Abstract
Most DNA in human sperm is bound to highly basic proteins called protamines, but a small proportion is complexed with histones similar to those found in active chromatin. This raises the intriguing possibility that histones in sperm are marking sets of genes that will be preferentially activated during early development. We have examined the chromatin structure of members of the β-globin gene family, which are expressed at different times in development, and the protamine 2 gene, which is expressed in spermatids prior to the widespread displacement of histones by transition proteins. The genes coding for ɛ and γ globin, which are active in the embryonic yolk sac, contain regions which are histone associated in the sperm. No histone-associated regions are present at the sites tested within the β- and δ-globin genes which are silent in the embryonic yolk sac. The trends of histone or protamine association are consistent for samples from the same person, and no significant between-subject variations in these trends are found for 13 of the 15 fragments analyzed in the two donors. The results suggest that sperm chromatin structures are generally similar in different men but that the length of the histone-associated regions can vary. The association of sperm DNA with histones or protamines sometimes changes within as little as 400 bp of DNA, suggesting that there is fine control over the retention of histones.
Complex rearrangements of the sperm chromatin occur during mammalian spermatogenesis. In the elongating spermatids, the histones associated with the DNA are displaced by transition proteins, which in turn are replaced by protamines. As a result of this process, the chromatin in the spermatozoa is highly condensed (17). In human sperm, approximately 15% of the DNA remains associated with histones in a sequence-specific manner (12, 25). The histones in sperm chromatin are a subset of the histones found in somatic chromatin (13), and they form nucleosomes which are more closely packed than those found in somatic cells (4). Histone H1 is absent, histone H2 takes the form of two minor variants, called H2A.X and H2A.Z, and the histones H3 and H4 are extensively acetylated (13). Absence of histone H1 and acetylation of histones are both features of active chromatin (28, 29). This has led to the suggestion that histones in sperm could influence which genes are first transcribed after fertilization (12).
To explore the impact of sperm chromatin on temporal gene regulation, we analyzed the β-globin family of genes which are transcribed at different times during development. The β-globin locus from 5′ to 3′ consists of the locus control region (LCR) and the ɛ-, Gγ-, Aγ-, δ-, and β-globin genes. Only the ɛ- and γ-globin genes are transcribed in the primitive erythroblasts in the embryonic yolk sac which differentiates at 3 weeks of gestation. The expression of the γ-globin gene predominates during the fetal period when the site of erythropoiesis shifts to the definitive erythroblasts in the fetal liver. The δ-globin gene product is a minor variant produced after birth in bone marrow. The β-globin gene is expressed to a small extent in the fetus but predominates after birth in bone marrow (21, 23). In this complex process of globin switching, it is currently not clear what roles are played by chromatin structures (2, 10) and by interactions between the genes and the LCR and transcription factors (5). The protamine 2 gene was studied as an example of a gene which is transcribed at the stage when histones are being displaced from spermatids but thereafter remains silent in the embryo (19).
Our study employed a technique which selectively removes histones from the sperm chromatin and then assesses the accessibility of specific sites to restriction enzymes. We identified regions which have conserved patterns of histone or protamine association in sperm samples from the same person and between two individuals. We have related the presence of these regions to expression patterns during development. The detection of histone-associated regions in the ɛ- and γ-globin genes suggests that the presence of histones may mark these genes for early expression in the embryo.
MATERIALS AND METHODS
Fractionation of sperm chromatin.
Liquefied semen samples from two donors were collected and stored at −20°C until analysis. Collection and utilization of the sperm for the present study was approved by the Central Sydney Area Health Service Ethics Review Committee. Sperm DNA was separated into fractions enriched for histone-associated DNA (HDNA) and protamine-associated DNA (PDNA) by a modification of the method of Gatewood et al. (12). Disulfide bonds between protamine molecules were reduced by dithiothreitol, and the tails of the sperm were disrupted by cetyltrimethylammonium bromide (3). All subsequent steps were carried out very gently in the presence of 0.05% digitonin to minimize clumping of nuclei. The nuclei were washed five times by centrifugation at 3,000 × g for 5 min and suspension in Tris saline buffer. Histones were selectively removed from the chromatin by treatment with a solution containing 0.65 M NaCl, 1 mM EDTA, and 10 mM Tris HCl, pH 8.0 (12). The nuclei were washed once with a solution containing 100 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, and 10 mM Tris HCl, pH 8.0. To release the HDNA fraction, the chromatin was cleaved with BamHI and DraI restriction enzymes (Boehringer Mannheim) in the above buffer for 1.5 h at 37°C with occasional rocking. The nuclei from two to four ejaculates (about 5 × 108 cells) were incubated with 2,000 U of enzyme. The chromatin was centrifuged at 3,000 × g for 2 min. The resulting supernatant contained the HDNA fraction, and the pellet contained the PDNA fraction. The supernatant was centrifuged twice at 9,000 × g to remove any contaminating PDNA.
For a fragment to be released into the HDNA fraction, sites at both ends of the fragment must be accessible to the enzyme. In cases where one site is bound to histone and the other site is bound to protamine, the DNA will remain in the PDNA fraction. DNA classed as histone associated may also include some DNA free of both histones and protamines.
Isolation of DNA.
The HDNA fraction was incubated with 200 μg of proteinase K per ml and 0.5% sodium dodecyl sulfate (SDS) at 55°C overnight and extracted twice with phenol-chloroform (1:1 [vol/vol]). The HDNA was precipitated with ethanol and suspended in TE (10 mM Tris [pH 8.0], 0.1 mM EDTA). Protamines were removed from the pellet containing the PDNA fraction by a modified version of the method of Gatewood (13a). The pellet was solubilized in a solution containing 8 M urea, 0.6 M NaCl, 0.2 M dithiothreitol, and 10 mM Tris, pH 8.0, and passed through 18-, 21-, and 25-gauge needles, successively. The solubilized chromatin was then bound for 20 min at 4°C to an equal volume of preequilibrated SP Sephadex C-25 (Pharmacia Biotech). PDNA was purified from the unbound fraction by at least four extractions with phenol-chloroform followed by ethanol precipitation and suspension in TE. Total DNA from sperm was isolated by the same method as for the PDNA with the omission of the histone removal and restriction enzyme digestion steps.
Protein analysis.
Following the 0.65 M NaCl treatment of the nuclei to selectively release histones, the supernatant containing the extracted proteins was collected. Acid-insoluble proteins were removed as described by Gatewood et al. (12), and the remaining proteins were desalted and concentrated by using Centricon 3 microconcentrators (Amicon). The proteins were then analyzed on gels prepared with 15% acrylamide–0.1% bisacrylamide–0.9 N acetic acid–2.5 M urea and visualized by Coomassie blue staining (24) (Fig. 1).
FIG. 1.
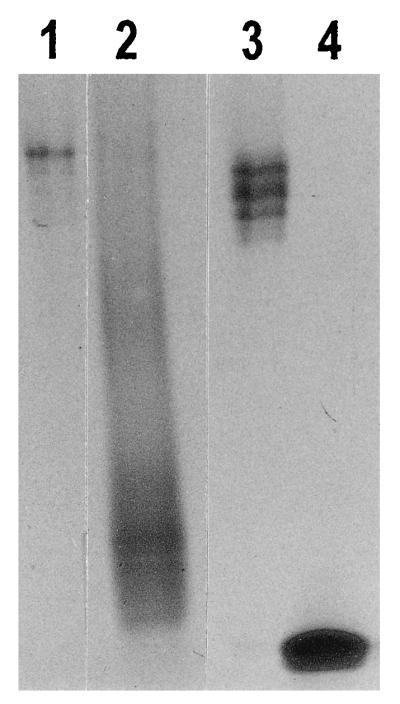
Selective extraction of histones from the sperm chromatin by 0.65 M NaCl treatment. This procedure has released most of the histones from the sperm chromatin, leaving the protamines still associated with the DNA. Lane 1 contains 1 μg of protein extracted from human sperm nuclei and consists primarily of histones with no detectable protamines. Lane 2 contains 3.0 μg of protein which remained associated with the sperm chromatin after extraction. The fraction consists mostly of protamines with a trace amount of histones. Lane 3 contains 1 μg of calf thymus histones for comparison with lane 1. Calf thymus histones are different from human sperm histones but are of comparable size. Lane 4 contains 1 μg of salmon sperm protamines for comparison with lane 2. Salmon sperm protamines are smaller (∼30 amino acids) than human protamines (∼50 amino acids). The pattern of fractionation of histones and protamines is similar to that reported by Gatewood et al. (Fig. 2 of reference 12).
The proteins which remained associated with the chromatin during the 0.65 M NaCl treatment were solubilized and bound to SP Sephadex C-25, as described above. The proteins were then released from the Sephadex in a 2 M NaCl–10 mM Tris solution (pH 8) and, in the same manner as the 0.65 M NaCl-extractable proteins, were purified and analyzed on acid-urea-polyacrylamide gels.
Southern blot analysis.
Genomic clones of the human genes coding for protamine 2, ɛ globin, and Gγ, Aγ, δ, and β globin were kindly provided by W. Engel, M. Baron, and R. Trent, respectively. DNA probes incorporating [α-32P]dCTP, illustrated in Fig. 2 to 5 and 7, were generated by using a Gigaprime DNA labelling kit (Bresatec). DNA was fractionated on 1.5% agarose superfine resolution gels (Amresco) and transferred to Hybond N+ filters (Amersham) by the standard procedure (20). Hybridization with DNA probes was performed in a mixture of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt’s solution, 0.5% SDS, and 0.1 mg of herring sperm DNA per ml overnight at 68°C. Blots were washed twice in 2× SSC–0.1% SDS at 68°C and twice in 0.5× SSC–0.1% SDS at 68°C for at least 30 min each time. Results were visualized and quantitated with a PhosphorImager (Storm 860; Molecular Dynamics).
FIG. 2.
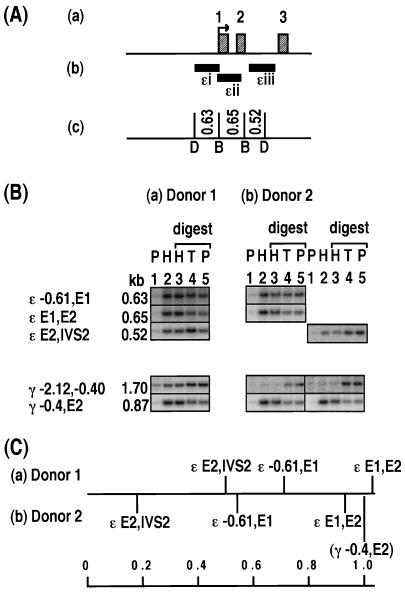
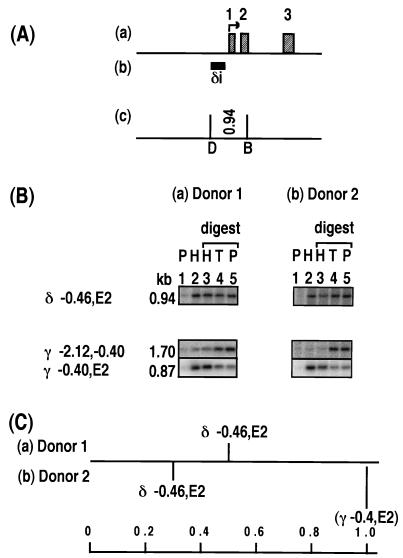
Relative levels of histone association of restriction enzyme sites in the ɛ-globin gene. (A) Maps of the ɛ-globin gene showing the locations of the transcription start site (arrow) and exons (labelled hatched boxes) (a), DNA probes (ɛi to ɛiii (b), and restriction enzyme sites (DraI [D] and BamHI [B]) and the lengths of DNA fragments (in kilobases) (c). (B) Southern blot analysis of HDNA and PDNA fractions for two sperm donors, donor 1 and donor 2. Lanes 1 and 2 contain PDNA (P) and HDNA (H). Lanes 3, 4, and 5 contain HDNA (H), total DNA (T), and PDNA (P) digested with BamHI (lanes 3) and DraI (lanes 5) restriction enzymes. Each lane contains 2 μg of DNA. The positions of the sites examined, on the border of each fragment, are marked as follows: E, exon; IVS, intron; negative values, distance upstream of the mRNA start site (in kilobases). Each column shows the same Southern blot probed with multiple DNA probes. The results for two γ-globin fragments, γ −2.12,−0.40 and γ −0.4,E2 are provided as negative and positive controls for enrichment in the HDNA fraction, respectively. The locations of these γ-globin fragments are illustrated in Fig. 3. The images were generated by using Molecular Dynamics ImageQuant and were compiled without alteration by using Microsoft Powerpoint and Adobe Photoshop. (C) Level of enrichment of a fragment in the HDNA fraction relative to that of the γ −0.4,E2 control fragment. For donor 1, the relative level of enrichment is displayed as an average for each fragment. The number of samples (in parentheses) and the values or the range of values are as follows: ɛ −0.61,E1 (2) 0.71 and 0.72; ɛ E1,E2, (2) 0.99 and 1.06; and ɛ E2,IVS2, (3) 0.41 to 0.66. For donor 2, the result of the sperm sample displayed in panel B is plotted. The other samples from donor 2 gave results similar to those presented, but the quantitation was less accurate as the signals were obtained using DNA probes labelled with digoxigenin-dUTP instead of [α-32P]dCTP, and the results were visualized on film rather than a phosphorimager screen. Figures 3 to 7 are set up like this figure and show different globin or protamine genes.
FIG. 5.
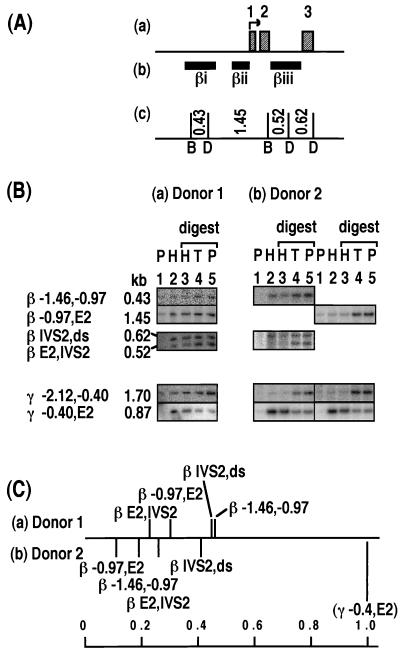
Relative levels of histone association of sites within the β-globin gene. The DNA probes are named βi to βiii. For donor 1 in panel C, the number of samples (in parentheses) and the values are as follows: β −1.46,−0.97, (2) 0.45 and 0.47; β −0.97,E2, (2) 0.30; β E2,IVS2, (2) 0.21 and 0.25; and β IVS2,ds, (2) 0.41 and 0.48. See the legend to Fig. 2.
FIG. 7.
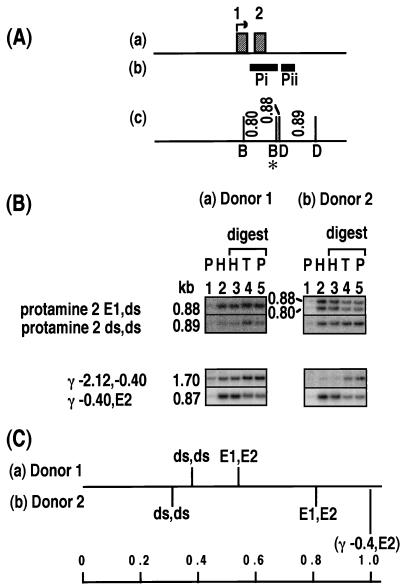
Relative levels of histone association of sites within the protamine 2 gene. The DNA probes are named Pi and Pii. The polymorphic BamHI site is marked with an asterisk. For donor 1 in panel C, the number of samples (in parentheses) and the range of values are listed as follows: protamine 2 E1,ds, (4) 0.33 to 0.67; and protamine 2 ds,ds, (3) 0.21 to 0.53. See the legend to Fig. 2.
RESULTS
Human sperm chromatin was separated into fractions enriched for either HDNA or PDNA at BamHI and DraI restriction enzyme sites. During fractionation, the HDNA and PDNA chromatin was fully or partially digested with restriction enzymes, respectively (see Materials and Methods). In order to obtain a quantitative comparison of DNA fragments in the two fractions, protein-extracted HDNA and PDNA (Fig. 2B to 7B, lanes 1 and 2) were redigested with BamHI and DraI restriction enzymes and termed HDNA or PDNA digest, respectively (Fig. 2B to 7B, lanes 3 and 5). Southern blots of equal amounts of DNA were then hybridized with DNA probes from genes of interest. The enrichment of a particular fragment in the HDNA digest lane compared to the TDNA and PDNA digest lanes indicates that, relative to the bulk of sperm DNA, the enzyme sites that were cleaved to generate the fragment were histone associated.
In order to standardize the results for different sperm preparations and different Southern blots, all blots were hybridized to the γi probe which detects the γ-globin 1.7-kb (γ −2.12,−0.40) and 0.87-kb (γ −0.4,E2) fragments (see Fig. 3A). The γ −2.12,−0.40 fragment was consistently depleted in the HDNA fraction, whereas the γ −0.4,E2 fragment was 2.1 ± 0.15 times enriched in the HDNA digest lane compared to the TDNA and PDNA digest lanes (Fig. 2B to 5B and 7B) and was the most histone associated of the fragments analyzed. For each fragment analyzed, the relative intensity of hybridization signal in the HDNA digest lane compared to that in PDNA digest lane was calculated and this ratio was then divided by the equivalent ratio for the γ −0.4,E2 control fragment on the same gel. The relative levels of enrichment compared to the γ −0.4,E2 control fragment were expressed on a scale from 0 to 1 (Fig. 2C to 5C and 7C).
FIG. 3.
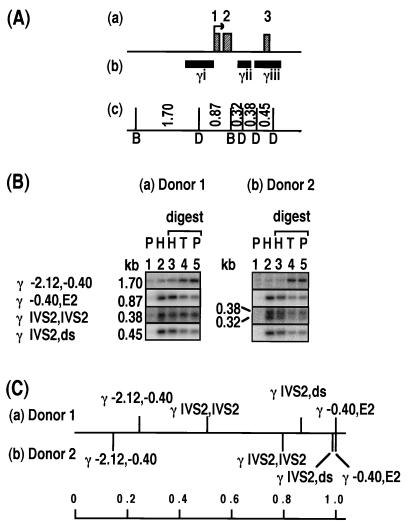
Relative levels of histone association of sites within the γ-globin gene. The DNA probes (γi to γiii), derived from the Gγ-globin gene, detect similar-sized fragments in both the Gγ- and Aγ-globin genes. The term ds denotes a site downstream of the gene. For donor 1 in panel C, the number of samples (in parentheses) and the values or the range of values are as follows: γ −2.12,−0.40, (5) 0.14 to 0.32; γ −0.40,E2, (5) 1.0; γ IVS2,IVS2, (3) 0.38 to 0.75; and γ IVS2,ds, (2) 0.86 and 0.87. See the legend to Fig. 2.
ɛ globin.
For the gene coding for ɛ globin, three BamHI/DraI fragments were assayed by using probes ɛi to ɛiii (Fig. 2A). The 0.65-kb fragment released from the body of the gene (ɛ E1,E2) was highly histone associated (Fig. 2B). The ratio of signal in HDNA compared to the PDNA digest tracks was similar to those observed for the γ −0.4,E2 positive-control fragment (Fig. 2C). The 0.63-kb fragment surrounding the promoter region of the gene (ɛ −0.61,E1) showed either an even distribution between the PDNA and HDNA fractions or a slight enrichment in the HDNA fraction (Fig. 2B). The level of enrichment was on average only about 0.6 times that observed for the ɛ E1,E2 fragment and the γ −0.4,E2 positive-control fragment (Fig. 2C). The 0.52-kb fragment derived from sites in the second exon and second intron of the ɛ-globin gene (ɛ E2,IVS2) was depleted in the HDNA fraction compared to the total DNA and PDNA fractions (Fig. 2B). Therefore, the order of relative association with histones is ɛ E1,E2 > ɛ −0.61,E1 > ɛE2,IVS2 for both donors (Fig. 2C). These results suggest that only the BamHI sites in E1 and E2 (Fig. 2A) are strongly histone associated.
γ globin.
For the gene coding for γ globin, five DNA fragments were examined by using probes γi to γiii (Fig. 3A); the smallest fragment, 0.32 kb, was at the size limit of detection and was not visible on all Southern blots. The 1.7-kb fragment, located entirely upstream of the gene (γ −2.12,−0.40), was very depleted in the HDNA fraction (Fig. 3B). The 0.87-kb fragment surrounding the promoter region (γ −0.40,E2), in contrast, was highly enriched in the HDNA fraction (Fig. 3B). This is the fragment used as a positive control on all Southern blots, as discussed above. The 0.45-kb fragment derived from a site in the second intron and another just downstream of the γ-globin gene (γ IVS2,ds) was also highly histone associated (Fig. 3B). Results for the 0.38- and 0.32-kb fragments in the second intron (γ IVS2,IVS2) were variable. For donor 1, in three of four sperm samples, these fragments showed no enrichment in the HDNA fraction (Fig. 3B). In the other preparation from donor 1, however, the fragments were enriched in the HDNA fraction to a level 75 to 80% that of the γ −0.4,E2 positive-control fragment (not shown). In two samples from donor 2, the 0.38- and 0.32-kb fragments were also enriched to a level 75 to 80% that of the γ −0.4,E2 positive-control fragment (Fig. 3B and C). Despite the variability in the intron fragments, the human γ-globin gene displays histone association in the order γ −0.40,E2 and γ IVS2,ds > γ IVS2,IVS2 > γ −2.12,−0.40 in both donors (Fig. 3C). The results suggest first that the site at −2.12-kb (Fig. 3A) was protamine associated in both donors. Second, in donor 1, the sites at −0.40, E2, IVS2 (3′ half), and ds (Fig. 3A) were all histone associated, whereas the extent of histone association of the DraI site in the first half of IVS2 varied between sperm samples. Third, in donor 2, all five sites in the vicinity of the gene from −0.40 to ds were consistently histone associated, suggesting that the whole gene is localized within a histone-associated region in sperm.
δ globin.
For the δ-globin gene, the 0.94-kb fragment obtained by cleavage of a site at −0.46 kb upstream and a site in the second exon (δ −0.46,E2) (Fig. 4A) was not enriched in the HDNA fraction (Fig. 4B and C). This result suggests that at least one of these two restriction enzyme sites was protamine associated.
FIG. 4.
Relative levels of histone association of sites with the δ-globin gene. The DNA probe is named δi. For donor 1 in panel C, the value displayed for δ −0.46,E2 is the average of two samples with values of 0.37 and 0.62, respectively. See the legend to Fig. 2.
β globin.
For the β-globin gene, four fragments were examined by using probes βi to βiii, a 0.43-kb fragment spanning from −1.46 to −0.97 kb upstream (β −1.46,−0.97), a 1.45-kb fragment from −0.97 kb upstream to the second exon (β −0.97,E2), a 0.52-kb fragment from the second exon to the second intron (β E2,IVS2), and a 0.62-kb fragment from the second intron to just downstream of the gene (β IVS2,ds) (Fig. 5A). None of the fragments was enriched in the HDNA fraction (Fig. 5B and C). Of the two fragments which border on the same DraI site in the second intron, the 0.52-kb fragment was consistently more depleted than the 0.62-kb fragment in the HDNA fraction (Fig. 5B and C). This result suggests that the BamHI site in the second exon displays stronger protamine association than the downstream DraI site. It can be inferred from these results that at least one of the two sites bordering each fragment was protamine associated.
In summary, the ɛ- and γ-globin genes, which are expressed in the embryo, are associated with histones in human sperm. In contrast, at the sites examined, no histone-associated regions were detected in the human δ- and β-globin genes. The pattern of histone and protamine association was constant in different samples from the same person and between the two donors, with the exception of one site in the intron of the γ-globin gene.
Comparison between β-globin family members.
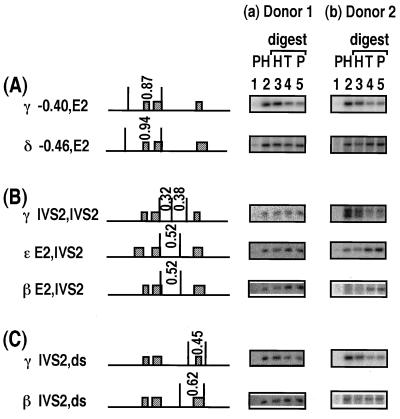
Further comparisons between the members of the β-globin family were made for sites which were located in very similar regions in the different genes (Fig. 6). First, the 0.87-kb (γ −0.40,E2) fragment surrounding the 5′ end of the γ-globin gene was histone associated, whereas the corresponding 0.94-kb (δ −0.46,E2) fragment in the δ-globin gene was not enriched in the HDNA fraction (Fig. 6A). Second, in all sperm preparations from donor 2 and in one from donor 1 (not shown), the 0.32- and 0.38-kb fragments in IVS2 of γ globin (γ IVS2,IVS2) were histone associated, whereas the similarly located 0.52-kb (ɛ E2,IVS2 and β E2,IVS2) fragments in the ɛ- and β-globin genes were depleted in the HDNA fraction (Fig. 6B); in the other three sperm preparations from donor 1, the γ-globin intron fragments were distributed equally in the HDNA and PDNA fractions and the β-globin E2,IVS2 fragment was depleted in the HDNA fraction (Fig. 6B). Third, the 0.45-kb γ-globin (IVS2,ds) fragment encompassing the third exon of the γ-globin gene was histone associated, whereas the corresponding 0.62-kb (β IVS2,ds) fragment in the β-globin gene was not enriched in the HDNA fraction (Fig. 6C). Thus, where direct comparisons can be made, sites in the γ-globin gene are generally more histone associated than those in the β- and δ-globin genes.
FIG. 6.
Comparison between different members of β-globin family. For each set of data, the same Southern blot was probed with multiple DNA probes. See the legend to Fig. 2.
Protamine 2.
The gene coding for protamine 2 is expressed during spermatogenesis prior to the general shutdown of transcription but is silent in the embryo. In donor 1, three fragments were assayed using probes Pi and Pii (Fig. 7A). Due to a heterozygous BamHI site (Fig. 7A), two overlapping fragments 0.80 and 0.88 kb long (protamine 2 E1,ds) are obtained from sites in exon 1 and just downstream of the gene. A 0.89-kb DraI fragment (protamine 2 ds,ds) is located completely downstream of the gene. In donor 2, the BamHI site is homozygous, so only the 0.80-kb (protamine 2 E1,ds) fragment is present. For the 0.80 (0.88)-kb (protamine 2 E1,ds) fragment(s), donor 1 consistently showed no enrichment in the HDNA fraction, whereas donor 2 always showed a significant enrichment, over 80% of the level of enrichment of the γ −0.4,E2 positive-control fragment (Fig. 7B and C). Therefore, for donor 1, one or both of the sites is protamine associated, whereas for donor 2, both sites are histone associated. The 0.89-kb fragment located downstream of the gene, in contrast, was clearly not enriched in the HDNA fraction of either donor (Fig. 7B and C), suggesting that the DraI site furthest from the gene is consistently protamine associated. We suggest that active transcription in spermatids may tend to cause retention of histones in a transcribed region. If this is the case for the protamine 2 gene, then it is probable that the E1 site is histone associated in both donors and that the variable levels of enrichment of the E1,ds fragment are due to the downstream site. Despite its association with histones in sperm, the protamine 2 gene is not expressed in the embryo, as it requires testis-specific transcription factors for its activation (16). The observed variation in the E1 and/or 0.36-kb downstream sites would have no effect on development.
DISCUSSION
The observation that sperm chromatin contains a distinct set of histones with features similar to histones found in active chromatin has led to the hypothesis that the association of these histones with specific gene sequences may be related to the epigenetic control of differential gene expression during embryonic development (12, 13). An alternative hypothesis is that all DNA sequences associated with gene transcription are histone associated in sperm, since the histone-associated portion of the sperm genome is sufficiently large to include all genes. In this study, we examined the histone-DNA association for defined gene sequences in human sperm and discovered that only some gene regions are histone associated in sperm. This finding is consistent with the first hypothesis and contrary to the second hypothesis. Of the 15 DNA fragments analyzed, 14 showed a consistent trend of either histone or protamine association in multiple samples from the same individual and 13 showed similar trends between two donors. There is, therefore, a high degree of conservation in the chromatin structures in human sperm. We also found that sperm DNA can vary from histone to protamine association within a distance as small as 400 bp (Fig. 3), suggesting that the physical configuration of the chromatin within the gene is subject to very fine control.
It is likely that critical chromatin structures are set up in the male germ cells prior to the meiotic divisions, since viable offspring can be produced from eggs fertilized with stages as early as secondary spermatocytes in mice and spermatids in humans (1). Premeiotic germ cells may contain gene regions which are consistently associated with either hyperacetylated or hypoacetylated histones. The regions associated with hyperacetylated histones may retain histones in the mature sperm and remain hyperacetylated in the male pronuclei. Because histone acetylation patterns can be inherited through cell division (29), the hyperacetylated histone structures in male pronuclear DNA could be passed to daughter cells as the embryo develops so that these hyperacetylated regions have the potential for activation. In contrast, the DNA associated with hypoacetylated histones in premeiotic germ cells may complex with protamines in sperm and become reassociated with hypoacetylated histones in the male pronuclei, thus maintaining the gene in a transcriptionally inactive state in the embryo.
Our study has focussed primarily on the human β-globin locus. In the ɛ-globin gene which is expressed in the embryonic yolk sac, two sites in exons 1 and 2, respectively, are consistently histone associated. A site in IVS2 was consistently protamine associated. Another site upstream of the gene at position −0.61 kb, which is located between the ɛ-PREIV and ɛ-PREII elements identified as being important in ɛ-globin expression (27), consistently shows no predilection for histone or protamine association, suggesting that individual sperm cells differ in their chromatin conformation at this site. In the γ-globin gene, which is expressed in the embryonic yolk sac and in the fetus, four sites located at −0.40 kb upstream, E2, IVS2, and 0.12 kb downstream of the gene, respectively, are consistently histone associated, suggesting that a large proportion of the gene retains its histones in sperm. Another site in IVS2, however, varies in its level of histone association, both between samples and between individuals. The results for the ɛ- and γ-globin genes suggest that there are critical regions which are consistently histone associated and thus are important for gene regulation. Other flanking regions which display variations in histone association are presumably of less importance. We consider it likely that the chromatin structure of a particular site will influence development only if the structure is the same in all fertile sperm. Other regions of the gene may acquire an active or inactive chromatin structure later under the influence of the critical regions.
In the δ- and β-globin genes, which are not expressed in the embryo, no histone-associated sites were detected. The two sites examined in the δ-globin gene were at positions −0.46 and E2 which are at similar locations in relation to the histone-associated sites at position −0.41 and E2 of the γ-globin gene. This finding is consistent with the delayed commencement of δ-globin transcription compared to the time of γ-globin transcription. In the case of the β-globin gene, the site at −0.97 kb was too far upstream to allow conclusions about the chromatin structure of the promoter region. Several sites, however, were examined in the body of the gene. Sequences in intron 2 and exon 3 are known to be very important in β-globin regulation (6, 8, 15). A β-globin gene fragment from IVS2 to 13 bp downstream is depleted in the HDNA fraction, whereas a γ-globin gene fragment in a similar position, from IVS2 to 120 bp downstream, is consistently enriched in the HDNA fraction. This result is consistent with the difference in temporal expression of the β- and γ-globin genes.
Switching of β-globin family expression involves mechanisms which (i) enable ɛ and γ globin to be expressed in the embryonic yolk sac but prevent β- and δ-globin expression at this early stage, (ii) silence ɛ-globin expression and initiate low-level β-globin expression in the fetal liver, and (iii) down-regulate γ-globin expression postnatally as β- and δ-globin expression increases. A published study of globin switching focussed on the latter two phenomena (5). Our results suggest that sperm chromatin structure might contribute to the first phenomenon by causing the formation of a chromatin structure permissive for ɛ- and γ-globin transcription but not for β- and δ-globin transcription during early development.
Consistent with this proposal, studies of the behavior of human fetal chromosomes in cell hybrids, formed by the fusion of human fetal liver cells and Friend virus-transformed mouse erythroleukemia (MEL) cells, suggest that the γ- and β-globin genes in the fetus differ by epigenetic modifications which make the γ-globin gene permissive and the β-globin gene relatively nonpermissive for transcription (10, 22). To date, similar studies have not been performed on the embryo. When human fetal erythroblasts were examined in terms of DNase I sensitivity, the Gγ- and Aγ-globin genes were very sensitive, the β-globin gene was moderately sensitive, and the δ-globin gene was insensitive (2). The difference in expression of the genes, however, is not linked to the presence of hypersensitive sites which are found both in the promoter regions of the Gγ- and Aγ-globin genes and in the δ- and β-globin genes at the fetal stage (15).
The mechanism which prevents histones from being displaced from certain regions of the ɛ- and γ-globin genes during spermatogenesis is still a matter for conjecture. As all the genes within the β-globin family have a similar G+C content, we can rule out overall base composition as an influence. There may, however, be certain DNA motifs which preferentially retain histones. The LCR with its potential chromatin-opening activity (9) is located closer to the ɛ- and γ-globin genes than the δ- and γ-globin genes, so the tendency to retain histones may increase with the proximity to the LCR. Hypomethylation and/or active transcription of a DNA sequence in spermatids as the chromatin condensation begins may lead to preferential retention of histones. The methylation and transcriptional statuses of the β-globin locus during human spermatogenesis are not known.
The protamine 2 gene was studied as an example of a gene which is transcribed in spermatids and is presumed to be in an open chromatin conformation when histones begin to be displaced. In another study where a transgene encompassing the human protamine 1, protamine 2, and transition protein 2 genes was transferred into the mouse germ line, the 28.5-kb segment containing the gene cluster was observed to reside in a DNase I-sensitive domain in sperm leading to the conclusion that this entire 28.5-kb segment was histone associated in sperm (7). In contrast, we have studied the endogenous human protamine 2 gene and have observed that in one of the donors, histone-associated sites are present in E1 and just downstream of the gene, while a site further downstream is protamine associated. In the other donor, either the site in E1 or the site just downstream of the gene is also protamine associated. These results cannot be reconciled with the gene being part of a totally open domain in human sperm.
If sperm chromatin has an influence on development, then the maternal chromatin must achieve a chromatin structure very similar to that of the paternal chromatin in the cleavage stage embryo. We suggest that either the paternal genome influences the maternal genome by allelic cross-talk (14) or the oocyte sets up patterns in the chromatin, which are functionally equivalent to those in the sperm, by means such as differential acetylation of histones. In gene regions where the paternal genome has inherited a different chromatin structure than that of the maternal genome, one can envisage that genomic imprinting could readily occur.
What types of DNA sequences tend to retain histones in sperm? We have demonstrated that some, but not all, gene sequences in human sperm are histone associated. Our study of the β-globin gene family supports the theory that genes expressed in early development tend to be histone associated. We suggest that an additional candidate for histone retention is the class of DNA sequences called CpG islands, as these regions are unmethylated in sperm and are associated with hyperacetylated histones in somatic cells (26). Centric heterochromatin is another candidate, since in mice, these regions are hypomethylated in sperm compared to the adult (11) and they are found hyperacetylated in embryonic stem cells, with deacetylation occurring only after differentiation (18). Further studies of sperm chromatin structure of genes, including imprinted genes and centric heterochromatin, should increase our understanding of the transmission of information from the gamete to the embryo.
ACKNOWLEDGMENTS
We are grateful to David Tremethick for helpful discussions and to Peter Rowe, Merlin Crossley, David Tremethick, and Emma Whitelaw for comments on the manuscript.
This work was supported by project grant 940497 from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Aitken R J, Irvine D S. Fertilization without sperm. Nature. 1996;379:493–495. doi: 10.1038/379493a0. [DOI] [PubMed] [Google Scholar]
- 2.Arapinis C, Elion J, Labie D, Krishnamoorthy R. Differences in DNase I sensitivity and methylation within the human β-globin gene domain and correlation with expression. Eur J Biochem. 1986;156:123–129. doi: 10.1111/j.1432-1033.1986.tb09556.x. [DOI] [PubMed] [Google Scholar]
- 3.Balhorn R, Gledhill B L, Wyrobek A J. Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry. 1977;16:4074–4080. doi: 10.1021/bi00637a021. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee S, Smallwood A, Hultén M. ATP-dependent reorganization of human sperm nuclear chromatin. J Cell Sci. 1995;108:755–765. doi: 10.1242/jcs.108.2.755. [DOI] [PubMed] [Google Scholar]
- 5.Baron M H. Transcriptional control of globin gene switching during vertebrate development. Biochim Biophys Acta. 1997;1351:51–72. doi: 10.1016/s0167-4781(96)00195-9. [DOI] [PubMed] [Google Scholar]
- 6.Behringer R R, Hammer R E, Brinster R L, Palmiter R D, Townes T M. Two 3′ sequences direct adult erythroid-specific expression of human β-globin genes in transgenic mice. Proc Natl Acad Sci USA. 1987;84:7056–7060. doi: 10.1073/pnas.84.20.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhary S K, Wykes S M, Kramer J A, Mohamed A N, Koppitch F, Nelson J E, Krawetz S A. A haploid expressed gene cluster exists as a single chromatin domain in human sperm. J Biol Chem. 1995;270:8755–8762. doi: 10.1074/jbc.270.15.8755. [DOI] [PubMed] [Google Scholar]
- 8.Collis P, Antoniou M, Grosveld F. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J, Tan-Un K C, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 10.Enver T, Brice M, Karlinsey J, Stamatoyannopoulos G, Papayannopoulou T. Developmental regulation of fetal to adult globin gene switching in human fetal erythroid × mouse erythroleukemia cell hybrids. Dev Biol. 1991;148:129–137. doi: 10.1016/0012-1606(91)90323-u. [DOI] [PubMed] [Google Scholar]
- 11.Feinstein S I, Racaniello V R, Ehrlich M, Gehrke C W, Miller D A, Miller O J. Pattern of undermethylation of the major satellite DNA of mouse sperm. Nucleic Acids Res. 1985;13:3969–3978. doi: 10.1093/nar/13.11.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatewood J M, Cook G R, Balhorn R, Bradbury E M, Schmid C W. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 13.Gatewood J M, Cook G R, Balhorn R, Schmid C W, Bradbury E M. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–20666. [PubMed] [Google Scholar]
- 13a.Gatewood, J. M. Personal communication.
- 14.Goldman M A. Executive decision: chromatin structure and gene regulation. Trends Genet. 1997;13:387–388. doi: 10.1016/s0168-9525(97)01272-9. [DOI] [PubMed] [Google Scholar]
- 15.Groudine M, Kohwi-Shigematsu T, Gelinas R, Stamatoyannopoulos G, Papayannopoulou T. Human fetal to adult hemoglobin switching: changes in chromatin structure of the β-globin gene locus. Proc Natl Acad Sci USA. 1983;80:7551–7555. doi: 10.1073/pnas.80.24.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha H, van Wijnen A J, Hecht N B. Tissue-specific protein-DNA interactions of the mouse protamine 2 gene promoter. J Cell Biochem. 1997;64:94–105. [PubMed] [Google Scholar]
- 17.Hecht N B. Regulation of ’haploid expressed genes’ in male germ cells. J Reprod Fertil. 1990;88:679–693. doi: 10.1530/jrf.0.0880679. [DOI] [PubMed] [Google Scholar]
- 18.Keohane A M, O’Neill L P, Belyaev N D, Lavender J S, Turner B M. X-inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 19.Kleene K C. Patterns of translational regulation in the mammalian testis. Mol Reprod Dev. 1996;43:268–281. doi: 10.1002/(SICI)1098-2795(199602)43:2<268::AID-MRD17>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 21.Mavilio F, Giampaolo A, Carè A, Migliaccio G, Calandrini M, Russo G, Pagliardi G L, Mastroberardino G, Marinucci M, Peschle C. Molecular mechanisms of human hemoglobin switching: selective undermethylation and expression of globin genes in embryonic, fetal, and adult erythroblasts. Proc Natl Acad Sci USA. 1983;80:6907–6911. doi: 10.1073/pnas.80.22.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papayannopoulou T, Brice M, Stamatoyannopoulos G. Analysis of human hemoglobin switching in MEL × human fetal erythroid cell hybrids. Cell. 1986;46:469–476. doi: 10.1016/0092-8674(86)90667-7. [DOI] [PubMed] [Google Scholar]
- 23.Peschle C, Mavilio F, Carè A, Migliaccio G, Migliaccio A R, Salvo G, Samoggia P, Petti S, Guerriero R, Marinucci M, Lazzaro D, Russo G, Mastroberardino G. Haemoglobin switching in human embryos: synchrony of ζ to α and ɛ to γ-globin switches in primitive and definitive erythropoietic lineage. Nature. 1985;313:235–238. doi: 10.1038/313235a0. [DOI] [PubMed] [Google Scholar]
- 24.Spiker S. A modification of the acetic acid-urea system for use in microslab polyacrylamide gel electrophoresis. Anal Biochem. 1980;108:263–265. doi: 10.1016/0003-2697(80)90579-5. [DOI] [PubMed] [Google Scholar]
- 25.Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp Cell Res. 1978;117:347–356. doi: 10.1016/0014-4827(78)90148-9. [DOI] [PubMed] [Google Scholar]
- 26.Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 27.Trepicchio W L, Dyer M A, Baron M H. Developmental regulation of the human embryonic β-like globin gene is mediated by synergistic interactions among multiple tissue- and stage-specific elements. Mol Cell Biol. 1993;13:7457–7468. doi: 10.1128/mcb.13.12.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 29.Wolffe A P. Inheritance of chromatin states. Dev Genet. 1994;15:463–470. doi: 10.1002/dvg.1020150604. [DOI] [PubMed] [Google Scholar]