Highlights
-
•
Automated immunopurification of monoclonal immunoglobulins.
-
•
Analysis of EXENT® system prepared eluates reflexed to LC-MS without further sample processing.
-
•
Using the molecular mass of monoclonal light chains to track a monoclonal immunoglobulin in serum samples from patients with multiple myeloma.
Keywords: Mass spectrometry, Monoclonal immunoglobulin, MALDI-TOF MS, LC-MS, Multiple myeloma
Abstract
Introduction
The EXENT® Solution, a fully automated system, is a recent advancement for identifying and quantifying monoclonal immunoglobulins in serum. It combines immunoprecipitation with MALDI-TOF mass spectrometry. Compared to gel-based methods, like SPEP and IFE, it has demonstrated the ability to detect monoclonal immunoglobulins in serum at lower levels. In this study, samples that tested negative using EXENT® were reflexed to LC-MS to determine if the more sensitive LC-MS method could identify monoclonal immunoglobulins missed by EXENT®.
Objectives
To assess whether monoclonal immunoglobulins that are not detected by EXENT® can be detected by LC-MS using a low flow LC system coupled to a Q-TOF mass spectrometer.
Methods
Samples obtained from patients confirmed to have multiple myeloma (MM) were diluted with pooled polyclonal human serum and analyzed using EXENT®. If a specific monoclonal immunoglobulin was not detected by EXENT®, the sample was then subjected to analysis by LC-MS. For the LC-MS analysis, the sample eluate, obtained after the MALDI-TOF MS spotting step, was collected and transferred to an autosampler tray for subsequent analysis using LC-MS.
Conclusion
LC-MS has the capability to detect monoclonal immunoglobulins that are no longer detected by EXENT®. Reflexing samples to LC-MS for analysis does not involve additional sample handling, allowing for a faster time-to-result compared to current approaches, such as Next-Generation Sequencing, Next-Generation Flow, and clonotypic peptide methods. Notably, LC-MS offers equivalent sensitivity in detecting these specific monoclonal immunoglobulins.
Introduction
For over two decades LC-MS has been used extensively in the development of therapeutic monoclonal antibodies (t-mAbs) [1]. The technology is ideally suited for the characterization of t-mAbs since monoclonality is defined as polypeptides with identical amino acid sequences, resulting in a single molecular mass. Recent studies have shown the utility of mass spectrometry in confirming correct t-mAb expression by analyzing intact immunoglobulin molecular mass, reduced light chain and heavy chain molecular masses, and tryptic peptide molecular masses [2], [3]. Initial investigations describing the use of LC-MS to characterize endogenous immunoglobulins in circulation were conducted in parallel with t-mAb characterization work, focusing on immunoglobulins with an IgG isotype. These studies explored aspects such as heavy chain Fc region oxidation, allelic variants, and post-translational modifications, such as glycosylation [4], [5]. Research into the analysis of endogenous monoclonal immunoglobulins in serum began with methods that combined enzymatic digestion and LC-MS/MS to monitor constant region peptides common to specific isotypes [6] and, later, variable region clonotypic peptides [7], [8]. Subsequently, LC-MS methods with higher throughput were developed to monitor endogenous monoclonal immunoglobulins. Initially, these methods involved either no purification or enrichment using Melon™ Gel prior to LC-MS. These approaches yielded significant findings including, the ability to isotype kappa and lambda light chains using top-down MS/MS [9], direct detection of monoclonal light chains from urine [10], insights into the origins of light chain polyclonal molecular mass distributions [11], [12], methods designed to quantify a t-mAb while monitoring oligoclonal immunoglobulin patterns [13], and comparison of oligoclonal immunoglobulins in cerebrospinal fluid (CSF) and serum [14].
The studies mentioned previously highlight the usefulness of LC-MS to identify and quantify endogenous monoclonal immunoglobulin. However, incorporating an LC-MS system in a laboratory that primarily employs gel-based analytical methods was challenging due to the associated costs of instrumentation and infrastructure. As an alternative, researchers explored the use of MALDI-TOF MS due to its lower cost, smaller footprint, reduced infrastructure requirements, and the availability of instruments bearing a Class I medical device rating, a prerequisite for in vitro diagnostic (IVD) applications. A proof-of-concept study demonstrated that MALDI-TOF MS could be utilized to identify endogenous monoclonal immunoglobulins, potentially replacing LC-MS, despite it being typically less sensitive [15]. Following this, alternative methods for isotype-specific immunoprecipitation were developed along with he optimization of MALDI-TOF MS sample preparation and acquisition parameters. This resulted in a viable methodology for detecting monoclonal immunoglobulin in a clinical laboratory setting [16], [17], [18].
EXENT® is a comprehensive clinical analyzer that consists of three main components: an automated liquid handler, EXENT-iP® 500, a MALDI-TOF mass spectrometer, EXENT-iX ® 500, and instrument control and data analysis software, EXENT-iQ®. Its primary purpose is to detect and quantify monoclonal immunoglobulins in serum, making it a valuable device for clinical laboratories seeking IVD compliance. One notable advantage of using the MALDI-TOF MS technology employed by EXENT® is its requirement for only a small sample volume for analysis. This ensures that a sufficient amount of the sample remains available for further analysis by LC-MS in instances where EXENT® yields a negative result. In this study, we present the results obtained from serum samples that were initially analyzed using EXENT® and subsequently subjected to reflex analysis by LC-MS. Our findings demonstrate that the eluates purified by EXENT® can be directly analyzed by LC-MS without the need for any additional sample manipulation. This approach provides complementary data to that obtained from EXENT®, enhancing comprehensive sample characterization.
2. Materials and methods
Monoclonal Immunoglobulins and Serum: Serum samples from patients with a confirmed disease diagnosis of multiple myeloma were used for this study. As only fully-anonymized patient samples were used that were not obtained specifically for use in this study through an interaction or intervention with living individuals, neither informed consent nor IRB review were required. An IRB approved informed consent letter associated with each sample approved the use of each sample for this type of study. The intact M-protein isotype and concentrations determined by EXENT® for undiluted baseline samples are listed in Table 1. The concentration of the baseline sample was used to determine the dilutions necessary to make a sample with a calculated concentration of 0.100 g/L (Dilution 1) and 0.001 g/L (Dilution 2) of each M-protein. A stock solution was made by first diluting the baseline sample in pooled normal human serum to a nominal concentration of 1.0 g/L. The 1.0 g/L stock was then diluted 10-fold using pooled normal human serum (total IgG concentration = 8.79 g/L, total IgA concentration = 1.67 g/L, and total IgM concentration = 0.070 g/L) to make Dilution 1. Dilution 2 was then made by diluting Dilution 1 by 100-fold using pooled normal human serum. All concentrations are reported in grams per liter (g/L).
Table 1.
M-protein isotype and concentration data for patient samples analyzed by the EXENT® system. Samples that were found to be negative for the specific M-protein from each patient were reflexed to LC-MS.
| Sample ID | M-protein Isotype | Baseline Sample (g/L) | Dilution 1 | Dilution 2 |
|---|---|---|---|---|
| GK223 | IgG Kappa | 35.6 | 0.241 | Neg. |
| GK227 | IgG Kappa | 37.9 | Neg. | Neg. |
| GK229 | IgG Kappa | 29.2 | Neg. | Neg. |
| GK233 | IgG Kappa | 38.2 | Neg. | Neg. |
| GL239 | IgG Lambda | 32.5 | Neg. | Neg. |
| GL241 | IgG Lambda | 69.1 | Neg. | Neg. |
| GL251 | IgG Lambda | 31.9 | Neg. | Neg. |
| AK347 | IgA Kappa | 22.8 | 0.223 | Neg. |
| AK349 | IgA Kappa | 22.9 | 0.135 | Neg. |
| AK353 | IgA Kappa | 24.2 | 0.136 | Neg. |
| AL359 | IgA Lambda | 35.7 | 0.458 | 0.132 |
| AL367 | IgA Lambda | 48.5 | 0.094 | Neg. |
| MK97 | IgM Kappa | 36.9 | 0.106 | Neg. |
| MK101 | IgM Kappa | 31.5 | 0.102 | Neg. |
| ML103 | IgM Lambda | 27.1 | 0.086 | Neg. |
| ML107 | IgM Lambda | 33.1 | 0.102 | Neg. |
| ML109 | IgM Lambda | 40.6 | 0.130 | Neg. |
2.1. Reagents
Immunoprecipitation (IP) of serum samples was performed using the EXENT-iP® 500 automated liquid handler. Reagents used for the IP were packaged and shipped by The Binding Site, part of ThermoFisher Scientific, as manufactured kits. Kits included, paramagnetic beads coated with sheep polyclonal anti-human antibodies against human IgG, IgA, IgM, total Kappa, and total Lambda, wash buffers (PBS with 0.1 % TWEEN-20), and elution buffer (5 % acetic acid + 20 mM TCEP in water). Reagents kits for the EXENT-IX® 500 MALDI-TOF mass spectrometer included CHCA matrix with 1:1 v/v water (saturated solution with an estimated final concentration of 10 mg/mL) and acetonitrile containing 0.1 % acetic acid and protein standard mixture for mass calibration consisting of trypsinogen and cytochrome C. Mass spectrometry grade reagents used for the LC mobile phase include the following: water, acetonitrile and 2-propanol (Honeywell, Charlotte, NC), and formic acid (Thermo Fisher Scientific, Waltham, MA). Instrument optimization was performed using Tuning Solution containing cesium iodide and a synthetic peptide while within run auto calibration was performed using APCI Positive Calibration Solution (SCIEX, ON, CA).
2.2. EXENT and LC-MS methods
Eluates containing the purified immunoglobulins were prepared as previously described [19]. Briefly, in a 96-well plate diluted sample (10 µL of serum + 90 µL buffer) was placed into separate wells containing a different bead designed to purify IgG, IgA, IgM, total Kappa, and total Lambda (5 wells total). After binding (15 min) and bead washing (3 repeated washings using a plate washer), purified immunoglobulins were eluted off the beads by adding 35 µL of 5 % acetic acid in water also containing 20 mM TCEP as the reducing agent. The acid is present to disrupt the binding of isotype specific intact immunoglobulins to the beads (i.e., IgG immunoglobulins bind to the anti-IgG beads). At the same time the TCEP reduces the disulfide bonds of the intact immunoglobulins resulting in the dissociation of heavy chains to light chains. The result is an acidic solution that contains light chains and heavy chains unbound to each other in solution. In addition, the TCEP breaks any J-chain to heavy chain bonds which are responsible for generating dimers in the case of IgA immunoglobulins and pentamers in the case of IgM. A mixture of matrix and sample (2 µL) were spotted onto a MALDI plate and analyzed using automated acquisition on a Bruker Sirius one linear MALDI-TOF mass spectrometer. The instrument was run in positive ion mode using a Smartbeam™ solid state laser with a 200 Hz repetition rate and 2,000 shots were acquired per spot. The m/z range acquired was 5,000 to 32,000 using an acceleration voltage of approximately 18 kV, 450 ns pulsed ion extraction, and a detector gain of roughly 2,900 V. The time to first result for 96 spots was 80 min, after which samples were analyzed for the presence of a monoclonal immunoglobulin. EXENT® eluates were transferred to an autosampler plate to be analyzed by LC-MS after it was determined if the sample was negative for a specific monoclonal immunoglobulin. Data from the heavy chain eluates was used to generate the data presented here since the M-protein isotype had been previously identified by EXENT®.
A volume of 5 µL of the eluate was injected onto a 2 µL loop to be separated using an Eksigent M5 microflow LC system coupled to the mass spectrometer. The LC separation conditions consisted of; mobile phase A: 100 % water + 0.1 % v/v formic acid, and mobile phase B: 90 % acetonitrile + 10 %2-propanol + 0.1 % v/v formic acid. Separation of the heavy and light chains was performed using a 1.0 x 50 mm MAbPac™ RP column with polystyrene divinylbenzene stationary phase with 4 µm particle size (ThermoFisher Scientific, Waltham, MA) heated to 60 °C and flowing at 25 µL/minute. The gradient used for separations started at 80 % A 20 % B where it was held for 30 s, then ramped to 75 % A / 25 % B over 1 min, then ramped to 61 % A / 39 % B over 7.5 min, then ramped to 5 % A / 95 % B over 8 min, then held for 1 min, then ramped to the starting percentage of 80 % A / 20 % B over 30 s, then re-equilibrated at 80 % A / 20 % B for 2.5 min for a total analysis time of 12 min. Mass spectra were acquired on a SCIEX Triple-TOF™ 6600 quadrupole time-of-flight mass spectrometer in ESI positive mode with a Turbo V dual ion source (SCIEX, ON, CA). The source conditions were, ion spray voltage (IS): 5,500, Temp: 500 °C, Curtain gas: 45, GS1 gas: 35, GS2 gas: 30, collision energy: 10. TOF MS scans were acquired from m/z 600–2,500 with an acquisition time of 250 ms in high sensitivity mode. Analyst TF version 1.7.1 was used for instrument control. Raw data were viewed using PeakView® version 2.2.0.11391 without smoothing. Once a positive sample is identified by EXENT®, these data can be used to calculate the expected multiply charged ion series in the LC-MS analysis. Utilizing the prominent and well resolved doubly charged ion, an estimation of the mass of the uncharged light chain (assuming protonation using one signification figure for the proton) can be calculated as follows: ((m/z of [M+2H+]2+) × 2) − 2 = M (molecular mass, Da). The m/z of the theoretical 11+ charge state for the same molecule, a comparatively low charge state chosen due to the lower level of polyclonal background compared to a higher m/z region, can then be calculated as follows: (M + 11)/11 = [M + 11H+]11+. However, in cases where the S/N of the +11 charge state is <3, an alternative higher charge state with a S/N > 3 is selected to generate an XIC. Mass spectra were summed over the retention time of the peak observed in the XIC to create an ESI mass spectrum with multiple charge states each one originating from the monoclonal light chain with a different number of protons attached to the light chain.
Deconvolution of unsmoothed raw data containing the multiply charged light chain ions was performed using the Bayesian Protein Reconstruct program in Bio Tool Kit version 2.2.0.11391 based on the algorithm first defined by Mann et. al [20]. The deconvolution settings for the light chain mass spectra were, Input m/z range 600–2,500, Output mass range 20,000–30,000, and Step mass: 1.00 with the Input Spectrum Isotope Resolution: Low and Charge agent: H+. No.
smoothing was performed on deconvoluted mass spectra. Once a mass spectrum was deconvoluted, the molecular mass (along with the peak area) was determined for the monoclonal light chain. The limit of detection criteria for calling a deconvoluted monoclonal light chain peak as a positive were, 1) S/N ≥ 3 for the deconvoluted peak intensity, 2) ≤ 3.5 Da mass difference between the calculated molecular mass from EXENT® and the deconvoluted peak molecular mass, 3) ≥ 5 multiply charged peaks each with a S/N ≥ 3 used to generate the deconvoluted peak. If a sample was run previously by LC-MS, then a retention time tolerance of ≤0.1 min was used in combination with the other criteria.
3. Results
The purpose of the data presented in this study is to simulate a scenario where a sample that previously tested positive for a monoclonal immunoglobulin using EXENT® later tests negative. This negativity may occur when a patient undergoes therapy aimed at reducing the number of malignant clonal plasma cells. As these plasma cells die off, the amount of monoclonal immunoglobulin originating from the malignant clonal plasma cells is reduced. When a sample becomes negative using a specific assay like the EXENT® assay, clinicians may request the use of a more sensitive assay, such as LC-MS, to assess minimal residual disease (MRD), i.e., the presence of malignant clonal plasma cells. If the more sensitive assay still detects the monoclonal immunoglobulin originally observed at the time of diagnosis, it suggests that malignant clonal plasma cells are still present, putting the patient at risk of relapse. The decision to reflex a negative sample from one assay to a higher sensitivity assay to look for MRD is typically managed by the clinician.
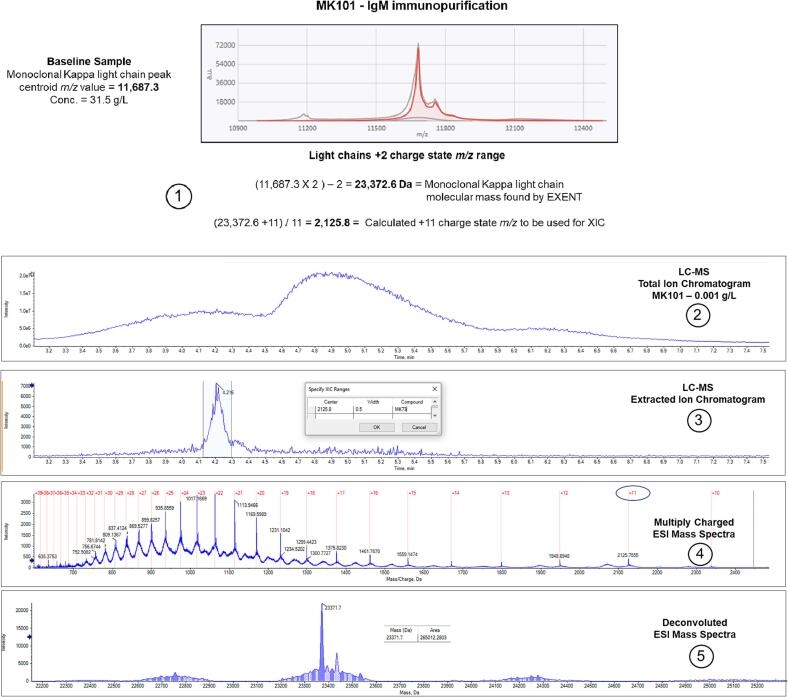
For example, Fig. 1 shows the steps involved in running a sample on EXENT® and then reflexing that sample to the higher sensitivity LC-MS assay. The transfer of the sample from an EXENT® 96-well sample elution plate to a 96-well LC-MS autosampler plate can be done manually or through a separate, stand-alone automated liquid handler. Fig. 2 shows the steps involved in LC-MS data acquisition and interpretation. The data were acquired for sample MK101, which contains an IgM Kappa M-protein, diluted to a calculated concentration of 0.001 g/L in pooled normal human serum. Step 1 of the process utilizes information from a sample positive by EXENT® to determine the molecular mass of the intact monoclonal kappa light chain. That information is then employed to calculate the +11 charge state m/z. In Step 2, this value is used to derive an extracted ion chromatogram (XIC) from the total ion chromatogram (TIC), with the y-axis in units of minutes. Step 3 involves summing the individual mass spectra comprising the peak associated with the monoclonal kappa light chain to produce the multiply charged ESI mass spectrum containing multiple charge states. The y-axis is given in units of Mass/Charge. Step 4 requires the deconvolution of the summed ESI mass spectrum to create the molecular mass spectrum with units in mass (Da). Step 5 involves recording the calculated molecular mass and peak area associated with the monoclonal kappa light chain. Table 2 shows the results from LC-MS analysis of the samples in Table 1, each at a concentration of 0.001 g/L. This includes the calculated molecular masses, peak areas, and signal-to-noise (S/N) values from the deconvoluted mass spectra observed for the monoclonal light chains. Supplementary materials Fig. 1 shows the method used to determine the S/N value for a peak in the deconvoluted mass spectrum.
Fig. 1.
Steps 1 through 5 in the process of taking a sample that becomes negative by EXENT® and then reflexing it to LC-MS.
Fig. 2.
The top panel shows mass spectra acquired using the EXENT® system run on sample MK101 containing an IgM Kappa M-protein and the bottom panels show LC-MS data. The figure steps through the calculations and operations done to generate LC-MS data on a sample reflexed from EXENT® to LC-MS.
Table 2.
M-protein molecular mass data observed for each patient specific M-protein analyzed using the EXENT® system and LC-MS. Peak areas and S/N values observed for samples diluted to 0.001 g/L that were reflexed to LC-MS are also listed.
| Sample ID | M-protein Isotype | EXENT Molecular Mass (Da) | LC-MS Molecular Mass (Da) | LC-MS Peak Area 0.001 g/L | LC-MS S/N 0.001 g/L |
|---|---|---|---|---|---|
| GK223 | IgG Kappa | 23,350.6 | 23,353.6 | 9.03E+04 | 6 |
| GK227 | IgG Kappa | 23,545.0 | 23,545.2 | Neg. | Neg. |
| GK229 | IgG Kappa | 23,614.0 | 23,614.3 | 2.00E+04 | 5 |
| GK233 | IgG Kappa | 24,305.6 | 24,303.0 | 3.16E+04 | 25 |
| GL239 | IgG Lambda | 22,711.0 | 22,709.0 | 1.18E+04 | 20 |
| GL241 | IgG Lambda | 22,722.8 | 22,720.1 | 3.58E+04 | 22 |
| GL251 | IgG Lambda | 22,973.2 | 22,973.0 | 1.25E+04 | 19 |
| AK347 | IgA Kappa | 23,775.0 | 23,777.5 | 1.84E+04 | 50 |
| AK349 | IgA Kappa | 23,736.2 | 23,739.7 | 7.71E+04 | 102 |
| AK353 | IgA Kappa | 23,167.6 | 23,166.5 | 4.65E+04 | 49 |
| AL359 | IgA Lambda | 23,075.2 | 23,075.1 | 3.43E+05 | 120 |
| AL367 | IgA Lambda | 22,264.6 | 22,265.4 | 1.10E+04 | 21 |
| MK97 | IgM Kappa | 23,489.2 | 23,486.1 | 5.70E+04 | 17 |
| MK101 | IgM Kappa | 23,372.6 | 23,371.8 | 2.65E+05 | 42 |
| ML103 | IgM Lambda | 23,012.6 | 23,007.6 | 5.83E+04 | 1,300 |
| ML107 | IgM Lambda | 22,682.8 | 22,683.2 | 1.69E+05 | 28 |
| ML109 | IgM Lambda | 23,120.6 | 23,119.5 | 1.62E+05 | 19 |
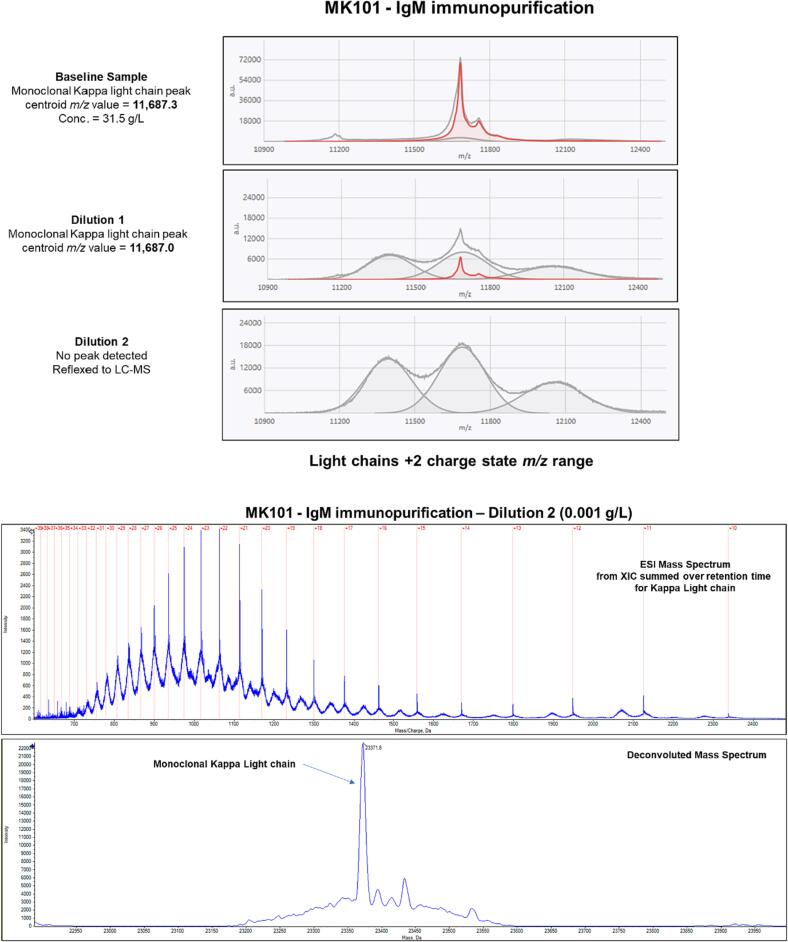
Fig. 3, Fig. 4, Fig. 5 show examples of positive and negative results for baseline, Dilution 1, and Dilution 2 samples using EXENT®. They also depict the results obtained for negative samples that were further tested using LC-MS. The top part of each figure presents the EXENT® mass spectra, while the bottom of each figure shows the LC-MS data. The data in Fig. 3 was derived from sample GK223, which contains an IgG Kappa M-protein. The monoclonal kappa light chain from the M-protein is illustrated by the red trace in the baseline sample and in Dilution 1, but not in Dilution 2. Dilution 2 was then reflexed to LC-MS, and the results are shown in the panel on the bottom of Fig. 3. After deconvolution, the monoclonal kappa light chain associated with sample GK223 is clearly visible with a calculated molecular mass of 23,353.6 Da. Each of the different charge states used to compile the deconvolution peak is highlighted with red lines in the ESI mass spectrum. Fig. 4 represents the results for sample AK337, which holds an IgA Kappa M-protein, while Fig. 5 demonstrates the results for sample MK101, which includes an IgM Kappa M-protein.
Fig. 3.
The top panel shows mass spectra acquired using the EXENT® system run on sample GK223 containing an IgG Kappa M-protein. Dilution 2 at 0.001 g/L was negative by EXENT® and was reflexed to LC-MS. The deconvoluted mass spectrum in the bottom panel shows the monoclonal kappa light chain matching the light chain modeled in the EXENT® MALDI-TOF MS mass spectrum at 23,353.6 Da. The polyclonal background is evident in mass spectra of diluted samples and is observed as a grey shaded normal distribution. These distributions represent the polyclonal light chains originating from Lambda (lower m/z distribution), Kappa (middle m/z distribution), and heavy Kappa (upper m/z distribution).
Fig. 4.
The top of panel shows mass spectra acquired using the EXENT® system run on sample AK337 containing IgA Kappa M-protein. Dilution 2 at 0.001 g/L was negative by EXENT® and was reflexed to LC-MS. The deconvoluted mass spectrum in the bottom panel shows the monoclonal kappa light chain matching the light chain modeled in the EXENT® MALDI-TOF MS mass spectrum at 23,777.5 Da. The polyclonal background is evident in mass spectra of diluted samples and is observed as a grey shaded normal distribution. These distributions represent the polyclonal light chains originating from Lambda (lower m/z distribution), Kappa (middle m/z distribution), and heavy Kappa (upper m/z distribution).
Fig. 5.
The top panel shows mass spectra acquired using the EXENT® system run on sample MK101 containing IgM Kappa M-protein. Dilution 2 at 0.001 g/L was negative by EXENT® and was reflexed to LC-MS. The deconvoluted mass spectrum at the bottom of the figure shows the monoclonal kappa light chain matching the light chain modeled in the EXENT® MALDI-TOF MS mass spectrum at 23,371.8 Da. The polyclonal background is evident in mass spectra of diluted samples and is observed as a grey shaded normal distribution. These distributions represent the polyclonal light chains originating from Lambda (lower m/z distribution), Kappa (middle m/z distribution), and heavy Kappa (upper m/z distribution).
The polyclonal background in the pooled serum used for dilutions contained the following concentrations of total Ig’s, as determined by turbidimetry: IgG = 8.79 g/L, IgA = 1.67 g/L, and IgM = 0.700 g/L. This polyclonal background is apparent in the mass spectra of diluted samples and is seen as a grey-shaded normal distribution. These distributions represent the polyclonal light chains originating from Lambda (lower m/z distribution), Kappa (middle m/z distribution), and heavy Kappa (upper m/z distribution) and have been described in detail previously [11], [12], [16]. The results summarized in Table 1 suggest that with EXENT®, higher levels of polyclonal light chain signal result in more negative results. This is illustrated by the Dilution 1 data, where only one of seven IgG samples was detectable at Dilution 1 levels, while all IgA and IgM M-proteins were detected. However, Table 2 shows that LC-MS can detect monoclonal light chains in a polyclonal background at a concentration 100-fold lower than the previously mentioned. Of the seventeen samples analyzed, only one was negative by LC-MS at 0.001 g/L. These observations suggest that higher levels of polyclonal background in samples immunopurified for IgG isotype don’t significantly impact the ability of LC-MS to detect monoclonal immunoglobulins. This differs from their impact on MALDI-TOF MS due to the added dimension of separation that the LC system provides prior to ESI-MS analysis.
To compare peak intensities independently of the peak area calculations performed by the deconvolution software, and to allow future comparisons between different LC-MS platforms, signal-to-noise (S/N) values were calculated. The S/N values in Table 2 display a broad range for Dilution 2, spanning from 5 for GK229 to 1,300 for ML103. This discrepancy was further evaluated by comparing the deconvoluted peak areas for Dilution 1 in the two IgM Lambda samples, ML103 and ML107. The values observed were exceedingly high, with sample ML103 displaying a value of 650,000 and sample ML107 having a value of 1,100,000. This finding suggests that the high S/N value of 1,300 for Dilution 2 from ML103 is a function of the deconvolution algorithm. Supplementary materials Fig. 2 shows LC-MS data for sample ML103 Dilution 2, which includes the ESI mass spectrum and deconvoluted mass spectrum. The deconvoluted mass spectrum at the bottom of the figure demonstrates how the deconvolution algorithm tends to decrease the background in m/z regions where the polyclonal background is lower. The figure also shows that two monoclonal peaks are present in the sample at the expected molecular mass of the monoclonal Lambda light chain. With an m/z difference of 17.0 Da, EXENT® did not resolve these two peaks. It is assumed that the mass difference of 5 between the calculated molecular mass by EXENT® and the observed molecular mass by LC-MS, as listed in Table 2, is likely a result of the centroid being modeled between the two peaks. Furthermore, the mass difference of 17.0 Da observed by LC-MS suggests that one set of monoclonal lambda light chains has an N-terminal glutamine converted to pyroglutamic acid. This is a commonly observed post-translational modification (PTM) for monoclonal immunoglobulins with N-terminal glutamines, which is typical in lambda light chains [21].
The peak areas calculated from deconvoluted mass spectra over a range of concentrations were performed using a set of serial dilutions, starting with Dilution 1. Supplementary materials Table 1 shows the LC-MS results from sample ML103 diluted serially into pooled normal human serum. The findings confirm that the deconvolution algorithm produces a linear set of peak area values, ranging from 0.100 g/L (100 mg/L) down to 0.00078 g/L (0.78 mg/L). The average peak area from four replicate injections was used to generate a linear regression plot shown under Table 1, with an r2 = 0.995. The %CV values for the replicate injections ranged from 4.2 for Dilution 1 at 0.100 g/L to 16 for the 0.00078 g/L serial dilution.
Supplementary materials Tables 2 and 3 present the results from serial dilutions of samples AK347 and GK223, respectively. The linear regression analysis of sample AK347, taken from the average peak areas of four replicate injections, resulted in an r2 = 0.992. The %CV values for the replicate injections ranged from 3.3 for Dilution 1 at 0.100 g/L to 23.4 for the 0.00078 g/L serial dilution. The linear regression analysis of sample GK223 taken from the average peak area of four replicate injections resulted in an r2 = 0.994. The %CV values for the replicate injections ranged from 7.3 for Dilution 1 at 0.100 g/L to 36.8 for the 0.00156 g/L serial dilution. The serial dilution at a calculated concentration of 0.00078 g/L was below the level of detection. Also, the m/z used for the extracted ion chromatogram was the +23 charge state at 1,016.2.
To evaluate the intra-day precision of the LC-MS system independent of an IP step with no polyclonal background, daratumumab, a monoclonal IgG Kappa t-mAb, was diluted in 5 % acetic acid + 20 mM TCEP to a concentration of 0.004 g/L and injected 20 times. The +11 charge state at m/z 2,126.6 was used to generate XIC peaks. Deconvoluted mass spectra were generated in the same manner as the samples containing an M-protein in pooled normal human serum. The peak areas found for the monoclonal kappa light chain had an average value of 1.9 x 106, a standard deviation of 1.2 x 105, and a %CV of 5.92. The LC retention time for the daratumumab kappa light chain peak had an average value of 4.04 min, a standard deviation of 0.01 min, and a %CV of 0.21. The average mass of the daratumumab light chain from the 20 replicates was 23,381.14 Da with a standard deviation of 0.07 Da (48 ppm mass measurement error from the known molecular mass of the Kappa light chain) obtained over the course of the four hour acquisition time.
4. Discussion
The data presented here demonstrates the usefulness of EXENT®, and the ability to reflex immunopurified samples that test negative with EXENT® directly to LC-MS. EXENT® is designed as a fully automated high-throughput in vitro diagnostic device, while the LC-MS method is a lower throughput, more sensitive, laboratory-developed test. The LC-MS method described here is similar to the previously reported microLC-ESI-Q-TOF MS method (i.e., miRAMM), and is simply referred to as LC-MS in this study. Furthermore, the results shown here focus only on the light chains' signal, and do not address the additional data acquired for heavy chains observed by LC-MS. Improved performance of 1 mm diameter microbore LC columns, the more sensitive Q-TOF mass spectrometer (utilizing a SCIEX 6600 in this study as opposed to a SCIEX 5600 used in previous publications), and new immunopurification components have resulted in a more sensitive LC-MS method compared to earlier studies [9], [10], [11], [12], [14], [24].
When reflexing to LC-MS from EXENT® the +2 charge state m/z data is used to track a specific monoclonal light chain. Utilizing the primary principle of molecular mass as a means of monitoring a monoclonal immunoglobulin avoids ambiguity when switching between EXENT® and LC-MS. When examining EXENT® +2 charge state mass spectra and LC-MS mass spectra, the entire collection of intact light chains can be visualized, allowing for the identification of specific monoclonal light chain(s) relative to the set of polyclonal light chains. This distinctive and informative perspective is independent of enzyme efficiency, amino acid sequence, or peptide ionization efficiency, which are potential issues associated with the clonotypic enzyme approach. Moreover, it is much faster to perform, as immunopurified samples can be analyzed directly by LC-MS without additional sample processing. Monitoring all intact light chains by EXENT® and by LC-MS ensures that any new clones or modifications to existing clones are accounted for. Additionally, certain post-translational modifications (PTMs), such as glycosylation, can be more readily assigned to a specific monoclonal light chain by monitoring the intact molecular mass. Methods that depend on the enzymatic digestion of immune-purified immunoglobulin pools can't use a single peptide to identify a specific light chain if the PTM is contained within an unmutated germline sequence, as multiple monoclonal light chains will have the same sequence.
The deconvolution software used to generate the data shown here employs a Bayesian deconvolution algorithm. This algorithm reconstructs the multiply charged ESI mass spectrum to an uncharged mass spectrum based on specific user inputs such as m/z input range, the charge agent (in this case +H), and molecular mass output range. It is clear that in certain cases the S/N of the deconvoluted peak of a monoclonal light chain is higher than expected given the processing steps used by the Bayesian deconvolution algorithm. This is illustrated in Supplementary materials Fig. 2 for sample ML103. This observation is linked to the fact that the m/z position of a monoclonal light chain peak (S) relative to the polyclonal background (N) impacts the final S/N calculation. In this particular scenario, the Bayesian deconvolution algorithm smoothed out the polyclonal background in the final deconvoluted mass spectrum, substantially decreasing the noise (N). Regardless, the manually determined S/N values and the software-derived peak areas for deconvoluted monoclonal light chains, irrespective of the m/z position, exhibit a linear response as demonstrated in Supplementary materials Tables 1–3. A comprehensive review of various deconvolution algorithms is discussed in the paper by Rolland and Prell [22]. The impact of raw data quality on the deconvolution process is discussed in the paper by Marty [23]. Nonetheless, the proposed unified scoring of raw and deconvoluted data suggested by Marty has yet to be widely implemented.
The purpose of this study was to demonstrate the process of reflexing a sample from the EXENT® system to an LC-MS system in scenarios where a previously identified monoclonal light chain from a monoclonal immunoglobulin is no longer detectable. In such situations, a more sensitive instrument is utilized to determine if the monoclonal immunoglobulin is still present. The data presented here exemplifies that samples reflexed to LC-MS can detect a specific monoclonal immunoglobulin by identifying the specific monoclonal light chain from the monoclonal immunoglobulin at levels achieved using clonotypic peptide-based methods, without the need for enzymatic digestion [24]. Additionally, it shows that analyzing eluates from EXENT® using LC-MS yields results at a substantially faster rate than methods that require a bone marrow biopsy, such as Next Generation Flow (NGF) and Next Generation Sequencing (NGS). Moreover, it often provides equal or better sensitivity [25].
Resolution and mass measurement accuracy / precision are analytical performance metrics that significantly influence a mass spectrometer's ability to monitor a specific monoclonal immunoglobulin light chain in a sample. In this study, the LC-MS system was operated in maximum sensitivity mode, which results in lower resolution (roughly 5,000). Other LC-MS platforms with higher resolution and mass measurement accuracy / precision are currently being used to monitor endogenous and therapeutic monoclonal immunoglobulins, both intact and reduced, without digestion. These platforms employ additional metrics in the gas-phase, allowing for unprecedented specificity and sensitivity that far surpasses what can be achieved using clonotypic peptides [26], [27], [28]. It is certain that LC-MS-based methods will continue to evolve, enabling the collection of additional phenotypic data that goes beyond merely monitoring the abundance of a monoclonal light chain. This will utilize as much data as possible to aid in tracking minimal residual disease.
5. Conclusion
Reflexing EXENT® negative samples to LC-MS to monitor monoclonal immunoglobulins is a straightforward, sensitive, and logical sequence in the progressive use of mass spectrometry to aid in the diagnosis of plasma cell dyscrasias.
CRediT authorship contribution statement
David Barnidge: Conceptualization, Data curation, Writing – original draft, Formal analysis, Methodology. Derek Troske: Data curation, Writing – review & editing. Simon North: Writing – review & editing, Methodology. Gregg Wallis: Data curation, Writing – review & editing, Methodology, Formal Analysis, Project administration. Mark Perkins: Writing – review & editing, Methodology. Stephen Harding: Writing – review & editing, Methodology, Resources, Project administration.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: DB, DT, SN, GW, MP, SH declare that they have potential competing interests relating to the potential commercial deployment of EXENT® as employees of The Binding Site, part of ThermoFisher. DB declares a potential competing interest with patents and royalties associated with the methodologies presented.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmsacl.2024.02.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lewis D.A., Guzzetta A.W., Hancock W.S., Costello M. Characterization of humanized anti-TAC, an antibody directed against the interleukin 2 receptor, using electrospray ionization mass spectrometry by direct infusion, LC/MS, and MS/MS. Anal. Chem. 1994;66(5):585–595. doi: 10.1021/ac00077a003. [DOI] [PubMed] [Google Scholar]
- 2.Srebalus Barnes C.A., Lim A. Applications of mass spectrometry for the structural characterization of recombinant protein pharmaceuticals. Mass Spectrom. Rev. 2007;26(3):370–388. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- 3.Beck A., Wagner-Rousset E., Ayoub D., Van Dorsselaer A., Sanglier-Cianférani S. Characterization of therapeutic antibodies and related products. Anal. Chem. 2013;85(2):715–736. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- 4.Goetze A.M., Zhang Z., Liu L., Jacobsen F.W., Flynn G.C. Rapid LC-MS screening for IgG Fc modifications and allelic variants in blood. Mol. Immunol. 2011 Oct;49(1–2):338–352. doi: 10.1016/j.molimm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Huhn C., Selman M.H., Ruhaak L.R., Deelder A.M., Wuhrer M. IgG glycosylation analysis. Proteomics. 2009;9(4):882–913. doi: 10.1002/pmic.200800715. [DOI] [PubMed] [Google Scholar]
- 6.Remily-Wood E.R., Benson K., Baz R.C., Chen Y.A., Hussein M., Hartley-Brown M.A., Sprung R.W., Perez B., Liu R.Z., Yoder S.J., Teer J.K., Eschrich S.A., Koomen J.M. Quantification of peptides from immunoglobulin constant and variable regions by LC-MRM MS for assessment of multiple myeloma patients. Proteomics Clin. Appl. 2014;8(9–10):783–795. doi: 10.1002/prca.201300077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnidge D.R., Tschumper R.C., Theis J.D., Snyder M.R., Jelinek D.F., Katzmann J.A., Dispenzieri A., Murray D.L. Monitoring M-proteins in patients with multiple myeloma using heavy-chain variable region clonotypic peptides and LC- MS/MS. J. Proteome Res. 2014;13(4):1905–1910. doi: 10.1021/pr5000544. [DOI] [PubMed] [Google Scholar]
- 8.Bergen H.R., 3rd, Dasari S., Dispenzieri A., Mills J.R., Ramirez-Alvarado M., Tschumper R.C., Jelinek D.F., Barnidge D.R., Murray D.L. Clonotypic light chain peptides identified for monitoring minimal residual disease in multiple myeloma without bone marrow aspiration. Clin. Chem. 2016;62(1):243–251. doi: 10.1373/clinchem.2015.242651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnidge D.R., Dasari S., Botz C.M., Murray D.H., Snyder M.R., Katzmann J.A., Dispenzieri A., Murray D.L. Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. J. Proteome Res. 2014;13(3):1419–1427. doi: 10.1021/pr400985k. [DOI] [PubMed] [Google Scholar]
- 10.Botz C.M., Barnidge D.R., Murray D.L., Katzmann J.A. Detecting monoclonal light chains in urine: microLC-ESI-Q- TOF mass spectrometry compared to immunofixation electrophoresis. Br. J. Haematol. 2014;167(3):437–438. doi: 10.1111/bjh.13003. [DOI] [PubMed] [Google Scholar]
- 11.Barnidge D.R., Dasari S., Ramirez-Alvarado M., Fontan A., Willrich M.A., Tschumper R.C., Jelinek D.F., Snyder M.R., Dispenzieri A., Katzmann J.A., Murray D.L. Phenotyping polyclonal kappa and lambda light chain molecular mass distributions in patient serum using mass spectrometry. J. Proteome Res. 2014 7;13(11):5198–5205. doi: 10.1021/pr5005967. [DOI] [PubMed] [Google Scholar]
- 12.Barnidge D.R., Lundström S.L., Zhang B., Dasari S., Murray D.L., Zubarev R.A. Subset of kappa and lambda germline sequences result in light chains with a higher molecular mass phenotype. J. Proteome Res. 2015;14(12):5283–5290. doi: 10.1021/acs.jproteome.5b00711. [DOI] [PubMed] [Google Scholar]
- 13.Mills J.R., Cornec D., Dasari S., Ladwig P.M., Hummel A.M., Cheu M., Murray D.L., Willrich M.A., Snyder M.R., Hoffman G.S., Kallenberg C.G., Langford C.A., Merkel P.A., Monach P.A., Seo P., Spiera R.F., St Clair E.W., Stone J.H., Specks U., Barnidge D.R. Using mass spectrometry to quantify rituximab and perform individualized immunoglobulin phenotyping in ANCA-associated vasculitis. Anal. Chem. 2016;88(12):6317–6325. doi: 10.1021/acs.analchem.6b00544. [DOI] [PubMed] [Google Scholar]
- 14.Barnidge D.R., Kohlhagen M.C., Zheng S., Willrich M.A., Katzmann J.A., Pittock S.J., Murray D.L. Monitoring oligoclonal immunoglobulins in cerebral spinal fluid using microLC-ESI-Q-TOF mass spectrometry. J. Neuroimmunol. 2015;15(288):123–126. doi: 10.1016/j.jneuroim.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Barnidge D.R., Krick T.P., Griffin T.J., Murray D.L. Using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to detect monoclonal immunoglobulin light chains in serum and urine. Rapid Commun. Mass Spectrom. 2015;29(21):2057–2060. doi: 10.1002/rcm.7314. [DOI] [PubMed] [Google Scholar]
- 16.Mills J.R., Kohlhagen M.C., Dasari S., Vanderboom P.M., Kyle R.A., Katzmann J.A., Willrich M.A., Barnidge D.R., Dispenzieri A., Murray D.L. Comprehensive assessment of M-proteins using nanobody enrichment coupled to MALDI- TOF mass spectrometry. Clin. Chem. 2016;62(10):1334–1344. doi: 10.1373/clinchem.2015.253740. [DOI] [PubMed] [Google Scholar]
- 17.Kohlhagen M.C., Barnidge D.R., Mills J.R., Stoner J., Gurtner K.M., Liptac A.M., Lofgren D.I., Vanderboom P.M., Dispenzieri A., Katzmann J.A., Willrich M.A., Snyder M.R., Murray D.L. Screening method for M-proteins in serum using nanobody enrichment coupled to MALDI-TOF mass spectrometry. Clin. Chem. 2016;62(10):1345–1352. doi: 10.1373/clinchem.2015.253781. [DOI] [PubMed] [Google Scholar]
- 18.Milani P., Murray D.L., Barnidge D.R., Kohlhagen M.C., Mills J.R., Merlini G., Dasari S., Dispenzieri A. The utility of MASS-FIX to detect and monitor monoclonal proteins in the clinic. Am. J. Hematol. 2017;92(8):772–779. doi: 10.1002/ajh.24772. [DOI] [PubMed] [Google Scholar]
- 19.El-Khoury H., Lee D.J., Alberge J.B., Redd R., Cea-Curry C.J., Perry J., Barr H., Murphy C., Sakrikar D., Barnidge D., Bustoros M., Leblebjian H., Cowan A., Davis M.I., Amstutz J., Boehner C.J., Lightbody E.D., Sklavenitis-Pistofidis R., Perkins M.C., Harding S., Mo C.C., Kapoor P., Mikhael J., Borrello I.M., Fonseca R., Weiss S.T., Karlson E., Trippa L., Rebbeck T.R., Getz G., Marinac C.R., Ghobrial I.M. Prevalence of monoclonal gammopathies and clinical outcomes in a high-risk US population screened by mass spectrometry: a multicentre cohort study. Lancet Haematol. 2022;9(5):e340–e349. doi: 10.1016/S2352-3026(22)00069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthias. Mann, Chin Kai. Meng, John B. Fenn, Interpreting mass spectra of multiply charged ions. Anal. 21. Liu YD, Goetze AM, Bass RB, Flynn GC. N-terminal glutamate to pyroglutamate conversion in vivo for human IgG2 antibodies, J. Biol. Chem. 286(13) (2011) 11211–11217. [DOI] [PMC free article] [PubMed]
- 21.Liu Z., Valente J., Lin S., Chennamsetty N., Qiu D., Bolgar M. Cyclization of N-terminal glutamic acid to pyro-glutamic acid impacts monoclonal antibody charge heterogeneity despite its appearance as a neutral transformation. J. Pharm. Sci. 2019;108(10):3194–3200. doi: 10.1016/j.xphs.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 22.Rolland A.D., Prell J.S. Approaches to heterogeneity in native mass spectrometry. Chem. Rev. 2022;122(8):7909–7951. doi: 10.1021/acs.chemrev.1c00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marty M.T. A universal score for deconvolution of intact protein and native electrospray mass spectra. Anal. Chem. 2020;92(6):4395–4401. doi: 10.1021/acs.analchem.9b05272. [DOI] [PubMed] [Google Scholar]
- 24.C. Wijnands, S. Noori, N.W.C.J.V. Donk, M.M. VanDuijn, J.F.M. Jacobs, Advances in minimal residual disease monitoring in multiple myeloma, Crit. Rev. Clin. Lab. Sci. 26 (2023) 1–17. Chem. 61(15) (1989) 1702–1708. [DOI] [PubMed]
- 25.Derman B.A., Stefka A.T., Jiang K., McIver A., Kubicki T., Jasielec J.K., Jakubowiak A.J. Measurable residual disease assessed by mass spectrometry in peripheral blood in multiple myeloma in a phase II trial of carfilzomib, lenalidomide, dexamethasone and autologous stem cell transplantation. Blood Cancer J. 2021;11(2):19. doi: 10.1038/s41408-021-00418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deighan W.I., Winton V.J., Melani R.D., Anderson L.C., McGee J.P., Schachner L.F., Barnidge D., Murray D., Alexander H.D., Gibson D.S., Deery M.J., McNicholl F.P., McLaughlin J., Kelleher N.L., Thomas P.M. Development of novel methods for non-canonical myeloma protein analysis with an innovative adaptation of immunofixation electrophoresis, native top- down mass spectrometry, and middle-down de novo sequencing. Clin. Chem. Lab. Med. 2020;59(4):653–661. doi: 10.1515/cclm-2020-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhenin J., Dupré M., Druart K., Krick A., Mauriac C., Chamot-Rooke J. A multiparameter optimization in middle- down analysis of monoclonal antibodies by LC-MS/MS. J. Mass Spectrom. 2023;58(3) doi: 10.1002/jms.4909. [DOI] [PubMed] [Google Scholar]
- 28.Wei B., Lantz C., Liu W., Viner R., Ogorzalek Loo R.R., Campuzano I.D.G., Loo J.A. Added value of internal fragments for top-down mass spectrometry of intact monoclonal antibodies and antibody-drug conjugates. Anal. Chem. 2023;95(24):9347–9356. doi: 10.1021/acs.analchem.3c01426. Epub 2023 Jun 6 PMID: 37278738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.