Abstract
Genomic imprinting is an epigenetic process that results in the preferential silencing of one of the two parental copies of a gene. Although the precise mechanisms by which genomic imprinting occurs are unknown, the tendency of imprinted genes to exist in chromosomal clusters suggests long-range regulation through shared regulatory elements. We characterize a 800-kb region on the distal end of mouse chromosome 7 that contains a cluster of four maternally expressed genes, H19, Mash2, Kvlqt1, and p57Kip2, as well as two paternally expressed genes, Igf2 and Ins2, and assess the expression and imprinting of Mash2, Kvlqt1, and p57Kip2 during development in embryonic and extraembryonic tissues. Unlike Igf2 and Ins2, which depend on H19 for their imprinting, Mash2, p57Kip2, and Kvlqt1 are unaffected by a deletion of the H19 gene region, suggesting that these more telomeric genes are not regulated by the mechanism that controls H19, Igf2, and Ins2. Mutations in human p57Kip2 have been implicated in Beckwith-Wiedemann syndrome, a disease that has also been associated with loss of imprinting of IGF2. We find, however, that a deletion of the gene has no effect on imprinting within the cluster. Surprisingly, the three maternally expressed genes are regulated very differently by DNA methylation; p57Kip2 is activated, Kvlqt1 is silenced, and Mash2 is unaffected in mice lacking DNA methyltransferase. We conclude that H19 is not a global regulator of imprinting on distal chromosome 7 and that the telomeric genes are imprinted by a separate mechanism(s).
In mammals, a subset of genes are preferentially expressed according to their parent of origin. This phenomenon, variously termed genomic, parental, or gametic imprinting, has been shown for approximately 20 autosomal genes in mice and humans (4). A fundamental question about imprinting involves the mechanism used for distinguishing the maternal and paternal alleles of a gene. The leading candidate is DNA methylation that is established in different patterns in the male and female germ lines and is maintained throughout embryogenesis to regulate the imprinted state. There are other epigenetic differences between the parental alleles of imprinted genes, including differential sensitivity of chromatin to nuclease digestion, asynchronous replication, and differential frequencies of meiotic recombination (5, 13, 18, 24, 25, 39), but these are thought to be the consequences of the primary epigenetic mark, not the causes.
A striking feature of imprinted genes is the frequency with which they are found in close proximity to another imprinted gene, often one that is imprinted in the opposite direction. Four clusters have been characterized, and each contains both maternally and paternally expressed genes (22, 23, 29, 37, 50, 56, 58, 61). The importance of clustering in imprinting remains unclear, but it suggests a role for a cis-regulatory element(s) that acts over a distance to permit the proper imprinting of genes in the cluster. In the case of Prader-Willi and Angelman syndromes, two human diseases that are associated with a cluster of imprinted genes on chromosome 15, deletions of a small region that spans the promoter of one of the paternally expressed genes, SNRPN, result in a disruption of the imprinting of genes hundreds of kilobases away. These observations imply the existence of an “imprint control element” acting on the entire cluster (8, 10, 49). Alternatively, clustering could arise if genes within an imprinted cluster interact functionally; for example, one gene could act in cis to silence a neighboring gene in much the same way that the Xist RNA is thought to be required for silencing the genes on the inactive X chromosome (36, 40). Finally, the integrity of imprinted clusters may also prove to be important for their regulation. In the case of another human disease associated with an imprinted gene cluster, Beckwith-Wiedemann syndrome (BWS), chromosomal rearrangements and translocations on chromosome 11 appear to be causative factors of the disease, in part by disrupting the imprinting of the insulin-like growth factor II (IGF2) gene (7, 20, 55).
In mice, evidence for the importance of imprinted gene clustering comes from studies of H19 and Igf2. These genes lie on distal chromosome 7, in a region syntenic to human chromosome 11p15.5, and the maternal silencing of Igf2 requires the presence of the H19 gene 90 kb away (31, 44). The role of H19 in the silencing of Igf2 is thought to arise from its ability to compete with Igf2 for a common set of endoderm-specific enhancers located downstream of the H19 gene (31). On the maternal chromosome, H19, because of its position relative to the enhancers, prevents enhancer activation of Igf2. On the paternal chromosome, however, allele-specific methylation suppresses the H19 promoter, allowing activation of Igf2 transcription (5, 13, 32). A similar explanation involving promoter competition has now been offered for the imprinting of the Igf2r gene on mouse chromosome 17 (3, 58).
In the last 3 years, several new imprinted genes have been mapped close to Igf2 and H19, including the placenta-specific gene Mash2 and the cyclin-dependent kinase inhibitor gene p57Kip2, both of which are maternally expressed (16, 19). A targeted disruption of Mash2 leads to embryonic death from placental failure in homozygous mutants and in heterozygous mutants inheriting the null allele maternally (17). Disruption of p57Kip2 leads to embryonic or early neonatal death when inherited in the same manner (60, 62). Interestingly, p57Kip2 mutant mice exhibit some features of BWS, including macroglossia and omphalocele.
Recently, another maternally expressed imprinted gene, KvLQT1, has been identified on human chromosome 11p15.5. KvLQT1 is imprinted in most human fetal tissues in which it is expressed, except for the heart (28). Mutations in KvLQT1 cause long-QT syndrome, a heart defect that often leads to sudden death (53). Consistent with the lack of KvLQT1 imprinting in the heart, this syndrome is not inherited in a parent-of-origin-specific manner.
This well-characterized cluster of imprinted genes provides an ideal opportunity to test experimentally the significance of linkage of imprinted genes. Toward that goal, we have generated a genetic and physical map of the region in the mouse, on which we have accurately placed eight genes. We show that a mutation at the H19 locus that disrupts imprinting of Igf2 and Ins2 has no effect on the imprinting of Mash2, Kvlqt1, and p57Kip2. Likewise, deletion of p57Kip2 does not affect the imprinting or expression of the other genes. In contrast, a mutation in the maintenance DNA methyltransferase gene (Dnmt) has different effects on imprinting depending on the gene in question.
MATERIALS AND METHODS
Genetic and physical mapping.
A total of 78 progeny of an interspecific (BTBR × M. spretus)F1 × BTBR backcross between Mus spretus and Mus domesticus were scored by PCR for the MIT markers D7Mit12 and D7Mit47 (Research Genetics). Restriction fragment length polymorphisms between the parental strains were detected with the 1.8-kb Mash2 cDNA fragment, a 2-kb SpeI p57Kip2 fragment, and a 4-kb EcoRI-SalI H19 fragment. Genomic DNA from the N2 progeny was digested with XbaI (Mash2 and p57Kip2) or SphI (H19), separated on a 1% agarose gel, transferred to a nitrocellulose membrane (Millipore), and hybridized to the appropriate radiolabeled fragment. The membranes were washed and visualized by autoradiography.
The Princeton and MIT YAC libraries were screened by PCR with Mash2-specific primers 5′-CTC TAC GTC TCC GTC CCG-3′ (forward) and 5′-CCA CCA CGT GTC TCC CTT AC-3′ (reverse) and p57Kip2-specific primers 5′-GCC GGG TGA TGA GCT GGG AA-3′ (forward) and 5′-AGA GAG GCT GGT CCT TCA GC-3′ (reverse). The BAC library (Research Genetics) was screened by hybridization of radiolabeled probes to library filters and visualization by autoradiography. The probes were inverse PCR products for the left arms of the YACs FDK.D2, FDI.G4, and D43.H9, a 600-bp BamHI-NcoI fragment located 3′ of Mash2, the 1.8-kb p57Kip2 cDNA, the 2-kb Kvlqt1 cDNA, the 1.8-kb Tyrosine hydroxylase (Th) cDNA, and a 3-kb EcoRI-SalI fragment containing H19. To construct a physical map, the BAC DNAs were digested with rare-cutting restriction enzymes, separated on pulsed-field gels, transferred to nylon membranes (Hybond), and probed with the same probes used to screen the BAC library.
Mice.
BTBR mice were obtained from William Dove, and C57BL/6J and 129/Sv mice were purchased from the Jackson Laboratory. The BTBR(SPR H19-p57) congenic strain containing distal chromosome 7 sequences from M. spretus and a similar strain which was a hybrid of C57BL/6J and Mus castaneus, B6(CAST H19-p57), were created by continuous backcrossing of F1 hybrids to BTBR and C57BL/6J and selection, respectively, for M. spretus and M. castaneus alleles of H19 and p57Kip2. The Dnmt mutant mice harboring the s allele were obtained from R. Jaenisch (33), and the p57Kip2 mutant mice were obtained from S. Elledge (62).
Mutant genotyping.
Dnmt genotyping was accomplished by a PCR that detected both the wild-type and targeted loci with primers 5′-CCT TCA GTG TGT ACT GCA GTC G-3′ (forward), 5′-AAT GAG ACC GGT GTC GAC AG-3′ (reverse), and 5′-CTT GTG TAG CGC CAA GTG C-3′ (reverse). A 20-μl reaction volume containing 100 ng of genomic DNA was prepared, and the conditions for amplification were 90°C for 30 s, 53°C for 30 s, and 72°C for 30 s for 35 cycles followed by 4 min at 72°C for 1 cycle. H19Δ13 genotyping used primers 5′-CAG TGT GGG AAA CAG CCT CG-3′ (forward) and 5′-CTT GTG TAG CGC CAA GTG C-3′ (reverse, same as the Dnmt genotyping primer) under the same conditions. p57Kip2 genotyping was performed as described by Zhang et al. (62).
RNA analysis.
Total RNA was isolated from embryonic day 6.5 to 9.5 (e6.5 to e9.5) embryos and ectoplacental cones by guanidine thiocyanate extraction and from e12.5 embryos and fetal and adult organs by LiCl-urea extraction (1, 2). The RNA was treated with DNase I (Stratagene) for 30 min and then extracted with phenol-chloroform (1:1), precipitated with 2 volumes of ethanol, and reverse transcribed by use of Superscript II (Gibco/BRL) with oligo(dT) as the primer as specified by the manufacturer. Analogous reactions were performed without reverse transcriptase (RT) to control for DNA contamination. Imprinting of H19 in Dnmt−/− embryos was assayed by single-strand conformational polymorphism analysis as described previously (51). H19 and Igf2 expression in p57Kip2-deficient mice was detected by allele-specific RNase protection assays (6, 30). For Mash2, cDNA was amplified by PCR in the presence of [33P]dCTP with Mash2-specific primers spanning intron 2, 5′-TTA GGG GGC TAC TGA GCA TC-3′ (forward) and 5′-AAG TCC TGA TGC TGC AAG GT-3′ (reverse). The conditions for amplification were 94°C for 1 min, 55°C for 2 min, and 72°C for 2 min for 35 cycles followed by 4 min at 72°C for 1 cycle. The products were digested with BstNI for 1 h at 60°C and run on a 40-cm 8% acrylamide gel at 50 W for 2 h. The gel was dried and visualized by autoradiography on BioMax film (Kodak). CD81 cDNA was amplified by PCR with primers 5′-AGC CAT TGT GGT AGC TGT C-3′ (forward) and 5′-CAT TGA AGG CAT AAC AGG GCT TAC-3′ (reverse). The conditions for amplification were 94°C for 30 s, 55°C for 60 s, and 72°C for 90 s for 35 cycles followed by 4 min at 72°C for 1 cycle. The products were digested with RsaI for 1 h at 37°C and analyzed on a 10% polyacrylamide gel. Kvlqt1 cDNA was amplified by PCR with primers 5′-GAT CAC CAC CCT GTA CAT TGG-3′ (forward) and 5′-CCA GGA CTC ATC CCA TTA TCC-3′ (reverse). On the basis of the structure of the human gene (28), these primers amplify sequences that span four introns. The conditions for amplification were 94°C for 30 s, 55°C for 60 s, and 72°C for 90 s for 35 cycles followed by 4 min at 72°C for 1 cycle. The product was digested with PvuII for 1 h at 37°C and analyzed on a 10% 1× TBE polyacrylamide gel. p57Kip2 cDNA was amplified by PCR with primers spanning intron 2, 5′-TTC AGA TCT GAC CTC AGA CCC-3′ (forward) and 5′-AGT TCT CTT GCG CTT GGC-3′ (reverse). The conditions for amplification were 94°C for 1 min, 57°C for 2 min, and 72°C for 2 min for 35 cycles followed by 4 min at 72°C for 1 cycle. The products were digested with AvaI for 1 h at 37°C and analyzed on a 10% polyacrylamide gel.
RESULTS
Genetic and physical mapping of the imprinted gene cluster on chromosome 7.
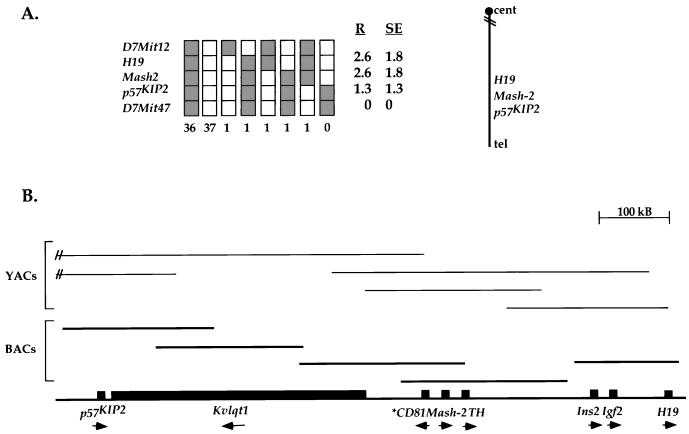
To characterize the imprinted domain at distal chromosome 7, we first constructed a genetic and physical map of the region. Previous linkage analysis had positioned p57Kip2 centromeric to H19 on mouse distal chromosome 7, analogous to the orientation of these genes on human chromosome 11p15.5 (19). To confirm this, we carried out a linkage analysis with 78 progeny of the interspecific backcross (BTBR × M. spretus)F1 × BTBR. In contrast to the previous report, our mapping places H19 at the centromeric end and p57Kip2 at the telomeric end of the cluster (Fig. 1A, right). Our gene order is based on results with three recombinant animals, whereas the other study found only one such animal. The recombination frequencies (expressed as mean genetic distance in centimorgans [cM] ± standard error) are D7Mit12-2.6 ± 1.8-H19-2.6 ± 1.8-Mash2-1.3 ± 1.3-p57KIP2-0-D7Mit47.
FIG. 1.
Genetic and physical mapping of genes on distal chromosome 7. (A) Haplotype analysis showing the segregation patterns of all loci in the interspecific backcross. Each column represents the haplotype of a chromosome that was inherited from the (BTBR × M. spretus)F1 parent. Shaded boxes represent BTBR alleles, whereas open boxes represent M. spretus alleles. The number of offspring inheriting each type of chromosome is shown at the bottom of each column. The percent recombination (R) between adjacent loci with the standard error (SE) is shown at the right of the grid. On the far right is shown the orientation of the imprinted genes relative to the centromere (cent) and telomere (tel). (B) Physical mapping of the imprinted genes on distal chromosome 7. The YAC contig is represented by thin lines, and the BAC contig is represented by thicker lines. The placement of the genes along the contig is indicated at the bottom. Arrows beneath the genes represent their transcriptional orientation. The asterisk indicates that the orientation of CD81 is based on that of the human. The 5′ and 3′ ends of Kvlqt1 cDNA are at least 250 kb apart, but its intron-exon structure remains unknown.
To determine the physical distances between the genes in the cluster, we used primers specific to p57Kip2 and Mash2 to screen the Princeton and MIT YAC libraries. Using both YAC ends and probes specific to p57Kip2, Mash2, and Kvlqt1, we also screened the Research Genetics BAC library. Restriction enzyme digest analysis of the resulting YAC and BAC clones placed H19 and p57Kip2 about 800 kb apart (Fig. 1B). Three additional genes were localized to the region: Kvlqt1, CD81 (Tapa1), and Th. In addition, we determined the transcriptional orientation of each gene by restriction mapping with 5′ and 3′ gene-specific probes and found that all are transcribed toward the centromere, with the exception of Kvlqt1 and possibly CD81, whose current orientation is based on that of the human gene (Fig. 1).
The physical distances between H19 and Mash2 (∼250 kb) and Mash2 and p57Kip2 (∼550 kb) are considerably shorter than those predicted from the genetic distances (∼5 and 2.5 Mb), respectively. The presence of a high rate of recombination during female meiosis is unexpected, given studies with humans that suggested that female meiotic recombination is suppressed in imprinted regions (39). Whether this reflects a species difference or whether it is specific to the interspecific cross we analyzed is unknown.
Imprinting of genes within the cluster.
To determine the imprinting profile of p57Kip2, Kvlqt1, CD81, and Mash2 during development, we generated progeny from reciprocal crosses between strains of M. domesticus and BTBR(SPR H19-p57), a congenic BTBR strain containing sequence from M. spretus at distal chromosome 7. We identified polymorphisms between alleles from the two species and developed RT-PCR assays to determine which parental allele was expressed in the offspring. For each assay, mixing controls were used to verify that there was no allelic bias in amplification (data not shown). Furthermore, all assays were done with primers that spanned at least one intron, to eliminate the possibility of amplification of genomic DNA.
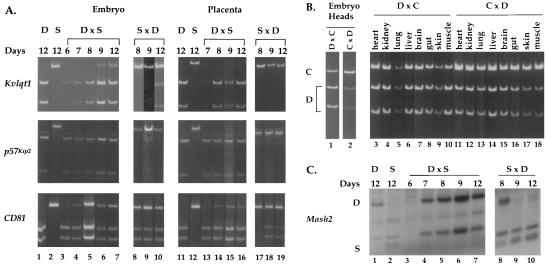
As shown in Fig. 2A, Kvlqt1 is maternally expressed in extraembryonic tissues at all stages of development analyzed but begins to lose its imprint in embryos after e9.5. This finding is in contrast to studies of human KvLQT1, which showed imprinting in all fetal tissues tested except for the heart (28). Interestingly, there appears to be a 1-day difference in the acquisition of paternal Kvlqt1 expression in 129/Sv M. domesticus × BTBR(SPR H19-p57) and BTBR(SPR H19-p57) × 129/Sv M. domesticus hybrids, suggesting that the M. spretus allele is more readily activated by e9.5. To determine whether the expression pattern of Kvlqt1 in mouse embryos past e8.5 was skewed by biallelic expression in the heart, we compared the expression of Kvlqt1 in the heads with that in the bodies of e13.5 embryos derived from crosses between C57BL/6 mice and a B6(CAST H19-p57) congenic strain (Fig. 2B and data not shown). Fortuitously, the M. castaneus Kvlqt1 allele possesses the same polymorphism as the M. spretus allele. Our results confirmed that Kvlqt1 is biallelically expressed in both embryo heads and bodies at e13.5, suggesting that the biallelic expression we detected in whole embryos was not solely attributable to contamination from heart RNA.
FIG. 2.
Temporal regulation of Kvlqt1, p57Kip2, CD81, and Mash2 imprinting during embryogenesis. (A) For Kvlqt1, p57Kip2, and CD81, RT-PCR analysis was performed on F1 progeny from a reciprocal interspecific cross between 129/Sv M. domesticus (D) and BTBR(SPR H19-p57) (S) mice dissected at e6.5, e7.5, e8.5, e9.5, and e12.5 (labeled 6, 7, 8, 9, and 12, respectively). The e6.5 embryo and extraembryonic tissues were dissected out together (lane 3); otherwise, the embryo was separated from the ectoplacental cone or placenta for analysis (lanes 4 to 10 and 13 to 19). 129/Sv or BTBR(SPR H19-p57) embryo (lanes 1 and 2) and placenta (lanes 11 and 12) at e12.5 was analyzed to show the parental alleles. (B) Kvlqt1 expression in e13.5 head and in neonatal tissues. RT-PCR analysis of e13.5 head RNA and postnatal day 4 tissues from F1 progeny of a reciprocal cross between 129/Sv M. domesticus (D) and B6(CAST H19-p57) (C). (C) Mash2 RT-PCR analysis of F1 progeny from a reciprocal interspecific cross between BTBR (D) and BTBR(SPR H19-p57) (S) mice. Dissections were of the whole decidua at e6.5, e7.5, and e8.5, of extraembryonic tissue at e9.5, and of placenta alone at e12.5.
To rule out tissue-specific imprinting that would be obscured by examination of whole embryos, we examined RNAs isolated from tissues of 4-day-old neonates derived from C57BL/6 × B6(CAST H19-p57) reciprocal crosses. As shown in Fig. 2B, Kvlqt1 was biallelically expressed in all neonatal tissues examined. Thus, we conclude that mouse Kvlqt1 imprinting is specific to extraembryonic tissues except at early stages of development.
Consistent with previous reports of p57Kip2 imprinting (19), we observed that p57Kip2 was imprinted at all developmental stages in both placental and embryonic tissues (Fig. 2A). By in situ hybridization, Guillemot et al. (16) had observed the expression of paternal Mash2 mRNA in a fraction of trophoblast cells at e6.5 and e7.5, suggesting that the imprinting of Mash2 is temporally regulated. To confirm this finding in wild-type animals, where there would be no selective pressure for inappropriate Mash2 expression, we used allele-specific RT-PCR; however, we were unable to detect Mash2 mRNA at e6.5 (Fig. 2C, lane 3). By e7.5, the paternal allele of Mash2 was silent when it was inherited from BTBR(SPR H19-p57) (lane 4). When paternal Mash2 was inherited from M. domesticus, however, its expression was still evident at e8.5 (lane 8) and was not extinguished until e9.5 (lanes 9 and 10). Thus, as suggested from the Mash2 gene disruption, Mash2 imprinting is developmentally acquired, at least when the paternal allele is derived from M. domesticus. Furthermore, the more rapid silencing of the M. spretus allele of Mash2 is an example of an allelic difference in the timing and expression of imprinting in mice.
Prior studies of CD81 (35) and Th (63) gene disruptions in mice did not indicate a parent-of-origin phenotype typical of imprinted genes. Th homozygous mutant embryos die of cardiovascular failure between e11.5 and e15.5, whereas heterozygotes are fully viable. The CD81 mutant phenotype is a subtle delay in the humoral response of B cells, and hence its imprinting might have been overlooked. Therefore, we used RT-PCR to examine its imprinting during development. Although CD81 expression shows a strong maternal bias early in development, by e8.5, it is expressed well from both parental alleles in embryonic and extraembryonic tissues (Fig. 2A). Thus, Mash2 is closely flanked by two predominantly nonimprinted genes. This observation is reminiscent of X chromosome inactivation in humans, where genes that escape inactivation are interspersed among genes that are inactivated (57). To date, L23MRP and NAP2, two nonimprinted genes that lie immediately telomeric and centromeric, respectively, to this region of human chromosome 11p15.5, have been viewed as defining the limits of the imprinting cluster (21, 52). The discovery of other nonimprinted genes embedded within the cluster, however, suggests that this notion should be reconsidered.
Effect of cis mutations on expression and imprinting in the cluster.
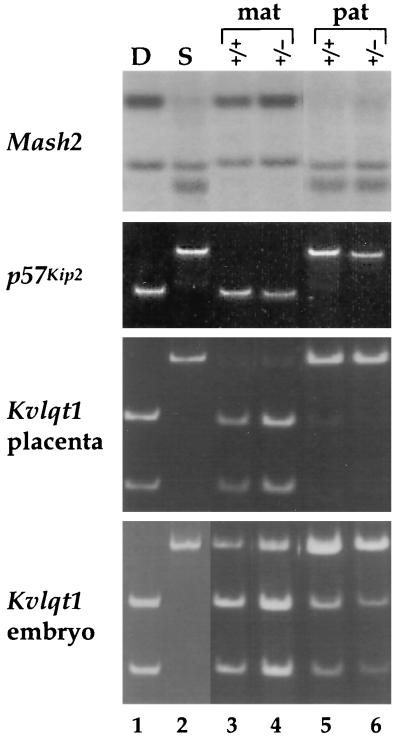
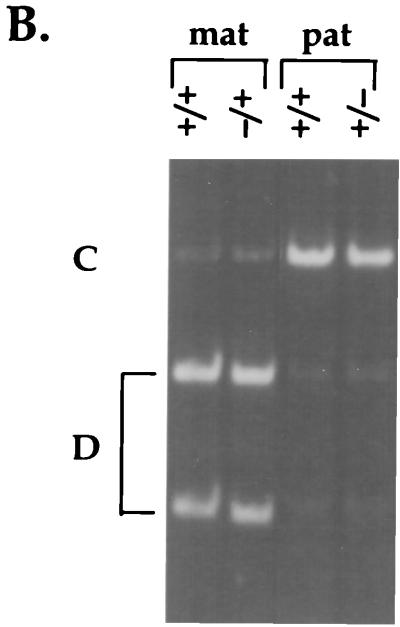
Because Igf2 and Ins2 are known to depend on the H19 gene for their imprinting, we were interested in whether H19 could exert its effect further along distal chromosome 7. The distal genes are expressed on the same chromosome as H19, and therefore we would not expect a role for H19 in promoter competition with Mash2, Kvlqt1, and p57Kip2. However, there is a precedent in the Prader-Willi imprinted gene cluster for a deletion of SNRPN, a paternally expressed gene, affecting the expression of linked paternal genes many kilobases away (45). To address this question, we assayed the imprinting status of Mash2, Kvlqt1, and p57Kip2 in mice lacking the H19 gene plus 10 kb of its 5′-flanking DNA (H19Δ13) (30). As shown in Fig. 3, all three genes showed a normal, imprinted expression profile in H19Δ13 heterozygous mice irrespective of whether the mutation was inherited maternally or paternally, suggesting that Mash2, Kvlqt1, and p57Kip2 are not regulated by H19.
FIG. 3.
Effect of an H19 deletion on the imprinting of Mash2, p57Kip2, and Kvlqt1: RT-PCR analysis of e12.5 placenta (Mash2 and Kvlqt1) and e12.5 embryo (Kvlqt1 and p57Kip2). Females heterozygous for the H19 gene body deletion (H19Δ13) (31) were crossed to BTBR(SPR H19-p57) males, and the F1 wild-type (+/+) and heterozygous (+/−) littermates (mat) were subjected to allele-specific assays (lanes 3 and 4). The reciprocal cross resulting in paternal inheritance of H19Δ13 (pat) was also analyzed (lanes 5 and 6). 129/Sv (D) or BTBR(SPR H19-p57) (S) embryo or placenta (lanes 1 and 2) at e12.5 was analyzed to show the parental alleles.
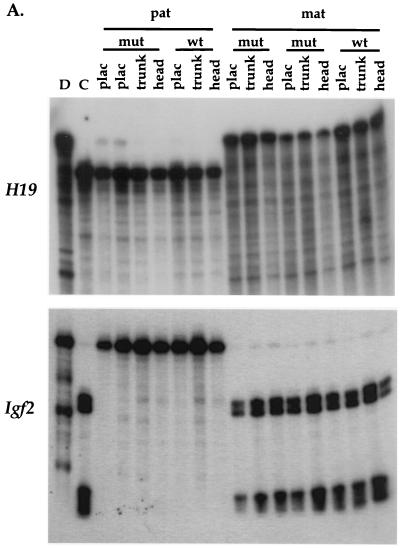
In humans, mutations in p57Kip2 have been associated with a minority of patients with BWS, and mice homozygous for an inherited p57Kip2 null allele have been put forward as a potential mouse model for the disease (27, 60, 62). Because the somatic overgrowth associated with BWS is often attributed to overexpression of Igf2 and because we have observed somatic overgrowth in e16.5 maternal heterozygous embryos (9a), we asked whether loss of p57Kip2 affected the expression or imprinting of Igf2 and/or H19. p57Kip2 heterozygous null mice (lacking exons 1 and 2 [87% of the coding region]), obtained from S. Elledge (Baylor College of Medicine), were crossed to B6(CAST H19-p57) mice to obtain e13.5 embryos inheriting the p57Kip2 null allele from either parent. With RNA derived from these embryos, we carried out allele-specific RNase protection assays to assess the effects on H19 and Igf2 RNAs, as well as RT-PCR analysis to examine the effects on imprinting and expression of Kvlqt1. As shown in Fig. 4, H19, Igf2, and Kvlqt1 imprinting and expression are not affected by the p57Kip2 deletion when it is present on the maternal chromosome, the chromosome from which p57Kip2 is normally expressed. Furthermore, the deletion of p57Kip2 DNA encompassing exons 1 and 2 did not disrupt local imprinting, since transcripts of the neomycin resistance (Neor) gene that replaces p57Kip2 were detected only upon maternal inheritance (9a). Given these results, we conclude that loss of p57Kip2 function has no effect on imprinting and expression of other genes in the region.
FIG. 4.
Effect of a p57Kip2 mutation on the imprinting of H19, Igf2, and Kvlqt1. (A) Females heterozygous for the p57Kip2 deletion (mat) (62) were crossed to B6(CAST H19-p57) males, and the F1 wild-type (wt) and heterozygous (mut) littermates were analyzed by use of allele-specific RNase protection assays of H19 and Igf2 RNAs in head, trunk, and placenta (plac) RNAs. The reciprocal cross resulting in paternal inheritance of the deletion allele (pat) was also analyzed. C57BL/6 (D) or B6(CAST H19-p57) (C) embryo or placenta at e12.5 was analyzed to show the parental alleles. (B) The same placental samples from the p57Kip2 deletion progeny were analyzed by RT-PCR for Kvlqt1 expression.
Effect of DNA methylation on imprinting in the cluster.
DNA methylation has different effects on imprinted genes. For H19 and Snrpn, methylation is required to maintain the silence of the genes, a finding that is consistent with the substantial methylation at their promoters (5, 13, 32, 46). In contrast, Igf2 and Igf2r, both of which are methylated on their expressed allele, are silenced in the absence of DNA methylation (32). It has been suggested that this classification of imprinted genes on the basis of the response to the loss of methylation is a useful way to distinguish genes that are the direct targets for DNA methylation (i.e., H19 and Snrpn) from those that are responding to methylation changes elsewhere (i.e., Igf2 and Igf2r) (3).
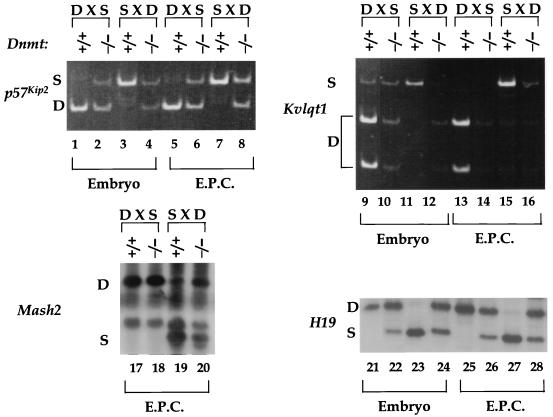
To classify the telomeric genes on distal chromosome 7 with regard to their response to methylation, we analyzed their imprinting in mice lacking the DNA methyltransferase gene (Dnmt), whose product is responsible for the maintenance of methylation in the genome (33). To allow us to distinguish the parental alleles of the genes in question, we bred the Dnmts null allele onto a BTBR(SPR H19-p57) background. Dnmt−/− mice die just after e9.5. Therefore, we studied embryonic and extraembryonic tissues from pools of e9.5 progeny of reciprocal crosses between heterozygous Dnmt+/− and Dnmt+/− BTBR(SPR H19-p57) mice. As shown in Fig. 5, in the absence of maintenance methylation, p57Kip2 is biallelically expressed (lanes 2, 4, 6, and 8), suggesting that DNA methylation is acting directly on p57Kip2 to repress its expression. In contrast, the maternal allele of Kvlqt1 is repressed in the absence of methylation (lanes 10, 12, 14, and 16), suggesting that this gene is an indirect target of DNA methylation and that methylation is required for Kvlqt1 expression.
FIG. 5.
Effect of a Dnmt mutation on the imprinting of p57Kip2, Kvlqt1, Mash2, and H19. Heterozygous Dnmts mice (33) were crossed to BTBR(SPR H19-p57) mice that were also heterozygous for Dnmts and wild-type (+/+) or mutant (−/−) progeny were dissected into embryonic, ectoplacental cone (E.P.C.), and yolk sac samples at e9.5. Yolk sac DNA was used to genotype the samples. Wild-type or mutant embryonic or ectoplacental cone samples were pooled for RT-PCR imprinting analyses. Abbreviations are the same as in Fig. 2.
The most surprising observation of the methylation study was made when the imprinting of Mash2 was examined and found to be unaffected by the loss of DNA methylation. This result is most striking in [129/Sv × BTBR(SPR H19-p57)]F1 hybrids (Fig. 5; compare lanes 17 and 18). In [BTBR(SPR H19-p57) × 129/Sv]F1 wild-type hybrids, the expression of the gene exhibits a strong maternal bias by e9.5 whereas the Dnmt−/− embryos are biallelic (compare lanes 19 and 20). This difference in the two F1 hybrids most probably reflects the fact that the DNA methyltransferase mutants are developmentally delayed about 1 day at e9.5 (33). Thus, when the expression of Mash2 at e8.5 is used as the appropriate comparison (Fig. 2C, lane 8), once again there is no impact of the Dnmt mutation. Given this finding, we wanted to confirm that DNA methylation had been affected in the Dnmt−/− embryos. Therefore, we used the same samples to examine the imprinting status of the H19 gene, which had previously been shown to become biallelic in the absence of Dnmt (32). As Fig. 5 illustrates, H19 RNA was detected from both alleles (lanes 22, 24, 26, and 28), confirming that Dnmt-dependent methylation is reduced in these tissues. Thus, this experiment provides no evidence for methylation playing a role in regulating the imprinting of Mash2.
DISCUSSION
Although the precise mechanisms by which imprinting occurs are unknown, the conserved localization of the imprinted genes on distal chromosome 7 in mice and humans suggests that clustering may be important for mechanistic or functional reasons. Our results show that the linkage of eight genes is conserved between mice and humans, consistent with the integrity of the region being important for proper imprinting of the genes contained therein (28, 41, 42). The synteny among imprinted genes in this region probably extends beyond the region. Recently, another maternally expressed imprinted gene, IPL/Ipl, has been characterized in humans and mice (41). In humans, this gene has been physically mapped centromeric to p57Kip2, and in mice, its genetic linkage places it in an analogous position. We have identified one major difference between the organization of this region in humans and mice in the positions of Th and CD81 relative to Mash2 and Ins2. In humans, TH is within 12 kb of INS (34), whereas in mice, the gene is just 25 kb centromeric of Mash2. In addition, a human P1 clone of the syntenic region of chromosome 11p15.5 (GenBank accession no. AC002536) places CD81 106 kb away from HASH2 (the human homolog of Mash2), whereas we detected CD81 sequences within 24 kb of Mash2. Another difference is the orientation of the cluster relative to the centromere. In humans, H19 is the most telomeric gene at 11p15.5, whereas our genetic analysis in mice places p57Kip2 closest to the telomere.
For the most part, the imprinting of the genes in this cluster is conserved between humans and mice. One difference we uncovered is in the maintenance of imprinting of Kvlqt1 during embryogenesis. In humans, the gene is imprinted in all fetal tissues except the heart (28), whereas in mice, the imprint is lost in all neonatal tissues examined. Species-specific differences in imprinting have been detected for the Igf2r gene as well, but in that case imprinting is relaxed in humans (47, 59).
Gene linkage has clearly been shown to be important for the imprinting of Igf2, H19, and Ins2. The mechanism is probably a transcriptional one, in which the genes require a common set of enhancers (31). DNA methylation on the paternal chromosome, the only epigenetic mark that has been identified, silences the H19 gene and thereby permits Igf2 and Ins2 expression (5, 13, 32). On the maternal chromosome, it is the position of the H19 gene, relative to the enhancers, that determines the preference for H19 transcription (54). This mechanism, however, does not extend to the telomeric genes in the cluster, since mutations that affect Igf2, H19 and Ins2 have no effect on these genes. Therefore, if a single element regulates distal chromosome 7 imprinting, that element does not appear to be the H19 gene.
The most compelling evidence in favor of a mechanistic link between the imprinting of genes throughout this cluster comes from observations in human patients with BWS. Approximately 80% of BWS patients exhibit biallelic IGF2 expression, and overexpression of IGF2 is thought to be responsible for most of the BWS phenotype, particularly the somatic overgrowth (43). Two recent mouse models of BWS, in which overexpression of Igf2 is achieved through transgenesis or genetic manipulation, lend strong support to this conclusion (12, 48). Some BWS patients have chromosomal abnormalities including balanced translocations whose breakpoints map to two regions of chromosome 11p15.5 (20). The first cluster of breakpoints lies in the 3′ end of the KvLQT1 gene, and one patient with such a translocation was shown to exhibit biallelic IGF2 expression (7). If this finding holds up with other BWS translocation patients, it strongly suggests that IGF2 imprinting requires linkage not just to H19 but also to sequences downstream of KvLQT1. The other cluster of translocation breakpoints is at least 4 Mb centromeric to p57Kip2, but the allelic expression of IGF2 has not been examined in any of these patients.
One reason for caution in interpreting the human translocations as implying a mechanistic linkage between the two domains of the cluster is that a small percentage of BWS patients have point mutations in the p57Kip2 gene itself (27, 38). It is unknown whether these rare patients display biallelic IGF2. If they do not, it is possible that the translocations are disrupting only p57Kip2 expression. As we have shown in this report, a loss-of-function mutation of p57Kip2 in mice does not result in biallelic Igf2 expression. The mice do exhibit some BWS-like symptoms, such as omphalocele, renal dysplasia, and adrenal cytomegaly, but they lack other features (60, 62). Thus, BWS is very likely to be a genetically complex disorder. Finally, there is indirect evidence for linkage between the genes in the cluster from studies of patients with Wilms’ tumor, where a general correlation between the expression of H19 and p57Kip2 has been observed (9).
Since H19 does not appear to be the global regulator of imprinting of the telomeric genes, we considered the possibility that these genes are regulated by a common mechanism involving DNA methylation. By analogy to the paternally expressed genes in the Prader-Willi complex, which are coordinately expressed on the unmethylated paternal chromosome and silenced on the methylated maternal chromosome (for reviews, see references 15 and 26), we expected Mash2, Kvlqt1, and p57Kip2 to respond in the same way to the absence of DNA methylation. Instead, each gene responded differently.
The imprinting of p57Kip2 in all tissues, coupled with the activation of its paternal allele in Dnmt−/− embryos, makes it a good candidate for a direct target of DNA methylation silencing. Indeed, Hatada and Mukai (19) had identified paternally specific methylation of a single HhaI site within the p57Kip2 gene itself. That site cannot be required for p57Kip2 imprinting, however, because it is deleted in p57Kip2 mutant mice, where the Neor gene retains imprinted expression (9a). Nevertheless, by analogy to other genes like H19 and Snrpn, our findings predict that there should be an imprint control region very close to the p57Kip2 gene. They also predict that the imprinting of p57Kip2 may not require the other genes in the cluster.
Kvlqt1, on the other hand, exhibits characteristics of a gene that is an indirect target of methylation. Like Igf2 and Igf2r, the expression of the active allele is extinguished in Dnmt−/− embryos. By analogy to those genes, we would expect that there is a yet-to-be-identified paternally expressed transcript in the locus that competes with Kvlqt1 for expression in the placenta. It would be that gene whose expression is directly silenced by DNA methylation. This is the first suggestion that maternally specific methylation might exist at this cluster.
An indirect mechanism for Kvlqt1 imprinting is also consistent with its tissue-specific imprinting. Tissue-specific imprinting can best be explained by considering the case of the Ins2 gene, which is imprinted in extraembryonic tissues but not in the pancreas (14). It has been proposed that the tissue specificity is a consequence of the position of transcriptional enhancers relative to the epigenetic mark at the H19 gene (4, 54). In extraembryonic tissues, Ins2 expression requires the same 3′ distal transcriptional enhancers that govern Igf2 and H19 expression, and thus its expression depends on the transcriptional status of the H19 gene. In the pancreas, an enhancer that lies 5′ of the gene is activated, and by virtue of its position, it escapes the influence of imprinting (11). For Kvlqt1, the target of the competition would be a placenta-specific enhancer.
The gene whose imprinting does not fit into one of these two categories of imprinted genes is Mash2, which is imprinted and expressed only in the placenta but appears to be unaffected by a loss in DNA methylation. It could be that Mash2 needs only a small amount of methylation to be imprinted. Li et al. (32) had noted that the Igf2r gene was more resistant than H19 to demethylation in mice carrying a hypomorphic allele of Dnmt; however, the gene was affected in animals carrying a null allele. Furthermore, even in mice with a null allele of Dnmt, such as the animals we used in this study, there is residual genomic DNA methylation at a level approximately 5 to 10% of that in wild-type embryos (33). Thus, it is formally possible that another DNA methylase provides the signal for Mash2 imprinting. No differentially methylated sites associated with Mash2 have been detected to date, however (8a). Moreover, we have observed that a 105-kb P1 clone encompassing the Mash2 locus displays biallelic expression in transgenic mice, arguing against local controls governing its imprinting (8a). If methylation is not involved in Mash2 imprinting, we must invoke an entirely novel imprinting control mechanism, such as heritable changes in chromatin structure.
In conclusion, our results with mice did not uncover long-range effects among the genes on distal chromosomes by known imprinting mechanisms as would be expected if the evolutionary conservation of the entire region is being maintained for regulatory reasons. Furthermore, a single mechanism whereby methylation spreads along the chromosome from a nucleating center can be argued against, since methylation is predicted to be on the paternal chromosome at p57Kip2, as it is for Igf2 and H19, but is expected to be on the maternal chromosome to affect Kvlqt1. The question that remains is whether there is any mechanistic link between p57Kip2, Kvlqt1, and Mash2 imprinting. Their common imprinting in the placenta is consistent with such a connection; however, the distinct ways in which they respond to the loss of DNA methylation cannot be readily reconciled. Thus, it is possible that distal chromosome 7 does not contain a single cluster of imprinted genes but, rather, contains multiple clusters, regulated by individual mechanisms.
ACKNOWLEDGMENTS
We thank Steve Elledge and Pumin Zhang, Baylor College of Medicine, for p57Kip2 mutant mice and the sequence of the p57Kip2 locus, and we thank Rudolph Jaenisch and En Li for the Dnmt mutant mice. We also thank R. S. Ingram for DNA sequencing, B. K. Jones for developing the Dnmt genotyping assay, and members of the laboratory for critical discussion.
This work was supported by a grant from the National Institute for General Medical Sciences (GM 51460).
T.C. and M.A.C. contributed equally to this work.
REFERENCES
- 1.Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 3.Barlow D P. Competition—a common motif for the imprinting mechanism? EMBO J. 1997;16:6899–6905. doi: 10.1093/emboj/16.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 5.Bartolomei M S, Webber A L, Brunkow M E, Tilghman S M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 6.Bartolomei M S, Zemel S, Tilghman S M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown K W, Villar A J, Bickmore W, Clayton-Smith J, Catchpole D, Maher E R, Reik W. Imprinting mutation in the Beckwith-Wiedemann syndrome leads to biallelic IGF2 expression through an H19-independent pathway. Hum Mol Genet. 1996;5:2027–2032. doi: 10.1093/hmg/5.12.2027. [DOI] [PubMed] [Google Scholar]
- 8.Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls R D, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 8a.Caspary, T., and S. M. Tilghman. Unpublished data.
- 9.Chung W-Y, Yuan L, Feng L, Hensle T, Tycko B. Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms’ tumors. Hum Mol Genet. 1996;5:1101–1108. doi: 10.1093/hmg/5.8.1101. [DOI] [PubMed] [Google Scholar]
- 9a.Cleary, M. A., and S. M. Tilghman. Unpublished data.
- 10.Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls R D, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- 11.Edlund T, Walker M D, Barr P J, Rutter W J. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science. 1985;230:912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- 12.Eggenschwiler J, Ludwig T, Fisher P, Leighton P A, Tilghman S M, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental-origin-specific epigenetic modifications of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 14.Giddings S J, King C D, Harman K W, Flood J F, Carnaghi L R. Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet. 1994;6:310–313. doi: 10.1038/ng0394-310. [DOI] [PubMed] [Google Scholar]
- 15.Glenn C C, Driscoll D J, Yang P, Nicholls R D. Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes. Mol Hum Reprod. 1997;3:321–332. doi: 10.1093/molehr/3.4.321. [DOI] [PubMed] [Google Scholar]
- 16.Guillemot F, Caspary T, Tilghman S M, Copeland N G, Gilbert D J, Jenkins N A, Anderson D J, Joyner A L, Rossant J, Nagy A. Genomic imprinting of Mash-2, a mouse gene required for trophoblast development. Nat Genet. 1995;9:235–241. doi: 10.1038/ng0395-235. [DOI] [PubMed] [Google Scholar]
- 17.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner A L. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 18.Gunaratne P H, Nakao M, Ledbetter D H, Sutcliffe J S, Chinault A C. Tissue-specific and allele-specific replication timing control in the imprinted human Prader-Willi syndrome region. Genes Dev. 1995;9:808–820. doi: 10.1101/gad.9.7.808. [DOI] [PubMed] [Google Scholar]
- 19.Hatada I, Mukai T. Genomic imprinting of p57/KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat Genet. 1995;11:204–206. doi: 10.1038/ng1095-204. [DOI] [PubMed] [Google Scholar]
- 20.Hoovers J M, Kalikin L M, Johnson L A, Alders M, Redeker B, Law D J, Bliek J, Steenman M, Benedict M, Wiegant J. Multiple genetic loci within 11p15 defined by Beckwith-Wiedemann syndrome rearrangement breakpoints and subchromosomal transferable fragments. Proc Natl Acad Sci USA. 1995;92:12456–12460. doi: 10.1073/pnas.92.26.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu R-J, Lee M P, Johnson L A, Feinberg A P. A novel human homologue of yeast nucleosome assembly protein, 65 kb centromeric to the p57KIP2 gene, is biallelically expressed in fetal and adult tissues. Hum Mol Genet. 1996;5:1743–1748. doi: 10.1093/hmg/5.11.1743. [DOI] [PubMed] [Google Scholar]
- 22.Kay G F, Penny G D, Patel D, Ashworth A, Brockdorff N, Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- 23.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 24.Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll D J, Nicholls R D, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- 25.Knoll J H, Cheng S D, Lalande M. Allele specificity of DNA replication timing in the Angelman/Prader-Willi syndrome imprinted chromosomal region. Nat Genet. 1994;6:41–46. doi: 10.1038/ng0194-41. [DOI] [PubMed] [Google Scholar]
- 26.Lalande M. Parental imprinting and disease. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 27.Lee M P, DeBaun M, Randhawa G, Reichard B A, Elledge S J, Feinberg A P. Low frequency of p57KIP2 mutation in Beckwith-Wiedemann syndrome. Am J Hum Genet. 1997;61:304–309. doi: 10.1086/514858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee M P, Hu R, Johnson L A, Feinberg A P. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 29.Leff S E, Brannan C I, Reed M L, Ozcelik T, Francke U, Copeland N G, Jenkins N A. Maternal imprinting of the mouse Snrpn gene and conserved linkage homology with the human Prader-Willi syndrome region. Nat Genet. 1992;2:259–264. doi: 10.1038/ng1292-259. [DOI] [PubMed] [Google Scholar]
- 30.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 31.Leighton P A, Saam J R, Ingram R S, Stewart C L, Tilghman S M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 32.Li E, Beard C, Jaenisch R. The role of DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 33.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 34.Lucassen A M, Julier C, Beressi J P, Boitard C, Froguel P, Lathrop M, Bell J I. Susceptibility to insulin dependent diabetes mellitus maps to a 4.1 kb segment of the DNA spanning the insulin gene and associated VNTR. Nat Genet. 1993;4:305–310. doi: 10.1038/ng0793-305. [DOI] [PubMed] [Google Scholar]
- 35.Maecker H T, Levy S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J Exp Med. 1997;185:1505–1510. doi: 10.1084/jem.185.8.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 37.Matsuura T, Sutcliffe J S, Fang P, Galjaard R-J, Jiang Y, Benton C S, Rommens J M, Beaudet A L. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 38.O’Keefe D, Dao D, Zhao L, Sanderson R, Warburton D, Weiss L, Anyane-Yeboa K, Tycko B. Coding mutations in p57KIP2 are present in some cases of Beckwith-Wiedemann syndrome but are rare or absent in Wilms tumors. Am J Hum Genet. 1997;61:295–303. doi: 10.1086/514854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paldi A, Gyapay G, Jami J. Imprinted chromosomal regions of the human genome display sex-specific meiotic recombination frequencies. Curr Biol. 1995;5:1030–1035. doi: 10.1016/s0960-9822(95)00207-7. [DOI] [PubMed] [Google Scholar]
- 40.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdroff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 41.Qian N, Frank D, O’Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B. The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum Mol Genet. 1997;6:2021–2029. doi: 10.1093/hmg/6.12.2021. [DOI] [PubMed] [Google Scholar]
- 42.Reid L H, Davies C, Cooper P R, Crider-Miller S J, Sait S N J, Nowak N J, Evans G, Stanbridge E J, deJong P, Shows T B, Weissman B E, Higgins M J. A 1-Mb physical map and PAC contig on the imprinted domain in 11p15.5 that contains TAPA1 and the BWSCR1/WT2 region. Genomics. 1997;43:366–375. doi: 10.1006/geno.1997.4826. [DOI] [PubMed] [Google Scholar]
- 43.Reik W, Maher E R. Imprinting in clusters: lessons from Beckwith-Wiedemann syndrome. Trends Genet. 1997;13:330–334. doi: 10.1016/s0168-9525(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 44.Ripoche M-A, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 45.Saitoh S, Buiting K, Rogan P K, Buxton J L, Driscoll D J, Arnemann J, Konig R, Malcolm S, Horsthemke B, Nicholls R D. Minimal definition of the imprinting center and fixation of a chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci USA. 1996;93:7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shemer R, Birger Y, Riggs A D, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA. 1997;94:10267–10272. doi: 10.1073/pnas.94.19.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smrzka O W, Fae I, Stoger R, Kurzbauer R, Fischer G F, Henn T, Weith A, Barlow D P. Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum Mol Genet. 1995;4:1945–1952. doi: 10.1093/hmg/4.10.1945. [DOI] [PubMed] [Google Scholar]
- 48.Sun F-L, Dean W, Kelsey G, Allen N D, Reik W. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 49.Sutcliffe J S, Nakao M, Christian S, Orstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 50.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 51.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 52.Tsang P, Gilles F, Yuan L, Kuo Y H, Lupu F, Samara G, Moosikasuwan J, Goye A, Zelenetz A D, Selleri L, Tycko B. A novel L23-related gene 40 kb downstream of the imprinted H19 gene is biallelically expressed in mid-fetal and adult human tissues. Hum Mol Genet. 1995;4:1499–1507. doi: 10.1093/hmg/4.9.1499. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Curran M E, Splawski I, Burn T C, Millholland J M, VanRaay T J, Shen J, Timothy K W, Vincent G M, de Jager T, Schwatz P J, Towbin J A, Moss A J, Atkinson D L, Landes G M, Connors T D, Keating M T. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 54.Webber A, Ingram R I, Levorse J, Tilghman S M. Location of enhancers is essential for imprinting of H19 and Igf2. Nature. 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- 55.Weksberg R, Shen D R, Fei Y L, Song Q L, Squire J. Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- 56.Wevrick R, Kerns J A, Francke U. Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- 57.Willard H F, Brown C J, Carrel L, Hendrich B, Miller A P. Epigenetic and chromosomal control of gene expression: molecular and genetic analysis of X chromosome inactivation. Cold Spring Harbor Symp Quant Biol. 1993;58:315–322. doi: 10.1101/sqb.1993.058.01.037. [DOI] [PubMed] [Google Scholar]
- 58.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y, Goodyer C G, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- 60.Yan Y, Frisén J, Lee M-H, Massagué J, Barbacid M. Ablation of the CDK inhibitor p57KIP2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 61.Zemel S, Bartolomei M S, Tilghman S M. Physical linkage of two mammalian imprinted genes. Nat Genet. 1992;2:61–65. doi: 10.1038/ng0992-61. [DOI] [PubMed] [Google Scholar]
- 62.Zhang P, Liégeois N J, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, DePinho R A, Elledge S J. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature. 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Q-Y, Quaife C J, Palmiter R D. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature. 1995;374:640–643. doi: 10.1038/374640a0. [DOI] [PubMed] [Google Scholar]