Abstract
Oncostatin M (OSM) is a member of a family of cytokines that includes ciliary neurotrophic factor, interleukin-6, interleukin-11, cardiotrophin-1, and leukemia inhibitory factor (LIF). The receptors for these cytokines consist of a common signaling subunit, gp130, to which other subunits are added to modify ligand specificity. We report here the isolation and characterization of a cDNA encoding a subunit of the mouse OSM receptor. In NIH 3T3 cells (which endogenously express gp130, LIF receptor β [LIFRβ], and the protein product, c12, of the cDNA described here), mouse LIF, human LIF, and human OSM signaled through receptors containing the LIFRβ and gp130 but not through the mouse OSM receptor. Mouse OSM, however, signaled only through a c12-gp130 complex; it did not use the LIF receptor. Binding studies demonstrated that mouse OSM associated directly with either the c12 protein or gp130. These data highlight the species-specific differences in receptor utilization and signal transduction between mouse and human OSM. In mouse cells, only mouse OSM is capable of activating the mouse OSM receptor; human OSM instead activates the LIF receptor. Therefore, these data suggest that all previous studies with human OSM in mouse systems did not elucidate the biology of OSM but, rather, reflected the biological actions of LIF.
Oncostatin M (OSM) is structurally and functionally related to a family of cytokines that includes the leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNTF), interleukin-11 (IL-11), cardiotrophin-1 (CT-1), and interleukin-6 (IL-6). All of the members of this family bind to, and signal through, a receptor complex that includes the glycoprotein gp130 (4, 8, 9, 12, 16). Most functional receptors for this group of cytokines consist of multimeric complexes of α chains, nonsignal-transducing receptor components required for high-affinity cytokine binding, and signal-transducing β chains. Known α components include CNTF receptor α (CNTFRα), IL-11 receptor α (IL-11Rα), and IL-6 receptor α (IL-6Rα); the known β chains are gp130 and the LIF receptor β (LIFRβ). This cytokine family can be further divided based upon specific β-chain usage. Functional receptors for human OSM (hOSM), CNTF, LIF, and CT-1 contain heteromultimers of LIFRβ and gp130 (in addition to specific α components), whereas the functional IL-6 and IL-11 receptors contain multimers of gp130 along with IL-6Rα and IL-11Rα, respectively. A subfamily of receptors is thus defined, based on the use of a LIFRβ-gp130 heteromultimer core, to which various α components are added to modify ligand binding specificity. LIF signals through the LIFRβ-gp130 complex alone, while CNTF signaling requires the addition of a specific α chain (CNTFRα) to the functional LIFR. CT-1 and hOSM can signal through the functional LIFR alone, but certain evidence suggested the existence of a second, OSM-specific, receptor in human cells (19). An hOSMRβ was recently reported (13) and demonstrated to interact functionally with gp130. We report here the cloning and characterization of a mouse OSMRβ (mOSMRβ) which we believe to be orthologous to the hOSMRβ. Characterization of this receptor revealed distinct modes of receptor activation by OSM in the mouse and human systems. Unexpectedly, mOSM and hOSM utilize different receptors on mouse cells.
MATERIALS AND METHODS
Materials.
Reagents for PCR were purchased from Boehringer Mannheim (Indianapolis, Ind.). The TA cloning kit and the pCRII vector were purchased from Invitrogen (San Diego, Calif.). The RNA transcription kit was purchased from Ambion (Austin, Tex.). Unless otherwise indicated, all other DNA modification enzymes were purchased from Boehringer Mannheim and all chemical reagents were purchased from Sigma (St. Louis, Mo.). Antiphosphotyrosine (anti-P.Tyr) antibodies were from UBI (Lake Placid, N.Y.). Anti-gp130 (M20), anti-STAT5B (C17), anti-STAT3 (C20), and anti-LIFR (C19) were from Santa Cruz Biotechnology (Santa Cruz, Calif.). Recombinant mouse and human OSM were from R & D Systems (Minneapolis, Minn.). Recombinant mouse and human LIF were from Genzyme (Cambridge, Mass.).
cDNA cloning.
The initial clone, cm5-00013-c12, was a 2,125-bp cDNA isolated from a mouse colon cDNA library. A pair of oligonucleotide primers were synthesized from the putative cytoplasmic domain of clone cm5-00013-c2, the sense oligonucleotide (5′-AGACACAGCACACCAACTTGG-3′, 1031-75) and the antisense oligonucleotide (5′-GCGCAATTAACCCTCACTAAAGCAGATCTTGTGCTGCTGGTGTTTACTG-3′, 1031-74). The antisense oligonucleotide encoded a T3 RNA polymerase recognition site before the region of complementarity to cm5-00013-c12. A 345-bp PCR product was generated with these primers from a mouse skeletal muscle library (Clontech, Palo Alto, Calif.) and cloned into the pCRII vector. A radiolabeled riboprobe was synthesized from the template thus prepared and used to screen 106 phage plaques, in duplicate, from a mouse skeletal muscle library (Clontech), as described below. A total of 120 primary positive plaques were isolated and subjected to further analysis.
To facilitate the cloning of the novel coding region sequence, anchored PCR was used. An antisense oligonucleotide (5′-GCGCAATTAACCCTCACTAAAGCAGATCTCTTCCACTGCAAATCACAGCG-3′; 1031-73), complementary to the 5′ end of the original clone, was combined with either of two vector arm-specific anchor oligonucleotides (left-arm oligonucleotide [5′-CCTTTTGAGCAAGTTCAGCCTGGTTAAGTCC-3′; 1065-30] and right-arm oligonucleotide [5′-CAGAGGTGGCTTATGAGTATTTCTTCCAGGG-3′; 1065-31]) in PCR amplification on the original 120 primary plaque pools. Twelve of the original pools allowed amplification with one, but not the other, of the two vector arm-specific oligonucleotides combined with oligonucleotide 1031-73. These PCR products were subcloned into the pCRII vector and sequenced. Eight of these contained novel sequence compared to the original clone, and all overlapped one another to some extent. The longest of these clones extended 1,708 bp 5′ of the original clone, defining a 2,949-bp open reading frame (ORF), but did not encode an in-frame stop codon in the putative 5′-untranslated region that would delineate the complete ORF.
A second round of anchored PCR on the original plaque pools was used with a new oligonucleotide (5′-GATGCCCTCAGGGACAGCAC-3′; 1091-36), complementary to a region in common between the previous extension products, combined with either of oligonucleotides 1065-30 and 1065-31 as before. Extension products were obtained from two independent plaque pools, whose sequences were identical to each other, overlapping and extending the previous sequence information by 142 bp. These clones still did not encode an in-frame stop codon, but sequencing from mouse genomic DNA revealed the presence of a stop codon 15 bp upstream of our consensus cDNA sequence and no additional methionine residues in frame with the predicted start methionine.
Library screening.
A 32P-labelled riboprobe was generated from the template prepared as described above, by transcription from the antisense T3 promoter with an in vitro transcription kit (Ambion). The 345-bp riboprobe was used to screen 106 plaques lifted from a mouse skeletal muscle cDNA library (Clontech). Plaques were lifted in duplicate onto nitrocellulose filters (Schleicher & Schuell, Keene, N.H.). Hybridization was performed in Stark’s buffer (50% formamide, 50 mM potassium phosphate [pH 6.5], 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% sodium dodecyl sulfate [SDS], 5× Denhardt’s solution, 0.05% sodium sarcosyl, 300 μg of salmon sperm DNA per ml) at 42°C overnight. The filters were washed to a final stringency of 0.1× SSC–0.5% SDS at 63°C and exposed to X-ray film (X-Omat AR; Eastman Kodak, Rochester, N.Y.) overnight at −70°C with intensifying screens. The films were developed, and 120 plaques hybridizing on both pairs of duplicate lifts were identified and isolated. Phages were eluted from agarose plugs in SM buffer (17) and stored at 4°C (primary plaque pools). Secondary and tertiary plaque purifications were performed in similar fashion to that for the primary pools on dilutions from the primary pools until single plaques could be isolated.
DNA sequence analysis.
DNA sequencing was performed on both strands of each template with the Taq Dye Terminator cycle-sequencing kit (Applied Biosystems, Foster City, Calif.) and primers appropriate to the cloning vector on an automated DNA sequencer (model 373A or 377; Applied Biosystems) as recommended by the manufacturer.
Northern analysis.
Mouse multiple-tissue Northern blots (Clontech) were probed with the 345-bp riboprobe in Stark’s buffer overnight at 42°C. The filters were washed to a final stringency of 0.1× SSC–0.1% SDS at 68°C for 90 min. They were then exposed to X-ray film for 2 days.
Antibody production.
Antipeptide antibodies were raised against the C-terminal 9 amino acids predicted by the cDNA c12. The peptide was synthesized with a cysteine on the N terminus, coupled to keyhole limpet hemocyanin with m-maleimidobenzoic acid-N-hydroxysuccinimide ester, and used as an antigen to make polyclonal rabbit antisera by standard procedures (6). These antibodies were affinity purified on a peptide column, as suggested by the manufacturer (SulfoLink kit; Pierce, Inc., Rockford, Ill.).
Production of mOSM.
The mOSM cDNA was amplified by PCR from first-strand adult mouse spleen cDNA (Clontech) with sense (5′-GGGAATTCGTATGCAGACACGGCTTCTAAGAACAC-3′) and antisense (5′-GGAGATCTCTAGGCCCTGGTCGTCGGGCTCTGGG-3′) oligonucleotides designed according to the published sequence (22). The amplified cDNA was subcloned into the expression vector pBJ5 (21). To transiently express mouse OSM, 5 μg of PBJ5-OSM plasmid was transfected as a calcium phosphate precipitate into 106 293T cells that had been plated on fibronectin-treated plates (2). At 18 h later, the medium was changed to serum-free Iscove’s modified Dulbecco’s medium. At 48 h after transfection, the medium from transfected and mock-transfected cells was harvested.
Expression of recombinant receptor.
The c12 ORF was subcloned into expression vector pBJ5. The insert of one construct was verified by sequence analysis and was used to express the full-length protein in COS7 cells. DNA was transfected into 1.5 × 106 cells by the DEAE-dextran technique (5). The next day the medium was changed, and the following day the medium was replaced by serum-free medium; 24 h later, the cells were treated and lysed. The cells were lysed in NP40 lysis buffer (50 mM Tris [pH 8.0], 150 mM sodium chloride, 1% Nonidet P-40 [NP-40], 10 mg of aprotinin per ml, 5 mM EDTA, 200 mM sodium orthovanadate). The lysates were centrifuged, and the supernatants were either boiled directly in sample buffer or used for immunoprecipitation. Immunoprecipitates were collected on protein G-Sepharose beads and boiled in sample buffer. After electrophoresis and electroblotting to polyvinylidene difluoride membranes, the immunoprecipitates were probed with various antibodies. Immune complexes were detected with horseradish peroxidase-conjugated secondary reagents by enhanced chemiluminescence, as described by the manufacturer (Amersham, Arlington Heights, Ill.).
Binding and cross-linking analysis.
Purified, carrier-free recombinant mouse (rmOSM; R&D Systems) was radiolabeled with Iodobeads (Pierce, Rockford, Ill.) as specified by the manufacturer. SDS-polyacrylamide gel electrophoresis (PAGE) analysis of the iodinated product revealed a single labeled protein of the expected size (21 kDa) (data not shown). The final specific activity was approximately 34 cpm/pg. COS7 cells were either mock transfected or transfected with c12 or gp130. Transfected cells were removed from the plates by scraping and were washed twice in RPMI containing 25 mM HEPES (pH 7.6) and 1 mg of bovine serum albumin per ml (binding buffer). Cells (∼1.5 × 106 cells/ml) were incubated with either 0.5 or 5 nM 125I-rmOSM for 2 h at room temperature in a final volume of 200 μl. Nonspecific binding was determined from parallel reaction mixtures containing a 100-fold molar excess of unlabeled rmOSM. Unbound ligand was separated from cell-bound ligand by centrifugation through a sucrose cushion, as described previously (21), and cell-associated counts were measured. The results shown in Fig. 7B correspond to the specifically bound counts per minute (cpm) after subtraction of nonspecifically bound cpm.
Cross-linking analysis was performed on similar sets of cells incubated with 5 nM 125I-rmOSM in the absence or presence of a 100-fold molar excess of unlabeled rmOSM. Cells were incubated for 2 h with labeled ligand in a volume of 1 ml, and then disuccinimidyl suberate (DSS; Pierce) was added to a final concentration of 0.2 mM and the mixture was incubated for 30 min at room temperature. Cells were washed four times with PBS containing 10 mM Tris (pH 7.5); lysed with a solution of PBS, 1 mM EDTA, and 0.5% NP-40; and incubated at 4°C for 10 min. Cellular debris was pelleted by centrifugation at 5,000 × g for 10 min at 4°C, and the supernatant was analyzed by SDS-PAGE on 4 to 20% gels (Novex). The gels were dried under vacuum, and the radioactivity was detected with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
OSM signaling in NIH 3T3 cells.
Subconfluent NIH 3T3 cells were serum starved overnight in medium containing 0.5% calf serum. These cells were treated for 15 min at 37°C with 100 ng of recombinant factors per ml or 1× conditioned medium containing mOSM. The cells were washed in PBS and lysed in NP-40. The lysates were analyzed as described above. The results were the same with commercially obtained rmOSM.
Nucleotide sequence accession number.
The cDNA sequence reported in this paper has been deposited into GenBank under accession no. AF058805.
RESULTS
cDNA cloning and characterization.
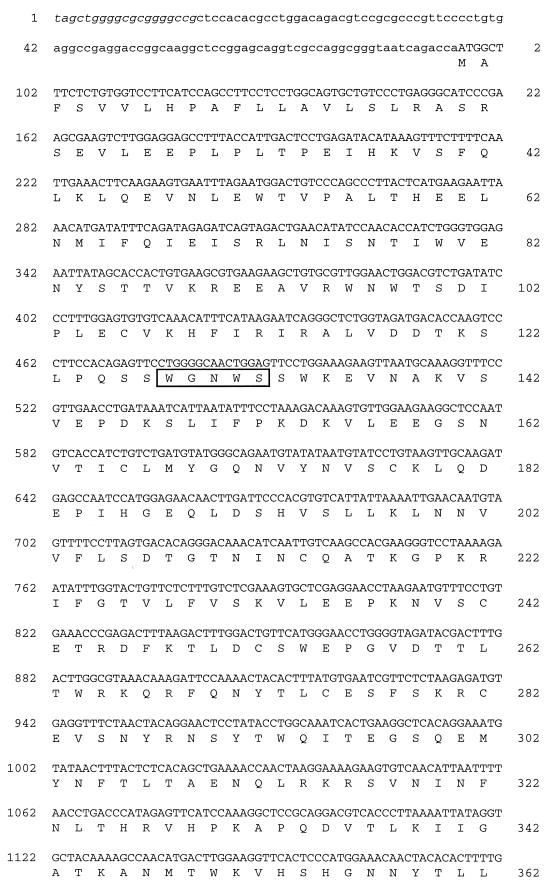
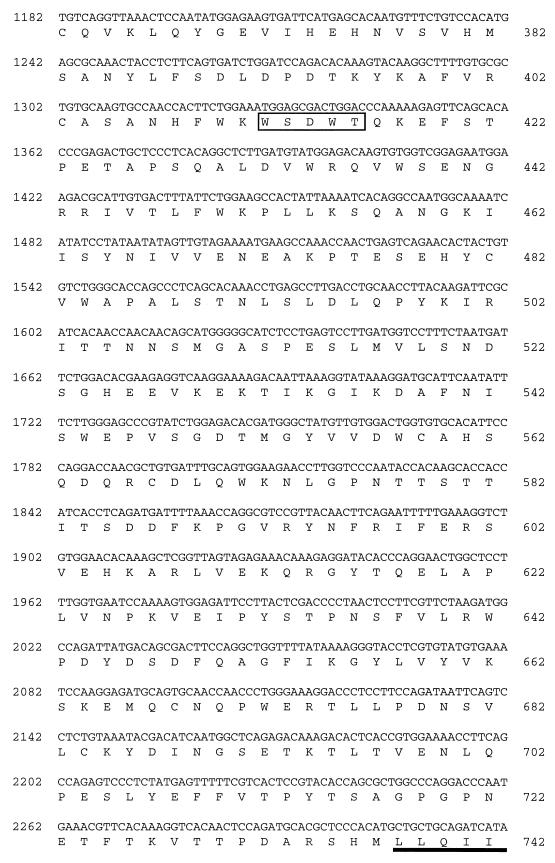
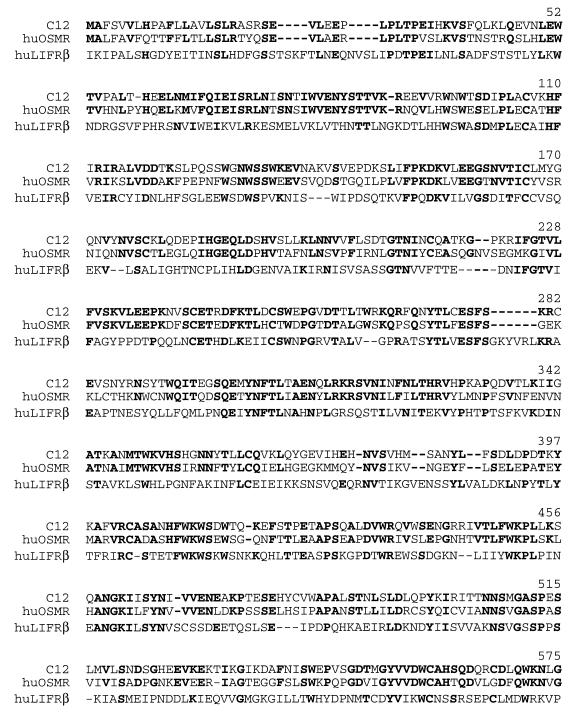
A partial cDNA (cm5-00013-c12) encoding the cytoplasmic portion of a predicted novel hematopoietin receptor was isolated by random sequence analysis from a mouse colon cDNA library. Recognition of a consensus sequence conserved in hematopoietin receptors (cytoplasmic box 1 [15]) in this clone led to further characterization. Northern blot analysis was used to assess the tissue distribution of mRNA expression. Multiple-tissue Northern blots of mouse poly(A)+ mRNA were hybridized with a 345-bp antisense riboprobe derived from the partial cDNA sequence described in Materials and Methods. A hybridizing band of ∼5 kb was detected in heart, brain, spleen, lung, liver, skeletal muscle, and kidney tissue but not in testis tissue (data not shown). An additional ∼6-kb band was detected only in the heart and skeletal muscle. Additional cDNAs were isolated by plaque hybridization of a mouse skeletal muscle cDNA library with the riboprobe, and several rounds of anchored PCR, with antisense oligonucleotides complementary to existing sequence paired with a vector-specific sense oligonucleotide, were performed on primary plaque pools. Discrete PCR products were amplified from nine primary plaque pools and characterized further. Sequencing of these clones allowed the construction of a long ORF containing both the initiation methionine and 3′-untranslated region but not an upstream in-frame stop codon that would delineate the complete ORF. Direct sequencing from a bacterial artificial chromosome containing the mouse genomic locus was used to identify an in-frame stop codon (TAG) 19 bp upstream of the end of the deduced cDNA sequence (114 bp upstream of the predicted translational initiation methionine). A contiguous cDNA sequence (c12) 4,891 bp long was identified from the overlapping clones and was verified by PCR amplification of the entire predicted ORF from mouse skeletal muscle first-strand cDNA (data not shown). A single ORF of 3,030 bp was identified, which encoded a putative protein of 971 amino acid (aa) residues (Fig. 1). The protein was predicted to contain an extracellular domain of 737 aa, a membrane-spanning domain of 20 aa, and a 214-aa cytoplasmic domain. The protein is most closely related to the hOSMRβ (13) followed by the hLIFRβ (3) (Fig. 2). Although only 55% identical to the hOSMRβ, the c12 protein exhibited many features conserved in members of the class I cytokine receptor family (reviewed in reference 7).
FIG. 1.
Nucleotide and predicted protein sequences of c12. The nucleotide sequence of mouse c12 cDNA is shown. Sequence information derived from the genomic DNA, which includes the 5′ stop codon upstream of the deduced cDNA sequence, is shown in italics. The nucleotide numbers of the cDNA are shown in the left margin, and the amino acid numbers, beginning with the predicted initiation methionine, are shown in the right margin. The WSXWS motifs are boxed, the transmembrane domain is underlined, and the cytoplasmic box 1 motif is highlighted in boldface type.
FIG. 2.
Amino acid sequence alignment of hLIFRβ, hOSMRβ, and mouse c12 proteins. The alignment of the three translated coding regions is a composite of pairwise comparisons made with the GAP tool from the Genetics Computer Group, Inc., package. The alignments are numbered from the predicted translational initiation methionine of c12. The translational initiation methionines of c12 and the hOSMRβ were coincident, and the coding region of the hLIFRβ upstream of the aligned sequences is not shown. Amino acid residues found to be identical between c12 and either one or both of the other proteins are in boldface. The single-letter amino acid code is used throughout.
The extracellular domain consists of a complete juxtamembrane hematopoietin domain, as well as a partial distal hematopoietin domain, similar to the hOSMRβ, but the WSXWS motifs are variant. The first motif (WGNWS; aa 128 to 132), contained within the membrane-distal partial hematopoietin domain, is more similar to the WGPWS motif found in the first hematopoietin domain of the related receptor encoded by c-mpl (20); while the second motif (WSDWT; aa 412 to 416), found in the complete membrane-proximal hematopoietin domain, is unlike any other example in this family of receptors. Neither of these completely conforms to the consensus sequence (WSXWS), but analysis of multiple independent cDNA clones confirmed the nucleotide sequence in these regions. The hematopoietin domains are followed by additional fibronectin III repeats in the extracellular region and then by transmembrane (aa 738 to 757) and cytoplasmic (aa 758 to 971) domains. The cytoplasmic domain encodes a box 1 motif (aa 767 to 774), a general box 2 motif (14), and two STAT3 binding motifs (aa 911 to 914 and 939 to 942) (18).
The c12 cDNA encodes a 180-kDa protein when expressed in COS cells.
To characterize the predicted protein product of c12, we expressed the cDNA transiently in COS7 cells. Lysates from c12-transfected and mock-transfected cells were analyzed by immunoblotting with an affinity-purified antiserum raised against a synthetic nonapeptide corresponding to the carboxy terminus of the predicted c12 protein (Fig. 3A). The protein expressed from c12 was heterogeneous in size and migrated at approximately 180 kDa. This heterogeneity was probably due to the differences in glycosylation that resulted as an artifact of the expression system and has been seen previously with other recombinant proteins. The native protein expressed by mouse cell lines was not as heterogeneous in size (see below).
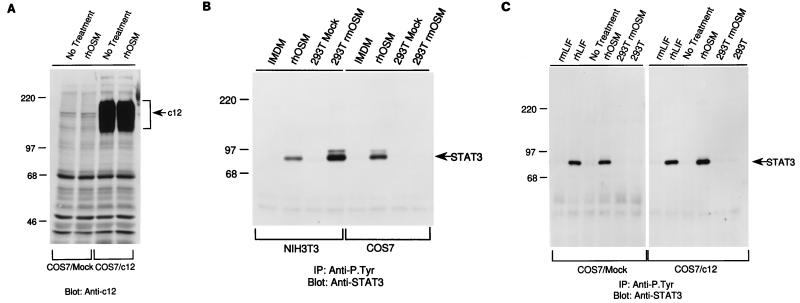
FIG. 3.
c12 cDNA directs expression of a 180-kDa protein in COS7 cells. (A) COS7 cells were transiently transfected, treated with hOSM, lysed, and analyzed for c12 protein expression. (B) rmOSM was expressed in 293T cells, and conditioned medium was collected. This medium, as well as medium from mock-transfected cells, control medium (Iscove modified Dulbecco medium), and rhOSM, was used to treat NIH 3T3 and COS7 cells, and signaling was monitored by the association of the STAT3 protein with the phosphotyrosine fraction after anti-P.Tyr immunoprecipitation. (C) COS7 cells expressing c12 or mock-transfected COS7 cells were treated with mLIF, hLIF, mOSM, and hOSM or control medium and were assayed for STAT3 signal transduction, as described for panel B.
Having expressed the c12 receptor, we were interested in determining the ligand for this protein. Based on the cDNA sequence homologies, it seemed possible that c12 was a receptor for mOSM. Since rmOSM was not commercially available at the time, we cloned and expressed rmOSM based on the published cDNA sequence (22). To verify that active rmOSM was being produced, the level of STAT3 protein in anti-P.Tyr immunoprecipitations was measured in mouse and primate cell lines that were treated with mock-transfected conditioned medium, rmOSM-containing conditioned medium, or rhOSM. Treatment of NIH 3T3 cells with the rmOSM-containing medium resulted in signaling through STAT3, as evidenced by increased association of STAT3 with the anti-P.Tyr fraction (Fig. 3B), confirming that the rmOSM was active. Conditioned medium from mock-transfected COS cells was negative, and rhOSM signaled in both cell types. We tested OSM and LIF from both species for signaling in COS7 cells expressing c12 (Fig. 3C). There was no change in signaling in as measured by STAT3 levels in anti-P.Tyr immunoprecipitations. Furthermore, recombinant c12 did not become phosphorylated on tyrosine in COS7 cells after any of these treatments (data not shown). These results show that, although primate LIFRβ and gp130 are present, mOSM does not signal in COS7 cells, regardless of the presence of murine c12 protein.
mOSM can bind to either c12 or gp130.
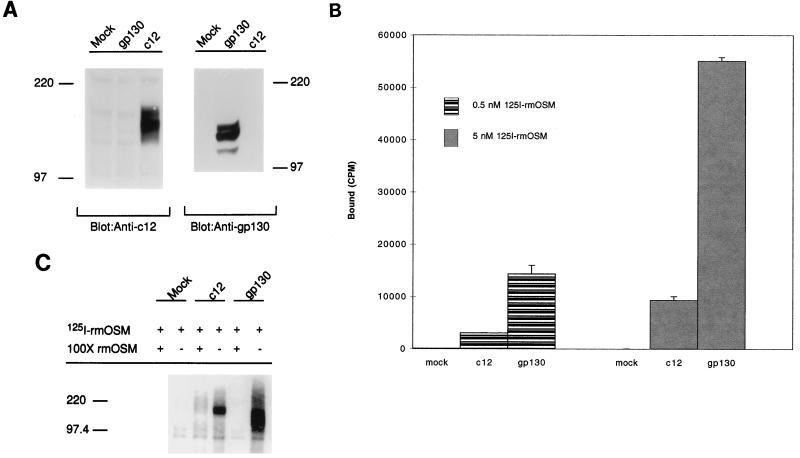
The previous analyses indicated that rmOSM signals in NIH 3T3 cells, but it was unclear with which receptor(s) it was interacting. Binding analyses were performed to determine whether iodinated rmOSM associated with either the mouse c12 protein or mouse gp130 (mgp130). COS cells were either mock transfected or transfected with the c12 cDNA or the mgp130 (Fig. 4). Parallel transfections were analyzed for protein expression levels by Western blotting with antibodies to either mgp130 or c12. Transfection with either mgp130 or c12 resulted in the expression of a protein of the anticipated size (Fig. 4A). Mock-transfected cells were unable to bind rmOSM (Fig. 4B), despite the presence of endogenous primate gp130 (see above), a result consistent with the lack of rmOSM-directed STAT3 signaling in COS7 cells. COS7 cells expressing either c12 or mgp130 independently bound rmOSM (Fig. 4B), while similar binding experiments performed with rhOSM indicated that binding to COS7 cells expressing c12 was not different from binding to mock-transfected COS7 cells (data not shown).
FIG. 4.
mOSM binds directly to either c12 or gp130. (A) Characterization of recombinant receptors produced in COS7 cells. COS7 cells were either mock transfected or transfected with gp130, c12, or a combination of c12 and gp130. Recombinant proteins were detected by Western blot analysis with antisera specific for either c12 (left) or gp130 (right). Molecular mass standards (in kilodaltons) are shown on the left and right. (B) COS cells transfected with either c12 or gp130 bind 125I-rmOSM. Parallel plates of transfected COS cells, as described in panel A, were tested for their ability to bind 125I-rmOSM. Shown are specifically bound cpm of 125I-rmOSM when tested at either 0.5 or 5 nM. Nonspecific binding represented 10 to 27% of total binding, depending on which receptor subunit was transfected and the concentration of the radiolabeled ligand. (C) 125I-rmOSM can be directly cross-linked to either c12 or gp130. Parallel plates of transfected COS cells, as described in panel A, were incubated with 125I-rmOSM in the presence (+) or absence (−) of a 100-fold molar excess of unlabeled rmOSM. The bound ligand was cross-linked to the receptors with DSS, the proteins were separated by SDS-PAGE, and the labeled complexes were visualized with a PhosphorImager. Molecular mass standards (in kilodaltons) are shown on the left.
The binding analyses indicated that COS7 cells expressing either c12 or mgp130 were able to bind rmOSM, but it was still formally possible that one or more endogenous COS7 cell proteins contributed to the binding. Therefore, cross-linking experiments were performed to determine if mOSM could interact directly with either c12 or mgp130. After treatment with 125I-rmOSM, a 210-kDa complex was detected in COS7 cells transfected with the c12 cDNA but not in the untransfected cells or when the c12-transfected cells were incubated with a 100-fold excess of unlabeled rmOSM prior to cross-linking (Fig. 4C). Analysis of COS7 cells transfected with only the mgp130 cDNA, and incubated with 125I-rmOSM revealed a different, 190-kDa complex specific for rmOSM. Therefore, the protein product of each mouse cDNA was individually able to bind rmOSM directly.
mOSM and hOSM use different receptors in mouse cell lines.
To study the receptor subunit composition of the mOSMR and the signaling by mOSM and hOSM in mouse cells, we chose a mouse cell line, NIH 3T3, that endogenously expresses gp130, LIFRβ, and c12. Immunoprecipitation with antibodies specific to the three receptor subunits demonstrated that all three were present and distinguishable by size (Fig. 5A). NIH 3T3 cells were treated with rmOSM-containing cell supernatants, rhOSM, rmLIF, or hLIF, and individual lysates were immunoprecipitated with anti-P.Tyr antibodies. Immunoprecipitates were individually immunoblotted with antibodies to STAT3, LIFRβ, or c12 (Fig. 5B). All four cytokine treatments resulted in the association of STAT3 with the phosphotyrosine fraction in NIH 3T3 cells, suggesting the activation of competent receptor complexes. As judged by receptor subunit phosphorylation, the c12 protein was used by rmOSM but not by rhOSM or LIF. The gp130 receptor was phosphorylated in response to all four cytokines. The LIFRβ subunit, however, was phosphorylated in response to LIF or rhOSM but not to rmOSM. Similar results were obtained with the TM-3 mouse testicular cell line (data not shown).
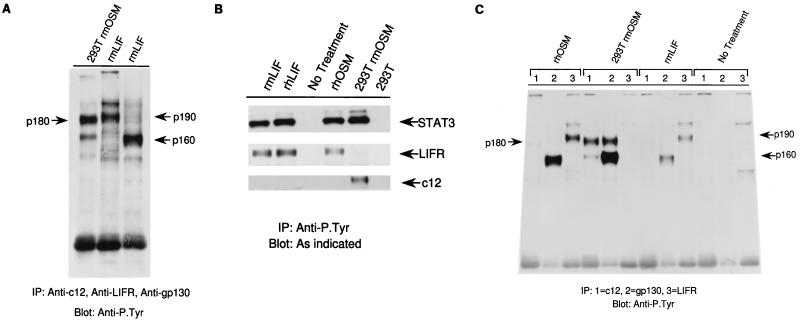
FIG. 5.
hOSM and mOSM use different receptors in mouse cells. (A) NIH 3T3 cells were treated with cytokines to allow the detection of receptors present with anti-P.Tyr antibody. Immunoprecipitation (IP) of rmOSM-treated cells with antibody to c12 or of rmLIF-treated cells with antibodies to LIFRβ or gp130 revealed that all three were present as p180, p190, and p160 proteins, respectively. (B) NIH 3T3 cells were treated, lysed, and immunoprecipitated with anti-P.Tyr antibodies. The immunoprecipitates were analyzed by immunoblotting for the presence of STAT3, LIFR, or c12. (C) Phosphorylation and coimmunoprecipitation of all three receptor subunits were tested by treating NIH 3T3 cells with the indicated cytokine and immunoprecipitating each separately with antibodies to the indicated receptor subunit, followed by immunoblotting with anti-P.Tyr antibodies.
It was of interest to determine which subunits might be found as multimeric complexes after cytokine treatment. Accordingly, coimmunoprecipitation studies were performed with antibodies to c12, LIFRβ, or gp130. As before, treatment with rmOSM resulted in phosphorylation of c12 and gp130 but not LIFRβ (Fig. 5C). The c12 protein was detected in anti-gp130 immunoprecipitations, and gp130 was detected in anti-c12 immunoprecipitations, indicating that the two receptor subunits were associated after ligand binding. Treatment with rhOSM and rmLIF resulted in tyrosine phosphorylation of LIFRβ and gp130, but they did not coimmunoprecipitate with antibodies directed against either receptor alone (Fig. 5C). The c12 protein was not phosphorylated under these conditions. The signaling by mOSM and hOSM or LIF on mouse (NIH 3T3) and primate (COS7) cells is summarized in Table 1. In the mouse cells, mOSM treatment resulted in the phosphorylation of c12 and gp130 but not LIFRβ. hOSM signaled through a receptor different from that for mOSM in mouse cells; the former used a receptor that included LIFRβ and gp130, while the latter used a receptor that included c12 and gp130. In the primate cells, hOSM and LIF both used LIFRβ whereas mOSM did not signal. We repeated many of the experiments for hOSM and mOSM in human cell lines (293 cells), and the results were the same as in the monkey cells (data not shown).
TABLE 1.
Activation of receptor subunits and signaling molecules by OSM and LIF in primate and mouse cells
| Cytokine | Phosphorylation in cell typea:
|
|
|---|---|---|
| COS7 cells | NIH 3T3 cells | |
| hOSM | STAT3(+) | STAT3(+) |
| gp130(+) | gp130(+) | |
| LIFRβ(+) | LIFRβ(+) | |
| c12(−) | ||
| mOSM | STAT3(−) | STAT3(+) |
| gp130(−) | gp130(+) | |
| LIFRβ(−) | LIFRβ(−) | |
| c12(+) | ||
| hLIF | STAT3(+) | STAT3(+) |
| gp130(+) | gp130(+) | |
| LIFRβ(+) | LIFRβ(+) | |
| c12(−) | ||
| mLIF | STAT3(−) | STAT3(+) |
| gp130(−) | gp130(+) | |
| LIFRβ(−) | LIFRβ(+) | |
| c12(−) | ||
+, phosphorylation on tyrosine residues in response to cytokine; −, no tyrosine phosphorylation.
DISCUSSION
It is clear that in human cells, in addition to the LIFR, OSM utilizes a specific receptor consisting of an OSMRβ-gp130 heteromultimer for signaling (13). It was assumed that in mouse cells, OSM also used the LIFRβ-gp130 heteromultimer, and indeed, this was the case with hOSM treatment. Recently, however, the mOSM cDNA was cloned (22), allowing subsequent studies that suggested that in mouse cells, mOSM does not bind (with high affinity) the same receptor as mLIF (11). These authors, however, did not identify the receptor for mOSM. We report here a cDNA clone that encodes a subunit of the mOSM receptor, which allowed the characterization of mOSM signaling in the mouse. The predicted protein encoded by c12 is most similar in primary sequence to the hOSMRβ (13). The domain structures of the two proteins are identical: both have a membrane-proximal hematopoietin domain and a partial membrane-distal hematopoietin domain. Both the WSXWS motifs in the c12 protein are unusual but comply with the general rules defined by mutational studies of the growth hormone and erythropoietin receptors (1, 10). The membrane-proximal WSXWS box is remarkable, however, in that the fifth residue is threonine rather than serine. This is the only example of this substitution in all known type I cytokine receptors, but mutagenesis of this site to a threonine in the erythropoietin receptor was functionally tolerated (10), indicating that this uncharacteristic motif can be functional. The cytoplasmic domains of c12 and OSMRβ are also completely homologous with respect to signaling motifs. Since c12 has all of the cytoplasmic characteristics required of a signaling molecule, it should be considered an mOSM receptor β chain, despite the lack of certain orthology with the hOSMRβ.
Analysis of the tissue distribution of the c12 mRNA indicated that it was broadly expressed. As determined by Northern blot analysis, every tissue tested except for testis tissue showed c12 expression. Reverse transcriptase PCR analysis of the hOSMRβ determined that it also was expressed in several human tissues and a wide variety of cell lines (13). Although further studies are required to determine the exact cellular distribution of c12 mRNA and protein expression, the present studies suggest a wide role(s) for mOSM in a variety of mouse tissues.
hOSM was shown previously to bind gp130 with low affinity; subsequent recruitment of either LIFRβ or OSMRβ formed a high-affinity signaling receptor (13). mOSM was only recently reported (22), and only one publication addressed receptor binding of mOSM (11). It was concluded that mOSM binds mgp130 with low affinity but that the addition of LIFRβ does not convert mOSM/mgp130 to a high-affinity complex. Since NIH 3T3 cells had high-affinity sites, it was speculated that there must be an OSMR on the NIH 3T3 cells and that mOSM does not use LIFRβ as a receptor subunit. The present data identified the specific molecular component responsible for the previous observations on the binding of mOSM and also revealed species-specific differences in OSM receptor-ligand interaction. Although previous reports have focused on binding affinities, more relevant data are the biological effects, in this case, signal transduction. We show that rmOSM bound directly to the c12 protein and gp130 and resulted in the tyrosine phosphorylation and association of both receptor subunits. In total, the binding, signaling, and coimmunoprecipitation data provide strong evidence that a complex of c12 and gp130, but not LIFRβ, constitutes the functional mOSMR complex.
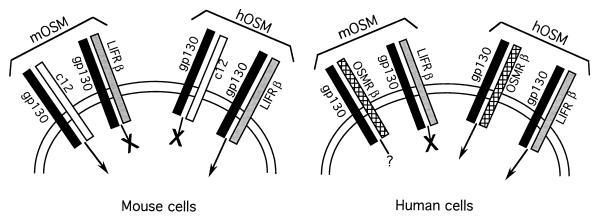
The present results, taken together with previous data, have allowed the construction of a model for OSM signalling in mice and humans (Fig. 6). The model contains two surprising elements. First, mOSM does not signal through the mLIFR or hLIFR (as hOSM does on human cells), but signals only through the specific mOSMR. As such, the classification of OSM as a member of the subfamily of cytokines which use LIFRβ and gp130 is called into question. For hOSM the association holds; for mOSM it does not. Second, hOSM, while competent to signal in mouse cells, does so through the mLIFR rather than the mOSMR. This apparent species specificity in OSM signaling mechanisms is important and implies that the biological activities of hOSM and mOSM are not equivalent. The fact that hOSM signals through a receptor distinct from that used by mOSM affects the experimental results that have been obtained in studies with hOSM or bovine OSM on mouse cell lines, as well as all in vivo transgenic and injected protein data. It seems likely that data from such experiments reflect the biology of LIF rather than that of OSM. With mOSM and the specific mOSMR now cloned, the biology of OSM in mice can be elucidated.
FIG. 6.
Model showing signaling by OSM receptors in mouse and human cells. Receptor dimers that are signaling competent in response to mOSM or hOSM are indicated by an intracellular arrow; an X indicates no detectable signaling. Signaling was determined by receptor subunit tyrosine phosphorylation and STAT3 tyrosine phosphorylation.
ACKNOWLEDGMENTS
We acknowledge the Amgen Genomics Project team for isolating the original clone cm5-000130c12, and we also thank Leif Selander and Laarni Antonio for assistance with the sequence analysis.
REFERENCES
- 1.Baumgartner J W, Wells C A, Chen C-M, Waters M J. The role of the WSXWS equivalent motif in growth hormone receptor function. J Biol Chem. 1994;269:29094–29101. [PubMed] [Google Scholar]
- 2.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gearing D P, Thut C J, Vandenbos T, Gimpel S D, Delaney P B, King J, Price V, Cosman D, Beckmann M P. Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J. 1991;10:2839–2848. doi: 10.1002/j.1460-2075.1991.tb07833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gearing D P, Comeau M R, Friend D J, Gimpel S D, Thut C J, McGourty J, Brasher K K, King J A, Gillis S, Mosley B, Ziegler S F, Cosman D. The IL-6 signal transducer, gp130: an oncostatin M receptor and affinity converter for the LIF receptor. Science. 1992;255:1434–1437. doi: 10.1126/science.1542794. [DOI] [PubMed] [Google Scholar]
- 5.Gorman C. DNA cloning: a practical approach. Vol. 2. Washington, D.C: IRL; 1985. [Google Scholar]
- 6.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 7.Heldin C-H. Dimerization of cell surface receptors in signal transduction. Cell. 1995;180:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 8.Hibi M, Murakami M, Saito M, Hirano T, Taga T, Kishimoto T. Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell. 1990;63:1149–1157. doi: 10.1016/0092-8674(90)90411-7. [DOI] [PubMed] [Google Scholar]
- 9.Hilton D J, Hilton A A, Raicevic A, Rakar S, Harrison-Smith M, Gough N M, Begley C G, Metcalf D, Nicola N A, Wilson T A. Cloning of a mouse IL-11 receptor α-chain: requirement for gp130 for high affinity binding and signal transduction. EMBO J. 1994;13:4765–4775. doi: 10.1002/j.1460-2075.1994.tb06802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilton D J, Watowich S S, Latz L, Lodish H F. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J Biol Chem. 1996;271:4699–4708. doi: 10.1074/jbc.271.9.4699. [DOI] [PubMed] [Google Scholar]
- 11.Ichihara M, Hara T, Kim H, Murate T, Miyajima A. Oncostatin M and leukemia inhibitory factor do not use the same functional receptor in mice. Blood. 1997;90:165–173. [PubMed] [Google Scholar]
- 12.Ip N Y, Nye S H, Boulton T G, Davis S, Taga T, Li Y, Birren S J, Yasukawa K, Kishimoto T, Anderson D J, Stahl N, Yancopoulos G D. CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell. 1992;69:1121–1132. doi: 10.1016/0092-8674(92)90634-o. [DOI] [PubMed] [Google Scholar]
- 13.Mosley B, De Imus C, Friend D, Boiani N, Thoma B, Park L S, Cosman D. Dual oncostatin M (OSM) receptors. J Biol Chem. 1996;271:32635–32643. doi: 10.1074/jbc.271.51.32635. [DOI] [PubMed] [Google Scholar]
- 14.Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguuchi M, Taga T, Kishimoto T. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci USA. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Neal K D, Yu-Lee L Y. The proline-rich motif (PRM): a novel feature of the cytokine hematopoietin receptor superfamily. Lymphokine-Cytokine Res. 1993;12:309–312. [PubMed] [Google Scholar]
- 16.Pennica D, Shaw K J, Swanson T A, Moore M W, Shelton D L, Zioncheck K A, Rosenthal A, Taga T, Paoni N F, Wood W I. Cardiotrophin-1: biological activities and binding to the leukemia inhibitory factor receptor/gp130 signaling complex. J Biol Chem. 1995;270:10915–10922. doi: 10.1074/jbc.270.18.10915. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Stahl N, Farruggella T J, Boulton T G, Zhong Z, Darnell J E, Yancopoulos G D. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 19.Thoma B, Bird T A, Friend D J, Gearing D P, Dower S K. Oncostatin M and leukemia inhibitory factor trigger overlapping and different signals through partially shared receptor complexes. J Biol Chem. 1994;269:6215–6222. [PubMed] [Google Scholar]
- 20.Vignon I, Mornon J-P, Cocault L, Mitjavila M-T, Tambourin P, Gisselbrect S, Souyri M. Molecular cloning and characterization of MPL, the human homolog of the v-mpl oncogene: identification of a member of the hematopoietic growth factor receptor superfamily. Proc Natl Acad Sci USA. 1992;89:5640–5644. doi: 10.1073/pnas.89.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welcher A A, Bitler C M, Radeke M J, Shooter E M. Nerve growth factor binding domain of the nerve growth factor receptor. Proc Natl Acad Sci USA. 1991;88:159–163. doi: 10.1073/pnas.88.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland N G, Gilbert D J, Jenkins N A, Hara T, Miyajima A. Mouse oncostatin M: an immediate early gene induced by multiple cytokines through the JAK-STAT5 pathway. EMBO J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]