While La bound at the 3′ end of pre-tRNA might theoretically interfere with the substrate’s ability to fit into the recognition site of RNase P (17, 40), the data suggest that this is not the case since La proteins that bind and protect the 3′ end of pre-tRNAiMet do not block processing by RNase P. This point is best made by comparing U-La and S-La, two proteins that differ only in their serine 366 phosphorylation status yet which differentially protect pre-tRNAiMet from RNase P (Fig. 8B).
General versus specific binding of La to RNA.
In the cell, La is presumably directed to its natural ligands, nascent Pol III transcripts (30, 34), by the La-containing Pol III holoenzyme (39). However, La protein can bind certain viral mRNAs. These include the leader sequence of vesicular stomatitis virus RNA (20), as well as the 5′ regions of TAR RNA (6), rubella virus RNA (28), poliovirus RNA (27), and hepatitis C virus RNA (1). Since these are not Pol III transcripts, the pathway(s) by which La interacts with them has remained a mystery. Our data indicate that La recognizes the 5′ triphosphate of newly synthesized RNA (Fig. 6 and 7). These findings suggest the possibility that La may interact with leader sequences through contacts made with their nascent 5′ triphosphate termini. Although the results with these leader sequences suggest that La may be involved in viral RNA biogenesis, we should also suspect that an RNA motif that is normally buried within a longer transcript but removed from its context by synthesis in vitro and therefore bearing a 5′ triphosphate might exhibit a higher affinity for La than it otherwise would. Moreover, templates with EcoRI-restricted 3′ ends artificially add 3′ U residues to the transcript, further increasing the apparent affinity of the RNA for La (1, 6, 26).
Bipartite binding by La and nascent RNA.
5′- and 3′-end-mediated binding of RNA to La can probably be accommodated by most Pol III nascent transcripts, since their 5′ and 3′ termini are often found in proximity to one another. The conserved structure of La in which two RRMs reside in the N-terminal domain and a basic region resides in the C-terminal domain (19) suggests that bipartite interactions with nascent RNA may be of fundamental importance to La function (see references 10 and 38 for alignments). It is also noteworthy that the 5′ triphosphate of the tRNA-like transcript of E. coli known as RNA I guards against a 3′ nuclease. In that case, a protein that mediates RNA I 3′-end metabolism also influences 5′ processing (41).
The present results help explain, in terms of an RNA-binding modality, recent findings that indicate that La interacts with the 5′ m7Gppp of certain selected synthetic RNAs that terminate in a 3′ U residue that lies in close proximity to the 5′ cap (15), findings which are consistent with the bipartite binding model described here. We, too, can readily UV cross-link 32pppG-labeled RNA to La (9, 15). Thus, La residues are in close contact with the 5′ triphosphate of a nascent RNA, and this interaction is apparently not blocked in the m7Gppp cap structure (15). This raises the possibility that La may also be able to recognize the hypermethylated caps of mRNAs. If La can block recognition of an m7Gppp cap by other proteins (15), this might explain in part why La protein can direct internal ribosome entry site-mediated initiation when added to in vitro translation reactions (1, 26, 27). It will also be interesting to examine whether the other cap structure, me-pppN, which is limited to a subset of Pol III transcripts, interferes with La binding and/or if me-ppp capping occurs on an La-RNA substrate (31, 32).
According to the bipartite binding model, La’s N-terminal domain interacts with the oligo(U) terminus of the pre-tRNA and La’s C-terminal domain interacts with the nascent triphosphate 5′ end of the pre-tRNA. Although it has been reasonable to assume that oligo(U) binding provides the primary interaction between La and a nascent RNA, we must now consider the possibility that La interacts first with the 5′ end of the nascent transcript, as might occur during the early elongation phase of transcription after La is brought to the template by the Pol III holoenzyme (39). This alternative, i.e., primary interaction with the 5′ end of the nascent transcript, may help explain why La appears to function as a Pol III initiation factor in some assays (23).
A role for La in coordinating RNA biogenesis and maturation.
Previous data indicate a role for La phosphorylation in regulating the recycling efficiency of Pol III transcription complexes (10). In view of the present results that indicate a role for La phosphorylation in posttranscriptional events, we must consider the possibility that La phosphorylation-dephosphorylation allows coordination of transcriptional and early posttranscriptional stages of RNA biogenesis.
Although it might appear paradoxical that La would first stimulate RNA synthesis and then prevent maturation of the same transcript, thereby acting positively and negatively in a single pathway, we believe that the net effect of La is to facilitate production of functional RNAs by acting as a quality control factor. By binding to the leader and trailer regions of a pre-tRNA, La may not interfere with tRNA recognition by other factors. La might serve as a platform, stabilizing nascent transcripts and ensuring that tRNA modifying and maturation enzymes have ample time to complete their activities, after which phosphorylation may signal release of the RNA. By contrast, uncontrolled processing might allow some transcripts to bypass important modifying enzymes and so fail to undergo efficient maturation to a fully functional tRNA. It will therefore be important to determine the extent to which La contacts pre-tRNA and whether modifications can occur on a pre-tRNA substrate that is bound to La.
La interacts transiently with most if not all nascent transcripts synthesized by Pol III, yet the mechanism by which these are released from La is unknown. The results reported here for pre-tRNAs may be extendable to Pol III transcripts in general. Phosphorylation may be used to control posttranscriptional handling of RNA at the level of dissociation of the nascent RNA-La complex, an issue that has received relatively little attention (16, 35, 36).
ACKNOWLEDGMENTS
We thank D. Kenan and J. Keene for generous and insightful discussions of La structure; J. Steitz and the reviewers who made helpful comments on the manuscript; and J. Anderson, A. Hinnebusch, and LMGR members for helpful discussions.
J.R.C. was a predoctoral fellow supported by NIH grant T32 GM07315. Work by J.R.C. and D.R.E. was supported by NIH grant GM34869. H.F. and J.L.G. were supported by the NICHD Visiting Fellow Program.
Footnotes
R.J.M. dedicates this research paper to the memory of Christopher P. Cully.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castano J G, Ornberg R, Koster J G, Tobian J A, Zasloff M. Eukaryotic Pre-tRNA 5′ processing nuclease: copurification with a complex cylindrical particle. Cell. 1986;46:377–387. doi: 10.1016/0092-8674(86)90658-6. [DOI] [PubMed] [Google Scholar]
- 3.Castano J G, Tobian J A, Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNAiMet (3′ pre-tRNase) J Biol Chem. 1985;260:9002–9008. [PubMed] [Google Scholar]
- 4.Chamberlain J R, Pagan-Ramos E, Kindelberger D W, Engelke D R. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan E K L, Tan E M. The small ribonucleoprotein SS-B/La binds RNA with a conserved protease-resistant domain of 28 kilodaltons. Mol Cell Biol. 1987;7:2588–2591. doi: 10.1128/mcb.7.7.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y-N, Kenan D J, Keene J D, Gatignol A, Jeang K-T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean N, Berk A J. Separation of TFIIIC into two functional components by sequence specific DNA affinity chromatography. Nucleic Acids Res. 1987;15:9895–9907. doi: 10.1093/nar/15.23.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher M P. tRNA processing nucleases. In: Soll D, RajBhandary U L, editors. tRNA: structure, biosynthesis and function. Washington, D.C: ASM Press; 1995. pp. 51–56. [Google Scholar]
- 9.Fan, H., and R. J. Maraia. 1998. Unpublished observations.
- 10.Fan H, Sakulich A L, Goodier J L, Zhang X, Qin J, Maraia R J. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 11.Fan H, Sugiura M. A plant basal in vitro system supporting accurate transcription of both RNA polymerase II- and III-dependent genes: supplement of green leaf component(s) drives accurate transcription of a light-responsive rbcS. EMBO J. 1995;14:1024–1031. doi: 10.1002/j.1460-2075.1995.tb07083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodier J L, Fan H, Maraia R J. A carboxy-terminal basic region controls the RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol. 1997;17:5823–5832. doi: 10.1128/mcb.17.10.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb E, Steitz J A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlieb E, Steitz J A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm C, Lund E, Dahlberg J E. In vivo selection of RNAs that localize in the nucleus. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guddat U, Bakken A H, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- 17.Harris M E, Nolan J M, Malhotra A, Brown J W, Harvey S, Pace N R. Use of photoaffinity crosslinking and molecular modeling to analyze the global archiecture of ribonuclease P RNA. EMBO J. 1994;13:3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inokuchi H, Yamao F. Structure and expression of prokaryotic tRNA genes. In: Soll D, RajBhandary U L, editors. tRNA: structure, biosynthesis and function. Washington, D.C: ASM Press; 1995. pp. 17–30. [Google Scholar]
- 19.Kenan D J. RNA recognition by the human La protein and its relevance to transcription, translation and viral infectivity. Ph.D. thesis. Durham, N.C: Duke University; 1995. [Google Scholar]
- 20.Kurilla M G, Keene J D. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1983;34:837–845. doi: 10.1016/0092-8674(83)90541-x. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Deutscher M P. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 22.Maraia R. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res. 1991;19:5695–5702. doi: 10.1093/nar/19.20.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maraia R J. Transcription termination factor La is also a reinitiation factor for RNA polymerase III. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maraia R J, Chang D-Y, Wolffe A P, Vorce R L, Hsu K. The RNA polymerase III terminator used by a B1-Alu element can modulate 3′ processing of the intermediate RNA product. Mol Cell Biol. 1992;12:1500–1506. doi: 10.1128/mcb.12.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maraia R J, Kenan D J, Keene J D. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;67:3798–3807. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 27.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogue G P, Hofmann J, Duncan R, Best J M, Etherington J, Sontheimer R D, Nakhasi H L. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruijn G J, Slobbe R L, van Venrooij W J. Analysis of protein-RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 1991;19:5173–5180. doi: 10.1093/nar/19.19.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinke J, Steitz J A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- 31.Shumyatsky G, Shimba S, Reddy R. Capping signals correspond to the 5′ end in four eukaryotic small RNAs containing gamma-monomethylphosphate cap structure. Gene Expr. 1994;4:29–41. [PMC free article] [PubMed] [Google Scholar]
- 32.Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci USA. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprague K. Transcription of eukaryotic tRNA genes. In: Soll D, RajBhandary U L, editors. tRNA: structure, biosynthesis and function. Washington, D.C: ASM Press; 1995. pp. 31–50. [Google Scholar]
- 34.Stefano J E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 35.Steitz J A, Berg C, Hendrick J P, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terns M P, Lund E, Dahlberg J E. 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol Cell Biol. 1992;12:3032–3040. doi: 10.1128/mcb.12.7.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topfer F, Gordon T, McCluskey J. Characterization of the mouse autoantigen La (SS-B) J Immunol. 1993;150:3091–3100. [PubMed] [Google Scholar]
- 38.Van Horn D J, Yoo C J, Xue D, Shi H, Wolin S L. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westhoff E, Altman S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5133–5137. doi: 10.1073/pnas.91.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu F, Cohen S N. RNA degradation in Escherichia coli is regulated by 3′ adenylation and 5′ phosphorylation. Nature (London) 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 42.Yoo C J, Wolin S L. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo C J, Wolin S L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 44.Yoshinaga S K, Boulanger P A, Berk A J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci USA. 1987;84:3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zasloff M, Santos T, Romeo P, Rosenberg M. Transcription and precursor processing of normal and mutant human tRNAiMet genes in a homologous cell-free system. J Biol Chem. 1982;257:7857–7863. [PubMed] [Google Scholar]