Abstract
Mammalian replication origins appear paradoxical. While some studies conclude that initiation occurs bidirectionally from specific loci, others conclude that initiation occurs at many sites distributed throughout large DNA regions. To clarify this issue, the relative number of early replication bubbles was determined at 26 sites in a 110-kb locus containing the dihydrofolate reductase (DHFR)-encoding gene in CHO cells; 19 sites were located within an 11-kb sequence containing ori-β. The ratio of ∼0.8-kb nascent DNA strands to nonreplicated DNA at each site was quantified by competitive PCR. Nascent DNA was defined either as DNA that was labeled by incorporation of bromodeoxyuridine in vivo or as RNA-primed DNA that was resistant to λ-exonuclease. Two primary initiation sites were identified within the 12-kb region, where two-dimensional gel electrophoresis previously detected a high frequency of replication bubbles. A sharp peak of nascent DNA occurred at the ori-β origin of bidirectional replication where initiation events were 12 times more frequent than at distal sequences. A second peak occurred 5 kb downstream at a previously unrecognized origin (ori-β′). Thus, the DHFR gene initiation zone contains at least three primary initiation sites (ori-β, ori-β′, and ori-γ), suggesting that initiation zones in mammals, like those in fission yeast, consist of multiple replication origins.
At least 22 replication origins have now been mapped in the chromosomes of flies, frogs, and mammals (references 14, 45, and 61 and references cited below) by using several different strategies (reviewed in reference 13). While all the data are consistent with bidirectional replication involving classical replication bubbles and forks, a complex and sometimes contradictory view of replication origins has emerged. While some studies conclude that most initiation events occur at specific sites analogous to those found in single-cell organisms such as yeast, Tetrahymena, and Physarum, other studies conclude that most initiation events are distributed throughout large DNA regions with no preference for one site over another. This paradox is best illustrated by studies on the mammalian rRNA and dihydrofolate reductase (DHFR) gene regions, where several different methods have been applied to the same genomic loci.
When nascent DNA is labeled with nucleotide precursors during its biosynthesis, the data suggest that most initiation events occur at specific DNA sequences referred to as origins of bidirectional replication (OBRs). In the rRNA gene region, the earliest labeled DNA fragments have been identified (8, 21) and the growth and relative abundance of nascent DNA chains have been determined (21, 29, 60). These studies reveal two primary initiation sites: a 1- to 6-kb locus upstream of the rRNA gene promoter that is >10-fold more active than distal sites and a ∼3-kb locus with weaker activity downstream of the 3′ end of the gene. The major site appears to be conserved among species (37). In the DHFR gene region in Chinese hamster ovary (CHO) cells, the earliest labeled DNA fragments have been identified (1, 4, 5, 34, 43), the polarity of replication forks has been determined by measuring both Okazaki fragment and leading-strand biases (6, 7, 30, 52), and the growth and relative abundance of nascent DNA chains have been determined (47, 53). These studies reveal two OBRs, a strong one (ori-β) located within a 0.5- to 3-kb sequence ∼17 kb downstream of the 3′ end of the DHFR gene, and a weaker one (ori-γ) located about 23 kb further downstream. The frequency of initiation at ori-β is >10-fold greater than at distal sites.
In contrast, when replicating intermediates are isolated from mammalian cells, fractionated by two-dimensional (2D) gel electrophoresis, and then hybridized with sequence-specific probes, initiation events appear to be distributed almost randomly throughout large DNA regions (initiation zones). Neutral-neutral 2D gel electrophoresis has detected replication bubbles throughout the 31-kb intergenic region in rRNA gene repeats and the 55-kb region between the DHFR and 2BE2121 genes, while neutral-alkaline 2D gel electrophoresis has detected equivalent numbers of replication forks within these regions, traveling in both directions (17, 44).
How might these two sets of data be reconciled? Analysis of newly synthesized DNA relies upon a quantitative comparison of results at different DNA sequences and thereby reveals sites where the frequency of initiation is greatest. However, the relative frequency of initiation events at different sites is difficult to quantify by 2D gel electrophoresis, and therefore most publications simply report their presence or absence. Nevertheless, the intensity of bubble arcs in the rRNA gene initiation zone varied ∼10-fold and was greatest in the regions corresponding to the two primary initiation sites (44). Visual inspection of neutral-neutral 2D gel electrophoresis data from the DHFR gene region suggests that the frequency of initiation bubbles is greatest within a 12-kb region containing ori-β (17, 18, 56). Thus, 2D gel electrophoresis may be unable to detect primary initiation sites. A relevant paradigm is provided by Schizosaccharomyces pombe. Initial studies by 2D gel electrophoresis suggested that initiation events in the ura4 region were distributed throughout a ∼5-kb initiation zone (62). However, when genetic analyses were coupled with 2D gel electrophoresis analyses (20), initiation events were discovered to occur coincident with three separate autonomously replicating sequence (ARS) elements. Thus, what at first appeared to be a continuous initiation zone is actually a cluster of three replication origins.
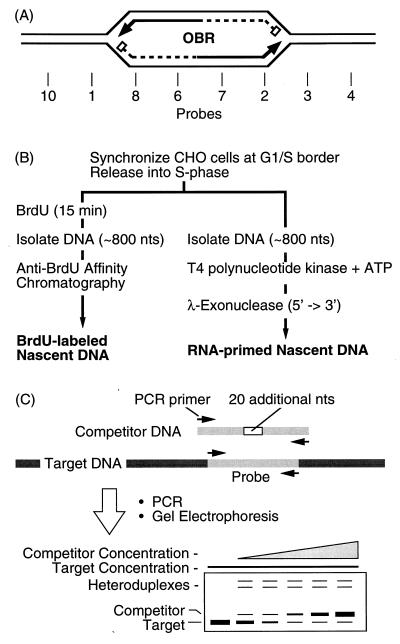
To determine whether additional, previously unrecognized replication origins exist in mammalian chromosomes at sites where 2D gel electrophoresis detects strong bubble arcs, the nascent-strand abundance assay was used to quantify the relative abundance of ∼0.8-kb nascent DNA fragments in and around the ori-β in the DHFR gene region of CHO cells. These nascent DNA fragments represent newly initiated replication bubbles. This assay and its close relative, the nascent-strand length assay, are based on the fact that sequences closest to an OBR are represented more frequently in nascent DNA strands than are sequences further away (Fig. 1A). They have been used to map at least 10 different replication origins in mammalian cells to sites as small as 0.5 kb (reviewed in references 24, 47, 51, and 60). In the present study, quantification was carried out at 26 sites by competitive PCR and nascent DNA was identified by two independent criteria, incorporation of bromodeoxyuridine (BrdU) in vivo and resistance to λ-exonuclease in vitro. Twenty of these probes were distributed throughout the ∼12.2 kb that contains the two EcoRI restriction fragments, F′ and F. Both fragments produce strong bubble arcs in 2D gel electrophoresis, but only fragment F′ has previously been shown to contain an OBR (ori-β).
FIG. 1.
Mapping replication origins by the nascent DNA strand abundance assay. (A) Early replication bubbles contain an OBR and two RNA-primed (box) nascent DNA strands (the arrow indicates the direction of growth). (B) Nascent DNA strands from early replication bubbles were isolated either as newly synthesized (BrdU-labeled) DNA or as RNA-primed DNA with an average length of ∼0.8 kb. (C) Competitive PCR was used to measure the relative abundance of specific probe sequences. nts, nucleotides.
The results reveal that EcoRI fragments F′ and F each contain one primary initiation site. The strongest initiation site was coincident with the ori-β OBR in fragment F′ originally identified in synchronized CHO cells by a transition from continuous to discontinuous DNA synthesis (6, 7, 30, 52) and subsequently confirmed by nascent-strand length (53) and abundance (47) assays in exponentially proliferating CHO cells. A second, previously unrecognized initiation site (ori-β′) of lower intensity was detected 5 kb further downstream in fragment F. Thus, when results from both the DHFR and rRNA gene regions are taken together, initiation zones in mammals appear to consist of one or more primary initiation sites (OBRs) as well as several low-frequency, secondary initiation sites that are detected only by 2D gel electrophoresis methods.
MATERIALS AND METHODS
Culture and synchronization of CHO cells.
CHO K1 cells were seeded in 150-mm tissue culture dishes at about 10% confluence and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and nonessential amino acids at 37°C under 5% CO2. Two different methods were used to synchronize cells at their G1/S boundary. The first was a double aphidicolin block. When the cell monolayers were about 20% confluent, they were cultured in the presence of 1 μg of aphidicolin per ml and 10 nCi of [14C]thymidine per ml for 24 h. The monolayers were washed twice with prewarmed medium to remove aphidicolin and the radiolabel, and the washed cells were cultured for 12 h to allow them to pass through the S phase. Aphidicolin (1 μg/ml) was then added to the culture medium for an additional 12 h to impose a second aphidicolin arrest as they again entered the S phase. The second method was an isoleucine deprivation-serum starvation-aphidicolin block. When the cell monolayers were 80 to 90% confluent, the cells were washed with prewarmed phosphate-buffered saline and then cultured in DMEM without isoleucine and supplemented with only 0.5% fetal calf serum (dialyzed) (GIBCO BRL) for 48 h to arrest the cells in the G1 phase. The cells were washed twice with prewarmed complete DMEM and then cultured for an additional 12 h in complete DMEM supplemented with 10% fetal calf serum and 10 μg of aphidicolin per ml to arrest the cells as they entered the S phase.
Isolation of nascent BrdU-labeled DNA from early replication bubbles.
Cells synchronized at their G1/S boundary were washed twice with prewarmed medium supplemented with serum and nonessential amino acids and then incubated for 15 min at 37°C with prewarmed fresh medium containing 1 μM [3H]dC and 100 μM BrdU. After incubation, the cells were washed twice with 20 ml of ice-cold phosphate-buffered saline containing 0.02% sodium azide, 5 ml of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA–100 mM NaCl was added, and the cells were collected by being scraped off the dish with a rubber policeman. All the subsequent steps were performed in a dark room or with an orange safety light to prevent photodamage to BrdU-substituted DNA (Br-DNA).
Total DNA (∼80 μg) was extracted from one dish containing ∼107 cells by the procedure described in reference 50. The DNA was suspended in 400 μl of 10 mM Tris-HCl (pH 7.8)–1 mM EDTA, denatured by incubation in boiling water for 5 min, and cooled immediately on ice. The DNA was divided in two aliquots and fractionated in 5 to 30% linear sucrose gradients (5 ml) in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–0.3 M NaCl with an SW50Ti rotor in a Beckman centrifuge (45,000 rpm for 5 h at 20°C) (24). Alternatively, the DNA was treated with 0.2 N NaOH at room temperature for 20 min and then fractionated in 5 to 30% linear sucrose gradients in 1 mM EDTA–0.2 N NaOH–0.3 M NaCl under the same conditions. DNA collected in either neutral or alkaline sucrose gradients gave the same results when ori-β was mapped. Sucrose gradient fractions (25) of 200 μl were collected, and the size of DNA in each fraction was determined by alkaline agarose gel electrophoresis as described in reference 50. Four fractions that contained DNA with a mean length of ∼0.8 kb were pooled, dialyzed against TBSE (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1 mM EDTA), and further purified by anti-BrdU affinity chromatography (9). At least 90% of these molecules were determined by electrophoresis in alkaline agarose gels (2) to be 800 ± 200 nucleotides.
Goat anti-mouse immunoglobulin G was coupled to CNBr-activated Sepharose 4B (Pharmacia, Uppsala, Sweden), and then mouse anti-BrdU monoclonal antibody was bound to the coupled Sepharose. The immunoaffinity beads (0.5 ml) were poured into a column and washed twice with 10 ml of TBSE before being incubated with the Br-DNA sample (1.5 ml). After 2 h at room temperature with slow agitation, unbound DNA was collected as the flowthrough fraction. The column was then washed twice with 4 ml of TBSE before eluting bound Br-DNA with 2 ml of 150 mM NaCl adjusted to pH 11.5 with NH4OH. The eluate was neutralized on an NAP 10 column (Pharmacia) and treated with proteinase K (200 μg/ml) overnight at 37°C before being extracted once with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and once with chloroform-isoamyl alcohol (24:1, vol/vol), and the DNA was precipitated in 0.1 M NaCl by the addition of 2 volumes of ethanol. The DNA pellet was resuspended in 1.5 ml of Tris-EDTA (TE) buffer. Nonreplicating DNA was prepared from G0 cells (a confluent culture arrested in 0.5% serum), and 30 μg of DNA in 200 μl of TE was sonicated for 13 s at 80% power with a microtip (Misonix). The products were fractionated by sucrose gradient sedimentation to isolate the same size population taken from Br-DNA.
Enrichment of 5′-RNA-DNA chains from early replication bubbles.
DNA from 107 synchronized cells was isolated as described above, except that the RNase treatment was omitted. DNA of ∼0.8 kb was isolated by neutral sucrose gradient sedimentation and precipitated with ethanol. The pellet was washed with 70% ethanol, air dried briefly, dissolved in 20 μl of water, and denatured for 2 min at 100°C. To this sample was added 4 μl of 500 μM ATP, 2 μl of T4 polynucleotide kinase (New England Biolabs), 4 μl of 10× T4 kinase buffer (New England Biolabs), 5 μl of water (RNase free), and 5 μl of EcoRI-digested, dephosphorylated, 2.7-kb pUC18 DNA (100 ng/μl; Pharmacia). The 40-μl mixture was incubated for 30 min at 37°C, and 10 μl of 1% Sarkosyl–0.1 M EDTA–0.25-μg/μl proteinase K was added. The sample was incubated for 30 min at 50°C and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and then with chloroform-isoamyl alcohol (24:1). DNA was precipitated and washed with ethanol, and the pellet was resuspended in 22 μl of water (RNase free). To 20 μl was added 16 μl of 2.5× λ-exonuclease buffer (167.5 mM glycine-KOH [pH 8.8], 6.25 mM MgCl2, 125 μg of bovine serum albumin per ml) plus 4 μl of λ-exonuclease (Gibco BRL), and the mixture was incubated for 12 h at 37°C. The remaining 2-μl sample of predigested DNA was compared with a 4-μl sample of λ-exonuclease-digested DNA by electrophoresis in 1% agarose to confirm that all of the pUC18 5′-phosphorylated-DNA control had been digested. λ-Exonuclease was then inactivated by heating at 75°C for 10 min, and the DNA was extracted, precipitated, and resuspended in 500 μl of Tris-EDTA buffer.
Competitive PCR.
For each of the 26 DNA target sites to be amplified, four oligonucleotides complementary to the target sequence were synthesized (Table 1), two external primers (primers 1 and 4) and two internal primers (primers 2 and 3). Each internal primer carries the same 20-nucleotide tail attached to its 5′ end: tail 1 (5′-GTCGACGGATCCCTGCAGGT-3′) and tail 2 (5′-ACCTGCAGGGATCCGTCGAC-3′) are unrelated to genome sequence and are complementary, so that the PCR products can be used to construct a competitor sequence that is identical to its corresponding target sequence except for an additional 20 nucleotides in the center. For the genomic positions of each primer set, see Fig. 3. Competitor DNAs (from 156 to 266 nucleotides) were constructed as previously described (19), and their concentrations were determined as follows. PCR was performed with 10 μl of diluted competitor DNA (<10 pg/μl) with the two external primers of each set. The PCR mixture contained the standard amount of dCTP (10 nmol) plus 0.4 μl (0.66 pmol) of [α-32P]dCTP (6,000 Ci/mmol, 10 mCi/ml; Amersham, Little Chalfont, United Kingdom), corresponding to 3.4 × 106 cpm. Amplification products were resolved by electrophoresis in 1.8% agarose gel (50), and the radioactive band was eluted in 100 μl of TE buffer. Residual [α-32P]dCTP was removed on a NICK column (Pharmacia). For example, 400 μl of competitor 6 was recovered from the NICK column, and 50 μl contained 5.2 × 103 cpm or 1.0 fmol of [α-32P]dCTP. Since the ratio of cold to hot dCTP in a PCR mixture was 1 × 104/0.66, 1.0 fmol [1 × 104/0.66] = 15.2 pmol of cytidine. Since competitor 6 contained 87 cytidines/molecule, there is 15.2/87 = 0.175 pmol of competitor 6 in 50 μl. The amount of unlabeled dCTP contributed by [α-32P]dCTP was insignificant and was therefore ignored.
TABLE 1.
PCR primer sets used in this studya
| Name | Sequence (5′ to 3′) | Map positions (bp) |
|---|---|---|
| H-1 | AGACAGAGCCTCTTCAGGAG | |
| H-2 | TCCTCAGAAGAAACTGCATT | |
| H-3 | TTCCAAATCTCAGGCTGATC | |
| H-4 | CCCACATAACCTTGAAGTAC | |
| I-1 | GGTTGGGTGATAACAAAGTC | |
| I-2 | TACAAGTACATGCCACCAAG | |
| I-3 | ATCCTATCAGTGGATCAGCA | |
| I-4 | GGTCTCTCGCTATGATCTGG | |
| J-1 | GAGAATGGATATTGCTTGAA | |
| J-2 | TGCTGAGATAGGAAAGGCAT | |
| J-3 | GGGTTTGTAGCATCAGGTCA | |
| J-4 | GAGGATATGAAGGTAGTGGA | |
| G-1 | GGAAGTGGTCTTGAGCATCT | |
| G-2 | CTCTCTCTCCCCAGACCTCG | |
| G-3 | GTGGGGCTTCCACCATAACG | |
| G-4 | AAGCTCCACCTGAGCTCCTA | |
| 10-1 | GCTTCCGTCCAGTAAGTGCT | 66–85 |
| 10-2 | GCTCAGTGCTTGTCTGTGGT | 102–121 |
| 10-3 | CAAGCTCCAGGAGTCCTGTT | 122–141 |
| 10-4 | GGTCAAGAGTCCATGAGCTC | 231–250 |
| 1-1 | ATCCTCCTAGCTCGGAGTCA | 1534–1553 |
| 1-2 | CATCTGAGACTTGGCTGGGA | 1611–1630 |
| 1-3 | AGTCACAGACCTGCATGGCA | 1631–1650 |
| 1-4 | GGCTTATCTGCATCCTATTC | 1677–1696 |
| 8-1 | GCCTTTGAGCTCAGACTAGA | 2020–2039 |
| 8-2 | CATTCATCAAGCTGGAAAGC | 2061–2080 |
| 8-3 | TCCATGGCAGTCTTCACACT | 2081–2100 |
| 8-4 | GTCCTCGGTATTAGTTCTCC | 2183–2202 |
| 6-1 | GGAAATGTTCTGAAACCAGC | 2464–2483 |
| 6-2 | AACCCACACCTACCTTACGA | 2508–2527 |
| 6-3 | ATAGCATAATCCCACACAAG | 2528–2547 |
| 6-4 | TTGTGTTTTGAGGCAGGGAC | 2600–2619 |
| 7-1 | CCTTCCTTCTCAGTGAGTCC | 2669–2688 |
| 7-2 | TAGTGCGTCTTTAAGACCTG | 2703–2722 |
| 7-3 | GATGCTGAACTTAACAGTAA | 2723–2742 |
| 7-4 | CTGAACTTTATCAGTGCAGT | 2786–2805 |
| 2-1 | CTACTGCCGTATTATAAGAC | 2842–2861 |
| 2-2 | GGTAGGGACTTCAGAAAAAC | 2977–2996 |
| 2-3 | AGGACACAACGCACCCTGGT | 2997–3016 |
| 2-4 | AGGTGACACCTTGCTTTTGT | 3069–3088 |
| 3-1 | ATAACTACTGTCTTAGCTGG | 3108–3127 |
| 3-2 | GACAATGGATTAAACCTCTG | 3208–3227 |
| 3-3 | ATGATTGCAGGAAGTATGGT | 3228–3247 |
| 3-4 | GGTGTGGCCTTGTTGGAAGA | 3310–3329 |
| 4-1 | GCCATTTTCATTCAAACCAC | 3353–3372 |
| 4-2 | GGTATAAGCTACCTTGTAGC | 3377–3396 |
| 4-3 | CCAGCTTGCTATTTCTGATG | 3397–3416 |
| 4-4 | GCTTCTTCCTAATTTGAACT | 3483–3502 |
| 5-1 | CAAGCAGTCCTCGTGGAGCT | 4102–4121 |
| 5-2 | GAAACCAGAGTTGCCATGGT | 4179–4198 |
| 5-3 | CTTGGGCAGCACCTGCTTTA | 4199–4218 |
| 5-4 | GTTTCATGAGCTGATTGGTC | 4232–4251 |
| 11-1 | GGACCTCAGCCTCTGAAACA | 5085–5104 |
| 11-2 | ATACTAAGCTCTCTTTATAG | 5167–5186 |
| 11-3 | TTTAAGAAATCTGTAGCTAT | 5187–5206 |
| 11-4 | CTTTCTCCACTCACTTCACC | 5261–5280 |
| 9-1 | ATCAGACTGGTCCCATATCC | 5955–5974 |
| 9-2 | AAATGTCTCCCTCAGTTGAT | 6005–6024 |
| 9-3 | TTGGATTGAAAGCATAATGC | 6025–6044 |
| 9-4 | ATGGCTTAAATGTGACTCCC | 6074–6093 |
| 16-1 | GAAGACCTGACTGTTCCATG | 6437–6456 |
| 16-2 | TTCAATTCTACACGCAGCTT | 6491–6510 |
| 16-3 | GAGCACAGCATTTGAGTGAC | 6511–6530 |
| 16-4 | CCCTGTTCTCTGCTAAGCAG | 6591–6610 |
| 12-1 | CCAGGACCAATGTGATACAA | 6921–6940 |
| 12-2 | AATGAGATGAGACCTTGGGA | 6986–7005 |
| 12-3 | TTGCGTGTGCCTTTGACACC | 7006–7025 |
| 12-4 | GCTTAAGGCTCACTTATGGA | 7086–7105 |
| 17-1 | TGAAAGTAGGTTAGAAGGGC | 7412–7431 |
| 17-2 | TTCATGTGTGTGTTCTGGGG | 7481–7500 |
| 17-3 | GGTAAACGATGGTTAAAGGA | 7501–7520 |
| 17-4 | GCCACAGCCTGTCACTTCAA | 7566–7585 |
| 13-1 | ACCAAGCACTAGCTAACCCC | 7917–7936 |
| 13-2 | AAGATTGTTCATACTCTTCT | 7995–8014 |
| 13-3 | AGCTTTTCTGTATATGTGTC | 8015–8034 |
| 13-4 | TTCACAGCCTTTCATGTTGC | 8075–8094 |
| 14-1 | TAGGAAGATGCTGGACTTCT | 8909–8928 |
| 14-2 | AACCAGAAGATTATATATGG | 8990–9009 |
| 14-3 | TATAAACAGATTGCTTTACC | 9010–9029 |
| 14-4 | ATGACACTCCAATCAGAGAC | 9081–9100 |
| 19-1 | ATAATCCAGGCAAGCCTGGC | 9429–9448 |
| 19-2 | TTGCCATGCACAGATGGGTG | 9481–9500 |
| 19-3 | GCAGTACCCTGACTCTGTAT | 9501–9520 |
| 19-4 | TGGCCTCCCTCCTCAGAGAA | 9578–9597 |
| 15-1 | TGGTTCCTCAGAAAGCAGTC | 9900–9919 |
| 15-2 | TAGTAAGTATACGGGATCTT | 9997–10016 |
| 15-3 | TTAAGATGTATATAAACCTC | 10017–10036 |
| 15-4 | ACCCTCTTCCTCTAATCAGG | 10102–10121 |
| F-1 | GGCTCTTATCCACACATATA | 10678–10697 |
| F-2 | AGTAGCGTCAAAGGCAGGGA | 10725–10744 |
| F-3 | CTGTTTCTCCAACCCTGCTT | 10745–10764 |
| F-4 | AGAGGTCCTGGGGTCAGAGT | 10806–10825 |
| K-1 | GGAGCTGCATTGGGAGATGG | |
| K-2 | AGTAAGTTAGACTTTCTGCC | |
| K-3 | TGAGAGTTAACTTGGAAATG | |
| K-4 | CATCTGTCAGGCTAACTTCT | |
| O-1 | AGAAAATCCCAAGTTGGTTT | |
| O-2 | GAGTCCAATGGTACACCTTT | |
| O-3 | ATTTCCCCAAATAGTATAC | |
| O-4 | CACTCACCCTTATGTTCTCC | |
| P-1 | ACTTTAGTCTCTTAAGGCAG | |
| P-2 | TTGAGATGGGAAGGAGTCAG | |
| P-3 | GCAGTATAAAGCCAATAGAG | |
| P-4 | GTTACACTTCTGTAGGAAAA |
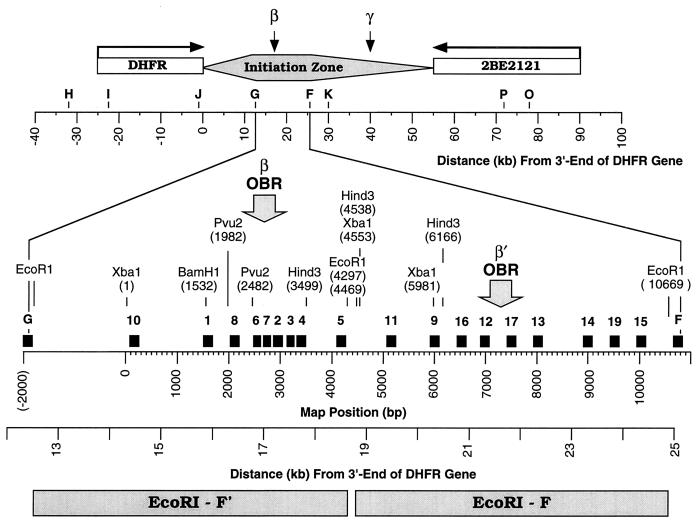
FIG. 3.
PCR primer sets used to map the ori-β region. Primer sets H, I, J, G, F, and K in this study are the same sites as A, B, C, D, E and F (30), respectively. Their DNA sequences were determined from plasmids pDGKS-H, pDGKS-I, pDGKS-J, pDGKS-G, pDGKS-F, and pDGKS-K (25), respectively, originally cloned by H. Cedar. DNA sequences of primer sets P and O were determined from plasmids pDGKS-P and pDGKS-O (25), respectively, originally cloned by J. Hamlin. Primer sets for probes 1 to 19 and F were taken from the 10,825 sequenced nucleotides [Map Position (bp)] now available (GenBank accession no. X94372 and AF028017). Primer sets H, I, J, G, K, P, and O are identical to probes of the same name in reference 25, and primer sets 6, 7, and 9 are similar to probes C, D, and R (6). Primer set 8 is similar to primer set 8 in reference 47. The direction of transcription of the DHFR and 2BE2121 genes is indicated with arrows. ori-β is defined by the data summarized in Fig. 7, ori-β′ is defined by the data in Fig. 5 and 6, and ori-γ is defined by the data in references 1, 30, 43, and 59. The initiation zone is defined by data in references 17, 18, and 56. The distance from the 3′ end of the DHFR gene is based on restriction fragment analyses of plasmids and cosmids. Primer sequences are provided in Table 1.
PCR was performed on 10-μl samples of size-fractionated cellular DNA in the presence of known amounts of competitor DNA. Fifty cycles were carried out with 5 U of AmpliTaq Gold Taq polymerase (Perkin-Elmer), which was activated at 95°C for 8 min before the PCR was started (total volume, 50 μl). Each cycle consisted of denaturation for 30 s at 94°C, annealing for 30 s at 60°C, and extension for 30 s at 72°C. Amplified DNA products were then fractionated by electrophoresis in 10% polyacrylamide gels (Bio-Rad Ready Gel, 100 V [constant voltage], 1.5 h at room temperature), stained with 0.5 μg of ethidium bromide per ml for 10 min at room temperature, and then quantified by densitometry with an Eagle Eye densitometer (Stratagene, La Jolla, Calif.). The ratio of competitor to target DNA was used to determine the number of target molecules, as described in Results.
RESULTS
Measuring the relative abundance of nascent DNA strands.
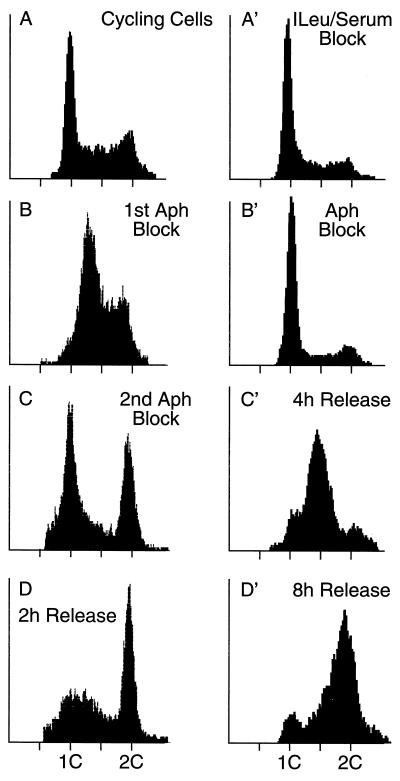
Most analyses of replication origins in the DHFR gene region have been carried out with cells synchronized at their G1/S boundary. Synchronized cells enrich for initiation events at replication origins that are activated at the beginning of the S phase and help to ensure that nascent DNA from specific loci results from initiation of replication at those loci and not from replication forks traveling through them from neighboring origins. To facilitate comparison between earlier studies and those reported here, CHO K1 cells were synchronized by two different methods and the efficacy of each method was evaluated by fluorescence-activated cell sorter (FACS) analysis. In the first method, randomly proliferating cells (Fig. 2A) were cultured for 24 h in low concentrations of aphidicolin, a specific inhibitor of replicative DNA polymerases. This caused the cells to accumulate in both the S and G2 phases (Fig. 2B). The cells were allowed to recover and then subjected to a second aphidicolin block that arrested about 40% of the cells at their G1/S border (Fig. 2C). Most of these cells entered S phase when the aphidicolin was removed (Fig. 2D). Cells that had arrested in G2 did not incorporate BrdU when aphidicolin was removed (data not shown). These cells presumably suffered DNA damage during the synchronization procedure (27) and therefore responded to checkpoint controls (11). The second synchronization method avoided the production of a large population of G2 cells. The cells were arrested first in G1 phase by depriving them of isoleucine and serum (Fig. 2A′) and then released from this block in the presence of a high concentration of aphidicolin to arrest them as they entered S phase (Fig. 2B′). Subsequent removal of aphidicolin allowed most (>95%) of these cells to enter (Fig. 2C′) and complete (Fig. 2D′) S phase. Both methods yielded identical origin-mapping results, although the second was more commonly used in previous studies and produced a larger population of G1/S-phase cells.
FIG. 2.
FACS analyses of CHO K1 cell synchronization procedures. CHO cell synchronization was evaluated by staining nuclei with Cycle Test Plus (Becton Dickinson) and measuring their DNA content by flow cytometry (FACScan; Becton Dickinson). Between 5,000 and 10,000 fluorescent events were counted. (A to D) Double aphidicolin block. (A) Randomly proliferating cells; (B) cells in the first aphidicolin block; (C) cells in the second aphidicolin block; (D) cells 2 h after release from the second aphidicolin block. (A′ to D′) Isoleucine-serum-aphidicolin block. (A′) Cells arrested by depletion of isoleucine and fetal calf serum; (B′) cells released into aphidicolin; (C′ and D′) cells released from aphidicolin for 4 h (C′) and 8 h (D′).
Two methods were used to map replication origins by the nascent-strand abundance assay (Fig. 1B). The first is that of Giacca et al. (23, 24). CHO cells were released into S phase in the presence of BrdU to label nascent DNA (Br-DNA). DNA strands isolated were 800 ± 200 nucleotides long, because they should be small enough to originate from newly formed replication bubbles (Fig. 1A) but large enough to exclude CHO cell Okazaki fragments, which have a mean length of 105 nucleotides (7). Br-DNA of this length was then purified by affinity chromatography to avoid contamination with fragments of nonreplicated DNA that resulted from DNA damage. In fact, prelabeled genomic [14C]DNA was never detected in the purified Br-DNA fraction.
Br-DNA was then analyzed by competitive PCR to determine the relative concentrations of specific DNA sequences present at 26 genomic sites distributed within a 110-kb region containing ori-β (Fig. 3). An average of one probe every 0.6 kb was constructed in the 10.8 kb of sequenced DNA around ori-β. Since the nascent DNA strands mapped in these experiments were ∼0.8 kb, all origins within this region should be detected. Competitive PCR (Fig. 1C) permits quantification of small amounts of DNA sequences by coamplifying the target DNA in the presence of known amounts of a competitor DNA that has the same primer recognition sites (28). The competitor DNA contains a 20-nucleotide insertion, so that its amplified products can be distinguished from those of the target. Since target and competitor DNAs compete for the same PCR primers, they are amplified with the same efficiency. Thus, when the amount of competitor DNA added to the reaction mixture equals the amount of target DNA present, the ratio of amplified competitor to target sequences is unity. Since this method is independent of the time course of amplification, small amounts of target can be amplified until the reaction is exhausted.
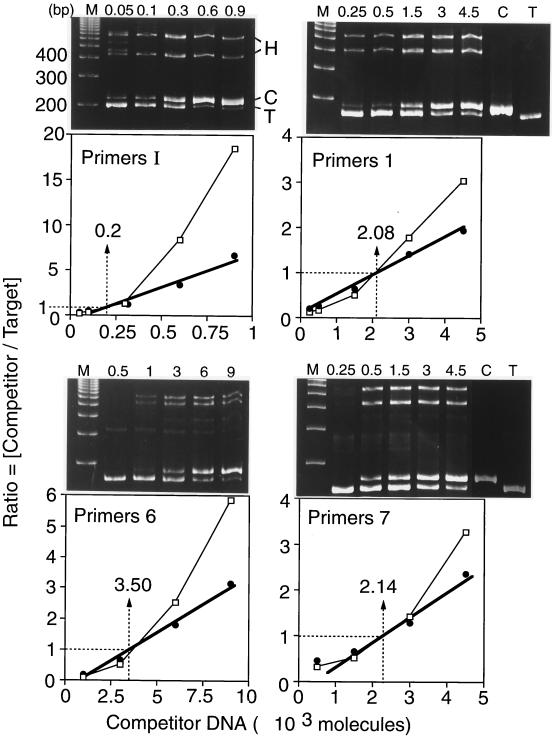
The results for primer sets I, 1, 6, and 7 from a single DNA preparation illustrate the method (Fig. 4). Two slower-migrating bands of DNA invariably appeared in our PCR products. These represented heteroduplexes between target and competitor DNA amplicons that formed during the final cycle of denaturation and renaturation. They were absent whenever either target or competitor DNA was omitted from the PCR mixture (Fig. 4, primer sets 1 and 7) and appeared when preamplified competitor and target DNAs were mixed, denatured, and then slowly renatured (data not shown). Since the ratio of competitor to target DNA is used to determine the actual amount of target DNA present, the amount of heteroduplex DNA formed must be taken into account. This was done as follows. Amplified target DNA is indicated by T, and amplified competitor DNA is indicated by C. Homoduplex DNAs are indicated by T · T or C · C, and heteroduplex DNAs are indicated by T · C or C · T. Total target DNA [T] = [T · T + (T · C + C · T)/2], and the total competitor DNA [C] = [C · C + (T · C + C · T)/2]. Using this formula, the ratio of [Total C] to [Total T] was linear as a function of the amount of competitor DNA added to the PCR mixture (Fig. 4). In contrast, when the ratio of [C · C] to [T · T] was plotted against the amount of competitor DNA added to the PCR mixture, a curved line resulted (Fig. 4). Thus, when the experimental ratio of [C · C] to [T · T] in the PCR mixture was ≈1, the effect of heteroduplex formation on the calculated value for [T] was negligible, but when [T] was determined by extrapolation between data points, the effect of heteroduplex formation could be significant.
FIG. 4.
Use of competitive PCR to quantify the amount of a specific sequence. A fixed amount of newly synthesized Br-DNA was amplified in the presence of increasing amounts of the indicated competitor molecules (primer sets I, 1, 6, and 7). DNA products were resolved by gel electrophoresis and stained with ethidium bromide (upper panels). The amount of competitor DNA added to the PCR mixture is indicated above each lane. Lanes M contain a 100-bp DNA ladder. Lanes C lacked target DNA. Lanes T lacked competitor DNA. Two heteroduplex DNA bands consisting of either T · C or C · T are indicated by H. The ratio of competitor DNA to target DNA was determined by quantifying the fraction of DNA in each of the four major bands by densitometry (lower panels). This ratio was plotted as a function of the amount of competitor DNA added to the PCR assay: C/T versus [competitor added] (open symbols, thin line), and [C + H/2] / [T + H/2] versus [competitor added] (solid symbols, heavy line; H = T · C + C · T). The thin lines with open symbols take into account heteroduplex DNA. The amount of target DNA determined in each example is given.
Mapping replication origins with newly synthesized BrdU-labeled DNA from early replication bubbles.
To identify primary initiation sites in the DHFR gene region, five independent samples of newly synthesized Br-DNA from CHO cells were assayed in parallel with five samples of nonreplicating DNA fragments of the same size prepared from G0-phase CHO cells. To present the results from these experiments on the same graph, the average value for probes H, I, and J in each Br-DNA sample was taken as unity and each of the probes used in that sample was normalized to this value. The results from five analyses of nascent Br-DNA were then averaged and are presented in Fig. 5A and B. The same procedure was carried out with the five analyses of nonreplicating G0 DNA. Since probes H, I, and J are located within or slightly upstream of the DHFR gene, a region in which initiation events have never been detected by any method including 2D gel electrophoresis (17, 18, 56), a value of unity indicated the absence of initiation events at that probe site. As expected, probes H, I, and J were consistently represented at the lowest levels in nascent Br-DNA.
FIG. 5.
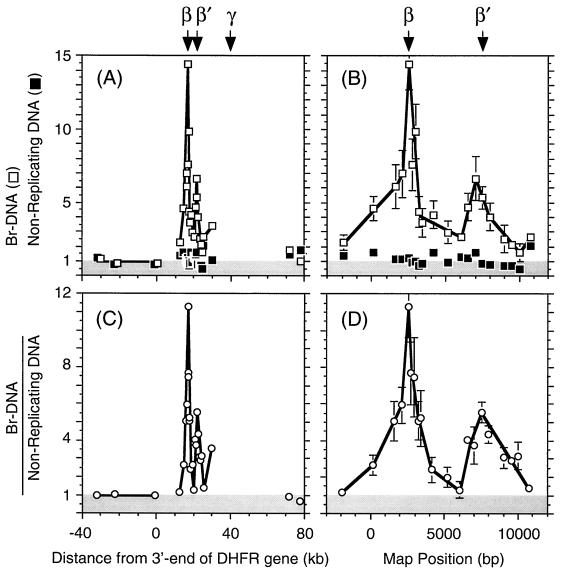
Mapping replication origins with newly synthesized BrdU-labeled DNA from early replication bubbles. (A) The relative abundance of 26 different probes (Fig. 3) was determined on five samples of nonreplicating DNA (▪) and on five samples of newly synthesized Br-DNA (□) chains with an average length of ∼0.8 kb by competitive PCR. The mean values and the standard errors of the mean are shown. The amount of each probe was normalized to the average of probes H, I, and J (the region where 2D gel electrophoresis has never detected replication bubbles). Therefore, numbers of unity or less (shaded area) indicate absence of initiation events. Since most of the region from −40 to +80 kb has not been sequenced (Fig. 3), the distance from the 3′ end of the DHFR gene was plotted. (B) Since most of the region from 12 to 27 kb downstream of the 3′-end of the DHFR gene has been sequenced (Fig. 3), the nucleotide map positions of these data in panel A were plotted. (C) The ratios of Br-DNA to nonreplicating DNA were calculated (○) in each of the five experiments and averaged. (D) Data in the region from 12 to 27 kb downstream of the 3′ end of the DHFR gene in panel C were replotted on their nucleotide map position. The positions of ori-β, ori-β′, and ori-γ OBRs are indicated.
In principle, the copy number for each probe in the G0 DNA samples should be the same, if each probe represents a single-copy locus. In practice, the mean copy number for 26 primer sets in five DNA samples was 1.14 ± 0.08 (standard error of the mean) (Fig. 5A and B). This variation reflected the degree of inaccuracy in our measurements of competitor DNA concentrations. In addition, competitor DNA, like any DNA stored at low concentrations, can deteriorate over time (41). Therefore, these variations were corrected in each experiment by assaying a sample of nascent Br-DNA in parallel with a sample of nonreplicated G0 DNA, and the ratio of Br-DNA to nonreplicating DNA was calculated for each probe in each experiment. The average ratio of Br-DNA to nonreplicating DNA at each probe was then determined (Fig. 5C and D).
In one experiment, a 10-μl sample of Br-DNA contained 0.2 × 103 copies of primer set I located in the DHFR gene and 3.5 × 103 copies of primer set 6 located close to the ori-β OBR (Fig. 4). Primer sets H and J gave results very similar to those obtained with primer set I. Therefore, in this experiment, primer set 6 was enriched about 17.5-fold over the DHFR gene region in nascent DNA. Primer sets 1 and 7, which flanked primer set 6, gave intermediate levels, consistent with an OBR at or near primer set 6.
The average enrichment in all five experiments for primer set 6 over the primer sites in the DHFR gene in Br-DNA was (14.4 ± 2.3)-fold (Fig. 5A and B). Since the region around primer set 6 produced a reasonably symmetrical peak of nascent DNA centered on primer set 6 that was absent in nonreplicating DNA (Fig. 5A and B), these results suggested bidirectional replication from a site at or close to primer set 6. This peak was even more symmetrical when the average ratio of Br-DNA to nonreplicating DNA was displayed (Fig. 5C and D). After correcting for experimental variation among the probes, the ratio of primer set 6 (peak of initiation activity) relative to the DHFR gene (mean value of primer sets H, I, and J) in nascent DNA was 11.3 ± 1.7 (Fig. 5C and D). Since primer set 6 is coincident with the ori-β OBR (see Discussion), these data demonstrate that the ori-β OBR is a primary initiation site for DNA replication.
A second, previously undetected initiation site (ori-β′) was discovered about 5 kb downstream of ori-β (Fig. 5B and D). The average ratio of Br-DNA to nonreplicating DNA at ori-β′ was 5.5 ± 1, representing about half as many initiation events as were observed at ori-β. The data also indicated a third initiation site that may correspond to the previously identified ori-γ locus located about 23 kb downstream of ori-β, but the absence of sequence information in this region precluded further analysis. Therefore, the DHFR gene initiation zone contains at least two (β and β′) and probably three (β, β′, and γ) primary initiation sites.
Mapping replication origins with RNA-primed DNA from early replication bubbles.
Gerbi and Bielinsky (22) showed that the 5′ to 3′ λ-exonuclease activity could be used to enrich for RNA-primed nascent DNA chains because they are resistant to digestion whereas DNA chains containing a 5′-terminal phosphate are degraded. RNA-primed DNA chains isolated in this manner were used to map the transition from continuous to discontinuous DNA synthesis at OBRs in simian virus 40 (SV40) and yeast to a few nucleotides (3). Therefore, to confirm the results obtained with Br-DNA (described above) and to eliminate possible artifacts from BrdU-induced DNA damage and repair, this strategy was adapted to mammalian cells.
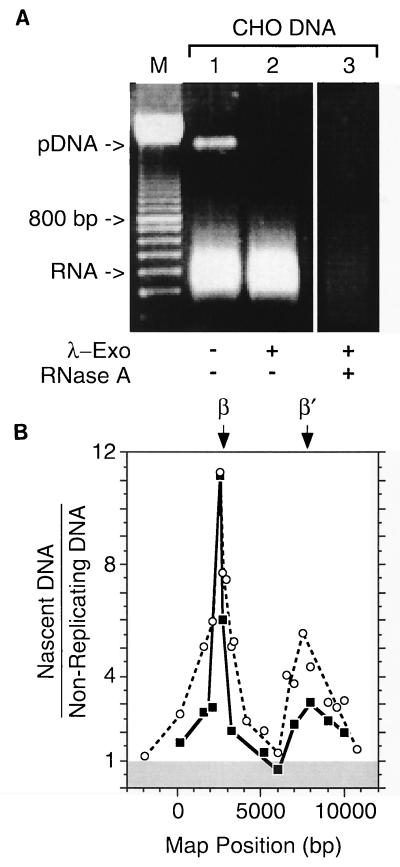
At least 90% of Okazaki fragments from CHO cells carry a 10-residue oligoribonucleotide (7). For the 3′ end of an Okazaki fragment to be ligated to the 5′ end of a long, growing nascent DNA chain, the RNA primer of the last Okazaki fragment joined to this chain must be excised. However, since this excision step is slow, a fraction of long, nascent DNA chains should still have one or more ribonucleotides attached to their 5′-ends (2, 16). Accordingly, a second method (Fig. 1B) for the nascent-strand abundance assay was used in which DNA with an average length of ∼0.8 kb was purified as before from CHO cells, except that the cells were not incubated with BrdU and the DNA was not treated with RNase A. A linear dephosphorylated plasmid DNA was added to provide an internal control. Under the conditions used, the plasmid DNA was completely phosphorylated by T4 polynucleotide kinase in the presence of ATP and then digested by λ-exonuclease, while residual cellular RNA remained unless RNase A was added to degrade it (Fig. 6A). These DNA samples were then analyzed by competitive PCR as described above.
FIG. 6.
Mapping replication origins with RNA-primed DNA from early replication bubbles. DNA with an average length of ∼0.8 kb was enriched for RNA-primed nascent DNA chains by treatment first with T4 polynucleotide kinase plus ATP to ensure that all 5′-DNA ends were phosphorylated and then with λ-exonuclease to degrade all 5′-phosphorylated-DNA chains (Fig. 1B). Linear, dephosphorylated pUC18 DNA was included as an internal control. (A) An aliquot of the sample was fractionated by electrophoresis in a 1% agarose gel before (lane 1) and after (lane 2) λ-exonuclease treatment. A sample of the λ-exonuclease-treated material was later digested with RNase A (lane 3). A 100-bp DNA ladder (Pharmacia Biotech) was run in parallel to provide size markers (M). (B) Two preparations of the T4 kinase/λ-exonuclease-treated material were used to determine the relative abundance of RNA-primed nascent DNA chains (▪) ∼0.8 kb long by competitive PCR. These results are compared with the relative abundance of BrdU-labeled nascent DNA chains (○) of the same size class.
Since these data were essentially the same as in Fig. 4 and 5, the ratios of RNA-primed nascent DNA to nonreplicating DNA were compared with the ratios of Br-DNA to nonreplicating DNA. The results revealed that the same replication origins, ori-β and ori-β′, were identified by either definition of nascent DNA (Fig. 6B). Digestion with λ-exonuclease was required to detect ori-β and ori-β′.
DISCUSSION
Visual inspection of the results of neutral-neutral 2D gel electrophoresis analyses of the DHFR gene region (17, 18, 56) suggests that a high frequency of initiation events occurs within each of two adjacent ∼6.1-kb EcoRI restriction fragments (F′ and F [Fig. 3]) but only fragment F′ appeared to contain an OBR (ori-β). The results presented here confirm previously published work that ori-β is a primary initiation site for DNA replication, and they extend it with the discovery of a second, previously unrecognized initiation site (ori-β′) 5 kb downstream in fragment F (Fig. 3). Thus, the DHFR initiation zone contains at least three primary initiation sites (ori-β, ori-β′, and ori-γ) within a 28-kb locus. Taken together with similar results from the rRNA gene region (see the introduction), these results show that initiation zones in mammalian chromosomes (like those in yeast) consist of multiple primary initiation sites (OBRs). In addition, there may be low-frequency initiation sites that escape detection by biolabeling methods but not by 2D gel electrophoresis methods.
These results support the hypothesis that while many potential initiation sites exist in metazoan DNA, in vivo some of these potential initiation sites are suppressed while others are activated (Jesuit model) (12, 14). Site-specific initiation of DNA replication at ori-β can be activated de novo in isolated nuclei by soluble Xenopus egg factors (25), but not until the nuclei have entered late G1 phase (43a, 58, 59), suggesting that assembly of prereplication complexes (46) is not complete in mammalian cells until late G1. Thus, to the extent that prereplication complexes form at alternative sites, initiation events may also occur at other, secondary initiation sites. Four parameters have been implicated in the selection of initiation sites: nuclear structure (25, 26, 58), chromatin structure (42), DNA sequences (14), and DNA methylation (48, 49). Developmental changes in one or more of these parameters could account for the observation that initiation of DNA replication in the Xenopus rRNA gene repeats changes from nonspecific to site specific following the midblastula transition (39).
ori-β is a primary initiation site for DNA replication.
The presence of a DNA replication origin downstream of the DHFR gene in CHO cells was first suggested by the discovery that two restriction fragments within a 28-kb region downstream of the DHFR gene incorporate radioactive DNA precursors in the first 20 min of the S phase (33). These fragments coincide with ori-β and ori-γ. The 12-kb fragment closest to the DHFR gene (ori-β) replicated within the first 10 min of the S phase (4). DNA labeled in vivo during the first 2 min of S phase (4) and putative replication forks from early S-phase cells labeled in vitro (5) hybridized preferentially to a 4.5-kb XbaI fragment, suggesting that this fragment contained an origin of replication (Fig. 7B, gray rectangle). This locus was later refined to a 2-kb BamHI-HindIII fragment (43) by using an in-gel renaturation method that reduced background (Fig. 7B, solid rectangle) and then to a 0.5-kb PvuII DNA fragment (Fig. 7F) by introducing DNA cross-links to prevent replication bubbles from expanding beyond their origin.
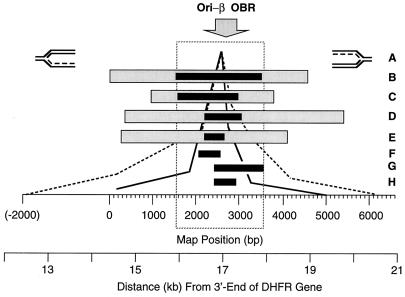
FIG. 7.
Comparison of results from the nascent DNA strand abundance assay with results from other nascent DNA strand origin-mapping strategies. Six different methods for mapping replication origins reveal a primary initiation site for DNA replication in CHO cells that can be localized to a 2-kb region encompassing the original ori-β OBR identified by the transition between leading- and lagging-strand synthesis ( ) (6). (A) Leading-strand distribution (7, 30); (B) earliest labeled DNA fragment (4, 5, 25, 34, 43, 59); (C) nascent DNA strand length (53); (D) nascent DNA strand abundance in unsynchronized cells (47); (E) nascent DNA strand abundance in synchronized cells (this paper) (–––, Br-DNA; ——, RNA-DNA); (F) replication bubble trap (1); (G and H) Okazaki fragment distribution (6, 52). Lightly shaded rectangles indicate the outer limits of the origin, while solid rectangles indicate the region of maximum origin activity. For example, the outer limits of the initiation site described here (E) are between primer sites 10 and 5, while the center is between primer sites 8 and 7 (Fig. 6). These data show that most initiation events occur within the 2-kb locus indicated by the dashed box. The highest Br-DNA/nonreplicating DNA ratios were routinely observed at probe 6 (nucleotides 2464 to 2619). The ori-β OBR is located between nucleotides 2431 and 2914, ∼17 kb downstream of the DHFR gene.
) (6). (A) Leading-strand distribution (7, 30); (B) earliest labeled DNA fragment (4, 5, 25, 34, 43, 59); (C) nascent DNA strand length (53); (D) nascent DNA strand abundance in unsynchronized cells (47); (E) nascent DNA strand abundance in synchronized cells (this paper) (–––, Br-DNA; ——, RNA-DNA); (F) replication bubble trap (1); (G and H) Okazaki fragment distribution (6, 52). Lightly shaded rectangles indicate the outer limits of the origin, while solid rectangles indicate the region of maximum origin activity. For example, the outer limits of the initiation site described here (E) are between primer sites 10 and 5, while the center is between primer sites 8 and 7 (Fig. 6). These data show that most initiation events occur within the 2-kb locus indicated by the dashed box. The highest Br-DNA/nonreplicating DNA ratios were routinely observed at probe 6 (nucleotides 2464 to 2619). The ori-β OBR is located between nucleotides 2431 and 2914, ∼17 kb downstream of the DHFR gene.
One caveat in these studies was the use of synchronized CHOC 400 cells containing ∼1,000 tandem integrated copies of the DHFR gene region. Either the process of cell synchronization or the presence of an excessive number of gene copies might artifactually induce origin specificity. Therefore, Vassilev et al. (53) used PCR to measure the lengths of nascent DNA strands in the ori-β region of unsynchronized, single-copy CHO cells containing a single copy of the DHFR gene region per haploid genome. They concluded that long nascent DNA strands expanded bidirectionally from ori-β, and they mapped the origin of this expansion to a 1.4- to 2.8-kb region of the 4.5-kb XbaI fragment identified in earlier studies (Fig. 7C). Later, Pelizon et al. (47) used competitive PCR to quantify the relative number of ∼1-kb BrdU-labeled nascent DNA strands at seven probes distributed over a 25-kb region (Fig. 7D and E). The fact that both of these studies were done with unsynchronized CHO cells shows that ori-β is a primary initiation site during cell proliferation. In the present study, the same nascent-strand abundance assay was applied to synchronized CHO cells at 20 probes in the 13-kb region containing ori-β. At least 95% of initiation events detected in this region (Fig. 5D) were localized to two primary initiation sites, ori-β and ori-β′. ori-β activity was confined to a 2- to 4-kb locus centered at the 786 bp between primer sets 8 and 7 (map positions 2020 to 2805), coincident with the OBR originally identified by analysis of the distribution of Okazaki fragments in this region (6). These results are in excellent agreement with those of Pelizon et al. (47). Recently, sites corresponding to ori-β plus ori-β′ and ori-γ have been identified by using fluorescence in situ hybridization analysis to map sites of BrdU incorporation relative to the DHFR gene in CHO cells (57a).
Compelling evidence that DNA replication in metazoan chromosomes originates at specific sites by using the classic replication fork mechanism comes from measurements of replication fork polarity. A transition from discontinuous to continuous DNA synthesis (the OBR) occurs on each DNA strand where bidirectional replication is initiated. When the leading-strand bias was measured by preferential inhibition of Okazaki fragment synthesis with emetine, at least 85% of replication forks 5 kb upstream and 9 kb downstream of ori-β were traveling away from ori-β in both CHO and CHOC 400 cells (Fig. 7A). This OBR was further refined by measuring the Okazaki fragment bias. At least 80% of the replication forks within a 27-kb segment emanated from an OBR located within a 0.45-kb region ∼17 kb downstream from the DHFR gene (Fig. 7G and H). The Okazaki fragment bias has been used to map an OBR to a few nucleotides in SV40 (15, 31), polyomavirus (15, 35, 36), and Saccharomyces cerevisiae (22). In each case, the OBR was coincident with the binding site for replication initiation proteins, suggesting that the ori-β OBR is a site where prereplication complexes assemble.
Comparison of results from all 12 studies revealed that the regions of maximum origin activity define a 2-kb region encompassing the original ori-β OBR (6) that was identified by the transition between leading- and lagging-strand synthesis (Fig. 7, dashed box). This 2-kb locus contains several features that may be related to its origin activity (14, 32, 49), but so far none have been proven to be required.
Interpretation of nascent DNA strand analyses.
Why was ori-β′ not detected in any of the previous nascent-strand analyses? In previous nascent-strand abundance assays (47), the probes were not located in sites that would have detected ori-β′. In early-labeled DNA fragment assays and nascent DNA strand length assays, ori-β′, which is three- to fourfold less active than and only 5 kb distant from ori-β, simply contributed to a broadening of the peak defining ori-β. In fork polarity assays, the bias in leading strands from asynchronous cells was lost at probes close to ori-β (30) and the Okazaki fragment bias close to ori-β was difficult to observe unless the cells were well synchronized (6), consistent with a second OBR close to ori-β. In a recent attempt to observe the transition from continuous to discontinuous DNA synthesis at the ori-β OBR, the expected strong Okazaki fragment bias was observed on the DHFR gene side of ori-β but not on the downstream side containing ori-β′ and ori-γ (56). Less than optimal cell synchronization could account for this problem, particularly when exacerbated by omission of the Br-DNA affinity purification step in the Okazaki fragment distribution method (56). Under these conditions, Okazaki fragments anneal to pieces of contaminating DNA in solution, which reduces their hybridization to filter-bound probes, preventing quantification with small probes (see e.g., Fig. 6 in reference 56). Originally, it was thought that the ori-β OBR could be mapped in exponentially growing CHOC 400 cells by the Okazaki fragment distribution method (6), but subsequent studies revealed that CHOC 400 cells were actually synchronized in early S phase under the conditions used (25). The importance of these considerations is evident from studies on SV40 and polyomavirus DNA replication, where even with a single origin, various parameters limited the observed strand bias to ∼6:1 (36).
A general problem in mapping replication origins is interference from DNA damage and repair. For example, λ-exonuclease was required to eliminate fragments of unreplicated DNA in order to map the ARS1 OBR in yeast (3). This problem is more pronounced in mammalian cells when they are synchronized at their G1/S boundary with inhibitors of DNA synthesis such as aphidicolin and mimosine that arrest replication forks after initiation of replication has occurred (10, 27, 38, 57). This results in damaged DNA, including breaks at replication forks (27, 38, 40), and could account for the low ratio of replication bubbles to forks observed at ori-β and ori-β′ 4 min after the cells were released from aphidicolin (56). Cells appear to recover from aphidicolin synchronization after 30 min (55) and from mimosine after 60 to 90 min into the S phase (17). Some cells accumulate in the G2 phase (Fig. 2), presumably as a result of checkpoint controls (11). Thus, while the Br-DNA affinity purification step can be omitted from the nascent-strand abundance assay with unsynchronized cells (23), we found that either affinity purification of BrdU-labeled nascent DNA fragments or elimination of broken DNA with λ-exonuclease was required to detect ori-β and ori-β′ in synchronized cells. Therefore, it is not surprising that the signal at ori-β relative to nearby sequences can vary from negligible (56) to 2- to 3-fold (25, 43) to 5- to 10-fold (1, 4, 5) in early-labeled fragment assays. Resolution in these assays is strongly dependent on cell synchrony, on allowing cells to recover from the synchronization protocol, and on blocking extensive elongation.
A major concern in the nascent-strand abundance assay is artifactual overrepresentation of nascent DNA at some sequences relative to others. In the experiments described here, this problem was eliminated as follows. (i) Repetitive sequence elements were avoided in the selection of PCR probes. This was verified by the production of a single major PCR product and by the equal representation of all 26 target sites in nonreplicating DNA from G0 cells (1.14 ± 0.08 copies/site [Fig. 5A and B). (ii) The ratio of nascent DNA to nonreplicating DNA was measured at each target site. This eliminated artifacts from errors in determining the concentration of different competitor DNAs and from overestimates of origin activity if some target sites were components of active origins elsewhere in the genome. (iii) The same peaks of origin activity were observed when two different definitions of nascent DNA were used (Fig. 5 and 6). Therefore, the high frequency of nascent DNA at ori-β and ori-β′ did not result from DNA labeled by damage and repair and did not result from preferential PCR amplification of BrdU-labeled DNA relative to the same, nonreplicated sequence.
If the ori-β OBR (transition from discontinuous to continuous DNA synthesis) occurred within a few base pairs (as it does in SV40, polyomavirus, and yeast), most of the nascent DNA chains should map within a ∼2-kb-wide peak (failure in Okazaki fragment ligation will produce some 1-kb chains that begin at the center of the replication bubble and extend in either direction [Fig. 1A]). This model is supported by the 5′-RNA-primed nascent DNA strand map (Fig. 6). The BrdU-labeled nascent DNA strands mapped within a ∼4-kb-wide peak (Fig. 5), suggesting that they were contaminated by broken pieces of Br-DNA from larger replication bubbles. Most of these broken pieces would lack a 5′-RNA primer. Both sets of data show that initiation events at the ori-β OBR are at least 10 to 12 times more frequent than at distal sequences, and they suggest that most initiation events at ori-β occur within a 2-kb locus centered at the OBR.
Interpretation of 2D gel analyses.
Why have 2D gel analyses failed to identify primary initiation events within the DHFR gene initiation zone? Although there are many technical differences among the various methods (reviewed in references 13 and 54), each method yields reproducible results at the same sites when carried out by different laboratories, as well as consistent results at other chromosomal sites, regardless of whether single-copy or multiple-copy cells are used, whether cells are synchronized or not, or whether DNA is labeled in vivo or in vitro. Therefore, neither specific OBRs nor broad initiation zones can be dismissed easily as experimental artifacts. Thus, the question whether the frequency of initiation events at an OBR differs significantly from the frequency of initiation events elsewhere revolves around quantification of data and resolution.
The following comments offer some insight. 2D gel electrophoresis patterns represent a photograph of all DNA structures that exist at a particular time, whereas analyses of DNA labeled during its biosynthesis reveal only the active replication structures. Moreover, 2D gel electrophoresis analyses on mammalian cells are generally carried out 1 to 2 h after cells are released into S phase, when the signal from replication bubbles is greatest, while nascent-strand analyses are generally carried out 4 to 20 min into S phase. Thus, while 2D gel electrophoresis analyses focus on the most prominent structures in early S phase, nascent-strand analyses focus on the most active ones at the beginning of S phase. However, since 2D gel electrophoresis, nascent-strand length, and nascent-strand abundance assays on nonsynchronized cells were essentially the same as those on synchronized cells, neither OBRs nor initiation zones can be considered unique to a particular portion of the S phase.
Perhaps of greater significance is the fact that 2D gel electrophoresis analyses of DNA replication in mammalian and frog genomes depends completely upon enriching for replication intermediates by their association with nuclear matrix and benzoylated-naphthoylated-DEAE cellulose prior to their analysis. These methods select for single-stranded sequences. In contrast, nascent-strand assays select only for newly synthesized DNA. Therefore, if the extent of single-stranded DNA varies as replication bubbles expand, representation of bubbles and forks in 2D gel electrophoresis will vary. Furthermore, if the tacit assumption that replication origins (prereplication complexes) as well as replication forks are attached to nuclear matrix is false, a finite time or distance will elapse before replication forks associate with matrix (i.e., before they form replication factories). This could account for the absence of replication intermediates on 2D gel electrophoresis of cells arrested in mimosine (17). This could also account for the fact that small replication bubbles are underrepresented in 2D gel electrophoresis patterns of mammalian and Xenopus DNA but not in 2D gel electrophoresis patterns either of plasmid DNA replication in Xenopus egg extract or of DNA amplification in flies, studies where the nuclear matrix enrichment step was omitted. Thus, while visual inspection of 2D gel electrophoresis data indicates that most replication bubbles occur in the vicinity of an OBR (see the introduction), the need to enrich for replication intermediates may reduce their resolution.
With these considerations in mind, the results reported here, together with those of 13 other studies involving six different origin-mapping methods (Fig. 7), strongly support a reconciliation between these data and those from 2D gel electrophoresis based on the concept that initiation zones consist of one or more primary initiation sites surrounded by several secondary initiation sites.
ACKNOWLEDGMENT
T.R. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Anachkova B, Hamlin J L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989;9:532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson S, DePamphilis M L. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J Biol Chem. 1979;254:11495–11504. [PubMed] [Google Scholar]
- 3.Bielinsky A K, Gerbi S A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- 4.Burhans W C, Selegue J E, Heintz N H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci USA. 1986;83:7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burhans W C, Selegue J E, Heintz N H. Replication intermediates formed during initiation of DNA synthesis in methotrexate-resistant CHOC 400 cells are enriched for sequences derived from a specific, amplified restriction fragment. Biochemistry. 1986;25:441–449. doi: 10.1021/bi00350a025. [DOI] [PubMed] [Google Scholar]
- 6.Burhans W C, Vassilev L T, Caddle M S, Heintz N H, DePamphilis M L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- 7.Burhans W C, Vassilev L T, Wu J, Sogo J M, Nallaseth F S, DePamphilis M L. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991;10:4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffman F D, Georgoff I, Fresa K L, Sylvester J, Gonzalez I, Cohen S. In vitro replication of plasmids containing human ribosomal gene sequences: origin localization and dependence on an aprotinin-binding cytosolic protein. Exp Cell Res. 1993;209:123–132. doi: 10.1006/excr.1993.1292. [DOI] [PubMed] [Google Scholar]
- 9.Contreas G, Giacca M, Falaschi A. Purification of BrdUrd-substituted DNA by immunoaffinity chromatography with anti-BrdUrd antibodies. BioTechniques. 1992;12:824–826. [PubMed] [Google Scholar]
- 10.Dai Y, Gold B, Vishwanatha J K, Rhode S L. Mimosine inhibits viral DNA synthesis through ribonucleotide reductase. Virology. 1994;205:210–216. doi: 10.1006/viro.1994.1636. [DOI] [PubMed] [Google Scholar]
- 11.Dasso M, Newport J W. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- 12.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 13.DePamphilis M L, editor. Identification and analysis of replication origins in eukaryotic cells. Vol. 13. San Diego, Calif: Academic Press, Inc.; 1997. [Google Scholar]
- 14.DePamphilis M L. Origins of DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- 15.DePamphilis M L, Martínez-Salas E, Cupo D Y, Hendrickson E A, Fritze C E, Folk W R, Heine U. Initiation of polyomavirus and SV40 DNA replication, and the requirements for DNA replication during mammalian development. In: Stillman B, Kelly T, editors. Eukaryotic DNA replication. Vol. 6. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 165–75. [Google Scholar]
- 16.DePamphilis M L, Wassarman P M. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- 17.Dijkwel P A, Hamlin J L. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol Cell Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dijkwel P A, Hamlin J L. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol Cell Biol. 1992;12:3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diviacco S, Norio P, Zentilin L, Menzo S, Clementi M, Biamonti G, Riva S, Falaschi A, Giacca M. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene. 1992;122:313–320. doi: 10.1016/0378-1119(92)90220-j. [DOI] [PubMed] [Google Scholar]
- 20.Dubey D D, Zhu J, Carlson D L, Sharma K, Huberman J A. Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J. 1994;13:3638–3647. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gencheva M, Anachkova B, Russev G. Mapping the sites of initiation of DNA replication in rat and human rRNA genes. J Biol Chem. 1996;271:2608–2614. doi: 10.1074/jbc.271.5.2608. [DOI] [PubMed] [Google Scholar]
- 22.Gerbi S A, Bielinsky A-K. Replication initiation point mapping. Methods. 1997;13:271–280. doi: 10.1006/meth.1997.0526. [DOI] [PubMed] [Google Scholar]
- 23.Giacca G, Pelizon C, Falaschi A. Mapping replication origins by quantifying the relative abundance of nascent DNA strands using the competitive polymerase chain reaction. Methods. 1997;13:301–312. doi: 10.1006/meth.1997.0529. [DOI] [PubMed] [Google Scholar]
- 24.Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci USA. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert D M, Miyazawa H, DePamphilis M L. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert D M, Miyazawa H, Nallaseth F S, Ortega J M, Blow J J, DePamphilis M L. Site-specific initiation of DNA replication in metazoan chromosomes and the role of nuclear organization. Cold Spring Harbor Symp Quant Biol. 1993;58:475–485. doi: 10.1101/sqb.1993.058.01.054. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert D M, Neilson A, Miyazawa H, DePamphilis M L, Burhans W C. Mimosine arrests DNA synthesis at replication forks by inhibiting deoxyribonucleotide metabolism. J Biol Chem. 1995;270:9597–9606. doi: 10.1074/jbc.270.16.9597. [DOI] [PubMed] [Google Scholar]
- 28.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gogel E, Langst G, Grummt I, Kunkel E, Grummt F. Mapping of replication initiation sites in the mouse ribosomal gene cluster. Chromosoma. 1996;104:511–518. doi: 10.1007/BF00352115. [DOI] [PubMed] [Google Scholar]
- 30.Handeli S, Klar A, Meuth M, Cedar H. Mapping replication units in animal cells. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- 31.Hay R T, DePamphilis M L. Initiation of SV40 DNA replication in vivo: location and structure of 5′ ends of DNA synthesized in the ori region. Cell. 1982;28:767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- 32.Heintz N H. DNA replication in mammals. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 983–1004. [Google Scholar]
- 33.Heintz N H, Hamlin J L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci USA. 1982;79:4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heintz N H, Stillman B W. Nuclear DNA synthesis in vitro is mediated via stable replication forks assembled in a temporally specific fashion in vivo. Mol Cell Biol. 1988;8:1923–1931. doi: 10.1128/mcb.8.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrickson E A, Fritze C E, Folk W R, DePamphilis M L. The origin of bidirectional DNA replication in polyoma virus. EMBO J. 1987;6:2011–2018. doi: 10.1002/j.1460-2075.1987.tb02465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrickson E A, Fritze C E, Folk W R, DePamphilis M L. Polyoma virus DNA replication is semi-discontinuous. Nucleic Acids Res. 1987;15:6369–6385. doi: 10.1093/nar/15.16.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez P, Martin-Parras L, Martinez-Robles M L, Schvartzman J B. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993;12:1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes T A, Cook P R. Mimosine arrests the cell cycle after cells enter S-phase. Exp Cell Res. 1996;222:275–280. doi: 10.1006/excr.1996.0035. [DOI] [PubMed] [Google Scholar]
- 39.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 40.Kalejta R F, Hamlin J L. The dual effect of mimosine on DNA replication. Exp Cell Res. 1997;231:173–183. doi: 10.1006/excr.1996.3444. [DOI] [PubMed] [Google Scholar]
- 41.Kohler T, Rost A K, Remke H. Calibration and storage of DNA competitors used for contamination-protected competitive PCR. BioTechniques. 1997;23:722–726. doi: 10.2144/97234st07. [DOI] [PubMed] [Google Scholar]
- 42.Lawlis S J, Keezer S M, Wu J-R, Gilbert D M. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. J Cell Biol. 1996;135:1207–1218. doi: 10.1083/jcb.135.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leu T H, Hamlin J L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989;9:523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Li, C.-J., and M. L. DePamphilis. Unpublished results.
- 44.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L, Tower J. A transcriptional insulator element, the su(Hw) binding site, protects a chromosomal DNA replication origin from position effects. Mol Cell Biol. 1997;17:2202–2206. doi: 10.1128/mcb.17.4.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 47.Pelizon C, Diviacco S, Falaschi A, Giacca M. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol Cell Biol. 1996;16:5358–5364. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rein T, Natale D A, Gartner U, Niggemann M, DePamphilis M L, Zorbas H. Absence of an unusual “densely methylated island” at the hamster dhfr ori-beta. J Biol Chem. 1997;272:10021–10029. doi: 10.1074/jbc.272.15.10021. [DOI] [PubMed] [Google Scholar]
- 49.Rein T, Zorbas H, DePamphilis M L. Active mammalian replication origins are associated with a high-density cluster of mCpG dinucleotides. Mol Cell Biol. 1997;17:416–426. doi: 10.1128/mcb.17.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Staib C, Grummt F. Mapping replication origins by nascent DNA strand length. Methods. 1997;13:293–300. doi: 10.1006/meth.1997.0528. [DOI] [PubMed] [Google Scholar]
- 52.Tasheva E S, Roufa D J. A mammalian origin of bidirectional DNA replication within the Chinese hamster RPS14 locus. Mol Cell Biol. 1994;14:5628–5635. doi: 10.1128/mcb.14.9.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vassilev L T, Burhans W C, DePamphilis M L. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol. 1990;10:4685–4689. doi: 10.1128/mcb.10.9.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilev L T, DePamphilis M L. Guide to identification of origins of DNA replication in eukaryotic cell chromosomes. Crit Rev Biochem Mol Biol. 1992;27:445–472. doi: 10.3109/10409239209082569. [DOI] [PubMed] [Google Scholar]
- 55.Vaughn J P, Dijkwel P A, Hamlin J L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- 56.Wang S, Dijkwel P A, Hamlin J L. Lagging-strand, early-labelling, and two-dimensional gel assays suggest multiple potential initiation sites in the Chinese hamster dihydrofolate reductase origin. Mol Cell Biol. 1998;18:39–50. doi: 10.1128/mcb.18.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Zhao J, Clapper J, Martin L D, Du C, DeVore E R, Harkins K, Dobbs D L, Benbow R M. Mimosine differentially inhibits DNA replication and cell cycle progression in somatic cells compared to embryonic cells of Xenopus laevis. Exp Cell Res. 1995;217:84–91. doi: 10.1006/excr.1995.1066. [DOI] [PubMed] [Google Scholar]
- 57a.Windle, B., and I. Parra. Personal communication.
- 58.Wu J R, Gilbert D M. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- 59.Wu J R, Gilbert D M. The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1-phase event that precedes restriction point control. Mol Cell Biol. 1997;17:4312–4321. doi: 10.1128/mcb.17.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon Y, Sanchez J A, Brun C, Huberman J A. Mapping of replication initiation sites in human ribosomal DNA by nascent-strand abundance analysis. Mol Cell Biol. 1995;15:2482–2489. doi: 10.1128/mcb.15.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Y, Tsutsumi R, Yamaki M, Nagatsuka Y, Ejiri S, Tsutsumi K. Initiation zone of DNA replication at the aldolase B locus encompasses transcription promoter region. Nucleic Acids Res. 1994;22:5385–5390. doi: 10.1093/nar/22.24.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu J, Brun C, Kurooka H, Yanagida M, Huberman J A. Identification and characterization of a complex chromosomal replication origin in Schizosaccharomyces pombe. Chromosoma. 1992;102:S7–S16. doi: 10.1007/BF02451780. [DOI] [PubMed] [Google Scholar]