Abstract
Background
Pleomorphic adenoma is a well-known benign salivary gland neoplasm characterized by the presence of varying proportions of three different components, including bi-layered ducts, myoepithelial cells, and admixed within a chondromyxoid/fibrous stroma.
Method
We report an interesting case of an adult male who presented with bleeding from an extensively degenerated parotid gland mass, concerning for a vascular neoplasm versus primary malignant tumor. Microscopically, majority of the viable tumor exhibited diffuse proliferation of spindle to epithelioid cells, with focal areas depicting cribriform glands, ducts, and scant chondromyxoid stroma.
Result
Next-generation sequencing (NGS) RNA-based fusion panel analysis identified a gene rearrangement involving the pleomorphic adenoma gene 1 (PLAG1), with a novel, cryptogenic fusion partner known as LINC01606; [LINC01606::PLAG1; inv(8;8)(8q12.1;8q12.1)].
Conclusion
To the best of our knowledge, this is the first documented case of a long non-coding RNA (lnc-RNA) serving as a rearrangement partner with the PLAG1 gene. We reviewed the molecular characteristics of this entity and explored the potential role of LINC01606::PLAG1 in the tumorigenesis of pleomorphic adenoma.
Keywords: Pleomorphic adenoma, PLAG1, Gene rearrangement, LINC01606
Introduction
Salivary gland neoplasms, whether benign or malignant, pose certain diagnostic challenges. Histopathologic examination and immunohistochemical (IHC) stains can reveal the diagnosis in most cases; however, molecular characterization is crucial to identify certain difficult cases [1, 2]. Pleomorphic adenoma (PA) is the most common benign salivary gland neoplasm, which usually presents as a slowly growing mass [3–5]. In certain instances, the presentation could be atypical and more concerning than just being a benign neoplasm. Pathologically, PA is characterized by the presence of varying proportions of three components, including bi-layered ducts, myoepithelial cells, and admixed within a chondromyxoid/fibrous stroma. It can be readily diagnosed by Hematoxylin & eosin (H&E) stain, without the need for IHC stains. However, when one of its three components predominates over the others, it can turn out to be a challenging diagnosis. Pleomorphic adenoma gene 1 (PLAG1) and high-mobility group AT-hook 2 (HMGA2) are hallmark genes showing rearrangement with various partner genes [6]. We present a challenging and unique case of a degenerated myoepithelial cell-rich PA, showing PLAG1 rearrangement with a novel partner gene.
Clinical and Radiologic Findings
A 77-year-old male presented to the emergency room (ER) with excessive bleeding and purulent discharge from a ruptured right cheek/neck mass. The mass had been progressively enlarging over a period of five years. Physical examination revealed a soft, “boggy” lateral neck mass adjacent to the submandibular region with a bleeding skin defect, worrisome for a vascular tumor. The bleeding was controlled promptly with Surgical and Tranexamic Acid. A contrast-enhanced computed tomography (CT) imaging study revealed a circumscribed, complex cystic mass with internal enhancement arising from the parotid gland (Fig. 1). No skin or bone invasion, regional lymphadenopathy, or involvement of any other site were noted. Previously, in the fine needle aspiration (FNA) clinic, the cyst was aspirated revealing degenerated fluid and a core biopsy was performed which yielded predominantly necrotic material. He was scheduled for an elective parotidectomy. However, he presented shortly again in the ER with excessive bleeding from the cutaneous defect, necessitating urgent parotidectomy along with right levels II to IV lymph node dissection. It is worth mentioning that the patient had no prior medical history of bleeding disorder or the administration of blood-thinning products.
Fig. 1.
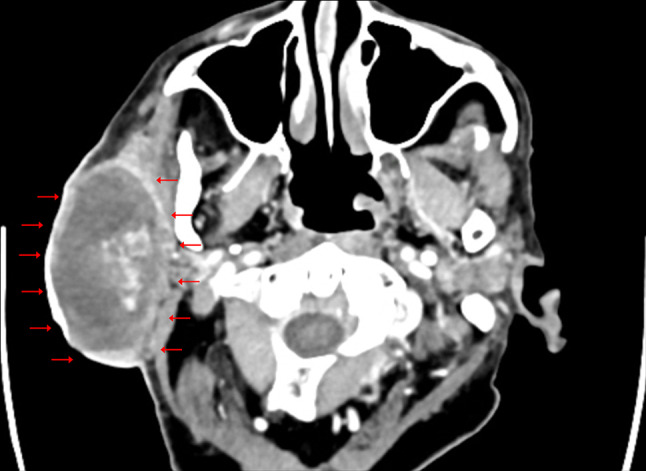
Computed tomographic image shows an 8.7-cm right parotid circumscribed complex cystic mass (arrows) with heterogeneous internal enhancement
Materials and Methods
Formalin-fixed, paraffin-embedded (FFPE) 4-µm-thick tissue sections were stained for H&E section. Immunohistochemical stains were performed on 3- to 5-µm-thick FFPE sections using the DAKO Envision system (DAKO, Glostrup, Denmark) in an automated immunostainer (DAKO, Autostainer plus). The utilized IHC antibodies are summarized in Table 1. Heat-induced antigen retrieval was used for all antibodies in appropriate buffers as per the manufacturer’s guidelines.
Table 1.
Immunohistochemical panel used
| Antibody | Clone | Manufacturer | Animal species | Dilution |
|---|---|---|---|---|
| CK7 | OV-T1 12/20 | Agilent | Mouse monoclonal | RTU |
| p40 | Anti-p40 | Biocare | Rabbit polyclonal | 1:200 |
| p63 | 4A4 | Biocare | Mouse monoclonal | RTU |
| S100 | Anti-S100 | Agilent | Rabbit polyclonal | RTU |
| SOX10 | BC34 | Biocare | Mouse monoclonal | 1:100 |
| SMA | 1A4 | Agilent | Mouse monoclonal | RTU |
| Ki-67 | MIB-1 | Agilent | Mouse monoclonal | RTU |
RTU Ready to use
Molecular Testing
Molecular testing was performed as a send-out test to NeoGenomics Laboratories, USA. Total nucleic acid was extracted from FFPE tissue from unstained slides. The Salivary Gland NGS Fusion Panel utilizes hybridization capture-based targeted next-generation RNA sequencing for the detection of fusions involving select exons of the following genes (ARID1A, ATF1, CRTC1, CRTC3, DDX3X, ETVS, EWSR1, HMGA2, MAML1, NAML2, MYB, MYBL1, MYH9, NCOA4, NFIB, NTRK1, NTRK2, NTRK3, PLAG1, PRKD1, PRKD2, RET, and USPS) was performed.
Histopathologic and Molecular Findings
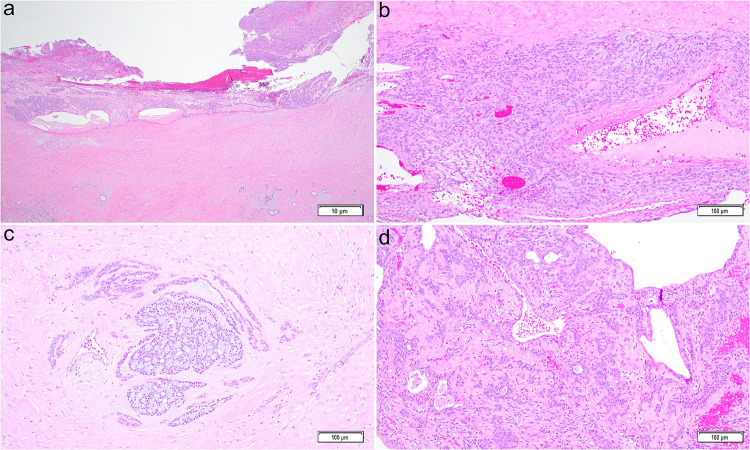
On gross examination, an 8.2-cm mass was detected within the right parotid gland which was adherent to the overlying skin with 5.2-cm hemorrhagic/necrotic cystic component. The mass with adjacent skin and salivary tissue was submitted in its entirety for microscopic evaluation. Histopathologic evaluation revealed a well-defined, circumscribed salivary gland neoplasm with extensive (~ 75% of the mass) areas of hemorrhage, infarction, and cystic degeneration (Fig. 2a). The viable (~ 25% of the tumor) areas showed diffuse proliferation of bland, spindle to epithelioid/plasmacytoid cells (Fig. 2b) with focal areas demonstrating cribriform/glandular nest formation (Fig. 2c-2d). In the background, scant areas of chondromyxoid stroma were noted. At the periphery, the lesion showed dense peritumoral fibrosis, granulation tissue, and chronic inflammation; the changes compatible with FNA procedure. No area of significant cellular atypia, increased mitotic activity (> 3 mitosis/10 high-power field), or definitive tumor necrosis were noted. No microscopic features of tumor invasion into surrounding parotid tissue were identified. No lymphovascular or perineural invasion was identified. A panel of IHC stains revealed that the tumor cells were diffusely positive for CK7, S100, and SOX10. CK7 stained more strongly in the epithelial component in the cribriform and glandular areas as compared to the background spindle and epithelioid myoepithelial cell proliferation. In addition to S100 and SOX10, the myoepithelial cells were also patchy positive for SMA, p63, and p40. The Ki-67 labeling index was variable from 1% to up to 10%; overall 5% in the lesional cells. Thirty lymph nodes, including peri-parotid and right lateral neck, were negative for metastatic carcinoma. Based on morphologic and IHC findings, a diagnosis of myoepithelial cell-rich cellular pleomorphic adenoma was rendered. Because of unusual clinical presentation, the tissue was sent for an RNA-based salivary gland NGS panel which revealed a chromosomal fusion with inversion, namely, LINC01606::PLAG1 [inv(8;8)(8q12.1;8q12.1)]. No other fusions detected.
Fig. 2.
Histologic section shows circumscribed mass with a thick fibrotic wall, hemorrhage, and cystic degeneration (2a, hematoxylin and eosin (H&E) stain, total magnification × 20). In viable areas, the lesional cells are spindle to epithelioid, without atypia, in the background of myxoid and hyalinized stroma (2b, H&E, × 100). In focal areas, cribriform nests and ducts/glands are identified (2c-d, H&E, × 100)
Discussion
Salivary gland neoplasms encompass a wide array of tumors and their classification has grown comprehensively with the progress of molecular techniques. Histopathologic features and IHC staining play a role in narrowing down the range of differential diagnoses by elucidating distinct tumor components [4]. Moreover, the patient’s clinical presentation and the specific salivary gland affected contribute to further refining the diagnostic process.
Despite being the most frequently encountered salivary gland neoplasm, PA can pose a considerable diagnostic challenge for several reasons. This begins with the uneven distribution of its constituent components and the variability in the histologic features of epithelial and stromal elements. The variations in the stromal component include mucoid, myxoid, hyalinized, or chondroid alterations. Additionally, the epithelial elements have the potential to undergo metaplastic changes, which can vary from squamous, oncocytic, and mucocytes to sebaceous transformations. In such instances, common potential differential diagnoses may include oncocytoma, myoepithelial neoplasms, basal cell adenoma/adenocarcinoma, adenoid cystic carcinoma, or polymorphous adenocarcinoma [4].
Our patient had a relatively large parotid mass (8.2 cm) with ulceration of the overlying skin and extensive cystic degeneration that raised the possibility of carcinoma ex pleomorphic adenoma (CEPA) in the differential diagnosis. However, the parotid swelling had been gradually increasing in size over five years without any pain or abrupt growth. The CT scan showed a well-circumscribed mass without any skin or bone invasion. Intraoperatively, the lesion was retracted from the facial nerve and its branches without difficulty. The development of cystic degeneration in our case likely followed after FNA which may be attributed to either manipulation of the blood supply during the procedure or the tumor demand of blood supply exceeding than the capacity of feeder vessels [5, 6]. Histologically, the tumor was well circumscribed, without any significant nuclear atypia, increased mitotic activity, or invasion into surrounding parotid tissue. All these features helped us to exclude malignancy such as CEPA in our patient. Thus, this case underscores the importance of a comprehensive clinicopathologic correlation as both PA and CEPA can show the same driver (PLAG1 and HMGA2) molecular rearrangement [6]. When cystic changes are present, other differential diagnoses, such as mucoepidermoid carcinoma, mucocele, or squamous cell carcinoma, can also be considered in an appropriate morphologic context [7].
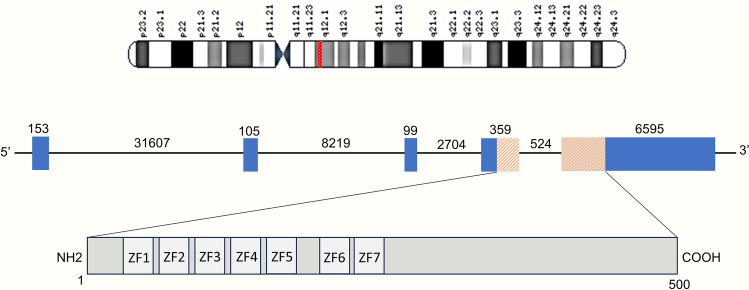
Pleomorphic adenoma is characterized by specific genomic abnormalities, involving translocations with breakpoints in the 8q12 and 12q13-15 regions. These translocations result in gene fusions with transcription factor genes, such as PLAG1 and HMGA2, respectively [8]. It is essential to note that PLAG1 and HMGA2 rearrangements are the primary driver gene mutations in PA, and they tend to be mutually exclusive [9]. The PLAG1 gene itself consists of five coding exons, with functional C2H2 zinc finger domains encoded by parts of exon 4 and exon 5 (Fig. 3). As a transcription factor, PLAG1 plays a significant role in upregulating the expression of insulin-like growth factor 2 (IGF2) and activating the PI3K/MAPK pathway, both of which contribute to the tumorigenic characteristics of pleomorphic adenoma [10, 11].
Fig. 3.
Schematic representation of the structure of the human PLAG1 gene and its protein. On the top, chromosome 8 is shown with a red-highlighted area of band 8q12.1. The introns are represented by a line and exons by boxes. The orange-shaded area of exon 4 and exon 5 shows the translated region. Numbers refer to the number of base pairs. In the protein structure at the bottom, numbers refer to the number of amino acids and lightly shaded boxes indicated by ‘ZF’ represent the seven zinc finger domains. Structure is based on the Ensembl sequences ENST00000316981.8
Rearrangement abnormalities on chromosome 8q12 involving PLAG1 represent the largest subgroup, accounting for 50–60% of PA. These rearrangements can occur between two different chromosomes (inter-chromosomal) or within the same chromosome (intra-chromosomal). Specifically, inter-chromosomal rearrangements involve PLAG1 fusion with CTNNB1 (beta-catenin), LIFR (leukemia inhibitory factor receptor), and CHCHD7 (Coiled-coil-helix-coiled-coil-helix domain-containing protein 7). In contrast, intra-chromosomal fusion occurs with TCEA1/SII (transcription elongation factor) [12, 13]. Furthermore, various other binding partners have been identified in these rearrangements, including FGFR1, TGFBR3, GEM, ACTA2, and ND4 [9]. In our patient, we identified a novel finding of LINC01606 as a rearrangement partner with PLAG1, which has not been previously reported.
The elevated expression of PLAG1 mRNA and protein can be attributed to promoter swapping or substitution, which places PLAG1 under the influence of a highly active promoter. This swapping primarily involves the promoters of translocation partner genes with constitutively active promoters, such as CTNNB1, leading to the upregulation of PLAG1 expression. In cases involving specific translocations, like t(3;8)(p21;q12), the 5' non-coding regions of both genes are translocated, signifying the swapping of regulatory elements. Similarly, fusion with CHCHD7 involves the rearrangement of non-coding exon 1 of CHCHD7 with non-coding exon 2 of PLAG1 through para-centric inversion. In the case of TCEA1 fusion, the exon 1- and 5'-flanking sequence of TCEA1 are rearranged with intron 1 of PLAG1, effectively swapping the regulatory elements [12]. All these instances underscore the notion that disturbances in the 5' untranslated regulatory regions result in the over-activation of PLAG1, leading to a gain of function.
In our case, a similar pattern of rearrangement was observed. An inversion event, inv(8;8)(8q12.1;8q12.1), led to the fusion of exons 1–2 of the LINC01606 gene with exons 3–5 of the PLAG1 gene. It is important to note that the coding exons responsible for the zinc finger domains (exon 4 and exon 5) remained intact, with the rearrangement occurring only in the upstream regions of these exons.
Long intergenic non-coding RNA (linc-RNAs) are subtypes of long non-coding RNAs (lnc-RNA). Lnc-RNA are longer than 200 nucleotides which are not translated into functional proteins and have been further defined by anatomic properties. For example, intronic lnc-RNAs are encoded within introns of a single protein-coding gene, and intergenic linc-RNAs are encoded within intergenic space, between two protein-coding loci. Lnc-RNAs affect stages of mRNA life, including splicing, turnover, and translation by different mechanisms [14, 15]. HOTAIR, MALAT1, MEG3, and H19 are some of the lnc-RNAs recorded in ≥ 16 cancer types [16]. LINC01606 consists of around 2900 base pairs (bp) and 7 non-coding exons. In our patient, exons 1 and 2 of LINC01606 were involved in rearrangement. So far, there is no data demonstrating the pathogenesis of LINC01606::PLAG1 in neoplastic proliferation. However, the translocation follows the theme of altering the regulatory elements in the 5’UTR of PLAG1. It is hard to speculate whether LINC01606 had any role in that chromatin remodeling in our case or not, without further investigation.
LINC01606 has been studied in some cancers for its carcinogenic roles. LINC01606 is reported to participate in the persistent activation of the Wnt/β-catenin pathway in colon cancer cells [17]. Other study reported its role in the invasion and migration of multiple myeloma cells [18]. It is shown to control the activity of micro-RNA (mi-RNA), another type of non-coding regulatory RNA, leading to tumor progression indirectly. There have been few studies about detection of non-coding RNA expression in salivary gland pleomorphic adenoma as well. Shen et al. evaluated for co-expression of coding RNA (mRNA) with non-coding RNAs (mi-RNA and lnc-RNA) and possible interaction between them [19]. Their research highlights the co-expression of a variety of lnc-RNAs along with tumorigenesis-causing genes like P53, PLAG1, beta-catenin, and IGF2 on microarray analysis which also demonstrate a complex interplay of ‘lincRNA – mi-RNA—mRNA’ in an ex vivo model. One of the lnc-RNA, T042819 was shown to increase the expression of the PLAG1 gene by sponge-like action on miR-195–5p, which otherwise inhibits PLAG1 expression [19].
In our case, the viable tumor was primarily composed of a spindle/epithelioid cell population. The potential correlation between LINC01606::PLAG1 fusion and the presence of myoepithelial cell-rich histomorphology in PA could be a subject for future investigation as more data become available.
While LINC01606 has been implicated in carcinogenesis, it has not been previously identified in benign tumors, possibly due to the limited use of NGS in studying benign entities. However, it raises the question of whether LINC01606 could potentially influence specific tumor behavior and clinical presentation even in benign cases.
At six months and one year of follow-up, our patient remains asymptomatic with no disease recurrence on imaging.
Conclusion
In summary, our case reinforces the importance of a comprehensive, clinicopathologic correlation for cases of pleomorphic adenoma which may have an atypical clinical presentation due to large size and show post-FNA degeneration/infarction. In addition, to our knowledge, this is the first report of pleomorphic adenoma showing gene rearrangement involving PLAG1 with a novel, cryptogenic fusion partner known as LINC01606; [LINC01606::PLAG1; inv(8;8)(8q12.1;8q12.1)].
Acknowledgements
No acknowledgements.
Authors Contribution
Udit Naik contributed to writing of the manuscript and data collection; Sara E Amin contributed to reviewing and editing of the manuscript and photo collection; Mahmoud Elsayad contributed to reviewing and editing of the manuscript; Karan Saluja contributed to conceptualization and reviewing and editing of the manuscript.
Funding
Not applicable. The authors did not receive support from any organization for the submitted work.
Data Availability
Not applicable.
Declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical Approval
Not applicable.
Research Involving Human/Animal Participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
For this type of study, informed consent is not required.
Consent for Publication
For this type of study, consent for publication is not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Skálová A, Stenman G, Simpson RHW, et al. (2018) The role of molecular testing in the differential diagnosis of salivary gland carcinomas. Am J Surg Pathol. doi:10.1097/PAS.0000000000000980. [DOI] [PubMed]
- 2.Todorovic E, Dickson BC, Weinreb I (2020) Salivary gland cancer in the era of routine next-generation sequencing. Head Neck Pathol 14:311–320. 10.1007/s12105-020-01140-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith CC, Schmitt AC, Little JL, Magliocca KR (2017) New developments in salivary gland pathology: clinically useful ancillary testing and new potentially targetable molecular alterations. Arch Pathol Lab Med 141:381–395. 10.5858/arpa.2016-0259-SA [DOI] [PubMed] [Google Scholar]
- 4.Hernandez-Prera JC, Skálová A, Franchi A et al (2021) Pleomorphic adenoma: the great mimicker of malignancy. Histopathology 79:279–290. 10.1111/his.14322 [DOI] [PubMed] [Google Scholar]
- 5.Chen YK, Lin CC, Lai S et al (2004) Pleomorphic adenoma with extensive necrosis: report of two cases. Oral Dis 10:54–59. 10.1046/j.1354-523x.2003.00966.x [DOI] [PubMed] [Google Scholar]
- 6.Antony J, Gopalan V, Smith RA, Lam AK (2012) Carcinoma ex pleomorphic adenoma: a comprehensive review of clinical, pathological and molecular data. Head Neck Pathol 6:1–9. 10.1007/s12105-011-0281-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khetrapal S, Jetley S, Hassan MJ, Jairajpuri Z. Cystic change in pleomorphic adenoma: a rare finding and a diagnostic dilemma. J Clin Diagn Res. 2015;. doi:10.7860/JCDR/2015/14101.6764. [DOI] [PMC free article] [PubMed]
- 8.Martins C, Fonseca I, Roque L et al (2005) PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol 18:1048–1055. 10.1038/modpathol.3800386 [DOI] [PubMed] [Google Scholar]
- 9.Kaur K, Mehta S, Vanik S et al (2022) The evolving role of molecular pathology in the diagnosis of salivary gland tumours with potential pitfalls. Eur Arch Otorhinolaryngol 279:3769–3783. 10.1007/s00405-022-07326-6 [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, Xu L, Hassan M et al (2020) Bayesian modeling identifies PLAG1 as a key regulator of proliferation and survival in rhabdomyosarcoma cells. Mol Cancer Res 18:364–374. 10.1158/1541-7786.MCR-19-076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Dyck F, Declercq J, Braem CV, Van de Ven WJ (2007) PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review). Int J Oncol 30:765–774 [PubMed] [Google Scholar]
- 12.Asp J, Persson F, Kost-Alimova M, Stenman G (2006) CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer 45:820–828. 10.1002/gcc.20346 [DOI] [PubMed] [Google Scholar]
- 13.Voz ML, Aström AK, Kas K, Mark J, Stenman G, Van de Ven WJ (1998) The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of PLAG1 gene expression under control of the LIFR promoter. Oncogene 16:1409–1416. 10.1038/sj.onc.1201660 [DOI] [PubMed] [Google Scholar]
- 14.Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinn JJ, Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17:47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, Huang S, Wei J et al (2022) Long noncoding RNA LINC01606 protects colon cancer cells from ferroptotic cell death and promotes stemness by SCD1-Wnt/β-catenin-TFE3 feedback loop signalling. Clin Transl Med 12:e752. 10.1002/ctm2.752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Fan X, Zhang B, Wu L, Wu X (2021) Expression of LINC01606 in multiple myeloma and its effect on cell invasion and migration. Am J Transl Res 13:8777–8786 [PMC free article] [PubMed] [Google Scholar]
- 19.Shen S, Lu H, Liu S, Yang W, Liu L, Xu W (2022) Integrative analysis of long non-coding RNAs and mRNAs associated with tumorigenesis of salivary gland pleomorphic adenoma. Arch Oral Biol 133:105303. 10.1016/j.archoralbio.2021.105303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.