Abstract
The U3 small nucleolar RNA participates in early events of eukaryotic pre-rRNA cleavage and is essential for formation of 18S rRNA. Using an in vivo system, we have developed a functional map of the U3 small nucleolar RNA from Saccharomyces cerevisiae. The test strain features a galactose-dependent U3 gene in the chromosome and a plasmid-encoded allele with a unique hybridization tag. Effects of mutations on U3 production were analyzed by evaluating RNA levels in cells grown on galactose medium, and effects on U3 function were assessed by growing cells on glucose medium. The major findings are as follows: (i) boxes C′ and D and flanking helices are critical for U3 accumulation; (ii) boxes B and C are not essential for U3 production but are important for function, most likely due to binding of a trans-acting factor(s); (iii) the 5′ portion of U3 is required for function but not stability; and, (iv) strikingly, the nonconserved hairpins 2, 3, and 4, which account for 50% of the molecule, are not required for accumulation or function.
The small nucleolar RNAs (snoRNAs) play essential roles in posttranscriptional maturation of rRNAs (reviewed in references 2, 17, 38, 40, 51, 65, and 69). A few snoRNAs are required for cleavage of rRNA precursors which encode the 18S, 5.8S, and 25S/28S rRNAs. Most and possibly all of the other snoRNAs serve as guide RNAs in nucleotide modification reactions, in particular ribose methylation and pseudouridine formation. Scores of snoRNAs have already been identified in protist, fungal, plant, and animal cells, and all are believed to exist as snoRNP particles. The various rRNA maturation reactions are thought to occur in a large, poorly defined complex called the processome (16; see also references 17 and 40).
U3, the subject of the present report, is one of a few phylogenetically conserved snoRNAs and appears to join the processome at an early, vital stage. In mammalian and yeast cells U3 is approximately 1 order of magnitude more abundant than other snoRNAs (58, 73). U3 is required for several early pre-rRNA cleavage reactions, including (i) an initial cleavage in the 5′ external transcribed spacer (5′ ETS) region (in yeast cells, mouse extracts, and Xenopus oocyte extracts [25, 29, 45]); (ii) two sites that flank the 18S rRNA coding region (in yeast cells [25]); and (iii) a site near the boundary of internal transcribed spacer-1 (ITS1) and 5.8S rRNA (Xenopus oocytes [60]). U3 appears to interact with pre-rRNA at a variety of sites. In yeast cells, U3 has been proposed to contact pre-rRNA at three sites: one in the 5′ ETS portion and two early in the 18S rRNA coding region. The site in the 5′ ETS was identified by in vivo cross-linking, and subsequent mutation analysis showed it to be essential for 18S rRNA production (7, 9). Additional support for this interaction came from a suppressor mutation in U3 (8; see also below). The two interactions within the 18S rRNA coding region are proposed to occur through base-pairing of a pseudoknot in U3 (24, 44). This suggestion is supported by strong conservation of the putative interacting sequences in both U3 and 18S RNA and by preliminary mutations in the corresponding U3 nucleotides which affect cell viability.
Progress in defining proteins associated with U3 is still at an early stage. For many years U3 RNAs have been known to be associated with the nucleolar protein fibrillarin, or its yeast homolog, Nop1p. This association, which is probably indirect, is now known to be characteristic of snoRNAs in one of two major snoRNA groups, i.e., the box C/D family; the second major family consists of box H/ACA snoRNAs (reviewed in references 65 and 69). Depletion of Nop1p from yeast cells causes severe defects in ribosome biogenesis, including cleavage and methylation of pre-rRNA and ribosome assembly (70, 71). More recently, other proteins associated with U3 have been identified. A U3 RNA-protein complex from CHO cells was shown to contain three previously uncharacterized proteins and seems likely to be a core particle of the native U3 snoRNP (37). Two proteins associated with yeast U3 have been identified, Sof1p and Mpp10p (15, 27). The gene for Sof1p was found in a search for suppressors of a temperature-sensitive growth phenotype caused by substituting the yeast NOP1 gene with its human homolog. The gene for Mpp10p was identified during a search for a structural homolog of a human protein (MPP10), which, in turn, had been detected with antibodies specific for proteins phosphorylated during mitosis. Neither yeast protein is known to be associated with other snoRNAs, as determined by limited hybridization screening of immunoprecipitated RNAs (Sof1p and Mpp10p) and analysis of immunoprecipitated and postlabeled RNAs (Mpp10p). It is not known if Sof1p and Mpp10p interact directly with U3. However, depletion of these proteins causes defects in rRNA processing similar to those resulting from loss or inactivation of U3 itself.
All known U3 RNAs contain a 5′ cap, and the nature of the cap correlates with the type of RNA polymerase used in transcription. Protist, fungal, and animal U3 RNAs are transcribed by RNA polymerase II and have a trimethyl guanosine (TMG) cap. In plants U3 is produced by RNA polymerase III and has a gamma-monomethyl phosphate cap (reviewed in reference 17). In all cases, known U3 is transcribed from independent genes. This is in contrast to most other snoRNAs in animals, which are encoded within introns of protein genes, and some plant snoRNAs, which are transcribed from polycistronic snoRNA operons (reviewed in reference 69). The U3 genes in several species of fungi have the remarkable distinction of being the only snoRNA coding sequences known which themselves contain introns (11, 47).
Phylogenetic comparison of the first U3 snoRNAs revealed conserved sequence elements, called boxes A to D (26, 28, 30, 52, 73, 77). Additional elements, called boxes A′ and C′ (or A°), have been defined more recently (39, 47, 74). Other snoRNAs were subsequently determined to contain boxes C and D, and these elements now define the box C/D family of snoRNAs. Most box C/D snoRNAs possess long (>12 nucleotides) sequences complementary to rRNA that are located immediately upstream of box D and/or an analog box D′ (reviewed in references 2, 65, and 69). RNAs with these features guide the formation of 2′-O-methylated nucleotides in rRNAs (14, 32, 48, 75). The U3 snoRNAs do not contain the methylation motif and thus are not believed to function in 2′-O-methylation.
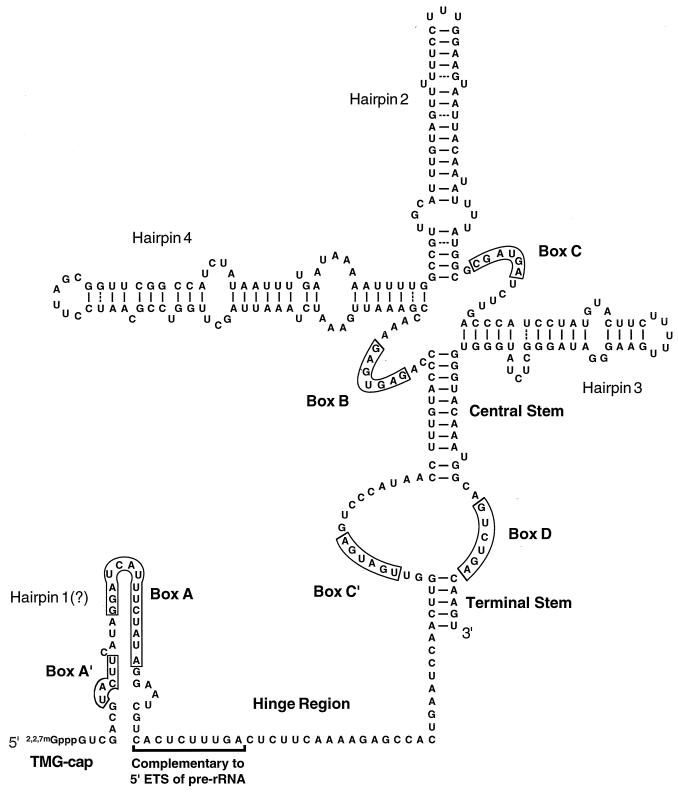
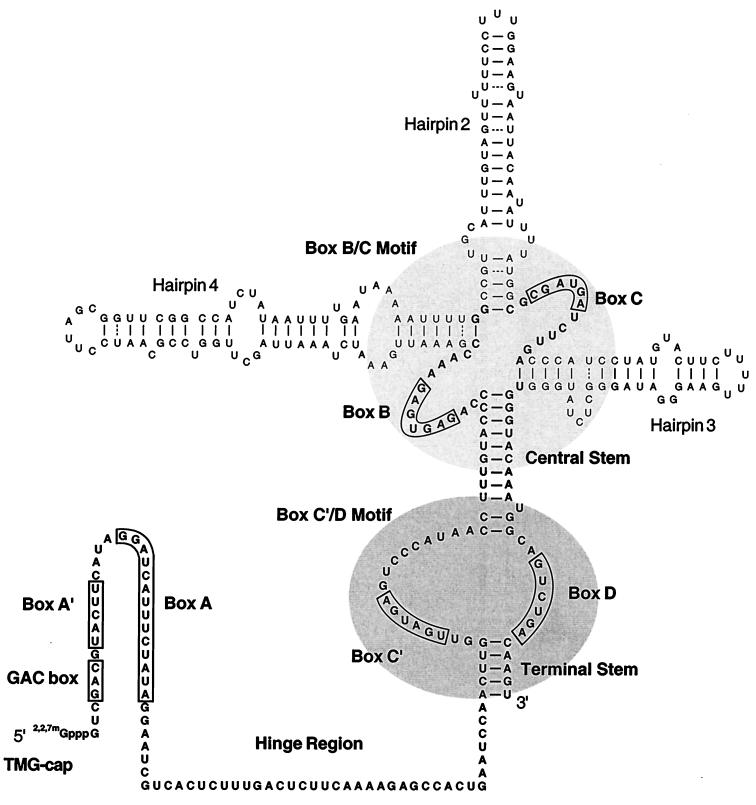
Secondary structure models have been proposed for a variety of U3 molecules, based on computer folding, phylogenetic sequence comparisons, and direct structure probing data (10, 18, 21, 22, 26, 28, 30, 31, 34, 39, 41–44, 46, 47, 49, 50, 52, 53, 61, 62). A consensus structure has not yet been determined, but several features occur consistently (Fig. 1). The conserved box A′ and A elements and adjoining nonconserved nucleotides, known as a hinge region, define the 5′ portion of the molecule. No unambiguous secondary structure has been proposed for this region: one or two hairpins were proposed to be formed in yeasts, one or no hairpins were proposed for higher eukaryotes, and no hairpins were proposed for protists. The rest of the molecule is believed to be highly folded and relatively consistent structures have been proposed for this portion. Boxes C′ and D occur between phylogenetically conserved terminal and central stems, juxtaposed next to each other. The regions surrounding boxes B and C are not conserved; however, in all cases boxes B and C are predicted to be in close proximity. Folding of the surrounding segments results in different numbers of hairpins: from none in protists to three in some fungal species.
FIG. 1.
The U3 snoRNA from the yeast S. cerevisiae. The model shown is derived from a compilation of structures proposed for U3 RNAs from different organisms. Several configurations have been suggested for the 5′ segment including (i) nonfolded (28, 49), (ii) possessing a single helix (hairpin 1 [50]), or (iii) containing two helices (hairpin 1a and 1b, not shown [44, 52]). The well-structured 3′ portion of S. cerevisiae U3 contains two highly conserved (central and terminal stems) and nonconserved helices (hairpins 2, 3, and 4 [26]). The number of hairpins in this region varies from none to three in different organisms. The relative positions of the phylogenetically conserved sequence elements, boxes A, A′, B, C, C′, and D are well preserved in all U3 snoRNAs known. The box A′ and A segments are believed to interact with pre-rRNA (see text). Box C influences association with the protein fibrillarin (6). Box D is required for formation of the 5′ TMG cap structure and nuclear retention of mature RNA (68). The primary sequence of the hinge region separating the conserved elements of the 5′ and 3′ portions is not well preserved phylogenetically. Ten nucleotides of the hinge segment in yeast U3 have been postulated to interact with the 5′ ETS of pre-rRNA through complementary base-pairing (8).
Functional mapping of U3 is still at an early stage. A few elements have been tied to different aspects of snoRNA synthesis, localization, or function. Box A of yeast U3 was shown to be essential for yeast cell viability, based on substitution and deletion mutations (24, 44). This is one of the two regions proposed to base-pair with pre-rRNA, based on cross-linking data obtained with yeast and animal U3 RNAs (66, 72). Box A′ has also been implicated in base-pairing with pre-rRNA, and mutation of this element has been found to affect yeast cell viability (44). The hinge region of yeast U3 has been proposed to interact directly with pre-rRNA through a segment of 10 complementary nucleotides (9). Consistent with binding, a mutation in this segment rescued a lethal rRNA mutation by providing complementarity to the mutant rRNA region (8). No functional information is yet available for box B and the central stem. Mutations in box C influence U3 association with fibrillarin in human cell extracts, and alterations in box D and the 3′ terminal stem block cap formation in Xenopus U3 transcripts injected into oocytes (6, 68). Mutations in the latter elements also impair retention of U3 in the nucleus (68). In yeast cells a large deletion covering most of the hinge region, the proximal part of the 3′ terminal stem, and box C′ has been shown to abolish U3 accumulation (44).
The present investigation was undertaken to develop a map of essential regions in this vital RNA. The major aims of our study were to evaluate the importance of the conserved and nonconserved elements for U3 production and function in S. cerevisiae. The elements featured in our analysis included (i) the 5′ segment encompassing conserved boxes A′ and A and the nonconserved hinge region; (ii) conserved boxes B, C, C′, and D; (iii) the conserved central and terminal stems, and (iv) the nonconserved hairpins in the highly structured 3′ domain.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae JH84 (αleu2-3,12 ura3-52 his3-Δ ade2-1 can1100 u3aΔ UASGAL:U3A::URA3 U3B::LEU2) was kindly provided by John Hughes and was used as the test strain (a similar strain, JH44, is described in reference 25). Yeast cells were grown at 30°C unless otherwise specified. Transformants were grown initially on selective solid 0.67% yeast nitrogen base (YNB) medium containing 2% galactose. This allowed maintenance of the introduced plasmid and expression of U3 from the galactose-dependent genomic cassette; hence, even cells containing nonfunctional U3 genes on the plasmid were able to grow.
To test the ability of mutant U3 genes to support growth, transformants were restreaked from galactose plates onto plates containing 5% glucose (in this condition the genomic U3 cassette is silent) and grown for 5 days. To distinguish unambiguously between residual growth and slow growth, cells were restreaked onto fresh glucose plates and incubated for another 5-day period.
In all, except some control experiments, production of RNA was analyzed with cells grown overnight in selective liquid medium (YNB) containing 2% galactose. In control experiments where the efficiency of repressing the galactose-dependent U3 allele was evaluated, RNA was isolated from cells grown overnight in selective liquid medium containing 5% glucose.
All cloning procedures were carried out with Escherichia coli DH5α (supE44 lacU169 [φ80 lacZ-ΔM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1) grown on either liquid or solid Luria-Bertani medium (0.5% yeast extract, 1% tryptone, 1% NaCl), supplemented with ampicillin (50 μg/ml) when necessary.
Bacterial and yeast strains were transformed with plasmids by slightly modified calcium chloride and lithium acetate methods, respectively (20, 54).
DNA and RNA manipulations.
Plasmid DNA was prepared from E. coli with a boiling miniprep procedure, and total yeast RNA was isolated with a hot-phenol–glass bead procedure (33, 59).
Northern hybridization assays were performed essentially as described previously (4, 55). Equity of RNA transfer to the nitrocellulose membrane was verified by hybridization of the blots with control oligonucleotides and/or ethidium bromide staining. Oligonucleotides for probing U3 RNA included C163 (CATAGGATGGGTCAAGATCATCGCGCC), which recognizes U3, U3*, and all derivatives except U3(del); SD14 (GCCGAACCGCTAAGGATTGCGGAC), which hybridizes to hairpin 4 of wild-type U3 but not to U3* and U3(del) variants; SD13 (CGGCTTAGGCTAAGCTAAGGCCAG), which binds to U3* and all its mutant derivatives except U3(del) but not to wild-type U3; and SD74 (see below), which recognizes all U3 forms including U3(del). Oligonucleotides used for controls were C164 (CGCCTTCCGCGCCGTATGTGTGTGTGACC), which is specific for U1 spliceosomal RNA, and C106 (CGATGGGTTCGTAAGCGTACTCCTACCGTGG), which is specific for U14 snoRNA.
Preparation of modified U3 coding sequences.
The original U3A gene was isolated from plasmid pRex4A (kindly provided by John Hughes) as a blunt-ended HindIII fragment and subcloned into the blunt-ended SalI site of the pBluescript IISK(−) vector with a previously removed BstXI site to generate plasmid pBU3. PCR strategies to alter the U3 gene were performed with this plasmid (Fig. 2). Plasmids used for yeast transformation were prepared by cloning EcoRI-XhoI fragments carrying the wild-type and mutated U3 RNA genes from the pBluescript IISK(−) vector into the yeast shuttle vector pRS313 (64). The final plasmids were as follows: pRU3, wild-type U3; pRU3*, tagged U3; pRU3*(C′:G→C), substitution of the first G in box C′ with C; pRU3*(C′:A→T), substitution of the first A in box C′ with T; pRU3*(C:G→C), substitution of the first G in box C with C; pRU3*(C:A→T), substitution of the first A in box C with T; pRU3*(D:A→T), substitution of the last A in box D with T; pRU3*(C:subst), substitution of the entire box C sequence CGATGA with GCTACT; pRU3*(B:subst), substitution of the entire box B sequence GAGTGAG with CTCACTC; pRU3*(t.st:P), substitution of the terminal stem proximal sequence ACTTG with TGGGC; pRU3*(t.st:D), substitution of the terminal stem distal sequence CAAGT with GCCCA; pRU3*(t.st:PD), substitution of both proximal and distal sequences of the terminal stem; pRU3*(c.st:P), substitution of the central stem proximal sequence CCTTTGTAGGG with GGAAACATGGG; pRU3*(c.st:D), substitution of the central stem distal sequence GGGTACAAATCC with CCCATGTTTTCC; pRU3*(c.st:PD), substitution of both proximal and distal sequences of the terminal stem; pRU3*(trunc), deletion from the 5′ end to the base of the terminal stem; and pRU3*(del), deletion of hairpins 2, 3, and 4.
FIG. 2.
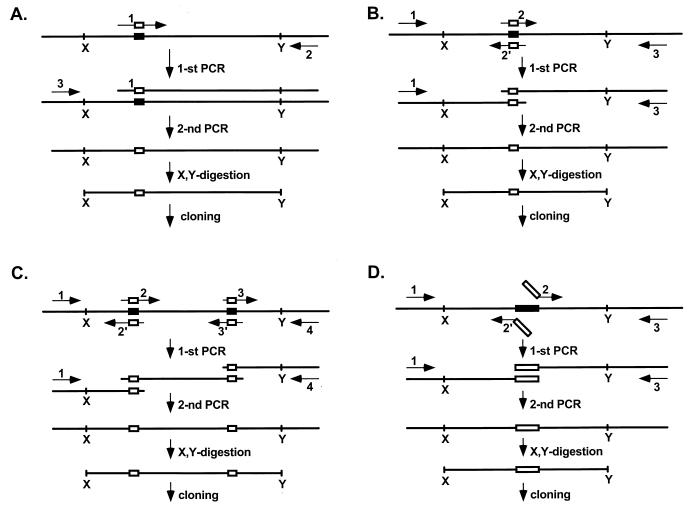
U3 mutagenesis strategies. Different PCR strategies were used for creating U3 mutations, depending on the position and length of the mutant sequence and the number of sequences to be substituted. In each case two PCR amplification steps were involved. To introduce point mutations or make a short sequence substitutions, the strategies presented in panels A or B were used. Multiple mutations were created with the strategy presented in panel C. Substitution or deletion of large regions were done as depicted in panel D. Arrows with numbers correspond to the oligonucleotide primers. Short vertical lines with letters correspond to restriction sites. Open and closed boxes designate mutant and wild-type sequences, respectively.
The oligonucleotides used for PCR mutagenesis were as follows: universal M13 forward (GTAAAACGACGGCCAGT) and reverse (GGAAACAGCTATGACCATG) primers for all mutations; SD10 and SD11 for U3* (CGGCTTAGGCTAAGCTAAGGCCAGCAAGCTAATTTAGATTCAA and CTGGCCTTAGCTTAGCCTAAGCCGCATCTATAATTTTGAATAA, respectively); SD19 (CCAACTTGGTTCATGAGTCCC) for U3*(C′:G→C); SD20 (CCAACTTGGTTGTTGAGTCCC) for U3*(C′:A→T); SD29 (GGATGGGTCAAGATCATGGCGCC) for U3*(C:G→C); SD30 (GGATGGGTCAAGATCAACGCGCC) for U3*(C:A→U); SD32 (GTGGTTAACTTGACAGACTGCC) for U3*(D:A→T); NS1 (CAATATTTTATGGCGGCTACTTCTTGACCCATCC) and NS2 (GGATGGGTCAAGAAGTAGCCGCCATAAAATATTG) for U3*(C:subst); NS3 (CTTAGGCTAAGCCAAGGCCAGCAAGCTAATTTAGATTTCAATTTCGGTTTGAGTCTCTGGGGTAC) for U3*(B:subst); SD34 (CACTGAATCCATGGGCGTTGATGAGT) and SD35 (GACTCATCAACGCCCATGGATTCAGTG) for U3*(t.st:P) and U3*(t.st:PD); SD33 (TGGCAGTCTGAGCCCATAACCACTTT) and D149 (AAAGTGGTTATGGGCTCAGACTGCCA) for U3*(t.st:D) and U3*(t.st:PD); SD71 (GGAAACATGGGCAGAGTGAGAAACCGAAATTG) and SD72 (CGGTTTCTCACTCTGCCCATGTTTCCTTATGGGACTCATCAACCAA) for U3*(c.st:P) and U3*(c.st:PD); SD69 (GGTCCCATGTTTTCCCAGTCTGACAAGTTAACCAC) and SD70 (CTTGTCAGACTGGGAAAACATGGGACCCATAGAGCCCTATCCCTTC) for U3*(c.st:D) and U3*(c.st:PD); SD65 (TGACTCTGTCGACAACTTGGTTGATGAGTCCC) for U3*(trunc); and SD73 (GTGAGAAACCGGCGCGATGATCTTGATGGGTACAAATGGCAGTCTGAC) and SD74 (ATCAAGATCATCGCGCCGGTTTCTCACTCTGGGGTAC) for U3(del).
Restriction endonucleases used for treating the final PCR fragments before cloning (see Fig. 2) were SalI and BstXI. All new constructs were verified by sequencing the mutated regions using the dsDNA Cycle Sequencing System Kit from BRL Life Technologies, Inc., and appropriate primers.
RESULTS
Genetic system for functional mapping of yeast U3 RNA.
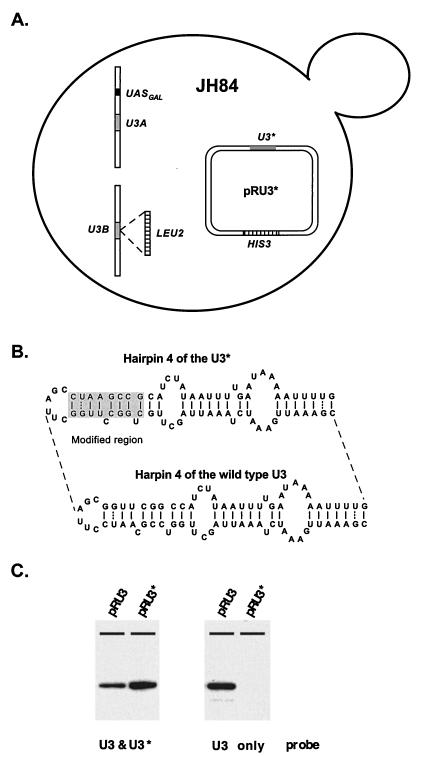
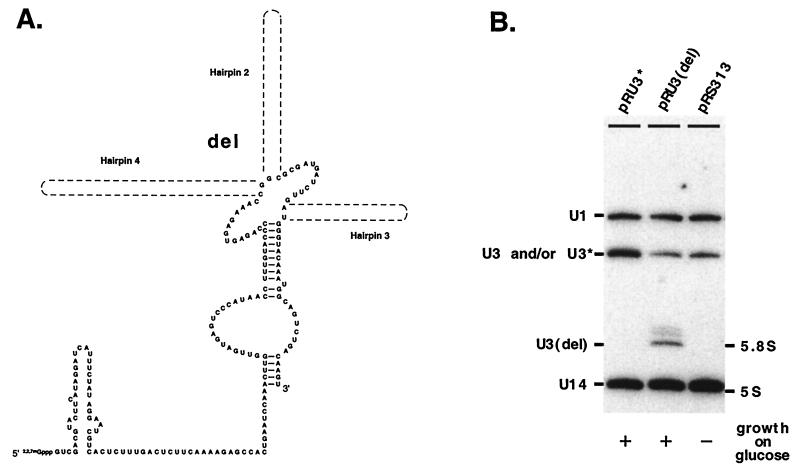
To assess the importance of U3 structural elements, we developed a genetic system that features a S. cerevisiae JH84 test strain containing a galactose-dependent U3 gene in the genome (UASGAL:U3A). All mutations were introduced into a constitutively expressed U3 gene on the plasmid, called U3* (Fig. 3A; see also Materials and Methods). The RNA expressed from the U3* gene contains a unique hybridization “tag,” which makes it possible to distinguish the products of this allele from RNA produced from the genomic locus. When designing this gene, we took into consideration the fact that hairpin 4 of the S. cerevisiae U3 RNA is the least phylogenetically conserved portion of the molecule and in most organisms (including the yeast Schizosaccharomyces pombe) is absent altogether and thus probably plays an unimportant role. Modifications were made in this stem-loop domain that changed the primary sequence slightly but left the hypothetical secondary structure intact (Fig. 3B).
FIG. 3.
Genetic system for functional mapping of yeast U3 RNA. (A) Mutational analyses were carried out with test strain JH84 (25). In this strain one of two genes encoding U3 RNA (U3B) is disrupted with a LEU2 marker gene, and the second (U3A) is under control of the UASGAL-regulated promoter. Cells grow well on medium containing galactose as the sole carbon source, since U3A RNA is transcribed from the promoter induced by galactose. In the presence of glucose the UASGAL-regulated promoter is repressed, and this leads to severe underaccumulation of U3 and subsequent cell death. Lethality can be avoided, however, if the cells also contain a plasmid with a functional U3 RNA gene under control of the normal U3 promoter. (B) In order to distinguish RNAs expressed from the chromosome and the plasmid, a modified U3 gene with a unique hybridization tag (U3*) was constructed. The U3* RNA has a different sequence in the nonconserved hairpin 4 region; however, the secondary structure of the hairpin is preserved. The importance of specific U3 elements for RNA function and production was evaluated by transforming test cells with the plasmid-encoded mutant genes and examining cell growth on glucose and the accumulation of mutant U3 RNA on galactose. (C) Depletion of wild-type U3 produced from the galactose-dependent chromosomal allele during cell growth on glucose was evaluated by Northern blotting analysis. The test strain was transformed with plasmids encoding wild-type (pRU3) or tagged (pRU3*) U3. Total RNA was prepared from transformants grown overnight in liquid selective medium containing 5% glucose. A blot containing these RNAs was first probed with a radiolabeled oligonucleotide (C163) that recognizes both wild-type (U3) and tagged U3 (U3*; left panel) RNAs. The blot was washed and then probed with an oligonucleotide (SD14) that recognizes exclusively wild-type U3 (right panel). Depletion of wild-type U3 is nearly complete in cells transformed with pRU3* and grown on glucose.
Our system allows the importance of specific U3 elements to be tested in two ways. The inability of mutant U3* alleles to support growth on glucose medium indicates that an essential nucleotide(s) has been mutated. By examining U3* RNA levels in cells grown on galactose, it is possible to distinguish mutations that influence RNA accumulation from those that impair U3* function without affecting production.
Before initiating the mapping analysis, we analyzed the behavior of the test strain itself. Growth and U3 RNA production from both genomic and plasmid alleles were evaluated on galactose and glucose medium. Cells were transformed with either a commonly used vector lacking an insert (pRS313 [64]) or a vector containing either the natural U3A gene (pRU3) or the tagged U3 gene (pRU3*). Transformants were first analyzed for the ability to grow on galactose and glucose. Cells containing plasmids with the natural or tagged U3 genes grew with the same efficiency on solid medium containing glucose; no growth was observed for cells containing the control plasmid. All three types of cells grew equally well on solid medium containing galactose. These results demonstrate that the galactose-dependent U3 allele (chromosome) is repressed on glucose and does not support cell growth; this allele does allow normal growth on galactose. The results also show that the tagged U3* gene supports wild-type growth on glucose, when the galactose-dependent allele is silent.
We next analyzed the patterns of U3 RNA produced by these same transformants grown in liquid galactose or glucose medium by using probes specific to natural or tagged U3 or one that recognizes both RNAs simultaneously. The results demonstrated that the tagged U3* RNA accumulates at a normal level, both in glucose- and in galactose-grown cells. The tagged (U3*) and natural (U3) RNAs were easily distinguished with the specific probes. Repression of the chromosomal galactose-dependent U3 gene on glucose was nearly complete. The RNA from this allele was barely detectable in cells with the plasmid-encoded U3* gene grown in liquid medium overnight; we estimate the amount of natural U3 at less than 5% of the normal level. Thus, it seems likely that little or no residual wild-type U3 is present in the test cells when the ability of the mutant U3* gene to support growth is examined. Much of the characterization data for the test system are not shown here since most control genes are included in the experiments that follow. Northern blotting demonstrated that wild-type U3 is essentially depleted from test cells grown in liquid glucose medium (Fig. 3C).
U3 contains a structural homolog of the U14 box C/D terminal stem motif.
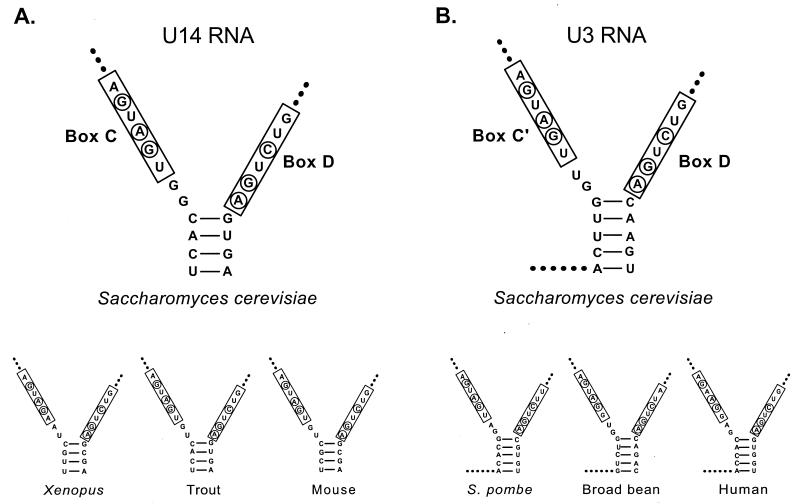
In box C/D snoRNAs, boxes C (UGAUGA) and D (GUCUGA) are usually located near the 5′ and 3′ ends of the mature molecule, respectively. Structure and phylogenetic studies of S. cerevisiae U14 RNA revealed that box C, box D, and an adjoining terminal stem form a box C/D stem structural motif (Fig. 4A; see also references 3 and 23). Identical structures occur in folding models of U14 molecules from other organisms (several examples are presented). In yeast and Xenopus sp., each component of the motif has been shown to be essential for U14 accumulation (23, 78). Certain of the box C and D nucleotides are particularly important (Fig. 4, circled).
FIG. 4.
Structural homologs of the box C/D stem motif in U3 and U14 snoRNAs. (A) Boxes C and D of U14 snoRNA together with adjoining inverted repeats form a conserved structure known as the box C/D stem motif. Each of the components of this motif is essential for U14 accumulation in S. cerevisiae (23). Box C and box D nucleotides important for RNA production are circled. (B) Boxes C′ and D of U3 can also be arranged in a secondary structure similar to that of the U14 box C/D stem motif. As in U14 this hypothetical structure is phylogenetically conserved in all U3 homologs. The nucleotides essential for U14 accumulation are also preserved in U3 (circled).
U3 differs from U14 and other box C/D snoRNAs in that it possesses two box C-like elements (boxes C and C′), both of which are in the interior of the molecule. Interestingly, box C′ (but not box C) and box D of U3 snoRNA and flanking 5-nucleotide inverted repeats can be arranged in a secondary structure similar to that of the U14 box C/D stem motif (Fig. 4B; see also references 18, 21, 28, 31, 34, 39, 41, 44, 49, 50, 52, and 62). As in U14 this hypothetical structure is phylogenetically conserved in U3 homologs (examples are shown). Furthermore, the nucleotides essential for U14 accumulation are also preserved (circled). The phylogenetic similarities suggest that the U3 box C′/D stem motif is both a structural and functional homolog of the box C/D stem motif in U14. This hypothesis was examined in the experiments described below.
Boxes C′ and D, but not box C, are essential for U3 accumulation.
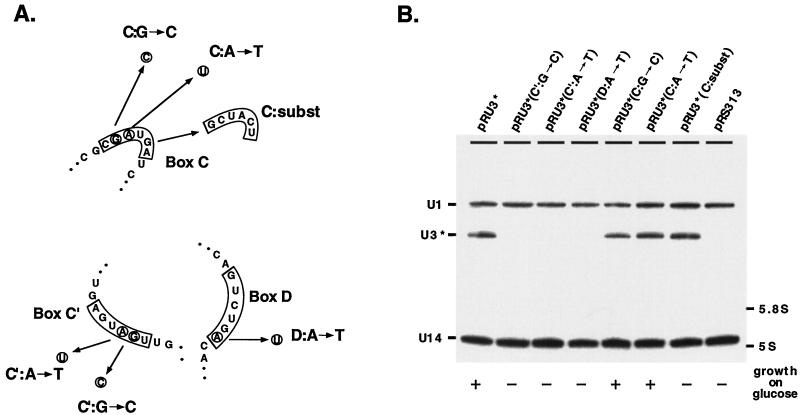
If the box C or C′ element (or both) of U3 is a functional homolog of U14 box C, the corresponding nucleotides vital for U14 accumulation should also be necessary for U3 RNA production. This prediction was examined by introducing two point mutations in boxes C and C′ of U3* which are known to cause severe underaccumulation of U14 (Fig. 5A; see also Fig. 4).
FIG. 5.
A box C/D stem motif containing box C′ rather than box C is important for U3 accumulation. (A) Single base substitutions were made in U3 boxes C and C′ at positions known to be essential for the box C stabilizing function in U14. A point mutation was created in box D, which in U14 is known to lead to underaccumulation of snoRNA (23). A substitution mutation which replaces the entire box C was also prepared. (B) Effects of mutations on U3 RNA accumulation were assessed by Northern blotting analysis of total RNA isolated from galactose-grown cells. Levels of mutant U3* RNA were estimated with a probe specific to a hybridization tag (upper portion of panel). RNA loading was assessed with hybridization probes for U14 and U1 RNAs. Mutation effects on cell viability were determined by growing transformants on glucose-containing plates (lower portion of panel). Growth rates are indicated as follows: + corresponds to wild-type growth, and − represents no growth.
Substitutions in box C had no effect on either cell growth on glucose or accumulation of the U3* RNA on galactose [Fig. 5B, lanes pRU3*(C:G→C) and pRU3*(C:A→T)]. In contrast, point mutations in box C′ impaired cell growth on glucose and accumulation of U3* in galactose-grown cells [lanes pRU3*(C′:G→C) and pRU3*(C′:A→T)]. To determine if box C plays a role in RNA production, a U3* allele was prepared in which all of box C was replaced with an unrelated sequence. No significant difference in mutant RNA levels was observed for transformants grown on galactose [lane pRU3*(C:subst)]. However, the mutant allele did not support growth on glucose, indicating that box C is required for U3* function. A point mutation in U3* box D was also created analogous to one in U14 box D that leads to severe underaccumulation of that RNA. This mutation led to impaired growth on glucose and a dramatic reduction in the amount of stable RNA [lane pRU3*(D:A→T)]. Taken together, these results argue that boxes C′ and D in U3 provide the critical accumulation function(s) first established for boxes C and D in yeast U14. The canonical box C in U3 is not required for RNA accumulation but it is needed for U3 function since substitution of this element leads to lethality.
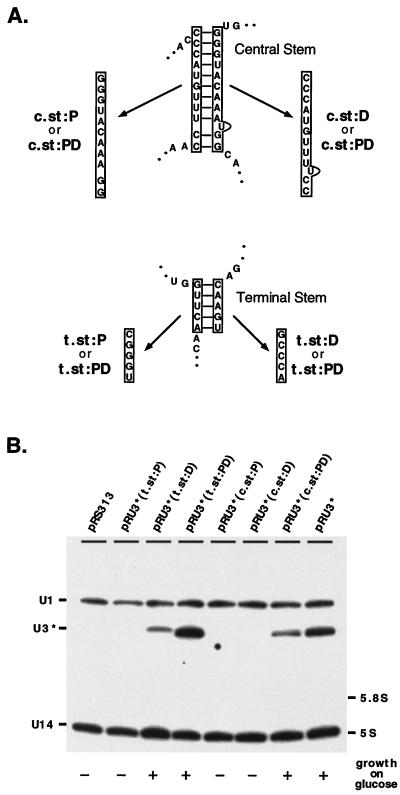
Both stems flanking boxes C′ and D are required for U3 production.
We propose the box C′ and D elements in U3 comprise a structure similar to the U14 box C/D stem motif. If true, the inverted repeats flanking boxes C′ and D must form a helix in vivo that is important for snoRNA accumulation. To test this, we separately substituted both the 5′ proximal [pRU3*(t.st:P)] and 3′ distal [pRU3*(t.st:D)] inverted repeats with noncomplementary sequences. An additional construct, pRU3*(t.st:PD), contained proximal and distal stem segments with reconstituted base-pairing possibilities but with different sequences (Fig. 6A). Transformants containing these alleles were analyzed for growth on glucose, and RNA was prepared from cells grown on galactose. RNA expressed from plasmid pRU3*(t.st:P) accumulated at a very low level on galactose and did not support normal growth on glucose medium [Fig. 6B, lane pRU3*(t.st:P)]. Although U3 expressed from the pRU3*(t.st:D) construct supported normal growth, the mutant RNA accumulated at a level lower than did the U3* control [lane pRU3*(t.st:D)]. Cells dependent on the mutant allele in which substitution mutations restored base-pairing grew on glucose as well as cells expressing wild-type U3*; however, the level of the mutant RNA expressed on galactose actually exceeded that of wild-type U3* [lanes pRU3*(t.st:PD) and pRU3*].
FIG. 6.
The terminal and central stems are required for stable RNA production. (A) The importance of the terminal and central stems flanking boxes C′ and D was evaluated with mutations that disrupt the complementary interactions. Mutations which change the primary but preserve the secondary structures of these stems were also prepared. (B) Substitutions in either proximal (P) or distal (D) parts of the terminal stem (t.st) or central stem (c.st) lead to underaccumulation of U3 on galactose medium and impair cell growth on glucose. Substitutions in both proximal and distal parts (PD) which preserve complementarity rescue these effects. Hybridization probes for the Northern blotting analyses and symbols indicating cell growth on glucose are the same as in Fig. 5.
To evaluate the importance of the central stem, we examined three U3* alleles containing substituted proximal [pRU3*(c.st:P)] or distal [pRU3*(c.st:D)] stem segments and a stem defined by novel complementarities [pRU3*(c.st:PD); Fig. 6A]. Characterization of transformants cultured on both glucose and galactose media clearly demonstrated the importance of the central stem structure for RNA accumulation (Fig. 6B). RNA produced from the galactose-inducible alleles containing disrupted base-pairings accumulated at much lower levels than RNA synthesized from the control U3* allele and did not support normal growth on glucose [compare lanes pRU3*(c.st:P) and pRU3*(c.st:D) and lane (pRU3*)]. RNA with restored complementary interactions accumulated during galactose growth at a level only slightly lower than that of U3* RNA [lanes pRU3*(c.st:PD) and pRU3*] and provided normal growth on glucose.
These results demonstrate that base-pairing in the terminal and central stems is necessary for U3 accumulation and cell growth and imply that these stems exist in vivo. Why RNA with a substituted distal part of the terminal stem [pRU3*(t.st:D)] still accumulates, though at a lower level, is not clear. It is possible that the new sequence creates an additional stabilizing signal or allows an alternative stable structure to be formed. Taken together, the results argue that U3 boxes C′ and D and flanking inverted repeats form a structure in vivo which is functionally homologous to the box C/D stem motif of U14. The internal inverted repeats, which in U3 comprise the central stem, are most likely also involved in the formation of this structure.
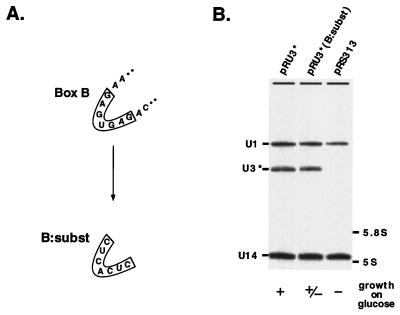
Box B is required for U3 function but not accumulation.
To assess the importance of the conserved box B, we prepared a construct in which the seven most phylogenetically stable nucleotides were substituted with an unrelated sequence [pRU3*(B:subst); Fig. 7A]. Expression of this construct in test cells cultured on galactose revealed no change in accumulation level, indicating that box B is not important for U3 production (Fig. 7B). While the RNA accumulated, it could not support normal growth of cells on solid glucose medium. To assess the effect of the mutation in more detail, we analyzed cell growth at different temperatures and in liquid medium. At 15°C and on solid glucose medium, cells expressing U3*(B:subst) grew more slowly than those expressing wild-type U3*. However, at 37°C both types of cells grew equally well. A growth curve analysis at 30°C in liquid glucose medium revealed that the onset of logarithmic growth was delayed 10 h (data not shown). The basis of this effect is unknown. In any case, the cold-sensitive growth indicates that box B is important for U3 RNA function but does not affect its accumulation.
FIG. 7.
Box B is required for U3 function but not RNA accumulation. (A) The importance of the highly conserved box B element was examined by substituting the entire box with an unrelated sequence. (B) Substitution of box B (B:subst) does not affect U3 accumulation on galactose but does cause a cs slow-growth phenotype for cells grown on glucose medium. Probes and growth symbols are the same as in Fig. 5, except that ± refers to partially impaired growth (see text).
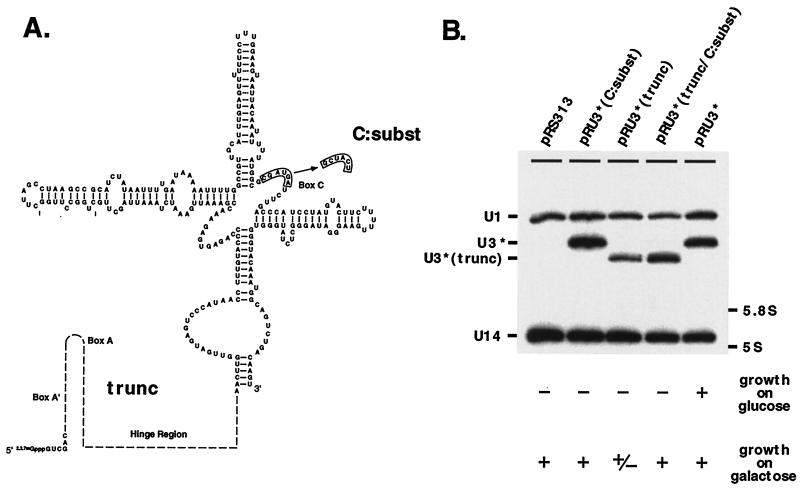
U3 lacking a 5′ segment is toxic, but mutation of box C rescues this effect.
The box A′ and A elements of the U3 5′ segment are known to be required for RNA function but not RNA stability (24, 44). We evaluated a construct in which nearly all of the 5′-end segment was removed, leaving only six native nucleotides at the very 5′ end of the molecule and one nucleotide preceding the proximal part of the terminal stem [pRU3*(trunc); Fig. 8A]. The truncated RNA accumulated normally on galactose but was unable to support growth on glucose medium [Fig. 8B, lane pRU3*(trunc)]. Interestingly, cells containing the abnormal RNA grew more slowly on galactose-containing plates than cells containing U3* or just the plasmid vector pRS313. The growth curve of cells cultured in liquid galactose medium revealed a pre-logarithmic-phase delay of 8 h (not shown). We wondered if the toxic effect of the truncated RNA might reflect competition of this molecule for an essential trans-acting factor(s). Results from this and earlier studies suggest that a factor(s), presumably a protein(s), binds to the central part of U3 where the conserved boxes B and C occur (5, 6, 37, 44, 50, 68). The possibility that the truncated U3 variant competes for such a factor(s) was examined with a mutant U3 containing the same deletion, as well as with a fully substituted box C mutation (Fig. 8A). Strikingly, the box C mutation rescued both the slow-growth phenotype on solid galactose medium and the delayed-growth phenotype in liquid medium (not shown). The mutant RNA was produced at a normal level [Fig. 8B, lane pRU3*(trunc/C:subst)].
FIG. 8.
U3 lacking the 5′ segment is toxic, but substitution of box C rescues this effect. (A) The importance of the entire 5′ segment for U3 RNA accumulation was examined by creating a deletion spanning the region between position +6 and a nucleotide preceding the terminal stem. (B) This variant, U3*(trunc), accumulates normally in cells grown in galactose but, as expected, does not support growth on glucose. Interestingly, cells expressing both the wild-type and truncated forms of U3 on galactose grow slower than cells which only express normal U3. Substitution of box C with an unrelated sequence, U3*(trunc/C:subst), rescues the toxic effect of the truncated molecule in cells grown on galactose. This fact suggests binding of the hypothetical trans-acting factor(s) with the portion of U3 where boxes B and C are located. Probes and growth symbols are as in Fig. 5 and 7.
The results from this series of functional mapping are summarized as follows: (i) the 5′ portion of U3 containing the hinge region and conserved box A′ and A elements is essential for RNA function but is not important for RNA production; (ii) U3 with a truncated 5′ end competes with wild-type U3 for a hypothetical trans-acting factor(s); and (iii) the hypothetical factor(s) most likely binds box C (and perhaps box B), which is located in the central part of the 3′ region.
The essential 5′ region contains a previously unidentified GAC box element.
To gain additional insight into the role of the 5′ region, we conducted a phylogenetic comparison of the 5′ segments from more than 20 U3 RNA species (Fig. 9). This analysis revealed a novel, absolutely conserved GAC box element which precedes box A′. The conserved nature of this box suggests that it is important for U3 function (see also Discussion).
FIG. 9.
Phylogenetic comparison of U3 5′ regions. Alignment of the 5′ segment sequences from diverse organisms reveals a novel conserved GAC box element. The length but not the primary structure of the hinge region appears well preserved. The U3 sequences analyzed (GenBank accession numbers are given in parentheses) are from protists Leptomonas collosoma (L32919), Trypanosoma brucei (M25776), Leishmania tarentolae (L20948), Tetrahymena thermophila (X71349), Euglena gracilis (U27297), and Dictyostelium discoideum (V00190); fungi Saccharomyces cerevisiae (M26648 and X05498), Hansenula wingei (X91005), and Schizosaccharomyces pombe (X56982 and X56189); plants Zea mays (Z29641), Oryza sativa (X79685), Triticum aestivum (X63065), Solanum lycopersicum (X14411), Solanum tuberosum (Z11883), and Arabidopsis thaliana (X52629, X52630, and X58068); and animals Xenopus laevis (X07318), Xenopus borealis (X07319), Mus musculus (X63743), Rattus norvegicus (J01884 and K00780), and Homo sapiens (M14061). The first nucleotide (R) follows the cap (either TMG or gamma-monomethyl phosphate). T.st(P) corresponds to the proximal region of the terminal stem. The G* nucleotide in the GAC box of mouse U3 has been shown to cross-link to the 5′ ETS of pre-rRNA (72).
Hairpins 2, 3, and 4 are not required for accumulation or function.
The functional importance of the nonconserved hairpins 2, 3, and 4 was evaluated with a deletion construct that removed all three stem-loop structures [pRU3*(del); Fig. 10A]. In this construct the hairpins were almost completely deleted, leaving only the nucleotides which comprise the first base pair of each hairpin. Surprisingly, the RNA expressed from the new allele was stable and also provided normal growth in the absence of wild-type U3 (Fig. 10B). Minor larger species present in the Northern blot correspond to U3 derivatives with a 3′ end extended by approximately 20 to 30 nucleotides, as demonstrated with hybridization probes specific for the 5′ or 3′ flanking regions in the U3 gene (not shown). These striking results indicate that in yeast cells a small U3 variant lacking most of the nonconserved sequence is able to accumulate normally and provide all of the essential functions. This mini-variant resembles the U3 RNAs from protists (18, 21, 49).
FIG. 10.
Hairpins 2, 3, and 4 together are not essential for U3 accumulation and function. (A) The importance of hairpins 2, 3, and 4 was evaluated by deleting all three stem-loop domains. (B) The ability of mutant U3(del) RNA to accumulate was assessed with cells grown in liquid galactose medium, in which natural U3 is also produced from the chromosomal gene. Since the mutant RNA lacks hairpin 4, which contains the unique hybridization tag, a different probe was used in this analysis. This probe (SD74) hybridizes to the U3(del) region corresponding to the central stem and boxes B and C, which are also present in the wild-type U3 produced from the genomic locus. Since U3(del) is much smaller than wild-type U3 it is easily identified. The mutant U3(del) accumulates at a good level on galactose and, remarkably, supports wild-type growth of cells on glucose medium, where the wild-type U3 gene is repressed. Probes for control RNAs and growth symbols are the same as in Fig. 5.
DISCUSSION
Our results provide several new insights into the structure and function elements of the essential U3 snoRNA from S. cerevisiae and should be relevant to other U3 homologs as well. The genetic system used allowed us not only to identify defective U3 genes but also to distinguish mutations that influence U3 production from others that affect only U3 function.
The unfolded 5′ segment of U3 is essential for activity but not RNA production.
Deleting most of the 5′ portion of U3 did not affect RNA accumulation; rather, a stable, but nonfunctional by-product was produced. Biochemical and genetic studies conducted previously with U3 RNAs from various organisms strongly suggest that the 5′ segment interacts directly with pre-rRNA (see the introduction). However, attempts to identify specific nucleotides involved in such interaction(s) have yielded ambiguous results. The primary candidates in U3 are the highly conserved nucleotides in the box A′/A region, and the proposed target sites in pre-rRNA include nucleotides in the noncoding region of the 5′ ETS and in an early portion of the 18S rRNA coding region (9, 24, 44, 66, 72). In reality U3 may interact with pre-rRNA at multiple sites, perhaps in an evolving dynamic fashion.
Our phylogenetic comparison of the 5′ segments from more than 20 U3 snoRNA species revealed a novel GAC box element. The absolutely conserved nature of this element suggests it is important for U3 function. This view is supported by experimental results showing that the C nucleotide of the GAC box was a site of cross-linking to the pre-rRNA 5′ ETS in mouse cell extracts (72). Recently, the GAC nucleotides have been proposed to base pair with pre-rRNA in the 18S rRNA portion (44).
Another interesting observation is that despite obvious differences in the primary structure of the hinge region in various U3 molecules, its length is well conserved (Fig. 10; see also electronic databases described in references 19 and 79). This property suggests that the hinge region serves as a spacer to separate the 5′ segment containing the GAC, A′, and A box elements and the highly folded, protein-bound 3′ portion of U3 (see also below). Interestingly, a mutation in the hinge region of yeast U3 has been identified that suppresses a lethal mutation in the 5′ ETS segment of pre-rRNA (8). This mutation provided base-pairing between U3 and the mutant pre-rRNA. However, the mutated region in U3 is not essential and is not conserved, suggesting that this segment may not normally interact with pre-rRNA. Nonetheless, this finding argues that yeast U3 and pre-rRNA do interact through base-pairing.
Computer folding and chemical probing analyses have not produced a consistent secondary structure for the 5′ segment. This situation indicates that the 5′ region might not be folded at all or only transiently so. Indeed, association with pre-rRNA might be complicated by stable secondary folding of the 5′ segment. This view is supported by different chemical modification patterns observed for yeast U3 analyzed as free RNA or a RNP complex (44).
Boxes C′ and D comprise a recognition motif characteristic of other box C/D snoRNAs.
Previous studies with other box C/D snoRNAs revealed that box C, box D, and an adjoining terminal stem are required for snoRNA accumulation and form a structural motif in mature RNA (3, 12, 13, 23, 76, 78). This box C/D stem motif is believed to be recognized by a hypothetical protein(s) which protects the mature RNA from exonucleolytic degradation. Our mutational results demonstrate that boxes C′ and D and flanking complementary elements are required for U3 production, as shown earlier for boxes C and D and flanking inverted repeats of yeast U14 (see also reference 23). These results argue that the box C′/D stem motif of U3 corresponds functionally to the box C/D motif of U14. Consistent with this view and a role in protein binding, the box C′/D stem segments were found to be protected in structure probing studies of U3 RNP complexes in human and yeast cell extracts and in intact yeast cells (44, 50). The canonical box C is located far from box D in the U3 secondary structure. This fact and our demonstration that box C is not required for RNA production indicate that box C in U3 is not homologous to box C from other box C/D snoRNAs.
Of particular interest is our demonstration that the central stem of U3 is also required for RNA accumulation, suggesting that this helix is important for the formation of the box C′/D stem recognition motif. This result is consistent with the finding that formation of the stabilizing box C/D stem signal in yeast U14 involves an additional complementary interaction as well. In U14 the enhanced structural stability is provided by a single complementary element that extends into the flanking precursor regions (56; see also reference 3). A role for internal folding to create the RNA stabilizing motif which contains boxes C and D is also favored by the knowledge that some box C/D snoRNAs lack an obvious terminal stem (see reference 76). A protein(s) specific for boxes C (C′ in U3) and D has not yet been identified, though results from competition analysis of RNAs injected into Xenopus oocytes indicate that the same trans-acting factor(s) stabilizes different box C/D snoRNAs (68).
In addition to contributing to RNA stabilization, box D is required for nuclear targeting of U3 injected into the cytoplasm of Xenopus oocytes and for nuclear retention when injected into the nucleus (5, 67, 68). In work to be described elsewhere, we have demonstrated that the box C/D motif is sufficient for nucleolar targeting of model box C/D snoRNAs expressed in yeast and mammalian cells (56). It is likely that stabilization-processing and targeting-retention functions are provided by the same protein(s). In many other box C/D snoRNAs known, the box D element is also a determinant in guiding the formation of 2′-O-methylated nucleotides in rRNAs (see Introduction). Because U3 does not have a discernible methylation motif, it is not believed to participate in that process.
Interestingly, box D has also been implicated in the methylation of U3 itself, specifically, in the hypermethylation of a monomethyl guanosine cap to yield TMG (68). In contrast to TMG cap formation for the spliceosomal RNAs, hypermethylation of U3 does not occur in the cytoplasm but in the nucleus (67). The TMG cap is believed to protect the 5′ segment of U3 from degradation by exonucleases. Indeed, when TMG is not formed, a shorter molecule, stabilized by the box C′/D stem motif accumulates. This shorter by-product was observed when U3 was encoded in an intron context or removed posttranscriptionally with a ribozyme (57, 76). Consistent with a role in protecting 5′ ends, in vivo addition of a TMG cap to the normally uncapped yeast U14 snoRNA yields a final product with a capped 5′ extension; the 5′ end is defined by the TMG cap (35; see also references 3 and 36). Whether protection of the 5′ end is the true and only function of the TMG cap in U3 and other capped snoRNAs remains to be determined.
Boxes B and C are predicted to form a recognition motif for a U3-specific protein(s).
The canonical box C element is required for normal function of U3; this is due, most likely, to interaction with some trans-acting factor(s). In the secondary structure, box B is always found in close proximity to box C, suggesting that boxes B and C may form a recognition motif and interact with the same factor(s). The importance of box B was demonstrated by a substitution mutation which impaired U3 function, although to a lesser degree than substitution of box C.
Box C is known to influence the association of mammalian U3 with fibrillarin (6). This effect was demonstrated by adding in vitro-transcribed U3 to cell extracts and assaying complex formation with antibodies to fibrillarin. It is not known if this association is direct or depends on other trans-acting factors. A region containing boxes B and C has been implicated in the binding of a 55-kDa protein from mammalian cells (37). This protein was isolated as part of a U3 RNP complex from CHO cells and subsequently shown to bind U3 in vitro. In another study, chemical and nuclease probing analyses with free Trypanosoma sp. and human U3 RNAs and the corresponding RNPs revealed that regions containing boxes B and C are most likely covered with protein (21, 50). Similar results have been also obtained with U3 RNP complexes in yeast cells and extracts (44). We speculate that boxes B and C form a conserved U3-specific box B/C motif and that this structure serves as a recognition signal for one or more protein factors. This putative interaction is essential for the formation of a functional U3 snoRNP.
Nearly 50% of wild-type U3 is dispensable.
Our results demonstrate that the nonconserved stem-loop domains in the highly structured 3′ portion are not required for U3 accumulation or function. Molecules lacking all three hairpins, corresponding to about 50% of the nucleotides, accumulate and provide normal function. This fact, though quite remarkable, is not completely surprising. For example, the yeast U2 spliceosomal RNA is almost six times larger than mammalian U2 and more than 80% of the molecule, corresponding to the nonconserved parts, can be removed without significantly affecting function (1, 63). The U3 from S. cerevisiae with 333 nucleotides is among the largest known U3 molecules. The smallest ones, from protists, are about 50% smaller and do not possess hairpins 2 to 4 (18, 21, 49). However, the latter RNAs contain all of the phylogenetically conserved U3 elements. Our functional mini-U3 molecule looks very similar to the protist homologs.
The nonconserved hairpins in S. cerevisiae U3 do not appear to be without function. In addition to the major expected species, cells expressing the mini-U3 accumulate two minor, larger variants with extra nucleotides at the 3′ end. We speculate that some or all of the nonconserved hairpins might be important for proper RNA folding and subsequent processing. Interestingly, internal deletions in the spliceosomal U2 snRNA can also result in extension of the 3′ end (63).
Structure-function map of U3.
The structural and functional information available for the U3 small nucleolar RNA from S. cerevisiae is summarized in Fig. 11. The 5′ segment containing the conserved GAC, A′, and A boxes is believed to be involved in direct interaction with pre-rRNA, which is critical for rRNA processing. The hinge region is proposed to serve a spacer function to divide the protein-covered 3′ portion of U3 and the 5′ segment, which interacts with pre-rRNA. The 5′ TMG cap is postulated to protect the 5′ region upstream of the box C′/D stem motif from exonucleolytic degradation. The box C′/D stem and box B/C motifs are proposed to be recognition signals for RNA binding proteins. A protein(s) associated with the box C′/D stem motif provides U3 stability and processing and allows nucleolar targeting functions. Protein(s) recognizing the box B/C motif is predicted to be important for U3 function. Finally, the nonconserved hairpins 2, 3, and 4 are dispensable but appear to facilitate RNA folding and processing, thus affecting the homogeneity of the final U3 products. The present map will be useful for studies aimed at identifying the precise role that U3 RNA plays in ribosome biogenesis and characterizing its mechanism of action.
FIG. 11.
Functional map of U3 snoRNA. Results from the present and earlier studies have been combined to yield a functional map of the U3 molecule from S. cerevisiae. The 5′ segment encompassing conserved boxes A and A′ and a novel conserved element, designated the GAC box, is involved in direct interaction with pre-rRNA and is essential for snoRNA function but not production. A hinge region is predicted to provide proper spacing between the 5′ portion which interacts with pre-rRNA and the 3′ segment complexed with RNA binding proteins. The 5′ and hinge segments are protected from degradation by a TMG (gamma-monomethyl phosphate in plants) cap. The box C′/D stem motif is homologous to the box C/D stem motifs in other box C/D snoRNAs and most likely serves as a recognition element for protein(s) which in U3 (i) protects the RNA from degradation, (ii) facilitates formation of the 5′ TMG-cap, and (iii) specifies nucleolar localization. Formation of the box C′/D stem structure requires pairing of the central stem. Boxes B and C are proposed to form a box B/C structure motif which serves as a recognition element for a trans-acting factor(s), presumably a protein(s). Interaction with this factor(s) is required for U3 function. Hairpins 2, 3, and 4 are dispensable for U3 accumulation and function. These latter domains may be considered a species-specific “signature.” The nucleotide regions corresponding to the minimal functional U3 are indicated by boldface letters. The shaded areas correspond to putative protein recognition sites.
ACKNOWLEDGMENTS
We thank Nandita Sharma for help in preparing box B and box C substitution mutations, Matthew Huang and Xiwei Wang for sharing unpublished data, Andrey Balakin for valuable discussions, and Elizabeth Furter-Graves for expert editorial work. We also thank Susan Baserga, Michael Terns, and anonymous reviewers for critical reading of the manuscript prior to publication and for helpful suggestions.
This research was supported by National Institutes of Health grant GM19351.
REFERENCES
- 1.Ares M., Jr U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986;47:49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie J P, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 3.Balakin A G, Lempicki R A, Huang G M, Fournier M J. Saccharomyces cerevisiae U14 small nuclear RNA has little secondary structure and appears to be produced by post-transcriptional processing. J Biol Chem. 1994;269:739–746. [PubMed] [Google Scholar]
- 4.Balakin A G, Schneider G S, Corbett M S, Ni J, Fournier M J. SnR31, snR32, and snR33: three novel, non-essential snRNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5391–5397. doi: 10.1093/nar/21.23.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baserga S J, Gilmore-Hebert M, Yang X W. Distinct molecular signals for nuclear import of the nucleolar snRNA, U3. Genes Dev. 1992;6:1120–1130. doi: 10.1101/gad.6.6.1120. [DOI] [PubMed] [Google Scholar]
- 6.Baserga S J, Yang X W, Steitz J A. An intact box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein L B, Mount S M, Weiner A M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983;32:461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- 11.Brule F, Venema J, Segault V, Tollervey D, Branlant C. The yeast Hansenula wingei U3 snoRNA gene contains an intron and its coding sequence co-evolved with the 5′ ETS of the pre-ribosomal RNA. RNA. 1996;2:183–197. [PMC free article] [PubMed] [Google Scholar]
- 12.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 13.Cavaille J, Bachellerie J P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 14.Cavaille J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 15.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier M J, Maxwell E S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993;18:131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- 17.Gerbi S A. Small nucleolar RNA. Biochem Cell Biol. 1995;73:845–858. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- 18.Greenwood S J, Schnare M N, Gray M W. Molecular characterization of U3 small nucleolar RNA from the early diverging protist, Euglena gracilis. Curr Genet. 1996;30:338–346. doi: 10.1007/s002940050142. [DOI] [PubMed] [Google Scholar]
- 19.Gu J, Reddy R. Small RNA database. Nucleic Acids Res. 1997;25:98–102. doi: 10.1093/nar/25.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Jessee J, Bloom F R. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 21.Hartshorne T, Agabian N. A common core structure for U3 small nucleolar RNAs. Nucleic Acids Res. 1994;22:3354–3364. doi: 10.1093/nar/22.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartshorne T, Agabian N. RNA B is the major nucleolar trimethylguanosine-capped small nuclear RNA associated with fibrillarin and pre-rRNAs in Trypanosoma brucei. Mol Cell Biol. 1993;13:144–154. doi: 10.1128/mcb.13.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang G M, Jarmolowski A, Struck J C R, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes J M X. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 25.Hughes J M X, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes J M X, Konings D A M, Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeppesen C, Stebbins-Boaz B, Gerbi S A. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 30.Kiss T, Solymosy F. Molecular analysis of a U3 RNA gene locus in tomato: transcription signals, the coding region, expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res. 1990;18:1941–1949. doi: 10.1093/nar/18.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiss T, Toth M, Solymosy F. Plant small nuclear RNAs. Nucleolar U3 snRNA is present in plants: partial characterization. Eur J Biochem. 1985;152:259–266. doi: 10.1111/j.1432-1033.1985.tb09192.x. [DOI] [PubMed] [Google Scholar]
- 32.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 33.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 34.Leader D J, Connelly S, Filipowicz W, Brown J W S. Characterization and expression of a maize U3 snRNA gene. Biochim Biophys Acta. 1994;1219:145–147. doi: 10.1016/0167-4781(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 35.Lempicki, R. A., and M. J. Fournier. 1994. Unpublished data.
- 36.Li H V, Fournier M J. U14 function in Saccharomyces cerevisiae can be provided by large deletion variants of yeast U14 and hybrid mouse-yeast U14 RNAs. EMBO J. 1992;11:683–689. doi: 10.1002/j.1460-2075.1992.tb05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubben B, Marshallsay C, Rottmann N, Luhrmann R. Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 1993;21:5377–5385. doi: 10.1093/nar/21.23.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maden T. Click here for methylation. Nature. 1996;383:675–676. doi: 10.1038/383675a0. [DOI] [PubMed] [Google Scholar]
- 39.Marshallsay C, Connelly S, Filipowicz W. Characterization of the U3 and U6 snRNA genes from wheat: U3 snRNA genes in monocot plants are transcribed by RNA polymerase III. Plant Mol Biol. 1992;19:973–983. doi: 10.1007/BF00040529. [DOI] [PubMed] [Google Scholar]
- 40.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 41.Mazan S, Bachellerie J P. Structure and organization of mouse U3B RNA functional genes. J Biol Chem. 1988;263:19461–19467. [PubMed] [Google Scholar]
- 42.Mazan S, Gulli M P, Joseph N, Bachellerie J P. Structure of the differentially expressed mouse U3A gene. Eur J Biochem. 1992;205:1033–1041. doi: 10.1111/j.1432-1033.1992.tb16871.x. [DOI] [PubMed] [Google Scholar]
- 43.Mazan S, Qu L H, Sri-Widada J, Nicoloso M, Bachellerie J P. Presence of a differentially expressed U3A RNA variant in mouse. Structure and evolutive implications. FEBS Lett. 1990;267:121–125. doi: 10.1016/0014-5793(90)80304-2. [DOI] [PubMed] [Google Scholar]
- 44.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 45.Mougey E B, Pape L K, Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5′ external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993;13:5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mougin A, Gregoire A, Banroques J, Segault V, Fournier R, Brule F, Chevrier-Miller M, Branlant C. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA. 1996;2:1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 47.Myslinski E, Segault V, Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 48.Nicoloso M, Qu L H, Michot B, Bachellerie J P. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 49.Orum H, Nielsen H, Engberg J. Sequence and proposed secondary structure of the Tetrahymena thermophila U3-snRNA. Nucleic Acids Res. 1993;21:2511. doi: 10.1093/nar/21.10.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker K A, Steitz J A. Structural analyses of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peculis B A, Mount S M. Ribosomal RNA: small nucleolar RNAs make their mark. Curr Opin Cell Biol. 1996;6:1413–1415. doi: 10.1016/s0960-9822(96)00745-2. [DOI] [PubMed] [Google Scholar]
- 52.Porter G L, Brennwald P J, Holm K A, Wise J A. The sequence of U3 from Schizosaccharomyces pombe suggests structural divergence of this snRNA between metazoans and unicellular eukaryotes. Nucleic Acids Res. 1988;16:10131–10152. doi: 10.1093/nar/16.21.10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy R, Henning D, Busch H. Nucleotide sequence of nucleolar U3B RNA. J Biol Chem. 1979;254:11097–11105. [PubMed] [Google Scholar]
- 54.Rose M D, Winston F, Heiter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 55.Samarsky D A, Balakin A G, Fournier M J. Characterization of three new snRNAs from Saccharomyces cerevisiae: snR34, snR35, and snR36. Nucleic Acids Res. 1995;23:2548–2554. doi: 10.1093/nar/23.13.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samarsky, D. A., E. Bertrand, R. H. Singer, and M. J. Fournier. Unpublished data.
- 57.Samarsky, D. A., G. Ferbeyre, E. Bertrand, R. H. Singer, R. Cedergren, and M. J. Fournier. Unpublished data.
- 58.Samarsky, D. A., and M. J. Fournier. 1996. Unpublished data.
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Savino R, Gerbi S A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segault V, Mougin A, Gregoire A, Banroques J, Branlant C. An experimental study of Saccharomyces cerevisiae U3 snRNA conformation in solution. Nucleic Acids Res. 1992;20:3443–3451. doi: 10.1093/nar/20.13.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selinger D A, Porter G L, Brennwald P J, Wise J A. The two similarly expressed genes encoding U3 snRNA in Schizosaccharomyces pombe lack introns. Mol Biol Evol. 1992;9:297–308. doi: 10.1093/oxfordjournals.molbev.a040721. [DOI] [PubMed] [Google Scholar]
- 63.Shuster E O, Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988;55:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- 64.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 66.Stroke I L, Weiner A M. The 5′ end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursor. J Mol Biol. 1989;210:497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- 67.Terns M P, Dahlberg J E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- 68.Terns M P, Grimm C, Lund E, Dahlberg J. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 70.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 72.Tyc K, Steitz J A. A new interaction between the mouse 5′ external transcribed spacer of pre-rRNA and the U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res. 1992;20:5375–5382. doi: 10.1093/nar/20.20.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tycowski K T, Shu M D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 75.Tycowski K T, Smith C M, Shu M D, Steitz J A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 77.Wise J A, Weiner A M. Dictyostelium small nuclear RNA D2 is homologous to rat nucleolar RNA U3 and is encoded by a dispersed multigene family. Cell. 1980;22:109–118. doi: 10.1016/0092-8674(80)90159-2. [DOI] [PubMed] [Google Scholar]
- 78.Xia L, Watkins N J, Maxwell E S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]
- 79.Zwieb C. The uRNA database. Nucleic Acids Res. 1996;24:76–79. doi: 10.1093/nar/24.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]