Abstract
The ubiquitous molecular chaperone Hsp90 acts in concert with a cohort of associated proteins to facilitate the functional maturation of a number of cellular signaling proteins, such as steroid hormone receptors and oncogene tyrosine kinases. The Hsp90-associated protein p23 is required for the assembly of functional steroid aporeceptor complexes in cell lysates, and Hsp90-binding ansamycin antibiotics disrupt the activity of Hsp90-dependent signaling proteins in cultured mammalian cells and prevent the association of p23 with Hsp90-receptor heterocomplexes; these observations have led to the hypotheses that p23 is required for the maturation of Hsp90 target proteins and that ansamycin antibiotics abrogate the activity of such proteins by disrupting the interaction of p23 with Hsp90. In this study, I demonstrate that ansamycin antibiotics disrupt the function of Hsp90 target proteins expressed in yeast cells; prevent the assembly of Sba1, a yeast p23-like protein, into steroid receptor-Hsp90 complexes; and result in the assembly of receptor-Hsp90 complexes that are defective for ligand binding. To assess the role of p23 in Hsp90 target protein function, I show that the activity of Hsp90 target proteins is unaffected by deletion of SBA1. Interestingly, steroid receptor activity in cells lacking Sba1 displays increased sensitivity to ansamycin antibiotics, and this phenotype is rescued by the expression of human p23 in yeast cells. These findings indicate that Hsp90-dependent signaling proteins can achieve a functional conformation in vivo in the absence of p23. Furthermore, while the presence of p23 decreases the sensitivity of Hsp90-dependent processes to ansamycin treatment, ansamycin antibiotics disrupt signaling through some mechanism other than altering the Hsp90-p23 interaction.
The 90-kDa heat shock protein (Hsp90) is a highly conserved, abundant protein chaperone. Hsp90 is essential for viability in Saccharomyces cerevisiae (7), Drosophila melanogaster (14), and Schizosaccharomyces pombe (1). Although the role of Hsp90 in maintaining cell viability is not well understood, it is notable that many Hsp90 target proteins are involved in cellular signaling (4), including such diverse signaling proteins as v-src family oncogene tyrosine kinases (9), steroid hormone receptors (6, 10, 47), the basic helix-loop-helix dioxin receptor (67), the sevenless and torso receptor tyrosine kinases (14), and the Wee1 tyrosine kinase (1). Recent experiments suggest that Hsp90 also plays a role in establishing the specific conformation of the p53 tumor suppressor protein (2). The mechanism of Hsp90 action in promoting the activity of these proteins is unknown.
Hsp90 is an abundant homodimer, representing up to 5% of total cell protein under non-stress conditions (37). A significant fraction of cellular Hsp90 exists in complex with other proteins, including Hsp70, p60 (a Sti1-like protein), immunophilins, and p23 (42, 57). Steroid hormone receptors have served as a useful model for the functional characterization of the Hsp90 protein complex. Steroid receptors, which act as ligand-regulated transcription factors, must interact with Hsp90 to bind ligands with high affinity and, thereby, transduce the hormonal signal (5, 8, 41, 55). In the unliganded, inactive state, several steroid receptor types exist in cells as components of aporeceptor complexes, composed of a single receptor protein, a dimer of Hsp90, and other associated proteins (45, 46, 57). After ligand binding, Hsp90 dissociates from the receptor and the liganded receptor translocates to the nucleus, binds specific sites on the chromosomes, and modulates the transcriptional activity of target genes (6, 20, 69).
Interestingly, functional aporeceptor complexes can be reconstituted onto free monomers of the glucocorticoid and progesterone receptors (GR and PR, respectively) in vitro in reticulocyte lysates (50, 56). This reconstitution assay has been used to identify and characterize many of the components of the aporeceptor complex; for instance, in addition to Hsp90, both Hsp70 and p23 are required for reconstitution of the aporeceptor complex in vitro (28, 29, 33), and drugs which abrogate the formation of these aporeceptor complexes also disrupt the p23-Hsp90 interaction (58, 65). Furthermore, genetic studies of yeast cells have demonstrated that mutations in HSP82 (an Hsp90 gene [3, 5]), YDJ1 (a DnaJ-like gene [34]), STI1 (a p60-like gene [12]), or CPR7 (a Cyp-40-like gene [18]) cause defects in steroid receptor and pp60v-src function. These studies indicate that the ability of Hsp90 to support the function of steroid receptors is dependent both on the structure of Hsp90 itself and on the presence of the proper cohort of associated proteins.
Signaling proteins display differing requirements for interaction with Hsp90 and associated proteins. On one end of the spectrum, the steroid hormone receptors appear to require contact with Hsp90 to be competent to bind and respond to ligand; bound Hsp90 appears to favor a high ligand affinity conformation that is unstable in the absence of interaction with Hsp90. This is in contrast to membrane-associated tyrosine kinases, such as pp60v-src, which interact with Hsp90 transiently following synthesis until the nascent pp60v-src protein is inserted in the membrane; pp60v-src appears to require Hsp90 to avoid nonproductive interactions and/or to facilitate proper association with the cell membrane (9, 68); once the “mature” conformation is achieved, pp60v-src appears to be functional without further need for Hsp90. A requirement for such a transient interaction with Hsp90 may also explain the observation that the retinoic acid receptor, which has not been isolated in stable complexes with Hsp90 (15, 20), displays decreased function and ligand affinity in yeast cells expressing reduced levels of Hsp90 (27). Thus, in some cases, Hsp90 may help proteins assume stable functional structures by transiently interacting with target proteins to prevent nonproductive interactions or degradation. However, in the case of steroid receptors, stable interaction with Hsp90 appears to be required to achieve a high ligand affinity conformation, such that Hsp90 is an obligatory component of the functional aporeceptor complex (8, 55).
Analysis of the role of Hsp90 in cell signaling has been facilitated by the characterization of quinone ansamycin antibiotics, such as the macbecins, geldanamycin, and herbimycin A; these compounds display potent antitumor activity in vitro (61) and in vivo (40, 48) and had been hypothesized to act directly as tyrosine kinase inhibitors (63). However, Whitesell et al. (66) demonstrated that these compounds specifically bind Hsp90 and inhibit Hsp90-pp60v-src complex formation, with a resulting loss of tyrosine kinase activity. Similarly, ansamycins have been shown to inhibit progesterone receptor complex assembly in vitro (58) and inhibit transcriptional activation by progesterone and glucocorticoid receptors in cultured mammalian cells (58, 65). Furthermore, it has been observed that ansamycins disrupt the interaction of p23 with Hsp90 (33) and prevent the formation of aporeceptor complexes containing p23 (58, 65). p23 was identified by Johnson et al. (31) as an Hsp90-associated protein and as a component of progesterone aporeceptor complexes. These investigators and others subsequently demonstrated that p23 is a required constituent of lysates that are competent to assemble high ligand affinity steroid aporeceptor complexes in vitro (29, 33). The intersection between studies of p23 activity and the mechanism of ansamycin action has led to the hypothesis that ansamycins block the participation of p23 in the maturation of Hsp90 target polypeptides and, thereby, prevent the accumulation of functional proteins (58, 60).
In this study, I test the hypothesis that ansamycin antibiotics abrogate the activity of Hsp90 target proteins by disrupting the interaction of p23 with Hsp90 in vivo. First, I undertake a pharmacologic and genetic analysis of the effects of quinone ansamycins on Hsp90 target proteins in S. cerevisiae. Guided by the results of this analysis, I exploit yeast genetics to investigate the role of p23, an Hsp90-associated protein, in signaling and ansamycin sensitivity.
MATERIALS AND METHODS
Chemicals and reagents.
Macbecins I and II (MI and MII, respectively) and geldanamycin were obtained from the Developmental Therapeutics Program, National Cancer Institute, Bethesda, Md. Herbimycin A (HA) was from Calbiochem. Both were stored at −20°C, protected from light, either in powder form or as a 10 mM stock solution in 100% dimethyl sulfoxide (DMSO; Sigma). Restriction enzymes and ligase were from Promega, New England Biolabs, and Gibco/BRL. Taq polymerase was from Boehringer Mannheim. [1H]dexamethasone ([1H]Dex) and [3H]Dex (38 Ci/mmol) were obtained from Sigma and Amersham, respectively. AEBSF was from Calbiochem. Aprotinin, leupeptin, and pepstatin A were from Boehringer Mannheim. Immobilon-P membranes were from Millipore Corp. Affi-prep protein A, Affi-gel 10, and alkaline phosphatase-conjugated goat anti-mouse and goat anti-rabbit immunoglobulin G were from Bio-Rad. Horseradish peroxidase-conjugated goat anti-mouse Fc-specific immunoglobulin G was from Sigma. 5-Bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium alkaline phosphatase substrate was from Kirkegaard & Perry. Luminescent horseradish peroxidase substrate was from Amersham. Antibodies were generously provided by the following scientists: rabbit anti-yeast Hsp82 antiserum was from Susan Lindquist (Howard Hughes Medical Institute, University of Chicago); rabbit anti-SSA and anti-SSB antisera were from Elizabeth Craig (University of Wisconsin, Madison); mouse monoclonal anti-Sti1 was from David Toft (Mayo Clinic, Rochester, Minn.); and rabbit anti-Ydj1 antiserum was from Avrom Caplan (Mt. Sinai Hospital, New York, N.Y.). Mouse monoclonal anti-Flag antibody was from IBI. Mouse monoclonal anti-v-src antibody, designated LA074, was from Quality Biotech, and 4G10 mouse monoclonal anti-phosphotyrosine antibody was from Upstate Biotechnology. Other reagents were from Sigma.
Yeast strains and plasmids.
The strain referred to as wild-type in this manuscript is YNK100 (MATα pdr5-101 [35]); Pdr5 is an ATP-binding-cassette transporter, and the pdr5-101 mutation results in increased glucocorticoid potency (36). YNK234 (MATα pdr5-101 sba1::HIS3) is derived from YNK100 and has a deletion of SBA1, a yeast p23-like gene (denoted YKL117w in reference 19; GenBank entry Z28117) by insertion of the HIS3 gene. A construct targeting the chromosomal SBA1 gene for replacement by the HIS3 gene via homologous recombination was made by PCR amplification of the −9 to −229 region of the SBA1 gene (with respect to the predicted translation start site) as a BamHI-SalI fragment with primers 5′-GCGGGATCCGTGGTTATTGGTACACATATACC-3′ and 5′-GCGCGTCGACGAATCGATGATCTTGGGAAC-3′; likewise, a 355-bp downstream fragment, including only 25 bp of the predicted SBA1 coding sequence with 330 bp of downstream chromosomal DNA, was amplified with flanking SalI and PstI sites by using primers 5′-CGCGGTCGACGGAAATAGAGCCGGAAGTGAAAGC-3′ and 5′-CGCCTGCAGGCTTACACCTGATAATCATCCAGC-3′. The amplified fragments were digested with SalI, ligated, and cloned into pUC19 as a BamHI-PstI fragment; the HIS3 gene was subsequently subcloned into the SalI site between the SBA1 flanking regions. The flanking regions and HIS3 gene were liberated from the plasmid as a BamHI-SphI fragment, agarose gel purified, and transformed into YNK100. Histidine prototrophs were selected, and single insertion of HIS3 into the predicted chromosomal location was confirmed by Southern blotting. In all experiments, yeast strains not protrophic for histidine or leucine were transformed with pRS313 (HIS3, low copy number) and/or pRS315 (LEU2, low copy number) to confer histidine and leucine prototrophy, respectively (54). Thus, all strains were grown in equivalent medium.
F620S mutant rat GR was expressed from a TRP1-marked, high-copy-number plasmid by using the glyceraldehyde-6-phosphate dehydrogenase (GPD) promoter (3, 21). F620S GR displays higher ligand affinity in yeast cells than does wild-type rat GR (21); Hsp90-dependent signal transduction by F620S GR has otherwise been shown to similar to that of wild-type GR (3). The human PR expression plasmid, YEphPR-B (64), and the N556 constitutive GR expression plasmid (5) have been described elsewhere. pΔS26X is a URA3-marked, high-copy-number plasmid containing the Escherichia coli lacZ gene driven by three glucocorticoid-progesterone response elements upstream of the CYC1 TATA (49) and was used as the reporter for assays of GR and PR activity. RAR and PR expression plasmids and the RAR reporter plasmid have been described elsewhere (26, 43). Yeast cells were transformed by a standard lithium acetate protocol (23).
Subcloning of SBA1 and human p23.
A search of the SwissProt and GenPept sequence databases with the BLAST program (22) identified several sequences of significant homology, including a putative polypeptide encoded by a single open reading frame (ORF) from the S. cerevisiae genome. This ORF, which I will call SBA1 for increased sensitivity to benzoquinone ansamycins, was designated YKL117w by Dujon et al. (19) and is predicted to encode a protein with an estimated molecular mass of 24.1 kDa; SBA1 was the only yeast ORF predicted to encode a protein with detectable similarity to human p23. The predicted Sba1 polypeptide sequence displays 27.5% identity and 54.4% similarity with the human p23 protein over 160 amino acids (see Fig. 4), as measured by the Gap program (22); Sba1 at 216 amino acids is significantly longer than human p23, which is 160 amino acids in length. Deletion of the yeast SBA1 gene is described above.
FIG. 4.
Comparison of yeast Sba1 and human p23 protein sequences. Protein sequences for yeast Sba1 and human p23 are shown. Sba1 and p23 sequences are 28% identical and 54% similar over the entire 160-amino-acid length of human p23. Sequences were aligned by the Gap program to demonstrate maximum similarity (22). Symbols: |, an exact amino acid match; :, a conserved substitution; …, gaps introduced into the amino acid sequence to optimize the pairing of regions of similarity.
A Flag-tagged Sba1 expression plasmid was created by PCR amplification of the Sba1 coding sequence from yeast genomic DNA with primers designed to add the Flag epitope, NH2-DYKDDDDK-COOH, to the carboxy terminus of the predicted Sba1 polypeptide sequence; the primer sequences were 5′-GCGGAATCCATGTCCGATAAAGTTATTAACCCTC-3′ and 5′-CGCTCTAGATTACTTGTCATCGTCGTCCTTGTAGTCAGCTTTCACTTCCGGCTCTATTTCCTC-3′. The amplified fragment was subcloned as a BamHI-XbaI fragment into a pRS315 (LEU2, low copy number [54])-based plasmid containing the GPD promoter as an EcoRI-BamHI fragment.
A plasmid for the expression of human p23 in yeast was created by PCR amplification of the human p23 coding sequence from a previously described (31) bacterial expression plasmid, provided by David Toft, Mayo Clinic; the primer sequences were 5′-GCGGAATCCATGCAGCCTGCTTCTGCAAAGTGG-3′ and 5′-CGCTCTAGATTACTCCAGATCTGGCATTTTTTCATCATCACTGTC-3′. The amplified fragment was subcloned as a BamHI-XbaI fragment into a pRS425 (LEU2, high copy number)-based plasmid (13) containing the GPD promoter as an EcoRI-BamHI fragment.
β-Gal assays.
The ligand response of GR in the context of ansamycin treatment and/or SBA1 mutation was determined by measuring the β-galactosidase (β-Gal) activity induced in response to various concentrations of Dex. Appropriate yeast strains were grown overnight at 30°C in selective medium (SD complete medium lacking His, Trp, Leu, and Ura [53]) and diluted 1:6 into fresh medium containing Dex in ethanol or ethanol only, as well as with the denoted ansamycin in dimethyl sulfoxide (DMSO) or with DMSO only; final concentrations of ethanol and DMSO were 1% each in all samples. Cultures were incubated overnight at 30°C. To measure β-Gal activity, cells were permeabilized by combining 50 μl of each culture with 50 μl of 2× Z buffer (1× Z buffer is 60 mM Na2HPO4 · 7H2O, 60 mM NaH2PO4 · H2O, 5 mM KCl, and 0.5 mM MgSO4 · 7 H2O at pH 7 with 0.025% β-mercaptoethanol, freshly added) and 50 μl of chloroform in a microcentrifuge tube and vigorously vortex mixing for 30 s. Assays were conducted at 30°C by adding 700 μl of a 2-mg/ml mixture of o-nitrophenyl-β-d-galactopyranoside (ONPG) in 1× Z buffer and incubating for 10 min. Reactions were stopped by adding 500 μl of 1 M Na2CO3. The optical density at 420 nm (OD420) of each reaction mixture and the OD600 of each culture were determined, and β-Gal units were calculated as 1,000 times the OD420 divided by the product of the OD600, the reaction time (in minutes), and the culture volume used (in milliliters).
In vivo ligand binding.
All binding studies were conducted on cells expressing F620S GR, which displays higher affinity for ligand than does wild-type GR when expressed in yeast cells. In vivo ligand binding assays were as described by Kralli et al. (35). Duplicate cultures of yeast cells expressing wild-type or mutant Hsp82 and F620S GR were inoculated into selective liquid medium and grown overnight at 30°C. Cells were diluted into fresh medium containing ansamycin at the denoted concentration or into DMSO only (1% DMSO in all samples) and were incubated for 2 h at 30°C. [3H]Dex (adjusted to 1 Ci/mmol with [1H]Dex) was added to a final concentration of 1 μM in the absence or presence of 100-fold excess [1H]Dex, and cultures were incubated 2 h longer at 30°C. Portions (1 ml) of each culture were harvested by centrifugation at 12,000 × g at 4°C, and cells were washed three times by resuspending and recentrifugation in 1 ml of ice-cold phosphate-buffered saline (PBS) plus 2% glucose. After the washes, cells were resuspended in 50 μl of PBS-glucose, and counts bound were measured by liquid scintillation. Specific counts bound were determined as counts bound in the absence of excess unlabeled ligand minus counts bound in its presence. No specific binding was detected in cells not expressing GR (data not shown).
Coimmunoprecipitations and immunoblotting.
Extracts were prepared essentially as previously described (3). Briefly, YNK234 expressing Sba1-Flag, containing the pΔS26X reporter plasmid, and expressing either F620S rat GR or human PR was grown at 30°C in SD complete medium lacking His, Trp, Leu, and Ura (53) in the presence or absence 10 μM Dex and/or 50 μM MI (ethanol and DMSO concentrations were normalized to 1% each). All subsequent manipulations were done at 4°C. Cells were collected by centrifugation, washed with 10 ml of wash buffer (PBS containing 2 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM AEBSF, and aprotinin, leupeptin, and pepstatin A [1 μg of each per ml, freshly added]). Cells were then resuspended in 1 ml of wash buffer, transferred to microcentrifuge tubes, pelleted, and resuspended in an equal volume (150 to 250 μl) of lysis buffer (10 mM Tris-HCl [pH 7.6]–50 mM NaCl–1 mM EDTA–10% [vol/vol] glycerol, 2 mM DTT, 1 mM PMSF, 1 mM AEBSF, and aprotinin, leupeptin, and pepstatin A [1 μg of each per ml, freshly added]). Tubes were filled with glass beads to the bottom of the meniscus, and cells were lysed by vortex mixing for 30 min in an Eppendorf model 5432 platform vortex mixer. Lysates were centrifuged for 5 min in a microcentrifuge, fresh PMSF and AEBSF were added (1 mM concentrations of each), and extracts were cleared by centrifugation for 30 min at 85,000 rpm in a 100.1 Ti rotor with a Beckman tabletop ultracentrifuge. Protein concentrations were normalized to 10 mg/ml with lysis buffer, and Triton X-100 was added to 0.2%. Extract was then incubated with mouse monoclonal anti-GR antibody BuGR2 covalently and cross-linked to Affi-prep protein A for 2 h with agitation to immunoprecipitate the GR. Immunoprecipitates were washed, twice with 1 ml of lysis buffer containing 0.2% Triton X-100 and once with lysis buffer only. Equal amounts of each immunoprecipitate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5 and 12.5% gels and blotted to Immobilon-P membranes. Proteins were detected with appropriate antibodies, as listed above, and blots were developed with alkaline phosphatase-conjugated secondary antibodies, except for Sba1-Flag blots, which were developed with horseradish peroxidase-conjugated secondary antibodies.
Assay of human and yeast Hsp90 binding to GA beads.
GA was covalently coupled to Affi-gel 10 resin (Bio-Rad) by the technique described by Whitesell et al. (66). Yeast cell lysates from strains YNK234 (for yeast Hsp-Hsc82) and HH1Kat6 (for human Hsp90) were prepared as described above. The HH1Kat6 strain expresses human Hsp90 as the sole Hsp90 protein (41). NaCl was added to the extracts to a final concentration of 200 mM, and whole yeast cell extract (1 mg of total protein per assay), with the prior addition of MI (at a final concentration of 10 μM) or vehicle only, was incubated with GA-coupled beads for 2 h at 4°C. Beads were subsequently washed three times with ice-cold lysis buffer with 200 mM NaCl and once with ice-cold lysis buffer. Bound proteins were eluted by heating in reducing loading buffer, separated by SDS-PAGE, and detected by silver staining.
Assay of pp60v-src expression and activity.
Strain W303 (MATa ade2-1 his3-11 his3-15 leu2-3 leu2-112 trp1-1 ura3-1) was used for all experiments involving pp60v-src. The pp60v-src gene was expressed from a URA3-marked, low-copy-number plasmid under the control of a galactose-inducible promoter from plasmid Y316 v-src (38). Control strains contained the parent plasmid, pRS316 (54), only. Production of pp60v-src was induced by growing cultures overnight at 30°C in SC medium lacking uracil with 2% raffinose and 0.1% sucrose as carbon sources. Cultures were centrifuged, and cell pellets were resuspended in SC lacking uracil with 2% galactose and 0.1% glucose as carbon sources. Cultures were split into equal fractions to which MI (50 μM) or vehicle only (1% DMSO) was added and were subsequently incubated at 30°C for 6 h.
Cell extracts were prepared as described above with the exception that cell pellets were washed with PBS containing 25 mM NaF, 5 mM Na2V2O5, and 1 mM sodium pyrophosphate, in addition to 2 mM DTT, 1 mM PMSF, 1 mM AEBSF, and aprotinin, leupeptin, and pepstatin A (1 μg per ml of each freshly added), and lysis buffer was as above with the addition of 25 mM NaF, 5 mM Na2V2O5, and 1 mM sodium pyrophosphate. An equal amount of total protein from each extract was separated on SDS–10% PAGE gels and then blotted, and pp60v-src and phosphotyrosine were detected with the appropriate antibodies.
RESULTS
MI inhibits steroid receptor activity in yeast.
The benzoquinone ansamycin geldanamycin has been shown to bind Hsp90 and to disrupt the activity of Hsp90-dependent signaling proteins in cultured mammalian cells (65, 66). Given that structurally similar compounds can display differing degrees of potency in yeast versus mammalian cells, I screened a panel of several benzoquinone ansamycins for the ability to disrupt the ligand-dependent activation of the glucocorticoid receptor in yeast cells. I found that MI (62) most potently disrupted GR signaling (Fig. 1), whereas geldanamycin is a relatively poor inhibitor of GR signaling, reducing ligand response by only 29% at a geldanamycin concentration of 100 μM (data not shown). An analysis of the structures of various benzoquinone ansamycins in the context of the potencies of these compounds in yeast cells suggests that the more hydrophobic compounds are more potent. This may reflect differences in the intracellular availability of these compounds; geldanamycin, though only weakly active in vivo in yeast cells, specifically binds yeast Hsp-Hsc82 in whole-cell extract in a manner similar to the binding of human Hsp90 and this binding is competed by MI (see below). Because of its high potency, I chose to utilize MI to analyze the effects of benzoquinone ansamycins on signaling in yeast cells.
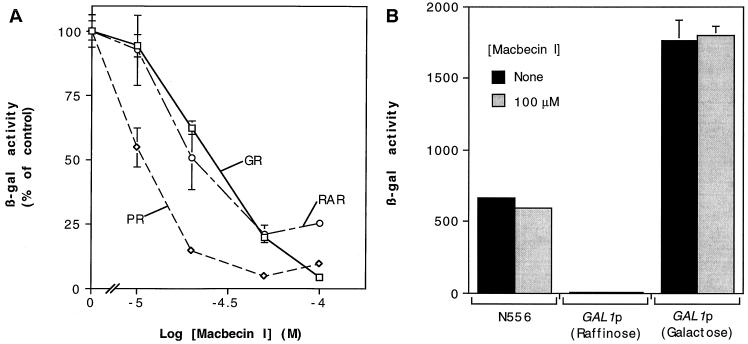
FIG. 1.
MI inhibits ligand response of steroid and nuclear receptors in a dose-dependent manner. (A) Yeast cells expressing GR, PR, or RAR and containing the appropriate β-Gal reporter plasmid were treated with 10 μM Dex, 100 nM progesterone, or 10 μM retinoic acid, respectively, in the presence of increasing concentrations of MI. (B) MI (100 μM) does not affect the activity of a constitutive form of GR (N556) or induction of the GAL1 promoter in response to galactose. Data are the means and ranges for two independent transformants and are representative of three or more experiments. β-Gal activity was determined as described in Materials and Methods. Data for F620S mutant rat GR are represented here and in subsequent experiments (see Materials and Methods); MI treatment has a similar effect on the ligand response of wild-type rat GR (data not shown). A 100% activity for GR, PR, or RAR was approximately 2,100, 1,400, or 45 U, respectively.
To assess the effects of MI on steroid receptor function in vivo, yeast cells containing plasmids expressing glucocorticoid, retinoic acid, or progesterone receptors (GR, RAR, and PR, respectively) were treated with agonist ligand and increasing concentrations of MI; the ability of the receptors to induce production of β-Gal from a reporter gene under control of the appropriate receptor response elements was then measured. MI inhibited the ligand-dependent induction of the reporter by the various receptors in a dose-dependent manner (Fig. 1A). In contrast, MI did not affect the induction of the GRE-linked β-Gal reporter by N556, a constitutive GR mutant lacking the carboxy-terminal signaling domain (24), or the induction of the GAL1 promoter in the presence of galactose (Fig. 1B). These findings indicate that decreased receptor activity in the presence of MI is not the result of nonspecific defects in transcription or translation; rather, the effects of MI appear to be restricted to the ability of these receptors to recognize and respond to cognate ligands. Furthermore, yeast Hsp90 in whole-cell extracts, like mammalian Hsp90 (65, 66), specifically binds benzoquinone ansamycins (data not shown), suggesting that the effects on MI in yeast cells are mediated through altering the activity of Hsp90.
MI inhibits pp60v-src tyrosine kinase activity and decreases pp60v-src protein level.
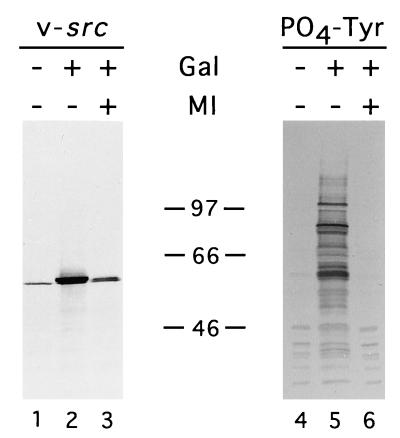
The quinone ansamycin, geldanamycin, has been shown to inhibit the transforming activity of the Hsp90 target protein pp60v-src in mammalian cells in culture by decreasing pp60v-src protein levels and activity (66). Similarly, pp60v-src levels and tyrosine kinase activity were decreased in yeast cells treated with MI (Fig. 2). Consistent with this finding, yeast cells treated with MI do not undergo growth arrest upon induction of pp60v-src expression, as do untreated yeast cells (data not shown). When the amount of lysate from different conditions is normalized for the pp60v-src protein level, tyrosine kinase activity is not detected in MI-treated extracts but is easily detected in untreated extracts, indicating that pp60v-src activity is affected independently of the protein level (data not shown). These findings indicate that MI abrogates the activity of diverse Hsp90-dependent signaling proteins in yeast cells.
FIG. 2.
Tyrosine kinase activity and protein level of pp60v-src are decreased in the presence of MI. Yeast cells containing a galactose-inducible expression vector were grown in glucose or galactose with or without MI (50 μM) as indicated. Samples of whole-cell extract were separated by SDS-PAGE; pp60v-src protein and phosphotyrosine were detected by immunoblotting with the appropriate antibodies. The amount of sample loaded for each blot was normalized to total protein. The approximate positions of the protein molecular weight markers are indicated. The band in lane 1 represents nonspecific cross-reactivity of the anti-pp60v-src antibody with a yeast protein and is seen in extracts of yeast cells lacking the pp60v-src expression plasmid (data not shown).
Yeast Hsp90 specifically binds benzoquinone ansamycins.
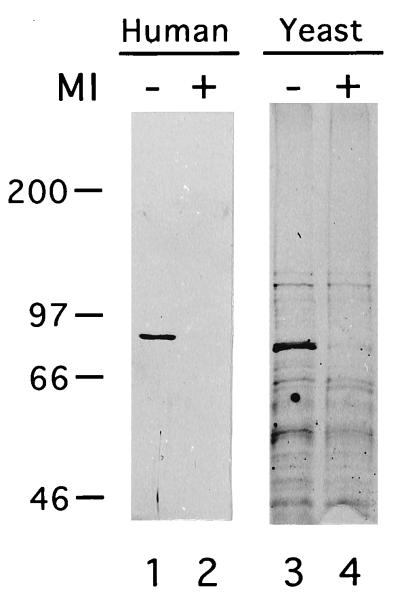
Prior to the demonstration by Whitesell et al. (66) that benzoquinone ansamycins specifically bind to Hsp90, these compounds were believed to act as tyrosine kinase inhibitors. To strengthen my hypothesis that the effects of MI in yeast cells are the result of alterations in Hsp90 function, I assessed the ability of the yeast Hsp90-like proteins Hsc82 and Hsp82 to bind to GA-coupled beads (66) and the ability of MI to compete for this binding. When GA-coupled beads are exposed to yeast whole-cell extract from cells expressing either human or yeast Hsp90, Hsp90 is the major protein retained by the beads (Fig. 3). MI competes for Hsp90 binding, indicating that this binding is specific. Thus, yeast Hsp90, like its human counterpart, specifically binds benzoquinone ansamycins, suggesting that the effects of MI in yeast are mediated through Hsp90.
FIG. 3.
Yeast Hsp-Hsc82 specifically binds immobilized geldanamycin. Whole-yeast-cell extracts of strains expressing human Hsp90 (lanes 1 and 2) or yeast Hsp82 and Hsc82 (lanes 3 and 4) were incubated with GA-coupled beads with (lanes 2 and 4) or without (lanes 1 and 3) the prior addition of MI (10 μM). GA-coupled beads were subsequently washed, and bound proteins were eluted, separated by SDS-PAGE, and detected by silver staining. Numbers at the left of the figure indicate the approximate positions of protein molecular weight markers. The identities of the major specifically bound proteins as human Hsp90 (lane 1) and yeast Hsp-Hsc82 (lane 3) were confirmed by immunoblotting (data not shown).
Identification of SBA1, a yeast gene encoding a p23-like protein.
The finding that inhibition of steroid receptor transcriptional activation and ligand binding by benzoquinone ansamycins correlates with a loss of p23 from the aporeceptor complex in mammalian cells has led to the hypothesis that p23 is required for the formation of receptor complexes capable of binding ligand with high affinity (58, 65). Furthermore, that p23 is required to render cell lysates competent for the assembly of aporeceptor complexes in vitro suggests a prominent role for p23, along with other chaperones, in the aporeceptor complex assembly (29, 32). To test whether benzoquinone ansamycins similarly alter the interaction of p23 with Hsp90-dependent signaling proteins in yeast, I sought to identify a yeast protein with significant amino acid sequence similarity to mammalian p23.
A sequence similarity search of the entire S. cerevisiae genome reveals a single ORF predicted to encode a protein with significant similarity to human p23; I will refer to this ORF as SBA1 for the increased sensitivity of steroid receptor signaling to benzoquinone ansamycin antibiotics in a strain with deletion of this gene (see below). SBA1 has been previously described as YKL117w (19) and is predicted to encode a 216-amino-acid polypeptide that shares 28% identity and 54% similarity over the 160 amino acids of the predicted human p23 polypeptide sequence (31; Fig. 4). The amino acid sequence similarity is evenly spread throughout the polypeptide sequences, with the notable exception of the 7-amino-acid sequence WPRLTKE beginning at amino acids 104 and 86 of Sba1 and human p23, respectively, which is identical in the two proteins.
MI alters aporeceptor complex composition in vivo.
Previous studies in vitro and in vivo have demonstrated that benzoquinone ansamycins alter the subunit composition of GR and PR aporeceptor complexes (17, 33, 58, 65). To assess the effects of MI treatment on a GR aporeceptor composition in yeast, I immunoprecipitated GR from yeast cells grown in the presence or absence of dexamethasone and/or MI with anti-GR monoclonal antibodies under nondenaturing conditions. GR and associated proteins were separated by SDS-PAGE and detected by immunoblotting (Fig. 5, left panel). GR aporeceptor complex composition in yeast cells is similar to that seen in mammalian cells or cell extracts (Fig. 5, lane 1); in addition to the Hsp90 family proteins, Hsp82 and Hsc82, the complexes contain a p60-like protein (Sti1), Hsp70 family proteins (Ssa and Ssb), a DnaJ-like protein (Ydj1), and Sba1, the yeast p23-like protein (see Fig. 5 and text below). The composition of GR aporeceptor complexes in yeast cells shown here is similar to that published by Kimura et al. (34), with the notable exception that the Hsp70-like Ssb protein coprecipitates with GR under the conditions used here (Fig. 5, lanes 1 to 4). In yeast cells, over 70% of Ssb protein is associated with translating ribosomes, and deletion of the genes encoding the Ssb proteins results in increased sensitivity to antibiotics that inhibit translation (39); thus, it is believed that the primary function of Ssb is to assist in the structural maturation of newly synthesized proteins (30, 39). Kimura et al. (34) epitope tagged the amino terminus of GR to facilitate immunoprecipitation; in contrast, I used a monoclonal antibody directed at an epitope within the GR protein sequence. It is possible that the amino-terminal epitope tag is not accessible to antibody until after the dissociation of the Ssb proteins, such that Kimura et al. isolated only the structurally mature receptor, whereas I am isolating both mature and nascent forms. Alternatively, the association of Ssb, whether functional or artifactual, may simply represent differences in the conditions used in performing these experiments. In any case, the association of the Hsp70-like proteins, Ssa and Ssb, does not appear to reflect alterations in the conformation and composition of the GR aporeceptor complex induced by ligand or MI (Fig. 5, lanes 1 to 4).
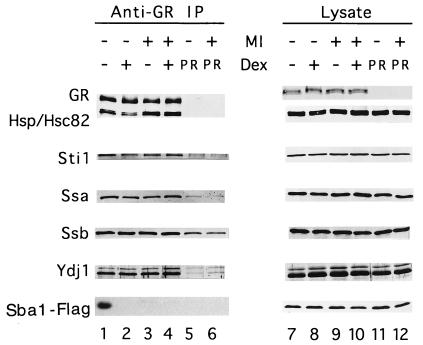
FIG. 5.
MI alters glucocorticoid aporeceptor complexes in yeast cells. GR was immunoprecipitated under nondenaturing conditions from whole-cell extracts of yeast cells expressing GR and epitope-tagged yeast Sba1 (Sba1-Flag) in the absence or presence of 10 μM Dex and/or 50 μM MI (lanes 1 to 4). GR and associated proteins in immunoprecipitates were resolved by SDS-PAGE and detected by immunoblotting with antibodies to specific proteins (as described in Materials and Methods). In parallel, extracts of yeast cells expressing human PR and epitope-tagged Sba1 in the absence or presence of 50 μM MI were exposed to anti-GR antibody resin, the resulting precipitates were resolved by SDS-PAGE, and specific proteins in these precipitates were detected to provide an estimate of nonspecific binding (lanes 5 and 6). The total amounts of proteins analyzed were similar in each type of extract (lanes 7 to 12). Whole-cell extract of each type was normalized for total protein concentration and separated by SDS-PAGE, and specific proteins were then detected by immunoblotting. Data for a given protein are from a single experiment and are representative of at least three independent experiments.
In the presence of ligand, a significant fraction of Hsp90 and all of the GR-associated Sba1 dissociate from the liganded receptor. This observation correlates with the ability of GR to regulate transcription in a ligand-regulated manner in yeast cells and confirms that aporeceptor complexes formed in yeast cells expressing GR are functional to the extent that the composition of such complexes is altered by ligand binding in a manner very similar to that observed in mammalian cells and cell extracts (Fig. 5, lane 2). While there are several possible explanations for the observation that only about 50% of GR-associated Hsp90 dissociates with ligand, the observation that there is little change in the levels of associated Sti1 protein suggests that a significant fraction of GR in yeast is complexed with Hsp90 and Sti1. Such complexes have been proposed to be low-affinity intermediates in the stepwise assembly of mature aporeceptor complexes (hypothesized to contain GR, Hsp90, immunophilin, and p23 [58]). Consistent with this model, all of the GR-associated Sba1 is displaced in the presence of ligand, indicating that the Sba1-containing aporeceptor complexes bind ligand and subsequently dissociate.
The addition of MI to yeast cultures results in two observed alterations in GR aporeceptor complex composition and behavior. First, ansamycin treatment displaces detectable Sba1 from aporeceptor complexes (Fig. 5, lane 3). A similar observation has been made with regard to the interaction of p23 with GR and PR aporeceptor complexes in cultured mammalian cells treated with geldanamycin (58, 65). However, in contrast to observations in mammalian cells, the interaction of GR with the other proteins examined in this study is apparently unaffected by MI. Second, Hsp90 does not dissociate from GR in response to Dex when MI is present (Fig. 5, lane 4), indicating that GR is not being activated by ligand and consistent with the observation that MI abrogates activation of transcription by GR in response to ligand. MI and/or Dex treatment had no effect on the level of GR or aporeceptor complex components present in whole-cell extracts (Fig. 5, right panel).
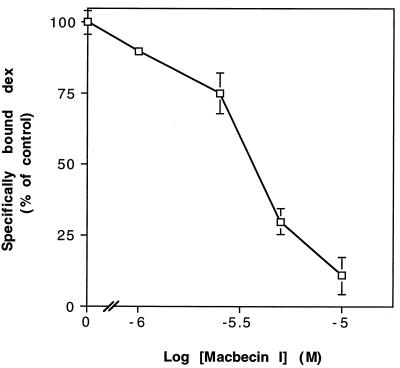
MI inhibits Dex binding by GR in a dose-dependent manner.
Two simple models could explain the finding that GR aporeceptor complexes do not dissociate in response to ligand in the presence of MI; through its interaction with Hsp90, MI may produce GR aporeceptor complexes that are unable to bind Dex, or GR aporeceptor complexes may bind ligand in the presence of MI, but MI may prevent dissociation of aporeceptor components subsequent to ligand binding. Whole-cell ligand-binding assays showed that Dex binding by GR is dramatically reduced in the presence of MI in a dose-dependent manner (Fig. 6), supporting the former hypothesis. Total GR protein levels are comparable in the absence or presence of MI (Fig. 5, lanes 7 to 10). Thus, while aporeceptor complexes form in the presence of MI, the resulting complexes are defective for ligand binding.
FIG. 6.
MI reduces Dex binding by yeast cells expressing GR in a dose-dependent manner. Yeast cells expressing GR were preincubated with various concentrations of MI or vehicle only for 2 h at 30°C, followed by a 2-h incubation with [3H]Dex with or without [1H]Dex competitor. Cells were washed, and the specific counts bound were determined (see Materials and Methods). Each datum point represents the mean ± range for two independent samples from a given experiment normalized to maximum specific counts bound and is representative of three experiments. A 100% binding is approximately 3,100 sites per cell. Yeast cells not expressing GR display no specific binding.
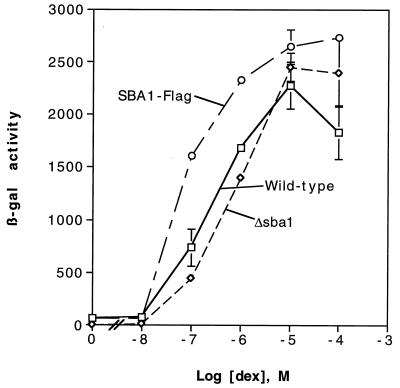
SBA1 deletion does not affect yeast cell growth, temperature sensitivity, or signaling protein function.
To test whether p23 is required for aporeceptor complex formation in vivo, I exploited the genetic techniques for yeast cells to delete the SBA1 gene. Deletion of the entire SBA1 gene via replacement of this chromosomal sequence with the HIS3 gene by homologous recombination resulted in a yeast strain with no appreciable growth defects at 30°C or altered sensitivity to growth at elevated or decreased temperature (deletion of SBA1 was confirmed by Southern blot analysis; data not shown). Furthermore, activation of a target promoter by GR in response to dexamethasone was unaffected by deletion of SBA1 (Fig. 7). Similarly, no effect on cell growth inhibition by pp60v-src or PR ligand response was noted as a result of SBA1 deletion (data not shown). These findings indicate that Sba1 function is not required in vivo in yeast cells either for viability or for steroid receptor function. No other p23-like ORFs were identified in the S. cerevisiae genome by computerized homology search, moderate stringency hybridization to yeast genomic DNA, or PCR amplification from genomic DNA with degenerate primers targeted to polypeptide domains conserved among several species (3a). Of note, there is a slight, but consistent, increase in ligand response of GR in cells expressing Sba1-Flag, as well as an approximately 50% increase in GR protein levels versus those in wild-type or SBA1 deletion strains, as measured by ligand binding (see Fig. 9) and immunoblotting (data not shown).
FIG. 7.
GR signaling in response to Dex is unaffected by SBA1 deletion. Yeast strains that are wild type, those with SBA1 deleted (Δsba1), or those with SBA1 deleted and with a plasmid expressing epitope-tagged Sba1 (SBA1-Flag) were incubated overnight with various concentrations of Dex. All cell types express GR and contain a GR-inducible lacZ reporter. Cells were harvested and β-Gal activities were measured (see Materials and Methods). Data, expressed as arbitrary β-Gal units, represent the means ± ranges of two independent samples from a given experiment and are representative of several experiments.
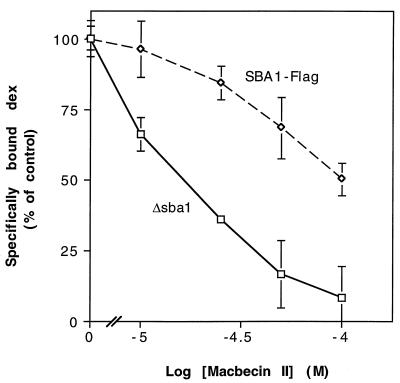
FIG. 9.
Dex binding in cells lacking Sba1 displays increased sensitivity to MII. Yeast strains with the chromosomal SBA1 gene deleted and containing plasmids expressing no Sba1 (Δsba1) or epitope-tagged Sba1 (SBA1-Flag) were preincubated with various concentrations of MII or vehicle only for 2 h at 30°C, followed by a 2-h incubation with [3H]Dex with or without [1H]Dex competitor. Cells were washed, and specific counts bound were determined (see Materials and Methods). Each datum point represents the mean ± range for two independent samples from a given experiment normalized to maximum specific counts bound, and results are representative of three experiments. Total binding is approximately 3,100 sites per cell for SBA1-Flag and 1,900 sites per cell for Δsba1.
SBA1 deletion results in increased sensitivity of GR activity to ansamycins; human p23 expression rescues this phenotype.
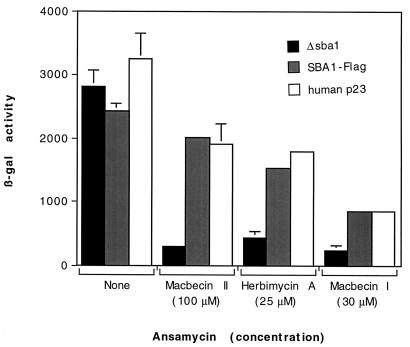
Given that p23 is a component of the aporeceptor complex and the compelling evidence from other experimental systems of a role for p23 in steroid receptor function, I assessed the effect of SBA1 deletion on the sensitivity of GR signaling to ansamycin treatment. MII and HA weakly inhibit GR signaling in wild-type yeast; however, an SBA1 deletion strain displays increased sensitivity to MII and HA (Fig. 8), an observation consistent with a role for p23 in antagonizing the effects of ansamycins on GR signaling. Similarly, that GR signaling in the SBA1 deletion strain displays increased sensitivity to MI (Fig. 8) suggests that the loss of Sba1 results in increased sensitivity of GR signaling to all ansamycins. Furthermore, the ability of the human p23 protein to rescue the increased sensitivity of GR signaling to benzoquinone ansamycins in yeast cells lacking Sba1 (Fig. 8) indicates that human p23 and yeast Sba1 are functionally interchangeable and suggests that yeast cells provide a valid system for studying the role of p23 in steroid receptor function in vivo.
FIG. 8.
GR signaling in SBA1 deletion strains displays increased sensitivity to ansamycins; expression of human p23 rescues this increased sensitivity. Yeast strains with the chromosomal SBA1 gene deleted and containing plasmids expressing no Sba1 (Δsba1), epitope-tagged Sba1 (Sba1-Flag), or human p23 were incubated overnight at 30°C with Dex (10 μM) with or without MII, HA, or MI. All strains express GR and contain a GR-inducible lacZ reporter. Cells were harvested, and β-Gal activities were measured (see Materials and Methods). Data, expressed as arbitrary β-Gal units, represent the means ± ranges of two independent samples from a given experiment and are representative of several experiments. The response of the parent yeast strain, with the chromosomal SBA1 gene intact, is comparable to those of the SBA1-Flag and human p23 strains (data not shown).
To confirm that the defect in GR signaling caused by MII treatment is a result of decreased ligand binding, I measured the effects of MII on Dex binding in whole cells lacking Sba1 or expressing Sba1-Flag (Fig. 9). As expected, MII caused a significant, dose-dependent decrease in Dex binding in SBA1 deletion strains versus a less severe decrease in the Sba1-Flag strain. Furthermore, GR and Hsp90 protein levels and the levels of GR-Hsp90 complexes are similar in wild-type and SBA1 deletion strains, whereas the GR protein level is about 50% higher (as mentioned above) and the Hsp90 level is similar in the Sba1-Flag strains (data not shown). Taken together, these findings suggest that, while p23 is not required for Hsp90 target protein function, the presence of p23 protects Hsp90 from the effects of ansamycins and, thereby, preserves the activity of proteins that are dependent on Hsp90 to achieve a mature, functional conformation.
DISCUSSION
A variety of signaling proteins, including nuclear hormone receptors (6, 10, 27, 47), tyrosine kinases (1, 9, 14), and the dioxin receptor (67), require Hsp90 to achieve normal cellular activity. The discovery that benzoquinone ansamycins bind to Hsp90 (66) and, in so doing, disrupt the activity of Hsp90-dependent signaling proteins (51, 58, 65, 66) provides a powerful reagent both for the identification of Hsp90 substrate proteins and for the characterization of Hsp90 function. In this study, I have demonstrated that ansamycins abrogate the activity of diverse Hsp90 substrates, specifically GR, PR, RAR, and pp60v-src, in yeast cells in a manner similar to that observed in cultured mammalian cells.
Hsp90 acts with a number of associated proteins (42, 57), including Hsp70, p60-Sti1, immunophilins, and p23-Sba1, to promote the maturation of substrate proteins. Several lines of evidence have converged on the hypothesis that p23 is required for Hsp90-dependent steroid receptor function. First, depletion of p23 from reticulocyte lysate renders the lysate incompetent to promote the conversion of free steroid receptors into high-ligand-affinity aporeceptor complexes (29, 32). Second, this loss of activity is rescued by supplementation with biochemically purified p23 (29, 32). Third, addition of purified p23 to wheat germ lysate, which does not normally support the assembly of aporeceptor complexes in vitro, renders this extract competent to assemble aporeceptor complexes that are able to bind ligand (29). Fourth, the loss of function and the decreased stability of steroid receptors following geldanamycin treatment in cultured mammalian cells correlates with disruption of the Hsp90-p23 interaction (58, 65). While these findings lend support for an obligate role for p23 in the conformational maturation of steroid receptors, the deletion of the yeast p23-like gene SBA1 had, surprisingly, no effect on GR function, indicating that p23 is not required for Hsp90 target protein function in yeast. Furthermore, while ansamycin antibiotics disrupt the interaction of p23 with steroid aporeceptor complexes, some effect of ansamycin treatment other than the displacement of p23 must lead to defects in the signaling activity of Hsp90 target proteins.
At first glance, my findings appear to contradict the results of experiments with animal cells and cell extracts. However, it is important to note that observations in cultured cells demonstrate only a correlation between the loss of steroid receptor function and the disruption of the Hsp90-p23 interaction in response to geldanamycin treatment (58, 65). In contrast, experiments with cell lysates suggest that p23 is required for complex assembly in in vitro systems (29, 32). One possible interpretation is that aporeceptor complex assembly in cell lysates may reflect only a p23-dependent subset of the routes that Hsp90 may take in vivo to form functional complexes with target proteins. This view would imply that Hsp90-associated proteins, other than p23, may be competent to assist Hsp90 in completing the maturation of target proteins to their functional form in vivo. If this were the case, such alternate pathways would remain intact in yeast cells lacking Sba1 and support the activity of Hsp90 target proteins. Alternatively, p23 may increase the stability of mature aporeceptor complexes without actually being required to achieve a high-ligand-affinity conformation. In findings published during the preparation of this manuscript, Dittmar et al. (16, 17) suggest that p23 is, in fact, not required to assemble mature aporeceptor complexes in vitro, but instead serves to stabilize such complexes once assembled; Segnitz and Gehring (52) found that in mammalian cells in culture, while geldanamycin inhibits the activity of various steroid receptors, the degree of receptor protein destabilization resulting from geldanamycin treatment varies in different cell types. Thus, the importance of p23 for receptor function may depend on the stability of aporeceptor complexes in a given cell type, which in turn would reflect as-yet-uncharacterized factors in the cellular milieu. Finally, despite the striking similarities between the aporeceptor complexes in yeast and animal cells (11, 57), it remains possible that the assembly of aporeceptor complexes in yeast cells may be fundamentally different than that in metazoans. In any case, my findings demonstrate that functional receptor-Hsp90 complexes can form in vivo in the absence of p23.
Although Sba1 is not required for the function of Hsp90 target proteins in yeast cells, it does have a role in steroid receptor signaling, as evidenced by the increased sensitivity of GR ligand response to ansamycins in the SBA1 deletion strain. This altered sensitivity to ansamycins suggests that p23 antagonizes the action of ansamycin antibiotics. It has been proposed that geldanamycin and p23 may directly compete for Hsp90 binding (58). This model is consistent with my findings; however, the properties of the geldanamycin-nucleotide binding domain of Hsp90, as revealed by the crystal structures of geldanamycin (59) or nucleotide (44) bound to an amino-terminal fragment of Hsp90, suggest that ansamycins compete with adenosine nucleotides for binding to a common site on Hsp90. Though it is unlikely that ansamycins and p23 compete directly for binding to Hsp90, ATP and Mg2+ are required for the formation of Hsp90-p23 complexes in vitro (33); furthermore, Sullivan et al. (60) have shown that ADP inhibits p23 binding to Hsp90 and that ATP alters some biochemical characteristics of Hsp90 without having a detectable effect on p23, suggesting that ATP acts through altering Hsp90. Furthermore, Grenert et al. (25) have demonstrated that nucleotide binding to Hsp90 is disrupted by geldanamycin. As has been proposed (25, 44, 60), these findings suggest that the ATP-bound form of Hsp90 is required at some point in the functional maturation of Hsp90 target proteins and that ansamycins may disrupt Hsp90 function by mimicking nucleotides and inhibiting nucleotide binding to Hsp90.
My findings expand upon these proposals by demonstrating that, while ATP may be required for Hsp90 to complete its chaperone function, bound p23 is not required for the maturation of Hsp90 target proteins to the functional form in vivo in yeast cells. However, the finding that receptor function is more sensitive to ansamycin activity in cells lacking Sba1 suggests that p23 may play some role in stabilizing complexes containing ATP-bound Hsp90. Thus, although p23 is not required in the contexts that I examined here, one can envision potential Hsp90 target proteins or cellular environments that require increased stability of complexes containing ATP-bound Hsp90 and, therefore, are dependent on the presence of p23 for Hsp90 target protein function. Finally, whatever the role of p23 in Hsp90 function, it would appear to be highly conserved, given that the expression of human p23 protein in yeast cells can rescue the phenotype caused by deletion of the SBA1 gene.
Ansamycin antibiotics have been demonstrated to be effective antitumor agents in vitro (61) and in mouse models (40, 48). Recent structural and biochemical analyses, as cited above, have contributed greatly to our understanding of the mechanism of ansamycin action. In an effort to even better understand the effects of ansamycins, yeast genetic analysis offers the opportunity to dissect the roles of Hsp90 and of associated proteins, such as p23-Sba1, in an easily manipulated in vivo system. The findings presented here compel significant modifications in our current understanding of the role of Hsp90 and associated proteins in cell signaling and offer insight into the mechanism of action of ansamycin antibiotics. Further investigation of the dependence of a variety of cell signaling systems on Hsp90 and other chaperones in yeast cells, mammalian cell culture, and cell lysates may identify new targets for pharmacological intervention and, in combination with structural analysis, will provide a basis for refinement of the therapeutic action of known Hsp90 binding compounds.
ACKNOWLEDGMENTS
I thank A. Kralli and B. Freeman for critically reading the manuscript. Thanks go to A. Caplan, E. Craig, S. Lindquist, and D. Toft for generously providing antibodies. My appreciation goes to K. Yamamoto, L. Neckers, E. Mimnaugh, D. Toft, C. Klee, D. Hursh, A. Clements, C. Wu, and members of the Wu laboratory for technical advice and helpful discussions, and thanks go to M. Singer for laboratory space and helpful discussions. Thanks go to S. Davis, Jr., for assistance with immunoprecipitations and ligand-binding assays.
S.B. was supported by a National Institutes of Health postdoctoral Intramural Research Training Award. This work was supported by the Intramural Research Program of the National Cancer Institute.
REFERENCES
- 1.Aligue R, Akhavan N H, Russell P. A role for Hsp90 in cell cycle control: Wee1 tyrosine kinase activity requires interaction with Hsp90. EMBO J. 1994;13:6099–6106. doi: 10.1002/j.1460-2075.1994.tb06956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blagosklonny M V, Toretsky J, Bohen S, Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci USA. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohen S P. Hsp90 mutants disrupt glucocorticoid receptor ligand binding and destabilize aporeceptor complexes. J Biol Chem. 1995;270:29433–29438. doi: 10.1074/jbc.270.49.29433. [DOI] [PubMed] [Google Scholar]
- 3a.Bohen, S. P. Unpublished observations.
- 4.Bohen S P, Kralli A, Yamamoto K R. Hold ’em and fold ’em: chaperones and signal transduction. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 5.Bohen S P, Yamamoto K R. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohen S P, Yamamoto K R. Modulation of steroid receptor signal transduction by heat shock proteins. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334. [Google Scholar]
- 7.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bresnick E H, Dalman F C, Sanchez E R, Pratt W B. Evidence that the 90-kDa heat shock protein is necessary for the steroid binding conformation of the L cell glucocorticoid receptor. J Biol Chem. 1989;264:4992–4997. [PubMed] [Google Scholar]
- 9.Brugge J S. Interaction of the Rous sarcoma virus protein pp60src with the cellular proteins pp50 and pp90. Curr Top Microbiol Immunol. 1986;123:4992–4997. doi: 10.1007/978-3-642-70810-7_1. [DOI] [PubMed] [Google Scholar]
- 10.Catelli M G, Binart N, Jung T I, Renoir J M, Baulieu E E, Feramisco J R, Welch W J. The common 90-kd protein component of non-transformed ‘8S’ steroid receptors is a heat-shock protein. EMBO J. 1985;4:3131–3135. doi: 10.1002/j.1460-2075.1985.tb04055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang H C, Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- 12.Chang H C, Nathan D F, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 14.Cutforth T, Rubin G M. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 15.Dalman F C, Sturzenbecker L J, Levin A A, Lucas D A, Perdew G H, Petkovitch M, Chambon P, Grippo J F, Pratt W B. Retinoic acid receptor belongs to a subclass of nuclear receptors that do not form “docking” complexes with hsp90. Biochemistry. 1991;30:5605–5608. doi: 10.1021/bi00236a038. [DOI] [PubMed] [Google Scholar]
- 16.Dittmar K D, Demady D R, Stancato L F, Krishna P, Pratt W B. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery: the role of p23 is to stabilize receptor-hsp90 heterocomplexes formed by hsp90-p60-hsp70. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- 17.Dittmar K D, Pratt W B. Folding of the glucocorticoid receptor by the reconstituted Hsp90-based chaperone machinery. The initial hsp90.p60.hsp70-dependent step is sufficient for creating the steroid binding conformation. J Biol Chem. 1997;272:13047–13054. doi: 10.1074/jbc.272.20.13047. [DOI] [PubMed] [Google Scholar]
- 18.Duina A A, Chang H C, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 19.Dujon B, Alexandraki D, Andre B, Ansorge W, Baladron V, Ballesta J P, Banrevi A, Bolle P A, Bolotin F M, Bossier P, et al. Complete DNA sequence of yeast chromosome XI. Nature (London) 1994;369:371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- 20.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garabedian M J, Yamamoto K R. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 23.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godowski P J, Rusconi S, Miesfeld R, Yamamoto K R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature (London) 1987;325:365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- 25.Grenert J P, Sullivan W P, Fadden P, Haystead T A J, Clark J, Mimnaugh E, Krutzsch H, Ochel H J, Schulte T W, Sausville E, et al. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 26.Hall B L, Smit M Z, Privalsky M L. Reconstitution of retinoid X receptor function and combinatorial regulation of other nuclear hormone receptors in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:6929–6933. doi: 10.1073/pnas.90.15.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holley S J, Yamamoto K R. A role for Hsp90 in retinoid receptor signal transduction. Mol Biol Cell. 1995;6:1833–1842. doi: 10.1091/mbc.6.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchison K A, Dittmar K D, Czar M J, Pratt W B. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J Biol Chem. 1994;269:5043–5049. [PubMed] [Google Scholar]
- 29.Hutchison K A, Stancato L F, Owens G J, Johnson J L, Krishna P, Toft D O, Pratt W B. The 23-kDa acidic protein in reticulocyte lysate is the weakly bound component of the hsp foldosome that is required for assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. J Biol Chem. 1995;270:18841–18847. doi: 10.1074/jbc.270.32.18841. [DOI] [PubMed] [Google Scholar]
- 30.James P, Pfund C, Craig E A. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 31.Johnson J L, Beito T G, Krco C J, Toft D O. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol. 1994;14:1956–1963. doi: 10.1128/mcb.14.3.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J L, Toft D O. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J Biol Chem. 1994;269:24989–24993. [PubMed] [Google Scholar]
- 33.Johnson J L, Toft D O. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–678. doi: 10.1210/mend.9.6.8592513. [DOI] [PubMed] [Google Scholar]
- 34.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 35.Kralli A, Bohen S P, Yamamoto K R. LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc Natl Acad Sci USA. 1995;92:4701–4705. doi: 10.1073/pnas.92.10.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kralli A, Yamamoto K R. An FK506-sensitive transporter selectively decreases intracellular levels and potency of steroid hormones. J Biol Chem. 1996;271:17152–17156. doi: 10.1074/jbc.271.29.17152. [DOI] [PubMed] [Google Scholar]
- 37.Lai B-T, Chin N W, Stanek A E, Keh W, Lanks K W. Quantitation and intracellular localization of the 85K heat shock protein by using monoclonal and polyclonal antibodies. Mol Cell Biol. 1984;4:2802–2810. doi: 10.1128/mcb.4.12.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy S M, Bergman M, Morgan D O. Suppression of c-Src activity by C-terminal Src kinase involves the c-Src SH2 and SH3 domains: analysis with Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5290–5300. doi: 10.1128/mcb.13.9.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson R J, Zuegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 40.Ono Y, Kozai Y, Ootsu K. Antitumor and cytocidal activities of a newly isolated benzenoid ansamycin, macbecin I. Gann. 1982;73:938–944. [PubMed] [Google Scholar]
- 41.Picard D, Khursheed B, Garabedian M J, Fortin M G, Lindquist S, Yamamoto K R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature (London) 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 42.Pratt W B, Scherrer L C, Hutchison K A, Dalman F C. A model of glucocorticoid receptor unfolding and stabilization by a heat shock protein complex. J Steroid Biochem Mol Biol. 1992;41:223–229. doi: 10.1016/0960-0760(92)90348-m. [DOI] [PubMed] [Google Scholar]
- 43.Privalsky M L, Sharif M, Yamamoto K R. The viral erbA oncogene protein, a constitutive repressor in animal cells, is a hormone-regulated activator in yeast. Cell. 1990;63:1277–1286. doi: 10.1016/0092-8674(90)90423-c. [DOI] [PubMed] [Google Scholar]
- 44.Prodromou C, Roe S M, O’Brien R, Ladbury J E, Piper P W, Pearl L H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 45.Rehberger P, Rexin M, Gehring U. Heterotetrameric structure of the human progesterone receptor. Proc Natl Acad Sci USA. 1992;89:8001–8005. doi: 10.1073/pnas.89.17.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rexin M, Busch W, Gehring U. Protein components of the nonactivated glucocorticoid receptor. J Biol Chem. 1991;266:24601–24605. [PubMed] [Google Scholar]
- 47.Sanchez E R, Toft D O, Schlesinger M J, Pratt W B. Evidence that the 90-kDa phosphoprotein associated with the untransformed L-cell glucocorticoid receptor is a murine heat shock protein. J Biol Chem. 1985;260:12398–12401. [PubMed] [Google Scholar]
- 48.Sasaki K, Yasuda H, Onodera K. Growth inhibition of virus transformed cells in vitro and antitumor activity in vivo of geldanamycin and its derivatives. J Antibiot. 1979;32:849–851. doi: 10.7164/antibiotics.32.849. [DOI] [PubMed] [Google Scholar]
- 49.Schena M, Yamamoto K R. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 50.Scherrer L C, Hutchison K A, Sanchez E R, Randall S K, Pratt W B. A heat shock protein complex isolated from rabbit reticulocyte lysate can reconstitute a functional glucocorticoid receptor-Hsp90 complex. Biochemistry. 1992;31:7325–7329. doi: 10.1021/bi00147a017. [DOI] [PubMed] [Google Scholar]
- 51.Schulte T W, Blagosklonny M V, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- 52.Segnitz B, Gehring U. The function of steroid hormone receptors is inhibited by the hsp90-specific compound geldanamycin. J Biol Chem. 1997;272:18694–18701. doi: 10.1074/jbc.272.30.18694. [DOI] [PubMed] [Google Scholar]
- 53.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith D F. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 56.Smith D F, Stensgard B A, Welch W J, Toft D O. Assembly of progesterone receptor with heat shock proteins and receptor activation are ATP mediated events. J Biol Chem. 1992;267:1350–1356. [PubMed] [Google Scholar]
- 57.Smith D F, Toft D O. Steroid receptors and their associated proteins. Mol Endocrinol. 1993;7:4–11. doi: 10.1210/mend.7.1.8446107. [DOI] [PubMed] [Google Scholar]
- 58.Smith D F, Whitesell L, Nair S C, Chen S, Prapapanich V, Rimerman R A. Progesterone receptor structure and function altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stebbins C E, Russo A A, Schneider C, Rosen N, Hartl F U, Pavletich N P. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri E S, Litwack G, Toft D. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 61.Supko J G, Hickman R L, Grever M R, Malspeis L. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother Pharmacol. 1995;36:305–315. doi: 10.1007/BF00689048. [DOI] [PubMed] [Google Scholar]
- 62.Tanida S, Hasegawa T, Higashide E. Macbecins I and II, new antitumor antibiotics. J Antibiot. 1980;33:199–212. doi: 10.7164/antibiotics.33.199. [DOI] [PubMed] [Google Scholar]
- 63.Uehara Y, Fukazawa H. Use and selectivity of herbimycin A as inhibitor of protein-tyrosine kinases. Methods Enzymol. 1991;201:370–379. doi: 10.1016/0076-6879(91)01033-x. [DOI] [PubMed] [Google Scholar]
- 64.Vegeto E, Allan G F, Schrader W T, Tsai M J, McDonnell D P, O’Malley B W. The mechanism of RU486 antagonism is dependent on the conformation of the carboxy-terminal tail of the human progesterone receptor. Cell. 1992;69:703–713. doi: 10.1016/0092-8674(92)90234-4. [DOI] [PubMed] [Google Scholar]
- 65.Whitesell L, Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 66.Whitesell L, Mimnaugh E G, De Costa B, Myers C E, Neckers L M. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilhelmsson A, Cuthill S, Denis M, Wikstrom A C, Gustafsson J A, Poellinger L. The specific DNA binding activity of the dioxin receptor is modulated by the 90 kd heat shock protein. EMBO J. 1990;9:69–76. doi: 10.1002/j.1460-2075.1990.tb08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci USA. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto K R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]