Abstract
To isolate and characterize proteins that interact with the unique domain and SH3 and SH2 domains of Src and potentially regulate Src activity, we used the yeast two-hybrid assay to screen a human lung fibroblast cDNA library. We identified RACK1, a receptor for activated C kinase and a homolog of the β subunit of G proteins, as a Src-binding protein. Using GST-Src fusion proteins, we determined that RACK1 binds to the SH2 domain of Src. Coimmunoprecipitation of Src and RACK1 was demonstrated with NIH 3T3 cells. Purified GST-RACK1 inhibited the in vitro kinase activity of Src in a concentration-dependent manner. GST-RACK1 (2 μM) inhibited the activities of purified Src and Lck tyrosine kinases by 40 to 50% but did not inhibit the activities of three serine/threonine kinases that we tested. Tyrosine phosphorylation on many cellular proteins decreased in 293T cells that transiently overexpressed RACK1. Src activity and cell growth rates decreased by 40 to 50% in NIH 3T3 cells that stably overexpressed RACK1. Flow cytometric analyses revealed that RACK1-overexpressing cells do not show an increased rate of necrosis or apoptosis but do spend significantly more time in G0/G1 than do wild-type cells. Prolongation of G0/G1 could account for the increased doubling time of RACK1-overexpressing cells. We suggest that RACK1 exerts its effect on the NIH 3T3 cell cycle in part by inhibiting Src activity.
The cellular gene c-src and its viral homolog v-src (the transforming gene of Rous sarcoma virus) encode 60-kDa, cytoplasmic, membrane-associated protein-tyrosine kinases (reviewed in reference 6). For the viral protein (v-Src) or for transforming mutants of the cellular protein (c-Src or Src), a close correlation exists between elevated specific kinase activity and cell transformation. At least four factors are known to influence Src activity: (i) mutation within the coding region of the src gene, (ii) phosphorylation on Tyr 527 and Tyr 416 of Src, (iii) subcellular localization of Src and its substrates, and (iv) association of Src with other cellular proteins.
Compelling evidence indicates that Src-binding proteins can regulate Src activity (reviewed in reference 6). A number of interacting proteins that upregulate Src activity have been identified. The prototype is middle T antigen (mT), the transforming protein of polyomavirus. Src complexed with mT has elevated specific activity due to dephosphorylation of Tyr 527 (5, 8, 11, 16). Activation of Src is required for the induction of mammary tumors in mT transgenic mice (24). Characterization of the Src-mT complex led to discovery of the fundamental mechanism by which the cellular Src protein is converted to a transforming protein and defined the requirement of Src for polyomavirus transformation. Thus, characterization of a single Src-binding protein contributed substantially to our understanding of both RNA and DNA tumor virology.
While a number of interacting proteins that upregulate Src activity have been identified, few that downregulate Src activity have been identified. Because it is the repression of c-Src activity rather than the elevation of v-Src activity that accounts for differences in their transforming abilities (9, 29, 53, 56), it is important to search for cellular mechanisms that inactivate c-Src. Recently, caveolin, a 22-kDa integral membrane protein that is the principal structural and regulatory component of caveolae, was shown to bind Src and suppress its tyrosine kinase activity (33–35).
Domains within Src kinases target the kinases to specific subcellular locations where they bind to regulatory and/or substrate proteins and are integrated into cell signaling pathways and cell cycle events (reviewed in reference 6). For example, the N-terminal unique domain (UD) confers the specificity of binding of Lck to CD4 and CD8 (64) and of Fyn to the zeta chain of the T-cell receptor (22), thus coupling intracellular tyrosine kinases to the signaling pathways of cell surface receptors. The SH3 domain binds to proline-rich motifs in proteins such as Sam68, an RNA-binding protein that binds to Src and becomes tyrosine phosphorylated during mitosis (20, 63). The SH2 domain of Src binds to tyrosine-phosphorylated proteins such as the platelet-derived growth factor (PDGF) receptor, and this interaction, which results in Src activation, is required for PDGF-induced DNA synthesis (57, 65). Thus, the UD and the SH3 and SH2 domains (UD/SH3/SH2) in Src are key binding sites for proteins that regulate Src activity and integrate Src into important signaling pathways and cell cycle events.
The purpose of this study was to isolate and characterize Src-interacting proteins that potentially regulate Src activity. We focused on protein interactions that involve UD/SH3/SH2 of Src. Using the yeast two-hybrid assay to screen a human lung fibroblast cDNA library, we identified RACK1, a receptor for activated C kinase and a homolog of the β subunit of G proteins, as a Src SH2-binding protein. We found that the overexpression of RACK1 inhibits the activity of Src tyrosine kinases and the growth of NIH 3T3 cells. RACK1 exerts its effect on growth during the G0/G1 phase of the cell cycle.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Mediatech, Herndon, Va.) supplemented with 10% calf serum (Sigma, St. Louis, Mo.). NIH 3T3 cells transfected with pneoMLV–wild-type c-src (3T3/c-Src cells; 9) or pcDNA3-HA-RACK1 were maintained in G418 (200 μg/ml) (Gibco-BRL, Life Technologies, Gaithersburg, Md.). 3T3/c-Src cells transfected with pTK-Hygro (Clontech, Palo Alto, Calif.) and pcDNA3-HA-RACK1 or pZeoSV-HA-RACK1 were maintained in G418 (200 μg/ml) and hygromycin (150 μg/ml). NIH 3T3 cells transfected with pZeoSV-RACK1 were maintained in Zeocin (Invitrogen, La Jolla, Calif.) (150 μg/ml). HeLa, 293T, and human colon carcinoma WiDr cell lines (American Type Culture Collection, Manassas, Va.) were cultured in DMEM supplemented with 10% fetal bovine serum (Sigma).
To analyze cell growth rates and doubling times, 104 cells were seeded in 35-mm-diameter plates. Cells from quadruplicate plates were counted with a hemocytometer at 24-h intervals from days 2 to 6. Cell number (log scale) was plotted versus time (linear scale), and doubling time was calculated from the slope of the line.
Plasmids.
pGEMsrc, a gift from Tony Hunter and Martin Broome (The Salk Institute, La Jolla, Calif.), was digested with NcoI and SmaI, and the src insert was subcloned into the yeast vector pAS2, creating pAS2-srcUD/SH3/SH2. The full-length, 1.2-kb human RACK1 cDNA (H12.3) was excised from the yeast two-hybrid vector pGAD-GL and subcloned into pcDNA3 (Invitrogen), pZeoSV (Invitrogen), and pGEX 4T3 (Pharmacia, Piscataway, N.J.) to create pcDNA3-RACK1, pZeoSV-RACK1, and pGEX-RACK1, respectively. The influenza virus hemagglutinin (HA) tag (YPYDVPDYA) was inserted into the BglII site of pcDNA3-RACK1 and pZeoSV-RACK1 to create pcDNA3-HA-RACK1 and pZeoSV-HA-RACK1, respectively. These plasmids were used for both transient and stable RACK1 expression. Hygromycin-resistant plasmid pTK-Hygro was cotransfected with RACK1-containing plasmids into 3T3/c-Src cells. Murine c-src cDNAs were subcloned into the pGEX 4T3 plasmid to create pGEXsrc (full length), pGEXsrc-SmaI (UD/SH3/SH2), pGEXsrc-UD, pGEXsrc-SH3, pGEXsrc-SH2, pGEXsrc-UD/SH3, and pGEXsrc-SH2/SH3. The pGEX-src constructs were used to generate glutathione S-transferase (GST)–Src fusion proteins. pGEMsrc was used for in vitro translations.
Antibodies, purified kinases, and synthetic peptides.
A peptide corresponding to amino acids 527 to 533 of Src was synthesized, coupled with glutaraldehyde to a carrier (keyhole limpet hemocyanin; Calbiochem, San Diego, Calif.), and injected into New Zealand White rabbits to produce antipeptide antibody R7 (26). Monoclonal antibodies (MAb) to RACK1 (Transduction Laboratories, Lexington, Ky.), the HA epitope (12CA5; Boehringer Mannheim Biochemicals, Indianapolis, Ind.), and phosphotyrosine (PY20; Transduction Laboratories) were commercially available. Src MAb 327 hybridoma cells (38) were used to generate MAb 327 ascites. Antipeptide antibody specific for Yes (Yes 3; 31) was a gift from Sara Courtneidge (Sugen, Redwood City, Calif.). Purified Src (expressed by recombinant baculovirus containing the human c-src gene in Sf9 insect cells; specific activity, 900 U/μg), Lck (purified from membrane fractions of bovine thymus), and protein kinase C (PKC; a mixture of α, β, and γ isoforms purified to >97% homogeneity from rat brain) were obtained from Upstate Biotechnology, Inc., Lake Placid, N.Y. Purified protein kinase A (PKA; the catalytic subunit of cyclic AMP-dependent protein kinase purified to homogeneity from bovine heart) and casein kinase II (CKII; purified from rat liver in the tetrameric form α α′ β2) were purchased from Promega, Madison, Wis. Src peptide substrate (K-V-E-K-I-G-E-G-T-Y-G-V-V-Y-K, corresponding to amino acids 6 to 20 of p34cdc2) was purchased from Upstate Biotechnology, Inc. Kemptide (L-R-R-A-S-L-G), CKII substrate (R-R-R-E-E-E-T-E-E-E) and biotinylated PKC substrate (Neurogranin28-43; A-A-K-I-Q-A-S-F-R-G-H-M-A-R-K-K) were purchased from Promega.
Yeast two-hybrid screen.
UD/SH3/SH2 of murine c-src was inserted into the plasmid vector pAS2 (which contains an HA epitope tag) as a GAL4 DNA-binding domain fusion and transformed into cycloheximide-resistant (Chxr) MATα Saccharomyces cerevisiae Y190 (3, 19). Expression of the Src-GAL4 DNA-binding domain fusion protein was confirmed by immunoblot analysis of yeast cell lysates with MAb 327, which binds to the SH3 domain of Src, and with MAb 12CA5, which recognizes the HA epitope (data not shown). A WI-38 human lung fibroblast cell line cDNA library fused to the GAL4 activation domain in the pGAD-GL vector (Clontech) was transformed into the Src-UD/SH3/SH2 Y190 strain, and 2 × 106 transformants were screened for interactors (19). Yeast plasmid DNA was isolated from His+ β-galactosidase-positive Chxr colonies and rescued into LeuB Escherichia coli HB101. False-positives were eliminated by (i) generating Leu+ Trp− transformants and assaying them for β-galactosidase activity; (ii) cotransforming pGAD-GL clones and a control bait, pAS2-lamin; and (iii) retransforming pGAD-GL clones into Y190 harboring pAS2-Src-UD/SH3/SH2 and reassaying for β-galactosidase activity and growth on Trp− Leu− His− medium with 50 mM 3-aminotriazole. Redundant clones were eliminated by analyzing insert sizes. Nonredundant, Src-specific cDNA clones derived from the two-hybrid screen were sequenced by the dideoxynucleotide chain termination method with Sequenase 2.0 (United States Biochemicals, Cleveland, Ohio). Nucleotide sequence databases (GenBank and EMBL) were searched for homologous sequences with FASTDB analysis (Intelligenetics, Santa Clara, Calif.).
In vitro translation and GST fusion protein-binding assays.
Circular plasmid DNAs (2 μg of pGEMsrc or pcDNA3-RACK1) were transcribed and translated in vitro with a TNT-coupled rabbit reticulocyte lysate system (Promega) as instructed by the manufacturer. In vitro-translated Src was shown in an in vitro kinase assay (see below) to have in vitro kinase activity (data not shown).
Cultures of E. coli DH5α containing pGEX-RACK1 or various pGEXsrc plasmids were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (United States Biochemicals) for 3 h at 30°C. Bacteria were harvested, resuspended in Tris-buffered saline (TBS) containing 1% Triton X-100 and 100 mM EDTA, and sonicated. After centrifugation at 12,000 × g for 10 min to remove debris, the supernatant was incubated with glutathione-agarose beads (Sigma) for 2 h at 4°C with agitation. Beads were washed three times with TBS. GST fusion proteins were eluted by the addition of 100 mM Tris (pH 8.0)–120 mM NaCl–20 mM glutathione and dialyzed four times against TBS.
PRO-MIX[35S] (70% l-[35S]methionine and 30% l-[35S]cysteine; >1,000 Ci/mmol; Amersham, Arlington Heights, Ill.)-labeled in vitro translation products were diluted in binding buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.2% Nonidet P-40 [NP-40]) and incubated with 1 μg of purified GST fusion protein for 3 h at 4°C. Protein complexes were collected with the addition of 30 μl of glutathione beads, washed four times in buffer containing 0.5% NP-40, 20 mM Tris (pH 8.0), 100 mM NaCl, and 1 mM EDTA, and boiled in sodium dodecyl sulfate (SDS) sample buffer (7–11). Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). A 1/10 or 1/20 quantity of the unbound translation reaction product was loaded directly on the gel as a measure of the amount of protein translated. The gel was stained with Coomassie brilliant blue G-250 (Sigma) and treated with Fluoro-Hance (Research Products International Corp., Mount Prospect, Ill.). Radiolabeled proteins were detected by fluorography.
Protein extractions, immunoprecipitations, and in vitro protein kinase assays.
Cells were washed three times with ice-cold TBS. For coimmunoprecipitation experiments, cells were lysed in ice-cold NP-40 buffer (0.5% NP-40, 20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 100 μM sodium vanadate, 50 mM sodium fluoride, 50 μM leupeptin, 1% aprotinin, 1 mM dithiothreitol [DTT]). For other experiments, cells were lysed in ice-cold modified radioimmunoprecipitation assay (RIPA) buffer (0.1% SDS, 1% NP-40, 1% sodium deoxycholate, 0.15 M sodium chloride, 10 mM sodium phosphate [pH 7.0], 100 μM sodium vanadate, 50 mM sodium fluoride, 50 μM leupeptin, 1% aprotinin, 2 mM EDTA, 1 mM DTT). Lysates were centrifuged at 14,000 × g for 1 h at 4°C. Protein concentrations were measured by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.), and samples were standardized to equal amounts of total cellular protein (7–11, 49, 50). Lysates were incubated for 3 h at 4°C with 1 μg of MAb 327, antipeptide antibody R7, or RACK1 MAb, and protein complexes were collected with the addition of 30 μl of protein A/G Sepharose beads (Pharmacia) for MAb 327 or R7 immunoprecipitates or with the addition of 30 μl of goat anti-mouse immunoglobulin M (IgM) conjugated to agarose beads (Sigma) for RACK1 MAb immunoprecipitates (7–11, 49, 50). Src kinase assays were performed by incubating MAb 327 or RACK1 MAb immunoprecipitates for 10 min at 30°C in 30 μl of kinase buffer, containing 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 7.0), 10 mM manganese chloride, 10 mM DTT, 1 μg of acid-denatured rabbit muscle enolase (Boehringer), 10 μM ATP, and 25 μCi of [γ-32P]ATP (4,000 Ci/mmol; ICN, Costa Mesa, Calif.) (7–11, 49, 50). In some cases, purified Src was preincubated for 15 min at 4°C with 2 μM GST or GST-RACK or with RACK1 MAb or rabbit anti-mouse IgG immunoprecipitates prior to the addition of kinase buffer. Proteins were resolved on SDS–7% polyacrylamide gels (acrylamide-bisacrylamide, 20:1) to achieve maximum separation among 60-kDa Src, 55-kDa IgG heavy chain, and 47-kDa enolase. Gels were stained with Coomassie brilliant blue G-250 to confirm that equivalent amounts of enolase were present in each lane. Radiolabeled proteins were detected with Kodak XAR or Fuji RX film and an intensifying screen at −70°C. 32P incorporation into proteins was quantified by Cerenkov counting of excised gel pieces and, for some autoradiograms, by scanning densitometry. Both methods gave similar results. Src or Yes in vitro protein kinase activity is linearly related to the concentration of total cellular protein (7–11, 49, 50).
Immunoblot analysis.
Src or RACK1 immunoprecipitates were resolved on SDS–10% polyacrylamide gels (acrylamide-bisacrylamide, 29:0.8). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, Mass.) in transfer buffer (25 mM Tris-HCl [pH 7.4], 192 mM glycine, 15% methanol) with a Trans-Blot apparatus (Bio-Rad, Hercules, Calif.) for 2 h at 60 V (7–11, 49, 50). Protein-binding sites on the membranes were blocked by incubating membranes overnight in TNT buffer (10 mM Tris-HCl [pH 7.5], 100 mM sodium chloride, 0.1% [vol/vol] Tween 20 [Sigma]) containing 3% nonfat powdered milk (blocking buffer). Membranes were incubated with RACK1 MAb (0.08 μg/ml), affinity-purified MAb 327 ascites (2 μg/ml), or antiphosphotyrosine MAb PY20 (0.08 μg/ml) for 1 h, washed in TNT buffer, with changes every 5 min for 30 min, and incubated with horseradish peroxidase-conjugated donkey anti-mouse IgM (Zymed, San Francisco, Calif.) for RACK1 MAb blots or goat anti-mouse IgG (Bio-Rad) for MAb 327 or PY20 blots (7–11, 49, 50). Proteins were detected by enhanced chemiluminescence (Amersham) according to the manufacturer’s protocol.
Soluble protein kinase assays with peptide substrates.
Purified PKA, PKC, CKII, Src, and Lck kinases were subjected to in vitro protein kinase assays with [γ-32P]ATP and specific peptide substrates (25). Attempts were made to use equivalent specific activities for the kinases (900 U/μg). The following kinase buffers and peptides were used for each reaction: (i) Src and Lck (13): 100 mM Tris-HCl (pH 7.2), 0.25 mM Na3VO4, 2 mM EGTA, 125 mM magnesium acetate, 25 mM MnCl2, 0.1 mM ATP, and 250 μM Src peptide; (ii) PKC (1): 20 mM morpholinepropanesulfonic acid (MOPS) (pH 7.2), 1 mM Na3VO4, 1 mM DTT, 25 mM β-glycerophosphate, 1 mM CaCl2 with 0.5 mg of phosphatidylserine per ml and 0.5 mg of diglycerides per ml as activators of PKC, 0.1 mM ATP, and 0.1 mM biotinylated PKC substrate; (iii) PKA (55): 40 mM Tris-HCl (pH 7.4), 20 mM magnesium acetate, 0.2 mM ATP, and 130 μM Kemptide; and (iv) CKII (30): 25 mM Tris-HCl (pH 7.4), 200 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, and 100 μM CKII peptide. Kinases were preincubated with 2 μM GST or GST-RACK1 at 4°C for 15 min prior to the addition of kinase buffer and [γ-32P]ATP (0.5 × 104 to 1.0 × 104 cpm/pmol). Kinase reactions were run for 5 or 10 min at room temperature (RT). Proteins were precipitated with trichloroacetic acid (40%), and aliquots of the supernatant containing the phosphorylated peptide were spotted on streptavidin discs (Promega) for PKC and on phosphocellulose P81 discs (Whatman, Hillsboro, Oreg.) for other kinases. Discs were washed five times with 0.75% phosphoric acid and once with acetone (25). 32P incorporation into peptides was quantified by counting in Ready Safe Liquid Scintillation Cocktail (Beckman Instruments Inc., Fullerton, Calif.).
Transfection assays.
NIH 3T3 cells were transfected with pZeoSV or pZeoSV-RACK1 by use of Lipofectamine (Gibco-BRL) according to the manufacturer’s protocol. Briefly, 2 × 105 cells were plated in DMEM containing 10% calf serum in six-well plates 24 h before transfection. Transfections were performed with serum-free DMEM containing 1 μg of pZeoSV or pZeoSV-RACK1 and 15 μl of Lipofectamine. After 24 h, the medium was replaced with fresh DMEM containing 10% calf serum. Colonies were isolated 14 to 17 days after the addition of 500 μg of Zeocin per ml and analyzed for stable RACK1 overexpression by immunoblotting with RACK1 MAb.
HA-RACK1 transfectants were produced by transfecting NIH 3T3 or 3T3/c-Src cells with 1 μg of pcDNA3-HA-RACK1 or pZeoSV-HA-RACK1. pTK-Hygro (0.1 μg) was cotransfected with RACK1-containing plasmids into 3T3/c-Src cells. Stable clones were obtained 10 to 14 days after the addition of 150 μg of hygromycin per ml and/or 400 μg of G418 per ml. Subclones were selected and analyzed for HA-RACK1 expression by immunoblotting with RACK1 MAb and anti-HA MAb 12CA5. Transient transfections in 293T cells were performed as described above, except that cells were harvested 30 h after transfection. For immunoblot analysis with antiphosphotyrosine MAb PY20, cells were treated with 100 μM sodium vanadate for 30 min prior to harvesting. Cells were harvested in trypsin, collected by centrifugation, and lysed in boiling SDS sample buffer.
Cell proliferation assays.
NIH 3T3 cells (5 × 104) stably expressing HA-RACK1 or those transfected with the vector alone were seeded in DMEM containing 10% calf serum (no hygromycin) on three replicate 96-well plates. Cell number was determined 72 h later by use of the CellTiter 96 nonradioactive cell proliferation assay (28, 54), which relies on the ability of a living cell to convert a tetrazolium salt into a formazan product; the assay was performed according to the manufacturer’s (Promega) protocol. Briefly, after 72 h of growth, 15 μl of tetrazolium salt (dye solution) was added and the cells were incubated for 4 h at 37°C. Stop/Solubilization Solution (100 μl) was added, and the cells were incubated overnight in a moist container at RT. The formazan product was quantified by measuring the end-point absorbance at a 595-nm wavelength with a ThermoMax microplate reader in conjunction with SOFTmax 2.32 software (Molecular Devices).
Cell cycle analysis.
Unsynchronized cells in the mid-log phase were seeded at a density of 2 × 106 cells/10-cm-diameter plate. After 24 h, cells were collected by mild trypsinization and gentle centrifugation, washed twice in phosphate-buffered saline, and incubated in 1 ml of a solution containing 50 μg of propidium iodide (PI; Sigma) per ml, 0.1% Triton X-100, 0.1% sodium citrate, and 1 mg of RNase A per ml for 30 min at 4°C in the dark (2, 17). The DNA content of 5 × 104 PI-stained cells was analyzed by flow cytometry (FACScan) at a 488-nm excitation, with gating set to exclude debris and cells containing less than diploid DNA. The PI signal was filtered through a 635-nm band-pass filter to remove scattered light. Linear amplification of fluorescence signals was used. Pulse-processing electronics (peak integral versus peak height) were used to analyze the DNA signals and to eliminate cell doublets from the analysis (2, 17). LYSIS II (Becton Dickinson) was used to analyze flow cytometry data.
Analysis of cell death and apoptosis.
Unsynchronized cells in the mid-log phase were seeded at a density of 2 × 106 cells/10-cm-diameter plate. After 24 h, cells were collected by mild trypsinization and gentle centrifugation, washed twice in phosphate-buffered saline, and resuspended in 0.5 ml of buffer containing 10 mM HEPES-NaOH (pH 7.4), 140 mM NaCl, and 2.5 mM CaCl2. Fluorescein-conjugated anti-Annexin V (Pharmingen, La Jolla, Calif.) (10 μl) and 50 μg of PI per ml were added to the buffer, and cells were gently vortexed and incubated for 15 min at RT in the dark (2, 17). Stained cells were analyzed by flow cytometry. Necrotic cells stained with both fluorochromes, apoptotic cells stained with anti-Annexin V alone, and live cells did not stain with either fluorochrome (2, 17).
RESULTS
Isolation of RACK1 as a Src-interacting protein.
To isolate proteins that interact with UD/SH3/SH2 of Src and potentially regulate its activity, we used the yeast two-hybrid assay to screen a human lung fibroblast cDNA library. Of 2 × 106 colonies cotransformed with pAS2-Src-UD/SH3/SH2 and a pGAD-GL fibroblast library, 48 were Trp+ Leu+ His+ LacZ+. After elimination of false-positives by cycloheximide selection and recloning of the plasmids in E. coli, 12 clones that interacted with the Src fusion protein and not a negative control fusion protein, pAS2-lamin, were identified. Redundant clones were eliminated by analysis of insert sizes, and the eight remaining clones were sequenced. One clone had 100% homology at the nucleotide sequence level with human H12.3, a homolog of the heterotrimeric G-protein β subunit (23), and 100% homology at the amino acid sequence level with rat brain RACK1, a receptor for activated C kinase (23, 60, 61).
RACK1 associates with the SH2 domain of Src in vitro.
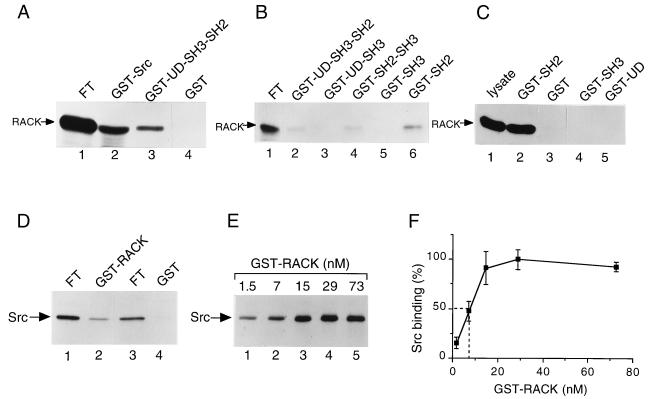
To confirm the results obtained with the yeast two-hybrid assay, we initially studied the Src-RACK1 interaction with in vitro binding experiments. Full-length Src or UD/SH3/SH2 was expressed as a bacterial fusion protein with GST, purified to homogeneity on glutathione-agarose beads, and tested for the ability to bind [35S]methionine-labeled, in vitro-translated RACK1 (Fig. 1A). One-tenth of the unbound translation reaction product (flowthrough) was loaded on the gel as a measure of the amount of protein translated (Fig. 1A, lane 1). We observed that GST-Src (Fig. 1A, lane 2) and GST-UD/SH3/SH2 (lane 3) bound RACK1, whereas the GST control (lane 4) did not. We estimated (by scanning densitometry of RACK1 bands) that 4% of in vitro-translated RACK1 bound GST-Src.
FIG. 1.
Association of RACK1 and Src in vitro. (A) Binding of in vitro-translated RACK1 to GST-Src fusion proteins. [35S]methionine- and [35S]cysteine-labeled RACK1 was synthesized in rabbit reticulocyte lysates and incubated with 1 μg of purified GST-Src (lane 2), GST-UD/SH3/SH2 (lane 3), or GST (lane 4) for 3 h at 4°C. Protein complexes were collected on glutathione-agarose beads, washed, and boiled in SDS sample buffer. Proteins were resolved by SDS-PAGE. For each reaction, 1/10 of the unbound translation reaction product (flowthrough [FT]) was loaded directly on the gel as a measure of the amount of protein translated (lane 1). 35S-labeled proteins were visualized by autoradiography and quantified by scanning densitometry. (B) Binding of in vitro-translated RACK1 to additional GST-Src fusion proteins. In vitro-translated RACK1 (lane 1) was assessed for binding to GST-UD/SH3/SH2 (lane 2), GST-UD/SH3 (lane 3), GST-SH2/SH3 (lane 4), GST-SH3 (lane 5), or GST-SH2 (lane 6) as described for panel A. (C) Binding of RACK1 from HeLa cell lysates to GST-Src fusion proteins. HeLa cell lysates containing 500 μg of total cellular protein were incubated with 5 μg of GST-SH2 (lane 2), GST alone (lane 3), GST-SH3 (lane 4), or GST-UD (lane 5) for 3 h at 4°C. Proteins bound to GST or GST-Src fusion proteins were recovered, resolved by SDS-PAGE, transferred to PVDF membranes, and subjected to immunoblot analysis with anti-RACK1 MAb. Lysate containing 20 μg of total cellular protein was loaded directly on the gel prior to transfer and immunoblotting with anti-RACK1 MAb (lane 1). (D) Binding of in vitro-translated Src to GST-RACK1. In vitro-translated Src was assessed for binding to GST-RACK1 (lane 2) or GST alone (lane 4) as described in panel A. For each reaction, 1/20 of the FT was loaded directly on the gel (lanes 1 and 3). (E) Concentration-dependent binding of GST-RACK1 to in vitro-translated Src. A constant amount of in vitro-translated Src was assessed for binding to increasing concentrations of GST-RACK1 (1.5 to 73 nM). Data are representative of four independent experiments. (F) Quantitation of data on concentration-dependent binding of GST-RACK1 to in vitro-translated Src. Data represent average values from four independent experiments and are expressed as a percentage of maximal Src binding. Error bars indicate standard errors. Broken lines show the amount of GST-RACK1 required for half-maximal Src binding.
To further define the binding site on Src for RACK1, we generated GST-Src fusions containing the UD, the SH3 domain, and/or the SH2 domain, and tested their abilities to bind in vitro-translated RACK1 (Fig. 1B). We observed that all fusions containing the SH2 domain bound RACK1 (Fig. 1B, lanes 2, 4, and 6), whereas those that lacked the SH2 domain did not (lanes 3 and 5). Similarly, we found that a GST-Src fusion containing the SH2 domain bound RACK1 in HeLa cell lysates (Fig. 1C, lane 2), whereas fusions containing the SH3 domain (lane 4), UD (lane 5), or GST alone (lane 3) did not. These in vitro binding studies demonstrated that RACK1 binds to the SH2 domain of Src.
In reverse experiments, GST-RACK1 was tested for its ability to bind [35S]methionine-labeled, in vitro-translated Src (Fig. 1D). Here, 1/20 of the flowthrough was loaded on the gel (Fig. 1D, lanes 1 and 3). We observed that GST-RACK1 bound Src (Fig. 1D, lane 2), whereas GST alone (lane 4) and GST fused to a protein unrelated to RACK1 (elongation factor Tu; data not shown) did not. We estimated that 2% of the in vitro-translated Src bound GST-RACK1. Additional experiments showed that GST fusions of two Src-related kinases, Lck and Fyn, also bound RACK1 (data not shown). Together, these studies demonstrated that Src family kinases associate with RACK1 in vitro.
To determine whether the binding of RACK1 and Src is concentration dependent and saturable, we measured the binding of increasing amounts of GST-RACK1 to a constant amount of [35S]methionine-labeled, in vitro-translated Src (Fig. 1E and F). We observed concentration-dependent, saturable binding of GST-RACK1 to Src, with ≈8 nM GST-RACK1 being required for half-maximal Src binding.
To examine the specificity of the interaction of RACK1 with Src family kinases, we tested other nonreceptor tyrosine kinases and phosphatases for binding to RACK1. We did not detect binding of RACK1 to the focal adhesion kinase (FAK) or to the C-terminal Src kinase (Csk) after immunoprecipitation of RACK1 and immunoblotting with MAb specific for FAK or Csk (data not shown). Similarly, we did not detect binding of GST-RACK1 to the Shp-2 tyrosine phosphatase (data not shown). Thus, for the tyrosine kinases and phosphatases tested, RACK1 interacted specifically with Src family kinases and not with other nonreceptor tyrosine kinases or phosphatases.
RACK1 associates with Src in vivo.
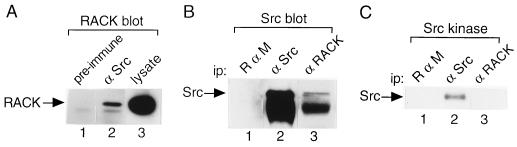
To determine whether RACK1 associates with Src in vivo, proteins from NP-40 lysates of NIH 3T3 cells or proteins precipitated from lysates with preimmune serum or excess Src polyclonal antibody R7 were resolved by SDS-PAGE, transferred to PVDF membranes, and immunoblotted with an MAb specific for RACK1 (Fig. 2A). RACK1 protein was detected in lysate (Fig. 2A, lane 3) and in Src immunoprecipitates (lane 2) but not in control immunoprecipitates (lane 1). A nonspecific immunoreactive band running below RACK1 was present in all lanes. We estimated (by scanning densitometry of RACK1 bands) that 1% of total cellular RACK1 bound Src (compare RACK1 protein levels in lanes 2 and 3 of Fig. 2A).
FIG. 2.
Association of RACK1 and Src in vivo. (A) RACK1 coimmunoprecipitates with Src. Proteins were precipitated with preimmune serum (lane 1) or excess Src polyclonal antibody R7 (lane 2) from NP-40 lysates of NIH 3T3 cells containing 800 μg of total cellular protein, resolved by SDS-PAGE, transferred to PVDF membranes, and immunoblotted with anti-RACK1 antibody. Lysate containing 20 μg of total cellular protein was loaded directly on the gel prior to transfer and immunoblot analysis with anti-RACK1 antibody (lane 3). (B) Src coimmunoprecipitates with RACK1. Proteins were immunoprecipitated (ip) with rabbit anti-mouse IgG (R α M, lane 1), Src MAb 327 (lane 2), or RACK1 MAb (lane 3) from NP-40 lysates of 3T3/c-Src cells containing 500 μg of total cellular protein and subjected to immunoblot analysis with Src MAb 327. The band below Src is mouse IgG heavy chain. (C) In vitro protein kinase activity of Src bound to RACK1. Immunoprecipitates parallel to those shown in panel B were incubated with [γ-32P]ATP and MnCl2 for 10 min at 30°C in an in vitro protein kinase assay. 32P-labeled proteins were resolved by SDS-PAGE and visualized by autoradiography.
To determine whether Src coimmunoprecipitates with RACK1, proteins from NP-40 lysates of NIH 3T3 cells overexpressing Src (3T3/c-Src cells; 8) were immunoprecipitated with MAb specific for RACK1 or Src (327) or rabbit anti-mouse IgG and subjected to immunoblot analysis with MAb 327 (Fig. 2B). Src protein (≈60 kDa) was detected in Src (Fig. 2B, lane 2, upper band) and RACK1 (lane 3, upper band) immunoprecipitates but not in control immunoprecipitates (lane 1). The ≈55-kDa band running below Src in Fig. 2B, lanes 2 and 3, is mouse IgG heavy chain (7–11, 49, 50). We estimated that 3% of immunoprecipitable Src bound RACK1 (compare Src protein levels in lanes 2 and 3 of Fig. 2B). Together, these findings indicated that Src and RACK1 associate in vivo.
RACK1 inhibits Src activity in vitro.
To assess the effect of RACK1 on Src protein kinase activity, we incubated IgG, Src, or RACK1 immunoprecipitates (identical to those used for immunoblotting in Fig. 2B) with [γ-32P]ATP and MnCl2 and measured the phosphorylation of Src in an in vitro protein kinase assay (Fig. 2C). As expected, we observed autophosphorylation of Src in Src immunoprecipitates (Fig. 2C, lane 2). Surprisingly, we did not detect Src autophosphorylation in RACK1 immunoprecipitates (Fig. 2C, lane 3) that contained detectable levels of Src protein (Fig. 2B, lane 3). Because autophosphorylation of Src in a kinase assay is usually more sensitive than immunoblot analysis for detecting Src, this finding suggested that RACK1 inhibits Src activity.
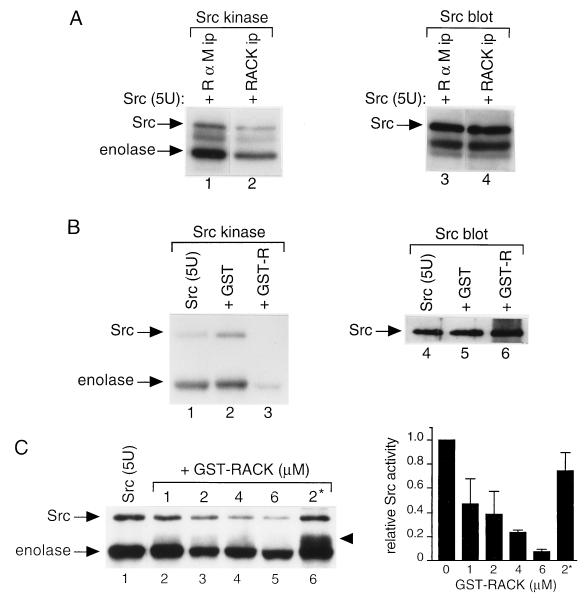
To investigate this finding further, we assessed the effect of immunoprecipitable RACK1 on the activity of purified Src kinase (Fig. 3A). We observed that the kinase activity of purified Src, as measured either by Src autophosphorylation or by enolase phosphorylation, decreased 60 to 70% more with the addition of RACK1 immunoprecipitates than with the addition of IgG immunoprecipitates (compare lanes 1 and 2 of Fig. 3A). Immunoblotting with Src antibody confirmed that equivalent amounts of purified Src protein were present in both reaction mixtures (compare lanes 3 and 4 of Fig. 3A). Thus, the addition of RACK1 immunoprecipitates to purified Src inhibits the specific activity of Src.
FIG. 3.
Effect of RACK1 on Src activity in vitro. (A) Effect of RACK1 immunoprecipitates on the activity of purified Src. Proteins were immunoprecipitated (ip) with excess anti-RACK1 antibody (lanes 2 and 4) or rabbit anti-mouse IgG (lanes 1 and 3) from NP-40 lysates of NIH 3T3 cells containing 100 μg of total cellular protein, incubated with 5 U of purified Src, and subjected to an in vitro protein kinase assay (lanes 1 and 2) or immunoblot analysis with Src MAb 327 (lanes 3 and 4). The band below Src is IgG heavy chain. (B) Effect of purified GST-RACK1 on the activity of purified Src. In vitro protein kinase assays (lanes 1 and 3) or immunoblot analysis with MAb 327 (lanes 4 to 6) was performed with 5 U of purified Src alone (lanes 1 and 4) or with the addition of 2 μM GST (lanes 2 and 5) or GST-RACK1 (GST-R) (lanes 3 and 6). (C) Concentration-dependent inhibition of Src activity by GST-RACK1. (Left panel) In vitro protein kinase assays were performed with 5 U of purified Src together with 2 μM GST (lane 1), 1 to 6 μM GST-RACK1 (lanes 2 to 5), or 2 μM boiled GST-RACK1 (2*) (lane 6). 32P-labeled proteins were resolved by SDS-PAGE and visualized by autoradiography. The arrowhead indicates a 55-kDa phosphorylated protein that comigrated with Coomassie-blue stained GST-RACK1. Data are representative of four independent experiments. (Right panel) Quantitation of data on concentration-dependent inhibition of Src activity by GST-RACK1. 32P incorporation into Src was quantified by scanning densitometry. Data represent average values from four independent experiments and are expressed relative to those for purified Src with the addition of GST alone. Error bars indicate standard errors.
To determine whether the inhibition of Src activity was due to RACK1 itself or to proteins or lipids associated with RACK1 in immunoprecipitates, we assessed the effect of GST-RACK1 that had been purified to homogeneity from bacteria on the activity of purified Src kinase (Fig. 3B). We observed that Src activity was slightly increased with the addition of 2 μM GST alone (compare lanes 1 and 2 of Fig. 3B) and was decreased by more than 80% with the addition of 2 μM GST-RACK1 (compare lanes 2 and 3). Immunoblotting with Src antibody confirmed that equivalent amounts of purified Src protein were present in all reaction mixtures (compare lanes 4 to 6 of Fig. 3B). To confirm that the inhibition of Src by GST-RACK1 was due to RACK1 and not to the GST portion of the fusion protein, we proteolytically cleaved RACK1 from GST-RACK1 by using thrombin, purified RACK1 by dialysis, and measured the effect of purified RACK1 on the activity of purified Src. We observed a similar inhibition of Src activity with thrombin-cleaved, purified RACK1 as we did with GST-RACK1 (data not shown). Together, these results suggested that RACK1 inhibits Src directly and that other proteins or lipids associated with RACK1 and/or Src are not responsible for the inhibition.
To determine whether RACK1 inhibits Src in a concentration-dependent manner, we added increasing amounts of purified GST-RACK1 fusion protein to a constant amount (5 U) of purified Src kinase and measured Src activity (Fig. 3C and D). We observed concentration-dependent inhibition of Src activity with the addition of increasing amounts of GST-RACK1. Interestingly, the addition of 2 μM GST-RACK1 inhibited Src activity (as measured by autophosphorylation) by ≈60%, whereas the addition of the same amount of boiled GST-RACK1 inhibited Src activity by <30%. These results revealed that RACK1 inhibits Src in a concentration-dependent manner and suggested that the conformation of RACK1 is important for its ability to inhibit Src activity.
Incidentally, when boiled GST-RACK1 was added to the kinase reaction, we observed an ≈55-kDa phosphorylated protein running between Src and enolase (Fig. 3C, left panel, lane 6, arrowhead). We determined that the protein was GST-RACK1 because it comigrated with Coomassie blue-stained GST-RACK1. Thus, boiled GST-RACK1 can be phosphorylated in vitro, probably because phosphorylation sites that are inaccessible in the folded protein are exposed in the denatured protein.
RACK1 inhibits other Src family kinases but not serine/threonine kinases in vitro.
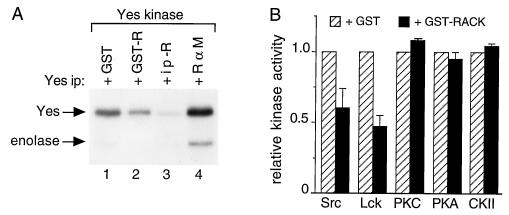
To determine whether RACK1 affects the activities of other Src family members, we measured Yes kinase activity in Yes immunoprecipitates after the addition of 2 μM GST or GST-RACK1 or after the addition of immunoprecipitates of RACK1 or IgG (Fig. 4A). We observed that Yes activity (as measured by autophosphorylation) decreased 55% more with the addition of GST-RACK1 than with the addition of GST alone (compare lanes 1 and 2 of Fig. 4A) and decreased 95% more with the addition of immunoprecipitated RACK1 than with the addition of IgG (compare lanes 3 and 4). We observed more phosphorylation of Yes than of enolase, probably because Yes is a better substrate for itself than is enolase (48, 49). These results indicated that RACK1 inhibits the activity of another member of the Src kinase family, Yes, and that of Src to similar degrees.
FIG. 4.
Effect of RACK1 on the activities of Src family and serine/threonine protein kinases. (A) Effect of RACK1 on Yes kinase activity. Proteins were precipitated with excess antipeptide antibody specific for Yes from RIPA lysates of WiDr colon carcinoma cells containing 500 μg of total cellular protein. Yes immunoprecipitates were incubated with 2 μM GST (lane 1) or GST-RACK1 (GST-R) (lane 2) or with anti-RACK1 antibody (R) (lane 3) or rabbit anti-mouse (R α M) IgG (lane 4) immunoprecipitates (ip) of WiDr cell lysates containing 100 μg of total cellular protein for 15 min at 4°C and subjected to an in vitro kinase assay with enolase as an exogenous substrate. (B) Effect of GST-RACK1 on the activities of Src family and serine/threonine protein kinases. Purified kinases were incubated with 2 μM GST or GST-RACK1 and assayed for in vitro protein kinase activity with specific peptides as substrates. All assays were performed with equivalent specific activities for each kinase. Data represent average values from three independent measurements of Src or Lck activity and two independent measurements of PKC, PKA, or CKII activity. Data on kinase activity measured after the addition of GST-RACK1 (solid bars) are expressed relative to those measured after the addition of GST alone (hatched bars). Error bars indicate standard errors.
To determine whether RACK1 affects the activities of additional Src family members or serine/threonine kinases, purified kinases were incubated with specific peptide substrates and 2 μM GST or GST-RACK1 and assayed for in vitro protein kinase activity (Fig. 4B). GST-RACK1 inhibited the activity of Src by ≈40% and inhibited the activity of the Src-related kinase Lck by ≈50%. GST-RACK1 had no significant effect on the activities of three serine/threonine kinases: PKC, PKA, and CKII. Thus, of the protein kinases that we tested and at the concentrations of GST-RACK1 that we used, RACK1 inhibited the activities of Src tyrosine kinases but not those of serine/threonine kinases.
The data on the ability of RACK1 to inhibit Src activity, as measured by phosphorylation of a Src-specific peptide (Fig. 4B), are internally consistent with those obtained by measurement of the phosphorylation of enolase (Fig. 3C); both show that 2 μM purified GST-RACK1 inhibits the ability of purified Src to phosphorylate an exogenous substrate by 40 to 50%.
RACK1 overexpression results in decreased tyrosine phosphorylation on proteins in vivo.
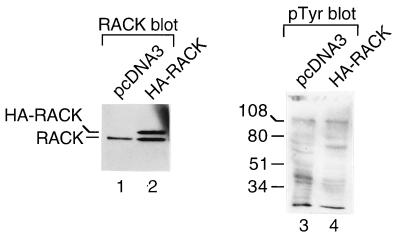
To examine the effect of RACK1 on protein tyrosine phosphorylation in vivo, we transiently expressed HA-tagged RACK1 in 293T cells, measured HA-RACK1 expression by immunoblotting with anti-HA MAb 12CA5 (data not shown) and an anti-RACK1 MAb (Fig. 5, lanes 1 and 2), and measured tyrosine phosphorylation on proteins by immunoblotting with antiphosphotyrosine MAb PY20 (Fig. 5, lanes 3 and 4). We observed that HA-RACK1 (Fig. 5, lane 2, upper band) was approximately 1 kDa larger than endogenous RACK1 (lane 2, lower band, and lane 1). The apparent molecular mass of HA-RACK was consistent with the predicted molecular mass of a RACK1 protein containing nine additional amino acids from the HA tag. We observed that the amount of transiently expressed HA-RACK1 was similar to that of endogenous RACK1. The antiphosphotyrosine MAb PY20 blot revealed that while tyrosine phosphorylation of several proteins in RACK1-overexpressing cells was similar to or higher than that in vector-transfected cells, tyrosine phosphorylation of many other proteins in RACK1-overexpressing cells was lower than that in control cells (compare lanes 3 and 4 of Fig. 5). In four additional RACK-1 overexpressing clones that we studied, we observed a similar decrease in tyrosine phosphorylation on many proteins (data not shown). Coomassie blue-stained gels showed that the levels of cellular proteins expressed in RACK1-transfected clones were similar to those expressed in vector-transfected clones (data not shown). Thus, the changes in tyrosine phosphorylation observed on the antiphosphotyrosine blots appeared to be due to changes in the levels of phosphotyrosine and not to changes in the levels of protein expression. Src activity was decreased in cells transiently overexpressing HA-RACK1 (data not shown). These findings showed that transient overexpression of RACK1 results in decreased tyrosine phosphorylation on many proteins in vivo.
FIG. 5.
Effect of RACK1 overexpression on protein tyrosine phosphorylation in vivo. 293T cells were transiently transfected with 1 μg of pcDNA3 (lanes 1 and 3) or pcDNA3-HA-RACK1 (lanes 2 and 4). After 30 h, cells were treated with 100 μM sodium vanadate for 30 min and lysed. Lysates were resolved by SDS-PAGE, transferred to PVDF membranes, and incubated with anti-RACK1 antibody (lanes 1 and 2) or antiphosphotyrosine antibody PY20 (pTyr) (lanes 3 and 4). Protein sizes are indicated in kilodaltons.
RACK1 overexpression results in inhibition of Src activity.
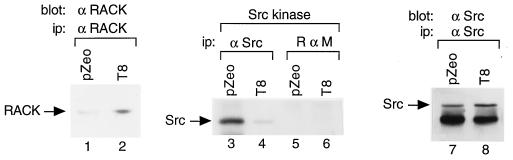
To determine whether stable overexpression of RACK1 affects Src kinase activity, we transfected NIH 3T3 cells with pZeoSV or pZeoSV-RACK1, screened isolated clones for stable expression of RACK1 by immunoblotting with an anti-RACK1 MAb, and assayed clones for Src activity by immunoprecipitating proteins with MAb 327 and performing in vitro kinase reactions. We observed that 2 of 12 isolated clones overexpressed RACK1 (data not shown). One clone, T8, expressed approximately threefold-higher levels of RACK1 protein than did a clone transfected with the vector alone (compare lanes 1 and 2 of Fig. 6). The activity of Src isolated from T8 cells was 90% lower than that of Src isolated from cells transfected with the vector alone (compare lanes 3 and 4 of Fig. 6). Immunoblotting with an anti-Src MAb confirmed that equivalent amounts of Src protein were present in both immunoprecipitates (compare lanes 7 and 8 of Fig. 6). The T8 clone and the only other RACK1-overexpressing clone that we succeeded in isolating by using this approach died after a few weeks in culture. Thus, a clone of NIH 3T3 cells that expressed threefold-higher levels of RACK1 than did control cells and showed 90% inhibition of Src activity did not survive long in culture.
FIG. 6.
Effect of RACK1 overexpression on Src activity. NIH 3T3 cells were transfected with pZeoSV (pZeo) or pZeoSV-RACK1, and isolated clones that stably overexpressed RACK1 were assayed for Src kinase activity. The results from one clone, T8, are shown. Proteins were immunoprecipitated (ip) from lysates containing 500 μg of total cellular protein with excess anti-RACK1 MAb (lanes 1 and 2), Src MAb 327 (lanes 3, 4, 7, and 8) or rabbit anti-mouse (R α M) IgG (lanes 5 and 6) and subjected to immunoblot analysis with anti-RACK1 MAb (lanes 1 and 2) or MAb 327 (lanes 7 and 8) or to in vitro protein kinase assays (lanes 3 to 6). Data are representative of two independent experiments.
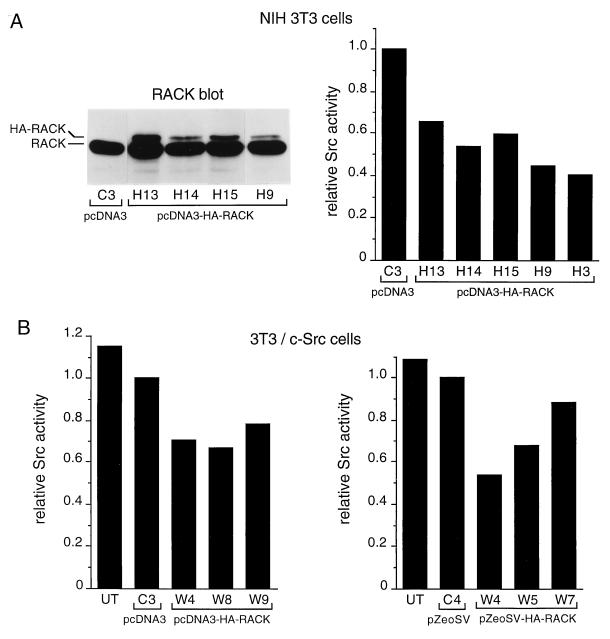
Along with the above-described experiments, we transfected NIH 3T3 cells with pcDNA3 or pcDNA3-HA-RACK1, screened isolated clones for HA-RACK1 expression by immunoblotting with an anti-RACK1 MAb and anti-HA antibody 12CA5, and assayed clones for Src activity by immunoprecipitating proteins with MAb 327 and performing in vitro kinase reactions (Fig. 7A). Using HA-tagged RACK1 allowed us to distinguish introduced RACK1 from endogenous RACK1 (Fig. 5, lane 2). Among cells transfected with pcDNA3-HA-RACK1, 6 of 24 isolated clones expressed detectable levels of HA-RACK1 (data on 4 of the 6 clones are shown in Fig. 7A, left panel). While the level of HA-RACK1 expression varied somewhat between clones, overall it was lower than that of endogenous RACK1 or transiently expressed HA-RACK1 (Fig. 5, lane 2). Src activity in the six HA-RACK1-overexpressing clones was 30 to 60% lower than that in the clone transfected with the pcDNA3 vector alone (data on five of the six clones is shown in Fig. 7A, right panel). There were no significant differences in levels of Src activity between the various clones that overexpressed HA-RACK1. Additional experiments showed that HA-RACK1 was present in Src immunoprecipitates (data not shown).
FIG. 7.
Effect of HA-RACK1 overexpression on Src activity. (A) (Left panel) NIH 3T3 cells were transfected with pcDNA3 or pcDNA3-HA-RACK1, subclones were isolated, and lysates containing 20 μg of total cellular protein were assayed for stable expression of HA-RACK1 by immunoblotting with anti-RACK1 antibody. (Right panel) Clones that expressed detectable levels of HA-RACK1 were assayed for Src activity by immunoprecipitating proteins with MAb 327 and performing in vitro kinase reactions. 32P incorporation into enolase was quantified by Cerenkov counting of excised gel bands. Data represent average values from two independent experiments. Data on Src activity in RACK1-overexpressing clones are expressed relative to those on Src activity in the vector-transfected clone. (B) 3T3/c-Src cells were transfected with pcDNA3-HA-RACK1 (left panel) or pZeo-HA-RACK1 (right panel), and isolated subclones were assayed for expression of HA-RACK1 and Src activity as described in panel A. UT, untransfected 3T3/c-Src cells.
We also examined the effect of the stable expression of HA-RACK1 on Src activity in cells that overexpressed wild-type c-Src (3T3/c-Src cells). 3T3/c-Src cells were transfected with pcDNA3, pcDNA3-HA-RACK1, pZeoSV, or pZeoSV-HA-RACK1, screened for HA-RACK1 expression by immunoblotting with an anti-RACK1 MAb, and assayed for Src activity by immunoprecipitation of proteins with MAb 327 and in vitro kinase reactions (Fig. 7B). Among cells transfected with pcDNA3-HA-RACK1, 3 of 11 isolated clones expressed detectable levels of HA-RACK1 (data not shown). Src activity in the three clones was 20 to 35% lower than that in a clone transfected with the pcDNA3 vector alone (Fig. 7B, left panel). Similarly, among cells transfected with pZeoSV-HA-RACK1, 3 of 14 isolated clones expressed HA-RACK1, and Src activity in the 3 clones was 15 to 45% lower than that in a clone transfected with the pZeoSV vector alone (Fig. 7B, right panel). Thus, in six clones stably overexpressing both c-Src and RACK1, Src activity was inhibited by 20 to 40%.
Together, these studies demonstrated that RACK1, when stably overexpressed from either a simian virus 40 or a cytomegalovirus promoter, results in the inhibition of c-Src activity in both NIH 3T3 cells and NIH 3T3 cells that overexpress c-Src. The inhibition is greater in NIH 3T3 cells, presumably because they contain less c-Src. Moreover, the inhibition is greater in cells transiently expressing RACK1 than in cells stably expressing RACK1, probably because the former express higher levels of RACK1 protein.
RACK1 overexpression inhibits the growth of NIH 3T3 cells.
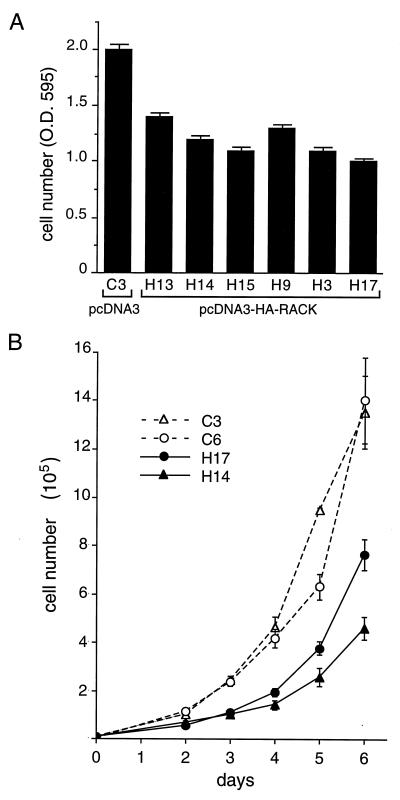
We observed that NIH 3T3 clones that stably overexpressed HA-RACK1 and had inhibited Src kinase activity (Fig. 7) appeared to grow more slowly than clones containing the vector alone. To analyze this result further, we seeded equivalent numbers of the six NIH 3T3 clones that overexpressed HA-RACK1 (the HA-RACK1 expression levels for four of the six clones and the Src activities for five of the six clones are shown in Fig. 7A) and a clone expressing the vector alone and measured cell numbers after 3 days of growth. We observed that all six NIH 3T3 clones that overexpressed HA-RACK1 and had inhibited Src kinase activity grew more slowly than the clone containing the vector alone (Fig. 8A). After 3 days of growth, there were, on average, 30 to 50% fewer RACK1-overexpressing cells than control cells.
FIG. 8.
Effect of RACK1 overexpression on the growth of NIH 3T3 cells. NIH 3T3 transfectants that stably overexpressed HA-RACK1 and showed inhibited Src activity (Fig. 7A) were assayed for cell growth rates. (A) Cell number (optical density at 595 nm [O.D. 595]) after 3 days of growth was measured with the CellTiter 96 assay. Data are representative of three independent experiments. Data represent means ± standard errors from three plates of each clone. (B) Six-day growth curves. Cells were plated at 104 cells/dish (35-mm diameter) and counted daily from days 2 to 6. Data are representative of two independent experiments. Data represent means ± standard errors from four plates at each time point.
Measurement of cell growth rates over 6 days (Fig. 8B) confirmed that RACK1-overexpressing clones grew more slowly than wild-type clones. These data are internally consistent with those from the proliferation assay (Fig. 8A); both showed that after 3 days of growth, there were 40 to 50% fewer RACK1-overexpressing cells than wild-type cells. The doubling time for wild-type cells during exponential growth was 20 to 21 h, whereas that for RACK1-overexpressing cells was 23 to 27 h (Table 1). Thus, RACK1 overexpression inhibits the growth of NIH 3T3 cells.
TABLE 1.
Doubling time and cell cycle analysis of wild-type and RACK-1-overexpressing cells
| Cell line | Doubling time (h)a | Mean ± SEM length (h) of cell cycle phaseb
|
||
|---|---|---|---|---|
| G0/G1c | S | G2/M | ||
| C3 | 20 | 12.5 ± 1.9*† | 1.8 ± 0.6 | 4.5 ± 1.4 |
| C6 | 21 | 13.3 ± 2.3‡§ | 1.9 ± 0.1 | 4.3 ± 1.7 |
| H14 | 27 | 20.8 ± 1.2*§ | 1.4 ± 0.3 | 3.7 ± 0.7 |
| H17 | 23 | 17.5 ± 0.4†‡ | 1.4 ± 0.1 | 3.2 ± 0.3 |
Doubling time was determined from cell growth rates (Fig. 8B); cell number (log scale) was plotted versus time (linear scale), and doubling time was calculated from the slope of the line.
Cells in the mid-log phase were stained with propidium iodide. The percentage of cells in each phase was determined by flow cytometry and was multiplied by the doubling time to calculate the length of each cell cycle phase. Data represent the means ± standard errors of the mean (H14, n = 5; H17 and C3, n = 4; C6, n = 3).
Statistical analysis was performed by analysis of variance. Only the length of G0/G1 was significantly different among the four cell lines as a group (P < 0.01). Further analysis with an independent t test identified pairs with significant differences (∗, P < 0.005; † and ‡, P < 0.05; §, P < 0.01).
RACK1 overexpression prolongs the G0/G1 phase of the NIH 3T3 cell cycle.
We used flow cytometry to analyze in greater detail the mechanism(s) by which RACK1 affects NIH 3T3 cell proliferation. Two general mechanisms could account for the longer doubling time of RACK1-overexpressing cells: (i) RACK1-overexpressing cells die at a higher rate than wild-type cells and/or (ii) RACK1-overexpressing cells proliferate more slowly than wild-type cells. To distinguish between these possibilities, we used Annexin V and propidium iodide staining to estimate necrotic and apoptotic cell death in wild-type and RACK1-overexpressing cells grown under normal conditions. The results showed that the proportions of dead RACK1-overexpressing cells in the mid-log phase (13% of H14 cells and 9% of H17 cells) were similar to those of dead wild-type cells (14% of C3 cells and 9% of C6 cells).The fractions of apoptotic RACK1-overexpressing cells (<1% of H14 cells and 2% of H17 cells) were also similar to those of apoptotic wild-type cells (1% of C3 and C6 cells). These findings implied that the increased doubling time of RACK1-overexpressing cells does not reflect increased cell death; therefore, we inferred that RACK1-overexpressing cells proliferate more slowly than wild-type cells.
To analyze cell proliferation in greater detail, we used flow cytometry to determine whether RACK1 exerts its effect at a particular phase of the cell cycle. Analysis of unsynchronized populations during logarithmic growth revealed no significant difference between wild-type and RACK1-overexpressing cells in the fraction traversing the S phase or the G2/M phase; however, the fraction of RACK1-overexpressing cells in the G0/G1 phase was significantly higher than the fraction of wild-type cells in the G0/G1 phase (data not shown). Combining these flow cytometric data with the observed doubling times for wild-type and RACK1-overexpressing cells allowed us to estimate the time that each cell type spends in each phase of the cell cycle (Table 1). Our data indicate that there is no significant difference between wild-type and RACK1-overexpressing cells in the time that they spend in the S phase (1.5 to 2 h) or in the G2/M phase (3 to 4.5 h). In contrast, RACK1-overexpressing cells spend significantly more time in the G0/G1 phase (18 to 21 h) than do wild-type cells (13 h). Thus, G0/G1 is prolonged by 5 to 8 h in RACK1-overexpressing cells. For RACK1-overexpressing clone H14, the prolongation of G0/G1 could account for the prolongation of the doubling time (≈7 h). For RACK1-overexpressing clone H17, the prolongation of G0/G1 was greater than the prolongation of the doubling time (≈3 h), suggesting, but not proving, that compensatory mechanisms are at work in the cell to override the effect of RACK1 on G0/G1. Taken together, our results suggest that RACK1 influences NIH 3T3 cell growth at least in part by altering the length of the G0/G1 phase of the cell cycle.
DISCUSSION
Because it is the repression of c-Src activity rather than the elevation of v-Src activity that accounts for differences in the transforming abilities of the two proteins (9, 29, 53, 56), it is important to identify mechanisms that inactivate c-Src in normal cells. While a number of interacting proteins that upregulate Src activity have been identified, few that downregulate Src activity have been identified. Here we report the identification of a protein, RACK1, which appears to be an inhibitor of Src activity.
RACK1 is one of a group of proteins collectively termed RACKs (reviewed in references 42 and 43). RACK1 was cloned from a rat brain cDNA expression library (60) and shown to be 100% identical (at the protein level) to human H12.3, which was previously identified as a homolog of the heterotrimeric G-protein β subunit (23). Overall, human RACK1 has 44% homology with human Gβ (61). RACK1 and Gβ are both members of an ancient family of regulatory proteins made up of highly conserved repeating units usually ending with Trp-Asp (WD) (reviewed in references 46 and 47). WD repeat proteins are functionally diverse, although all seem to be regulatory and few are enzymes. The WD repeats in RACK1 are conserved from Chlamydomonas spp. to humans (reviewed in reference 47). Thus, the function of RACK1 was probably fixed before the evolutionary divergence of plants and animals.
While RACK1 binds to both PKC and Src, there are clear differences in its interactions with the two kinases. Most notably, RACK1 does not appear to inhibit PKC activity (60) (Fig. 4B) as it does Src activity (Fig. 3 to 7). RACK1, which is located in the cell particulate fraction, appears to be required for the translocation of cytosolic βPKC to the plasma membrane, where isozyme-specific substrates are located (reviewed in references 43 and 59). In this way, RACK1 serves as an anchoring protein for PKC (reviewed in references 18, 42, and 43). The interaction of RACK1 and PKC closely resembles that of the βγ subunits of G proteins and the β-adrenergic receptor kinase (βARK). βγ subunits appear to be required for the translocation of βARK to the plasma membrane, where it phosphorylates the agonist-occupied β-adrenergic receptor, which then becomes desensitized to further stimulation (51). The fact that RACK1, which is also a member of the G-protein β-subunit family, is an anchor for PKC and possibly Src is interesting in view of the role of βγ subunits of G proteins as anchors for βARK. Recently, βγ subunits of G proteins were shown to mediate Src-dependent phosphorylation of the epidermal growth factor receptor and to serve as a scaffold for G protein-coupled receptor-mediated Ras activation (39). Thus, a common theme is emerging for signaling via protein kinases: membrane-anchoring proteins, particularly subunits of G proteins such as βγ and RACK1, appear to be important for targeting specific nonreceptor protein kinases (before or after activation) to their specific substrates. In this regard, it will be important to determine the effect of RACK1 on subcellular targeting of Src to its substrates. Preliminary immunofluorescence and subcellular fractionation studies show colocalization of RACK1 and Src in the cytoplasm of 3T3/c-Src cells (12).
RACK1 resembles the membrane-anchored scaffolding protein caveolin in its interaction with Src. Caveolin, like RACK1, inhibits Src activity (35). Caveolin also interacts with H-Ras and G-protein α subunits and downregulates the GTPase activity of the latter (35). In a broader analysis, caveolin and RACK1 functionally resemble another family of scaffolding proteins, the AKAPs (A-kinase anchoring proteins). One family member, AKAP79, interacts with PKA, PKC (α and β isoforms), and protein phosphatase 2B. Another family member, AKAP250 (gravin), interacts with PKA and PKC (45). Both AKAPs suppress the activities of the enzymes with which they interact (14, 15, 18, 27, 45), just as caveolin and RACK1 suppress the activity of Src. Thus, a broader theme is emerging for signaling via intracellular protein kinases and phosphatases: membrane-anchored scaffolding proteins, such as AKAPs, caveolin, and RACK1, not only may restrict the subcellular movement and localization of kinases and phosphatases but also may restrict their activity. It will be important to determine whether RACK1, PKC, and Src function in a trimolecular complex and, if so, how they regulate each other’s activity. Interestingly, in v-Src-transformed cells, PKC δ associates with Src, becomes phosphorylated on tyrosine, and is downregulated (67).
How does RACK1 inhibit Src activity? While the specific mechanism is unknown, the recently identified three-dimensional structures of G-protein β subunits and Src kinases may provide clues. G-protein β subunits fold into a highly symmetric, seven-bladed, β propeller containing seven structurally similar WD repeats (21, 32). This Gβ structure provides a rigid scaffold that serves as an anchor for interacting proteins. In the inactive state, Src folds up with phosphorylated Tyr 527 in the C-terminal tail binding to the SH2 domain. The ligand-binding surfaces of the SH2 and SH3 domains are tucked inside, thus presenting an inert surface to the outside environment (40, 62, 66). Thus, it is possible that RACK1 inhibits Src activity by clamping down on Src and holding it in the closed, inactive, conformational state. Preliminary studies suggest that there are multiple sites in RACK1 that bind to Src and multiple sites in Src that bind to RACK1 (12). Once the precise binding sites on Src and RACK1 have been identified, we may better understand the mechanism by which RACK1 inhibits Src activity.
A related mechanism by which RACK1 may inhibit Src activity is by acting as a molecular chaperone, a protein that assists other proteins in folding (reviewed in reference 4). Newly synthesized v-Src appears to interact with one chaperone, Hsp90, to hold it in the inactive state and later with another chaperone, Ydj1 (a member of the DnaJ chaperone family), to dissociate it from Hsp90 or to achieve the final activated state (reviewed in reference 4). Thus, it seems possible that RACK1 serves as a chaperone for c-Src by assisting it in folding into the inactive state.
How can the significant amount of Src inhibition observed in HA-RACK1-transfected NIH 3T3 cells be explained by the fairly low levels of HA-RACK1 that are stably expressed in the cells (Fig. 7) and the small quantities of Src and RACK1 molecules that appear to interact in vivo (Fig. 2)? One explanation, which accounts for the latter finding, is that, in addition to directly binding to Src in cells, RACK1 indirectly influences Src activity through its interaction with other regulators of Src. For example, RACK1 could inactivate tyrosine phosphatases which dephosphorylate Src at Tyr 527, activate tyrosine kinases that phosphorylate Tyr 527 (like Csk), or recruit known inhibitors of Src (like caveolin). Thus, RACK1, like other scaffolding proteins, could regulate enzymes or recruit adaptor proteins that, in turn, downregulate Src. However, our in vitro data showing that 1 μM purified GST-RACK1 is sufficient to inhibit the activity of purified Src by ≈50% (Fig. 3C) argue against the involvement of other downregulators of Src. A more likely explanation is that we are underestimating the amounts of Src and RACK1 that interact. Most of the estimates are based on postlysis, immunoprecipitable analyses of RACK1, which is a fairly insoluble protein. We estimate that only 5% of total cellular RACK1 is immunoprecipitable from cell lysates (data not shown). Historically, the detection of the RACK1-PKC complex by coimmunoprecipitation analysis has been difficult, and overlay assays have been used instead to detect the complex (41, 58–60).
One possible explanation for the fairly low levels of stable HA-RACK1 expression observed in NIH 3T3 cells (Fig. 7A), the gradual loss of HA-RACK1 expression in these cells over 2 to 3 months in culture (data not shown), and the failure of cells that initially express high levels of RACK1 to survive more than a few weeks in culture (like clone T8) is that RACK1 inhibits both Src activity (Fig. 3 to 7) and the growth of cells (Fig. 8 and Table 1); thus, there is a survival advantage for cells that do not overexpress RACK or that do so but at very low levels. A similar decrease in the expression of the growth-inhibitory protein SSeCKs (Src-suppressed C kinase substrate; see below) was seen with serial passage of cells in culture (36, 37).
Our in vitro data show that nanomolar amounts of GST-RACK1 are sufficient to obtain maximal binding of in vitro-translated Src (Fig. 1F), yet micromolar amounts of GST-RACK1 are required to obtain maximal inhibition of baculovirus-expressed Src (Fig. 3C). One possible explanation is that these two forms of Src have different posttranslational modifications and that specific posttranslational modifications on Src are important for binding and/or inhibition by RACK1. For example, baculovirus-expressed Src is important for binding and/or inhibition by RACK1. For example, baculovirus-expressed Src has a higher level of specific activity (because of increased phosphorylation on Tyr 416 and decreased phosphorylation on Tyr 527; 52) than does in vitro-translated Src; therefore, baculovirus-expressed Src may require higher concentrations of GST-RACK1 for inhibition. Curiously, when we added increasing concentrations of GST-RACK1 and measured Src activity by autophosphorylation, we observed linear inhibition of Src activity and nearly complete inhibition with the addition of 6 μM GST-RACK1 (Fig. 3C). In contrast, when we measured Src activity by phosphorylation of the exogenous substrate enolase, we observed only 50% inhibition of Src activity with the addition of 6 μM GST-RACK1. Although the reason for these results is unknown, one possibility is that RACK1 binds to the autophosphorylation site on Src (Tyr 416), thereby inhibiting phosphorylation at this site but not entirely inhibiting the enzymatic activity of Src.
The finding that RACK1 inhibits both Src family kinases (Fig. 3 to 7) and the growth of NIH 3T3 cells (Fig. 8 and Table 1) suggests a role for RACK1 in Src kinase-mediated mitogenic signaling. A clear correlation exists between the suppression by RACK1 of Src kinase activities and its suppression of cell growth. Thus, it is tempting to suggest that the two are linked and that it is in part through the repression of Src kinases that RACK1 inhibits cell growth. It is also tempting to suggest that RACK1 exerts its influence on Src activity at the G1/S boundary, where the activation of Src is required for the PDGF-induced G1/S transition and DNA synthesis (57, 65). However, the inhibition of Src activity is only one of many possible mechanisms by which RACK1 could influence cell growth.
Interestingly, the deduced amino acid sequence of the product of the cpc-2 gene of Neurospora crassa reveals 70% homology with RACK1 (44). cpc-2 upregulates the activity of amino acid biosynthetic enzymes in response to amino acid starvation and is required for the formation of the female sexual organs, protoperithecia (44). Mutations which drastically reduce cpc-2 expression reduce the rate of growth of N. crassa in exponential cultures by 50% and result in female infertility. Moreover, mutants of both cpc-2 and the related gene cpc-1 become temperature-sensitive synthetic lethal mutants. Thus, cpc-2, a gene closely related to the RACK1 gene, appears to regulate the growth of N. crassa.
The 322 gene product, which is closely related to AKAP-250, is growth inhibitory when overexpressed in NIH 3T3 cells (36). In addition, 322 is transcriptionally suppressed in cells transformed by src and ras (36) and encodes a protein which is a substrate of PKC (thus its name, SSeCKs (37). Therefore, SSeCKs, a close relative of AKAPs, a substrate of PKC, and a protein whose expression is suppressed in Src-transformed cells, may be another example of a membrane-anchored scaffolding protein that is growth inhibitory.
In summary, we have shown that RACK1 inhibits the activities of Src tyrosine kinases and inhibits the growth of NIH 3T3 cells. In this way, RACK1 resembles other membrane-anchored scaffolding proteins that restrict the activities of associated kinases and phosphatases and that are growth inhibitory. Thus, anchors containing WD repeats may direct the assembly and regulation of intracellular kinases and phosphatases involved in mitogenic signaling pathways. Understanding the mechanisms by which these anchors regulate protein phosphorylation may ultimately lead to strategies for selectively regulating cell growth.
ACKNOWLEDGMENTS
We thank Rachel Harte for assistance with data analysis and figure preparation. We thank Joosang Park and Annette Walter for generating pGEXsrc plasmids and Salvador Gallardo for help with binding studies. We are grateful to Robert Beatty, Sheri Krams, and Olivia Martinez for help with flow cytometric analyses. We thank Sara Courtneidge for Yes3 antibody and Tony Hunter and Martin Broome for pGEMsrc. We are grateful to Daria Mochly-Rosen, Tony Hunter, Joe Bolen, Bishr Omary, and Anson Lowe for helpful discussions. We thank Daria Mochly-Rosen, Joe Bolen, and Jim Whitlock for critical review of the manuscript.
This work was supported by grants to C.A.C., initially from the American Cancer Society (BE-246) and subsequently from the National Institutes of Health (R01 DK43743). B.Y.C. is a recipient of a National Research Service award from the National Cancer Institute.
REFERENCES
- 1.Allen B G, Katz S. Isolation and characterization of the calcium and phospholipid dependent protein kinase (protein kinase C) subtypes from bovine heart. Biochemistry. 1991;30:4334–4340. doi: 10.1021/bi00231a032. [DOI] [PubMed] [Google Scholar]
- 2.Altmeyer A, Simmons R C, Krajewski S, Reed J C, Bornkamm G W, Chen-Kiang S. Reversal of EBV immortalization precedes apoptosis in IL-6 induced human B cell terminal differentiation. Immunity. 1997;7:667–677. doi: 10.1016/s1074-7613(00)80387-8. [DOI] [PubMed] [Google Scholar]
- 3.Bartel P L, Chien C T, Stemglanz R, Fields S. Using the two hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 4.Bohen S P, Kralli A, Yamamoto K R. Hold’em and fold’em: chaperones and signal transduction. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. [DOI] [PubMed] [Google Scholar]
- 5.Bolen J B, Thiele C J, Israel M A, Yonemoto W, Lipsich L A, Brugge J S. Enhancement of cellular src gene product associated tyrosyl kinase activity following polyoma virus infection and transformation. Cell. 1984;38:767–777. doi: 10.1016/0092-8674(84)90272-1. [DOI] [PubMed] [Google Scholar]
- 6.Brown M T, Cooper J A. Regulation, substrates and function of Src. Biochim Biophys Acta. 1996;128:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 7.Cartwright C A, Meisler A I, Eckhart W. Activation of the pp60c-src protein kinase is an early event in colonic carcinogenesis. Proc Natl Acad Sci USA. 1990;87:558–562. doi: 10.1073/pnas.87.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartwright C A, Kaplan P L, Cooper J A, Hunter T, Eckhart W. Altered sites of tyrosine phosphorylation in pp60c-src associated with polyomavirus middle tumor antigen. Mol Cell Biol. 1986;6:1562–1570. doi: 10.1128/mcb.6.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartwright C A, Eckhart W, Simon S, Kaplan P L. Cell transformation by pp60c-src mutated in the carboxyl-terminal regulatory domain. Cell. 1987;49:83–91. doi: 10.1016/0092-8674(87)90758-6. [DOI] [PubMed] [Google Scholar]
- 10.Cartwright C A, Kamps M P, Meisler A I, Eckhart W. pp60c-src activation in human colon carcinoma. J Clin Invest. 1989;83:2025–2033. doi: 10.1172/JCI114113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartwright C A, Hutchinson M, Eckhart W. Structural and functional modifications of pp60c-src associated with polyomavirus middle tumor antigen from infected or transformed cells. Mol Cell Biol. 1985;5:2647–2652. doi: 10.1128/mcb.5.10.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, B. Y., and C. A. Cartwright. Unpublished data.
- 13.Cheng H C, Nishio H, Hatase U, Ralph S, Wang J H. A synthetic peptide derived from p34cdc2 is a specific and efficient substrate of Src family tyrosine kinases. J Biol Chem. 1992;267:9248–9256. [PubMed] [Google Scholar]
- 14.Coghlan V M, Perrino B A, Howard M, Langeberg L K, Hicks J B, Gallatin W M, Scott J D. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 15.Coghlan V M, Hausken Z E, Scott J D. Subcellular targeting of kinases and phosphatases by association with bifunctional anchoring proteins. Biochem Soc Trans. 1995;23:592–596. doi: 10.1042/bst0230592. [DOI] [PubMed] [Google Scholar]
- 16.Courtneidge S A, Smith A E. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature (London) 1983;303:435–439. doi: 10.1038/303435a0. [DOI] [PubMed] [Google Scholar]
- 17.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz M A, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 18.Faux M C, Scott J D. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- 19.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 20.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 21.Gaudet R, Bohm A, Sigler P B. Crystal structure at 2.4Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 22.Gauen L K T, Kong A N T, Samelson L E, Shaw A S. p59fyn tyrosine kinase associates with multiple T-cell receptor subunits through its unique amino-terminal domain. Mol Cell Biol. 1992;12:5438–5466. doi: 10.1128/mcb.12.12.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillemot F, Billault A, Auffray C. Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc Natl Acad Sci USA. 1989;86:4594–4598. doi: 10.1073/pnas.86.12.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy C T, Muthuswamy S K, Cardiff R D, Soriano P, Muller W J. Activation of the c-Src tyrosine kinase is required for the induction of mammary tumors in transgenic mice. Genes Dev. 1994;8:23–32. doi: 10.1101/gad.8.1.23. [DOI] [PubMed] [Google Scholar]
- 25.Hardie D G, editor. Protein phosphorylation: a practical approach. Oxford, England: IRL Press; 1993. [Google Scholar]
- 26.Harlow E, Lane D, editors. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 27.Klauck T M, Faux M C, Labudda K, Langeberg L K, Jaken S, Scott J D. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 28.Klemke R L, Yebra M, Bayna E M, Cheresh D A. Receptor tyrosine kinase signaling required for integrin α v beta 5-directed cell motility but not adhesion on vitronectin. J Cell Biol. 1994;127:859–866. doi: 10.1083/jcb.127.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kmiecik T E, Shalloway D. Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell. 1987;49:65–73. doi: 10.1016/0092-8674(87)90756-2. [DOI] [PubMed] [Google Scholar]
- 30.Kuenzel E, Krebs E. A synthetic peptide substrate specific for casein kinase II. Proc Natl Acad Sci USA. 1985;82:737–741. doi: 10.1073/pnas.82.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kypta R M, Goldberg Y, Ulug E T, Courtneidge S A. Association between the PDGF receptor and members of the src family of tyrosine kinases. Cell. 1990;62:481–492. doi: 10.1016/0092-8674(90)90013-5. [DOI] [PubMed] [Google Scholar]
- 32.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Okamoto T, Chun M, Sargiacomo M, Casanova J E, Hansen S H, Nishimoto I, Lisanti M P. Evidence of a regulated interaction between heterotrimeric G proteins and caveolin. J Biol Chem. 1996;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Seitz R, Lisanti M P. Phosphorylation of caveolin by Src tyrosine kinases. J Biol Chem. 1996;271:3863–3868. [PubMed] [Google Scholar]
- 35.Li S, Couet J, Lisanti M P. Src tyrosine kinases, Gα subunits, and H-ras share a common membrane-anchored scaffolding protein, caveolin. J Biol Chem. 1996;271:29182–29190. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin X, Nelson P J, Frankfort B, Tombler E, Johnson R, Gelman I H. Isolation and characterization of a novel mitogenic regulatory gene, 322, which is transcriptionally suppressed in cells transformed by src and ras. Mol Cell Biol. 1995;15:2754–2762. doi: 10.1128/mcb.15.5.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin X, Tombler E, Nelson P J, Ross M, Gelman I H. A novel src- and ras-suppressed protein kinase C substrate associated with cytoskeletal architecture. J Biol Chem. 1996;271:28430–28438. doi: 10.1074/jbc.271.45.28430. [DOI] [PubMed] [Google Scholar]
- 38.Lipsich L A, Lewis A J, Brugge J S. Isolation of monoclonal antibodies that recognize the transforming proteins of avian sarcoma viruses. J Virol. 1983;48:352–360. doi: 10.1128/jvi.48.2.352-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luttrell L M, Della Rocca G J, van Biesen T, Luttrell D K, Lefkowitz R J. Gβγ subunits mediate Src-dependent phosphorylation of the epidermal growth factor receptor. J Biol Chem. 1997;272:4637–4644. doi: 10.1074/jbc.272.7.4637. [DOI] [PubMed] [Google Scholar]
- 40.Mayer B J. Signal transduction: clamping down on Src activity. Curr Biol. 1997;7:R295–R298. doi: 10.1016/s0960-9822(06)00141-2. [DOI] [PubMed] [Google Scholar]
- 41.Mochly-Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for protein kinase C. Proc Natl Acad Sci USA. 1991;88:3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mochly-Rosen D, Smith B L, Chen C H, Disatnik M H, Ron D. Interaction of protein kinase C with RACK1, a receptor for activated C-kinase: a role in β protein kinase C mediated signal transduction. Biochem Soc Trans. 1995;23:596–560. doi: 10.1042/bst0230596. [DOI] [PubMed] [Google Scholar]
- 43.Mochly-Rosen D. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- 44.Muller F, Kruger D, Sattlegger E, Hoffmann B, Ballario P, Kanaan M, Barthelmess I B. The cpc-2 gene of Neurospora crassa encodes a protein entirely composed of WD-repeat segments that is involved in general amino acid control and female fertility. Mol Gen Genet. 1995;248:162–173. doi: 10.1007/BF02190797. [DOI] [PubMed] [Google Scholar]
- 45.Nauert J B, Klauck T M, Langeberg L K, Scott J D. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 46.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 47.Neer E J, Smith T F. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 48.Park J, Meisler A I, Cartwright C A. c-Yes tyrosine kinase activity in human colon carcinomas. Oncogene. 1993;8:2627–2635. [PubMed] [Google Scholar]
- 49.Park J, Cartwright C A. Src activity increases and Yes activity decreases during mitosis of human colon carcinoma cells. Mol Cell Biol. 1995;15:2374–2382. doi: 10.1128/mcb.15.5.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Z-Y, Cartwright C A. Regulation of the Src tyrosine kinase and Syp tyrosine phosphatase by their cellular association. Oncogene. 1995;11:1955–1962. [PubMed] [Google Scholar]
- 51.Pitcher J A, Inglesse J, Higgins J B, Arriza J L, Casey P J, Kim C, Benovic J L, Kwatra M M, Caron M G, Lefkowitz R J. Role of βγ subunits of G proteins in targeting the β-adrenergic receptor kinase to membrane-bound receptors. Science. 1992;257:1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- 52.Piwnica-Worms H, Kaplan D R, Whitman M, Roberts T M. Retrovirus shuttle vector for study of kinase activities of pp60c-src synthesized in vitro and overproduced in vivo. Mol Cell Biol. 1986;6:2033–2040. doi: 10.1128/mcb.6.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piwnica-Worms H, Saunders K B, Roberts T M, Smith A E, Cheng S H. Tyrosine phosphorylation regulates the biochemical and biological proerties of pp60c-src. Cell. 1987;49:75–82. doi: 10.1016/0092-8674(87)90757-4. [DOI] [PubMed] [Google Scholar]
- 54.Preito A L, Edelman G M, Crossin K L. Multiple integrins mediate cell attachment to cytotactin/tenacin. Proc Natl Acad Sci USA. 1993;90:10154–10158. doi: 10.1073/pnas.90.21.10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rannels S R, Beasley A, Corbin J D. Regulatory subunits of bovine heart and rabbit skeletal muscle cAMP dependent protein kinase isozymes. Methods Enzymol. 1983;99:55–62. doi: 10.1016/0076-6879(83)99040-7. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds A B, Vila J, Lansing T J, Potts W M, Weber M J, Parsons J T. Activation of the oncogenic potential of the avian cellular src protein by specific structural alteration of the carboxyl terminus. EMBO J. 1987;6:2359–2364. doi: 10.1002/j.1460-2075.1987.tb02512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roche S, Koegl M, Barone M V, Roussel M F, Courtneidge S A. DNA synthesis induced by some but not all growth factors requires Src family tyrosine kinases. Mol Cell Biol. 1995;15:1102–1109. doi: 10.1128/mcb.15.2.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ron D, Mochly-Rosen D. Agonists and antagonists of protein kinase C function, derived from its binding proteins. J Biol Chem. 1994;269:21395–21398. [PubMed] [Google Scholar]
- 59.Ron D, Luo J, Mochly-Rosen D. C2 region-derived peptides inhibit translocation and function of β protein kinase C in vivo. J Biol Chem. 1995;270:24180–24187. doi: 10.1074/jbc.270.41.24180. [DOI] [PubMed] [Google Scholar]
- 60.Ron D, Chen C H, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of the intracellular receptor for protein kinase C: a homolog of the β subunits of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shan X, Luo H, Houle B, Wu J. Expression of a G-protein β subunit-related gene during lymphocyte activation. Int Immunol. 1994;6:739–749. doi: 10.1093/intimm/6.5.739. [DOI] [PubMed] [Google Scholar]
- 62.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 63.Taylor S J, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 64.Turner J M, Brodsky M H, Irving B A, Levin S D, Perlmutter R M, Littman D R. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 1990;60:755–765. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 65.Twamley-Stein G M, Pepperkok R, Ansorge W, Courtneidge S A. The Src family tyrosine kinases are required for PDGF mediated signal transduction in NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:7696–7700. doi: 10.1073/pnas.90.16.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu W, Harrison S C, Eck M J. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 67.Zang Q, Lu Z, Curto M, Barile N, Shalloway D, Foster D A. Association between v-Src and protein kinase C δ in v-Src transformed fibroblasts. J Biol Chem. 1997;272:13275–13280. doi: 10.1074/jbc.272.20.13275. [DOI] [PubMed] [Google Scholar]