Abstract
The transcription factor TFII-I was initially isolated as a factor that can bind to initiator elements in core promoters. Recent evidence suggests that TFII-I may also have a role in signal transduction. We have found that overexpression of TFII-I can enhance the response of the wild-type c-fos promoter to a variety of stimuli. This effect depends on the c-fos c-sis–platelet-derived growth factor-inducible factor binding element (SIE) and serum response element (SRE). There is no effect of cotransfected TFII-I on the TATA box containing the c-fos basal promoter. Three TFII-I binding sites can be found in c-fos promoter. Two of these overlap the c-fos SIE and SRE, and another is located just upstream of the TATA box. Mutations that distinguish between serum response factor (SRF), STAT, and TFII-I binding to the c-fos SIE and SRE suggest that the binding of TFII-I to these elements is important for c-fos induction in conjunction with the SRF and STAT transcription factors. Moreover, TFII-I can form in vivo protein-protein complexes with the c-fos upstream activators SRF, STAT1, and STAT3. These results suggest that TFII-I may mediate the functional interdependence of the c-fos SIE and SRE elements. In addition, the ras pathway is required for TFII-I to exert its effects on the c-fos promoter, and growth factor stimulation enhances tyrosine phosphorylation of TFII-I. These results indicate that TFII-I is involved in signal transduction as well as transcriptional activation of the c-fos promoter.
TFII-I is a transcription factor that was initially characterized as a factor that binds to initiator sites (Inr) of various promoters (48). It has been implicated in the initiation of transcription of TATA-less promoters and in cell-type-specific transcription as well (31, 48). Deletions of TFII-I are closely associated with neurodevelopmental Williams-Beuren syndrome in humans (41). TFII-I can also bind to E-box elements and can interact with upstream regulatory factors, including USF1 and c-myc (46, 47). In addition, TFII-I can associate with Bruton’s tyrosine kinase (Btk), and its phosphorylation on tyrosine is stimulated by it (69). The activity of TFII-I is regulated by phosphorylation, and one of the potential phosphorylation sites is a mitogen-activated protein (MAP) kinase phosphorylation site (40). These observations suggest that TFII-I may play a role in signal transduction as well as in transcriptional initiation. In addition, Grueneberg et al. (13) have shown that TFII-I associates with the serum response factor (SRF) and the Phox1 protein, which are both involved in the regulation of the c-fos promoter.
The c-fos promoter is the best-studied immediate-early gene promoter and is well known for being responsive to a variety of extracellular ligands (2, 12, 30, 36). The c-fos promoter is a TATA box-containing promoter with several upstream elements, including a calcium cyclic AMP response element (CRE), a serum response element (SRE), and a c-sis–platelet-derived growth factor (PDGF)-inducible factor element (SIE) (1, 52, 68). The CREB transcription factor has been associated with calcium and cyclic AMP induction of the c-fos promoter through the CRE, and the SIE element binds to STAT proteins and can regulate the responses to PDGF and tyrosine kinases (9, 34, 50, 56–59). The SRE binds to SRF and can form complexes with a number of other transcription factors, most notably ternary complex factors (TCFs), including Elk-1 and SAP-1, which are responsive to MAP kinase and related pathways (11, 20, 24, 32, 38, 65).
Although the c-fos promoter has been extensively studied, there are still a number of outstanding issues remaining in the understanding of its regulation. SRF can form a ternary complex with the various TCFs and can respond to signals from MAP kinase and MAP kinase-related kinases (3, 24, 55). However, there is substantial evidence of another pathway that leads to the activation of the SRE that is activated by small GTP-binding proteins (G proteins), such as RhoA (23, 27, 44). This pathway appears not to require the TCFs. The mechanism by which this pathway stimulates the c-fos promoter through the SRE is unclear. In addition, results from the introduction of the c-fos promoter into transgenic animals indicate a high degree of interdependence between c-fos promoter elements (45). The mechanism that accounts for this interdependence is also not well understood.
Here, we report that TFII-I can enhance the transcriptional activity of the c-fos promoter in a manner that is dependent on upstream elements, including both the SIE and SRE. TFII-I can bind three different sites in the c-fos promoter, including the SIE and SRE, in a sequence-specific manner, and these TFII-I binding sites are required to be intact for the optimal activity of the c-fos promoter. Moreover, we find that critical promoter binding proteins SRF, STAT1, and STAT3 form complexes with TFII-I in vivo. TFII-I requires a functioning ras pathway for its activity, and its tyrosine phosphorylation is enhanced after epidermal growth factor (EGF) stimulation. These results suggest that TFII-I plays an important role in the activation and regulation of the c-fos promoter.
MATERIALS AND METHODS
Plasmids.
For reporter assays, pSVOAΔ5′ containing a 379-bp murine c-fos promoter was used for the wild-type c-fos luciferase construct (4, 17), and other mutant promoters were generated by PCR and cloned into the same vector. All insertions were made into the HinDIII sites of the vector. The structures of the newly generated constructs were verified by sequencing. Thymidine kinase (TK) luciferase constructs were described previously (17, 63). For transient overexpression of human TFII-I, the pEBG-His6-TFII-I construct was used (1a). Mouse STAT1 and human STAT3 expression plasmids were previously described (9, 71). Human SRF full-length cDNA was excised from the pG3.5 plasmid (38) and cloned into pCDNA3 vector (Invitrogen) for mammalian expression. pCMV.Elk-1 was obtained from Peter Shaw (29). The pRL-TK luciferase construct was obtained from Promega for standardization of transfection. pMT3.N17Ras was obtained from Larry Feig and was previously described (8).
The following top strand primers were used in the PCR mutagenesis (lowercase letters indicate nucleotides that differ from the wild-type sequence): ΔSIE, GGGGAAGCT TCTGCAGTCCT T TACACAGGATGTCCATAT TAGGACA; m34SIE, GGGGAAGCTTCTGCAGCCGGCGAGCTGTTCaCGTCAATCCCTCCCTCCTTTACACAGGATGTCCATATTAGGACATCT; m67SIE, GGGGAAGCT TCTGCAGCCGGCGAGCatT TCCCGTaAATCCCTCCCTCCT T TACACAGGATGTCCATATTAGGACATCT; mTCF, GGGGAAGCTTCTGCAGCTGTTCCCGTCAATCCCTCCCTCCTTTACAactGATGTCCATATTAGGACATCTGCGTCAGCAGGTTTCCACGG; mSRE-S, GGGGAAGCTTCTGCAGCTGT TCCCGTCAATCCCTCCCTCCT T TACACAGGATGTCCATATTAccACATCTGCGTCAGCAGGTTTCCACGG; mSRE-T, GGGGAAGCTTCTGCAGCTGTTCCCGTCAATCCCTCCCTCCT T TACACAGGATGTCCtTATaAGGACATCTGCGTCAGCAGGTTTCCACGG; mSRE-ST, GGGGAAGCTTCTGCAGCTGTTCCCGTCAATCCCTCCCTCCT T TACACAGGATGTggATATTAccACATCTGCGTCAGCAGGTTTCCACGG; Basal, GGGGAAGCTTCTGCAG TAGGAAGTCCATCCATTCACAGCGC T TC TATAAAGGCGCCAGCTGAGGCGCCTACTA; mBasal, GGGGAAGCTTCTGCAGCCGGCGAGCTG T TCCCG TCAATCCCTCCCTCCT T TACACAGGATGTCCATATTAGG; and m67SIE/mSRE-T, GGGGAAGCTTCTGCAGCCGGCGAGCatT TCCCG TaAATCCCTCCCTCC T T TACACAGGATG TCCAaAT TtGGACATCTGCGTCAGCAGGTTTCCACGG.
The following bottom strand primer (except mBasal) was also used in the PCR mutagenesis: GGGGAAGCTTCCCGGGAGTAGTAGGCGCCTCAGCTGGCGCCTTTATA. The bottom strand primer for mBasal was GGGGAAGCTTCCCGGGAGTAGTAGGCGCCTCAGCTGGCCGTTTATAGAAGCGCTGTGcccGGATGGACTTCCTACGTCACTGGGCGGAAC.
Antibodies.
Anti-STAT1 polyclonal rabbit antiserum was made against the synthetic peptide containing the unique C-terminal 37 amino acids of murine STAT1 protein. Anti-STAT3 antibody was a polyclonal rabbit antiserum made against another unique sequence in the C-terminal region of human STAT3 (amino acids 688 to 700). Anti-TFII-I polyclonal antibody was made against the synthetic peptide (amino acids 301 to 321) and affinity purified as previously described (39). The anti-CDK4, -Elk-1, and -SRF antibodies were obtained from Santa Cruz Biotechnology. The anti-phosphotyrosine antibody (4G10) was obtained from UBI. The anti-GST antibody was obtained from Sigma.
Cell culture and transfections.
Murine NIH 3T3 fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% calf serum (CS). COS-1 cells were cultured in DMEM with 10% fetal CS (FCS). For transient transfection assays, the calcium phosphate method was used with a calcium phosphate transfection kit from 5 Prime→3 Prime Co. (Boulder, Colorado). For reporter assays, NIH 3T3 cells were maintained for 30 to 40 h in medium containing 0.5% CS following transfection and stimulated with various reagents (CS, 10%; lipophosphatidic acid (LPA), 10 μM; tetradecanoyl phorbol acetate (TPA), 50 ng/ml; PDGF, 25 ng/ml) for 2.5 to 4 h before harvest. Four micrograms of reporter construct, 3 μg of pEBG-TFII-I (or pEBG) expression plasmid, and 1 μg of pRL-TK normalization plasmid were used per 60-mm-diameter dish. pMT3.N17Ras (0.5 μg) was included where indicated. A dual-luciferase assay was carried out according to the manufacturer’s recommendation (Promega). All transfection experiments were performed in duplicate, and results were normalized to the expression of the Renilla luciferase transfection control. For mammalian overexpression of TFII-I, SRF, Elk-1, STAT1, and STAT3, COS-1 cells were maintained before harvest for 36 h in medium containing 10% FCS following transfection. Seven micrograms of pEBG-TFII-I or pEBG or 10 μg of the SRF, Elk-1, STAT1, or STAT3 constructs was used per 100-mm-diameter plate. The total amount of DNA was kept at 17 μg per 100-mm-diameter dish, with pEBG added to adjust total amounts of DNA where necessary. For EGF stimulation, COS-1 cells were maintained for 40 h in medium containing either 0.5 or 10% FCS following transfection and stimulated with 20 ng of EGF per ml for 20 min before harvest for Western blotting analysis.
Isolation of overexpressed TFII-I.
Transfected COS-1 cells were lysed in buffer A (20 mM Tris [pH 7.8], 500 mM KCl, 10% glycerol, 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 1 μg each of leupeptin, antipain, pepstatin, and chymostatin per ml), and the lysate was then centrifuged for 10 min at 10,000 × g at 4°C. Supernatants were incubated with Ni-nitrilotriacetic acid (NTA) agarose beads (Qiagen) for 1 h at 4°C to pull down His6–TFII-I. The beads were collected by quick spin with a table top microcentrifuge and washed with buffer A supplemented with 20 mM imidazole four times, and the bound fraction was eluted from the column with 250 mM imidazole. The excess imidazole was removed from the sample by gel filtration with a PD10 prepacked desalting column (Pharmacia). The TFII-I–SRF complex for Fig. 2B was purified similarly from TFII-I–SRF cotransfected COS-1 cells by using buffer B (20 mM Tris [pH 7.8], 100 mM KCl, 10% glycerol, 0.2% Triton X-100, 0.5 mM PMSF, and 1 μg each of leupeptin, antipain, pepstatin, and chymostatin per ml).
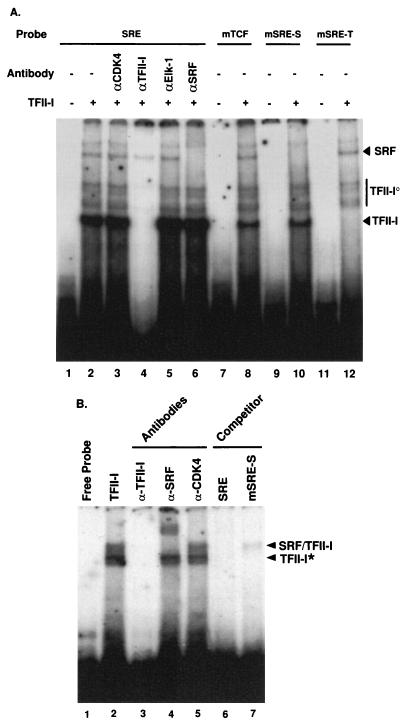
FIG. 2.
Binding of TFII-I to the c-fos SRE. (A) TFII-I protein purified from TFII-I-transfected COS-1 cells was incubated with the indicated 32P-labeled oligonucleotide probes and processed for EMSA as described in Materials and Methods. The specified antibodies were preincubated with affinity-purified TFII-I for 15 min at room temperature where indicated. (B) TFII-I purified from TFII-I–SRF-cotransfected COS-1 cells was incubated with the c-fos SRE 32P-labeled oligonucleotide probes and processed for EMSA as described in Materials and Methods. EDTA was removed from the gel and the running buffer, and 3 mM MgCl2 was added as a supplement. The specified antibodies were preincubated with the extracts for 15 min at room temperature where indicated.
Preparation of nuclear extracts.
NIH 3T3 cells were starved for 48 h with 0.5% CS in DMEM and stimulated with 25 ng of PDGF per ml for 20 min. The cells were rinsed three times with ice-cold phosphate-buffered saline (PBS). PBS containing 1 mM Na3VO4 and 5 mM NaF was added to each plate, and the cells were scraped from the dish and pelleted at 1,500 × g for 10 min at 4°C. Cells were resuspended in the same buffer and pelleted as before. The pellet was then resuspended in 0.8 ml of ice-cold hypotonic buffer, transferred to microcentrifuge tubes, and allowed to swell on ice for 15 to 30 min. The lysate was vortexed vigorously for 1 min, and the nuclei were pelleted (10,000 × g for 30 s). The nuclear pellets were resuspended in 100 to 150 μl of high-salt buffer and rotated at 4°C for 30 min. The extracted proteins were separated from residual nuclei (10,000 × g for 10 min), and the supernatant was quick-frozen in a dry ice-methanol bath. The buffer compositions were as follows. (i) Hypotonic buffer contained 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM Na4P2O7, 20 mM NaF, 1 mM dithiothreitol (DTT), 0.5 mM PMSF, and 1 μg each of leupeptin, antipain, pepstatin, and chymostatin per ml. (ii) High-salt buffer contained 20 mM HEPES (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM Na4P2O7, 20 mM NaF, 1 mM DTT, 0.5 mM PMSF, and 1 μg each of leupeptin, antipain, pepstatin, and chymostatin per ml.
EMSA.
Binding reactions for electrophoretic mobility shift assays (EMSAs) with affinity-purified recombinant TFII-I (see Fig. 2A and 3) or TFII-I–SRF complex (Fig. 2B) were done in a mixture of 20 mM Tris (pH 7.5), 100 mM KCl, 3 mM MgCl2, 5 mM DTT, 5% glycerol, 250 μg of bovine serum albumin (BSA) per ml, and 5 μg of poly(dG-dC) per ml. EMSA with nuclear extract (see Fig. 9) was done in a mixture of 20 mM Tris (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 5 mM DTT, 10% glycerol, 250 μg of BSA per ml, and 50 μg of poly(dG-dC) per ml. Four micrograms of nuclear extract was used with or without purified TFII-I. Reaction mixtures were incubated at room temperature for 15 min prior to addition of labeled probes and for another 15 min after the addition of various 32P-labeled probes (50,000 cpm). All of the EMSA probes were labeled by Klenow reaction with α-32P-dCTP after annealing of two complementary oligonucleotides. If necessary, antibodies or cold competitors (50- or 200-fold excess) were preincubated before the addition of various 32P-labeled probes. Binding reaction mixtures, except for those used for Fig. 2B (TFII-I–SRF complex EMSA), were electrophoresed through 5% polyacrylamide gels (39:1 acrylamide-bisacrylamide) containing 2.5% glycerol in 0.5× Tris-borate-EDTA buffer at room temperature. The binding reaction mixtures with the TFII-I–SRF complex (Fig. 2B) were electrophoresed through a 3.5% polyacrylamide gel (39:1 acrylamide-bisacrylamide containing 1.25% glycerol in 89 mM Tris-borate buffer without EDTA and supplemented with 3 mM MgCl2 at room temperature. The gel was then fixed, dried, and exposed to X-ray film.
FIG. 3.
Binding of TFII-I to the c-fos SIE. (A) TFII-I, prepared as described for Fig. 2A, was incubated with 32P-labeled c-fos SIE probe and processed for EMSA as described in Materials and Methods. Anti-TFII-I antibody (α-TFII-I) or the indicated unlabelled oligonucleotide competitor DNAs were preincubated with affinity-purified TFII-I for 15 min at room temperature where indicated. (B) Affinity-purified recombinant TFII-I was incubated with 32P-labeled c-fos basal probe (−60 to +18) and processed for EMSA as described in Materials and Methods. Anti-TFII-I antibody or the indicated unlabelled oligonucleotide competitor DNAs were preincubated with the extracts for 15 min at room temperature where indicated.
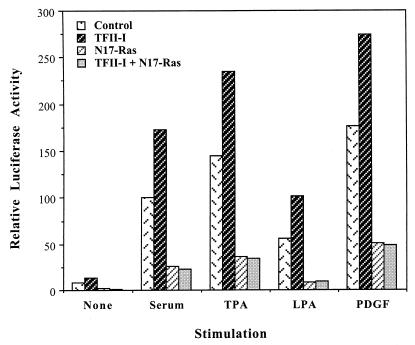
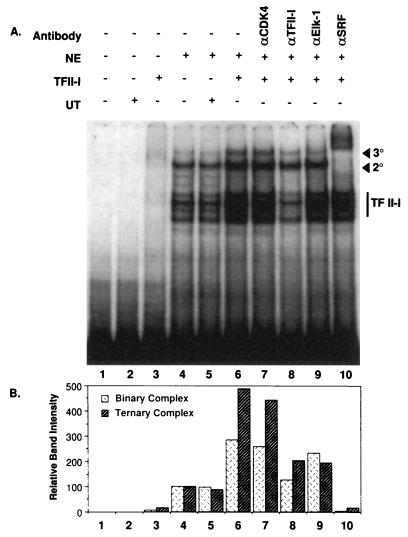
FIG. 9.
N17-Ras inhibits TFII-I enhancement of the c-fos promoter. NIH 3T3 cells were transfected with the wild-type c-fos/Luc reporter constructs and TFII-I (or empty vector) in the absence or presence of pMT3.N17Ras plasmid. Cells were serum starved for 36 h in 0.5% calf serum and then stimulated for 4 h with 10% CS, 50 ng of TPA per ml, 10 μM LPA, or 25 ng of PDGF-BB per ml in DMEM. Cell extracts were then processed for luciferase activity. Relative luciferase activities shown here are from a representative experiment, and similar results were obtained from three further independent experiments. All transfections were performed in duplicate for each experiment, and the values between the duplicates were within 10% of the mean.
The following oligonucleotides were used in EMSAs: SIE, AATTCCTGTTCCCGTCAATCCCTCCCC; m34SIE, AATTCCTGTTCaCGTCAATCCCTCCCC; m67SIE, AATTCCatTTCCCGTaAATCCCTCCCC; mTCF, AATTCTCCTTTACAactGATGTCCATATTAGGACATCTC; SRE, AATTCTCCTTTACACAGGATGTCCATATTAGGACATCTC; mSRE-S, AATTCTCCTTTACACAGGATGTCCATATTAccACATCTC; mSRE-T, AATTCTCCTTTACACAGGATGTCCtTATaAGGACATCTC; SIE/SRE, AATTCCTGTTCCCGTCAATCCCTCCCTCCTTTACACAGGATGTCCATATTAGGACATCTC; c-fos basal, AAT TCG TAGGAAG TCCATCCATTCACAGCGC T TC TATAAACGGCCAGCTGAGGCGCCTACTACTCCAACCGCGACTGCAC; −47/II-I, AATTCGTAGGAAGTCCATCCATTCACAGCGCTTCC; TATA, AATTCCACAGCGCTTCTATAAACGGCCAGCTGAG; −5/II-I, AATTCAGGCGCCTACTACTCCAACCGCGACTGCAC; m −47/II-I, AATTCGTAGGAAGTCCgggCATTCACAGCGCTTCC; and E-box, GGGCCCCCACCACGTGGTGCCTGA.
GST pulldown and Western analysis.
Transfected COS-1 cells were lysed in buffer B (20 mM Tris [pH 7.8], 100 mM KCl, 10% glycerol, 0.2% Triton X-100, 0.5 mM PMSF, and 1 μg each of leupeptin, antipain, pepstatin, and chymostatin per ml), and the lysate was then centrifuged for 10 min at 10,000 × g at 4°C. Supernatants were incubated with glutathione-Sepharose beads (Pharmacia) for 1 h at 4°C to pull down either glutathione S-transferase (GST) or GST–TFII-I. The beads were collected by quick spin with a table top microcentrifuge, washed with buffer B four times, and finally resuspended in 2× Laemmli sample buffer. Western blot analyses with anti-SRF, -Elk-1, -STAT1, -STAT3, -GST, or -phosphotyrosine (4G10) antibodies were carried out under standard conditions.
RESULTS
TFII-I binds to the c-fos SRE and forms a complex with SRF in vivo.
TFII-I binds to a subset of consensus Inr sequences (49). Comparison of the Inr consensus-binding sequence (YYAN[A/T]YY) (26) to that of the c-fos promoter indicates four possible TFII-I binding sites, three of which are shown in Fig. 1. Two of these potential binding sites overlap the c-fos SIE and SRE, and the other two (−47 and −5) are located in the basal region of the c-fos promoter. To determine whether TFII-I can bind to the c-fos SRE, recombinant TFII-I protein affinity purified from His6–TFII-I-transfected COS-1 cells was incubated with 32P-labeled SRE probe and analyzed by EMSA. The results are shown in Fig. 2A. Several bands are shifted with this probe. To determine which of these bands contain TFII-I, anti-TFII-I antibody was added to the binding reaction mixture prior to electrophoresis. From lane 4, the anti-TFII-I antibody appeared to disrupt most efficiently the lower, heavier band and a series of slightly higher bands (indicated as TFII-I and TFII-Io, respectively, in Fig. 2A). The multiple TFII-I-containing bands (TFII-Io) are presumably due to processing, differential phosphorylation, or formation of complexes with other factors. Interestingly, one of the weak upper bands that did not react with the TFII-I antibody ran at a position that was consistent with the position of SRF. To determine whether this band in fact contained SRF, an anti-SRF antibody was added to the binding reaction mixture prior to electrophoresis. From lane 6, it can be seen that the anti-SRF antibody specifically disrupted one of the upper bands (indicated as SRF in Fig. 2A), indicating that this weak band does contain SRF. These data demonstrate that endogenous SRF or SRF-related protein from COS-1 cells forms an in vivo complex with TFII-I and copurifies with it. SRF and TFII-I also form a complex that can be coprecipitated from transfected cells (see Fig. 7). The interaction between TFII-I and SRF is relatively strong in solution, since His6–TFII-I was purified over the Ni-NTA agarose column in the presence of 1% Triton X-100 and 500 mM KCl, which are relatively stringent conditions. Control extracts from cells without transfected His6–TFII-I run over the Ni-NTA agarose column under the same conditions showed neither TFII-I nor SRF binding activity (data not shown). Anti-Cdk4 and anti-Elk-1 antibodies failed to react with any of the shifted bands (lanes 3 and 5). Likewise, anti-STAT1 and anti-STAT3 antibodies also failed to shift any of the bands (data not shown). Neither TFII-I nor Elk-1 appears to be in the SRF complex observed on the band shift gel in Fig. 2A run in the presence of EDTA. Thus, despite the fact that the TFII-I complex copurifies with SRF in solution over the Ni-NTA agarose beads, the complex dissociates under the conditions of this EMSA. This behavior has been noted for other transcription factors that interact with TFII-I (35).
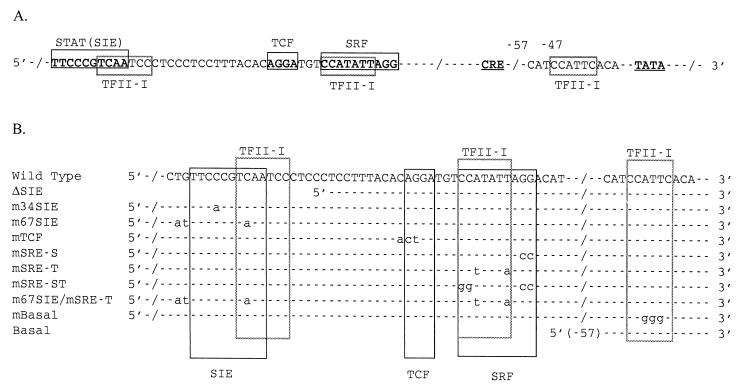
FIG. 1.
Diagram of c-fos promoter sequences and binding sites. (A) Major elements and TFII-I binding sites in the c-fos promoter. (B) Mutant c-fos promoters. (The wild-type sequence is given for comparison.)
FIG. 7.
TFII-I forms in vivo complexes with SRF, STAT1, and STAT3. SRF (A), Elk-1 (B), STAT1 (C), and STAT3 (D) expression plasmids were cotransfected into COS-1 cells with pEBG (lanes 1 and 3) and pEBG-TFII-I (lanes 2 and 4) expression plasmids. Transfected COS-1 cells were lysed and subjected to GST pulldown assay as described in Materials and Methods. Ten micrograms of each total lysate (lanes 1 and 2) and glutathione (GSH)-Sepharose bead-bound proteins (lanes 3 and 4) were fractionated by SDS-PAGE (12% polyacrylamide). Proteins associating with GST or GST–TFII-I were detected by Western blot analyses with anti-SRF (A), anti-Elk-1 (B), anti-STAT1 (C), or anti-STAT3 antibody (Ab).
mSRE-S and mSRE-T mutants dissociate SRF and TFII-I binding to the SRE.
To determine whether the binding of SRF and TFII-I to the SRE could be discriminated, two mutant SRE probes were designed and synthesized. The mSRE-S probe has mutations at the 3′ end of the CArG box, which is outside of the TFII-I binding site (Fig. 1B), and the mSRE-T probe has mutations in the internal core region of the CArG box that leave the exterior C’s and G’s intact (Fig. 1B). It has been shown that SRF binding can tolerate these internal core base changes (42), but the mSRE-T mutation should eliminate the binding of TFII-I (26). Therefore, the mSRE-S mutant was predicted to be specifically defective for SRF binding, leaving TFII-I binding intact and vice versa for the mSRE-T mutant. When each of these oligonucleotides was used as a probe in the binding assay, the results were as predicted. The mSRE-S oligonucleotide fails to form the upper band which contains SRF (Fig. 2A, lane 10), and the mSRE-T probe has diminished binding of the lower band which contains TFII-I (Fig. 2A, lane 12). The band patterns in the EMSA are not significantly affected by introduction of the mutations in the TCF site of the SRE probe (Fig. 2A, lane 8).
The TFII-I–SRF complex can bind to c-fos SRE.
In order to demonstrate that TFII-I and SRF can bind to the c-fos SRE as a complex, modified EMSA conditions were used. TFII-I was purified through the Ni-NTA agarose column from TFII-I- and SRF-co-overexpressing COS-1 cells to maximize the formation of TFII-I–SRF complex. EDTA was removed from both the EMSA polyacrylamide gel and running buffer, and 3 mM MgCl2 was used as a supplement to stabilize the DNA-protein complexes. The results are shown in Fig. 2B. Two bands are shifted under these conditions (lane 2), and both of these bands are diminished by the addition of anti-TFII-I antibody, indicating that both bands contain TFII-I (lane 3). The upper band (indicated as TFII-I/SRF on the figure) in addition can be shifted with anti-SRF antibody (lane 4), but not with an anti-Cdk4 antibody (lane 5). Thus, the upper band contains both TFII-I and SRF. It also can be concluded that TFII-I DNA binding contributes to the binding of the TFII-I–SRF complex to the SRE, because the mSRE-S competitor oligonucleotide, which is specifically defective for SRF binding while leaving TFII-I binding intact, efficiently competes out the binding (lane 7). If the binding of the TFII–SRF complex were primarily determined by SRF binding to c-fos SRE, then the mSRE-S oligonucleotide should not have competed out the binding.
m34SIE and m67SIE mutants dissociate STAT and TFII-I binding to the SIE.
To determine whether TFII-I can bind to the c-fos SIE, the same affinity-purified, recombinant TFII-I as that shown in Fig. 2A was incubated with 32P-labeled SIE probe and analyzed by EMSA. The results are shown in Fig. 3A. A major band (indicated as TFII-I in Fig. 3) can be seen in lane 2 and is abolished by the addition of anti-TFII-I antibody (lane 3), suggesting that TFII-I does bind to c-fos SIE. Competition experiments with the 32P-labeled SIE probe and 50- or 200-fold excess cold SIE, m34SIE, m67SIE, and SRE oligonucleotides as competitors were also carried out, and the results are shown in lanes 4 to 11. Both the wild-type SIE and the m34SIE (Fig. 1B), which is defective for STAT binding (68), can compete for TFII-I binding (lanes 4 to 7). Interestingly, the high-affinity mutant SIE (m67SIE), which binds STAT proteins with greater affinity (68), has a mutation that disrupts the consensus binding motif for TFII-I (Fig. 1B) and does not compete efficiently for TFII-I binding, even in the presence of 200-fold excess unlabeled probe (lanes 8 and 9). Thus, consistent with the results of Grueneberg et al. (13), the m34SIE mutant is specifically defective for STAT binding, leaving TFII-I binding intact and vice versa for the m67SIE mutant. Moreover, the finding that the SRE oligonucleotide also competes very effectively for TFII-I binding against the SIE (lanes 10 and 11) suggests that the same DNA binding domain of TFII-I is utilized for binding of both SRE and SIE.
TFII-I can bind to c-fos basal promoter through the −47 TFII-I site.
Two possible TFII-I binding sites can be found in the basal region of murine c-fos promoter at −47 (−50 catccattcaca −39) and −5 (−8 ctactactccaac +3) (underlined sequences match the TFII-I consensus sequence). (Note that the putative +1 site is taken from the GenBank murine c-fos entry, accession no. V00727.) To determine whether TFII-I can bind these sites, purified recombinant TFII-I was incubated with 32P-labeled c-fos basal probe (−60 to +18) and analyzed by EMSA. The results are shown in Fig. 3B. TFII-I can bind to the c-fos basal probe (lane 2), and this band is abolished either by anti-TFII-I antibody (lane 3) or by self competitor (lane 4), showing the specificity of this binding. SRE unlabeled competitor can also compete this binding (lane 6), suggesting that TFII-I utilizes the same DNA binding domain for c-fos basal probe binding as well as SRE binding. This domain is distinct from that used in E-box binding (lane 5) to which TFII-I also binds (41, 42). This observation is consistent with the notion that TFII-I has two different DNA binding domains.
In order to locate the functional TFII-I binding site in the basal probe, three different regions of c-fos basal probe, including the −47 TFII-I site (indicated as −47/II-I; −60 to −32), TATA box (indicated as TATA; −42 to −14), and −5 TFII-I site (indicated as −5/II-I; −11 to +18) were used as unlabeled competitor DNAs (lanes 7 to 9). The results demonstrate that only the sequence at −47 is capable of competing for TFII-I binding (lane 7). Moreover, an oligonucleotide (indicated as m−47/II-I in lane 10) which has mutations in the consensus sequence for TFII-I binding (see mBasal in Fig. 1) was not able to compete for TFII-I binding, demonstrating that the binding between TFII-I and c-fos basal probe is mediated by the TFII-I binding motif present at the −47 location. Thus, TFII-I can bind the basal region of c-fos promoter as well as upstream enhancers, including the SIE and SRE. However, mutation of this site had no significant effect on c-fos promoter activation by serum in a transient assay (data not shown). In addition, TFII-I appears to be unable to bind to the putative −5 site, which is the location nearest the initiation site. Moreover, this putative Inr sequence is not conserved between the mouse (CTACTC) and human (GTACTC) c-fos promoters (change underlined), though the other 3 TFII-I binding sites are (43). Thus, it is unlikely that c-fos has a functional Inr element, and since TFII-I does not bind near the initiation site as well, it is highly unlikely that TFII-I functions as a classical initiator factor in the c-fos promoter.
TFII-I enhances c-fos promoter activation in vivo in a manner that is dependent on upstream elements.
The fact that TFII-I can bind to the c-fos SRE and SIE and can form a complex with SRF suggests that TFII-I may be involved in c-fos promoter regulation. To determine whether TFII-I can functionally affect c-fos induction, TFII-I was cotransfected with a wild-type c-fos promoter driving a luciferase reporter gene into NIH 3T3 fibroblasts, and luciferase activity was measured before and after simulation with various agents. The results are shown in Fig. 4A. TFII-I cotransfection enhanced the activity of the wild-type c-fos promoter two- to fourfold. This enhancement occurred in response to a variety of c-fos stimulators, including serum, TPA, LPA, and PDGF. Since these agents activate a variety of c-fos signal transduction pathways, this result suggests that TFII-I has an impact on more than one signal transducer in the c-fos promoter and may be involved in enhancing the cooperativity between them.
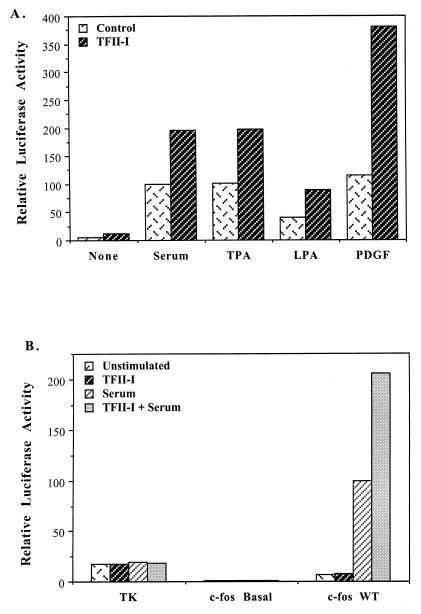
FIG. 4.
TFII-I enhances c-fos promoter activation. (A) NIH 3T3 cells were transfected with the wild-type (WT) c-fos/Luc reporter constructs and TFII-I. For control cultures, the empty vector of TFII-I expression plasmid was cotransfected with reporter constructs. Cells were serum starved for 36 h in 0.5% CS and then stimulated for 4 h with 10% CS, 50 ng of TPA per ml, 10 μM LPA, or 25 ng of PDGF-BB per ml in DMEM. Cell extracts were then processed for luciferase activity. Relative luciferase activities shown here are from a representative experiment, and similar results were obtained from four further independent experiments. All transfections were performed in duplicate for each experiment, and the values between the duplicates were within 10% of the mean. (B) TFII-I fails to transactivate the TK and c-fos basal promoters. The TK/Luc, c-fos basal (−57)/Luc, and wild-type c-fos/Luc reporter constructs were transfected into NIH 3T3 cells with or without TFII-I. Unstimulated or 10% CS-stimulated cells were then analyzed for luciferase activity. The data shown are the average of three independent experiments, and the standard deviations between experiments were on average 30% of the values shown here. All transfections were performed in duplicate for each experiment, and the values between the duplicates were within 10% of the mean.
Since TFII-I was originally isolated as a basal transcription initiator factor, the effects of TFII-I on the activation of truncated (−57) c-fos basal (Fig. 1B) or TK promoter by serum were also examined. The results are shown in Fig. 4B. c-fos promoter activity in the absence of upstream elements is not affected by TFII-I cotransfection. In addition, ectopic TFII-I fails to enhance the activity of the Inr-less TK promoter under either serum-stimulated or unstimulated conditions. Thus, the enhancement effect of TFII-I on the c-fos promoter requires the presence of upstream elements which are not present in either the truncated c-fos basal promoter or the TK promoter.
To determine which c-fos upstream elements are required for enhancement by TFII-I, a variety of reporter constructs containing point mutations or small deletions in the wild-type c-fos promoter were generated and tested for their response to ectopically expressed TFII-I during serum induction of c-fos promoter. The mutations introduced into the reporters include ΔSIE, m34SIE, m67SIE, mTCF, mSRE-S, mSRE-T, mSRE-ST, and each mutation is denoted in Fig. 1B. The results are summarized in Fig. 5 and 6.
FIG. 5.
Effect of mutations of the c-fos SRE on TFII-I transactivation. NIH 3T3 cells were transfected with the indicated c-fos/Luc reporter constructs with or without TFII-I. Cells were serum starved for 36 h in 0.5% calf serum and then stimulated for 2.5 h with 10% CS in DMEM. Cell extracts were then processed for luciferase activity. Data shown in panel A are from a representative experiment, and similar results were obtained from three qualitatively similar and independent experiments. All transfections were performed in duplicate for each experiment, and the values between the duplicates were within 10% of the mean. WT, wild type. Data shown in panel B are the comparison of the relative enhancement of each mutant reporter gene by TFII-I cotransfection to the fold enhancement of the wild-type c-fos reporter activation by TFII-I.
FIG. 6.
Effect of mutations of the c-fos SIE on TFII-I transactivation. NIH 3T3 cells were transfected with the indicated c-fos/Luc reporter constructs with or without TFII-I. Cells were serum starved for 36 h in 0.5% CS and then were stimulated for 2.5 h with 10% CS in DMEM. Cell extracts were then processed for luciferase activity. Data shown in panel A are from a representative experiment, and similar results were obtained from three qualitatively similar and independent experiments. All transfections were performed in duplicate for each experiment, and the values between the duplicates were within 10% of the mean. Data shown in panel B are the comparison of the relative enhancement of each mutant reporter gene by TFII-I cotransfection to the fold enhancement of the wild-type (WT) c-fos reporter activation by TFII-I.
Grueneberg et al. (13) found that TFII-I can cooperate with Phox1 to enhance the activity of a minimal SRE reporter construct. To study the effects of TFII-I by itself on the SRE in the context of the wild-type c-fos promoter, different SRE mutations, including mSRE-S and mSRE-T, which preferentially diminish binding of either TFII-I or SRF to the SRE, were examined. Figure 5 shows the results. Interestingly, the mSRE-T mutant reporter, which is specifically defective for TFII-I binding while leaving SRF binding intact, caused significant loss of promoter activity (40% level of the wild type) upon serum stimulation. This result suggests that the binding of endogenous TFII-I to the SRE may be necessary for maximal induction of the c-fos promoter. The mSRE-T mutant could not be further stimulated by cotransfection of TFII-I, suggesting that SRF–TFII-I protein interactions cannot overcome the loss of the TFII-I DNA binding site. The mSRE-S mutant reporter, which is specifically defective for SRF binding, leaving TFII-I binding intact, substantially lowered the activity of the promoter (15% of that of the wild type) as expected. However, in contrast to mSRE-T, a small but significant enhancement of promoter activity by TFII-I cotransfection was still observed with the mSRE-S mutation. One interpretation of this result could be that TFII-I can facilitate the recruitment of SRF to a weak SRE through SRF–TFII-I protein-protein interaction. A similar effect has been found for the Phox1 protein (15). The TFII-I enhancement effect on mSRE-S was completely eliminated in the mSRE-ST mutant reporter, which is defective in both SRF and TFII-I binding (Fig. 1B), confirming again that the functional effect of TFII-I requires its own intact binding site.
Next we sought to determine whether the TCF site might influence the effect of TFII-I on c-fos induction. Under the conditions we used, TCF was not copurified with TFII-I (see Fig. 2 and Fig. 7). As shown in Fig. 5, mTCF reporter, which is defective for TCF binding, exhibited a decreased response (50%) to the serum stimulation. Luciferase expression from the mTCF construct could still be enhanced by TFII-I overexpression, albeit with less efficiency (50%) than with the wild-type c-fos reporter. These results indicate that the activity of TFII-I is not strictly dependent on the TCF site, but this site contributes to the maximum effect of TFII-I.
Since we have found that TFII-I can bind to the SIE, we examined the effects of mutations in the SIE on TFII-I-mediated transactivation of the c-fos promoter. As described previously, deletion of the SIE from the promoter (ΔSIE) caused a significant reduction in the promoter response to serum stimulation (30% of the that of the wild type) (21). In addition, the ΔSIE reporter completely lost the ability to respond to TFII-I cotransfection, demonstrating that the intact SIE region of the promoter is required for the effect of TFII-I even in the presence of a wild-type SRE (Fig. 6). In order to dissociate the role of the STAT factors and TFII-I at the c-fos SIE, the activation of m34SIE and m67SIE reporters by serum was investigated (Fig. 6). The m67SIE mutation, which binds poorly to TFII-I, but retains binding to the STAT proteins, reduced the serum response of the promoter to 50% of the wild-type levels. This result suggests that the loss of endogenous TFII-I binding to the promoter is significant for c-fos induction even in the presence of STAT binding and SRF binding. The expression of the m67SIE reporter could not be further enhanced by overexpression of TFII-I, indicating that the binding of TFII-I to the SIE site is critical for the maximum effect of TFII-I on the c-fos promoter (Fig. 6). Interestingly, elimination of STAT binding by the m34SIE mutation also abolished promoter enhancement by TFII-I overexpression, even though the TFII-I binding site is left intact. In the absence of TFII-I overexpression, the m34SIE mutation, which is specifically defective for STAT binding, showed a level of serum inducibility similar to that of the wild-type c-fos promoter. This was not unexpected, since the STAT factors are not strongly induced over the basal levels by serum stimulation of NIH 3T3 cells (18, 68). This result suggests that some binding of STAT proteins to the SIE is required for the maximal activity of TFII-I. This observation prompted us to look more carefully for STAT–TFII-I interactions.
SRF, STAT1, and STAT3 can form complexes with TFII-I in vivo.
To attempt to detect protein complexes in solution between TFII-I and the STAT factors, GST–TFII-I (or GST) and STAT1 (or STAT3) expression plasmids were cotransfected into COS-1 cells. TFII-I complexes were isolated by glutathione-Sepharose pulldown assay, and the resulting complexes were separated by SDS-PAGE and Western blotted with anti-STAT1 (or anti-STAT3) antibodies. STAT1 and STAT3 were selected to investigate, since they are known to bind to the c-fos SIE (34, 51, 52, 71). As can be seen in Fig. 7C, significant complex formation was observed between STAT1 and TFII-I and between STAT3 and TFII-I in Fig. 7D. No interaction between Elk-1 and TFII-I could be detected under the same experimental conditions (Fig. 7B) or with cotransfected GST, indicating that this is a specific association.
To further confirm that our previously observed interaction between TFII-I and SRF can occur in vivo, GST–TFII-I (or GST) and SRF expression plasmids were also cotransfected into COS-1 cells and GST pulldown followed by Western blotting with an anti-SRF antibody was carried out. As can be seen in lane 4 of Fig. 7A, a specific interaction between SRF and TFII-I was observed.
TFII-I enhances the formation of binary and ternary complexes in vitro.
To examine the effects of TFII-I on the formation of complexes to the c-fos promoter, a probe containing both the SIE and SRE was labeled and allowed to bind to nuclear extract from NIH 3T3 fibroblasts in the absence and presence of TFII-I. The results are shown in Fig. 8. Overexpressed TFII-I purified from COS-1 cells was selectively added to the reaction mixtures. A limited amount of TFII-I, which was not itself sufficient to give detectable DNA binding, as shown in lane 3, was added to the nuclear extract in vitro and incubated with the c-fos probe. This resulted in a threefold-enhanced formation of an SRF binary complex (indicated as 2o) and a fivefold increase in SRF-TCF ternary complex (indicated as 3o) as shown in lane 6 (compared to lane 4). Interestingly, TFII-I binding to DNA was itself enhanced by the presence of the nuclear extract. Addition of an untransfected control lysate eluted from an Ni-NTA agarose column (indicated as UT) did not have either DNA binding activity (lane 2) or a complex enhancement effect (lane 5). Anti-Elk-1 antibody weakened primarily the ternary complex (lane 9) by as much as 60%, and anti-SRF antibody shifted both the binary and ternary complexes (lane 10) as expected. Anti-TFII-I antibody diminished the intensities of TFII-I band and, more importantly, reduced the intensity of the binary and ternary complex bands to control levels (lane 8). This observation confirms that the enhanced complex formation requires TFII-I. Anti-Cdk4 antibody was used as a negative control and had no effect (lane 7).
FIG. 8.
TFII-I enhances the protein-DNA complex formation to the c-fos SRE. A 32P-labeled c-fos SIE-SRE probe was incubated with PDGF-stimulated nuclear extracts (NE) in the presence (+) or absence (−) of TFII-I. TFII-I was affinity purified from transiently transfected COS-1 cells. UT (+) lanes contained the 250 mM imidazole-eluted fraction of vector-transfected COS-1 cell extracts passed over Ni-NTA agarose columns. Antibodies (αCDK4, αTFII-I, αElk-1, and αSRF) were preincubated with the extracts for 15 min at room temperature where indicated. Final reaction mixtures were analyzed by EMSA as described in Materials and Methods. Band intensities for binary and ternary complexes in control nuclear extract (lane 4) were considered as 100%, and the relative band intensities in other samples are represented as compared to that of lane 4.
Regulation of TFII-I by signal transduction pathways.
Since MAP kinase phosphorylation of bacterial TFII-I can activate its transcriptional activity in vitro (40a), we examined whether ras activation is required for the TFII-I activity. To do this, dominant negative N17-ras was cotransfected along with the wild-type c-fos reporter in the absence or presence of TFII-I and assayed for luciferase activity after stimulation by serum, TPA, LPA, or PDGF. As can be seen in Fig. 9, dominant negative ras not only diminished the induction of the c-fos promoter significantly but also completely abolished the ability of TFII-I to enhance the c-fos promoter. This result indicates that the ras pathway must be functioning for TFII-I to exert its effect on c-fos promoter. It is noteworthy that N17-ras does not completely abolish c-fos induction, but does completely abolish TFII-I enhancement of the promoter. This suggests that TFII-I does not simply enhance the activity of other transcription factors, but requires ras signaling for its own function.
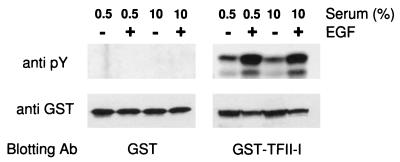
TFII-I is phosphorylated on both serine and tyrosine residues (40). In B cells, TFII-I can associate with Btk and become tyrosine phosphorylated by it (69). To determine whether TFII-I can be a direct target of signaling pathways stimulated by growth factors, GST–TFII-I (or GST) expression plasmid was transfected into COS-1 cells, and the phosphorylation of TFII-I on tyrosine residues was examined with or without EGF stimulation. Cell lysates were subjected to GST pulldown assay and analyzed by Western blotting with antiphosphotyrosine antibody (4G10) or anti-GST antibody. As can be seen in Fig. 10, EGF stimulation caused a significant increase in the phosphorylation of TFII-I on tyrosine residues. The anti-GST blot shows that the expression levels of transfected protein are comparable between samples. In parallel experiments with COS-1 cells, we found that TFII-I enhanced the activity of the c-fos promoter in response to EGF (data not shown). Thus, tyrosine phosphorylation may also regulate the transcriptional activity of TFII-I.
FIG. 10.
EGF stimulates TFII-I tyrosine phosphorylation. GST (left panels) and GST–TFII-I (right panels) expression plasmids were transfected into COS-1 cells. Transfected COS-1 cells were maintained in DMEM supplemented with either 0.5 or 10% FCS for 36 h and stimulated with 20 ng of EGF per ml for 20 min. Cells were lysed and subjected to GST pulldown assay as described in Materials and Methods. GSH-Sepharose bead-bound proteins were fractionated by SDS-PAGE (12% polyacrylamide). The tyrosine phosphorylation or protein level was detected by Western blot analyses with antiphosphotyrosine antibody (Ab) or anti-GST antibody.
DISCUSSION
The c-fos promoter is upregulated by a variety of extracellular ligands, and a number of transcription factors have been found to play a role in the regulation of this promoter. These transcription factors respond to pathways initiated by calcium, cyclic AMP, tyrosine kinases, phorbol esters, and serum (66). Three of these transcription factors—CREB, STAT, and p62TCF—have been shown to be phosphorylated in response to specific stimulation and have their activities regulated by this phosphorylation (22). SRF has been shown to bind cooperatively with members of the Ets family (Elk-1, SAP-1, and NET1) whose transcriptional activity is regulated by MAP kinase-related pathways (67). However, there is considerable evidence that this is not the only mechanism by which the c-fos SRE regulates the c-fos promoter. Several other transcription factors have been found either to bind to the SRE independently or to interact with SRF, including Phox1, YY1, ATF6, RAP74, and C/EBP (16, 28, 33, 37, 60, 72). Moreover, serum appears to induce c-fos through the SRE in a manner that involves the small G proteins, such as RhoA, Rac1, and CDC42, and in a way that is independent of the TCF binding site and of the TCF transcription factors (23, 27). Although RhoA, Rac1, and CDC42 upregulate c-fos through the SRE and presumably SRF, the molecular mechanisms underlying this signal transduction cascade are poorly understood. In addition, experiments with introduction of c-fos promoter constructs into transgenic mice have indicated that there may be more functional interdependence between c-fos promoter elements than has sometimes been apparent from tissue culture experiments (45).
Consistent with Grueneberg et al. (13), we find that the transcription factor TFII-I associates with the SRF in solution. In addition, we find that the TFII-I–SRF complex can bind to the SRE. Moreover, we find that TFII-I can enhance transcriptional activation of the c-fos promoter in a manner that is dependent on the SRE and SIE upstream elements. Interestingly, cotransfection of TFII-I with the basal c-fos promoter (−57) or with the TK promoter resulted in no enhancement. Previously, TFII-I had been notable for its ability to direct initiation at promoters which contained initiator elements in place of or in addition to the TATA box (48). However, TFII-I also interacts with various gene-specific activators, including c-myc, USF1, and NF-κB and can function as an upstream activator in the absence of a functional Inr element (47). Consistent with the latter observations, here we show that TFII-I appears to function as an upstream activator of the c-fos promoter. Moreover, we have found that the Inr-related sequence at −5 of the c-fos promoter fails to bind to TFII-I in vitro. Although it is unclear whether the c-fos promoter contains a functional Inr, the latter data indicate that TFII-I is not functioning as an initiator binding protein in the c-fos promoter.
Recent experiments have found that phosphorylation of TFII-I by MAP kinase regulates its initiator function in vitro (40a). Moreover, in B cells, TFII-I forms a complex with the Btk tyrosine kinase, and its phosphorylation on tyrosine is stimulated by it (69). Recent data (40a) also indicate that the kinase activity of Btk can regulate TFII-I transcriptional activity in vivo. These observations suggest that TFII-I is a mediator of multiple signal transduction pathways and thus could well be involved in signal transduction to the c-fos promoter. Consistent with these experiments, we find that EGF enhances phosphorylation of TFII-I on tyrosine and that the enhancement of the c-fos promoter activity by TFII-I is inhibited by cotransfection with dominant negative ras. Since dominant negative ras completely inhibits the effect of TFII-I, but not all c-fos induction, this suggests that TFII-I function itself is regulated by a ras-dependent pathway. This effect could be mediated through the inhibition of phosphorylation of TFII-I by one of the MAP kinase family proteins. Dominant negative ras can inhibit MAP kinase activation by EGF as well as activation of RhoA and Rac. (See the reference by Hunter [25] for review.) Thus, any of several signal transduction pathways could be regulating TFII-I, including the RhoA-mediated pathway. Future experiments will be directed toward delineating the ras-dependent pathways as well as the effects of tyrosine phosphorylation on TFII-I function.
It is interesting to note that the role of TFII-I in c-fos activation resembles that of Ste12p in the mating pheromone response in yeast (Saccharomyces cerevisiae). We have shown that the a-mating factor can stimulate gene expression in yeast through the Mcm1p and MATα1p transcription factors bound to the PQ box of α-cell-specificity genes (53, 54). MCM1 has homologies to the serum response factor, and MATα1 is in many ways analogous to c-fos TCF (19). We previously demonstrated that the Mcm1p-MATα1p complex has activity that is completely dependent on the STE12 gene (53). Although Ste12p can bind to specific DNA sequences and can mediate the pheromone response at genes such as FUS1, at the PQ box, it appears to be participating through protein-protein interactions. Despite the genetic requirement for STE12, Ste12p is difficult to find in complexes with Mcm1p and MATα1p in a bandshift assay (70). Overexpression of STE12 can enhance expression from the PQ box just as TFII-I enhances expression from the SRE (5). Ste12p itself is regulated by the MAP kinase-dependent pathway in response to pheromone (62). TFII-I also appears to be dependent on a ras-dependent pathway. Although sequence homologies between TFII-I and STE12 are minimal, it is possible that these proteins have conserved functions. It is also of interest that the consensus DNA binding site for Ste12p closely resembles the consensus binding site for TFII-I (6). These analogies are made even more intriguing by the fact that Ste12p is one of the few proteins in the yeast pheromone response pathway that does not yet have a mammalian homolog. This pathway includes the yeast homologs of CDC42 and MAP kinase (7, 10, 61). Further experiments are necessary to determine whether there may be functional conservation between STE12 and TFII-I.
Although TFII-I can enhance the activity of the c-fos promoter in a cotransfection experiment, it remains an open question as to whether endogenous TFII-I has an essential function in c-fos regulation in vivo. Several experiments presented here suggest that, in fact, it does. Although the effects of cotransfected TFII-I on the c-fos promoter are only two- to fourfold, there is endogenous TFII-I in the transfected cells. Therefore, it is entirely possible that endogenous TFII-I has exerted much of its effect in the absence of overexpression of the exogenous gene. Moreover, endogenous SRF copurifies with transfected TFII-I, suggesting an in vivo functional role for TFII-I in SRF activity. In addition, the mSRE-T mutation, which shows reduced TFII-I binding but wild-type levels of SRF binding, is significantly impaired in its ability to activate the c-fos promoter. This result suggests that binding of endogenous TFII-I to the SRE is required for full transcriptional activity of the promoter. Although it could be argued that the mSRE-T mutation may also impair the binding of factors other than TFII-I at the SRE, a similar effect is observed by mutation of the TFII-I binding site at the SIE in conjunction with mutation of the STAT binding site. It is also relevant that the effects of TFII-I on the c-fos promoter are impaired by mutations of the STAT and SRF binding sites. This result indicates that the enhancement observed is not simply due to enhanced binding of TFII-I to the promoter, but requires cooperation with these other factors.
The mechanism by which TFII-I enhances c-fos promoter activation is uncertain at present. The enhancement effect is dependent on the TFII-I binding sites in the promoter as well as the SRF and STAT binding sites. Consistent with this, we have found that binding of the TFII-I–SRF complex to the SRE is competed away by the mSRE-S oligonucleotide that binds TFII-I, but not SRF. This indicates that TFII-I DNA binding activity contributes to the binding of the complex to DNA. It is possible in addition that TFII-I facilitates SRF binding to DNA. This would be consistent with our finding that limited amounts of TFII-I enhance formation of SRF-DNA complexes. A mechanism such as this has been proposed for the ability of the paired-like homeodomain protein Phox1 to stimulate SRF binding and function (14). Interestingly, it has been shown recently that TFII-I can also interact with the Phox1 protein (13). It is not clear whether the Phox1 protein is expressed in NIH 3T3 or COS-1 cells or whether it is present in the affinity-purified TFII-I used in our experiments. Since the TFII-I used in our experiments was generated by overexpression in COS-1 cells, it is unlikely that Phox1, even if it is expressed endogenously, would be present stoichiometrically with TFII-I. However, it is also possible that other paired-like homeodomain proteins are present in our extracts and could have similar activities to Phox1 (15).
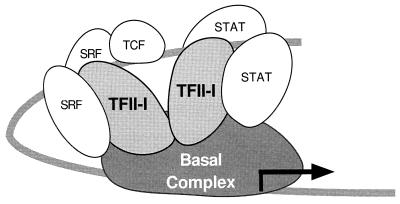
The fact that mutation of the TFII-I site in the SIE reduces the response of the promoter to TFII-I even in the presence of the wild-type SRE and vice versa suggests that TFII-I may play an important role in the mediating the functional interdependence of these elements. Further indication of cooperativity between the TFII-I binding sites is that serum induction of either the m67SIE or mSRE-T was not further repressed by double mutation of both sites simultaneously (data not shown). Since TFII-I can form complexes with both STATs and SRF as well as bind to multiple DNA sites, it would be well positioned to play an important role in the formation of a higher-order enhanceosome structure as HMG I(Y) does for the beta interferon gene (64). A possible model of this is shown in Fig. 11. However, it is uncertain whether the cooperativity between the SIE and SRE is mediated through the formation of a higher-order complex of SRE and SIE binding proteins or through independent contacts with the basal transcription machinery. TFII-I might act as an adapter protein connecting the upstream transcriptional activators to basal promoter elements (47). Since it is known that TFII-I has basal transcription activity and can interact with TATA box binding protein, this would be plausible (48). Moreover, there is a TFII-I binding site at −47 of the c-fos basal promoter in addition to the TATA box. Thus, TFII-I could directly couple the SIE and SRE to the c-fos basal promoter through a combination of protein-protein and/or protein-DNA interactions. Further experiments are necessary to test this hypothesis.
FIG. 11.
Model of the role of TFII-I on the c-fos promoter. After c-fos induction, activated TFII-I facilitates the formation of a putative enhanceosome structure consisting of SRF, TCF, STATs, and TFII-I. This results in the formation of a surface in which each activation domain can form optimal contacts with the basal transcription machinery.
ACKNOWLEDGMENTS
We are grateful to Beth Harvat, Kip Wharton, Peter Shaw, Larry Feig, and Richard Treisman for providing some of the plasmids used in these experiments. We thank Deepa Bhavsar for preparing the antisera to the STAT proteins and Daniel Ortiz for helpful advice. We thank Dorre Grueneberg and Michael Gilman for sharing unpublished results.
This work was supported by awards from the Concern Foundation for Cancer Research and the American Cancer Society (RPG-98-104-01-TBE) to A.L.R. and by NIH grant R01-GM51551 to B.H.C.
REFERENCES
- 1.Berkowitz L A, Riabowol K T, Gilman M Z. Multiple sequence elements of a single functional class are required for cyclic AMP responsiveness of the mouse c-fos promoter. Mol Cell Biol. 1989;9:4272–4281. doi: 10.1128/mcb.9.10.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Cheriyath, V., C. D. Novina, and A. L. Roy. Submitted for publication.
- 2.Cochran B H, Zullo J, Verma I M, Stiles C D. Expression of the c-fos gene and of a fos-related gene is stimulated by platelet-derived growth factor. Science. 1984;226:1080–1082. doi: 10.1126/science.6093261. [DOI] [PubMed] [Google Scholar]
- 3.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 4.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolan J W, Fields S. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 1990;4:492–502. doi: 10.1101/gad.4.4.492. [DOI] [PubMed] [Google Scholar]
- 6.Dolan J W, Kirkman C, Fields S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc Natl Acad Sci USA. 1989;86:5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elion E A, Grisafi P L, Fink G R. FUS3 encodes a cdc2+/CDC28− related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 8.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X-Y, Zhang J-J. Transcription factor p91 interacts with the epidermal growth factor receptor and mediates activation of the c-fos gene promoter. Cell. 1993;74:1135–1145. doi: 10.1016/0092-8674(93)90734-8. [DOI] [PubMed] [Google Scholar]
- 10.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 11.Gilman M Z. The c-fos serum response element responds to protein kinase C-dependent and -independent signals but not to cyclic AMP. Genes Dev. 1988;2:394–402. doi: 10.1101/gad.2.4.394. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–442. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 13.Grueneberg D A, Henry R W, Brauer A, Novina C D, Cheriyath V, Roy A L, Gilman M. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev. 1997;11:2482–2493. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grueneberg D A, Natesan S, Alexandre C, Gilman M Z. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 15.Grueneberg D A, Simon K J, Brennan K, Gilman M. Sequence-specific targeting of nuclear signal transduction pathways by homeodomain proteins. Mol Cell Biol. 1995;15:3318–3326. doi: 10.1128/mcb.15.6.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gualberto A, lePage D, Pons G, Mader S L, Park K, Atchison M L, Walsh K. Functional antagonism between YY1 and the serum response factor. Mol Cell Biol. 1992;12:4209–4214. doi: 10.1128/mcb.12.9.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvat B L, Wharton W. Serum response element and flanking sequences mediate the synergistic transcriptional activation of c-fos by 12-O-tetradecanoylphorbol-13-acetate and cholera toxin in AKR-2B cells. Cell Growth Differ. 1995;6:955–964. [PubMed] [Google Scholar]
- 18.Hayes T E, Kitchen A M, Cochran B H. Inducible binding of a factor of the c-fos regulatory region. Proc Natl Acad Sci USA. 1987;84:1272–1276. doi: 10.1073/pnas.84.5.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes T E, Sengupta P, Cochran B H. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 1988;2:1713–1722. doi: 10.1101/gad.2.12b.1713. [DOI] [PubMed] [Google Scholar]
- 20.Hill C S, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- 21.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–212. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 23.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 24.Hipskind R A, Rao V N, Mueller C G F, Reddy E S P, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- 25.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 26.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansen F-E, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joliot V, Demma M, Prywes R. Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature. 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 29.Kortenjann M, Thomae O, Shaw P E. Inhibition of v-raf-dependent c-fos expression and transformation by a kinase-defective mutant of the mitogen-activated protein kinase Erk2. Mol Cell Biol. 1994;14:4815–4824. doi: 10.1128/mcb.14.7.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruijer W, Cooper J A, Hunter T, Verma I M. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984;312:711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- 31.Manzano-Winkler B, Novina C D, Roy A L. TFII-I is required for transcription of the naturally TATA-less but initiator-containing Vbeta promoter. J Biol Chem. 1996;271:12076–12081. doi: 10.1074/jbc.271.20.12076. [DOI] [PubMed] [Google Scholar]
- 32.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 33.Metz R, Ziff E. The helix-loop-helix protein rE12 and the C/EBP-related factor rNFIL-6 bind to neighboring sites within the c-fos serum response element. Oncogene. 1991;6:2165–2178. [PubMed] [Google Scholar]
- 34.Meyer D J, Campbell G, Cochran B H, Argetsinger L, Larner A C, Finbloom D S, Carter-Su C, Schwartz J. Growth hormone induces a DNA-binding factor related to the interferon-stimulated 91-kDa transcription factor. J Biol Chem. 1993;269:4701–4704. [PubMed] [Google Scholar]
- 35.Montano M A, Kripke K, Norina C D, Achacoso P, Herzenberg L A, Roy A L, Nolan G P. NF-kappa B homodimer binding within the HIV-1 initiator region and interactions with TFII-I. Proc Natl Acad Sci USA. 1996;93:12376–12381. doi: 10.1073/pnas.93.22.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller R, Bravo R, Burckhardt J, Curran T. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 1984;312:716–720. doi: 10.1038/312716a0. [DOI] [PubMed] [Google Scholar]
- 37.Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol Cell Biol. 1995;15:5975–5982. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 39.Novina C D, Cheriyath V, Denis M C, Roy A L. Methods for studying the biochemical properties of an Inr element binding protein: TFII-I. Methods. 1997;12:254–263. doi: 10.1006/meth.1997.0477. [DOI] [PubMed] [Google Scholar]
- 40.Novina, C. D., V. Cheriyath, and A. L. Roy. Regulation of TFII-I activity by phosphorylation. Submitted for publication. [DOI] [PubMed]
- 40a.Novina, C. D., and A. L. Roy. Unpublished observations.
- 41.Perez Jurado L A, Wang Y K, Peoples R, Coloma A, Cruces J, Francke U. A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet. 1998;7:325–334. doi: 10.1093/hmg/7.3.325. [DOI] [PubMed] [Google Scholar]
- 42.Pollock R, Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990;18:6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renz M, Neuberg M, Kurz C, Bravo R, Muller R. Regulation of c-fos transcription in mouse fibroblasts: identification of DNase I-hypersensitive sites and regulatory upstream sequences. EMBO J. 1985;4:3711–3716. doi: 10.1002/j.1460-2075.1985.tb04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera V M, Sheng M, Greenberg M E. The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum induction. Genes Dev. 1990;4:255–268. doi: 10.1101/gad.4.2.255. [DOI] [PubMed] [Google Scholar]
- 45.Robertson L, Kerppola T, Vendrell M, Luk D, Smeyne R, Bocchiaro C, Morgan J, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 46.Roy A L, Carruthers C, Gutjahr T, Roeder R G. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature. 1993;365:359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- 47.Roy A L, Du H, Gregor P D, Novina C D, Martinez E, Roeder R G. Cloning of an Inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy A L, Malik S, Meisterernst M, Roeder R G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 49.Roy A L, Meisterernst M, Pognonec P, Roeder R G. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 50.Ruff-Jamison S, Chen K, Cohen S. Induction by EGF and interferon-γ of tyrosine phosphorylated DNA binding proteins in mouse liver nuclei. Science. 1993;261:1733–1736. doi: 10.1126/science.8378774. [DOI] [PubMed] [Google Scholar]
- 51.Ruff-Jamison S, Zhong Z, Wen Z, Chen K, Darnell J J, Cohen S. Epidermal growth factor and lipopolysaccharide activate Stat3 transcription factor in mouse liver. J Biol Chem. 1994;269:21933–21935. [PubMed] [Google Scholar]
- 52.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 53.Sengupta P, Cochran B H. MATα1 can mediate gene activation by a-mating factor. Genes Dev. 1991;5:1924–1934. doi: 10.1101/gad.5.10.1924. [DOI] [PubMed] [Google Scholar]
- 54.Sengupta P, Cochran B H. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol Cell Biol. 1990;10:6809–6812. doi: 10.1128/mcb.10.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw P E, Schroter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- 56.Sheng M, Dougan S T, McFadden G, Greenberg M E. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheng M, Thompson M A, Greenberg M E. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 58.Shuai K, Ziemiecki A, Wilks A F, Harpur A G, Sadowski H B, Gilman M Z, Darnell J E. Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature. 1993;366:580–583. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- 59.Silvennoinen O, Schindler C, Schlessinger J, Levy D E. Ras-independent growth factor signaling by transcription factor tyrosine phosphorylation. Science. 1993;261:1736–1739. doi: 10.1126/science.8378775. [DOI] [PubMed] [Google Scholar]
- 60.Simon K J, Grueneberg D A, Gilman M. Protein and DNA contact surfaces that mediate the selective action of the Phox1 homeodomain at the c-fos serum response element. Mol Cell Biol. 1997;17:6653–6662. doi: 10.1128/mcb.17.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon M N, De Virgilio C, Souza B, Pringle J R, Abo A, Reed S I. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 62.Song O, Dolan J W, Yuan Y O, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- 63.Subramaniam M, Schmidt L J, Crutchfield C E D, Getz M J. Negative regulation of serum-responsive enhancer elements. Nature. 1989;340:64–66. doi: 10.1038/340064a0. [DOI] [PubMed] [Google Scholar]
- 64.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 65.Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987;6:2711–2727. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 68.Wagner B J, Hayes T H, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang W, Desiderio S. BAP-135, a target for Bruton’s tyrosine kinase in response to B cell receptor engagement. Proc Natl Acad Sci USA. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan Y O, Stroke I L, Fields S. Coupling of cell identity to signal response in yeast: interaction between the alpha 1 and STE12 proteins. Genes Dev. 1993;7:1584–1597. doi: 10.1101/gad.7.8.1584. [DOI] [PubMed] [Google Scholar]
- 71.Zhong Z, Wen Z, Darnell J J. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 72.Zhu C, Johansen F-E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]