Fassl and Sicinski highlight work from Pang and colleagues that shows that the RNA-binding protein PC4 modulates cyclin D1 levels in cancer cells by regulating mRNA stability.
Abstract
The expression of cyclin proteins is tightly regulated during the cell cycle, to allow precise activation of cyclin-dependent kinases. In this issue, Pan et al. (https://doi.org/10.1083/jcb.202308066) identify an RNA-binding protein, PC4, as a regulator of cyclin D1 mRNA stability in hepatocellular carcinoma cells. This study provides a new mechanism regulating the levels of a key cell cycle protein, cyclin D1, in human cells.
Cyclin D1 is a regulator of mammalian cell cycles that links extracellular growth-controlling (mitogenic and anti-mitogenic) signals to the cell core cell cycle machinery (1). Growth-promoting signals acting through receptor tyrosine kinases (e.g., EGFR, HER2) or nuclear receptors (e.g., estrogen receptor, androgen receptor) lead to the induction of cyclin D1 expression, which constitutes the “go” signal for cell proliferation. Cyclin D1 binds and activates cyclin-dependent kinases CDK4 and CDK6. Cyclin D–CDK4/6 complexes subsequently phosphorylate the retinoblastoma protein RB1. In its hypophosphorylated state, RB1 binds the E2F transcription factors and blocks their activity. Phosphorylation of RB1 weakens and subsequently abolishes the interaction of RB1 with E2Fs, thereby allowing initiation of S phase through transcription of genes necessary for DNA replication.
Given the prominent role of cyclin D1 in cell cycle initiation, it is not surprising that cyclin D1 can act as an oncogene. Indeed, cyclin D1 is one of the most frequently overexpressed proteins in cancer. The oncogenic function of cyclin D1 in vivo was first demonstrated using transgenic mice engineered to overexpress cyclin D1 in their mammary epithelium; these mice are prone to develop mammary tumors (2). In contrast, mice lacking expression of cyclin D1 are fully resistant to the development of HER2-driven breast cancers (3). These observations led to the idea of using inhibitors of cyclin D1–CDK4/6 as a therapeutic strategy for breast cancer treatment. As the result of clinical trials, CDK4/6 inhibitors have been approved for the treatment of women with hormone receptor-positive breast cancers and are in clinical trials for several other cancer types.
Given the oncogenic potential of cyclin D1, its activity must be tightly controlled. Indeed, expression, activation, stabilization, degradation, and localization of cyclin D1 are strictly regulated throughout the cell cycle to ensure its precise activation (and inactivation) at the correct time. During G1 phase, cyclin D1 becomes induced, binds its kinase partners CDK4 and CDK6, and accumulates in cell nucleus. Upon S phase entry, cyclin D1 undergoes nuclear export and subsequent degradation of the majority, but not all protein, which plays additional roles beyond the G1 phase. Recently, the ubiquitin ligase AMBRA1 was added to the portfolio of proteins controlling cyclin D1 levels (4–6).
In this issue, Pan et al. (7) report that regulation of mRNA stability represents another mechanism controlling cyclin D1 expression. Through unbiased analysis of the expression of RNA-binding proteins in hepatocellular carcinoma (HCC), the authors identified positive cofactor 4 (PC4) as a protein that is overexpressed in HCC and correlates with poor prognosis. Depletion of PC4 had a strong growth inhibitory effect in HCC cells. PC4 RNA-immunoprecipitation sequencing revealed that RNAs bound to PC4 are enriched for cell cycle–related transcripts, in particular the ones encoding genes implicated in G1/S phase transition. Among them, cyclin D1, SKP1, SKP2, and E2F2 mRNAs bound to PC4. Depletion of PC4 decreased the stability of its bound mRNAs, while PC4 overexpression increased their half-life. These observations indicate that PC4 functions as a posttranscriptional regulator of cell cycle mRNAs’ stability.
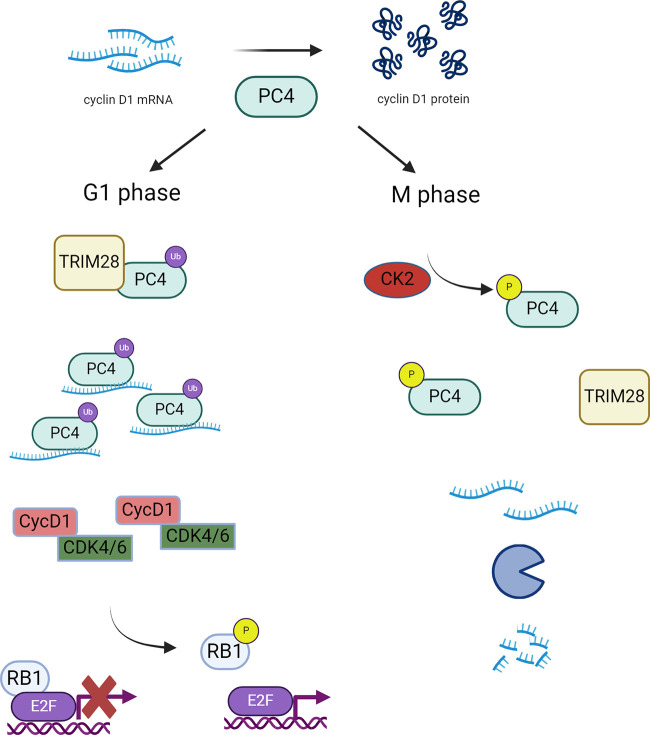
In their molecular analyses, the authors focused on cyclin D1 transcripts. The levels of cyclin D1 mRNA fluctuate during the cell cycle. Strikingly, these fluctuations were abolished in cells depleted of PC4, suggesting that PC4 is responsible for cell cycle–dependent control of cyclin D1 transcript levels. Since the levels of PC4 protein remain constant throughout the cell cycle, the authors hypothesized that the activity of PC4 must be regulated by some posttranslational modifications. Through mass spectrometry analysis of PC4 immunoprecipitated from HCC cells synchronized in different cell cycle phases, the authors identified two periodically changing modifications on PC4 with opposing patterns—ubiquitination of lysine 68 (K68) (present during the interphase) and phosphorylation of serine 17 (S17) during mitosis. The authors found that K68 ubiquitination strengthens the binding between PC4 and cyclin D1 mRNA, thereby promoting cyclin D1 mRNA stability. In contrast, S17-phosphorylated PC4 binds cyclin D1 transcripts very weakly and is unable to stabilize them.
The authors found that the RING domain-containing E3 ubiquitin ligase TRIM28 is responsible for K68 ubiquitination of PC4, while casein kinase 2 (CK2) phosphorylates S17 of PC4. Intriguingly, S17-phosphorylated PC4 is unable to interact with TRIM28. Hence, this modification of PC4 prevents ubiquitination at K68 and renders PC4 unable to stabilize cyclin D1 mRNA. According to the model proposed by Pan et al. (7), CK2 activity acts as a molecular switch regulating cyclin D1 mRNA stability. TRIM28-mediated ubiquitination of PC4 confers stabilization of cyclin D1 mRNA during interphase, and this function of PC4 is switched off by CK2 as the cells enter mitosis (Fig. 1). This results in degradation of cyclin D1 mRNA upon mitotic entry.
Figure 1.
PC4 regulates cyclin D1 mRNA stability in HCC cells. PC4 is regulated through a ubiquitination (Ub)/phosphorylation (P) switch. In G1 phase, TRIM28 K68-ubiquitinated PC4 binds and stabilizes cyclin D1 mRNA, leading to increased cyclin D1 protein expression. In M phase, PC4 becomes a target of CK2 phosphorylation. S17-phosphorylated PC4 is unable to bind TRIM28 and hence cannot become ubiquitinated. This reduces cyclin D1 transcript stability, resulting in lower cyclin D1 protein levels. Created with http://BioRender.com.
In addition to a mechanism physiologically regulating cyclin D1 levels across the cell cycle, aberrant ubiquitination/phosphorylation of PC4 may contribute to deregulated expression of cyclin D1 in cancer. Consistent with this notion, Pan et al. (7) demonstrated that in human HCC samples, K68 ubiquitination of PC4 is increased while S17 phosphorylation of PC4 is decreased compared to normal tissue.
It remains to be seen whether the mechanism described by Pan et al. (7) is specific to the liver, or if it is broadly employed in different cellular compartments. It is also unknown what is the contribution of deregulated cyclin D1 mRNA stability to driving uncontrolled tumor cell proliferation. Since PC4 may regulate transcript stability of multiple cell cycle–promoting genes, deregulated PC4 activity would be expected to promote cell division by upregulating the levels of several pro-proliferative transcripts.
Interestingly, Hirayama et al. (8) recently reported a fat mass and obesity-associated (FTO) –mediated mechanism of cyclin D1 mRNA stability regulation during cell cycle progression. This mechanism centers around N6-methyladenosine (m6A) modification of the cyclin D1 transcripts. According to Hirayama et al. (8), nuclear FTO removes m6A modification of cyclin D1 mRNA in the G1 phase, thereby stabilizing cyclin D1 transcripts. After the S phase entry, FTO is shuttled to the cytoplasm, thereby losing control over m6A modification and stability of cyclin D1 transcripts. An obvious question is whether the PC4- and FTO-dependent pathways are interconnected or independent of each other. Does m6A methylation status influence PC4 binding to cyclin D1 mRNA? Strikingly, nuclear export of FTO is controlled by CK2-mediated phosphorylation. Hence, for both FTO- and PC4-dependent mechanisms, CK2-driven phosphorylation—although occurring at different phases of the cell cycle (S phase for FTO, M phase for PC4)—blocks RNA stabilizing function, making CK2 a central player of cyclin D1 mRNA stability control. This is especially important as CK2 inhibition is considered as an anti-cancer strategy (9) and the first CK2 inhibitor (silmitasertib) has entered clinical trials (10). Based on the studies of Pan et al. (7) and Hirayama et al (8), one might expect that CK2 inhibition would cause stabilization of cyclin D1 mRNA and the resulting upregulation of cyclin D1 protein levels, thereby contributing to resistance or ineffectiveness of CK2 inhibition. Combination of CK2 inhibition with CDK4/6 inhibition might help to mitigate the undesired effect (11).
Treatment with CDK4/6 inhibitors represents the standard of care for women with advanced, hormone receptor-positive breast cancer and are being tested in clinical trials for several other cancer types (1). From the therapeutic standpoint, a particularly interesting outcome of the study of Pan et al. (7) is the demonstration that co-treatment of HCC xenografts with nanoparticles carrying PC4 siRNA together with a CDK4/6 inhibitor, palbociclib, reduced tumor burden, improved survival, and inhibited lung metastasis. Upregulation of several cell cycle proteins, including CDK6, cyclin E, or CDK2, represent frequent mechanisms of the acquired resistance of tumor cells to CDK4/6 inhibition (12, 13). Since PC4 depletion is expected to dampen the expression of multiple cell cycle–promoting genes, combining inhibition of CDK4/6 with depletion of PC4 may represent an attractive treatment option for HCC, as it may delay or prevent the acquisition of resistance mechanisms. If the observation of Pan et al. (7) could be confirmed in further preclinical models of HCC and maybe other solid tumor models, a combined inhibition of PC4 and CDK4/6 might represent a powerful treatment strategy.
Acknowledgments
A. Fassl is supported by the Mildred Scheel Career Center Frankfurt (Deutsche Krebshilfe). The writing of this article was supported by National Institutes of Health grant P01CA250959 to P. Sicinski.
References
- 1.Fassl, A., et al. 2022. Science. 10.1126/science.abc1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, T.C., et al. 1994. Nature. 10.1038/369669a0 [DOI] [Google Scholar]
- 3.Yu, Q., et al. 2001. Nature. 10.1038/35082500 [DOI] [Google Scholar]
- 4.Chaikovsky, A.C., et al. 2021. Nature. 10.1038/s41586-021-03474-7 [DOI] [Google Scholar]
- 5.Maiani, E., et al. 2021. Nature. 10.1038/s41586-021-03422-5 [DOI] [Google Scholar]
- 6.Simoneschi, D., et al. 2021. Nature. 10.1038/s41586-021-03445-y [DOI] [Google Scholar]
- 7.Pan, et al. 2024. J. Cell Biol. 10.1083/jcb.202308066 [DOI] [Google Scholar]
- 8.Hirayama, M., et al. 2020. Cell Rep. 10.1016/j.celrep.2020.03.028 [DOI] [Google Scholar]
- 9.Borgo, C., et al. 2021. Signal Transduct. Target. Ther. 10.1038/s41392-021-00567-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Amore, C., et al. 2020. Cell Oncol. 10.1007/s13402-020-00566-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padgaonkar, A., et al. 2018. Oncotarget. 10.18632/oncotarget.26514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner, N.C., et al. 2019. J. Clin. Oncol. 10.1200/JCO.18.00925 [DOI] [Google Scholar]
- 13.Cornell, L., et al. 2019. Cell Rep. 10.1016/j.celrep.2019.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]