Abstract
In Saccharomyces cerevisiae, PHO85 encodes a cyclin-dependent protein kinase (Cdk) with multiple roles in cell cycle and metabolic controls. In association with the cyclin Pho80, Pho85 controls acid phosphatase gene expression through phosphorylation of the transcription factor Pho4. Pho85 has also been implicated as a kinase that phosphorylates and negatively regulates glycogen synthase (Gsy2), and deletion of PHO85 causes glycogen overaccumulation. We report that the Pcl8/Pcl10 subgroup of cyclins directs Pho85 to phosphorylate glycogen synthase both in vivo and in vitro. Disruption of PCL8 and PCL10 caused hyperaccumulation of glycogen, activation of glycogen synthase, and a reduction in glycogen synthase kinase activity in vivo. However, unlike pho85 mutants, pcl8 pcl10 cells had normal morphologies, grew on glycerol, and showed proper regulation of acid phosphatase gene expression. In vitro, Pho80-Pho85 complexes effectively phosphorylated Pho4 but had much lower activity toward Gsy2. In contrast, Pcl10-Pho85 complexes phosphorylated Gsy2 at Ser-654 and Thr-667, two physiologically relevant sites, but only poorly phosphorylated Pho4. Thus, both the in vitro and in vivo substrate specificity of Pho85 is determined by the cyclin partner. Mutation of PHO85 suppressed the glycogen storage deficiency of snf1 or glc7-1 mutants in which glycogen synthase is locked in an inactive state. Deletion of PCL8 and PCL10 corrected the deficit in glycogen synthase activity in both the snf1 and glc7-1 mutants, but glycogen synthesis was restored only in the glc7-1 mutant strain. This genetic result suggests an additional role for Pho85 in the negative regulation of glycogen accumulation that is independent of Pcl8 and Pcl10.
In the budding yeast Saccharomyces cerevisiae, the PHO85 gene encodes a cyclin-dependent protein kinase (Cdk) with roles in both cell cycle and metabolic controls (43, 54). PHO85 was originally discovered because of its function in inorganic phosphate scavenging by nonspecific acid phosphatases such as Pho5 (76, 79). Pho85 regulates acid phosphatase gene expression when complexed with the cyclin Pho80 (44, 80). The Pho80-Pho85 kinase phosphorylates and negatively regulates Pho4, a transcription factor required for expression of PHO5 (38, 53). When phosphate is abundant, Pho4 is phosphorylated by Pho80-Pho85 and is mostly cytoplasmic, thus causing repression of PHO5 expression. When cells are starved for inorganic phosphate, Pho80-Pho85 is inhibited by the Cdk inhibitor Pho81 and transcription of PHO5 is activated (10, 11, 63). Thus, the Pho80 cyclin appears to specify the participation of Pho85 in phosphate metabolism.
In addition to causing constitutive PHO5 expression, deletion of PHO85 leads to a number of other phenotypic alterations. For example, pho85 strains grow poorly on glucose, have aberrant morphologies, and are larger than wild-type cells (47). They grow very slowly, compared to wild-type cells, on glycerol, ethanol, and acetate (21, 75) and, relevant to the present investigation, they hyperaccumulate the storage polysaccharide glycogen (33, 75). Diploid homozygous pho85 mutants do not sporulate. Not all of these phenotypes are associated with loss of PHO80, implying the involvement of other genes in discharging PHO85 functions. Indeed, nine other Pho85 cyclins, or Pcl’s, have been identified in addition to Pho80 (45, 47). Although the overall sequence identity is low among these proteins, all 10 Pcl proteins have a cyclin box, and phylogenetic analysis based on sequence alignment of this region has placed the Pcl’s into two families, the Pho80 family (Pho80, Pcl6, Pcl7, Pcl8, and Pcl10) and the Pcl1,2 family (Pcl1, Pcl2, Pcl5, Pcl9, and Clg1).
Pho85 is involved in regulation of the G1 phase of the cell cycle when complexed with the related cyclins Pcl1 and Pcl2 (14, 46). Entry into the cell cycle in late G1 phase is mainly controlled by the cyclin-dependent kinase Cdc28 and its associated G1 cyclins, Cln1, -2, and -3 (reviewed in references 51 and 52). Although cells lacking Pho85 or Pcl1 and Pcl2 are viable, Pcl1,2-Pho85 complexes are required for G1 progression in the absence of Cln1 and Cln2, suggesting a role for these kinases during G1 phase (14, 46). Recent work has also shown that expression of a related cyclin, PCL9, is cell cycle regulated, with peak transcript levels in late mitosis and early G1 phase (1, 71). Deletion of members of the Pcl1,2 subfamily of Pho85 cyclins leads to morphological abnormalities and budding defects, consistent with a role for these kinase complexes in proper cell morphogenesis during G1 phase (47, 71). The relevant substrates of Pcl1,2 subfamily kinases are unknown. In fact, except for Pho80, specific functions have not been ascribed to the Pho85 cyclins. We report here that two members of the Pho80 subfamily of Pho85 cyclins, Pcl8 and Pcl10, are involved in control of glycogen accumulation.
Glycogen is a storage carbohydrate synthesized prior to entry into stationary phase (18). Its biosynthesis requires several proteins, including the self-glucosylating initiator proteins Glg1 and Glg2 (6), the branching enzyme Glc3 (61, 74), and glycogen synthase (15, 16), which is generally considered to be the rate-limiting enzyme for glycogen accumulation. Yeast glycogen synthase is encoded by two closely related genes, GSY1 and GSY2, of which GSY2 appears to encode the dominant form, accounting for 90% of glycogen synthase activity at stationary phase (15, 16). Like its mammalian counterpart (65), yeast glycogen synthase is controlled by multisite phosphorylation that inactivates the enzyme (24). Three COOH-terminal residues, Ser-650, Ser-654, and Thr-667, have been implicated in control of Gsy2 activity in vivo (24). Full activity is restored to phosphorylated Gsy2 in the presence of the allosteric activator glucose-6-phosphate (glucose-6-P) so that the −/+ glucose-6-P activity ratio is often used as an index of the phosphorylation state of glycogen synthase. Dephosphorylation of glycogen synthase is thought to be mediated by a type I protein phosphatase (4, 17, 24, 57) encoded by GLC7, associated with the Gac1 (19, 68) or possibly the Pig1 (5) regulatory subunit.
Information regarding the protein kinase(s) responsible for modifying the three phosphorylation sites of Gsy2 is only now emerging. Cyclic AMP-dependent protein kinase can phosphorylate glycogen synthase in vitro, but it is not certain that this reaction is physiologically important (24, 57). More recently, two distinct Gsy2 kinase activities were partially purified from yeast extracts, and one of these contained a species that cross-reacted with antibodies to Pho85 (33). Deletion of PHO85 causes hyperaccumulation of glycogen and a significant reduction in the Gsy2 kinase activity measurable in yeast cell extracts (33, 75). Also, mutation of PHO85 suppresses the glycogen storage defect of a snf1 strain. SNF1 encodes a protein kinase which is required for the expression of glucose-repressible genes (37, 60) and which, in a separate pathway, regulates glycogen metabolism (4, 23, 73). Cells defective in Snf1 cannot accumulate glycogen synthase due to the inactivation and presumed hyperphosphorylation of glycogen synthase (23). Disruption of PHO85 in snf1 cells restores glycogen synthase activity to wild-type levels and allows normal glycogen accumulation (33, 75). We therefore suggested that Pho85 was a constituent of a major Gsy2 kinase that phosphorylated Ser-654 and Thr-667, two of the three phosphorylation sites in Gsy2 (33). We additionally postulated that there might be particular Pho85 cyclins that would specify this function. Based on both biochemical and genetic evidence, we now propose that Pcl8 and Pcl10 fulfill this role. We found that, in vitro, a Pcl10-Pho85 complex phosphorylated Gsy2 much more effectively than Pho4, whereas a Pho80-Pho85 complex selectively phosphorylated Pho4. In vivo, deletion of PCL8 and PCL10 caused glycogen hyperaccumulation but did not result in other phenotypic defects associated with disruption of PHO85. Thus, the in vivo specificities of Pho80 and Pcl10 were reflected in the substrate specificity that they imparted to Pho85 in vitro.
MATERIALS AND METHODS
Strains, media, and methods.
The S. cerevisiae strains used are listed in Table 1. In our experience, there is strain-to-strain variation in glycogen storage so that some backgrounds have more pronounced accumulation, which makes study of glycogen storage easier. We therefore preferred to analyze the glycogen-accumulating phenotype in strains related to EG328-1A. In addition, the pink or red coloration of strains with ade2 mutations affects the appearance of colonies after iodine staining, a common method for the semiquantitative analysis of glycogen accumulation. Standard rich medium (yeast extract-peptone-dextrose [YPD]) and supplemented minimal medium (SD) were used for most experiments. YP-glycerol has 3% glycerol instead of the dextrose used in YPD. Plasmids were maintained in Escherichia coli DH5α. Standard methods for yeast transformation and culture were used (22).
TABLE 1.
Yeast strainsa
| Strain(s) | Genotype | Reference or source |
|---|---|---|
| EG328-1A | MATα trp1 leu2 ura3-52 | K. Tatchell |
| DH4-101 | MATa trp1 ura3-52 thr4 MAL+ | 32 |
| DH28 | MATα trp1 leu2 ura3-52 pho85::URA3 | 33 |
| DH35-64 | MATa ura3-52 thr4 pho85::URA3 | This study |
| DH93-81, DH93-82 | MATα trp1 leu2 ura3-52 pcl10::URA3 | This study |
| DH96-11, DH96-52 | MATα trp1 leu2 ura3-52 pcl8::TRP1 | This study |
| DH97-13 | MATα trp1 leu2 ura3-52 pcl8::TRP1 pcl10::URA | This study |
| DH97-33 | MATa trp1 ura3-52 thr4 pcl8::TRP1 pcl10::URA3 | This study |
| EG353-1C | MATα trp1 leu2 ura3-52 snf1::URA3 | K. Tatchell |
| DH102-43, DH102-82 | MATα trp1 leu2 ura3-52 snf1::LEU2 pcl8::TRP1 | This study |
| DH103-31, DH103-33 | MATa trp1 leu2 ura3-52 thr4 snf1::LEU2 pcl10::URA3 | This study |
| DH104-104, DH104-163 | MATα trp1 leu2 ura3-52 snf1::LEU2 pcl8::TRP1 pcl10::URA3 | This study |
| EG327-1D | MATα trp1 leu2 ura3-52 glc7-1 | K. Tatchell |
| DH98-14 | MATa trp1 leu2 ura3-52 thr4 glc7-1 pcl8::TRP1 | This study |
| DH98-43 | MATa trp1 ura3-52 thr4 glc7-1 pcl8::TRP1 | This study |
| DH99-32, DH99-44 | MATα trp1 ura3-52 thr4 glc7-1 pcl10::URA3 | This study |
| DH100-61 | MATα trp1 leu2 ura3-52 glc7-1 pcl8::TRP1 pcl10::URA3 | This study |
| DH100-82 | MATα trp1 ura3-52 thr4 glc7-1 pcl8::TRP1 pcl10::URA3 | This study |
| BY546 | MATa ura3 trpl-1 ade2-1 his3-11,15 leu2-3,112 can1-100 GAL+ [psi+] pho85::hisG | D. Stillman |
| BY467 | MATa ura3 trp1-1 ade2-1 his3-11,15 leu2-3,112 can1-100 GAL+ [psi+] | B. Andrews |
| Y153 | MATa trp1 leu2 ura3 his3 ade2 gal4 gal80 URA3::GAL-lacZ LYS2::GAL-HIS3 | 12 |
| Y187 | MATα trp1 leu2 ura3 his3 ade2 gal4 gal80 URA3::GAL-lacZ | 25 |
| WW7 | MATatrp1LEU2ura3-52thr4 | This study |
| MATα trp1 leu2 ura3-52 THR4 | ||
| WW8 | MATaTRP1LEU2ura3-52thr4pho85::URA3 | This study |
| MATα trp1 leu2 ura3-52 THR4 pho85::URA3 | ||
| WW9 | MATatrp1LEU2ura3-52thr4pcl8::TRP1pcl10::URA3 | This study |
| MATα trp1 leu2 ura3-52 THR4 pcl8::TRP1 pcl10::URA3 |
EG328-1A, EG327-1D, and EG353-1C are isogenic. The DH and WW strains are all related to EG328-1A.
Gene disruptions and strain construction.
For disruption of PCL10, PCR was used to generate a DNA fragment from primers that contained 45 bp of the PCL10 sequence followed by 21 bp that matched pBluescript sequences straddling the chosen marker gene in an appropriate pRS plasmid. The URA3 gene on vector pRS306 (64) was used as the template for PCR. The primers used were CCA CAC CAC TGA CAC AGA GGA GTT TGA TGA TGG TGA TAT ACG TCC AGC AGA TTG TAC TGA GAG TGC (sense) and GGG AAG CTC TGA AGT TTT CTC CAA TAT CGA GTA CAG GTT TCC ACC CAT CTG TGC GGT ATT TCA CAC (antisense). The resulting PCR product contained the 5′ sequence (+18 to +63, referred to the open reading frame) and the 3′ sequence (+1439 to +1484) of PCL10 at each end of a 1.1-kb sequence containing the URA3 gene. This DNA fragment was then used to transform strain EG328-1A, to yield the PCL10-disrupted strain DH93 (Table 1). A similar strategy was employed for disruption of PCL8. The primers for gene disruption were CCA ACA AGT CTC TCA TTA ATG ACG CTT TGA CTC GGA GTA CGT CCTG AGC AGA TTG TAC TGA GAG TGC (sense) and GTT GCA GAC GGA GTT GTC GGG TAA TGC GGG CGT ATA CGA TAT AAT CAT CTG TGC GGT ATT TCA CAC (antisense). The TRP1 gene on vector pRS304 (64) was used as the template. The PCR products were used to transform the pcl10 strain DH93 to yield the pcl8::TRP1 pcl10::URA3 double-mutant strain DH97. The double mutant gave a strong glycogen-hyperaccumulating phenotype. To confirm the phenotype and to generate pcl8 single mutants, we crossed the double mutant DH97-33 with a wild-type strain, DH4-101. Tetrad analysis of the pcl8::TRP1 pcl10::URA3 heterozygote indicated a segregation ratio of 1:3 for the hyperaccumulation of glycogen for most of the tetrads. All spore clones with elevated glycogen levels were both Trp+ and Ura+. We generated pcl8 and/or pcl10 mutants in an snf1 or glc7-1 background in a similar manner, by crossing the double mutant DH97-33 with EG353-1C or EG327-1D, respectively.
Mutagenesis of Gsy2.
A two-step PCR protocol (27) was used to introduce mutations into GSY2 coding sequences to code for Asp instead of Ser-650 (AGT→GAT), Ser-654 (TCA→GAC), or Thr-667 (ACC→GAC). Three double mutants were constructed, S650D S654D, S650D T667D, and S654D T667D. The first double mutant was made by a single round of mutagenesis, whereas the other two were made by two sequential rounds, each mutating one of the phosphorylation sites. The outside primers for the secondary PCR contained a 5′ SacI site and a 3′ SmaI site, so that a 0.3-kb cassette could be excised from the secondary PCR product by digestion with SacI and SmaI and cloned into the GSY2 coding sequence. Gsy2 was contained in the pET28a-GSY2 vector (33), which was digested with SacI/XhoI, so as to cut at the SacI site noted above and at a XhoI site in the pET28a polylinker region. After filling in the XhoI site, the mutant SacI-SmaI cassette was ligated into the digested pET28a-GSY2 to introduce a mutated COOH terminus. All mutants were confirmed by sequencing of the region of the 0.3-kb cassette.
Two-hybrid analysis.
A yeast two-hybrid test (8, 12) was used to assay interactions between Gsy2 and various Pcl proteins. Yeast (Y153) bearing a plasmid expressing a GSY2 fusion (pGSY2 [6]) or the Gal4 DNA binding domain alone (pAS1) were mated to a strain (Y187) transformed with plasmids encoding Pcl fusion proteins identified in previous two-hybrid screens (47). Filter (6) or liquid (48) β-galactosidase assays were performed as previously described. A plasmid encoding a fusion of the Gal4 activation domain (AD) with the COOH-terminus of Glg2, which is known to give a positive signal in the assay (6), was used as a positive control.
Glycogen, glycogen synthase, and glycogen synthase kinase assays.
Yeast cells were grown in YPD and harvested in late log phase. Cells were resuspended in a homogenization buffer consisting of 50 mM Tris HCl, 1 mM EDTA, 3 mM dithiothreitol (DTT), 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone, 5 mM benzamidine hydrochloride, 0.25 μg of leupeptin per ml, and 0.5 μg of aprotinin per ml (pH 7.4). The cells were broken by shaking with glass beads, as described previously (24).
Glycogen synthase was assayed by the method of Thomas et al. (72), as described by Hardy et al. (23). The total activity of glycogen synthase is that measured in the presence of 7.2 mM glucose-6-P and is essentially proportional to the amount of protein present. The −/+ glucose-6-P activity ratio is defined as the activity measured in the absence of glucose-6-P divided by the activity measured in its presence. Glycogen was determined in extracts of cells as described by Hardy and Roach (24).
For assay of glycogen synthase kinase activity in cell extracts, the direct phosphorylation of added, purified recombinant Gsy2p was determined by analyzing the incorporation of 32P into Gsy2p from [γ-32P]ATP (36). The cells were resuspended in a homogenization buffer containing 50 mM Tris-HCl (pH 7.4), 0.1% (vol/vol) Triton X-100, 2 mM benzamidine hydrochloride, 1 mM PMSF, 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone, and 1 mM β-mercaptoethanol and broken with glass beads. Yeast extract (5 μl, diluted to ∼2.5 mg of protein/ml with homogenization buffer) was combined with 2.5 μg of His6Gsy2p and 100 nM okadaic acid in a final volume of 20 μl. The reaction was initiated by the addition of 5 μl of 1 mM [γ-32P]ATP mix (∼1,200 cpm/pmol) and 25 mM MgCl2. After incubation at 30°C for 15 min, 25 μl of a 1:1 slurry of Ni-nitrilotriacetic acid-agarose in wash buffer (50 mM Tris-HCl [pH 7.9], 0.1% [vol/vol] Triton X-100, 500 mM NaCl, 50 mM NaF, 50 mM imidazole, and protease inhibitors as above) was added, followed by 500 μl of ice-cold wash buffer, and the incubation was continued on ice for a further 30 min with occasional gentle agitation. The Ni-nitrilotriacetic acid-agarose was collected by centrifugation, and the pellet was washed four times with 500 μl of wash buffer. Bound His6Gsy2p was eluted by using 25 μl of wash buffer with the imidazole concentration increased to 500 mM. The eluted material was analyzed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE), followed by autoradiography.
Assay of Gsy2 and Pho4 phosphorylation by immunoprecipitated Pho85.
To analyze the activity of Pcl10-Pho85 complexes, cells were transformed with plasmid pAD-PCL10, which carries an AD-PCL10 fusion gene expressed from the ADH promoter (47). The encoded protein (AD-Pcl10) contains the Gal4 AD fused to the NH2 terminus of Pcl10. The first 27 NH2-terminal amino acids of Pcl10 are deleted in the fusion protein. As a control, cells were transformed with control vector (pACTII [2]), which carries a hemagglutinin (HA)-tagged Gal4 AD expressed from the ADH promoter. This vector is referred to as pAD. To analyze the activity of Pho80-Pho85 complexes, cells were transformed with pHA-PHO80, which encodes an HA-tagged Pho80 protein (HA-Pho80), as previously described (38). A URA3-based vector, pRS426 (9), was used as a control in the HA-Pho80 immunoprecipitation experiments.
Wild-type yeast cells (BY467) or cells of an isogenic pho85 disruptant (BY546) were transformed with appropriate plasmids and grown in selective minimal media to an optical density at 600 nm of approximately 1. Cells were collected by centrifugation, washed in cold sterile distilled H2O, and then stored at −80°C. Lysates were prepared essentially as described by Tyers et al. (78). Cell pellets were resuspended in 1 to 2 volumes of lysis buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1 mM DTT, 0.1% Nonidet P-40, 50 mM NaF, 5 mM EDTA and the following protease inhibitors: 1 mM PMSF, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml, 10 μg of soybean trypsin inhibitor per ml, 10 μg of N-tosyl-l-phenylalanine chloromethyl ketone per ml, and 0.6 mM dimethylaminopurine). Cells were broken with acid-washed glass beads (five to eight 60-s bursts) and centrifuged at 13,000 × g for 10 min, and the supernatant was used for immunoprecipitations. In a given experiment, samples were normalized to total protein content; about three times as many of pho85 cells as wild-type cells were needed to obtain sufficient protein. Lysates were incubated with 0.25 μl of 12CA5 ascitic fluid (monoclonal antibody to the influenza HA peptide) or 0.5 μl of 8CL-11 ascitic fluid (monoclonal antibody to the AD of Gal4) for 1 to 3 h on ice and then rocked in the presence of protein A-Sepharose beads for 1 to 3 h at 4°C. Beads were collected by centrifugation and washed four times in lysis buffer followed by two times in “kinase buffer” (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM DTT, and 10 μM ATP). The remaining liquid was removed by aspiration, and kinase reactions were initiated by adding 10 μl of kinase buffer containing 10 μCi of [γ-32P]ATP (0.1 Ci/μmol) and either 100 ng or 1 μg of purified Gsy2 or Pho4, as indicated in the legend to Fig. 5. In experiments where mutant forms of Gsy were used, 0.5 μg of protein substrate were present. Reactions were terminated after 20 min at 30°C with 10 μl of 2× SDS sample buffer and heated to 95°C for 2 min before analysis by SDS–PAGE (8% acrylamide). To quantitate the relative phosphorylation of Gsy2 and Pho4, gels were exposed on a Molecular Dynamics screen, scanned, and analyzed with a Molecular Dynamics PhosphorImager and Imagequant (version 4.2a) software.
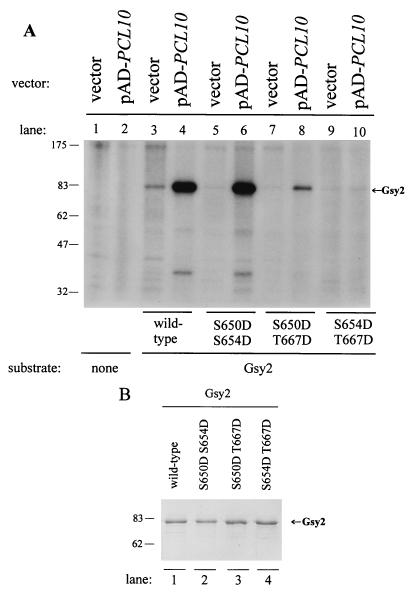
FIG. 5.
Substrate specificities of Pcl10- and Pho80-associated protein kinases in vitro. Fusion proteins were expressed from plasmids pAD-PCL10 or pHA-PHO80 in wild-type (wt) or pho85 mutant strains, as indicated, and immunoprecipitated from cell extracts; protein kinase activity was measured as described in Materials and Methods. Purified Gsy2 (lanes 1 through 6) or Pho4 (lanes 7 through 12) was added at 100 ng/reaction (1×) or 1 μg/reaction (10×) as a substrate, as indicated. Reactions were analyzed by SDS-PAGE, and corresponding autoradiograms (25-min exposures) are shown. (A) AD-Pcl10 (lanes 3 to 6 and 9 to 12) was immunoprecipitated from wt (BY467) or pho85 mutant (BY546) strains transformed with the pAD-PCL10 plasmid, as indicated. The control for Pcl10 immunoprecipitation (vector) used extracts from strains transformed with pAD, which expresses only the Gal4 AD. Immunoprecipitations utilized anti-Gal4 AD antibodies (17). (B) HA-Pho80 (lanes 3 to 6 and 9 to 12) was immunoprecipitated from wt or pho85 mutant strains transformed with the pHA-PhO80 plasmid, as indicated. The control for Pho80 immunoprecipitation (lanes 1, 2, 7, and 8) used extracts from strains transformed with the corresponding empty vector. Immunoprecipitations utilized 12CA5 anti-HA antibodies. The positions of migration of phosphorylated Gsy2 and Pho4 are shown to the right of relevant panels. The asterisk marks the position of an endogenous protein that is specifically phosphorylated in AD-Pcl10 immunoprecipitates and migrates slightly faster than exogenous Gsy2. Positions of migration of molecular size markers are indicated to the left of both panels, in kilodaltons.
Other materials and methods.
Protein was measured by the method of Bradford (3) with bovine serum albumin used as the standard. The SDS-PAGE protocol was a modification of the method of Laemmli (42). Synthesis of UDP-[U-14C]glucose was modified from the method of Tan (69). Recombinant His6Gsy2 with an NH2-terminal poly-His tag, as well as the three phosphorylation-site mutants, were produced in E. coli and purified essentially as previously described (33). Recombinant Pho4 was produced in E. coli and purified following a modification of the procedure of Kaffman et al. (38). Acid phosphatase activity was measured as described by Huang et al. (33) according to established methods (28, 77). The 12CA5 monoclonal antibodies were produced in mouse ascitic fluid by the monoclonal antibody facility of the Faculty of Medicine, University of Toronto. Monoclonal antibody 8CL-11 was raised to the activation domain of Gal4 and was a kind gift of I. Sadowski, University of British Columbia.
RESULTS
PCL8 and PCL10 control glycogen storage.
Having identified Pho85 as a constituent of a glycogen synthase kinase, we considered the possibility that a specific cyclin might be associated with this function. We initially tested whether deletion of the Pho85 cyclin genes known at the time, PHO80, PCL1, and PCL2, affected glycogen accumulation similarly to disruption of PHO85. No combination of disruptions, including a pho80 pcl1 pcl2 triple mutant, caused a significant increase in glycogen accumulation (34). When Measday et al. (47) identified an additional seven Pho85 cyclins, these Pcl’s also became candidates to target Pho85 to the control of glycogen synthesis. Reasoning that the relevant Pcl protein might interact physically with glycogen synthase, we screened several Pcl’s (Pcl2, Clg1, Pcl5, Pcl7, Pcl8, and Pcl10) for the ability to interact with Gsy2 by the two-hybrid assay. Only with Pcl10 was there a significant, albeit weak, signal indicative of Gsy2 binding, by a filter β-galactosidase assay (data not shown). Quantitative assays confirmed an almost fivefold elevation in β-galactosidase activity over the control when the pAD-PCL10 plasmid (PIP7.2 in reference 47) was expressed together with pGSY2 (Table 2). The signal was substantially weaker than that seen with the positive control provided by the pAD-GLG2 plasmid, which encodes a portion of Glg2 and which is known to interact with Gsy2 (6).
TABLE 2.
Interaction of Gsy2 and Pcl10 in the yeast two-hybrid systema
| Plasmid | β-Galactosidase activity (Miller unitsb)
|
Fold increasec | |
|---|---|---|---|
| pAS1 | pGSY2 | ||
| pAD | 0.18 | 0.05 | 0 |
| pAD-GLG2 | 0.20 | 39.5 | 198 |
| pAD-PCL10 | 0.63 | 2.9 | 4.6 |
Yeast strains transformed with a plasmid expressing a GSY2 fusion to the Gal4 DNA binding domain (pGSY2) or the Gal4 DNA binding domain alone (pAS1) were mated with strains transformed with Gal4 AD constructs (pAD) to allow coexpression of plasmid-encoded fusion proteins. pAD, AD alone; pAD-GLG2, positive control expressing an AD fusion to the 48 COOH-terminal residues of Glg2, which are known to interact with Gsy2; pAD-PCL10 (PIP7.2 in reference 47), expresses an AD fusion to all but the NH2-terminal 27 residues of the Pcl10 protein. Data are averages of two (for pAD and pAD-GLG2) or four (for pAD-PCL10) independent transformants.
As defined by Platt et al. (59).
Relative to the pAS1 control.
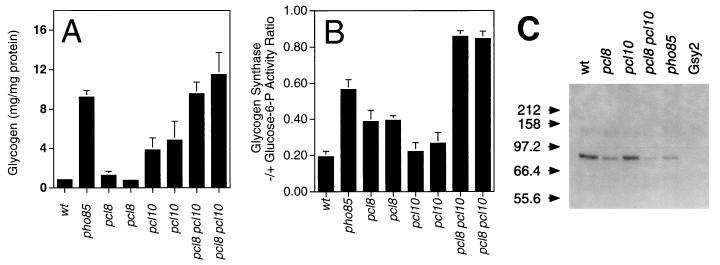
Pcl10 was thus a candidate to mediate Pho85 control of glycogen synthase, and we made a targeted disruption of the PCL10 gene. A pcl10 disruptant overaccumulated glycogen, though to a level short of that seen in pho85 mutants, and had only marginally increased glycogen synthase activity (Fig. 1A). Of the other nine Pcl proteins, Pcl10 most closely resembles Pcl8 in sequence, with regions of similarity outside the cyclin box not shared by other Pcls (47). Therefore, we considered the possibility that Pcl8 and Pcl10 might be functionally redundant. Deletion of PCL8 alone caused activation of glycogen synthase but had little effect on glycogen levels. However, in a pcl8 pcl10 double mutant, glycogen synthase was strongly activated and glycogen accumulation matched that observed in cells lacking PHO85 (Fig. 1B). Disruption of PCL8 and PCL10, alone or in combination, had little effect on the total glycogen synthase activity (data not shown), suggesting that restoration of glycogen levels was due to activation of the glycogen synthase and not alterations in the level of the protein. To test whether glycogen synthase kinase activity in yeast cells required the presence of the PCL8 and PCL10 genes, we assayed the ability of cell extracts to phosphorylate purified recombinant glycogen synthase. Gsy2 kinase activity, readily detectable in wild-type extracts, was significantly reduced in a pho85 strain, by 70 to 80% based on densitometric analysis of autoradiograms (Fig. 1C). The residual PHO85-independent activity may be due to a second Gsy2 kinase activity that has been previously described (33). A slight decrease in glycogen synthase kinase activity was seen after deletion of PCL10, and this decrease was more pronounced in pcl8 strains. In a pcl8 pcl10 double mutant, Gsy2 kinase activity was almost abolished. From densitometry, Gsy2 phosphorylation was decreased by 85 to 95%, to a level below that in the pho85 strain. Together, these results show that Pcl8 and Pcl10 are required for the major Gsy2 kinase activity in yeast under the conditions studied.
FIG. 1.
Effects of deletion of the PCL8 and/or PCL10 gene on yeast glycogen metabolism. Wild-type (wt), pho85, pcl8, pcl10, and pcl8 pcl10 yeast strains, as indicated, were analyzed as follows. (A) Glycogen levels were measured as described in Materials and Methods. The strains used were EG328-1A (wt), DH28 (pho85), DH96-11 and DH96-52 (pcl8), DH93-81 and DH93-82 (pcl10), and DH97-13 and DH97-33 (pcl8 pcl10). Averages and standard errors of three independent experiments are shown. (B) Glycogen synthase −/+ glucose-6-P activity measured in extracts from the strains represented in panel A. Averages and standard errors of three independent experiments are shown. (C) Glycogen synthase kinase activity was measured by the ability of cell extracts to transfer 32Pi from ATP to added purified Gsy2, which was then subjected to SDS-PAGE. Shown is an autoradiogram from one of three experiments yielding similar results. The relevant genotypes are indicated, and the strains analyzed were EG328-1A (wt), DH96-52 (pcl8), DH93-82 (pcl10), DH97-13 (pcl8 pcl10), and DH35-64 (pho85). Gsy2 refers to a control lacking added yeast extract. The molecular masses of standards are indicated, in kilodaltons.
Mutations in PCL8 and PCL10 are specific to glycogen accumulation.
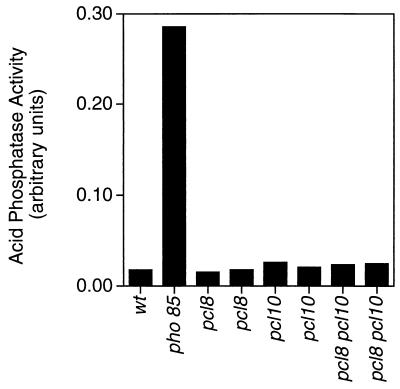
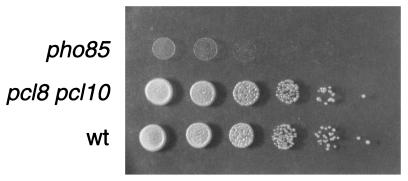
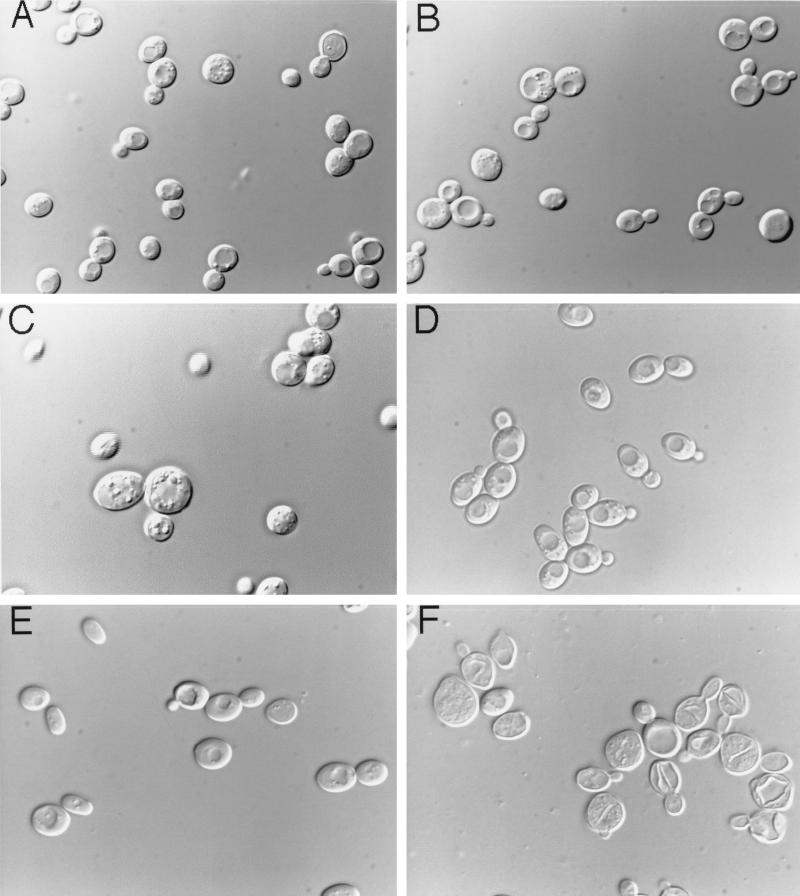
Disruption of PCL8 and PCL10 did not cause the other phenotypic alterations associated with deletion of PHO85. For example, deletions of PCL8 and PCL10 had no effect on the expression of acid phosphatase in a high-phosphate medium, indicating that their absence did not affect Pho4 phosphorylation (Fig. 2). The pcl8, pcl10, and pcl8 pcl10 mutants grew as well as wild-type cells on glycerol, glucose, galactose, acetate, and ethanol (Fig. 3 and data not shown). None of the morphological defects of pho85 mutants were observed with pcl8 pcl10 mutants (Fig. 4), and diploids homozygous for pcl8 and pcl10 sporulated similarly to wild-type controls (not shown). These results suggest that PCL8 and PCL10 are involved specifically with glycogen metabolism rather than other cellular functions of PHO85.
FIG. 2.
Effects of deletion of the PCL8 and/or PCL10 gene on yeast acid phosphatase activity. Wild-type (wt), pcl8, pcl10, and pcl8 pcl10 yeast strains, as indicated, were analyzed for acid phosphatase activity under repressed (high-phosphate) conditions, as described in Materials and Methods. The strains used were EG328-1A (wt), DH28 (pho85), DH96-11 and DH96-52 (pcl8), DH93-81 and DH93-82 (pcl10), and DH97-13 and DH97-33 (pcl8 pcl10). Values are normalized for the number of cells, and averages of two independent experiments are shown.
FIG. 3.
Growth of pho85 and pcl8 pcl10 mutants on glycerol. Liquid cultures of wild-type (wt) (EG328-1A), pho85 (DH28), and pcl8 pcl10 (DH97-13) strains were grown to saturation in rich medium (YPD). Serial 10-fold dilutions were made to generate cell densities from 103 to 108 cells/ml (right to left), and aliquots (2 μl) were spotted onto the surface of a YP-glycerol plate and incubated for 3 days at 30°C.
FIG. 4.
Morphologies of pho85 and pcl8 pcl10 mutants. Cells were grown to late logarithmic phase in rich medium (YPD). Aliquots were taken and concentrated approximately twofold by centrifugation and resuspension. Cells were observed and photographed at a magnification of ×600 with Nomarski optics mounted on a Nikon Microphot-FXA system. (A) Haploid wild-type (EG328-1A) cells. (B) Haploid pcl8 pcl10 (DH97-13) cells. (C) Haploid pho85 (DH28) cells. (D) Diploid wild-type (WW7) cells. (E) Diploid pcl8 pcl10 (WW9) cells. (F) Diploid pho85 (WW8) cells.
Pcl10 is specifically associated with Gsy2 kinase activity.
To test the substrate specificity conferred by different cyclins, we compared Pho4 and Gsy2 as substrates for Pho80-Pho85 and Pcl10-Pho85 complexes. We used antibodies to the AD of Gal4 to immunoprecipitate a Gal4-Pcl10 fusion protein (AD-Pcl10) from yeast extracts. The AD-Pcl10 fusion protein, which consists of the activation domain of Gal4 NH2-terminal to the Pcl10 sequence, was able to complement the glycogen hyperaccumulation defect of a pcl8 pcl10 mutant, indicating that the fusion protein retained function in vivo, at least when expressed from a high-copy-number plasmid (data not shown). Substantial Gsy2 kinase activity was detected in AD-Pcl10 immunoprecipitates from wild-type cell extracts, whereas the same immunoprecipitates phosphorylated Pho4 very poorly (Fig. 5A, lanes 3 and 9 and lanes 5 and 11). The activity of the AD–Pcl10-associated kinase was dependent on the presence of Pho85. A background kinase activity toward Gsy2 was associated with the protein A-Sepharose beads, as has previously been described (see below and reference 38). Huang et al. (33) have described a Gsy2 kinase distinct from Pho85, but there is no evidence that it associates with Pcl10. A species of Mr ∼80,000 that migrated slightly faster than Gsy2 was also phosphorylated in the AD-Pc110 kinase reaction (Fig. 5A, lanes 9 and 11). The predicted size of the AD-Pcl10 fusion protein is ∼70 kDa, and so one possibility is that this species becomes phosphorylated. Immunoprecipitated HA-tagged Pho80 was associated with significant Pho4 kinase activity but had relatively little activity toward Gsy2 (Fig. 5B; compare lanes 3 and 9 or 5 and 11). As has been previously reported, Pho4 kinase activity was strongly dependent on the presence of Pho85; background Pho4 phosphorylation was observed in extracts from pho85 cells, also consistent with previous reports (38, 71). Western analysis confirmed the expression of HA-Pho80 and AD-Pcl10 in the relevant cells (data not shown). Quantitation with a Molecular Dynamics PhosphorImager indicated that, under the conditions of these experiments, Pho80-Pho85 phosphorylated Pho4 about 10-fold more effectively than Gsy2. Phosphorylation of Pho4 by Pcl10-Pho85 was scarcely detectable above the vector control, indicating that this protein kinase exhibited very high specificity toward Gsy2. The relative substrate specificity of the Pho80-Pho85 and Pcl10-Pho85 complexes was retained over a 10-fold range of substrate concentrations (Fig. 5A and B, lanes 3 and 5 and lanes 9 and 11). We attempted similar types of experiments using reagents to isolate Pcl8-Pho85 complexes but did not succeed in measuring kinase activity towards any substrate. The results may well reflect a technical problem with the assay and, in the absence of a positive control for kinase activity, we are unable to judge whether Pcl8-Pho85 can phosphorylate Gsy2. For example, it may be that expression of Pcl8 is significantly lower than Pcl10, or else the protein may be less stable or become inactivated in cell extracts. However, we can conclude that Pcl10 is part of a highly specific Gsy2 kinase and that Pcl10 and Pho80 specify Pho85 to phosphorylate glycogen synthase and Pho4, respectively.
Three sites in Gsy2 are phosphorylated in vivo (24); two of these, Ser-654 and Thr-667, were inferred from in vivo studies to be involved in Pho85-mediated controls (33). We were interested in determining whether the same sites were phosphorylated in vitro by the Pcl10-Pho85 complex. We took advantage of a set of mutant forms of recombinant Gsy2 in which pairs of the three phosphorylation sites had been mutated to Asp. These mutants were originally constructed to test whether an acidic side chain could mimic Gsy2 phosphorylation, but the introduction of Asp had little effect Gsy2 activity (7). We found that Pcl10-Pho85 phosphorylated the S650D S654D and, more weakly, S650D T667D mutant proteins (Fig. 6). This result suggests that both Thr667 and Ser654 are phosphorylated by the Pcl10-Pho85 kinase. The S654D T667D mutant was not detectably phosphorylated in the same experiment. We conclude that only Thr-667 and Ser-654 are phosphorylated by Pcl10-Pho85 in vitro.
FIG. 6.
Phosphorylation of Gsy2 mutants by Pcl10-Pho85 kinase in vitro. (A) AD-Pcl10 fusion protein was expressed from plasmid pAD-PCL10 in a wild-type strain (BY467) and immunoprecipitated from cell extracts by using anti-Gal4 AD antibodies, as described in Materials and Methods and the legend to Fig. 5 (lanes 2, 4, 6, 8, and 10). The control for the kinase reaction used extracts from wild-type strains transformed with pAD, which expresses only the Gal4 activation domain (lanes 1, 3, 5, 7, and 9). Protein kinase activity was measured as described in Materials and Methods. Substrates (at 500 ng per reaction) were added to the kinase assay mixtures as indicated below the gel. The migration of phosphorylated Gsy2 is indicated. (B) Analysis of substrates used in kinase assays. One microgram (each) of purified wild-type Gsy2, S650D S654D, S650D T667D, and S654D T667D were analyzed by SDS-PAGE and visualized by Coomassie blue staining. Migrations of molecular size markers are shown to the left of both panels, in kilodaltons.
Interactions of PCL8 and PCL10 with GLC7 and SNF1 in determining glycogen accumulation.
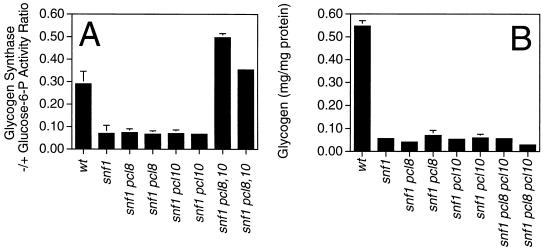
One result that implicated Pho85 in the control of glycogen metabolism was the observation that deletion of PHO85 activates glycogen synthase and restores glycogen accumulation to snf1 cells that are otherwise defective in glycogen synthesis (33). If the role of Pho85 in glycogen metabolism is solely dependent on it being complexed with Pcl8 and Pcl10, one would predict that deletion of PCL8 and PCL10 should also suppress the defective glycogen storage phenotype of snf1 strains. To test this idea, we made pcl8 pcl10 snf1 triple mutants as well as pcl8 snf1 and pcl10 snf1 double mutants. The total activity of glycogen synthase, and by inference the protein level, was not appreciably altered in any of these mutant strains (data not shown). The glycogen synthase activity ratio in snf1 cells is significantly lower than in wild-type cells (23; see also Fig. 7), and we had previously suggested that the glycogen synthesis defect of snf1 mutants could be explained by this observation. Deletion of neither PCL8 nor PCL10 alone had much effect on the glycogen synthase activity ratio. In pcl8 pcl10 snf1 triple mutants, the activity ratio was restored to the level observed in wild-type cells. However, none of the single or double mutations of PCL8 or PCL10 suppressed the glycogen accumulation defect of snf1 (Fig. 7). Although these results further support a role for Pcl8 and Pcl10 in determining the activity—and by inference, the phosphorylation—of glycogen synthase, they also imply that both SNF1 and PHO85 must control some other aspect of the glycogen biosynthetic pathway.
FIG. 7.
Effects of deletion of the PCL8 and/or PCL10 gene on yeast glycogen metabolism in snf1 mutant strains. The relevant genotype of each strain analyzed is shown on the abscissa; wt, wild type. The strains analyzed were EG328-1A (wt), EG353-1C (snf1), DH102-43 and DH102-82 (snf1 pcl8), DH103-31 and DH103-33 (snf1 pcl10), and DH104-104 and DH104-163 (snf1 pcl8 pcl10). (A) Glycogen synthase −/+ glucose-6-P activity measured in extracts from the indicated strains. Averages and standard errors of three independent experiments are shown. (B) Glycogen levels in the indicated strains measured as described in Materials and Methods. Averages and standard errors of three independent experiments are shown.
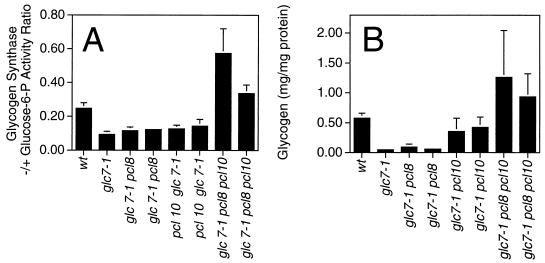
Certain alleles of GLC7, the gene encoding the type I protein phosphatase catalytic subunit in yeast, cause hyperphosphorylation of glycogen synthase and defective glycogen storage (4, 17, 57). Deletion of PHO85 in a glc7-1 strain suppressed the glycogen storage defect of cells grown on rich medium (33), and we asked whether mutations of PCL8 and PCL10 could elicit a similar result. Deletion of PCL8 or PCL10 alone caused minimal increases in the glycogen synthase activity ratio, but mutation of both genes resulted in values as high as or higher than that of wild-type cells (Fig. 8). Corresponding to this increased activity was the restoration of glycogen storage in pcl8 pcl10 glc7-1 cells. These results are consistent with Pcl8 and Pcl10 counteracting the activation of glycogen synthase by a Glc7-containing protein phosphatase.
FIG. 8.
Effects of deletion of the PCL8 and/or PCL10 gene on yeast glycogen metabolism in glc7-1 mutant strains. The relevant genotype of each strain analyzed is shown on the abscissa; wt, wild type. The strains analyzed were EG328-1A (wt), EG327-1D (glc7-1), DH98-14 and DH98-43 (glc7-1 pcl8), DH99-32 and DH99-44 (glc7-1 pcl10), and DH100-61 and DH100-82 (glc7-1 pcl8 pcl10). (A) Glycogen synthase −/+ glucose-6-P activity ratio measured in extracts from the indicated strains. Averages and standard errors of three independent experiments are shown. (B) Glycogen levels in the indicated strains measured as described in Materials and Methods. Averages and standard errors of three independent experiments are shown.
DISCUSSION
PHO85 encodes a multifunctional cyclin-dependent kinase whose activity depends on association with cyclin regulatory subunits. Although 10 Pho85 cyclins (Pcl’s) are known, a physiological substrate had only been identified for the Pho80-Pho85 kinase complex. In this study, we identify the Pcl8 and Pcl10 cyclins as proteins that direct Pho85 to control the phosphorylation state of glycogen synthase in vivo. Our results are relevant to understanding nutritional controls of metabolism but also have a broader importance for substrate selection by protein kinases in general. Deletion of PCL8 and PCL10 mimics mutation of PHO85 in the resulting effects on glycogen synthase activity and, in most cases, glycogen accumulation. At the same time, other defects associated with pho85 mutants, such as growth on nonfermentable carbon sources, morphological defects, and constitutive acid phosphatase expression, are unaffected, consistent with the idea that different cyclins direct Pho85 to different cellular tasks. This in vivo specificity is reflected in vitro, at least in the case of Pcl10, for which we have direct evidence that Pcl10-Pho85 specifically phosphorylates Gsy2. In addition, we showed that the sites modified in vitro, Ser-654 and Thr-667, are precisely those sites inferred from in vivo experiments to mediate Pho85-dependent control of glycogen synthase activity (33).
Elimination of both PCL8 and PCL10 had an even greater impact on glycogen metabolism than deletion of PHO85 alone. This observation may imply that Pcl8 and Pcl10 also influence glycogen synthase and glycogen synthesis via a pathway that does not involve Pho85. The Gsy2 kinase activity in the pcl8 pcl10 strain was lower than in pho85 cells, suggesting that such a pathway might involve another protein kinase. However, at this stage, it is difficult to exclude other types of control.
While we are confident in assigning Pcl8 and Pcl10 to roles in controlling glycogen metabolism, we do not yet know the extent to which their properties differ. Single deletions of PCL8 or PCL10 did not have identical effects on the parameters measured. In some instances, the quantitative effects on glycogen synthase activation did not match the effects on glycogen levels. However, the pcl8 pcl10 double mutant always gave a clear phenotype that was stronger than that associated with deletion of either PCL8 or PCL10 individually. This observation is consistent with the two cyclins having overlapping functions in vivo. We had initially inferred this possibility from the fact that Pcl8 and Pcl10 resemble each other in primary sequence more than they resemble other Pcl’s. Nonetheless, the overall sequence identity between Pcl8 and Pcl10 is only ∼36%, and we cannot exclude the possibility that the two proteins exhibit some unique and distinguishing properties.
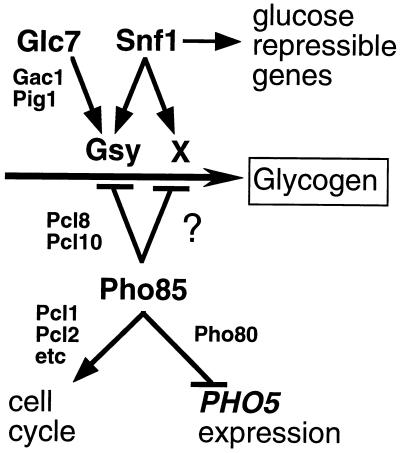
Our genetic experiments also suggest the existence of a hitherto unappreciated control of glycogen synthesis. The glycogen accumulation defects of both snf1 and glc7-1 mutants had been attributed to the presence of hyperphosphorylated, and thus inactivated, glycogen synthase (4, 17, 23, 24, 57). Consistent with this idea, the glycogen storage defect, in either strain, can be corrected by deletion of PHO85, which causes an increase in the glycogen synthase activity ratio. However, we found that deletion of PCL8 and PCL10 in these strains restored glycogen synthase activity to wild-type levels or beyond, but only in a glc7-1 background was glycogen storage rescued. Thus, in snf1 strains, although the defect in glycogen synthase activity was corrected by deletion of PCL8 and PCL10, the cells still did not accumulate glycogen. Therefore, snf1 cells have a second deficiency, besides the lack of glycogen synthase activation, that disables glycogen synthesis. These genetic results suggest an additional control over glycogen synthesis that requires Snf1 (Fig. 9). Normally, this pathway would not be rate determining for glycogen synthesis, and modulation of glycogen synthase activity would dictate the extent of glycogen accumulation. The pathway may involve a novel protein or a known protein whose regulation by Snf1 has not yet been appreciated. Some obvious candidates among known proteins would be Glg1 and Glg2, the self-glucosylating initiator proteins, or Glc3, the branching enzyme, but there is no evidence to date supporting control of these proteins by either Snf1 or Pho85. How Pho85 would interact with the pathway is unclear but presumably should involve a Pcl. Several Pcl’s still have unassigned functions, including Pcl6 and Pcl7 from the Pho80 subfamily. Deletion of PCL6 and PCL7, alone or together, did not cause a significant change in glycogen accumulation (35), but if the novel pathway is not limiting in wild-type cells, this result does not exclude the involvement of Pcl6 or Pcl7.
FIG. 9.
Model for Pho85 control of diverse cellular functions. The Pho85 catalytic subunit is targeted to different cellular functions via its association with different cyclins. Pcl1 and Pcl2 specify participation in cell cycle progression although the relevant substrates are not yet identified. Pho80 targets Pho4 phosphorylation and hence controls acid phosphatase (e.g., PHO5) expression. Pcl8 and Pcl10 specify control of glycogen metabolism via glycogen synthase phosphorylation (Gsy). However, we postulate the presence of another input, denoted “X,” into glycogen metabolism. X is required for glycogen synthesis and becomes limiting in snf1 mutants, presumably due to negative regulation by Pho85; however, this negative regulation is not mediated by Pcl8 or Pcl10. X may be a protein not known to be involved in this pathway, or it may be a known protein whose control by Pho85 and Snf1 has not yet been appreciated.
The present work is also significant to the broader issue of substrate selection by Cdk’s. Over the last several years, the idea has developed that the cyclins play an important role in defining the functional specificity of a Cdk (45, 50, 52), even though in relatively few cases can the in vivo function be correlated with the in vitro specificity of individual cyclin-Cdk complexes for physiological targets. In vitro, the concept that cyclins determine the substrate specificity of Cdk’s is supported by studies with a number of mammalian enzymes (13, 29, 39, 56, 70, 81). For example, both the p107 and Rb proteins are phosphorylated by mammalian cyclin A-Cdk complexes, but not by cyclin B-Cdk complexes, in vitro (56, 70, 81). Conversely, the Xenopus Nap1 protein (39) and the Cdc25-C phosphatase (29) are phosphorylated by their cognate cyclin B-Cdc2 kinase but not by cyclin A-Cdc2 kinase. Other examples of substrates that show preferential phosphorylation by specific cyclin-Cdk complexes in vitro include the E2f transcription factor (13), lamin B (31), and HSS-B (20). A few studies have addressed the relative specificity of different Cdk-cyclin complexes toward alternate peptide or other model substrates and have recorded rather modest differences in substrate specificities (26, 30, 41, 81). The novelty of our work lies in the fact that we have examined two bona fide substrates for the same Cdk catalytic subunit and have analyzed, by biochemical and genetic manipulations, the roles of the cyclin partners both in vivo and in vitro.
Although a more detailed enzyme kinetic study will be necessary for more quantitative assessments, we can conclude that Pcl10 and Pho80 confer considerable selectivity to Pho85 in terms of substrate preference. Switching from Pcl10 to Pho80 converts Pho85 from a Gsy2 kinase into a protein kinase that preferentially phosphorylates Pho4. Pcl10 and Pho80 may influence substrate selection in two different ways, either by modifying the inherent specificity of the catalytic site of the kinase or by providing additional contacts between the kinase and the substrate. The substrate specificity of protein Ser/Thr kinases has largely been discussed in terms of residues, usually close to the phosphate acceptor, that are likely to interact with the catalytic site of the kinase. From examination of known phosphorylation sites in proteins or the phosphorylation of synthetic peptides and, most recently, from the application of peptide library techniques (66, 67), meaningful consensus recognition motifs can sometimes be identified for protein Ser/Thr kinases (40, 55, 58), presumably reflecting critical interactions between the substrate and the catalytic site. Thus, one model would be that Pcl10 and Pho80 allosterically alter the geometry of the catalytic site of Pho85 such that its specificity is modified. The Pho85 sites in Pho4 have the sequence S-P-X-I/L (53), while the two putative Pho85 sites in Gsy2 have a similar but distinct sequence, S/T-P-X-D-L (33). The cyclin may dictate which of the two sequences is preferred. The second possible mechanism, which is not mutually exclusive of the first, is that the cyclin acts as a targeting subunit and is involved directly in substrate binding. The positive two-hybrid interaction between Pcl10 and Gsy2 supports this hypothesis. Comparison of the crystal structures of cyclin A and cyclin H reveals 10-helix core structures that are remarkably similar even though sequence identities are quite limited (see reference 49 for a review). The first five helices of this core form the “cyclin box,” which is the most conserved sequence among different cyclins and which is involved in binding to the catalytic subunit. There is, therefore, ample opportunity for cyclin-specific residues to participate in interactions with substrates, and this is particularly true for Pcl8 and Pcl10, which are relatively large cyclins. Thus, substrate recognition by Cdk’s may resemble interactions with the p27/Kip1 family of Cdk inhibitors, which, as seen in the crystal structure of the p27-cyclin A-Cdk2 complex, involves contacts with both the catalytic subunit and the cyclin (62). Substrate sites must conform to the local recognition requirements of kinase catalytic subunits, but in some manner a substantial proportion of the specificity may in fact derive from the presence of separate targeting subunits or domains.
ACKNOWLEDGMENTS
We are grateful to Mark Goebl, Ron Wek, Lawrence Quilliam, Mike Tyers, and Michael Moran for critical comments regarding the manuscript.
This work was supported in part by grant DK42576 from the National Institute of Diabetes and Digestive and Kidney Diseases (P.J.R.) and by the National Cancer Institute with funds from the Canadian Cancer Society (B.A.) and an Apotex, Inc./MRC industry grant. B.A. is a scientist of the MRC of Canada.
REFERENCES
- 1.Aerne B L, Johnson A L, Toyn J H, Johnston L H. Swi5 controls a novel wave of cyclin synthesis in late mitosis. Mol Biol Cell. 1998;9:945–956. doi: 10.1091/mbc.9.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai C, Elledge S. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cannon J F, Pringle J R, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng C, Huang D, Roach P J. Yeast PIG genes: PIG1 encodes a putative type 1 phosphatase subunit that interacts with the yeast glycogen synthase Gsy2p. Yeast. 1997;13:1–8. doi: 10.1002/(SICI)1097-0061(199701)13:1<1::AID-YEA49>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 6.Cheng C, Mu J, Farkas I, Huang D, Goebl M G, Roach P J. Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6632–6640. doi: 10.1128/mcb.15.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, C., and P. J. Roach. 1998. Unpublished results.
- 8.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Coche T, Prozzi D, Legrain M, Hilger F, Vandenhaute J. Nucleotide sequence of the PHO81 gene involved in the regulation of the repressible acid phosphatase gene in Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:2176. doi: 10.1093/nar/18.8.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creasy C L, Madden S L, Bergman L W. Molecular analysis of the PHO81 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:1975–1982. doi: 10.1093/nar/21.8.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 13.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 14.Espinoza F H, Ogas J, Herskowitz I, Morgan D O. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- 15.Farkas I, Hardy T A, DePaoli-Roach A A, Roach P J. Isolation of the GSY1 gene encoding yeast glycogen synthase and evidence for the existence of a second gene. J Biol Chem. 1990;265:20879–20886. [PubMed] [Google Scholar]
- 16.Farkas I, Hardy T A, Goebl M G, Roach P J. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J Biol Chem. 1991;266:15602–15607. [PubMed] [Google Scholar]
- 17.Feng Z H, Wilson S E, Peng Z Y, Schlender K K, Reimann E M, Trumbly R J. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J Biol Chem. 1991;266:23796–23801. [PubMed] [Google Scholar]
- 18.François J M, Blazquez J, Arino J, Gancedo C. Storage carbohydrates in the yeast Saccharomyces cerevisiae. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism: biochemistry, genetics, biotechnology. Lancaster, Pa: Technomic; 1997. pp. 285–311. [Google Scholar]
- 19.Francois J M, Thompson-Jaeger S, Skroch J, Zellenka U, Spevak W, Tatchell K. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 1992;11:87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs E, Pan Z Q, Niu H, Hurwitz J. Studies on the in vitro phosphorylation of HSSB-p34 and -p107 by cyclin-dependent kinases. Cyclin-substrate interactions dictate the efficiency of phosphorylation. J Biol Chem. 1996;271:22847–22854. doi: 10.1074/jbc.271.37.22847. [DOI] [PubMed] [Google Scholar]
- 21.Gilliquet V, Berben G. Positive and negative regulators of the Saccharomyces cerevisiae ‘PHO system’ participate in several cell functions. FEMS Microbiol Lett. 1993;108:333–339. doi: 10.1111/j.1574-6968.1993.tb06124.x. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie C, Fink R. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press; 1991. [PubMed] [Google Scholar]
- 23.Hardy T A, Huang D, Roach P J. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- 24.Hardy T A, Roach P J. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268:23799–23805. [PubMed] [Google Scholar]
- 25.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 26.Higashi H, Suzuki-Takahashi I, Taya Y, Segawa K, Nishimura S, Kitagawa M. Differences in substrate specificity between Cdk2-cyclin A and Cdk2-cyclin E in vitro. Biochem Biophys Res Commun. 1995;216:520–525. doi: 10.1006/bbrc.1995.2653. [DOI] [PubMed] [Google Scholar]
- 27.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–83. [Google Scholar]
- 28.Hirst K, Fisher F, McAndrew P C, Goding C R. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann I, Clarke P R, Marcote M J, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2—cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes J K, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 31.Horton L E, Templeton D J. The cyclin box and C-terminus of cyclins A and E specify CDK activation and substrate specificity. Oncogene. 1997;14:491–498. doi: 10.1038/sj.onc.1200851. [DOI] [PubMed] [Google Scholar]
- 32.Huang D, Chun K T, Goebl M G, Roach P J. Genetic interactions between REG1/HEX2 and GLC7, the gene encoding the protein phosphatase type 1 catalytic subunit in Saccharomyces cerevisiae. Genetics. 1996;143:119–127. doi: 10.1093/genetics/143.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang D, Farkas I, Roach P J. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, D., V. Measday, P. J. Roach, and B. Andrews. 1997. Unpublished results.
- 35.Huang, D., and P. J. Roach. 1997. Unpublished results.
- 36.Huang D, Wilson W A, Roach P J. Glucose-6-P control of glycogen synthase phosphorylation in yeast. J Biol Chem. 1997;272:22495–22501. doi: 10.1074/jbc.272.36.22495. [DOI] [PubMed] [Google Scholar]
- 37.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 38.Kaffman A, Herskowitz I, Tjian R, O’Shea E K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80–PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 39.Kellogg D R, Kikuchi A, Fujii-Nakata T, Turck C W, Murray A W. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennelly P J, Krebs E G. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 41.Kitagawa M, Higashi H, Jung H K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Lenburg M E, O’Shea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 44.Madden S L, Creasy C L, Srinivas V, Fawcett W, Bergman L W. Structure and expression of the PHO80 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2625–2637. doi: 10.1093/nar/16.6.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Measday V, Andrews B. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 46.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 47.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore L, Andrews B. Mutational analysis of a DNA sequence involved in linking gene expression to the cell cycle. Biochem Cell Biol. 1992;70:1073–1080. doi: 10.1139/o92-152. [DOI] [PubMed] [Google Scholar]
- 49.Morgan D O. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–772. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- 50.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 51.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 52.Nigg E A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 53.O’Neill E M, Kaffman A, Jolly E R, O’Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 54.Oshima Y. Regulatory circuits for gene expression: the metabolism of galactose and phosphate. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces cerevisiae: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 159–180. [Google Scholar]
- 55.Pearson R B, Kemp B E. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 56.Peeper D S, Parker L L, Ewen M E, Toebes M, Hall F L, Xu M, Zantema A, van der Eb A J, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng Z Y, Trumbly R J, Reimann E M. Purification and characterization of glycogen synthase from a glycogen-deficient strain of Saccharomyces cerevisiae. J Biol Chem. 1990;265:13871–13877. [PubMed] [Google Scholar]
- 58.Pinna L A, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 59.Platt T, Muller-Hill B, Miller J H. Assay of β-galactosidase. In: Miller J H, editor. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 60.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 61.Rowen D W, Meinke M, LaPorte D C. GLC3 and GHA1 of Saccharomyces cerevisiae are allelic and encode the glycogen branching enzyme. Mol Cell Biol. 1992;12:22–29. doi: 10.1128/mcb.12.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 63.Schneider K R, Smith R L, O’Shea E K. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 64.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skurat A V, Roach P J. Regulation of glycogen biosynthesis. In: LeRoith D, Olefsky J E, Taylor S, editors. Diabetes mellitus: a fundamental and clinical text. J. B. Philadelphia, Pa: Lippincott Company; 1995. pp. 213–222. [Google Scholar]
- 66.Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 67.Songyang Z, Lu K P, Kwon Y T, Tsai L-H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuart J S, Frederick D L, Varner C M, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan A W. A simplified method for the preparation of pure UDP[14C]glucose. Biochim Biophys Acta. 1979;582:543–547. doi: 10.1016/0304-4165(79)90146-6. [DOI] [PubMed] [Google Scholar]
- 70.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 71.Tennyson, C., J. Lee, and B. Andrews. A role for the Pcl9-Pho85 cyclin-cdk complex at the M/G1 boundary in S. cerevisiae. Mol. Microbiol., in press. [DOI] [PubMed]
- 72.Thomas J A, Schlender K K, Larner J. A rapid filter paper assay for UDP glucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25:486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 73.Thompson-Jaeger S, Francois J, Gaughran J P, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thon V J, Vigneron-Lesens C, Marianne-Pepin T, Montreuil J, Decq A, Rachez C, Ball S G, Cannon J F. Coordinate regulation of glycogen metabolism in the yeast Saccharomyces cerevisiae. Induction of glycogen branching enzyme. J Biol Chem. 1992;267:15224–15228. [PubMed] [Google Scholar]
- 75.Timblin B K, Tatchell K, Bergman L W. Deletion of the gene encoding the cyclin-dependent protein kinase Pho85 alters glycogen metabolism in Saccharomyces cerevisiae. Genetics. 1996;143:57–66. doi: 10.1093/genetics/143.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toh-e A, Tanaka K, Uesono Y, Wickner R B. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 77.Torriani A. Influence of organic phosphate on the formation of acid phosphatases by Escherichia coli. Biochim Biophys Acta. 1960;38:460–469. doi: 10.1016/0006-3002(60)91281-6. [DOI] [PubMed] [Google Scholar]
- 78.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uesono Y, Tanaka K, Toh-e A. Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and structural characterization of PHO85. Nucleic Acids Res. 1987;15:10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uesono Y, Tokai M, Tanaka K, Toh-e A. Negative regulators of the PHO system of Saccharomyces cerevisiae: characterization of PHO80 and PHO85. Mol Gen Genet. 1992;231:426–432. doi: 10.1007/BF00292712. [DOI] [PubMed] [Google Scholar]
- 81.Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]